Abstract
Brg1 (Brahma-related gene 1) is a catalytic component of the evolutionarily conserved mammalian SWI/SNF ATP-dependent chromatin remodeling enzymes that disrupt histone-DNA contacts on the nucleosome. While the requirement for the SWI/SNF enzymes in cell differentiation has been extensively studied, its role in precursor cell proliferation and survival is not as well defined. Muscle satellite cells constitute the stem cell pool that sustains and regenerates myofibers in adult skeletal muscle. Here we show that deletion of Brg1 in primary mouse myoblasts derived from muscle satellite cells cultured ex vivo leads to a cell proliferation defect and apoptosis. We determined that Brg1 regulates cell proliferation and survival by controlling chromatin remodeling and activating transcription at the Pax7 promoter, which is expressed during somite development and is required for controlling viability of the satellite cell population. Reintroduction of catalytically active Brg1 or of Pax7 into Brg1-deficient satellite cells rescued the apoptotic phenotype and restored proliferation. These data demonstrate that Brg1 functions as a positive regulator for cellular proliferation and survival of primary myoblasts. Therefore the regulation of gene expression through Brg1-mediated chromatin remodeling is critical not just for skeletal muscle differentiation but for maintaining the myoblast population as well.
Keywords: Brg1, SWI/SNF, Pax7, myoblast, cell proliferation
Introduction
Mammalian SWI/SNF ATP-dependent chromatin remodeling enzymes disrupt histone-DNA contacts on the nucleosome using the energy released by ATP hydrolysis (Imbalzano et al., 1994; Kwon et al., 1994). These structural alterations result in increased or decreased chromatin accessibility for the binding of regulatory proteins that modulate replication, recombination, and transcription (Bartholomew, 2014; Clapier and Cairns, 2009; Mueller-Planitz et al., 2013). The SWI/SNF family is highly conserved in eukaryotes and its catalytic activity is provided by one of two mutually exclusive ATPase subunits called Brahma (Brm) or Brg1 (Khavari et al., 1993; Muchardt and Yaniv, 1993; Wang et al., 1996). The SWI/SNF complexes also include other proteins known as Brg1 and Brm-associated factors that may modulate the activities of Brm or Brg1 (Wang et al., 1996). Chromatin remodeling is an important feature for integrating different signal transduction pathways into specific transcriptional responses that will determine the cellular fate. In this regard, the SWI/SNF complex is associated with the control of cellular proliferation, differentiation, and the cell cycle (de la Serna et al., 2006; Ho and Crabtree, 2010; Wu, 2012). The role of the SWI/SNF complex in differentiation of different lineages has been studied extensively; its role in proliferation and cell survival of precursor cells is less well understood.
Previous studies have definitively shown that Brg1 is necessary for early embryogenesis (Bultman et al., 2000; Bultman et al., 2006; Ho et al., 2009; Kidder et al., 2009; Sumi-Ichinose et al., 1997) and for terminal differentiation of most tissues (de la Serna et al., 2006; Ho and Crabtree, 2010; Wu, 2012). In contrast, it appears that the requirement for Brg1 in the development of precursor cells or in the maintenance of differentiated cells is more variable. For instance, the development and proliferation of mouse keratinocyte precursors were not impacted by Brg1 depletion (Indra et al., 2005). Depletion of Brg1 in Xenopus did not impact neural induction or cell fate determination (Seo et al., 2005). However, others studying depletion of Brg1 in mouse neural stem cells reached the opposite conclusion because Brg1 was required for neural stem cell maintenance (Lessard et al., 2007; Matsumoto et al., 2006). Expression of a Brg1 protein mutated in the ATPase domain supported primitive erythropoiesis in the yolk sac and development of pro- and basophilic erythroblasts in the fetal liver but were deficient for definitive erythropoiesis (Bultman et al., 2005). Brg1 is required to complete the stages of T cell development (Chi et al., 2003; Gebuhr et al., 2003) but is not required survival of mature T lymphocytes (Gebuhr et al., 2003). Differentiation of precursor cells into osteoblasts using a cell culture model system proceeded normally in the presence of a dominant negative Brg1 (Cruzat et al., 2009). Brg1 conditional embryonic fibroblasts depleted for Brg1 ex vivo survived and proliferated as well as control cells (Bultman et al., 2000). In vivo electroporation resulting in short-term expression of an ATPase-deficient Brg1 in skeletal muscle resulted in inhibition of expression of the Myogenin regulatory protein (Ohkawa et al., 2007), but skeletal muscle-specific depletion of Myogenin post-myogenesis resulted in only modest effects (Knapp et al., 2006), suggesting that there may not be an absolute requirement for Brg1 in terminally differentiated skeletal muscle.
Muscle satellite cells are located under the basal lamina that surrounds each myofiber (Mauro, 1961). Satellite cells have the capability to proliferate and to differentiate in order to sustain basal physiological myofiber turnover and muscle regeneration (Brack and Rando, 2012; Chang and Rudnicki, 2014; Montarras et al., 2013; Motohashi and Asakura, 2014; Sambasivan and Tajbakhsh, 2015), highlighting the need for effective mechanisms to maintain the satellite cell pool. The Pax7 transcriptional regulator has been shown to have an important role in the proliferation of the muscle stem cell pool (Brack and Rando, 2012; Buckingham and Rigby, 2014; Chang and Rudnicki, 2014; Montarras et al., 2013; Motohashi and Asakura, 2014; Sambasivan and Tajbakhsh, 2015). Pax7−/− mice die within 2–3 weeks after birth, presumably due to the abnormal development of neural crest derivatives (Mansouri et al., 1996). Pax7 knockout mice have a diminished number of muscle satellite cells and were impaired for muscle regeneration, supporting the idea that Pax7 is required for the propagation and function of the satellite cell population (Oustanina et al., 2004; Seale et al., 2000). Moreover, deletion of Pax7 led to an extended G2/M phase of the cell cycle and the pool of satellite cells is progressively lost due to cell death (Relaix et al., 2006). The anti-apoptotic properties of Pax7 cannot be compensated by the closely related Pax3 protein, highlighting the importance of Pax7 in promoting cell survival and in controlling the stem cell populations of adult tissues (Relaix et al., 2006). More recent tissue-specific analyses have provided additional evidence for Pax7 function in the maintenance and regenerative capacity of satellite cells (Gunther et al., 2013; von Maltzahn et al., 2013). The onset of satellite cell differentiation leads to the down-regulation of Pax7 and triggers the expression of Myogenin (Zammit et al., 2004). Consistent with these observations, overexpression or constitutive expression of Pax7 inhibited or delayed the expression of Myogenin in cultured cells (Olguin and Olwin, 2004; Zammit et al., 2006), while Myogenin expression repressed Pax7 expression, suggesting a reciprocal inhibition of regulators controlling satellite cell maintenance and differentiation (Olguin et al., 2007).
We investigated the function of the chromatin remodeling enzyme Brg1 in proliferating primary myoblasts derived from muscle satellite cells. We identified a critical role for Brg1 in directly regulating the transcriptional regulator Pax7 and, consequently, a role in satellite cell proliferation and survival under non-differentiating conditions.
Materials and methods
Generation of plasmids
Full-length, FLAG-tagged wild type human Brg1 and a mutated version in the ATP binding site have been previously described (De la Serna et al., 2000; Khavari et al., 1993; Sif et al., 2001). Briefly, both the FLAG-tagged wildtype and ATP binding site mutant Brg1 alleles were excised from the pBluescript vector as a ClaI fragment, blunted, and cloned into the EcoRV site of pCR3.1 (Invitrogen). Plasmid EYFP-c1-Brg1 (a kind gift from Gordon Hager), encodes YFP fused to the N-terminus of Brg1 (Johnson et al., 2008). The NheI–BamHI fragment from this plasmid was cloned into AgeI-BamHI digested pCR3.1-Brg1 for both the wildtype and mutant Brg1 sequences to create YFP-Brg1 fusion proteins. The resulting plasmids were digested with AgeI and PmeI and the sequences encoding the YFP-Brg1 fusion proteins were cloned into AgeI-PmeI digested pBABE vector encoding mCherry flanked by Cre recombination sites. The murine Pax7 coding region was PCR amplified from cDNA prepared from C57BL/6 primary myoblasts using the primers indicated in Table 1 and cloned into the pBABE retroviral vector.
Table 1.
Primers used in this study
| Primer name | Sequence 5′-3′ | Use |
|---|---|---|
| Pax7 F | GAGCTCCACCGCGGTGGCGGCCGCTCTAGC | Cloning pax7 |
| Pax7 R | GATCGAATTCTCTAGACTATTATTTGTCGTCATCATCC | Cloning pax7 |
| Pax7 LM1 F | GAGATCTGAATTCCTGCGCCTTGC | REAA |
| Pax7 LM1 R | GATCGAGGGGAGGAGGGAGTCC | REAA |
| Pax7 LM2 F | CCGGGAGATCTGAATTCCTGGGTAAG | REAA |
| Pax7 LM2 R | TCCCCTCTCCCTCCCGAACTGG | REAA |
| Pax7 LM3 F | CCGGGAGATCTGAATTCCTGGAGCAA | REAA |
| Pax7 LM3 R | CTCTTTCTTGAACCCAGATAGCCTGCC | REAA |
| Pax7 LM4 F | CCGGGAGATCTGAATTCCTGAGGGTG | REAA |
| Pax7 LM4 R | GGCTGGACAGGATCTTCA | REAA |
| qPax7 F | GCAGCTGGAGGAGCTAGAGAAG | Gene expression |
| qPax7 R | GTCTCCTGGCTTGATGGAGTCG | Gene expression |
| qEF1α F | AGCTTCTCTGACTACCCTCCACTT | Gene expression |
| qEF1α R | GACCGTTCTTCCACCACTGATT | Gene expression |
| Pax7 ChIP F | GTGGCGACAAGGAAGTTCAAACAAAC | ChIP |
| Pax7 ChIP R | AAAGAAAGCCACTCCGCAACCTCTG | ChIP |
Western blots and antibodies
Proliferating myoblasts were washed with PBS and solubilized with RIPA buffer (10 mM piperazine-N,N′-bis(2-ethanesulfonic acid), pH 7.4, 150 mM NaCl, 2 mM ethylenediamine-tetraacetic acid (EDTA), 1% Triton X-100, 0.5% sodium deoxycholate, and 10% glycerol) containing protease inhibitors (Complete Mini; Roche). 100 μg of each sample was prepared for SDS-polyacrylamide gel electrophoresis by boiling in sample buffer. The resolved proteins were electrotransferred to a polyvinylidene difluoride membrane (Bio-Rad). The proteins of interest were then detected with the specific polyclonal or monoclonal antibodies indicated, followed by species-appropriate peroxidase-conjugated antibodies (Pierce) and chemiluminescent detection (ECL PLUS; GE Healthcare). Rabbit anti-Brg1 antibody (H-88; sc-10768); goat anti-Pax-3/7 antibody (N-19; sc-7749); rabbit anti-Caspase-3 antibody (H-277; sc-7148) were purchased from Santa Cruz Biotechnology, Inc. Rabbit anti-PI3 Kinase p85 antibody, N-SH2 domain (ABS233) was obtained from Millipore. Mouse anti-GAPDH-Peroxidase conjugated (G9295) was obtained from Sigma Aldrich.
Animal use and cell culture
Mice were housed in the University of Massachusetts Medical School animal care facility as specified by Institutional Animal Care and Use Committee guidelines. Satellite cells were isolated from total leg muscle from Brg1 conditional mice (Bultman et al., 2000) as described (Bischoff and Heintz, 1994). The cells were grown in culture on plates coated in 0.02% collagen (Advanced BioMatrix) in a 1:1 mix of DMEM and F-10 media supplemented with 20% fetal bovine serum, 2% Chick Embryo Extract (Sera Laboratories) and 25 ng/ml recombinant basic FGF (Millipore).
Retrovirus was produced as previously described (Morgenstern and Land, 1990). Cells were infected with retrovirus in the presence of 8 μg/ml polybrene and selected for 2 days with 1 μg/ml puromycin (Invitrogen). After 2 days of puromycin selection, cells were replated at low density and allowed to grow for one week under puromycin selection. mCherry expressing cells were transferred to collagen coated wells of 96 well plates. Media was changed every other day, and cells were transferred to a new collagen coated well every 5 days. Once there were sufficient cells to reach 50% confluence on a 60 mm dish, cells were tested for transgene expression by western blot. Adenovirus for expression of Cre-recombinase (Ad-Cre; University of Iowa Viral Vector Core) (Stec et al., 1999) was administered at a MOI of 50.
TUNEL staining
Cells were fixed in PBS, pH 7.4 containing 2% formaldehyde at 4 °C for 15 min, permeabilized with ice cold 70% ethanol, and washed 3 times with ice cold PBS. Incorporation of biotinylated dUTP onto the 3′ ends of fragmented DNA was performed with terminal deoxynucleotidyl transferase. Briefly, cells were incubated in TdT equilibration buffer (2.5 mM Tris-HCl, pH 6.6, 0.2 M potassium cacodylate, 2.5 mM CoCl2, 0.25 mg/mL bovine serum albumin (BSA)). The cell suspension was incubated at 37°C for 30 min with occasional gentle mixing. Then 0.5 U/μL of TdT enzyme and 40 pmol/μL biotinylated-dUTP were added (Roche Diagnostics Corp.). Cells were washed in PBS, collected by centrifugation, and incubated 30 min at RT in blocking solution containing 3% BSA and 20% normal goat serum in 1% (w/v) in 0.1 M Tris-buffered saline (TBS). Samples were subsequently treated for 1 h at RT with peroxidase-labeled anti-digoxigenin antibody followed by washing and developed with 0.05% (w/v) 3,3&-diaminobenzidine tetrahydrochloride (DAB).
Gene expression analysis
RNA was purified from three independent biological replicates of proliferating primary myoblasts with TRIzol following the manufacturer’s protocol (Invitrogen). The RNA samples (1 μg) were used as templates for cDNA synthesis with random primers and SuperScript III reverse transcriptase (Invitrogen). Quantitative RT-PCR was performed with Fast SYBR green master mix on the ABI StepOne Plus Sequence Detection System using the primers listed in Table 1 and normalized to the levels of EF1-alpha expression (Ohkawa et al., 2006). One-tailed t tests were performed for statistical analyses using Kaleidagraph software.
Chromatin immunoprecipitation assays
Chromatin immunoprecipitation (ChIP) assays using cultured mouse primary myoblasts were performed in triplicate from three independent biological samples as described (Dacwag et al., 2007; Hernandez-Hernandez et al., 2013). Proliferating cell lysates were incubated overnight with polyclonal rabbit antisera against Brg1 (De la Serna et al., 2000), or normal rabbit IgG (Santa Cruz Biotechnology, sc-3888). Cross-linking was reversed, and DNA was purified using ChIP DNA Clean and Concentrator Columns (Zymo Research). qPCR was performed using Fast SYBR green master mix on the ABI StepOne Plus Sequence Detection System. Primers used are listed in Table 1. Quantification was done using the comparative Ct method to obtain the percent of total input DNA pulled down by each antibody. Unpaired Student’s t tests (two-tailed) were performed using the Kaleidagraph software.
Restriction enzyme accessibility assay
Nuclei from three independent biological replicates of proliferating myoblasts were isolated by sucrose gradients. Briefly, the cells were trypsinized, resuspended in buffer containing 10 mM HEPES, ph 7.9, 10 mM KCl, 1.5 mM MgCl2, 0.5 mM DTT and homogenized with a dounce tissue homogenizer. Nuclei were separated from cytoplasm by centrifuging the samples 5 min at 218 × g at 4 °C, resuspended in 3 ml of Buffer S1 (0.25 M sucrose, 10 mM MgCl2), layered over 3 ml of Buffer S2 (0.35 M sucrose, 10 mM MgCl2) and centrifuged at 1430 × g at 4 °C. Restriction enzyme accessibility assays using cultured myoblasts derived from mouse satellite cells were performed as described (Ohkawa et al., 2012) using primers described in Table 1. Unpaired Student’s t tests (two-tailed) were performed using the Kaleidagraph software.
Results and Discussion
Depletion of Brg1 leads to cell death in satellite cell derived myoblasts
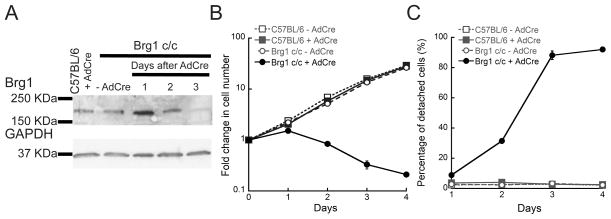
In order to understand the role of Brg1 in the proliferation and maintenance of the satellite cell population, we isolated primary satellite cells from mice homozygous for a conditional Brg1 allele (Brg1 c/c) and propagated them in culture. In this model (Bultman et al., 2000), the gene is rendered nonfunctional upon infection of the cells with adenovirus encoding the Cre recombinase (Ad-Cre). As a control for our experiments, we isolated primary satellite cells from wild type C57BL/6 mice. A representative western blot showed a decrease in the expression of Brg1 in the Ad-Cre-infected Brg1 conditional myoblasts 3 days after plating when compared to the C57BL/6 control cells. Depletion of Brg1 led to a decrease in the rate of myoblast proliferation (Fig. 1B). Moreover, a dramatic decrease in the myoblast population attached to the plate was observed upon Brg1 depletion (Fig. 1C). As early as one day after plating, approximately 10% of the Ad-Cre-infected Brg1 conditional cells detached from the plate, with approximately 95% of cells detached after 4 days of culture under proliferating conditions (Fig. 1C). In the case of control myoblasts derived from satellite cells from the wild type mice, ~5% of cells were detached from the plate upon Ad-Cre infection (Fig. 1C). Only 2–3% cell detachment was observed for the non-infected cells over the course of the experiment (Fig. 1C).
Fig. 1. Brg1 is necessary for proliferation and survival of satellite cell derived primary myoblasts.
(A) Representative western blot showing Brg1 expression in C57BL/6 and Brg1 c/c myoblasts infected or not with Ad-Cre virus. GAPDH was used as an internal loading control. (B) Effect of Brg1 depletion upon Ad-Cre infection on myoblast survival. (C) Percentage of detached cells upon Ad-Cre infection and Brg1 depletion. Data are the mean ± S.E. from three independent experiments.
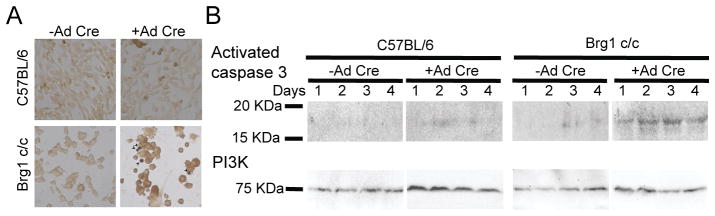
We sought to determine whether the decrease in cell number was due to cell death. First we analyzed the expression of apoptotic markers. Ad-Cre-infection of the Brg1 conditional myoblasts led to an increase in TUNEL staining, which indicates DNA fragmentation that results from apoptotic signaling cascades (Fig. 2A). This result was confirmed by western blots showing the activation of Caspase-3; only the myoblasts depleted for Brg1 showed elevated levels of activated Caspase-3 (Fig. 2B). It is interesting that even though some expression of Brg1 could still be detected 2 days post-infection (Fig. 1A), Caspase-3 was activated and detected as early as 1 day post-infection (Fig. 2B). The results suggest that in some cells the expression of Brg1 diminished soon after infection, which led to the rapid activation of the apoptotic pathway. These data also suggest that Brg1 serves as an anti-apoptotic factor in primary myoblasts. This finding is consistent with prior developmental studies indicating that Brg1 represses apoptosis at specific stages of T cell development (Gebuhr et al., 2003) and in neural crest cell proliferation that is necessary for subsequent events during cardiovascular development (Li et al., 2013). More broadly, this conclusion is consistent with observations that Brg1 suppresses apoptosis in response to UV and other DNA damaging agents (Gong et al., 2008; Kothandapani et al., 2012; Liu et al., 2014; Ondrusova et al., 2013; Park et al., 2009; Saladi et al., 2013) and that Brg1 represses E2F1-induced apoptosis (Liu et al., 2004).
Fig. 2. Myoblasts depleted for Brg1 undergo apoptosis.
(A) Representative light microscopy image of TUNEL stained C57BL/6 and Brg1 c/c myoblasts infected or not with Ad-Cre virus; arrowheads indicate apoptotic cells. (B) Representative western blot of activated Caspase-3 in myoblasts infected or not with Ad-Cre virus. PI3K levels were monitored as an internal loading control.
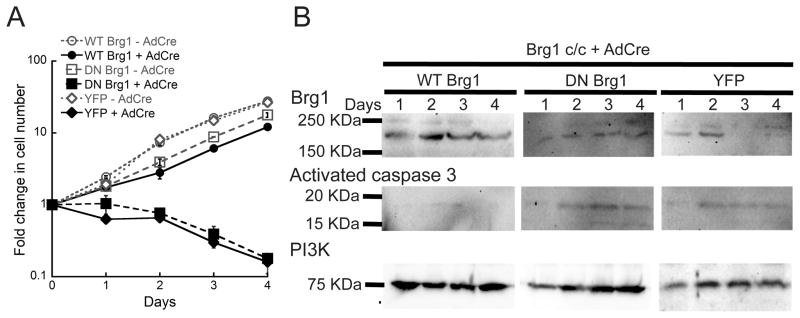
The catalytic activity of Brg1 is required to restore proliferation and to rescue apoptosis in myoblasts depleted for Brg1
We sought to investigate whether reintroduction of Brg1 or a catalytically inactive Brg1 into Brg1-depleted myoblasts would rescue the observed phenotypes. This experiment would demonstrate whether or not the ATPase activity of Brg1 was necessary to promote cell proliferation and prevent cell death. We generated a retrovirus that expresses mCherry flanked by Cre recombination sites upstream of a YFP-tagged wild-type Brg1 (WT-Brg1) or a YFP-tagged, ATPase deficient Brg1 mutant that contains a K-to-R point mutation at the ATP binding site. This mutant has previously been shown to act as a dominant negative (DN-Brg1) (De la Serna et al., 2000; de la Serna et al., 2005; Khavari et al., 1993). In the absence of Cre recombinase, only the mCherry is expressed. Upon introduction of Cre-recombinase, the conditional endogenous Brg1 alleles were deleted, the mCherry encoded by the retrovirus was deleted, and the YFP-Brg1 was expressed. The WT-Brg1 was able to rescue the proliferation phenotype in Brg1-depleted myoblasts, whereas the DN-Brg1 was not (Fig. 3A). Expression of both the WT-Brg1 and DN-Brg1 proteins was verified by western blot over the course of the experiment (Fig. 3B). Similarly, reintroduction of WT-Brg1, but not DN-Brg1, rescued Brg1 deficient myoblasts from apoptosis as shown by a decrease in the expression of activated Caspase-3 (Fig. 3B). We conclude that Brg1 is required for myoblast proliferation and viability and that these functions require a functional ATPase domain. We postulate that the requirement for the catalytic activity indicates tha- the mechanism of Brg1 function involves alterations in gene expression via its ATP-dependent chromatin remodeling function.
Fig. 3. Brg1 ATPase activity is required for myoblast proliferation and survival.
Brg1 c/c myoblasts were infected with an inducible pBABE vector encoding wild type (WT) or ATPase deficient (DN) Brg1 and with the Ad-Cre virus. YFP expressing retroviral vector was used as a control. (A) Complementation of Brg1 depleted myoblasts with WT but not DN Brg1 restored proliferation and survival. Data are the mean ± S.E. from three independent experiments. (B) Representative western blots showing expression of exogenous WT and DN Brg1 in Ad-Cre infected Brg1 c/c myoblasts. Levels of activated Caspase-3 were also evaluated. PI3K levels were monitored as an internal loading control.
Brg1 controls myoblast proliferation and survival by remodeling chromatin at the Pax7 promoter
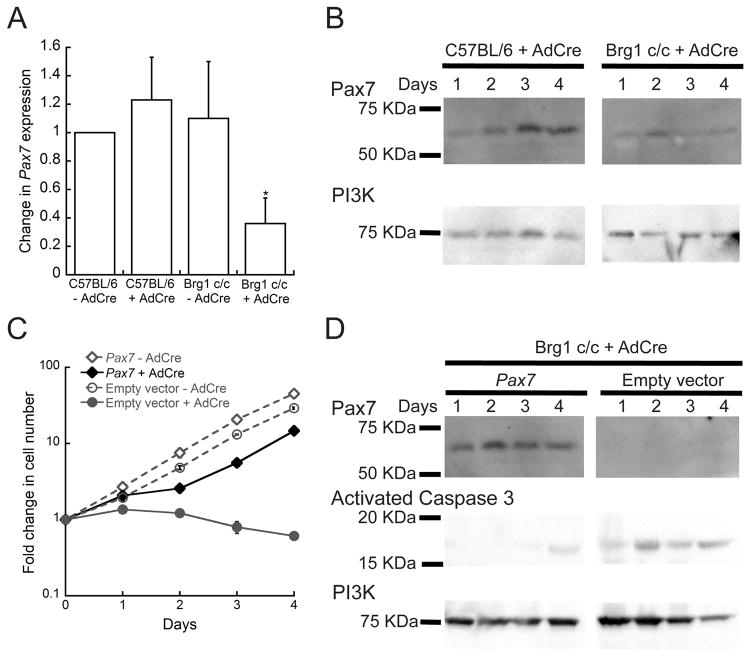
The decrease in proliferation and viability observed in the Brg1 depleted myoblasts suggested that expression of one or more genes required for the maintenance of myoblasts were perturbed. Pax7 is an important regulator of the proliferative state of muscle satellite cells (Gunther et al., 2013; Oustanina et al., 2004; Seale et al., 2000; von Maltzahn et al., 2013). Given the roles of Brg1 and Pax7 in myoblast proliferation, we tested the possibility that Brg1 might be regulating Pax7 mRNA and protein levels in proliferating satellite cell-derived myoblasts. We observed a decrease in Pax7 expression in proliferating Brg1-depleted myoblasts compared to the controls (Fig. 4A, B). This phenotype was partially rescued when Brg1-depleted myoblasts were co-infected with a retrovirus constitutively expressing Pax7 (Fig. 4C, D). These results suggest that a primary function of Brg1 in proliferating myoblasts is to facilitate the expression of Pax7 to maintain myoblast proliferation and survival. Interestingly, expression of Pax7 in non-infected control myoblasts caused a small but reproducible increase in the cell proliferation rate compared to the controls infected with the empty vector pBABE (Fig. 4D). These data indicate that overexpression of Pax7 accelerates the cell proliferation rate of myoblasts, highlighting the importance of Pax7 in the proliferation of satellite cells.
Fig. 4. Pax7 expression is altered in Brg1 depleted myoblasts.
(A) Depletion of Brg1 leads to a decrease in the expression of Pax7 mRNA. The individual mRNA levels were normalized to EF1α levels. The normalized expression levels of the control cells were set as 1. Data are the mean ± S.E. from three independent experiments. * p<0.05. (B) Representative western blot showing the expression of Pax7 in C57BL/6 and Brg1 c/c myoblasts infected with Ad-Cre virus. Samples were collected from day 1 to 4 after Ad-Cre infection. PI3K was used as an internal loading control. (C) Brg1 c/c myoblasts were infected with retrovirus encoding Pax7 or the empty retroviral vector and infected or not with Ad-Cre. A representative western blot shows the increased levels of Pax7 in the cells infected with pBABE-Pax7 compared to the pBABE empty vector. The levels of activated Caspase-3 were also evaluated, and PI3K was used as an internal loading control. (D) Proliferation assay on the cells described in (C). Data are the mean ± S.E. from three independent experiments.
Because of the prior work demonstrating the critical role of Pax7 on myoblast proliferation and maintenance, it was an obvious choice for a potential Brg1-regulated gene. qPCR confirmed that Pax7 expression was indeed dependent on Brg1. We were somewhat surprised that reintroduction of Pax7 into Brg1-depleted myoblasts rescued both viability and proliferation. This result suggests that the requirement for Brg1 may be limited to the expression of Pax7 and perhaps downstream Pax7 target genes, with modest contributions from other potential Brg1 target genes. It remains to be determined whether Brg1 regulates just one or a few genes in proliferating myoblasts or whether it is more widely required for gene expression, even if its contribution to the expression of many genes is not essential. Some precedent for the latter idea exists from an earlier study where it was determined that expression of a dominant negative Brg1 protein in MyoD reprogrammed fibroblasts inhibited differentiation and the expression of most myogenic genes. However, the extent of inhibition among the affected genes varied widely, with about one-third highly dependent on Brg1 and two-thirds showing only modest effects on gene expression in the presence of the catalytically inactive Brg1 (de la Serna et al., 2005).
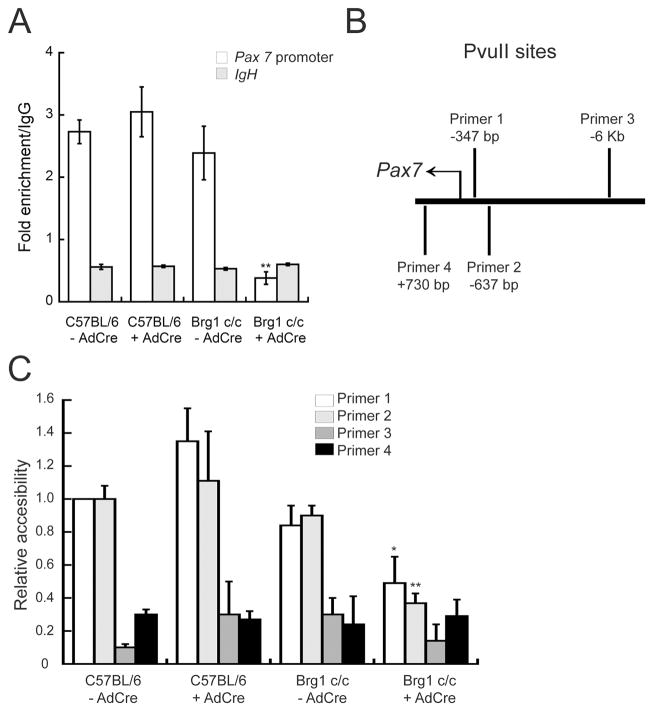
We sought to further explore the mechanism of Brg1-dependent Pax7 expression. Our data raised the possibility that Brg1 is capable of binding chromatin at the Pax7 promoter. Chromatin immunoprecipitation (ChIP) experiments indicated that Brg1 was bound to the Pax7 promoter in proliferating cells (Fig. 5A) as shown by a significant increase in pull-down compared to the IgG control. This interaction was abolished in the Brg1-depleted myoblasts. The IgH enhancer sequence was used as an additional specificity control; Brg1 binding was not observed at this sequence (Fig. 5A). Gene transcription is associated with alteration of nucleosome organization at regulatory sequences, which allows the binding of transcription factors to the chromatin and renders the chromatin more accessible to nuclease cleavage. Such changes are generally a consequence of ATP-dependent chromatin remodeling enzyme function. In order to determine whether or not Brg1 controls chromatin remodeling at the Pax7 promoter, we identified several PvuII cleavage sites in the promoter region of Pax7 (Fig. 5B) and assayed for changes in the accessibility of this enzyme upon Brg1 depletion. As shown in Figure 5C, a strong increase in PvuII accessibility was observed in the control cells at two nuclease sites (primer sets 1 and 2, Fig. 5B) in the promoter region of Pax7 that are 347 and 637 bp upstream of the transcription start site, respectively. In contrast, depletion of Brg1 by Ad-Cre infection in the conditional myoblasts led to a decrease in nuclease accessibility at the Pax7 promoter, resulting in a reduced level of PvuII cleavage (Fig. 5C). In contrast, PvuII sites located 6 kb upstream of the Pax7 transcription start site (primer set 3) or within the Pax7 coding region (primer set 4) showed no change in chromatin accessibility in any of the cells tested (Fig. 5C).
Fig. 5. Brg1 binds to and remodels chromatin at the Pax7 promoter in proliferating myoblasts.
(A) Chromatin immunoprecipitation assay of proliferating C57BL/6 and Brg1 c/c myoblasts infected or not with Ad-Cre virus. Brg1 binding to the Pax7 promoter region was shown by a significant increase in binding compared to IgG. Brg1 binding to the IgH enhancer was used as a control. Depletion of Brg1 led to a significant decrease in binding. Data are the mean ± S.E. of three independent experiments. ** p<0.01, comparing values for Brg1 c/c +/− Ad-Cre samples. (B) Schematic representation of the location of the PvuII sites and primers used for restriction enzyme accessibility assay (REAA). (C) REAA of C57BL/6 and Brg1 c/c myoblasts infected or not with Ad-Cre virus. The accessibility of PvuII sites in the Pax7 promoter was increased as measured by REAA relative to PvuII sites far upstream or in the coding sequence of the Pax7 gene. Depletion of Brg1 upon Ad-Cre infection led to a decrease in accessibility at promoter PvuII sites. All values are relative to the value for primer 1 in the uninfected C57BL/6 myoblasts, which was set to 1. Data are the mean ± S.E. from three independent experiments. * p<0.05; ** p<0.01.
The data suggest a model where Brg1, as the enzymatic subunit of the chromatin remodeling SWI/SNF complex, functions as a co-activator at the Pax7 promoter in proliferating myoblasts to drive Pax7 expression. This would be consistent with previous findings indicating that Polycomb family members repress Pax7 expression in satellite cells (Palacios et al., 2010) and the overall theme that Polycomb mediated repression is opposed by proteins of the Trithorax family, which includes Brg1 and related proteins (Kingston and Tamkun, 2014; Tamkun et al., 1992).
We suggest that Brg1 may function as a co-activator for the CCAAT/Enhancer Binding Protein beta (C/EBPβ) transcriptional activator protein. Recent evidence implicates C/EBPβ as a direct regulator of Pax7 expression in satellite cells and proliferating myoblasts (Fu et al., 2015; Marchildon et al., 2012). C/EBPβ is expressed in Pax7 expressing satellite cells and, like Pax7 itself, is down-regulated upon differentiation signaling. A putative C/EBPβ binding site was identified in the Pax7 promoter; ChIP experiments demonstrated C/EBPβ binding to promoter sequences containing this site and C/EBPβ stimulated the expression of Pax7 reporter genes. Knockdown of C/EBPβ resulted in premature myogenic differentiation in culture, even in the absence of differentiation signaling, and increased muscle fiber cross-sectional area and enhanced muscle repair after injury in vivo (Marchildon et al., 2012). Thus C/EBPβ is a critical regulator of Pax7 in satellite cells and myoblasts.
Multiple examples of Brg1-mediated co-activation of C/EBPβ exist. During bone differentiation, C/EBPβ and Brg1-based SWI/SNF chromatin remodeling enzymes cooperate to positively and negatively regulate tissue-specific gene expression and to ultimately drive differentiation (Aguilar et al., 2014; Grandy et al., 2011; Villagra et al., 2006). Vitamin D regulated transcription involves C/EBPβ and SWI/SNF enzymes (Christakos et al., 2007; Seth-Vollenweider et al., 2014; Zhong et al., 2009). Cooperativity between C/EBPβ and Brg1 has also been reported in the regulation of mammary-specific gene expression (Xu et al., 2007) and cooperativity between C/EBPβ and other SWI/SNF subunits indicate a role in adipogenic and myeloid differentiation (Caramel et al., 2008; Kowenz-Leutz and Leutz, 1999; Kowenz-Leutz et al., 2010). Given the evidence listed here, it seems likely that the requirement for Brg1 in Pax7 transcription in proliferating myoblasts is also dependent on C/EBPβ, though the evidence does not preclude a role for Brg1 in facilitating the function of other components of the transcriptional machinery present at the Pax7 promoter.
In conclusion, we report a direct and essential contribution for the Brg1 ATPase of the SWI/SNF chromatin remodeling enzyme in the proliferation and viability of satellite cell-derived myoblasts via a mechanism that promotes chromatin remodeling at and transcription of the Pax7 gene. The data extend the requirement for Brg1 in myogenesis from a role in facilitating differentiation-specific gene expression and differentiation to maintenance of the myoblast population.
Acknowledgments
Contract grant sponsor: NIH; Contract grant number: GM56244.
We thank G. Hager for plasmids, and Y-J. Hu for the critical reading of the manuscript. This work was funded by NIH grant GM56244.
References
- Aguilar R, Grandy R, Meza D, Sepulveda H, Pihan P, van Wijnen AJ, Lian JB, Stein GS, Stein JL, Montecino M. A functional N-terminal domain in C/EBPbeta-LAP* is required for interacting with SWI/SNF and to repress Ric-8B gene transcription in osteoblasts. Journal of cellular physiology. 2014;229(10):1521–1528. doi: 10.1002/jcp.24595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartholomew B. Regulating the chromatin landscape: structural and mechanistic perspectives. Annual review of biochemistry. 2014;83:671–696. doi: 10.1146/annurev-biochem-051810-093157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bischoff R, Heintz C. Enhancement of skeletal muscle regeneration. Developmental dynamics : an official publication of the American Association of Anatomists. 1994;201(1):41–54. doi: 10.1002/aja.1002010105. [DOI] [PubMed] [Google Scholar]
- Brack AS, Rando TA. Tissue-specific stem cells: lessons from the skeletal muscle satellite cell. Cell stem cell. 2012;10(5):504–514. doi: 10.1016/j.stem.2012.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckingham M, Rigby PW. Gene regulatory networks and transcriptional mechanisms that control myogenesis. Dev Cell. 2014;28(3):225–238. doi: 10.1016/j.devcel.2013.12.020. [DOI] [PubMed] [Google Scholar]
- Bultman S, Gebuhr T, Yee D, La Mantia C, Nicholson J, Gilliam A, Randazzo F, Metzger D, Chambon P, Crabtree G, Magnuson T. A Brg1 null mutation in the mouse reveals functional differences among mammalian SWI/SNF complexes. Molecular cell. 2000;6(6):1287–1295. doi: 10.1016/s1097-2765(00)00127-1. [DOI] [PubMed] [Google Scholar]
- Bultman SJ, Gebuhr TC, Magnuson T. A Brg1 mutation that uncouples ATPase activity from chromatin remodeling reveals an essential role for SWI/SNF-related complexes in beta-globin expression and erythroid development. Genes Dev. 2005;19(23):2849–2861. doi: 10.1101/gad.1364105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bultman SJ, Gebuhr TC, Pan H, Svoboda P, Schultz RM, Magnuson T. Maternal BRG1 regulates zygotic genome activation in the mouse. Genes Dev. 2006;20(13):1744–1754. doi: 10.1101/gad.1435106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caramel J, Medjkane S, Quignon F, Delattre O. The requirement for SNF5/INI1 in adipocyte differentiation highlights new features of malignant rhabdoid tumors. Oncogene. 2008;27(14):2035–2044. doi: 10.1038/sj.onc.1210847. [DOI] [PubMed] [Google Scholar]
- Chang NC, Rudnicki MA. Satellite cells: the architects of skeletal muscle. Current topics in developmental biology. 2014;107:161–181. doi: 10.1016/B978-0-12-416022-4.00006-8. [DOI] [PubMed] [Google Scholar]
- Chi TH, Wan M, Lee PP, Akashi K, Metzger D, Chambon P, Wilson CB, Crabtree GR. Sequential roles of Brg, the ATPase subunit of BAF chromatin remodeling complexes, in thymocyte development. Immunity. 2003;19(2):169–182. doi: 10.1016/s1074-7613(03)00199-7. [DOI] [PubMed] [Google Scholar]
- Christakos S, Dhawan P, Peng X, Obukhov AG, Nowycky MC, Benn BS, Zhong Y, Liu Y, Shen Q. New insights into the function and regulation of vitamin D target proteins. The Journal of steroid biochemistry and molecular biology. 2007;103(3–5):405–410. doi: 10.1016/j.jsbmb.2006.12.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clapier CR, Cairns BR. The biology of chromatin remodeling complexes. Annual review of biochemistry. 2009;78:273–304. doi: 10.1146/annurev.biochem.77.062706.153223. [DOI] [PubMed] [Google Scholar]
- Cruzat F, Henriquez B, Villagra A, Hepp M, Lian JB, van Wijnen AJ, Stein JL, Imbalzano AN, Stein GS, Montecino M. SWI/SNF-independent nuclease hypersensitivity and an increased level of histone acetylation at the P1 promoter accompany active transcription of the bone master gene Runx2. Biochemistry. 2009;48(30):7287–7295. doi: 10.1021/bi9004792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dacwag CS, Ohkawa Y, Pal S, Sif S, Imbalzano AN. The protein arginine methyltransferase Prmt5 is required for myogenesis because it facilitates ATP-dependent chromatin remodeling. Mol Cell Biol. 2007;27(1):384–394. doi: 10.1128/MCB.01528-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De la Serna IL, Carlson KA, Hill DA, Guidi CJ, Stephenson RO, Sif S, Kingston RE, Imbalzano AN. Mammalian SWI-SNF complexes contribute to activation of the hsp70 gene. Mol Cell Biol. 2000;20(8):2839–2851. doi: 10.1128/mcb.20.8.2839-2851.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Serna IL, Ohkawa Y, Berkes CA, Bergstrom DA, Dacwag CS, Tapscott SJ, Imbalzano AN. MyoD targets chromatin remodeling complexes to the myogenin locus prior to forming a stable DNA-bound complex. Mol Cell Biol. 2005;25(10):3997–4009. doi: 10.1128/MCB.25.10.3997-4009.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Serna IL, Ohkawa Y, Imbalzano AN. Chromatin remodelling in mammalian differentiation: lessons from ATP-dependent remodellers. Nat Rev Genet. 2006;7(6):461–473. doi: 10.1038/nrg1882. [DOI] [PubMed] [Google Scholar]
- Fu D, Lala-Tabbert N, Lee H, Wiper-Bergeron N. Mdm2 Promotes Myogenesis through the Ubiquitination and Degradation of CCAAT/Enhancer-binding Protein beta. The Journal of biological chemistry. 2015;290(16):10200–10207. doi: 10.1074/jbc.M115.638577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gebuhr TC, Kovalev GI, Bultman S, Godfrey V, Su L, Magnuson T. The role of Brg1, a catalytic subunit of mammalian chromatin-remodeling complexes, in T cell development. The Journal of experimental medicine. 2003;198(12):1937–1949. doi: 10.1084/jem.20030714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong F, Fahy D, Liu H, Wang W, Smerdon MJ. Role of the mammalian SWI/SNF chromatin remodeling complex in the cellular response to UV damage. Cell cycle. 2008;7(8):1067–1074. doi: 10.4161/cc.7.8.5647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandy R, Sepulveda H, Aguilar R, Pihan P, Henriquez B, Olate J, Montecino M. The Ric–8B gene is highly expressed in proliferating preosteoblastic cells and downregulated during osteoblast differentiation in a SWI/SNF- and C/EBPbeta-mediated manner. Mol Cell Biol. 2011;31(14):2997–3008. doi: 10.1128/MCB.05096-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunther S, Kim J, Kostin S, Lepper C, Fan CM, Braun T. Myf5-positive satellite cells contribute to Pax7-dependent long-term maintenance of adult muscle stem cells. Cell stem cell. 2013;13(5):590–601. doi: 10.1016/j.stem.2013.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez-Hernandez JM, Mallappa C, Nasipak BT, Oesterreich S, Imbalzano AN. The Scaffold attachment factor b1 (Safb1) regulates myogenic differentiation by facilitating the transition of myogenic gene chromatin from a repressed to an activated state. Nucleic acids research. 2013;41(11):5704–5716. doi: 10.1093/nar/gkt285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho L, Crabtree GR. Chromatin remodelling during development. Nature. 2010;463(7280):474–484. doi: 10.1038/nature08911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho L, Ronan JL, Wu J, Staahl BT, Chen L, Kuo A, Lessard J, Nesvizhskii AI, Ranish J, Crabtree GR. An embryonic stem cell chromatin remodeling complex, esBAF, is essential for embryonic stem cell self-renewal and pluripotency. Proc Natl Acad Sci U S A. 2009;106(13):5181–5186. doi: 10.1073/pnas.0812889106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imbalzano AN, Kwon H, Green MR, Kingston RE. Facilitated binding of TATA-binding protein to nucleosomal DNA. Nature. 1994;370(6489):481–485. doi: 10.1038/370481a0. [DOI] [PubMed] [Google Scholar]
- Indra AK, Dupe V, Bornert JM, Messaddeq N, Yaniv M, Mark M, Chambon P, Metzger D. Temporally controlled targeted somatic mutagenesis in embryonic surface ectoderm and fetal epidermal keratinocytes unveils two distinct developmental functions of BRG1 in limb morphogenesis and skin barrier formation. Development. 2005;132(20):4533–4544. doi: 10.1242/dev.02019. [DOI] [PubMed] [Google Scholar]
- Johnson TA, Elbi C, Parekh BS, Hager GL, John S. Chromatin remodeling complexes interact dynamically with a glucocorticoid receptor-regulated promoter. Molecular biology of the cell. 2008;19(8):3308–3322. doi: 10.1091/mbc.E08-02-0123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khavari PA, Peterson CL, Tamkun JW, Mendel DB, Crabtree GR. Brg1 Contains a Conserved Domain of the Swi2/Snf2 Family Necessary for Normal Mitotic Growth and Transcription. Nature. 1993;366(6451):170–174. doi: 10.1038/366170a0. [DOI] [PubMed] [Google Scholar]
- Kidder BL, Palmer S, Knott JG. SWI/SNF-Brg1 regulates self-renewal and occupies core pluripotency-related genes in embryonic stem cells. Stem cells. 2009;27(2):317–328. doi: 10.1634/stemcells.2008-0710. [DOI] [PubMed] [Google Scholar]
- Kingston RE, Tamkun JW. Transcriptional regulation by trithorax-group proteins. Cold Spring Harbor perspectives in biology. 2014;6(10):a019349. doi: 10.1101/cshperspect.a019349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knapp JR, Davie JK, Myer A, Meadows E, Olson EN, Klein WH. Loss of myogenin in postnatal life leads to normal skeletal muscle but reduced body size. Development. 2006;133(4):601–610. doi: 10.1242/dev.02249. [DOI] [PubMed] [Google Scholar]
- Kothandapani A, Gopalakrishnan K, Kahali B, Reisman D, Patrick SM. Downregulation of SWI/SNF chromatin remodeling factor subunits modulates cisplatin cytotoxicity. Experimental cell research. 2012;318(16):1973–1986. doi: 10.1016/j.yexcr.2012.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowenz-Leutz E, Leutz A. A C/EBP beta isoform recruits the SWI/SNF complex to activate myeloid genes. Molecular cell. 1999;4(5):735–743. doi: 10.1016/s1097-2765(00)80384-6. [DOI] [PubMed] [Google Scholar]
- Kowenz-Leutz E, Pless O, Dittmar G, Knoblich M, Leutz A. Crosstalk between C/EBPbeta phosphorylation, arginine methylation, and SWI/SNF/Mediator implies an indexing transcription factor code. Embo J. 2010;29(6):1105–1115. doi: 10.1038/emboj.2010.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon H, Imbalzano AN, Khavari PA, Kingston RE, Green MR. Nucleosome disruption and enhancement of activator binding by a human SW1/SNF complex. Nature. 1994;370(6489):477–481. doi: 10.1038/370477a0. [DOI] [PubMed] [Google Scholar]
- Lessard J, Wu JI, Ranish JA, Wan M, Winslow MM, Staahl BT, Wu H, Aebersold R, Graef IA, Crabtree GR. An essential switch in subunit composition of a chromatin remodeling complex during neural development. Neuron. 2007;55(2):201–215. doi: 10.1016/j.neuron.2007.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Xiong Y, Shang C, Twu KY, Hang CT, Yang J, Han P, Lin CY, Lin CJ, Tsai FC, Stankunas K, Meyer T, Bernstein D, Pan M, Chang CP. Brg1 governs distinct pathways to direct multiple aspects of mammalian neural crest cell development. Proc Natl Acad Sci U S A. 2013;110(5):1738–1743. doi: 10.1073/pnas.1218072110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu K, Luo Y, Lin FT, Lin WC. TopBP1 recruits Brg1/Brm to repress E2F1-induced apoptosis, a novel pRb-independent and E2F1-specific control for cell survival. Genes Dev. 2004;18(6):673–686. doi: 10.1101/gad.1180204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Tian X, Wang F, Ma Y, Kornmann M, Yang Y. BRG1 promotes chemoresistance of pancreatic cancer cells through crosstalking with Akt signalling. European journal of cancer. 2014;50(13):2251–2262. doi: 10.1016/j.ejca.2014.05.017. [DOI] [PubMed] [Google Scholar]
- Mansouri A, Stoykova A, Torres M, Gruss P. Dysgenesis of cephalic neural crest derivatives in Pax7−/− mutant mice. Development. 1996;122(3):831–838. doi: 10.1242/dev.122.3.831. [DOI] [PubMed] [Google Scholar]
- Marchildon F, Lala N, Li G, St-Louis C, Lamothe D, Keller C, Wiper-Bergeron N. CCAAT/enhancer binding protein beta is expressed in satellite cells and controls myogenesis. Stem cells. 2012;30(12):2619–2630. doi: 10.1002/stem.1248. [DOI] [PubMed] [Google Scholar]
- Matsumoto S, Banine F, Struve J, Xing R, Adams C, Liu Y, Metzger D, Chambon P, Rao MS, Sherman LS. Brg1 is required for murine neural stem cell maintenance and gliogenesis. Developmental biology. 2006;289(2):372–383. doi: 10.1016/j.ydbio.2005.10.044. [DOI] [PubMed] [Google Scholar]
- Mauro A. Satellite cell of skeletal muscle fibers. The Journal of biophysical and biochemical cytology. 1961;9:493–495. doi: 10.1083/jcb.9.2.493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montarras D, L’Honore A, Buckingham M. Lying low but ready for action: the quiescent muscle satellite cell. The FEBS journal. 2013;280(17):4036–4050. doi: 10.1111/febs.12372. [DOI] [PubMed] [Google Scholar]
- Morgenstern JP, Land H. Advanced mammalian gene transfer: high titre retroviral vectors with multiple drug selection markers and a complementary helper-free packaging cell line. Nucleic acids research. 1990;18(12):3587–3596. doi: 10.1093/nar/18.12.3587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motohashi N, Asakura A. Muscle satellite cell heterogeneity and self-renewal. Frontiers in cell and developmental biology. 2014;2:1. doi: 10.3389/fcell.2014.00001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muchardt C, Yaniv M. A Human Homolog of Saccharomyces-Cerevisiae Snf2/Swi2 and Drosophila-Brm Genes Potentiates Transcriptional Activation by the Glucocorticoid Receptor. Embo J. 1993;12(11):4279–4290. doi: 10.1002/j.1460-2075.1993.tb06112.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller-Planitz F, Klinker H, Becker PB. Nucleosome sliding mechanisms: new twists in a looped history. Nature structural & molecular biology. 2013;20(9):1026–1032. doi: 10.1038/nsmb.2648. [DOI] [PubMed] [Google Scholar]
- Ohkawa Y, Mallappa C, Vallaster CS, Imbalzano AN. An improved restriction enzyme accessibility assay for analyzing changes in chromatin structure in samples of limited cell number. Methods Mol Biol. 2012;798:531–542. doi: 10.1007/978-1-61779-343-1_32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohkawa Y, Marfella CG, Imbalzano AN. Skeletal muscle specification by myogenin and Mef2D via the SWI/SNF ATPase Brg1. Embo J. 2006;25(3):490–501. doi: 10.1038/sj.emboj.7600943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohkawa Y, Yoshimura S, Higashi C, Marfella CG, Dacwag CS, Tachibana T, Imbalzano AN. Myogenin and the SWI/SNF ATPase Brg1 maintain myogenic gene expression at different stages of skeletal myogenesis. The Journal of biological chemistry. 2007;282(9):6564–6570. doi: 10.1074/jbc.M608898200. [DOI] [PubMed] [Google Scholar]
- Olguin HC, Olwin BB. Pax-7 up-regulation inhibits myogenesis and cell cycle progression in satellite cells: a potential mechanism for self-renewal. Developmental biology. 2004;275(2):375–388. doi: 10.1016/j.ydbio.2004.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olguin HC, Yang Z, Tapscott SJ, Olwin BB. Reciprocal inhibition between Pax7 and muscle regulatory factors modulates myogenic cell fate determination. J Cell Biol. 2007;177(5):769–779. doi: 10.1083/jcb.200608122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ondrusova L, Vachtenheim J, Reda J, Zakova P, Benkova K. MITF-independent pro-survival role of BRG1-containing SWI/SNF complex in melanoma cells. PloS one. 2013;8(1):e54110. doi: 10.1371/journal.pone.0054110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oustanina S, Hause G, Braun T. Pax7 directs postnatal renewal and propagation of myogenic satellite cells but not their specification. Embo J. 2004;23(16):3430–3439. doi: 10.1038/sj.emboj.7600346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palacios D, Mozzetta C, Consalvi S, Caretti G, Saccone V, Proserpio V, Marquez VE, Valente S, Mai A, Forcales SV, Sartorelli V, Puri PL. TNF/p38alpha/polycomb signaling to Pax7 locus in satellite cells links inflammation to the epigenetic control of muscle regeneration. Cell stem cell. 2010;7(4):455–469. doi: 10.1016/j.stem.2010.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JH, Park EJ, Hur SK, Kim S, Kwon J. Mammalian SWI/SNF chromatin remodeling complexes are required to prevent apoptosis after DNA damage. DNA repair. 2009;8(1):29–39. doi: 10.1016/j.dnarep.2008.08.011. [DOI] [PubMed] [Google Scholar]
- Relaix F, Montarras D, Zaffran S, Gayraud-Morel B, Rocancourt D, Tajbakhsh S, Mansouri A, Cumano A, Buckingham M. Pax3 and Pax7 have distinct and overlapping functions in adult muscle progenitor cells. J Cell Biol. 2006;172(1):91–102. doi: 10.1083/jcb.200508044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saladi SV, Wong PG, Trivedi AR, Marathe HG, Keenen B, Aras S, Liew ZQ, Setaluri V, de la Serna IL. BRG1 promotes survival of UV-irradiated melanoma cells by cooperating with MITF to activate the melanoma inhibitor of apoptosis gene. Pigment cell & melanoma research. 2013;26(3):377–391. doi: 10.1111/pcmr.12088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambasivan R, Tajbakhsh S. Adult skeletal muscle stem cells. Results and problems in cell differentiation. 2015;56:191–213. doi: 10.1007/978-3-662-44608-9_9. [DOI] [PubMed] [Google Scholar]
- Seale P, Sabourin LA, Girgis-Gabardo A, Mansouri A, Gruss P, Rudnicki MA. Pax7 is required for the specification of myogenic satellite cells. Cell. 2000;102(6):777–786. doi: 10.1016/s0092-8674(00)00066-0. [DOI] [PubMed] [Google Scholar]
- Seo S, Richardson GA, Kroll KL. The SWI/SNF chromatin remodeling protein Brg1 is required for vertebrate neurogenesis and mediates transactivation of Ngn and NeuroD. Development. 2005;132(1):105–115. doi: 10.1242/dev.01548. [DOI] [PubMed] [Google Scholar]
- Seth-Vollenweider T, Joshi S, Dhawan P, Sif S, Christakos S. Novel mechanism of negative regulation of 1,25-dihydroxyvitamin D3-induced 25-hydroxyvitamin D3 24-hydroxylase (Cyp24a1) Transcription: epigenetic modification involving cross-talk between protein-arginine methyltransferase 5 and the SWI/SNF complex. The Journal of biological chemistry. 2014;289(49):33958–33970. doi: 10.1074/jbc.M114.583302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sif S, Saurin AJ, Imbalzano AN, Kingston RE. Purification and characterization of mSin3A-containing Brg1 and hBrm chromatin remodeling complexes. Genes Dev. 2001;15(5):603–618. doi: 10.1101/gad.872801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stec DE, Davisson RL, Haskell RE, Davidson BL, Sigmund CD. Efficient liver-specific deletion of a floxed human angiotensinogen transgene by adenoviral delivery of Cre recombinase in vivo. The Journal of biological chemistry. 1999;274(30):21285–21290. doi: 10.1074/jbc.274.30.21285. [DOI] [PubMed] [Google Scholar]
- Sumi-Ichinose C, Ichinose H, Metzger D, Chambon P. SNF2beta-BRG1 is essential for the viability of F9 murine embryonal carcinoma cells. Mol Cell Biol. 1997;17(10):5976–5986. doi: 10.1128/mcb.17.10.5976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamkun JW, Deuring R, Scott MP, Kissinger M, Pattatucci AM, Kaufman TC, Kennison JA. brahma: a regulator of Drosophila homeotic genes structurally related to the yeast transcriptional activator SNF2/SWI2. Cell. 1992;68(3):561–572. doi: 10.1016/0092-8674(92)90191-e. [DOI] [PubMed] [Google Scholar]
- Villagra A, Cruzat F, Carvallo L, Paredes R, Olate J, van Wijnen AJ, Stein GS, Lian JB, Stein JL, Imbalzano AN, Montecino M. Chromatin remodeling and transcriptional activity of the bone-specific osteocalcin gene require CCAAT/enhancer-binding protein beta-dependent recruitment of SWI/SNF activity. The Journal of biological chemistry. 2006;281(32):22695–22706. doi: 10.1074/jbc.M511640200. [DOI] [PubMed] [Google Scholar]
- von Maltzahn J, Jones AE, Parks RJ, Rudnicki MA. Pax7 is critical for the normal function of satellite cells in adult skeletal muscle. Proc Natl Acad Sci U S A. 2013;110(41):16474–16479. doi: 10.1073/pnas.1307680110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Cote J, Xue Y, Zhou S, Khavari PA, Biggar SR, Muchardt C, Kalpana GV, Goff SP, Yaniv M, Workman JL, Crabtree GR. Purification and biochemical heterogeneity of the mammalian SWI-SNF complex. Embo J. 1996;15(19):5370–5382. [PMC free article] [PubMed] [Google Scholar]
- Wu JI. Diverse functions of ATP-dependent chromatin remodeling complexes in development and cancer. Acta biochimica et biophysica Sinica. 2012;44(1):54–69. doi: 10.1093/abbs/gmr099. [DOI] [PubMed] [Google Scholar]
- Xu R, Spencer VA, Bissell MJ. Extracellular matrix-regulated gene expression requires cooperation of SWI/SNF and transcription factors. The Journal of biological chemistry. 2007;282(20):14992–14999. doi: 10.1074/jbc.M610316200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zammit PS, Golding JP, Nagata Y, Hudon V, Partridge TA, Beauchamp JR. Muscle satellite cells adopt divergent fates: a mechanism for self-renewal? J Cell Biol. 2004;166(3):347–357. doi: 10.1083/jcb.200312007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zammit PS, Relaix F, Nagata Y, Ruiz AP, Collins CA, Partridge TA, Beauchamp JR. Pax7 and myogenic progression in skeletal muscle satellite cells. Journal of cell science. 2006;119(Pt 9):1824–1832. doi: 10.1242/jcs.02908. [DOI] [PubMed] [Google Scholar]
- Zhong Y, Armbrecht HJ, Christakos S. Calcitonin, a regulator of the 25-hydroxyvitamin D3 1alpha-hydroxylase gene. The Journal of biological chemistry. 2009;284(17):11059–11069. doi: 10.1074/jbc.M806561200. [DOI] [PMC free article] [PubMed] [Google Scholar]