Abstract
Recent advances in VWD research have improved our understanding of the genotype and phenotype of VWD. The VWF gene is highly polymorphic, with a large number of sequence variations reported in healthy individuals. This can lead to some difficulty when attempting to discern genotype-phenotype correlations, as sequence variations may not represent disease. In type 1 VWD, mutations can be found throughout the VWF gene, but likely pathogenic sequence variations are found in only about 2/3 of type 1 VWD patients. Sequence variations in type 2 VWD are located in the region corresponding to the defect in the VWF protein found in each type 2 variant. In type 3 VWD, sequence variations are not confined to a specific region of the VWF gene, and also include large deletions which may not be picked up using conventional sequencing techniques. Use of genetic testing may be most helpful in diagnosis of type 2 VWD, where a larger number of known, well characterized mutations are present and demonstration of one of these may help confirm the diagnosis. Bleeding symptoms in general are more severe with decreasing VWF levels, and more severe in type 2 and type 3 VWD as compared to type 1 VWD. Prediction of phenotype for an individual patient, however, is still difficult, and the addition of genetic data most helpful in ascertaining the correct diagnosis for VWD patients.
Learner objective: to understand the relationship of genotype and phenotype as currently understood for VWD variants
Introduction
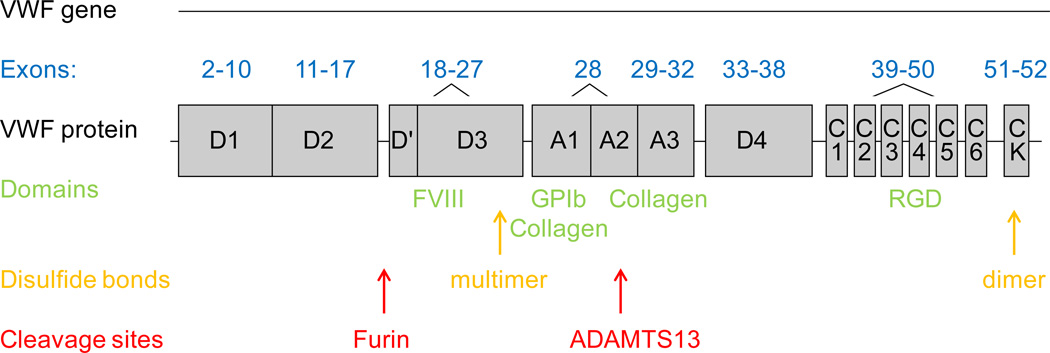
Von Willebrand factor (VWF) is the product of a large gene located on the short arm of chromosome 12, with 52 exons spread over about 178 kb of genomic DNA and 8439 bp of coding sequence.1 Apart from its size, the other factor contributing to genetic heterogeneity is the presence of a pseudogene on chromosome 22, which mimics VWF exons 23–34.2 An online database maintained by the VWD Scientific and Standardization Committee of the International Society on Thrombosis and Haemostasis (http://www.vwf.group.shef.ac.uk/variant.html) lists known sequence variations, both pathogenic and non-pathogenic.3 The VWF protein contains key functional domains that mediate binding to factor VIII (FVIII), platelet glycoprotein Ib (GPIb), and facilitate multimerization to form the final protein. The exons corresponding to key regions of the VWF protein are noted in figure 1. Common genetic mutation types, their location, and the clinical and laboratory phenotype for VWD type 1, type 3, and type 2 variants are listed in table 1.
Figure 1. VWF gene and protein structure correlation.
The exons comprising the coding sequence of the VWF gene are noted along with the corresponding regions of the VWF protein. Also noted are key functional domains with their ligands, location of disulfide bonds important in C-terminal dimerization and N-terminal multimerization, and enzyme cleavage sites.42
Table 1.
Genotype-phenotype correlations in VWD.
| VWD Type |
Mutation Type | Mutation Location | Inheritance Pattern |
Clinical Phenotype | Laboratory Phenotype |
|---|---|---|---|---|---|
| Type 1 | Missense, nonsense, deletion, insertion, splicing | Throughout VWF gene | Autosomal dominant | Mild-moderate bleeding | Proportionate decrease in VWF:Ag and VWF:RCo |
| Type 3 | Missense, nonsense, deletion, insertion, splicing | Throughout VWF gene | Autosomal recessive | Severe mucosal bleeding, may have joint bleeds | Undetectable VWF:Ag and VWF:RCo |
| Type 2A | Missense* | VWF A2 domain D1, D2, D′, D3 domains CK domain | Autosomal dominant | Moderately severe | ↓VWF:RCo/VWF:Ag ratio and loss of high molecular weight multimers |
| Type 2B | Missense | VWF A1 domain | Autosomal dominant | Moderately severe | ↓VWF:RCo/VWF:Ag ratio and loss of high molecular weight multimers spontaneous platelet binding ± thrombocytopenia |
| Type 2M | Missense* | VWF A1 and A3 domains | Autosomal dominant | Moderately severe | ↓VWF:RCo/VWF:Ag ratio and/or collagen binding defect |
| Type 2N | Missense* | D′ and D3 domains | Autosomal recessive | Moderately severe | Defect in FVIII binding |
Missense mutations are the most frequent, but deletions, insertions, and splicing mutations have been reported.3
Genotype-phenotype correlation: healthy individuals
The VWF gene is highly polymorphic. With the advent of relatively inexpensive whole exome and whole genome sequencing, a large number of sequence variants have been reported. The 1000 Genomes database demonstrated 2728 single nucleotide polymorphisms and 91 insertions/deletions in the VWF gene, with the highest degree of ethnic variability seen in Africans, followed closely by Asians.4
This variability can lead to some difficulty when attempting to discern genotype-phenotype correlations, as sequence variations may not represent disease. Indeed, one study of healthy controls demonstrated a high rate of variation in the VWF gene, particularly in African Americans. Increased genetic variability was observed in African Americans, with 80% of the novel sequence variations from the study, as compared to Caucasians, with only 20% of the novel sequence variations.5 Several sequence variations that had previously been reported in VWD were also found in the healthy control population. A number of variations were found at relatively high frequency (5–20%) in African American healthy controls.5 Caution is therefore required when evaluating pathogenicity of any novel sequence variation found in patients with VWD or who are undergoing workup for possible VWD.
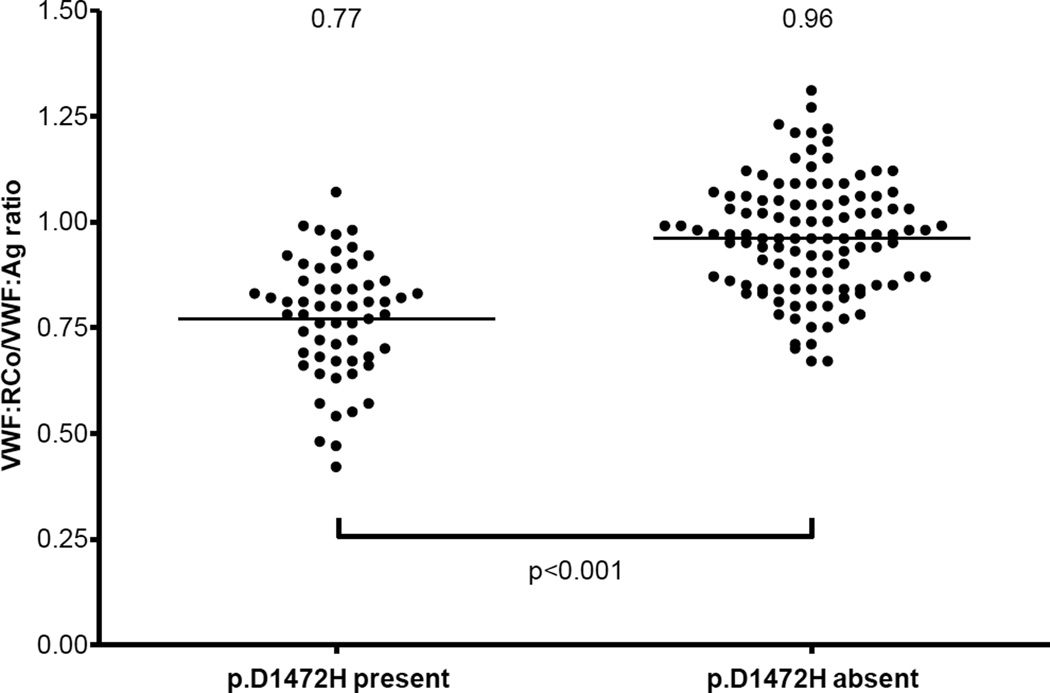
Some genetic variants do affect VWF levels. The NHLBI exome sequencing project identified a number of sequence variations associated with either increased (p.T789A and p.D1472H) or decreased (p.R2185Q) VWF antigen (VWF:Ag) in African Americans.6 The p.D1472H sequence variant has also been associated in healthy individuals with decreased VWF ristocetin cofactor activity (VWF:RCo)/VWF:Ag ratios, as depicted in figure 2.7 Presence of this variant could conceivably result in a mis-diagnosis of type 2M VWD. Polymorphisms in the CLEC4M gene have recently been associated with variations in VWF levels.8 Several other candidate genes have been discovered through genome-wide association studies and may also prove to have a significant role in modifying VWF:Ag.9 It is likely that other modifier genes will be discovered to affect expression or clearance of VWF in the future.
Figure 2. Decreased VWF:RCo/VWF:Ag ratio in healthy control subjects with p.D1472H.
This graph shows VWF:RCo/VWF:Ag ratios for healthy control subjects (59 African Americans and 113 Caucasians) enrolled in the US Zimmerman Program study, grouped by presence or absence of the p.D1472H sequence variation.7 Mean ratios for each group are shown at the top of the graph. There were 7 subjects with ratios <0.6, all either heterozygous or homozygous for p.D1472H. None of the subjects without p.D1472H had ratios <0.7.
Variability in VWF levels can also result from a number of extrinsic factors, some of which may also be genetic. Decreased VWF:Ag is seen in association with blood type O.10 Alterations in VWF glycosylation, which are also seen associated with blood type O, may affect VWF survival.11 African Americans have higher VWF levels as compared to Caucasians.12 Increased VWF:Ag may occur due to stress, age, exercise or pregnancy, or a variety of pro-inflammatory conditions.10,13,14 This can confound the diagnosis of VWD, as a single normal VWF:Ag does not necessarily exclude the diagnosis.
Genotype-phenotype correlation: type 1 VWD
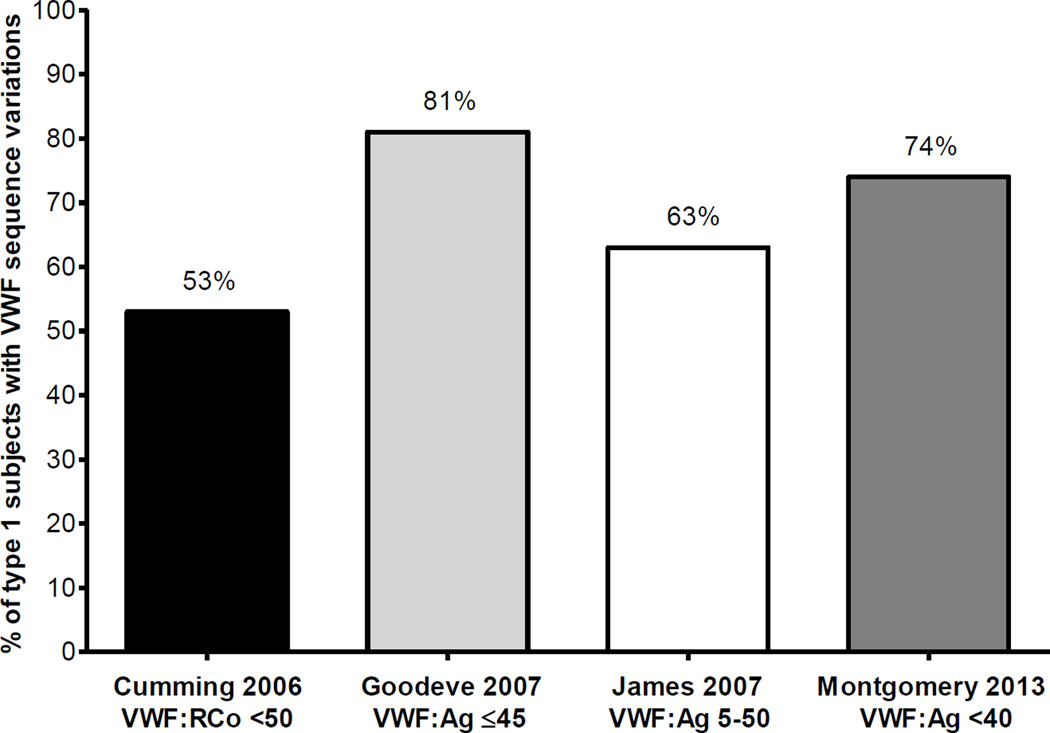
Several large studies of type 1 VWD have recently been performed, representing patients in Europe and North America, including the European Union MCMDM-1VWD study,15 Canada,16 the United Kingdom,17 and the United States.18 Data from these studies show that most (about 70%) mutations in type 1 VWD are missense mutations located in the VWF coding sequence, with splice site, transcription, small deletions, small duplications, and nonsense mutations each representing less than 10% of the mutations found.19 Defects in the VWF promoter have also been associated with type 1 VWD.20 However, only about 2/3 of index cases with type 1 VWD in these studies have a genetic mutation in VWF. Figure 3 summarizes the rate of sequence variations reported by the four recent studies noted above. The typical inheritance pattern for type 1 VWD is autosomal dominant, although variability in penetrance and symptoms have been noted in affected family members.
Figure 3. Rate of sequence variations in type 1 VWD studies.
This graph shows rates of sequence variations discovered in subjects with type 1 VWD in each of 4 large VWD studies. The first column (in black) represents the United Kingdom study subjects with VWF:RCo <50 IU/dL,17 the second column (in light grey) represents the MCMDM-1VWD study type 1 subjects with VWF:Ag ≤45 IU/dL,15 the third column (in white) represents the Canadian type 1 VWD study subjects with VWF:Ag between 5–50 IU/dL,16 and the fourth column (in dark grey) represents the US Zimmerman Program study type 1 subjects with VWF:Ag <40 IU/dL.18 The percent of each group with sequence variations is shown at the top of each column.
The likelihood of finding a causative sequence variation in type 1 VWD is directly proportional to the level of decrease in VWF:Ag; that is, the patients with the lowest VWF:Ag levels have the highest chance of a sequence variation in VWF. The precise cutoff varies by study, with VWF:Ag levels below 20–40 IU/dL most strongly correlated with presence of a VWF sequence variation. The MCMDM-1VWD study from the European Union reported that 88% of type 1 VWD subjects had sequence variations when the VWF:Ag was ≤30 IU/dL. This dropped to 53% in subjects with VWF:Ag between 31–45 IU/dL and 50% when the VWF:Ag was >45 IU/dL.15 In the Canadian type 1 VWD study, where subjects had VWF:Ag between 5 and 50 IU/dL, 63% were found to have sequence variations.16 A study of type 1 VWD from the United Kingdom demonstrated that 53% of subjects had sequence variations, all of whom met diagnostic criteria for type 1 VWD.17 The US Zimmerman Program type 1 VWD cohort demonstrated 74% of subjects to have VWF sequence variations when the VWF:Ag was less than 40 IU/dL.18 A recent Swedish study demonstrated the presence of sequence variations in 56% of their type 1 VWD subjects who underwent detailed sequence analysis.21
Correlation of genotype and phenotype for individual mutations is difficult due to the high degree of variability in type 1 VWD. There is some evidence that bleeding symptoms correlate with the degree of reduction in VWF:Ag.22 However, some patients have extensive bleeding symptoms, despite modest decreases in VWF levels. Other patients report minimal symptoms in the context of significant reductions in VWF. This heterogeneity can occur even within families, where the same mutation is present in each affected family member, yet manifests with different clinical symptoms. Some mutations have been characterized, as reported by Robertson and colleagues, with accelerated clearance and intracellular retention as reported mechanisms.23 Although in their study bleeding scores increased as VWF:RCo decreased, some variation in bleeding scores was observed among subjects who had the same mutation.23
Clearance defects represent a subset of type 1 VWD. In clearance defects (type 1C), there is an association of abnormal propeptide to VWF:Ag ratios with genetic variations, particularly in the D3 and D4 domains.24 The most common of these, p.R1205H or the Vicenza mutation, is caused by accelerated clearance of the VWF protein.25 VWF levels in type 1C do rise in response to desmopressin, but the increase in clearance results in a rapid fall back to the patient’s baseline.24 Genetic diagnosis may be helpful in this subtype, as VWF propeptide levels may not be readily available and a clear diagnosis will facilitate appropriate treatment.
Genotype-phenotype correlation: type 3 VWD
Type 3 VWD presents with undetectable VWF protein. Inheritance is autosomal recessive, although parents of type 3 VWD patients may or may not be symptomatic. Type 3 VWD is due to disruption of expression from both VWF alleles, typically either through point mutations (missense or null) or deletions involving the VWF gene. Some deletions may be small, affecting only one or two exons, while others encompass much larger segments of the VWF gene. The most common deletion found to date is a deletion of exons 4–5.26 Bleeding symptoms in type 3 VWD are on average more extensive than in type 1 or type 2 VWD, as expected given the absent VWF and low FVIII seen in this condition.27
Genetic diagnosis is not required for the diagnosis of type 3 VWD, although on occasion it may be useful in distinguishing severe type 1 VWD due to a clearance defect. Very low, but detectable, VWF:Ag may be seen in clearance defects (type 1C) but absent (undetectable) VWF:Ag is the hallmark of type 3 VWD. This differentiation relies on accurate quantification of VWF levels, which can be difficult when the VWF:Ag is very low. Knowing the causative mutations may also be helpful for prenatal testing. Not all cases of type 3 VWD have two mutations found in the VWF gene, with one recent study finding candidate mutations in only 91% of alleles28 and another study with mutations in only 17 of 19 subjects.29
Genotype-phenotype correlation: type 2A VWD
Type 2A VWD results from mutations that cause a defect in VWF activity through disruption of high molecular weight multimer formation. Inheritance follows an autosomal dominant pattern. These mutations may cause decreased secretion, increased ADAMTS13 proteolysis, defective protein storage, defective multimerization, or a combination of these.30 Mutations that result in increased clearance of high molecular weight multimers are typically found in exon 28, specifically in the A2 domain of VWF where the ADAMTS13 cleavage site is located. Mutations may also be present in either the N- or C-terminal multimerization domains. Since both type 2A and type 2B VWF present with decreased VWF:RCo/VWF:Ag ratios and lack of high molecular weight multimers, genetic diagnosis may be helpful in discriminating between these two type 2 variants.
The VWF A domains contain the majority of reported 2A mutations (82%), with 8% each in the D2 and CK domains and 1% in the D3 domain.31 Some of these variations have been well characterized, with known effects on function of the mutant VWF.30,32 Targeted VWF sequencing, encompassing only these regions, is offered by some laboratories. Unfortunately, novel sequence variations may be difficult to interpret, given the highly polymorphic nature of VWF.
Bleeding symptoms for patients with type 2 VWD are generally more severe than for type 1 VWD. One study reported an increase in bleeding for type 2A patients as compared with type 2M patients.33 This was attributed to an increase in GI bleeding, and did not seem to correlate with level of VWF:Ag. Interestingly, this study demonstrated a range of bleeding scores for subjects with the same mutation.33 Genetic diagnosis of type 2A VWD may be helpful in distinguishing from other type 2 variants, but the specific genotype may not necessarily predict future symptoms.
Genotype-phenotype correlation: type 2B VWD
Type 2B VWD represents a gain-of-function defect, where VWF has an increased affinity for platelet GPIb. This results in clearance of VWF-platelet complexes, loss of high molecular weight multimers, and frequently thrombocytopenia. Inheritance follows an autosomal dominant pattern. Since the pathogenic defect in type 2B is a problem with platelet binding, mutations in type 2B VWD are found exclusively in exon 28 of the VWF gene. There are a number of reported type 2B sequence variations, all in the VWF A1 domain where binding to platelet GPIb occurs.34 Although thrombocytopenia is a hallmark of classic type 2B VWD, recent studies have demonstrated that thrombocytopenia is not necessarily a universal feature.35 Several type 2B variants, specifically p.R1308C, p.P1266L, and p.P1266Q, seem to carry a lower likelihood of thrombocytopenia. Presence of thrombocytopenia has been associated with bleeding.35
Genetic diagnosis may be particularly helpful in determining whether type 2A or type 2B VWD is the cause of a decreased VWF:RCo/VWF:Ag ratio with a loss of high molecular weight multimers. While demonstration of increased VWF binding to platelets, either through elevated low-dose ristocetin-induced platelet aggregation or direct platelet binding assays, is considered to officially confirm the diagnosis of type 2B VWD, it is reasonable to consider demonstration of a known type 2B gene mutation in addition to or in lieu of platelet testing. Genetic diagnosis may also give additional insight into predicted phenotype, along with the platelet count.35
Platelet type, pseudo-VWD presents with similar laboratory findings to type 2B VWD (decreased VWF:RCo/VWF:Ag ratio, thrombocytopenia) but is due to a gain-of-function mutation in platelet GPIb. Missense mutations are the most common cause of platelet-type, pseudo-VWD but one deletion mutation has been reported.36 Failure to diagnose a defect in the VWF gene in a patient who has bleeding symptoms and laboratory findings otherwise consistent with type 2B VWD should trigger investigation of GP1BA.
Genotype-phenotype correlation: type 2M VWD
Type 2M VWD is characterized by a defect in the ability of VWF to bind platelet GPIb and thus a decrease in VWF:RCo/VWF:Ag ratio, but with a normal multimer distribution. This category has also been expanded to include collagen binding defects. Inheritance follows an autosomal dominant pattern. Genetic changes in type 2M VWD are therefore present in either the VWF A1 (platelet and collagen binding) domain or A3 (collagen binding) domain. There are numerous reported mutations in type 2M VWD, and at this point, no clear correlation between genotype and bleeding symptoms. As previously mentioned, one study demonstrated less bleeding in type 2M VWD as compared to type 2A VWD.33 There may be a suggestion of increased bleeding when both a platelet binding and a collagen binding defect are present.37
While most type 2M mutations predominantly affect platelet binding and thus the VWF:RCo activity, several reported variants predominantly or exclusively affect collagen binding. The first reported of these, p.S1731T, had borderline VWF:Ag and VWF:RCo (approximately 50–60 IU/dL) and reduced collagen binding (VWF:CB).38 A recently reported p.M1761K variant had normal VWF:Ag and VWF:RCo but VWF:CB of 10 IU/dL.39 Discovery of an A1 or A3 domain mutation may help confirm a diagnosis of type 2M VWD. However, genotype does not necessarily predict bleeding phenotype. For example, Castaman and colleagues report a series of 23 type 2M subjects who were all heterozygous for the p.R1374H sequence variation, but whose bleeding scores ranged from 1 to 18.33
Genotype-phenotype correlation: type 2N VWD
Type 2N VWD results from defects in the ability of VWF to bind FVIII. Sequence variations in type 2N are therefore found in the FVIII binding region of VWF spanning the D' and D3 domains. Exons 18–20 contain most of the reported type 2N mutations, but defects have been reported in exons 17–27.31 Type 2N VWD results when two mutations in the FVIII binding region are present, one on each allele. It is also possible to have a 2N phenotype resulting from the presence of one type 2N mutation and a type 1 VWF mutation, typically resulting in low VWF or a null allele. Therefore the inheritance pattern is autosomal recessive.
Bleeding in type 2N VWD is also variable. The p.R854Q mutation has been reported with a less severe phenotype,40 while p.R854W has been associated with a severe bleeding phenotype.41 More data is needed on the interaction of type 2N mutations with type 1 mutations and their effect on bleeding symptoms.
Summary
Genetic defects can be demonstrated in approximately 2/3 of type 1 VWD patients, and over 90% of type 2 and 3 VWD patients. However, use of genetic testing to predict phenotype above and beyond the knowledge available from standard VWF assays is limited. Part of the difficulty is the extreme heterogeneity of symptoms in VWD. Even in type 1 VWD, where the defect is a straightforward quantitative deficiency of VWF, bleeding symptoms do not always correlate with VWF:Ag levels. Additional regulatory genes, such as those involved in post-translational modification of VWF, may play a role. Alternately, the presence of modifiers in other coagulation factors may modulate symptoms, such as factor V Leiden creating a prothrombotic state to balance bleeding due to VWD. Prediction of bleeding phenotype based on genotype in VWD is therefore difficult at this time, beyond the general progression of bleeding in severity from type 1 to type 2 to type 3 VWD. Genetic testing can be quite useful, however, in diagnosis of type 2 variants, where mutations in specific regions correspond to the defect in VWF function.
Acknowledgments
The author would like to acknowledge research funding from the National Institutes of Health (HL102260).
Footnotes
Disclosures: No conflicts of interest to disclose.
Bibliography
- 1.Mancuso DJ, Tuley EA, Westfield LA, et al. Structure of the gene for human von willebrand factor. J Biol Chem. 1989;264:19514–19527. [PubMed] [Google Scholar]
- 2.Mancuso DJ, Tuley EA, Westfield LA, et al. Human von willebrand factor gene and pseudogene: Structural analysis and differentiation by polymerase chain reaction. Biochemistry. 1991;30:253–269. doi: 10.1021/bi00215a036. [DOI] [PubMed] [Google Scholar]
- 3.International Society on Thrombosis and Haemostasis Scientific and Standardization Committee. [May 1, 2014];ISTH-SSC VWF Online Database. http://www.vwf.group.shef.ac.uk. [Google Scholar]
- 4.Wang QY, Song J, Gibbs RA, Boerwinkle E, Dong JF, Yu FL. Characterizing polymorphisms and allelic diversity of von willebrand factor gene in the 1000 genomes. J Thromb Haemost. 2013;11:261–269. doi: 10.1111/jth.12093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bellissimo DB, Christopherson PA, Flood VH, et al. VWF mutations and new sequence variations identified in healthy controls are more frequent in the african-american population. Blood. 2012;119:2135–2140. doi: 10.1182/blood-2011-10-384610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Johnsen JM, Auer PL, Morrison AC, et al. Common and rare von willebrand factor (VWF) coding variants, VWF levels, and factor VIII levels in african americans: The NHLBI exome sequencing project. Blood. 2013;122:590–597. doi: 10.1182/blood-2013-02-485094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Flood VH, Gill JC, Morateck PA, et al. Common VWF exon 28 polymorphisms in african americans affecting the VWF activity assay by ristocetin cofactor. Blood. 2010;116:280–286. doi: 10.1182/blood-2009-10-249102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rydz N, Swystun LL, Notley C, et al. The C-type lectin receptor CLEC4M binds, internalizes, and clears von willebrand factor and contributes to the variation in plasma von willebrand factor levels. Blood. 2013;121:5228–5237. doi: 10.1182/blood-2012-10-457507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Smith NL, Chen MH, Dehghan A, et al. Novel associations of multiple genetic loci with plasma levels of factor VII, factor VIII, and von willebrand factor: The CHARGE (cohorts for heart and aging research in genome epidemiology) consortium. Circulation. 2010;121:1382–1392. doi: 10.1161/CIRCULATIONAHA.109.869156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gill JC, Endres-Brooks J, Bauer PJ, Marks WJ, Jr, Montgomery RR. The effect of ABO blood group on the diagnosis of von willebrand disease. Blood. 1987;69:1691–1695. [PubMed] [Google Scholar]
- 11.Castaman G, Tosetto A, Rodeghiero F. Reduced von willebrand factor survival in von Willebrand disease: pathophysiologic and clinical relevance. J Thromb Haemost. 2009;7(Suppl 1):71–74. doi: 10.1111/j.1538-7836.2009.03381.x. [DOI] [PubMed] [Google Scholar]
- 12.Miller CH, Haff E, Platt SJ, et al. Measurement of von willebrand factor activity: Relative effects of ABO blood type and race. J Thromb Haemost. 2003;1:2191–2197. doi: 10.1046/j.1538-7836.2003.00367.x. [DOI] [PubMed] [Google Scholar]
- 13.Hellgren M, Blomback M. Studies on blood coagulation and fibrinolysis in pregnancy, during delivery and in the puerperium. I. normal condition. Gynecol Obstet Invest. 1981;12:141–154. doi: 10.1159/000299596. [DOI] [PubMed] [Google Scholar]
- 14.Stakiw J, Bowman M, Hegadorn C, et al. The effect of exercise on von willebrand factor and ADAMTS-13 in individuals with type 1 and type 2B von willebrand disease. J Thromb Haemost. 2008;6:90–96. doi: 10.1111/j.1538-7836.2007.02790.x. [DOI] [PubMed] [Google Scholar]
- 15.Goodeve A, Eikenboom J, Castaman G, et al. Phenotype and genotype of a cohort of families historically diagnosed with type 1 von willebrand disease in the european study, molecular and clinical markers for the diagnosis and management of type 1 von willebrand disease (MCMDM-1VWD) Blood. 2007;109:112–121. doi: 10.1182/blood-2006-05-020784. [DOI] [PubMed] [Google Scholar]
- 16.James PD, Notley C, Hegadorn C, et al. The mutational spectrum of type 1 von willebrand disease: Results from a canadian cohort study. Blood. 2007;109:145–154. doi: 10.1182/blood-2006-05-021105.. [DOI] [PubMed] [Google Scholar]
- 17.Cumming A, Grundy P, Keeney S, et al. An investigation of the von willebrand factor genotype in UK patients diagnosed to have type 1 von willebrand disease. Thromb Haemost. 2006;96:630–641. [PubMed] [Google Scholar]
- 18.Montgomery RR, Christopherson PA, Bellissimo DB, et al. The complete type I VWD cohort of the zimmerman program for the molecular and clinical biology of VWD - phenotypic assignment, mutation frequency, and bleeding assessment. Blood. 2013;122:332. [Google Scholar]
- 19.Lillicrap D. Von willebrand disease: Advances in pathogenetic understanding, diagnosis, and therapy. Blood. 2013;122:3735–3740. doi: 10.1182/blood-2013-06-498303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Othman M, Chirinian Y, Brown C, et al. Functional characterization of a 13-bp deletion (c.-1522_-1510del13) in the promoter of the von willebrand factor gene in type 1 von willebrand disease. Blood. 2010;116:3645–3652. doi: 10.1182/blood-2009-12-261131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Johansson AM, Hallden C, Sall T, Lethagen S. Variation in the VWF gene in swedish patients with type 1 von willebrand disease. Ann Hum Genet. 2011;75:447–455. doi: 10.1111/j.1469-1809.2011.00652.x. [DOI] [PubMed] [Google Scholar]
- 22.Tosetto A, Rodeghiero F, Castaman G, et al. A quantitative analysis of bleeding symptoms in type 1 von willebrand disease: Results from a multicenter european study (MCMDM-1 VWD) J Thromb Haemost. 2006;4:766–773. doi: 10.1111/j.1538-7836.2006.01847.x. [DOI] [PubMed] [Google Scholar]
- 23.Robertson JD, Yenson PR, Rand ML, et al. Expanded phenotype-genotype correlations in a pediatric population with type 1 von willebrand disease. J Thromb Haemost. 2011;9:1752–1760. doi: 10.1111/j.1538-7836.2011.04423.x. [DOI] [PubMed] [Google Scholar]
- 24.Haberichter SL, Castaman G, Budde U, et al. Identification of type 1 von willebrand disease patients with reduced von willebrand factor survival by assay of the VWF propeptide in the european study: Molecular and clinical markers for the diagnosis and management of type 1 VWD (MCMDM-1VWD) Blood. 2008;111:4979–4985. doi: 10.1182/blood-2007-09-110940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gezsi A, Budde U, Deak I, et al. Accelerated clearance alone explains ultra-large multimers in von willebrand disease vicenza. J Thromb Haemost. 2010;8:1273–1280. doi: 10.1111/j.1538-7836.2010.03753.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sutherland MS, Cumming AM, Bowman M, et al. A novel deletion mutation is recurrent in von willebrand disease types 1 and 3. Blood. 2009;114:1091–1098. doi: 10.1182/blood-2008-08-173278. [DOI] [PubMed] [Google Scholar]
- 27.de Wee EM, Sanders YV, Mauser-Bunschoten EP, et al. Determinants of bleeding phenotype in adult patients with moderate or severe von willebrand disease. Thromb Haemost. 2012;108:683–692. doi: 10.1160/TH12-04-0244. [DOI] [PubMed] [Google Scholar]
- 28.Bowman M, Tuttle A, Notley C, et al. The genetics of canadian type 3 von willebrand disease: Further evidence for co-dominant inheritance of mutant alleles. J Thromb Haemost. 2013;11:512–520. doi: 10.1111/jth.12130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ahmad F, Budde U, Jan R, et al. Phenotypic and molecular characterisation of type 3 von willebrand disease in a cohort of indian patients. Thromb Haemost. 2013;109:652–660. doi: 10.1160/TH12-10-0737. [DOI] [PubMed] [Google Scholar]
- 30.Jacobi PM, Gill JC, Flood VH, Jakab DA, Friedman KD, Haberichter SL. Intersection of mechanisms of type 2A VWD through defects in VWF multimerization, secretion, ADAMTS-13 susceptibility, and regulated storage. Blood. 2012;119:4543–4553. doi: 10.1182/blood-2011-06-360875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Goodeve AC. The genetic basis of von willebrand disease. Blood Rev. 2010;24:123–134. doi: 10.1016/j.blre.2010.03.003. [DOI] [PubMed] [Google Scholar]
- 32.Brehm MA, Huck V, Aponte-Santamaria C, et al. Von willebrand disease type 2A phenotypes IIC, IID and IIE: A day in the life of shear-stressed mutant von willebrand factor. Thromb Haemost. 2014;112 doi: 10.1160/TH13-11-0902. in press. [DOI] [PubMed] [Google Scholar]
- 33.Castaman G, Federici AB, Tosetto A, et al. Different bleeding risk in type 2 A and 2 M von willebrand disease: A two-year prospective study in 107 patients. J Thromb Haemost. 2012;10:632–638. doi: 10.1111/j.1538-7836.2012.04661.x. [DOI] [PubMed] [Google Scholar]
- 34.Fujimura Y, Titani K, Holland LZ, et al. Von willebrand factor. A reduced and alkylated 52/48-kDa fragment beginning at amino acid residue 449 contains the domain interacting with platelet glycoprotein ib. J Biol Chem. 1986;261:381–385. [PubMed] [Google Scholar]
- 35.Federici AB, Mannucci PM, Castaman G, et al. Clinical and molecular predictors of thrombocytopenia and risk of bleeding in patients with von willebrand disease type 2B: A cohort study of 67 patients. Blood. 2009;113:526–534. doi: 10.1182/blood-2008-04-152280. [DOI] [PubMed] [Google Scholar]
- 36.Othman M, Kaur H, Emsley J. Platelet-type von willebrand disease: New insights into the molecular pathophysiology of a unique platelet defect. Semin Thromb Hemost. 2013;39:663–673. doi: 10.1055/s-0033-1353442. [DOI] [PubMed] [Google Scholar]
- 37.Flood VH, Gill JC, Christopherson PA, et al. Critical von willebrand factor A1 domain residues influence type VI collagen binding. J Thromb Haemost. 2012;10:1417–1424. doi: 10.1111/j.1538-7836.2012.04746.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ribba AS, Loisel I, Lavergne JM, et al. Ser968Thr mutation within the A3 domain of von willebrand factor (VWF) in two related patients leads to a defective binding of VWF to collagen. Thromb Haemost. 2001;86:848–854. [PubMed] [Google Scholar]
- 39.Keeling D, Beavis J, Marr R, Sukhu K, Bignell P. A family with type 2M VWD with normal VWF:RCo but reduced VWF:CB and a M1761K mutation in the A3 domain. Haemophilia. 2012;18:e33. doi: 10.1111/j.1365-2516.2011.02676.x. [DOI] [PubMed] [Google Scholar]
- 40.Mazurier C, Meyer D. Factor VIII binding assay of von willebrand factor and the diagnosis of type 2N von willebrand disease--results of an international survey. on behalf of the subcommittee on von willebrand factor of the scientific and standardization committee of the ISTH. Thromb Haemost. 1996;76:270–274. [PubMed] [Google Scholar]
- 41.Castaman G, Giacomelli SH, Jacobi P, et al. Homozygous type 2N R854W von willebrand factor is poorly secreted and causes a severe von willebrand disease phenotype. J Thromb Haemost. 2010;8:2011–2016. doi: 10.1111/j.1538-7836.2010.03971.x. [DOI] [PubMed] [Google Scholar]
- 42.Zhou YF, Eng ET, Zhu J, Lu C, Walz T, Springer TA. Sequence and structure relationships within von willebrand factor. Blood. 2012;120:449–458. doi: 10.1182/blood-2012-01-405134. [DOI] [PMC free article] [PubMed] [Google Scholar]