Abstract
Regulation of protein synthesis is of fundamental importance to cells. It has a critical role in the control of gene expression, and consequently cell growth and proliferation. The importance of this control is supported by the fact that protein synthesis is frequently upregulated in tumor cells. The major point at which regulation occurs is the initiation stage. Initiation of translation involves the interaction of several proteins to form the eIF4F complex, the recognition of the mRNA by this complex, and the subsequent recruitment of the 40S ribosomal subunit to the mRNA. This results in the formation of the 48S complex that then scans the mRNA for the start codon, engages the methionyl-tRNA and eventually forms the mature 80S ribosome which is elongation-competent. Formation of the 48S complex is regulated by the availability of individual initiation factors and through specific protein-protein interactions. Both of these events can be regulated by post-translational modification by ubiquitin or Ubls (ubiquitin-like modifiers) such as SUMO or ISG15. We provide here a summary of translation initiation factors that are modified by ubiquitin or Ubls and, where they have been studied in detail, describe the role of these modifications and their effects on regulating protein synthesis.
Keywords:
Abbreviations
- eIF
eukaryotic initiation factor
- PABP
poly(A) binding protein
- 4E-BP
eIF4E-binding protein
- mTORC
mechanistic target of rapamycin
- Ubl
ubiquitin-like protein
- HDAC
histone deacetylase
Introduction
Initiation of protein synthesis
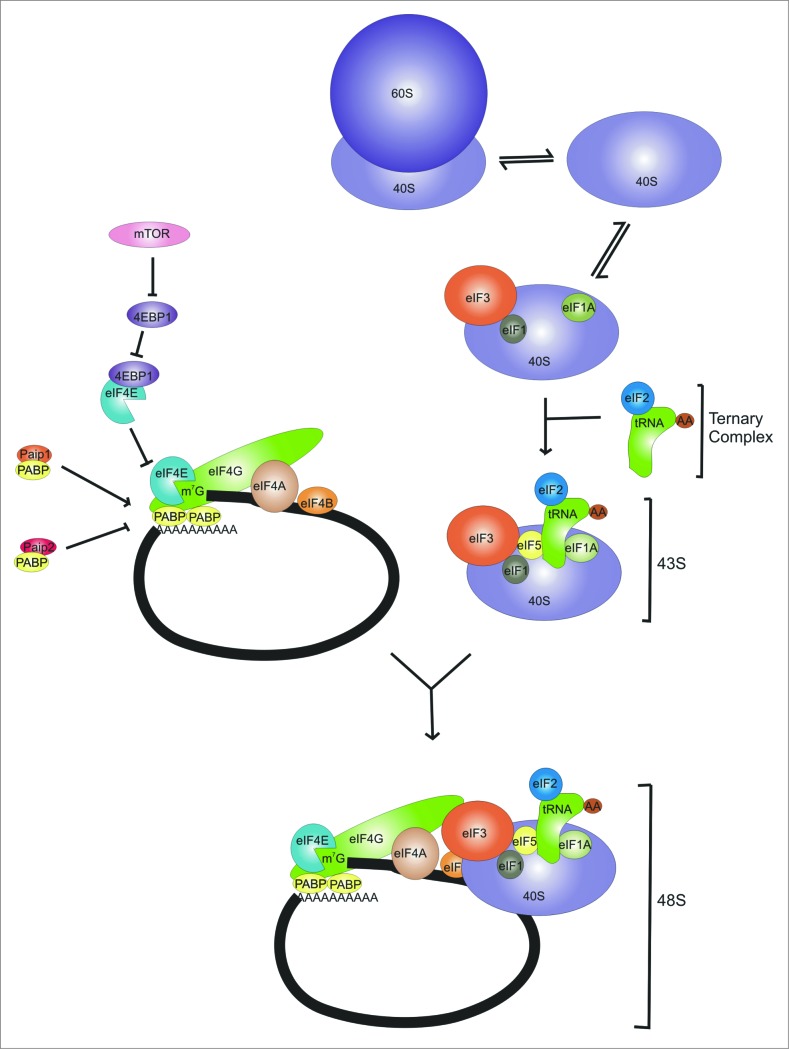
Protein synthesis is of fundamental importance in cells and its regulation is crucial for the continued viability of organisms. The process comprises 3 stages: initiation, elongation and termination. Of these, initiation is generally considered to be one of the major regulatory steps of gene expression in mammalian cells. Initiation requires the function of a number of translation initiation factors (Fig. 1 ), several of which have key roles in cell survival and oncogenesis. These proteins modulate the binding of mRNA to the ribosome, a process facilitated by the assembly of the cap binding protein (eIF4E), a helicase (eIF4A) and a scaffold protein (eIF4G), to form the eIF4F complex (eIF4E/eIF4A/eIF4G).1-3 The eIF4G scaffold protein possesses domains that interact with eIF4E, eIF4A, eIF3 and the poly(A) binding protein, PABP.1-4 PABP itself is regulated by interaction with other proteins; binding of Paip1 to PABP stimulates protein synthesis while interaction with Paip2 is inhibitory to translation.5,6 The activity of the eIF4F complex is regulated by a family of proteins, the eIF4E binding proteins (4E-BPs). Using a conserved motif, 4E-BPs compete with eIF4G for a common surface on eIF4E and inhibit eIF4F assembly. In mammalian cells, activation of the mechanistic target of rapamycin (mTORC1) leads to phosphorylation of 4E-BP1 in a hierarchical manner. This promotes protein synthesis by releasing eIF4E and enabling eIF4F complex assembly on the m7GTP cap of mRNA, mediating 40S ribosomal subunit binding by a bridging interaction between eIF4G and eIF3.1-3
Figure 1.
For figure legend, see page e959366-3. Figure 1. (See previous page). Formation of the 48S preinitiation complex. eIF1, 1A and 3 interact with the 40S ribosomal subunit. This then interacts with eIF5 and the ternary complex (eIF2-GTP-Met-tRNA) to form the 43S complex. In parallel, eIF4E and eIF4A are recruited by eIF4G to form the eIF4F complex. The availability of eIF4E is controlled by 4E-BP1, which in turn is regulated by phosphorylation by mTOR. The eIF4F complex binds to the cap on mRNA along with Poly(A)-binding protein (PABP) and eIF4B. PABP is regulated via interactions with 2 PABP proteins, PAIP1 and PAIP2. The 43S complex then binds close to the cap from where it can scan the mRNA for the start codon.
In most organisms there is more than one isoform of most of these translation initiation factors. For example, there are 3 isoforms of eIF4A, eIF4G and PABP.7-9 In some cases the functions of the isoforms are indistinguishable, in others there are indications that the different isoforms display mRNA-specific regulation.7-9 Further work will be required to uncover the full range of functions and specificities of these isoforms.
Ubiquitin like proteins
Ubiquitin-like proteins (Ubls) comprise a family of structurally related proteins. The different members of the family share sequence similarities, and in particular the proteins contain a conserved β-grasp fold consisting of 5 β sheets and one α helix.10 Ubiquitin is a 76 amino acid protein and is the most highly conserved member of the Ubl family, with 96% identity between yeast and human ubiquitin. SUMO (small ubiquitin-like modifier) is less conserved between species and contains a longer, more variable N-terminal extension than ubiquitin being around 100–110 amino acids in total length.11 ISG15, between 155–165 amino acids in length, contains 2 ubiquitin-like domains.12 It was the first member of the family to be identified and, unlike ubiquitin and SUMO, is present only in vertebrates. The gene was so named because it was observed to be an interferon stimulated gene encoding a 15 kDa protein.13 Most members of the Ubl family are synthesized as precursor proteins that need to be processed to a mature form to reveal a di-glycine motif at the C-terminus that is required for activation and subsequent conjugation of the Ubl to target proteins. The exception to this is ISG15 in fish and bovine species where the protein is synthesized in the mature form.14 Ubls are attached to one or more lysine residues in target proteins. There are no known consensus sequences for conjugation sites for ubiquitin and ISG15. However SUMO is frequently, although not always, attached to lysine residues present within the consensus sequence ψKxE, where ψ = a hydrophobic amino acid and x is any amino acid.11
Ubiquitylation
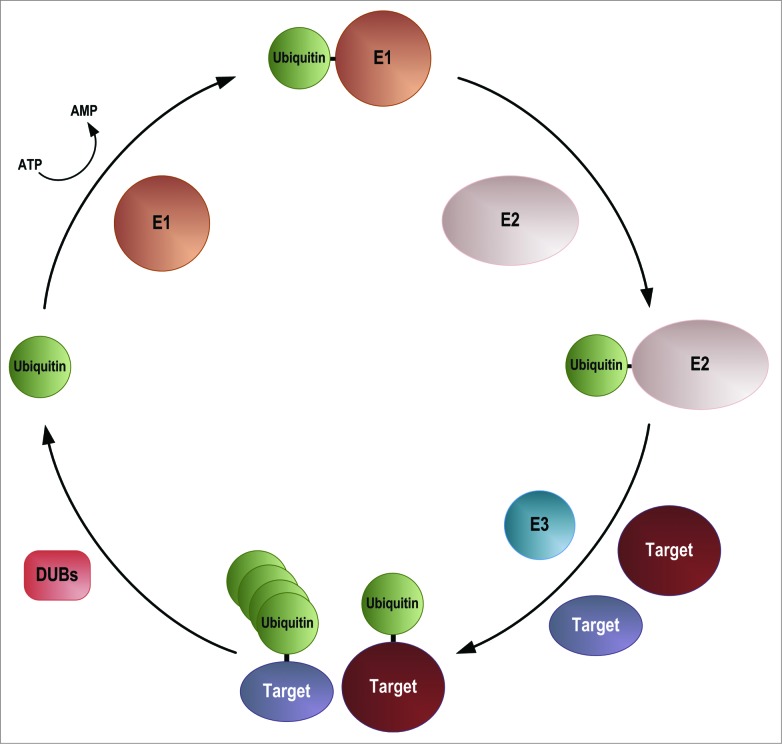
Ubiquitin can be covalently attached to lysine residues in target proteins as a monomer or in the form of chains. This occurs via the activity of a number of proteins, the E1 (ubiquitin activating enzyme), E2 (ubiquitin conjugating enzyme) and E3 (ubiquitin ligase) proteins (Fig. 2 ). In most organisms there is a single E1, around 40 E2s and hundreds of E3s (reviewed in15,16). Ubiquitin is produced as a precursor protein that is processed to the mature form by one of a small number of specific ubiquitin proteases, to reveal a diglycine motif at the C-terminus. Ubiquitin is then activated in an ATP-dependent manner, by the formation of a thioester bond between the C-terminal glycine residue and a cysteine residue on the E1 activating enzyme. From here it is passed to an E2 ubiquitin conjugating enzyme, again, via the formation of a thioester bond between the C-terminal glycine residue and a cysteine residue. Attachment of ubiquitin requires one of a large number of E3 ubiquitin ligases, which in many cases interact directly with target proteins, but which in some instances interact with targets via an adaptor protein. In the main, the E3s provide the specificity for the modification. Ubiquitin chain formation occurs via lysine residues within ubiquitin itself, and also requires the activities of the E1, E2 and E3 enzymes. The most common linkages are via K11, K48 and K63.17,18 Ubiquitin can be removed from targets by the actions of deubiquitinating enzymes (DUBs). Ubiquitylation has 2 main roles: targeting of proteins for proteolysis and modification of protein function. The best studied role of ubiquitylation is its targeting of proteins for proteasome-mediated degradation. This involves the recognition of K11- and K48-linked ubiquitin chains by the 26S proteasome.19 However, there is a rapidly expanding literature on other roles for ubiquitylation. For example ubiquitylation of PCNA is required for the recruitment of an error-prone polymerase to undertake translesion DNA synthesis e.g,20 while ubiquitylation of membrane proteins is required for endocytosis and ubiquitylation of PIN2 is required for vacuolar sorting (reviewed in21). In these cases the modification involves a single ubiquitin or K63-linked chains.
Figure 2.
Ubiquitylation pathway. E1 = Ubiquitin activating enzyme, E2 = ubiquitin conjugating enzyme, E3 = ubiquitin ligase, DUB = deubiquitylating enzyme. Ubiquitin is activated by the formation of a ubiquitin-adenylate before forming a thioester bond with a cysteine residue in the E1 ubiquitin activating enzyme. Ubiquitin is passed to an E2 ubiquitin conjugating enzyme, again forming a thioester bond. Target proteins are recognized by E3 ubiquitin ligases, either directly or via an adaptor, and ubiquitin is attached via the formation of an ε-amino bond. Ubiquitin can be attached to target proteins either as a monomer, or in the form of ubiquitin chains. Ubiquitin can be removed from target proteins by the action of one of a number of DUBs.
Sumoylation
The process of sumoylation is very similar to that of ubiquitylation, involving SUMO-specific E1 (SUMO activating enzyme), E2 (SUMO conjugating enzyme) and E3 (SUMO ligase) proteins.11 There is a single E1 (a heterodimer), a single E2 (Ubc9) and to date around 12 E3s have been identified. Unlike ubiquitylation, an E3 is not always required for modification, as the E2 is in some cases sufficient, and can provide a degree of target specificity.22 Like ubiquitin, SUMO can be attached to proteins either as a monomer or in the form of poly-SUMO chains.11 Sumoylation affects protein-protein interactions,23,24 protein activity25 and protein localization.26 In addition, SUMO chains interact with STUbLs (SUMO-targeted ubiquitin ligases) that bring about ubiquitylation of the target protein and associated SUMO chains, resulting in proteasome–mediated proteolysis.27
ISGylation
ISG15 is conjugated to target proteins in a manner similar to that of ubiquitin and SUMO.28 ISG15 expression and modification (ISGylation) are activated by Type I interferon (IFN), which is one of a number of critical cytokines in the innate immune response. As is the case for ubiquitin and SUMO, there are proteases that are specific for processing ISG15 and deconjugating it from target proteins (e.g., USP43,29) and a specific E1 enzyme for ISG15.29 However, some of the E2s (e.g., UbcH8) and E3s (e.g., Efp—the partner of UbcH8, and HHARI—the human homolog of Drosophila ariadne) involved in ISGylation also appear to be involved in ubiquitylation.30,31
Identification of Ubl Attachment Sites and the Roles of Modification
Early methods for the identification of modified sites involved site-directed mutagenesis of individual lysine residues in target proteins, followed by analysis in vitro or in vivo to determine whether modification still occurred. While this has been successful in some cases (e.g.,32) in many cases it has been problematic since other lysine residues are frequently used instead of the normal sites in the mutant proteins. More recently, mass spectrometry has been used successfully for site identification (e.g.,33). This involves the cleavage of modified proteins by trypsin or other suitable protease to release peptides from the target. This method is facilitated by having a protease cleavage site close to the C-terminal diglycine motif attached to the target, so that only a few extra amino acids remain attached to the modified site. Modification sites are thus detected by the identification of peptides that are increased in Mr by an amount dependent on the position of the cleavage site within the Ubl.
Analysis of the role of the modifications is hampered by the fact that frequently, only low levels of modified forms are observed in cells. The reason for this could be that the modifications are transient, are labile, or as in the case of poly-ubiquitylation and poly-sumoylation, are targeting the protein for proteasome-mediated destruction. It is also possible that modification may be confined to target molecules in a particular cellular location. Additionally, it is proposed that this form of post-translational modification is not like modifications such as phosphorylation—i.e., an on/off switch. For example, in the case of SUMO, it is proposed that in some cases modification results in a change in conformation of the target protein that is maintained even after desumoylation occurs. Thus analysis of the roles of these modifications has lagged behind analysis of the function of other types of modifications.
Identification of the roles of the modifications has been undertaken, in the main using in vitro assays to look at relative binding abilities of wild type and unsumoylatable mutant proteins for their binding partners e.g.,32 or by introduction of mutant coding sequences into cells to determine the effect of inability to modify a particular protein. This is relatively straightforward in yeast where a mutant copy can be integrated in the genome as the sole copy of the coding sequence e.g.,34 In mammalian cells, the mutant sequence can be introduced by transfection, but is dependent on having cells where the gene has been knocked out or where siRNA depletion is efficient. Depletion of the any of the enzymes in the conjugation pathway would be likely to affect multiple targets and would not be appropriate.
Role of Modification by Ubiquitin or Ubls in Translation Initiation Factors
A series of recent proteomic screens have identified numerous translation initiation factors that are modified by either ubiquitin or SUMO, or in many cases, by both (Table 1 ). Additionally, some of the screens have identified the lysine residues required for the modification. Early studies involved the overexpression of the modifier, but recently more refined methods using diGly capture techniques have been used to identify sites when the modifier is expressed at endogenous levels e.g.35,36 These studies use mass spectrometry to identify diGly-modified peptides obtained by trypsin digestion of cellular proteins. A list of modified sites can be found at PhosphoSitePlus37 (http://www.phosphosite.org/home). In many cases, individual lysine residues are identified as a single ‘hit’, making them less likely target sites than lysine residues that are highly represented, as for example is observed in eIF4A and eIF4G proteins.
Table 1.
Proteins identified in proteomic screens as being modified by ubiquitin or SUMO
| Initiation factor | Ubiquitin | SUMO | Reference |
|---|---|---|---|
| eIF1A | Hs Ubiquitin*Mm Ubiquitin | Rn SUMO-3 | 35,36,58,59 |
| eIF2A | Hs Ubiquitin*Mm Ubiquitin | Hs SUMO-2*At SUMO | 35,55,59,80 |
| eIF2α | Hs Ubiquitin*Mm Ubiquitin | Dm SUMO | 36,56,59 |
| eIF2B-β | Hs Ubiquitin*Mm Ubiquitin | Hs SUMO-1/2 | 36,59,81 |
| eIF2β | Hs Ubiquitin*Mm Ubiquitin | At SUMO*Sc SUMO-1 | 36,59,80,82 |
| eIF2 subunit 1 | Hs Ubiquitin*Mm Ubiquitin | Rn SUMO-3 | 35,58,59 |
| eIF2γ | Hs Ubiquitin*Mm Ubiquitin | Dm SUMO*Hs SUMO-1*Hs SUMO-2/3*Sc SUMO | 35,36,56,57,59,82,83, * |
| eIF5B* | Hs Ubiquitin*Mm Ubiquitin | Hs SUMO-2*Hs SUMO-1* | 55,59,61(A) |
| eIF3A | Hs Ubiquitin*Mm Ubiquitin*Rn Ubiquitin | Hs SUMO-2*Hs SUMO-1 | 36,55,59,61(B)* |
| eIF3B | Hs Ubiquitin*Mm Ubiquitin | Hs SUMO-2 | 35,55,59 |
| eIF3C | Hs Ubiquitin*Mm Ubiquitin | Hs SUMO 1/2 | 35,59,81 |
| eIF3D | Hs Ubiquitin*Mm Ubiquitin | Rn SUMO-3 | 35,58,59 |
| eIF3E | Hs Ubiquitin*Mm Ubiquitin | Hs SUMO-1/2 | 35,59,81 |
| eIF3F | Hs Ubiquitin*Mm Ubiquitin | 59,74(C) | |
| eIF3G | Hs Ubiquitin*Mm Ubiquitin | 35,36,59 | |
| eIF3H | Hs Ubiquitin*Mm Ubiquitin*Rn Ubiquitin | 35,36,59(D)* | |
| eIF3I | Hs Ubiquitin | Sc SUMO*Hs SUMO-1/2 | 35,36,59,60,82–84 |
| eIF3J | Hs Ubiquitin*Mm Ubiquitin | 36,59 | |
| eIF3K | Hs Ubiquitin*Mm Ubiquitin | 35,36* | |
| eIF3L | Hs Ubiquitin*Mm Ubiquitin | 35,59 | |
| eIF3M | Hs Ubiquitin*Mm Ubiquitin | Hs SUMO-1 | 35,59,83 |
| eIF3X | Hs SUMO-2 | 55 | |
| eIF4A1 | Hs Ubiquitin*Mm Ubiquitin*Rn Ubiquitin | Dm SUMO*Rn SUMO-3*Hs SUMO-1/2*At SUMO | 35,36,55–62(D) |
| eIF4A2 | Hs Ubiquitin*Mm Ubiquitin | Hs SUMO-1 | 35,59,61 |
| eIF4E | Hs Ubiquitin*Mm Ubiquitin | Hs SUMO-1 | 36,46,59 |
| eIF4GI | Hs Ubiquitin*Mm Ubiquitin | Hs SUMO-1/2 | 36,57,59,61 |
| eIF4GII | Hs Ubiquitin | 35 | |
| eIF4GIII | Hs Ubiquitin*Mm Ubiquitin | 35,36 | |
| eIF5A | Hs Ubiquitin*Mm Ubiquitin*Rn Ubiquitin | Hs SUMO-1/2 | 35,59,83(D) |
| PABP1 | Hs Ubiquitin*Mm Ubiquitin | Hs SUMO-2*Sc SUMO | 35,55,57,59,85 |
| PABP4 | Hs Ubiquitin*Mm Ubiquitin | Hs SUMO-2 | 35,55,59 |
Hs: human, Rn: rat, Mm: mouse, Sc: S. cerevisiae, At: Arabidospsis. (A) (2010) CST Curation Set: 9913; Year: 2010; SILAC: N; Biosample/Treatment: AMO-1(cell line)/Velcade; Disease: -; Specificity of Antibody Used to Purify Peptides prior to MS2: anti-UbK Antibody Used to Purify Peptides prior to MS2: Ubiquitin (D4A7A10) XP(R) Rabbit mAb Cat#: 3925, PTMScan(R) Ubiquitin Branch Motif (K-e-GG) Immunoaffinity Beads Cat#: 1990. (B) (2008) CST Curation Set: 3970; Year: 2008; SILAC: N; Biosample/Treatment: brain(tissue)/untreated; Disease: -; Specificity of Antibody Used to Purify Peptides prior to MS2: anti-UbK. (C) (2009) CST Curation Set: 8668; Year: 2009; SILAC: N; Biosample/Treatment: RPMI-8266(cell line)/Velcade; Disease: -; Specificity of Antibody Used to Purify Peptides prior to MS2: anti-UbK Antibody Used to Purify Peptides prior to MS2: Ubiquitin (D4A7A10) XP(R) Rabbit mAb Cat#: 3925, PTMScan(R) Ubiquitin Branch Motif (K-e-GG) Immunoaffinity Beads Cat#: 1990. (D) (2007) CST Curation Set: 3578; Year: 2007; SILAC: N; Biosample/Treatment: brain(tissue)/ischemia and Reperfusion; Disease: -; Specificity of Antibody Used to Purify Peptides prior to MS2: anti-UbK.
More detailed studies on the role of modification of a number of the individual proteins by ubiquitin, SUMO and in one case, ISG15 have also been reported. We summarize here what is known about the roles of these post-translational modifications and how they might affect translation rates in mammalian cells.
eIF4E
Regulation of eIF4E levels is important for normal cell growth, as disruption of its expression or its over-production leads to aberrant cell growth or oncogenesis.38 Additionally, eIF4E protein levels increase during differentiation e.g.,39 eIF4E is both mono- and poly-ubiquitylated40,41 and this has been demonstrated to occur mainly on K159.40 This modification is enhanced by the E3 ubiquitin ligase, Chip (carboxy terminus of Hsp-70 interacting protein) which is known to have a role in regulating protein quality control.42 A mutant form of eIF4E that is unable to interact with eIF4G or 4E-BP1 is more highly ubiquitylated than wild type eIF4E. This results in increased degradation by the proteasome of the mutant form, consistent with a role for ubiquitylation of eIF4E in a quality control process, removing inactive forms of the protein from the cell.40 A role for ubiquitylation in quality control is supported by a number of observations. First, that binding of eIF4E to 4E-BP1 (eIF4E binding protein that is also regulated by ubiquitylation—see below) suppresses ubiquitylation and degradation and that only non-ubiquitylated eIF4E binds eIF4G. Second, overexpression of 4E-BP1 prevents ubiquitin-mediated degradation of eIF4E. Third, heat shock (45°C 10 min, conditions that would result in a degree of protein misfolding) also induces ubiquitylation of eIF4E, as does exposure to another form of stress, cadmium chloride.41
While poly-ubiquitylation clearly has a role in targeted destruction of eIF4E, little work has been performed to determine whether there is a different role for mono-ubiquitylation in regulating levels or subcellular localization of eIF4E. In contrast, the biological significance of eIF4E phosphorylation and its effect on translation have been studied over many years; however, the role of phosphorylation in modulating the activity of the protein is still not completely understood, although enhanced levels of eIF4E phosphorylation are associated with a number of human tumors.43,44 Biophysical studies have suggested that phosphorylation of eIF4E decreases its affinity for the mRNA cap of mRNA, possibly allowing rapid recycling of eIF4E between competing mRNAs.45 However, it has also been suggested that phosphorylation of S209 causes a retractable salt bridge to form with K159 (the ubiquitylation site) which leads to increased binding of capped mRNA.40 Mutation of K159 to alanine but not arginine, reduces association with cap analogs, indicating that a positive charge is required at this position. Despite the fact that the K159R mutant cannot be ubiquitylated, it has been proposed that mono-ubiquitylation may stabilize the distance between S209 and K159, or that ubiquitin itself may form part of the bridge between S209 and K159.40
eIF4E is also modified by SUMO,32,46 in a process that is promoted by HDAC2 (histone deacetylase 2).46 Sumoylation occurs on several lysine residues, namely K36, 49, 162, 206 and 212. Interestingly, unlike what has been observed with a number of other proteins, such as IκBα and PCNA,34,47 sumoylation and ubiquitylation of eIF4E do not occur on the same lysine residues. Sumoylation of eIF4E is dependent on phosphorylation, but the reverse is not true: inability to sumoylate eIF4E does not affect its ability to be phosphorylated.32 Sumoylation results in the induction of translation of a subset of mRNAs required for cell proliferation and apoptosis. A mutant form of eIF4E that cannot be sumoylated is still able to bind m7GTP, indicating that cap-binding is unaffected. However, compared with wild type protein, the mutant form binds significantly better to 4E-BP1 than it does to eIF4G, and is unable to form stable eIF4F complexes. It has been suggested that sumoylation induces a conformational change in eIF4E producing a change in interaction surfaces resulting in release from 4E-BP1 and promoting interaction with eIF4G. The inability of the mutant protein to be sumoylated results in an increase in the amount of eIF4E interacting with 4E-BP1.32 While overexpression of wild type eIF4E in NIH-3T3 cells results in increased expression of eIF4E-regulated genes, this is not observed when unsumoylatable eIF4E is overexpressed.32 At this time is unclear whether sumoylation of eIF4E has any effect of global rates of translation or rates of export of specific mRNAs from the nucleus.
4EHP
4EHP, also known as eIF4E2, binds to the m7GTP cap in a manner similar to that of eIF4E. However, unlike eIF4E, it does not bind eIF4G and therefore does not allow ribosome recruitment. It thus competes with eIF4E for the mRNA and prevents translation.48 It is targeted for ubiquitylation49 and interestingly, also for modification with another Ubl, ISG15.50 Curiously, the E3 ligase HHARI, which has recently been shown to be a marker of cellular proliferation,51 stimulates both ubiquitylation and ISGylation of 4EHP.49,50 Proteomic studies have identified K239 as a ubiquitylation site, but this has not been verified in a detailed study. In contrast, ISGylation, which occurs on K134 and K222, has been analyzed in some detail.50 Binding studies indicate that ISGylated 4EHP has a higher affinity for m7GTP than the unmodified form. It has been proposed that this modification is used by cells to inhibit translation of specific mRNAs in innate immune responses. Interestingly, despite its similarity to 4EHP, eIF4E is not ISGylated.
4E-BP Family
The eIF4E binding proteins (4E-BPs) are key regulators of protein synthesis.1-3 As their name suggests, they function by interacting with eIF4E. This inhibits eIF4E function by preventing it from interacting with eIF4G to form the mature eIF4F complex. The 4E-BP proteins are phosphorylated following activation of mTORC1, in response to changes in growth conditions, and interaction of eIF4E with 4E-BP1 and -2 occurs with the hypophosphorylated form.1-5 A key factor in the regulation of translation initiation is that the relative levels of eIF4E and 4E-BP1 and -2 are highly controlled.52 The hypophosphorylated form, but not the hyperphosphorylated form, of 4E-BP1 is unstable if not bound to eIF4E. Under these conditions, 4E-BP1 is ubiquitylated and targeted for proteasome-mediated proteolysis.52,53 The role of ubiquitylation was identified following some rather unexpected results obtained when knockdown of eIF4E using shRNA was demonstrated to have no effect on protein synthesis.52 This was subsequently shown to be due to concomitant degradation of 4E-BP1, which resulted in the release of eIF4E molecules to compensate for the loss brought about by the reduced expression. K57, a lysine residue conserved between all 3 4E-BPs, was identified by the Sonenberg lab as the ubiquitylation site in 4E-BP1,52 and a screen of an siRNA library identified the KLHL25-CUL3 as the E3 ubiquitin ligase responsible for 4E-BP1 degradation. Knockdown of KLHL25 resulted in a decrease in translation, consistent with it having a role in controlling levels of 4E-BP1.52
Proteasome activity (presumed to be a result of poly-ubiquitylation) has also been demonstrated to be required for the formation of a truncated form of 4E-BP1 (tr4E-BP) in murine erythro-leukemia (MEL) cells containing activated p53.54 This truncated form is 3 kDa smaller than full-length protein, is unphosphorylated and relatively stable. It also binds to eIF4E in preference to the full-length protein. It has been proposed that the production of this p53-induced form may be contributing to the ability of p53 to regulate apoptosis and malignancy.
eIF4A
Two isoforms of eIF4A have been identified in proteomic screens as being modified by ubiquitin and SUMO.35,36,55-62 In contrast to what is observed with some of the other initiation factors, modified peptides from both eIF4A1 and eIF4A2 are highly abundant in the proteomic screens designed to identify ubiquitylation sites, implying that modification is likely to have a key role(s) in the regulation of the function of these 2 proteins. In the ubiquitin screens, most of the modified sites identified in the human eIF4A proteins were also observed in the mouse proteins, suggesting that they are likely to be true ‘hits’ and not false positives. Interestingly, eIF4A2 (but not eIF4A1) and translational repression have both been shown to be essential for miRNA-mediated gene regulation.63 However, the post-translational modification of these proteins by ubiquitin or Ubls has not been analyzed in detail and to date there are no reports on whether it affects the activity of the eIF4A protein or miRNA-mediated translational control.
In a role unrelated to its function in translation, ubiquitylation of Drosophila eIF4A has been shown to be linked with Decapentaplegic (Dpp) signaling.64 Additionally, rice DRM2 (required for RNA-directed DNA methylation) interacts with eIF4A via its ubiquitin associated (UBA) domain, (although whether this occurs with a ubiquitylated form has not been analyzed).65
eIF4G
There are 3 isoforms of the scaffold protein, eIF4G, eIF4GI-III. As observed with eIF4A, diGly-modified peptides from these proteins are abundant in proteomic screens designed to identify ubiquitylation sites,35,36,57,59,61 and again most are observed in both the human and mouse proteins. In eIF4GI these sites (6 in total, 4 common to both human and mouse) map to lysine residues occurring between amino acids 593–925 which map close to, or in the region of, the eIF4E and eIF4A/3 binding sites. The abundance of these modified tryptic fragments and their position in the protein suggests that this post-translational modification is likely to be important for regulating the functions of these proteins, possibly by affecting the interaction of eIF4G with other members of the eIF4F complex. Again, these modifications have not been analyzed in detail and to date there are no reports on whether they affect the activity of eIF4GI. In addition to this modification by ubiquitin, eIF4GI has been shown to be sumoylated in both fission yeast and human cells.66 Sumoylation of S. pombe eIF4G is increased following exposure of cells to 1 M KCl or arsenite, conditions which result in the formation of stress granules. In vitro sumoylation studies have identified 2 sumoylation sites in mammalian eIF4GI, K1368 and K1588, residing in the eIF3/4A binding site and the Mnk-binding domain, respectively. (Mnks (MAP kinase-interacting kinases) are kinases which bind to the C-terminus of eIF4G and phosphorylate eIF4E which is bound to the N-terminus of eIF4G.67) These data suggest that sumoylation may be affecting interactions of eIF4GI with associated proteins, e.g., eIF4E, and possibly the assembly of eIF4G into stress granules.
Paip2
Poly(A)-binding protein (PABP) is regulated through the interaction with 2 proteins, Paip1 and Paip2.5,6 Paip1, which also interacts with eIF3g, is eIF4G-like and is stimulatory for translation, while Paip2 represses PABP function by decreasing the affinity of PABP for polyadenylated mRNA, thus inhibiting translation. Paip1 and Paip2 both have 2 domains, PAM1 and PAM2 which interact with PABP. This interaction occurs through RRM-1 and PABC domains, respectively.68 Additionally, PAM2 is capable of interacting with EDD (a member of the HECT domain family of E3 ubiquitin ligases) which also contains a PABC domain.69 In cells where levels of PABP are depleted, Paip2A is ubiquitylated in an EDD-dependent manner prior to proteasome-mediated degradation.70 Interestingly, the affinity of the PAM2 domain of Paip2 for the PABC domain of PABP is greater than that of the affinity for the PABC of EDD. Thus, it is proposed that interaction of PABP with Paip2 competes with EDD for interaction with PAM2 on Paip2, and that this normally prevents ubiquitylation of Paip2.70 However, in apparently contradictory work, it has been demonstrated that during human cytomegalovirus infection PABP levels rise concomitantly with the levels of both Paip2 and EDD1. The reason for this is not known, but it has been proposed that it may provide cells with a process to allow rapid changes in protein levels if necessary.71 Paip2B is also polyubiquitylated, although at a somewhat lower level that Paip2, and is hence more stable.72
eIF3
Proteomic studies have identified many of the eIF3 subunits as targets for ubiquitylation and/or sumoylation. However, independent of these studies, eIF3f is the only eIF3 subunit where the function of these modifications has been studied in any detail. eIF3f is a non-core subunit of the eIF3 complex. It can act both as a repressor and as an enhancer of translation (reviewed in73). Its role as a translational enhancer came to light in a study on muscle atrophy.74 Here, eIF3f is ubiquitylated by the MAFbnx/Atrogin1 protein which is a muscle-specific F-box protein ubiquitin E3 ligase.75 This E3 is upregulated and essential for accelerated muscle protein loss in a number of disorders.76 Ubiquitylation of eIF3f occurs on multiple (6) lysines in the C-terminus74 and results in its ubiquitin-mediated proteolysis in myotubes undergoing atrophy. Under these conditions both MAFbnx and eIF3f are detected in the nucleus.75 It has been proposed that this ubiquitylation may be associated with the rapid downregulation of certain proteins during muscle atrophy. eIF3f also interacts with the ubiquitin E3 ligase TRC8 to inhibit protein synthesis. The mechanism by which this occurs is unknown, but it has been proposed that TRC8 targets an eIF3 subunit for ubiquitylation.77 Unrelated to its role in translation, eIF3f can act as a deubiquitylating enzyme (DUB). In this capacity it is capable of deubiquitylating, and thus contributing to the activation of, the Notch signaling receptor in Drosophila.78
Interestingly, recent work has shown that eIF3e is involved in eIF4E phosphorylation; Mnk1 binding to eIF4F is dependent on eIF3e, and eIF3e is sufficient to promote Mnk1-binding to eIF4G.79 As eIF3e is modified by both ubiquitylation and sumoylation, it would be interesting to know if these modifications of eIF3e also have a role in controlling eIF4E phosphorylation.
Summary
In conclusion, despite the fact that numerous translation initiation factors have been shown to be ubiquitylated and/or sumoylated in proteomic screens, relatively little is known about the effects of the modifications on the functions of individual proteins. In part this is due to the transient nature of these modifications, e.g., in many cases of sumoylation, less than 5% of a particular protein is modified at any one time, and the sumoylated species appear to be very labile in certain organisms due to highly active SUMO-specific proteases. Additionally, since ubiquitylation targets proteins for destruction, analysis of ubiquitylated proteins, other than in the presence of a proteasome inhibitor, is difficult.
The recent use of proteomic screens to identify modified proteins and the modified site(s) suggests that there are many more cases where post-translational modification by ubiquitin or Ubls is likely to affect translation initiation factors. For example, sumoylation of eIF4A1/2 might have a role in regulating both the unwinding of mRNA secondary structure and the ability of eIF4A2 to mediate miRNA-dependent gene expression in mammalian cells. Further work on these modifications is required to fully elucidate their effect on individual proteins and on translational control of gene expression as a whole.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- 1. Sonenberg N, Hinnebusch AG. Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell 2009; 136:731-45; PMID:19239892; http://dx.doi.org/10.1016/j.cell.2009.01.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Morley SJ, Coldwell MJ, Clemens MJ. Initiation factor modifications in the preapoptotic phase. Cell Death Differ 2005; 12:571-84; PMID:15900314; http://dx.doi.org/10.1038/sj.cdd.4401591 [DOI] [PubMed] [Google Scholar]
- 3. Jackson RJ, Hellen CU, Pestova TV. The mechanism of eukaryotic translation initiation and principles of its regulation. Nat Rev Mol Cell Biol 2010; 11:113-27; PMID:20094052; http://dx.doi.org/10.1038/nrm2838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Laplante M, Sabatini DM. mTOR signaling in growth control and disease. Cell 2012; 149:274-93; PMID:22500797; http://dx.doi.org/10.1016/j.cell.2012.03.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Craig AW, Haghighat A, Yu AT, Sonenberg N. Interaction of polyadenylate-binding protein with the eIF4G homologue PAIP enhances translation. Nature 1998; 392:520-3; PMID:9548260; http://dx.doi.org/10.1038/33198 [DOI] [PubMed] [Google Scholar]
- 6. Khaleghpour K, Svitkin YV, Craig AW, DeMaria CT, Deo RC, Burley SK, Sonenberg N. Translational repression by a novel partner of human poly(A) binding protein, Paip2. Mol Cell 2001; 7:205-16; PMID:11172725; http://dx.doi.org/10.1016/S1097-2765(01)00168-X [DOI] [PubMed] [Google Scholar]
- 7. Li Q, Imataka H, Morino S, Rogers GW, Jr., Richter-Cook NJ, Merrick WC, Sonenberg N. Eukaryotic translation initiation factor 4AIII (eIF4AIII) is functionally distinct from eIF4AI and eIF4AII. Mol Cell Biol 1999; 19:7336-46; PMID:10523622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Gorgoni B, Richardson WA, Burgess HM, Anderson RC, Wilkie GS, Gautier P, Martins JP, Brook M, Sheets MD, Gray NK. Poly(A)-binding proteins are functionally distinct and have essential roles during vertebrate development. Proc Natl Acad Sci U S A 2011; 108:7844-9; PMID:21518916; http://dx.doi.org/10.1073/pnas.1017664108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Sun F, Palmer K, Handel MA. Mutation of Eif4g3, encoding a eukaryotic translation initiation factor, causes male infertility and meiotic arrest of mouse spermatocytes. Development 2010; 137:1699-707; PMID:20430745; http://dx.doi.org/10.1242/dev.043125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Burroughs AM, Iyer LM, Aravind L. Structure and evolution of ubiquitin and ubiquitin-related domains. Methods Mol Biol 2012; 832:15-63; PMID:22350875; http://dx.doi.org/10.1007/978-1-61779-474-2_2 [DOI] [PubMed] [Google Scholar]
- 11. Ulrich HD. The SUMO system: an overview. Methods Mol Biol 2009; 497:3-16; PMID:19107407; http://dx.doi.org/10.1007/978-1-59745-566-4_1 [DOI] [PubMed] [Google Scholar]
- 12. Bogunovic D, Boisson-Dupuis S, Casanova JL. ISG15: leading a double life as a secreted molecule. Exp Mol Med 2013; 45:e18; PMID:23579383; http://dx.doi.org/10.1038/emm.2013.36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Reich N, Evans B, Levy D, Fahey D, Knight E, Jr, Darnell JE, Jr. Interferon-induced transcription of a gene encoding a 15-kDa protein depends on an upstream enhancer element. Proc Natl Acad Sci U S A 1987; 84:6394-8; PMID:3476954; http://dx.doi.org/10.1073/pnas.84.18.6394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Liu M, Reimschuessel R, Hassel BA. Molecular cloning of the fish interferon stimulated gene, 15 kDa (ISG15) orthologue: a ubiquitin-like gene induced by nephrotoxic damage. Gene 2002; 298:129-39; PMID:12426101; http://dx.doi.org/10.1016/S0378-1119(02)00932-0 [DOI] [PubMed] [Google Scholar]
- 15. Hershko A, Ciechanover A. The ubiquitin system. Annu Rev Biochem 1998; 67:425-79; PMID:9759494; http://dx.doi.org/10.1146/annurev.biochem.67.1.425 [DOI] [PubMed] [Google Scholar]
- 16. Lorenz S, Cantor AJ, Rape M, Kuriyan J. Macromolecular juggling by ubiquitylation enzymes. BMC Biol 2013; 11:65; PMID:23800009; http://dx.doi.org/10.1186/1741-7007-11-65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Pickart CM, Fushman D. Polyubiquitin chains: polymeric protein signals. Curr Opin Chem Biol 2004; 8:610-6; PMID:15556404; http://dx.doi.org/10.1016/j.cbpa.2004.09.009 [DOI] [PubMed] [Google Scholar]
- 18. Wickliffe KE, Williamson A, Meyer HJ, Kelly A, Rape M. K11-linked ubiquitin chains as novel regulators of cell division. Trends Cell Biol 2011; 21:656-63; PMID:21978762; http://dx.doi.org/10.1016/j.tcb.2011.08.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ciechanover A, Stanhill A. The complexity of recognition of ubiquitinated substrates by the 26S proteasome. Biochim Biophys Acta 2014; 1843:86-96; PMID:23872423; http://dx.doi.org/10.1016/j.bbamcr.2013.07.007 [DOI] [PubMed] [Google Scholar]
- 20. Ulrich HD. How to activate a damage-tolerant polymerase: consequences of PCNA modifications by ubiquitin and SUMO. Cell Cycle 2004; 3:15-8; PMID:14657656; http://dx.doi.org/10.4161/cc.3.1.623 [PubMed] [Google Scholar]
- 21. Tomanov K, Luschnig C, Bachmair A. Ubiquitin Lys 63 chains - second-most abundant, but poorly understood in plants. Front Plant Sci 2014; 5:15; PMID:24550925; http://dx.doi.org/10.3389/fpls.2014.00015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Knipscheer P, Flotho A, Klug H, Olsen JV, van Dijk WJ, Fish A, Johnson ES, Mann M, Sixma TK, Pichler A. Ubc9 sumoylation regulates SUMO target discrimination. Mol Cell 2008; 31:371-82; PMID:18691969; http://dx.doi.org/10.1016/j.molcel.2008.05.022 [DOI] [PubMed] [Google Scholar]
- 23. Pfander B, Moldovan GL, Sacher M, Hoege C, Jentsch S. SUMO-modified PCNA recruits Srs2 to prevent recombination during S phase. Nature 2005; 436:428-33; PMID:15931174 [DOI] [PubMed] [Google Scholar]
- 24. Papouli E, Chen S, Davies AA, Huttner D, Krejci L, Sung P, Ulrich HD. Crosstalk between SUMO and ubiquitin on PCNA is mediated by recruitment of the helicase Srs2p. Mol Cell 2005; 19:123-33; PMID:15989970; http://dx.doi.org/10.1016/j.molcel.2005.06.001 [DOI] [PubMed] [Google Scholar]
- 25. Hardeland U, Steinacher R, Jiricny J, Schär P. Modification of the human thymine-DNA glycosylase by ubiquitin-like proteins facilitates enzymatic turnover. EMBO J 2002; 21:1456-64; PMID:11889051; http://dx.doi.org/10.1093/emboj/21.6.1456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Joseph J, Tan SH, Karpova TS, McNally JG, Dasso M. SUMO-1 targets RanGAP1 to kinetochores and mitotic spindles. J Cell Biol 2002; 156:595-602; PMID:11854305; http://dx.doi.org/10.1083/jcb.200110109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Sriramachandran AM, Dohmen RJ. SUMO-targeted ubiquitin ligases. Biochim Biophys Acta 2014; 1843:75-85; PMID:24018209; http://dx.doi.org/10.1016/j.bbamcr.2013.08.022 [DOI] [PubMed] [Google Scholar]
- 28. van der Veen AG, Ploegh HL. Ubiquitin-like proteins. Annu Rev Biochem 2012; 81:323-57; PMID:22404627; http://dx.doi.org/10.1146/annurev-biochem-093010-153308 [DOI] [PubMed] [Google Scholar]
- 29. Jeon YJ, Yoo HM, Chung CH. ISG15 and immune diseases. Biochim Biophys Acta 2010; 1802:485-96; PMID:20153823; http://dx.doi.org/10.1016/j.bbadis.2010.02.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zou W, Zhang DE. The interferon-inducible ubiquitin-protein isopeptide ligase (E3) EFP also functions as an ISG15 E3 ligase. J Biol Chem 2006; 281:3989-94; PMID:16352599; http://dx.doi.org/10.1074/jbc.M510787200 [DOI] [PubMed] [Google Scholar]
- 31. Dao CT, Zhang DE. ISG15: a ubiquitin-like enigma. Front Biosci: J Virtual Libr 2005; 10:2701-22; PMID:15970528 [DOI] [PubMed] [Google Scholar]
- 32. Xu X, Vatsyayan J, Gao C, Bakkenist CJ, Hu J. Sumoylation of eIF4E activates mRNA translation. EMBO Rep 2010; 11:299-304; PMID:20224576; http://dx.doi.org/10.1038/embor.2010.18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Plant LD, Dementieva IS, Kollewe A, Olikara S, Marks JD, Goldstein SA. One SUMO is sufficient to silence the dimeric potassium channel K2P1. Proc Natl Acad Sci U S A 2010; 107:10743-8; PMID:20498050; http://dx.doi.org/10.1073/pnas.1004712107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Stelter P, Ulrich HD. Control of spontaneous and damage-induced mutagenesis by SUMO and ubiquitin conjugation. Nature 2003; 425:188-91; PMID:12968183; http://dx.doi.org/10.1038/nature01965 [DOI] [PubMed] [Google Scholar]
- 35. Kim W, Bennett EJ, Huttlin EL, Guo A, Li J, Possemato A, Sowa ME, Rad R, Rush J, Comb MJ, et al. Systematic and quantitative assessment of the ubiquitin-modified proteome. Mol Cell 2011; 44:325-40; PMID:21906983; http://dx.doi.org/10.1016/j.molcel.2011.08.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wagner SA, Beli P, Weinert BT, Nielsen ML, Cox J, Mann M, Choudhary C. A proteome-wide, quantitative survey of in vivo ubiquitylation sites reveals widespread regulatory roles. Mol Cell Proteomics: MCP 2011; 10(10):M111 013284; PMID:21890473; http://dx.doi.org/10.1074/mcp.M111.013284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Hornbeck PV, Kornhauser JM, Tkachev S, Zhang B, Skrzypek E, Murray B, Latham V, Sullivan M. PhosphoSitePlus: a comprehensive resource for investigating the structure and function of experimentally determined post-translational modifications in man and mouse. Nucleic Acids Res 2012; 40:D261-70; PMID:22135298; http://dx.doi.org/10.1093/nar/gkr1122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Mamane Y, Petroulakis E, LeBacquer O, Sonenberg N. mTOR, translation initiation and cancer. Oncogene 2006; 25:6416-22; PMID:17041626; http://dx.doi.org/10.1038/sj.onc.1209888 [DOI] [PubMed] [Google Scholar]
- 39. Walsh D, Meleady P, Power B, Morley SJ, Clynes M. Increased levels of the translation initiation factor eIF4E in differentiating epithelial lung tumor cell lines. Differentiation; Res Biol Diversity 2003; 71(2):126-34; PMID:12641566 [DOI] [PubMed] [Google Scholar]
- 40. Murata T, Shimotohno K. Ubiquitination and proteasome-dependent degradation of human eukaryotic translation initiation factor 4E. J Biol Chem 2006; 281:20788-800; PMID:16720573; http://dx.doi.org/10.1074/jbc.M600563200 [DOI] [PubMed] [Google Scholar]
- 41. Othumpangat S, Kashon M, Joseph P. Sodium arsenite-induced inhibition of eukaryotic translation initiation factor 4E (eIF4E) results in cytotoxicity and cell death. Mol Cell Biochem 2005; 279:123-31; PMID:16283521; http://dx.doi.org/10.1007/s11010-005-8284-2 [DOI] [PubMed] [Google Scholar]
- 42. Qian SB, McDonough H, Boellmann F, Cyr DM, Patterson C. CHIP-mediated stress recovery by sequential ubiquitination of substrates and Hsp70. Nature 2006; 440:551-5; PMID:16554822; http://dx.doi.org/10.1038/nature04600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Lee T, Pelletier J. Eukaryotic initiation factor 4F: a vulnerability of tumor cells. Future Med Chem 2012; 4:19-31; PMID:22168162; http://dx.doi.org/10.4155/fmc.11.150 [DOI] [PubMed] [Google Scholar]
- 44. McKendrick L, Morley SJ, Pain VM, Jagus R, Joshi B. Phosphorylation of eukaryotic initiation factor 4E (eIF4E) at Ser209 is not required for protein synthesis in vitro and in vivo. Eur J Biochem 2001; 268:5375-85; PMID:11606200; http://dx.doi.org/10.1046/j.0014-2956.2001.02478.x [DOI] [PubMed] [Google Scholar]
- 45. Scheper GC, van Kollenburg B, Hu J, Luo Y, Goss DJ, Proud CG. Phosphorylation of eukaryotic initiation factor 4E markedly reduces its affinity for capped mRNA. J Biol Chem 2002; 277:3303-9; PMID:11723111; http://dx.doi.org/10.1074/jbc.M103607200 [DOI] [PubMed] [Google Scholar]
- 46. Xu X, Vatsyayan J, Gao C, Bakkenist CJ, Hu J. HDAC2 promotes eIF4E sumoylation and activates mRNA translation gene specifically. J Biol Chem 2010; 285:18139-43; PMID:20421305; http://dx.doi.org/10.1074/jbc.C110.131599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Desterro JM, Rodriguez MS, Hay RT. SUMO-1 modification of IkappaBalpha inhibits NF-kappaB activation. Mol Cell 1998; 2:233-9; PMID:9734360; http://dx.doi.org/10.1016/S1097-2765(00)80133-1 [DOI] [PubMed] [Google Scholar]
- 48. Rom E, Kim HC, Gingras AC, Marcotrigiano J, Favre D, Olsen H, Burley SK, Sonenberg N. Cloning and characterization of 4EHP, a novel mammalian eIF4E-related cap-binding protein. J Biol Chem 1998; 273:13104-9; PMID:9582349; http://dx.doi.org/10.1074/jbc.273.21.13104 [DOI] [PubMed] [Google Scholar]
- 49. Tan NG, Ardley HC, Scott GB, Rose SA, Markham AF, Robinson PA. Human homologue of ariadne promotes the ubiquitylation of translation initiation factor 4E homologous protein, 4EHP. FEBS Lett 2003; 554:501-4; PMID:14623119; http://dx.doi.org/10.1016/S0014-5793(03)01235-3 [DOI] [PubMed] [Google Scholar]
- 50. Okumura F, Zou W, Zhang DE. ISG15 modification of the eIF4E cognate 4EHP enhances cap structure-binding activity of 4EHP. Genes Dev 2007; 21:255-60; PMID:17289916; http://dx.doi.org/10.1101/gad.1521607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Elmehdawi F, Wheway G, Szymanska K, Adams M, High AS, Johnson CA, Robinson PA. Human Homolog of Drosophila Ariadne (HHARI) is a marker of cellular proliferation associated with nuclear bodies. Exp Cell Res 2013; 319:161-72; PMID:23059369; http://dx.doi.org/10.1016/j.yexcr.2012.10.002 [DOI] [PubMed] [Google Scholar]
- 52. Yanagiya A, Suyama E, Adachi H, Svitkin YV, Aza-Blanc P, Imataka H, Mikami S, Martineau Y, Ronai ZA, Sonenberg N. Translational homeostasis via the mRNA cap-binding protein, eIF4E. Mol Cell 2012; 46:847-58; PMID:22578813; http://dx.doi.org/10.1016/j.molcel.2012.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Elia A, Constantinou C, Clemens MJ. Effects of protein phosphorylation on ubiquitination and stability of the translational inhibitor protein 4E-BP1. Oncogene 2008; 27:811-22; PMID:17653084; http://dx.doi.org/10.1038/sj.onc.1210678 [DOI] [PubMed] [Google Scholar]
- 54. Constantinou C, Elia A, Clemens MJ. Activation of p53 stimulates proteasome-dependent truncation of eIF4E-binding protein 1 (4E-BP1). Biol Cell / Under Auspices Eur Cell Biol Organ, 2008; 100(5):279-89; PMID:18021075 [DOI] [PubMed] [Google Scholar]
- 55. Blomster HA, Hietakangas V, Wu J, Kouvonen P, Hautaniemi S, Sistonen L. Novel proteomics strategy brings insight into the prevalence of SUMO-2 target sites. Mol Cell Proteomics 2009; 8:1382-90; PMID:19240082; http://dx.doi.org/10.1074/mcp.M800551-MCP200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Nie M, Xie Y, Loo JA, Courey AJ. Genetic and proteomic evidence for roles of Drosophila SUMO in cell cycle control, Ras signaling, and early pattern formation. PLoS One 2009; 4:e5905; PMID:19529778; http://dx.doi.org/10.1371/journal.pone.0005905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Bruderer R, Tatham MH, Plechanovova A, Matic I, Garg AK, Hay RT. Purification and identification of endogenous polySUMO conjugates. EMBO Rep 2011; 12:142-8; PMID:21252943; http://dx.doi.org/10.1038/embor.2010.206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Yang W, Thompson JW, Wang Z, Wang L, Sheng H, Foster MW, Moseley MA, Paschen W. Analysis of oxygen/glucose-deprivation-induced changes in SUMO3 conjugation using SILAC-based quantitative proteomics. J Proteome Res 2012; 11:1108-17; PMID:22082260; http://dx.doi.org/10.1021/pr200834f [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Wagner SA, Beli P, Weinert BT, Schölz C, Kelstrup CD, Young C, Nielsen ML, Olsen JV, Brakebusch C, Choudhary C. Proteomic analyses reveal divergent ubiquitylation site patterns in murine tissues. Mol Cell Proteomics 2012; 11:1578-85; PMID:22790023; http://dx.doi.org/10.1074/mcp.M112.017905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Shi Y, Chan DW, Jung SY, Malovannaya A, Wang Y, Qin J. A data set of human endogenous protein ubiquitination sites. Mol Cell Proteomics: MCP 2011;. 10(5):M110 002089; PMID:20972266; http://dx.doi.org/10.1074/mcp.M110.002089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Matafora V, D’Amato A, Mori S, Blasi F, Bachi A. Proteomics analysis of nucleolar SUMO-1 target proteins upon proteasome inhibition. Mol Cell Proteomics 2009; 8:2243-55; PMID:19596686; http://dx.doi.org/10.1074/mcp.M900079-MCP200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Mazur MJ, van den Burg HA. Global SUMO Proteome Responses Guide Gene Regulation, mRNA Biogenesis, and Plant Stress Responses. Front Plant Sci 2012; 3:215; PMID:23060889; http://dx.doi.org/10.3389/fpls.2012.00215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Meijer HA, Kong YW, Lu WT, Wilczynska A, Spriggs RV, Robinson SW, Godfrey JD, Willis AE, Bushell M. Translational repression and eIF4A2 activity are critical for microRNA-mediated gene regulation. Science 2013; 340:82-5; PMID:23559250; http://dx.doi.org/10.1126/science.1231197 [DOI] [PubMed] [Google Scholar]
- 64. Li J, Li WX. A novel function of Drosophila eIF4A as a negative regulator of Dpp/BMP signalling that mediates SMAD degradation. Nat Cell Biol 2006; 8:1407-14; PMID:17115029; http://dx.doi.org/10.1038/ncb1506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Dangwal M, Malik G, Kapoor S, Kapoor M. De novo methyltransferase, OsDRM2, interacts with the ATP-dependent RNA helicase, OseIF4A, in rice. J Mol Biol 2013; 425:2853-66; PMID:23732981; http://dx.doi.org/10.1016/j.jmb.2013.05.021 [DOI] [PubMed] [Google Scholar]
- 66. Jongjitwimol The S. pombe translation initiation factor eIF4G is sumoylated and assocaites with the SUMO protease Ulp2. PLoS ONE 2014; 40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Buxade M, Parra-Palau JL, Proud CG. The Mnks: MAP kinase-interacting kinases (MAP kinase signal-integrating kinases). Front Biosci 2008; 13:5359-73; PMID:18508592; http://dx.doi.org/10.2741/3086 [DOI] [PubMed] [Google Scholar]
- 68. Khaleghpour K, Kahvejian A, De Crescenzo G, Roy G, Svitkin YV, Imataka H, O’Connor-McCourt M, Sonenberg N. Dual interactions of the translational repressor Paip2 with poly(A) binding protein. Mol Cell Biol 2001; 21:5200-13; PMID:11438674; http://dx.doi.org/10.1128/MCB.21.15.5200-5213.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Deo RC, Sonenberg N, Burley SK. X-ray structure of the human hyperplastic discs protein: an ortholog of the C-terminal domain of poly(A)-binding protein. Proc Natl Acad Sci U S A 2001; 98:4414-9; PMID:11287654; http://dx.doi.org/10.1073/pnas.071552198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Yoshida M, Yoshida K, Kozlov G, Lim NS, De Crescenzo G, Pang Z, Berlanga JJ, Kahvejian A, Gehring K, Wing SS, et al. Poly(A) binding protein (PABP) homeostasis is mediated by the stability of its inhibitor, Paip2. EMBO J 2006; 25:1934-44; PMID:16601676; http://dx.doi.org/10.1038/sj.emboj.7601079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. McKinney C, Yu D, Mohr I. A new role for the cellular PABP repressor Paip2 as an innate restriction factor capable of limiting productive cytomegalovirus replication. Genes Dev 2013; 27:1809-20; PMID:23964095; http://dx.doi.org/10.1101/gad.221341.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Berlanga JJ, Baass A, Sonenberg N. Regulation of poly(A) binding protein function in translation: characterization of the Paip2 homolog, Paip2B. RNA 2006; 12:1556-68; PMID:16804161; http://dx.doi.org/10.1261/rna.106506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Marchione R, Leibovitch SA, Lenormand JL. The translational factor eIF3f: the ambivalent eIF3 subunit. Cell Mol Life Sci 2013; 70:3603-16; PMID:23354061; http://dx.doi.org/10.1007/s00018-013-1263-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Csibi A, Leibovitch MP, Cornille K, Tintignac LA, Leibovitch SA. MAFbx/Atrogin-1 controls the activity of the initiation factor eIF3-f in skeletal muscle atrophy by targeting multiple C-terminal lysines. J Biol Chem 2009; 284:4413-21; PMID:19073596; http://dx.doi.org/10.1074/jbc.M807641200 [DOI] [PubMed] [Google Scholar]
- 75. Lagirand-Cantaloube J, Offner N, Csibi A, Leibovitch MP, Batonnet-Pichon S, Tintignac LA, Segura CT, Leibovitch SA. The initiation factor eIF3-f is a major target for atrogin1/MAFbx function in skeletal muscle atrophy. EMBO J 2008; 27:1266-76; PMID:18354498; http://dx.doi.org/10.1038/emboj.2008.52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Sacheck JM, Hyatt JP, Raffaello A, Jagoe RT, Roy RR, Edgerton VR, Lecker SH, Goldberg AL. Rapid disuse and denervation atrophy involve transcriptional changes similar to those of muscle wasting during systemic diseases. FASEB J: Off Publ Fed Am Soc Exp Biol 2007; 21(1):140-55; PMID:17116744 [DOI] [PubMed] [Google Scholar]
- 77. Lee JP, Brauweiler A, Rudolph M, Hooper JE, Drabkin HA, Gemmill RM. The TRC8 ubiquitin ligase is sterol regulated and interacts with lipid and protein biosynthetic pathways. Mol Cancer Res 2010; 8:93-106; PMID:20068067; http://dx.doi.org/10.1158/1541-7786.MCR-08-0491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Moretti J, Chastagner P, Gastaldello S, Heuss SF, Dirac AM, Bernards R, Masucci MG, Israël A, Brou C. The translation initiation factor 3f (eIF3f) exhibits a deubiquitinase activity regulating Notch activation. PLoS Biol 2010; 8:e1000545; PMID:21124883; http://dx.doi.org/10.1371/journal.pbio.1000545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Walsh D, Mohr I. Coupling 40S ribosome recruitment to modification of a cap-binding initiation factor by eIF3 subunit e. Genes Dev 2014; 28:835-40; PMID:24736843; http://dx.doi.org/10.1101/gad.236752.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Miller MJ, Barrett-Wilt GA, Hua Z, Vierstra RD. Proteomic analyses identify a diverse array of nuclear processes affected by small ubiquitin-like modifier conjugation in Arabidopsis. Proc Natl Acad Sci U S A 2010; 107:16512-7; PMID:20813957; http://dx.doi.org/10.1073/pnas.1004181107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Westman BJ, Verheggen C, Hutten S, Lam YW, Bertrand E, Lamond AI. A proteomic screen for nucleolar SUMO targets shows SUMOylation modulates the function of Nop5/Nop58. Mol Cell 2010; 39:618-31; PMID:20797632; http://dx.doi.org/10.1016/j.molcel.2010.07.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Panse VG, Hardeland U, Werner T, Kuster B, Hurt E. A proteome-wide approach identifies sumoylated substrate proteins in yeast. J Biol Chem 2004; 279:41346-51; PMID:15292183; http://dx.doi.org/10.1074/jbc.M407950200 [DOI] [PubMed] [Google Scholar]
- 83. Becker J, Barysch SV, Karaca S, Dittner C, Hsiao HH, Berriel Diaz M, Herzig S, Urlaub H, Melchior F. Detecting endogenous SUMO targets in mammalian cells and tissues. Nat Struct Mol Biol 2013; 20:525-31; PMID:23503365; http://dx.doi.org/10.1038/nsmb.2526 [DOI] [PubMed] [Google Scholar]
- 84. Danielsen JM, Sylvestersen KB, Bekker-Jensen S, Szklarczyk D, Poulsen JW, Horn H, Jensen LJ, Mailand N, Nielsen ML, Mass spectrometric analysis of lysine ubiquitylation reveals promiscuity at site level. Mol Cell Proteomics: MCP 2011; 10(3):M110 003590; PMID:21139048; http://dx.doi.org/10.1074/mcp.M110.003590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Hannich JT, Lewis A, Kroetz MB, Li SJ, Heide H, Emili A, Hochstrasser M. Defining the SUMO-modified proteome by multiple approaches in Saccharomyces cerevisiae. J Biol Chem 2005; 280:4102-10; PMID:15590687; http://dx.doi.org/10.1074/jbc.M413209200 [DOI] [PubMed] [Google Scholar]