Abstract
The substitution of valine with phenylalanine at amino acid 617 of the Janus kinase 2 (JAK2) gene (JAK2 p.V617F) occurs in a high proportion of patients with myeloproliferative neoplasms (MPNs). The ability to accurately measure JAK2 p.V617F allele burden is of great interest given the diagnostic relevance of the mutation and the ongoing clinical evaluation of JAK inhibitors. A main hurdle in developing quantitative assays for allele burden measurement is the unavailability of accurate standards for both assay validation and use in a standard curve for quantification. We describe our approach to the validation of standards for quantitative assessment of JAK2 p.V617F allele burden in clinical MPN samples. These standards were used in two JAK2 p.V617F assays, which were used to support clinical studies of ruxolitinib (Jakafi®) in myelofibrosis, a real-time polymerase chain reaction assay for initial screening of all samples, and a novel single-nucleotide polymorphism typing (SNaPshot)-based assay for samples with less than 5% mutant allele burden. Comparisons of allele burden data from clinical samples generated with these assays show a high degree of concordance with each other and with a pyrosequencing-based assay used for clinical reporting from an independent laboratory, thus providing independent validation to the accuracy of these standards.
Introduction
Members of the hematopoietic receptor superfamily lack an intrinsic kinase activity and require members of the Janus kinase (JAK) family of nonreceptor tyrosine kinases for downstream signaling. There are four known JAK family members: JAK1, JAK2, JAK3, and tyrosine kinase 2 (TYK2). The JAK2 c.1849G>T (p.V617F) mutation (subsequently referred to as JAK2V167F) results in a valine to phenylalanine substitution at residue 617 in the JAK2 pseudokinase domain and is present in a high proportion of patients with myeloproliferative neoplasms (MPNs) (Levine and Wernig, 2006; Ihle and Gilliland, 2007). This mutation leads to growth factor hypersensitivity/independence through constitutive activation of the JAK2 kinase activity and increased downstream signaling. Increased signaling through the constitutive activation of JAK2 is believed to be an important component of the clinical manifestations of MPNs.
The prevalence of the JAK2V617F mutation varies among MPNs, ranging from 97% in polycythemia vera (PV) to ∼50% in essential thrombocythemia and primary myelofibrosis (Baxter et al., 2005; James et al., 2005; Kralovics et al., 2005; Levine et al., 2005). The ability to accurately measure JAK2V617F allele burden is of great interest given the diagnostic relevance of the mutation to MPNs as well as the ongoing clinical evaluation of JAK inhibitors. Numerous JAK2V617F assays have been described in the literature (Steensma, 2006). For nearly all assay formats, the accurate quantification of JAK2V167F allele burden requires comparison of unknowns to a standard curve containing different admixtures of wild-type (WT) and JAK2V167F DNA. Because of this, a robust and thoroughly validated set of standards is a key component of any quantitative assay. A critical issue with using JAK2V617F-positive and JAK2 WT cells for standards is the potential confounding effect of gene copy number and aneuploidy on the allele burden. Cell lines with amplified JAK2V617F can lead to artificially low allele burden measurements and overestimates of allele burden changes if the copy number is not accounted for in standards. Furthermore, using a cell line that is haploid for the WT JAK2 locus can have the same effect and even magnify the error when combined with a cell line with multiple copies of JAK2V617F.
We describe a process for the characterization and validation of standards prepared from the human erythroleukemic (HEL) 92.1.7 cell line and a PV patient's DNA sample for use in quantitative JAK2V617F assays. In addition, we describe our use of these standards in a two-tiered approach for assessing JAK2V617F status. All samples are assayed using a quantitative JAK2V617F real-time polymerase chain reaction (PCR) assay, with a confirmatory multiplex single-nucleotide polymorphism typing (SNaPshot) assay being performed on negative or low-percentage JAK2V617F samples identified by real-time PCR. The SNaPshot assay relies on the single-nucleotide extension of a JAK2-specific primer by fluorescent-labeled terminator dideoxynucleotides followed by capillary electrophoresis of the extension products for the determination of WT and mutant (MUT) JAK2 allele percentages. These assays and standards have been used to support Phase I/II and III ruxolitinib (Jakafi®) clinical studies in myelofibrosis (Verstovsek et al., 2010, 2012; Harrison et al., 2012).
Materials and Methods
Genomic DNA and standard curves
Genomic DNA for JAK2V617F standard curve development was obtained from the HEL 92.1.7 cell line from the American Type Culture Collection, commercially obtained samples from patients with PV (Asterand), and healthy volunteers. Genomic DNA was prepared from whole blood using PAXgene or QIAamp DNA Blood kits as recommended by the manufacturer (Qiagen). All patient samples were collected with informed consent (clinicaltrials.gov identifier NCT00509899).
The JAK2V617F status of the HEL 92.1.7 cell line was evaluated by standard dideoxy sequence analysis. A dilution series was prepared with DNAs isolated from the HEL 92.1.7 cell line and a PV patient sample obtained from Asterand (ID: MCV PV005) diluted in normal genomic DNA. The percentage JAK2V617F relative to total JAK2 sequences for these standards was assessed using Mutation Surveyor (Soft Genetics). The JAK2V617F copy number of the HEL92.1.7 was estimated by fitting the measured percentage JAK2V617F values of the dilution series to theoretical curves based on different JAK2V167F copy numbers. Based on these analyses, a dilution series using the PV patient sample, HEL 92.1.7, and control genomic DNA was generated for use in assay validation and standard curves for quantification. Samples that were less than 90% JAK2V167F were derived from PV patient sample DNA diluted in control DNA, whereas standards that were greater than 90% were derived from HEL 92.1.7 DNA appropriately diluted in control DNA.
Real-time PCR and pyrosequencing assay
Real-time PCR-based assays were performed essentially as described (Levine et al., 2006) with modifications to the primer and probe sequences (JAK2 forward GCAGCAAGTATGATGAGCAAGCT, JAK2 reverse GCTCTGAGAAAGGCATTAGAAAGC, WT probe: 6FAM-TCCACAGACACATACTC-MGBNFQ, and MUT probe: VIC-TCTCCACAGAAACATACTCCA-MGBNFQ). Twelve-point standard curves were run in duplicate and patient samples in triplicate. Standard curve values were plotted as changes in threshold cycle (ΔCt) values (WT Ct-MUT Ct) and %V617F values converted to log-fold change. The WT-V617F ΔCt values and log-fold change values were plotted as an X–Y scatter plot in MS Excel, with the trend line being calculated using a second-order polynomial. Unknown %V617F values were calculated from the resulting WT-V617F ΔCt values using the second-order polynomial equation from the standard curve, with the resulting log-fold change values being converted to percentage V617F. A pyrosequencing-based quantitative JAK2V167F assay was performed at the MD Anderson Cancer Center as a part of routine clinical testing as described (Jelinek et al., 2005).
Cell colony analysis
Colony-forming assays were performed by StemCell Technologies, using peripheral blood mononuclear cells (PBMCs) obtained from PV patients and normal volunteers. Individual colonies were picked from semisolid agar, washed in phosphate-buffered saline, and resuspended in 10 μL of water.
SNaPshot assay
PCR was performed using 20 ng of genomic DNA, or 2 μL colony cell suspension, a forward JAK2 primer (5′-GCTACATCCATCTACCTCAGTTTCC-3′), and a reverse JAK2 primer (5′-ATTCCAATGTTATGTTGAACCTGCC-3′). SNaPshot assays were performed using a JAK2 SNaPshot primer (5′-AGCAAGCTTTCTCACAAGCATTTGGTTTTAAATTATGGAGTATGT-3′) as recommended by the manufacturer (Applied Biosystems). GeneMapper® (Applied Biosystems) was used to determine the areas under the curve (AUC) for the MUT and WT peaks. A 16-point standard curve (run in duplicate) was fitted using a two-phase exponential association [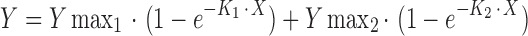 ]. Percentage V617F values for unknown samples (run in quadruplicate) were calculated in MS Excel from the MUT/WT AUCs using the above formula.
]. Percentage V617F values for unknown samples (run in quadruplicate) were calculated in MS Excel from the MUT/WT AUCs using the above formula.
Results
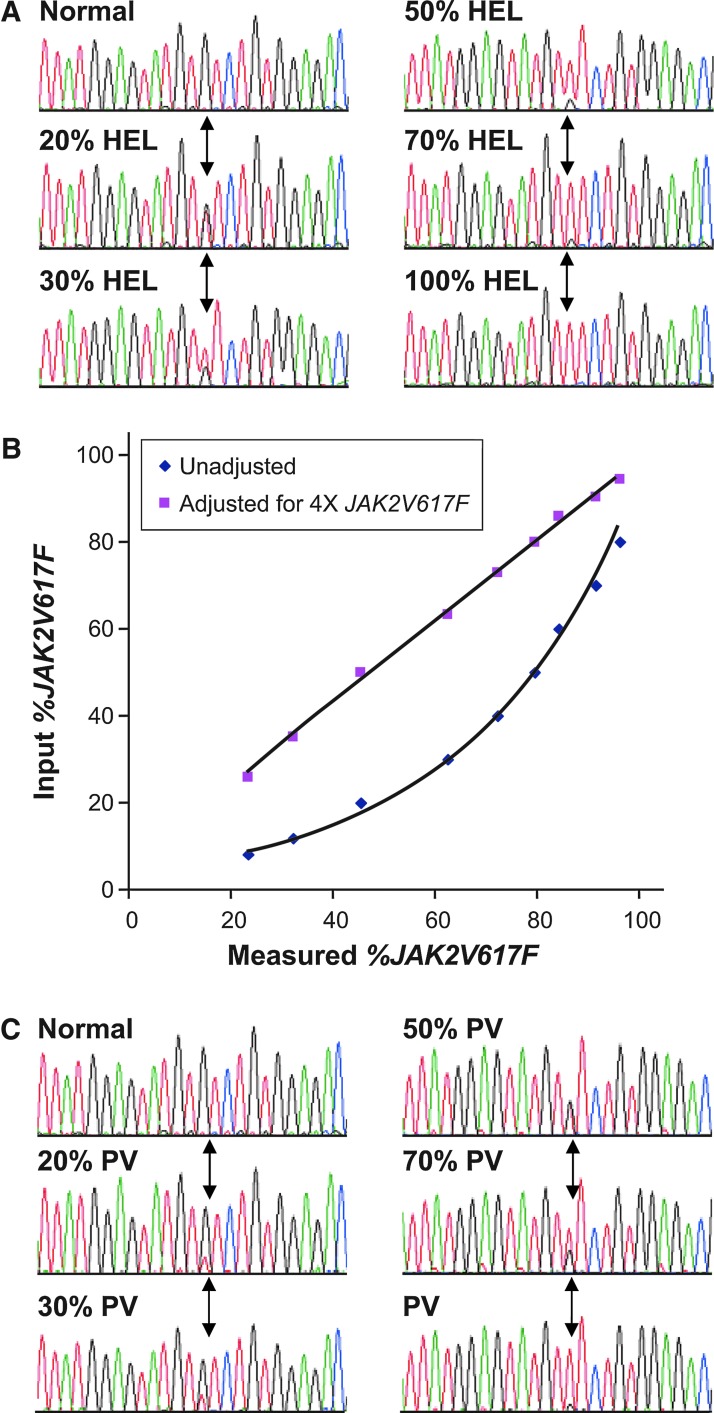
A key requirement for clinical JAK2V617F assays is a robust and well-calibrated set of standards for assay validation and standard curve generation. A dilution series of erythroleukemic HEL 92.1.7 cell line genomic DNA was prepared to explore assays for quantitatively assessing the allele burden of JAK2V617F in clinical samples. Although the HEL 92.1.7 cell line was shown to be homozygous for JAK2V617F by standard sequence-based methods (Fig. 1A), further sequence analysis with a DNA dilution series indicated a poor correlation between the HEL 92.1.7/control admixtures and the measured percentage JAK2V617F values (Fig. 1B). These data are consistent with the amplification of the JAK2 locus in the HEL 92.1.7 cell line, as has recently been described by others (Quentmeier et al., 2006; Voelkner et al., 2008; Lippert et al., 2009). An excellent correlation between the calculated and the measured percentage of JAK2V617F was obtained when the percentage JAK2V617F values of the HEL 92.1.7 dilution series were adjusted for a copy number of four copies of JAK2V617F per haploid genome (Fig. 1B). While the percent JAK2V617F of these standards was measured using the standard sequence analysis, this was performed on nine individual dilutions with the copy number estimate being based on the linear curve fitting of the nine samples (Fig. 1B). Using this approach, the overall accuracy of the copy number is far more precise than obtained with the standard sequence analysis of a single sample. If not accounted for, the amplification in the HEL cell line results in four times as much JAK2V617F allele being added to each standard assay point and an underestimation of JAK2V617F allele burden in the unknowns (Fig. 1B). A similar sequence analysis was performed on a reference PV sample obtained from a commercial vendor, with analysis of a DNA dilution series indicating that the DNA from the PV sample was ∼90% JAK2V617F (Fig. 1C). Genomic DNA from HEL 92.1.7 and PV samples were diluted in control genomic DNA to generate a JAK2V617F standard curve for use in assay validation and JAK2V617F quantification.
FIG. 1.
Analysis of JAK2V617F standards. (A) Human erythroleukemic (HEL) 92.1.7 genomic DNA was diluted in normal human peripheral blood DNA and analyzed by standard dideoxy sequence analysis. Position of the G to T transversion is highlighted by arrows. (B) Plot of the measured percentage JAK2V617F values obtained from diluted HEL 92.1.7 DNA versus expected percentage JAK2V617F values based on no copy number adjustment (blue) or with a copy number adjustment (pink). (C) Polycythemia vera (PV) patient peripheral blood DNA was diluted in normal human peripheral blood DNA and analyzed by standard dideoxy sequence analysis. Position of the G to T transversion is highlighted by arrows. The same normal control electropherogram is shown in (A) and (C).
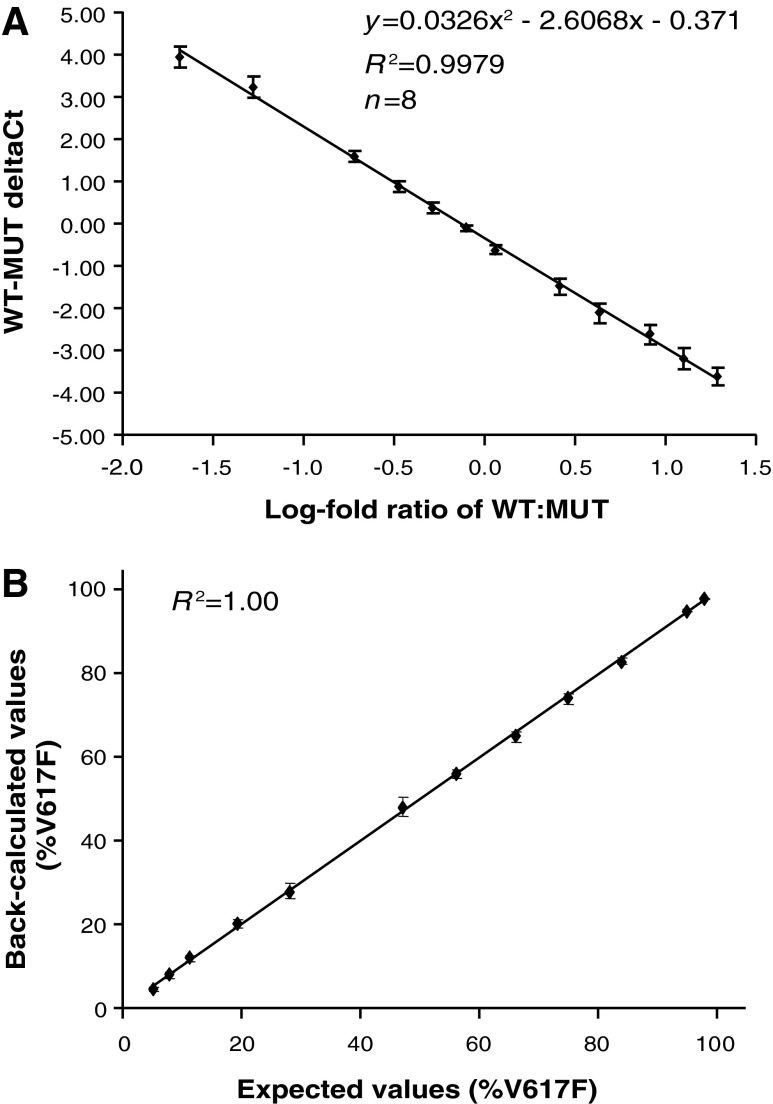
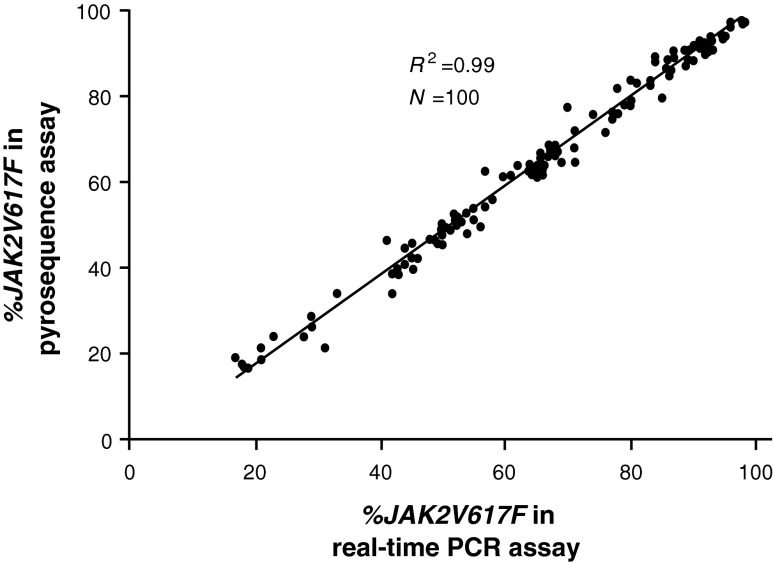
A real-time PCR assay was used as our primary assay for evaluating the JAK2V617F allele burden due to its throughput and lack of an upfront genomic PCR. Plotting the JAK2 WT-JAK2V617F (WT-MUT) ΔCt from the 12-point standard curve versus percentage JAK2V617F values (first converted to log-fold change) revealed a near-linear relationship (Fig. 2A). Analysis of the data in this manner allowed us to accurately calculate unknown V167F values from a standard curve using their WT-MUT ΔCt values. The reproducibility and accuracy of the assay were demonstrated in eight independent assays (Fig. 2B) with a sensitivity of 5% JAK2V617F. Importantly, the real-time PCR assay data are highly concordant with pyrosequencing data obtained from an independent clinical laboratory using 100 parallel clinical samples (Fig. 3), with the average percent difference for values between the two assays of 2%. This high concordance with the pyrosequencing assay, which semiquantitatively estimated the allele burden as a ratio of MUT and total JAK2 AUC, is a confirmation that the standard curve in the real-time PCR assay was correctly calibrated.
FIG. 2.
Reproducibility of real-time polymerase chain reaction (PCR) JAK2V617F assay. (A) Wild-type-mutant (WT-MUT) ΔCt values plotted against the log-fold ratio of JAK2V617F standards for eight independent experiments. Average values±standard deviation. (B) Percentage JAK2V617F values were determined for standards in eight independent experiments. Average values±standard deviation.
FIG. 3.
Comparison of clinical data in 100 paired samples using the real-time PCR assay and pyrosequencing analysis from an independent laboratory.
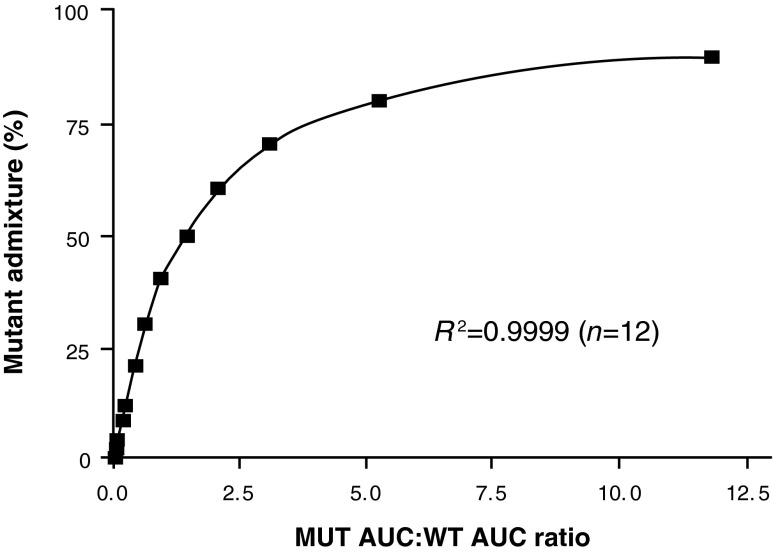
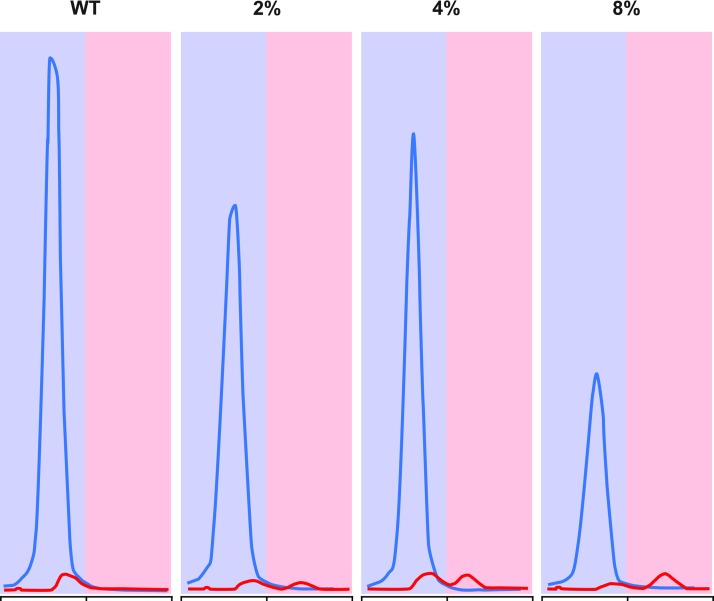
Although real-time PCR provided a rapid and accurate means to assess JAK2V167F allele burden, an additional assay was required to accurately differentiate samples with <5% JAK2V617F allele burden from JAK2V617F-negative samples, because this was below the lower limit of detection of both the real-time and the pyrosequencing-based assays. During our early evaluation of different assay formats, observable chromatogram peaks were seen in SNaPshot assays using 2% JAK2V617F controls, suggesting that the method may be more sensitive than real-time PCR and pyrosequencing. Analysis of JAK2V617F dilution curves in the SNaPshot assay indicated that the ratio of the AUC for the V617F allele to the WT allele reproducibly followed a two-phase exponential association, with R2 values consistently being greater than 0.99 (Fig. 4). Five replicates of a standard curve were run in the same assay, with the resulting equations from each curve being used to back-calculate the individual standards (Table 1). The calculated values for each standard matched the expected values well (R2≥0.99) for each set of standard curves in the assay. Interassay variability was similarly assessed using eight separate assays over the course of a 6-week period of time (Table 2 and Fig. 5). This latter analysis indicated that the assay had minimal interassay variation (Table 2 and Fig. 5). Although peaks could be seen for a 1% JAK2V617F standard, they could not be consistently quantitated by the GeneMapper software (data not shown). Because of this, 2% was set at the lower limit of quantification for the SNapShot assay. Chromatograms for 0–8% JAK2V617F samples, obtained by diluting the PV patient DNA (from above) in normal genomic DNA, are shown in Figure 6 and illustrate the sensitivity of the SNaPshot assay. Based on these data, the SNaPshot assay provides an accurate measure of JAK2V617F to 2% and correctly reported negative samples with high reproducibility between independent experiments. These data are consistent with the presence of measurable cross-reactivity of the MUT probe for the WT allele at low allele burdens, with JAK2V617F-negative samples scoring as 2–3% in the real-time PCR assay when calculating from the standard curve.
FIG. 4.
JAK2V617F SNaPshot assay. Relationship percentage JAK2V617F and area under the curve (AUC) ratios form MUT:WT peaks.
Table 1.
Intra-Assay Variability of Standard Curve Using Polycythemia Vera Patient DNA and Human Erythroleukemic 92.1.7 DNA Diluted in Wild-Type Genomic DNA
| Back-calculated JAK2V617F standards (%) | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| V6I7F standard (%) | Rep 1 | Rep 2 | Rep 3 | Rep 4 | Rep 5 | Mean | SD | CV | %Diff |
| 0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | N/A | N/A |
| 2 | 2.2 | 2.0 | 1.8 | 1.8 | 1.8 | 2 | 0.2 | 9.4 | −5.0 |
| 4 | 3.7 | 4.3 | 3.5 | 3.8 | 4.4 | 4 | 0.4 | 10.0 | −2.5 |
| 8 | 7.5 | 6.7 | 6.4 | 6.6 | 7.2 | 7 | 0.5 | 6.6 | −13.8 |
| 11 | 10 | 11 | 10 | 10 | 10 | 10 | 0.4 | 4.4 | −7.3 |
| 19 | 18 | 18 | 17 | 17 | 17 | 17 | 0.5 | 3.1 | −8.4 |
| 28 | 29 | 28 | 29 | 28 | 29 | 29 | 0.5 | 1.9 | 2.1 |
| 38 | 40 | 40 | 38 | 38 | 39 | 39 | 1.0 | 2.6 | 2.6 |
| 47 | 47 | 47 | 48 | 48 | 47 | 47 | 0.5 | 1.2 | 0.9 |
| 56 | 56 | 54 | 56 | 56 | 55 | 55 | 0.9 | 1.6 | −1.1 |
| 66 | 67 | 64 | 66 | 66 | 66 | 66 | 1.1 | 1.7 | −0.3 |
| 75 | 75 | 74 | 75 | 75 | 75 | 75 | 0.4 | 0.6 | −0.3 |
| 84 | 82 | 83 | 83 | 84 | 83 | 84 | 0.7 | 0.9 | −1.2 |
| 90 | 92 | 91 | 90 | 90 | 90 | 91 | 0.9 | 1.0 | 0.7 |
| 95 | 96 | 96 | 96 | 96 | 96 | 96 | 0.0 | 0.0 | 1.1 |
| 98 | 97 | 97 | 97 | 97 | 97 | 97 | 0.0 | 0.0 | −1.0 |
| R2 | 0.999 | 0.999 | 0.999 | 1.000 | 0.999 | 1.000 | |||
%Diff, percent difference of mean mutant V617F% from expected value; CV, coefficient of variation; Rep, replicate; SD, standard deviation; R2, R square calculation for back-calculated values when compared to expected values.
Table 2.
Inter-Assay Variability of Standard Curve Generated with Polycythemia Vera Patient DNA and Human Erythroleukemic 92.1.7 DNA diluted in Wild-Type Genomic DNA
| Back-calculated JAK2V617F standards (%) | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| V6I7F standard (%) | Run 1 | Run 2 | Run 3 | Run 4 | Run 5 | Run 6 | Run 7 | Run 8 | Mean | SD | CV | %Diff |
| 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | N/A | N/A |
| 2 | 2.0 | 1.9 | 2.2 | 2.4 | 1.9 | 2.4 | 1.7 | 1.9 | 2 | 0.3 | 15.0 | 0 |
| 4 | 3.7 | 4.2 | 3.5 | 4.1 | 4.2 | 4.3 | 3.6 | 3.5 | 4 | 0.4 | 10.3 | −3 |
| 8 | 7.1 | 7.5 | 6.9 | 7.2 | 7.5 | 7.5 | 6.5 | 7.4 | 7 | 0.4 | 5.6 | −10 |
| 11 | 10 | 12 | 11 | 11 | 12 | 11 | 10 | 11 | 11 | 0.7 | 6.4 | −1 |
| 19 | 17 | 18 | 17 | 18 | 18 | 19 | 19 | 20 | 18 | 1.1 | 6.0 | −4 |
| 28 | 28 | 28 | 27 | 27 | 28 | 29 | 27 | 27 | 28 | 0.7 | 2.5 | −1 |
| 38 | 40 | 38 | 39 | 40 | 38 | 40 | 40 | 40 | 39 | 1 | 2.5 | 3 |
| 47 | 49 | 49 | 49 | 47 | 49 | 46 | 49 | 47 | 48 | 1.2 | 2.5 | 3 |
| 56 | 55 | 55 | 55 | 54 | 55 | 54 | 54 | 53 | 54 | 0.7 | 1.3 | −3 |
| 66 | 65 | 65 | 65 | 67 | 65 | 66 | 66 | 67 | 66 | 0.8 | 1.2 | 0 |
| 75 | 75 | 76 | 74 | 75 | 76 | 76 | 75 | 76 | 75 | 0.6 | 0.8 | 1 |
| 84 | 84 | 84 | 84 | 83 | 84 | 84 | 84 | 83 | 84 | 0.4 | 0.5 | 0 |
| 90 | 90 | 90 | 90 | 90 | 90 | 90 | 90 | 90 | 90 | 0.1 | 0.1 | 0 |
| 95 | 95 | 96 | 96 | 96 | 96 | 96 | 96 | 95 | 96 | 0.3 | 0.3 | 1 |
| 98 | 98 | 97 | 97 | 97 | 97 | 98 | 97 | 98 | 97 | 0.3 | 0.3 | −1 |
| R2 | 0.999 | 0.999 | 0.999 | 0.999 | 0.999 | 0.999 | 0.999 | 0.999 | 1.000 | |||
FIG. 5.
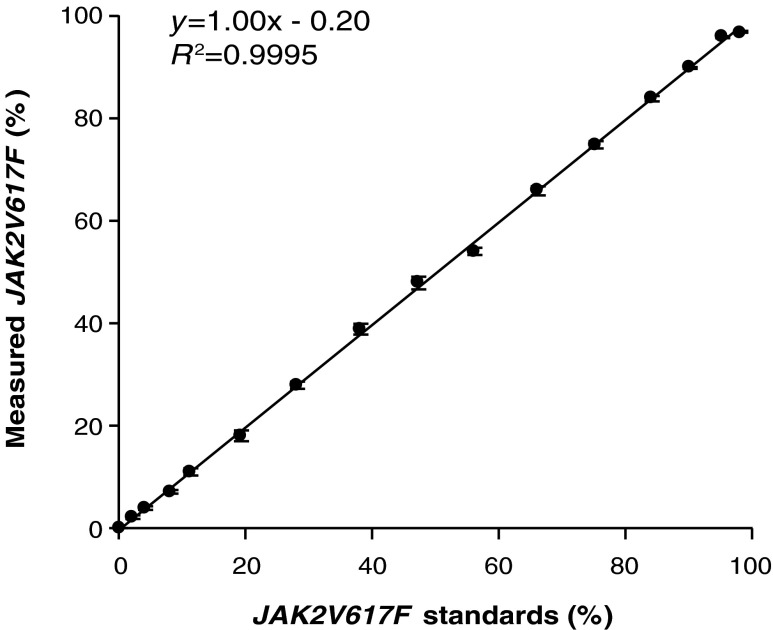
JAK2V617F SNaPshot assay. Comparison of expected and measured percentage JAK2V617F values from eight independent SNaPshot assays. Average values±standard deviation.
FIG. 6.
SNaPshot chromatograms showing WT, 2%, 4%, and 8% JAK2V617F samples. The WT JAK2 is shown in blue with red denoting the JAK2V617F.
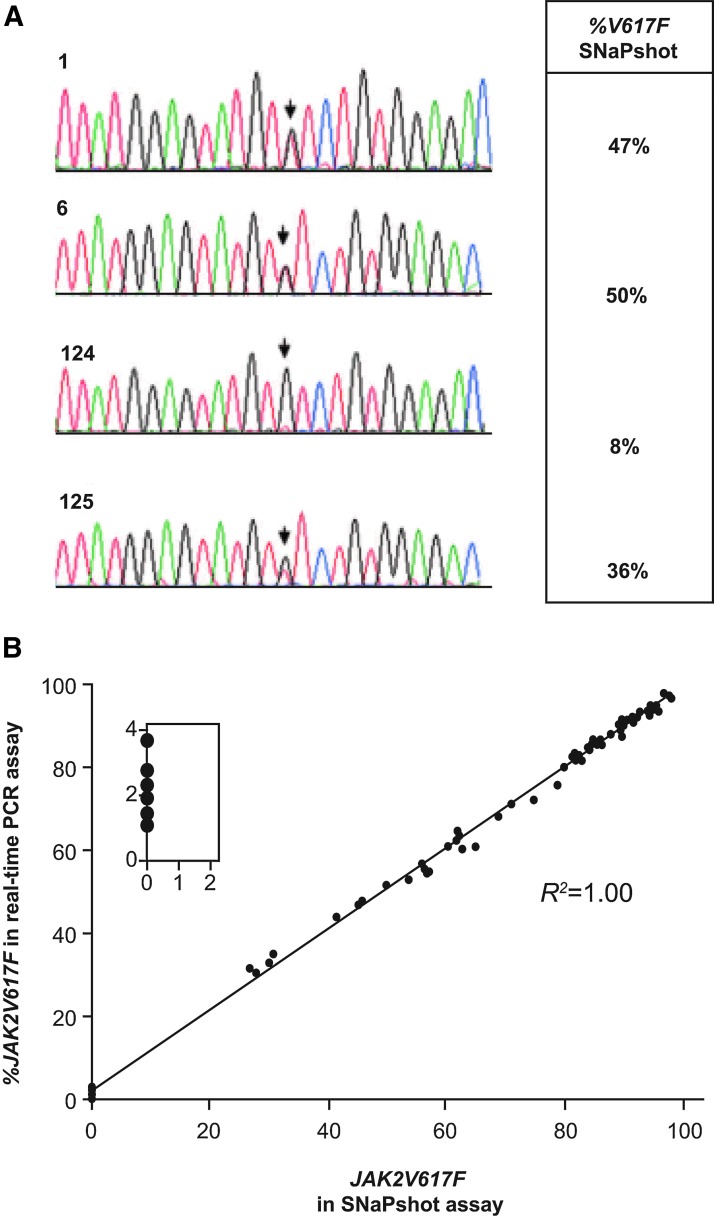
To confirm %V617F values reported by the SNaPshot assay, data from this assay were compared to standard dideoxy sequencing. For this analysis, we used PBMC DNAs from patients with PV and isolated colonies obtained from colony-forming assays. Representative data for two colonies that were called heterozygous are shown as well as data for two PBMC preparations used in the colony assays (Fig. 7A). In each case, the sequence chromatograms and SNaPshot data were consistent. The final validation of the V617F SNaPshot assay consisted of a comparison of 55 clinical samples that were run in both the SNaPshot and real-time PCR assays, with the two data sets being well correlated (Fig. 7B). Importantly, six JAK2V617F-negative samples that had an average percent JAK2V167F value of 2% in the real-time PCR assay were clearly negative in the SNaPshot assay (Fig. 7B, inset). This highlights the importance of the SNaPshot assay in our workflow since these could have been falsely called JAK2V617F-positive if relying solely on the real-time PCR. Because the SNaPshot assay is more labor-intensive and lower throughput (owing to the need for an upfront genomic PCR in addition to the SNaPshot PCR and phosphatase reactions), we reserved this assay for the quantitation of samples with less than 5% MUT allele burden as initially determined by real-time PCR.
FIG. 7.
Cross-platform validation of the JAK2V617F SNaPshot assay. (A) Dideoxy sequence analysis of two homozygous colonies from colony-forming assay performed with PV patient samples (1 and 6) and peripheral blood mononuclear cell DNA from the two PV patient samples used in the colony-forming assay (124 and 125). Corresponding percentage JAK2V617F data are shown from the SNaPshot assay. (B) Comparison of clinical data for 55 paired samples using the SNaPshot assay and pyrosequencing data from an independent laboratory. Inset shows samples that were <5% JAK2V617F in real-time PCR and negative in the SNaPshot assay.
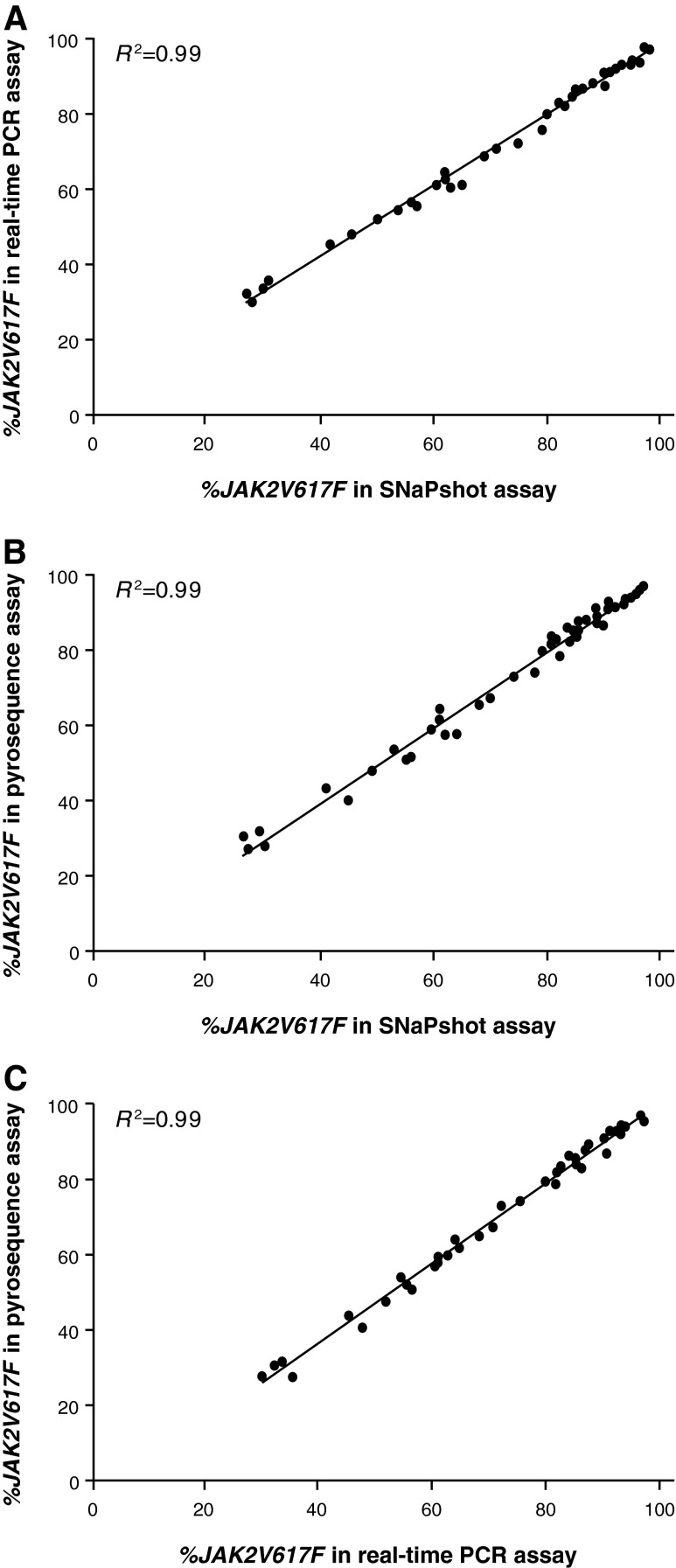
In subsequent analyses, we assessed data from 49 consecutive JAK2V617F-positive clinical samples that were run in the real-time PCR, pyrosequencing, and SNapShot assays with the samples not being preselected for JAK2V617F allele burden. There was an excellent correlation (R2=0.99) between all three assays (Fig. 8A–C). In addition, the average percent differences between separate measurements for the same sample were all below 3%. The excellent correlation between the pyrosequencing data, which is not dependent on a standard curve for measuring %V617F, and the SNaPshot data is an additional confirmation that we have correctly calibrated the standards used in these assays. Furthermore, these data confirm that %V617F data obtained with the SNaPshot assay have an excellent correlation with two widely used JAK2V617F assay formats.
FIG. 8.
Cross-comparison of three (A–C) assay formats. The same 49 clinical samples were compared across the real-time PCR, SNaPshot, and pyrosequencing assays.
Discussion
We described our approach for the characterization and cross-platform validation of a standard curve to enable the accurate quantitation of JAK2V617F MUT allele burden. Careful evaluation of the HEL 92.1.7 cell line indicated that there were approximately eight copies of the JAK2V617F allele per diploid genome, consistent with other reports (Quentmeier et al., 2006; Voelkner et al., 2008; Lippert et al., 2009). Furthermore, K562 cells are reported to have one copy of the WT JAK2 per diploid genome (Voelkner et al., 2008), an observation we have confirmed (data not shown). The use of HEL and K562 cells for standard curves without adjustment for copy number would lead to a calculated JAK2V617F allele burden that is artifactually eightfold lower. Furthermore, the practice of using uncorrected HEL/K562 standards in quantifying JAK2V617F changes during therapeutic intervention can dramatically overestimate changes in JAK2V671F allele burdens. Because of this potential pitfall, the importance of carefully evaluating one's standard curve samples by independent means and cross-validating with a method that does not rely on a standard curve cannot be overemphasized.
The real-time PCR assay for JAK2V617F was used as our primary screening assay for allele burden assessment. The assay is robust and requires one assay per sample without any upfront genomic PCR. When using a standard curve to calculate the percentage JAK2V617F of unknown samples, the assay is sensitive to a 5% MUT allele. We described the use of a second-tier SNaPshot assay for quantitatively measuring JAK2V617F allele burden in samples with less than 5% allele burden. Based on studies performed with a 16-point standard curve, the assay is quantitative to 2% V617F in a WT JAK2 background. SNaPshot assays are the basis for a number of qualitative genotyping assays; however, our data demonstrate that the assay is quantitative when used in conjunction with a standard curve. Although the SNaPshot assay is more sensitive than the real-time PCR assay, it requires two PCR reactions (genomic and SNaPshot) along with a phosphatase reaction after each PCR. These added steps increase the time requirements for the SNaPshot assay and contribute to its lower throughput and higher cost when compared with the real-time PCR assay.
A number of JAK2V617F genotyping assays have been previously described, including restriction length polymorphism analysis (Baxter et al., 2005), direct sequencing (Baxter et al., 2005; James et al., 2005; Kralovics et al., 2005; Levine et al., 2005), pyrosequencing (Jelinek et al., 2005; Jones et al., 2005), real-time PCR (James et al., 2006; Levine et al., 2006), allele-specific PCR (Jones et al., 2005; McClure et al., 2006), PCR melting curve analysis (Olsen et al., 2006), and alternately binding probe competitive PCR (Morishita et al., 2011). Although several investigators have described allele-specific PCR assays that are sensitive to less than 1% JAK2V617F (McClure et al., 2006; Lippert et al., 2009), these assay formats required two separate PCR assays for the MUT and WT alleles, which can double reagent costs for the assay. In addition, differences in PCR efficiencies between the two assays can potentially complicate data analysis.
Real-time PCR is widely used for JAK2V617F genotyping, and has the advantage that both WT and MUT can be assessed in a single reaction. However, careful upfront work is required for real-time PCR assays in the design of probes that differentiate the WT and MUT allele with minimal cross-reactivity. The reported sensitivity for the real-time PCR assays is 1–5% (Huijsmans et al., 2011); however, we found that with published as well as internally designed primer/probes, sufficient cross-reactivity of the probes prevented accurate measurement of JAK2V617F levels less than 5% when calculating unknowns using standard curve formulas. Recent work suggests that blocking WT amplification with a peptide nucleic acid oligonucleotide can improve the lower limit of detection (Huijsmans et al., 2011). The SNaPshot assay described here has a limit of quantification of 2% and can be performed on a standard capillary DNA sequencer found in nearly all molecular biology laboratories. In addition, the SNaPshot assay platform provides the ability to multiplex assays so that multiple mutations or alleles could be assessed in a single reaction. Although data from the real-time PCR and SNaPshot assay showed an excellent correlation with data on parallel samples assayed using a pyrosequencing-based assay, this latter assay requires dedicated instrumentation that may not be present in many molecular biology laboratories. In addition, the pyrosequencing-based assay has a sensitivity of 5–10% JAK2V617F (Jelinek et al., 2005), leading to an increased number of confirmatory assays when compared with the real-time PCR assay.
In conclusion, we described an approach to standard curve validation and calibration that yields highly concordant JAK2V617F allele burden measurements in MPN patient samples across three different assay formats: real-time PCR, SNaPshot, and pyrosequencing. Additionally, we have developed a two-tiered approach to clinical sample analysis using an upfront real-time PCR screening assay and a confirmatory SNaPshot assay for samples that are <5% JAK2V617F. This approach not only leverages the throughput of real-time PCR for efficiently analyzing allele burden in the majority of clinical samples, it also maximizes sensitivity using SNaPshot for the accurate assessment of JAK2V617F in the minority of samples with low abundance or absent MUT alleles. These assays and standards have been used to support Phase I/II and III ruxolitinib (Jakafi) clinical studies in myelofibrosis (Verstovsek et al., 2010, 2012; Harrison et al., 2012) and may lay the groundwork for future standardization in the field. As therapeutic interventions (JAK inhibitor combinations, interferon, bone marrow transplant, etc.) aimed at affecting the molecular aspects of disease are explored, robust, sensitive, and quantitative assays for V617F allele burden, such as those described herein, will play an important role in assessing molecular response and durability.
Acknowledgments
We thank Sheryl Weinerman of Evidence Scientific Solutions for editorial assistance. Editorial assistance was funded by Incyte Corporation.
Author Disclosure Statement
Keyur Patel, Rajylakshmi Luthra: No conflict of interests to disclose. Srdan Verstovsek reports receiving grant support through his institution from Incyte, Exelixis, Celgene, NS Pharma, Infinity Pharmaceuticals, SBIO, Lilly Oncology, AstraZeneca, Geron, Bristol-Myers Squibb, YM BioSciences, Gilead, and Roche. Paul Collier, Paul Waeltz, Mark Rupar, Phillip C.C. Liu, Gregory Hollis Reid Huber, and Timothy C. Burn are employees of Incyte Corporation.
References
- Baxter EJ. Scott LM. Campbell PJ, et al. Acquired mutation of the tyrosine kinase JAK2 in human myeloproliferative disorders. Lancet. 2005;365:1054–1061. doi: 10.1016/S0140-6736(05)71142-9. [DOI] [PubMed] [Google Scholar]
- Harrison C. Kiladjian JJ. Al-Ali HK, et al. JAK inhibition with ruxolitinib versus best available therapy for myelofibrosis. N Engl J Med. 2012;366:787–798. doi: 10.1056/NEJMoa1110556. [DOI] [PubMed] [Google Scholar]
- Huijsmans CJ. Poodt J. Savelkoul PH, et al. Sensitive detection and quantification of the JAK2V617F allele by real-time PCR blocking wild-type amplification by using a peptide nucleic acid oligonucleotide. J Mol Diagn. 2011;13:558–564. doi: 10.1016/j.jmoldx.2011.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ihle JN. Gilliland DG. Jak2: normal function and role in hematopoietic disorders. Curr Opin Genet Dev. 2007;17:8–14. doi: 10.1016/j.gde.2006.12.009. [DOI] [PubMed] [Google Scholar]
- James C. Delhommeau F. Marzac C, et al. Detection of JAK2 V617F as a first intention diagnostic test for erythrocytosis. Leukemia. 2006;20:350–353. doi: 10.1038/sj.leu.2404069. [DOI] [PubMed] [Google Scholar]
- James C. Ugo V. Le Couedic JP, et al. A unique clonal JAK2 mutation leading to constitutive signalling causes polycythaemia vera. Nature. 2005;434:1144–1148. doi: 10.1038/nature03546. [DOI] [PubMed] [Google Scholar]
- Jelinek J. Oki Y. Gharibyan V, et al. JAK2 mutation 1849G>T is rare in acute leukemias but can be found in CMML, Philadelphia chromosome-negative CML, and megakaryocytic leukemia. Blood. 2005;106:3370–3373. doi: 10.1182/blood-2005-05-1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones AV. Kreil S. Zoi K, et al. Widespread occurrence of the JAK2 V617F mutation in chronic myeloproliferative disorders. Blood. 2005;106:2162–2168. doi: 10.1182/blood-2005-03-1320. [DOI] [PubMed] [Google Scholar]
- Kralovics R. Passamonti F. Buser AS, et al. A gain-of-function mutation of JAK2 in myeloproliferative disorders. N Engl J Med. 2005;352:1779–1790. doi: 10.1056/NEJMoa051113. [DOI] [PubMed] [Google Scholar]
- Levine RL. Belisle C. Wadleigh M, et al. X-inactivation-based clonality analysis and quantitative JAK2V617F assessment reveal a strong association between clonality and JAK2V617F in PV but not ET/MMM, and identifies a subset of JAK2V617F-negative ET and MMM patients with clonal hematopoiesis. Blood. 2006;107:4139–4141. doi: 10.1182/blood-2005-09-3900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine RL. Wadleigh M. Cools J, et al. Activating mutation in the tyrosine kinase JAK2 in polycythemia vera, essential thrombocythemia, and myeloid metaplasia with myelofibrosis. Cancer Cell. 2005;7:387–397. doi: 10.1016/j.ccr.2005.03.023. [DOI] [PubMed] [Google Scholar]
- Levine RL. Wernig G. Role of JAK-STAT signaling in the pathogenesis of myeloproliferative disorders. Hematol Am Soc Hematol Educ Program. 2006;510:233–239. doi: 10.1182/asheducation-2006.1.233. [DOI] [PubMed] [Google Scholar]
- Lippert E. Girodon F. Hammond E, et al. Concordance of assays designed for the quantification of JAK2V617F: a multicenter study. Haematologica. 2009;94:38–45. doi: 10.3324/haematol.13486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClure R. Mai M. Lasho T. Validation of two clinically useful assays for evaluation of JAK2 V617F mutation in chronic myeloproliferative disorders. Leukemia. 2006;20:168–171. doi: 10.1038/sj.leu.2404007. [DOI] [PubMed] [Google Scholar]
- Morishita S. Komatsu N. Kirito K, et al. Alternately binding probe competitive PCR as a simple, cost-effective, and accurate quantification method for JAK2V617F allele burden in myeloproliferative neoplasms. Leuk Res. 2011;35:1632–1636. doi: 10.1016/j.leukres.2011.06.016. [DOI] [PubMed] [Google Scholar]
- Olsen RJ. Tang Z. Farkas DH, et al. Detection of the JAK2V617F mutation in myeloproliferative disorders by melting curve analysis using the LightCycler System. Arch Pathol Lab Med. 2006;130:997–1003. doi: 10.5858/2006-130-997-DOTJMI. [DOI] [PubMed] [Google Scholar]
- Quentmeier H. MacLeod RA. Zaborski M, et al. JAK2 V617F tyrosine kinase mutation in cell lines derived from myeloproliferative disorders. Leukemia. 2006;20:471–476. doi: 10.1038/sj.leu.2404081. [DOI] [PubMed] [Google Scholar]
- Steensma DP. JAK2 V617F in myeloid disorders: molecular diagnostic techniques and their clinical utility: a paper from the 2005 William Beaumont Hospital Symposium on Molecular Pathology. J Mol Diagn. 2006;8:397–411. doi: 10.2353/jmoldx.2006.060007. quiz 526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verstovsek S. Kantarjian H. Mesa RA, et al. Safety and efficacy of INCB018424, a JAK1 and JAK2 inhibitor, in myelofibrosis. N Engl J Med. 2010;363:1117–1127. doi: 10.1056/NEJMoa1002028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verstovsek S. Mesa RA. Gotlib J, et al. A double-blind, placebo-controlled trial of ruxolitinib for myelofibrosis. N Engl J Med. 2012;366:799–807. doi: 10.1056/NEJMoa1110557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voelkner MH. Ben-Ezra J. West AC, et al. Performance evaluation of a commercially available quantitative allele specific real time PCR assay for the Janus kinase 2 V617F (JAK-2) mutation in chronic myeloproliferative disorders (CMD) J Mol Diagn. 2008;10:557–626. Abstract H525. [Google Scholar]