Abstract
Two V-nitrogenase-based reaction systems were previously shown to reduce CO2 to hydrocarbons: (1) an enzyme-based system, which requires both components of V-nitrogenase for ATP-dependent reduction of CO2 to ≤C2 hydrocarbons; and (2) a cofactor-based system, which employs SmI2 to supply electrons to the isolated V-cluster for ATP-independent reduction of CO2 to ≤C3 hydrocarbons. Here, we report ATP-independent reduction of CO2 to hydrocarbons by a reaction system comprising Eu(II) DTPA and VFe protein. Combining features of both enzyme- and cofactor-based systems, this system demonstrates an improved activity of C-C coupling, as well as a widened product profile of ≤C4 hydrocarbons. The C-C coupling does not route via CO2-derived CO, and it is significantly enhanced in D2O. These observations afford the initial insights into the characteristics of this unique reaction and provide a potential template for future design of catalysts to recycle the greenhouse gas CO2 into useful products.
Keywords: V-nitrogenase, ATP-independent system, CO2 reduction, C-C coupling reactions, hydrocarbon formation
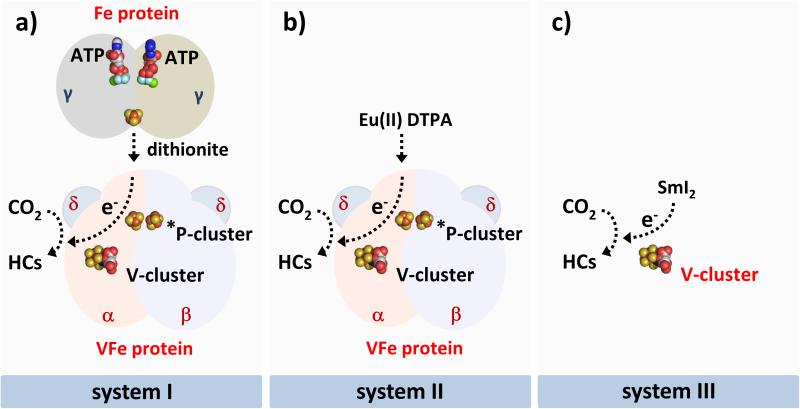
Nitrogenase plays a key role in the global nitrogen cycle, catalyzing the ATP-dependent reduction of atmospheric dinitrogen (N2) to bioavailable ammonia (NH3) under ambient conditions.[1,2] Other than N2, this enzyme is also capable of reducing carbon monoxide (CO) and carbon dioxide (CO2) to hydrocarbons;[3-10] in particular, the “alternative” vanadium (V)-nitrogenase displays considerably higher activities of CO- and CO2-reduction than the “conventional” molybdenum (Mo)-ntirogenase,[5,7] generating specific interest in exploring the catalytic characteristics of V-nitrogenase in these reactions. The V-nitrogenase from Azotobacter vinelandii consists of two component proteins: the γ2-homodimeric iron (Fe) protein, which contains a subunit-bridging [Fe4S4] cluster and a nucleotide-binding site per subunit;[11,12] and the α2β2δ4-octameric vanadium-iron (VFe) protein, which contains a P*-cluster (a [Fe4S4]-like cluster pair) at the α/β-subunit interface and a V-cluster (a [VFe7S9] cofactor) within each α-subunit (Figure 1a).[13,14] Two V-nitrogenase-based reaction systems were previously shown to reduce CO2 to hydrocarbons.[7,8] One of them utilized the complete enzymatic system of V-nitrogenase, namely, the Fe protein and the VFe protein, to enable ATP-dependent electron transfer from the [Fe4S4] cluster of the Fe protein to the V-cluster of the VFe protein for the reduction of CO2 to hydrocarbons in an aqueous buffer (Figure 1a);[7] the other employed the isolated cofactor of V-nitrogenase, namely, the V-cluster, to directly receive electrons from a strong reductant, samarium (II) iodide (SmI2; E0’ = −1.5 V), for ATP-independent reduction of CO2 to hydrocarbons in an organic solvent (Figure 1c).[8] While hydrocarbons of up to C2 and C3 in length were generated as the respective products of CO2 reduction by the enzyme- and cofactor-based reactions, the former consumed ATP as an energy source; whereas the latter was hampered by the instability of the V-cluster in the isolated state. These observations have prompted attempts to combine the desirable features of the enzyme- and cofactor-based reactions into an ATP-independent, reductant-driven reaction system, which utilizes the VFe protein-bound V-cluster as a stand-alone catalyst for improved production of hydrocarbons from CO2 reduction (Figure 1b).
Figure 1.
Three CO2-reducing reaction systems based on V-nitrogenase: a) V-nitrogenase plus dithionite (designated system I), which permit ATP-dependent transfer of electrons from the [Fe4S4] cluster of Fe protein to the V-cluster (or cofactor) of VFe protein in an aqueous buffer; b) VFe protein plus Eu(II) DTPA (designated system II), which permit ATP-independent electron transfer from Eu(II) DTPA to the V-cluster of VFe protein in an aqueous buffer; and c) V-cluster plus SmI2 (designated system III), which permit ATP-independent electron transfer from SmI2 to the isolated V-cluster in an organic solvent. For the purpose of simplicity, only one half of the VFe protein is shown. HCs, hydrocarbons.
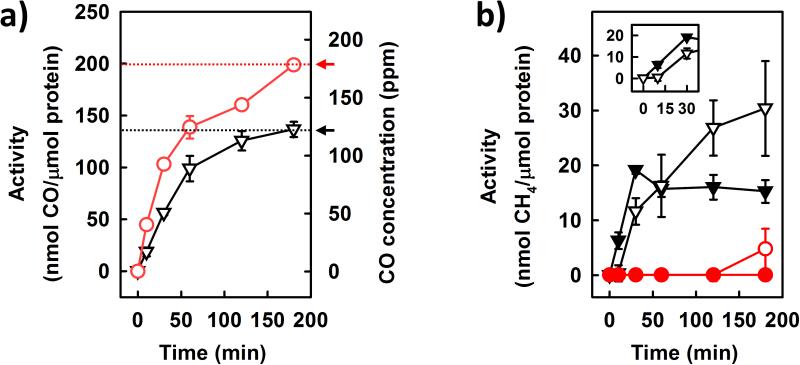
One such system could be generated by combining europium (II) DTPA [Eu(II) DTPA; E0’ = −1.1 V][15,16] with VFe protein in an aqueous buffer. Driven directly by Eu(II) DTPA, the VFe protein was capable of catalyzing ATP-independent reduction of CO2 to CO (Figure 2a) and CH4 (Figure 2b). The activity of the VFe protein to convert CO2 to CO was consistently higher in D2O (Figure 2a, ○) than that in H2O (Figure 2a,▽). In contrast, the activity of CH4 formation by VFe protein could not be detected in the presence of D2O (Figure 2b, ○); yet, it increased from 0 to 30 nmol/μmol protein over a time period of 180 minutes upon substitution of H2O for D2O (Figure 2b, ▽). Interestingly, when CO was supplied in a concentration simulating that achieved in from CO2 reduction by VFe protein at 180 minutes (see Figure 2a, arrows), no CD4 could be detected as a product in the presence of D2O (Figure 2b, •); whereas the activity of CH4 formation by VFe protein increased from 0 to a maximum of 18 nmol/μmol protein within the first 30 minutes of the reaction in the presence of H2O (Figure 2b, ▽). Such an increase in activity approximated the increase of 12 nmol/μmol protein between 10 and 30 minutes when CO2 was directly supplied to the reaction, following a delay of 10 minutes in which no CH4 could be detected (Figure 2b, ▼). Beyond 30 minutes, however, the activity of CH4 formation by VFe protein plateaued when CO was supplied as a substrate (Figure 2b, ▼); whereas it continued to increase when CO2 was supplied as a substrate (Figure 2b, ▽). The similarity between the initial phases of CO- and CO2-based CH4 formation by VFe protein (see Figure 2b, inset) suggests that a portion of CH4 is formed via CO2-derived CO, particularly given the 10-minute delay in the CO2-based reaction that could correlate with the time required to accumulate a sufficient amount of CO from CO2 reduction. The disparity between the two reactions beyond 30 minutes, on the other hand, implies that a certain portion of CH4 is generated directly from CO2 reduction, which could account for the different lineshapes of the time courses of CH4 formation, as well as the total amounts of CH4 generated in the CO- and CO2-based reactions.
Figure 2.
ATP-independent reduction of CO2 to C1 products by VFe protein. a) Time-dependent formation of CO from CO2-reduction in H2O (▽) or D2O (○). b) Comparison of time-dependent formation of CH4 from CO2-reduction in H2O (▽) or D2O (○) with that from CO-reduction in H2O (▼) or D2O (•).The amounts of CO supplied (in b) were 136 ppm (▼) and 178 ppm (•), respectively, which simulated the amounts of CO formed in H2O (▽) and D2O (○), respectively, from CO2-reduction at 180 min (see a, arrows). The inset (in b) is a blown-up of the first 30 min of CH4 formation in H2O where CO (▼) or CO2 (▽) was supplied as a substrate. Data are presented as mean ± SD (N = 3) after background correction.
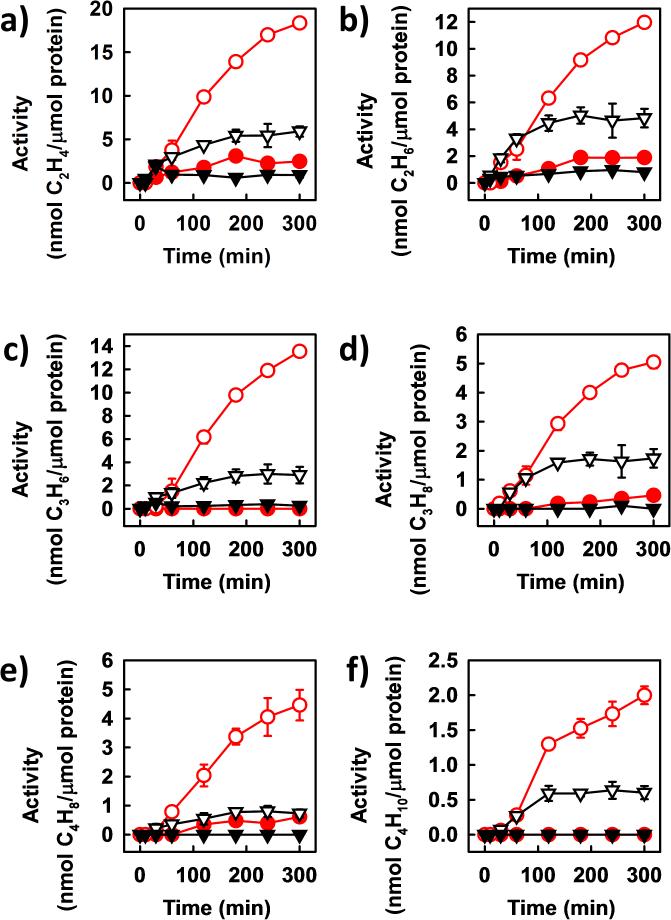
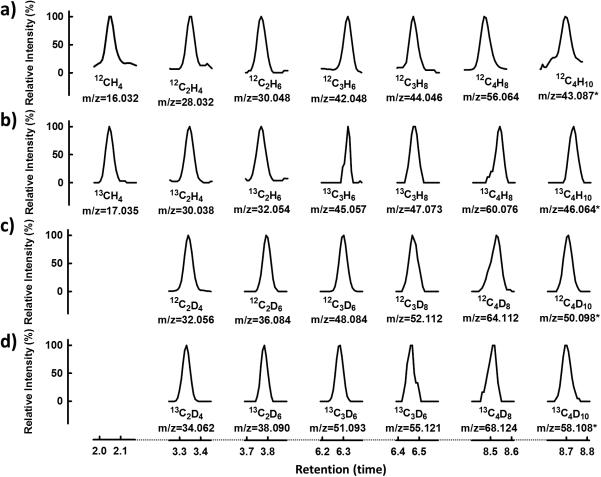
More excitingly, other than reducing CO2 to the C1 hydrocarbon product (i.e., CH4), the VFe protein could use CO2 as a substrate for ATP-independent C-C coupling into hydrocarbon products of up to C4 in length (Figure 3). Contrary to the formation of CH4, there was a dramatic increase in the activities of C2-C4 hydrocarbon formation by VFe protein upon substitution of D2O for H2O. In the presence of D2O, formation of C2D4, C2D6, C3D6, C3D8, C4D8 and C4D10 by VFe protein reached 18, 12, 14, 5, 4.5 and 2.0 nmol/μmol protein, respectively, over a time period of 300 minutes (Figure 3a-f, ○); whereas in the presence of H2O, formation of C2H4, C2H6, C3H6, C3H8, C4H8 and C4H10 started to plateau between 120 and 180 minutes at 6, 5, 3, 1.7, 0.8 and 0.6 nmol/μmol protein, respectively (Figure 3a-f, ▽). When CO was supplied in a concentration mimicking that achieved from CO2 reduction by VFe protein at 180 minutes (see Figure 2, arrows), little or no C2-C4 product could be detected in the presence of either D2O (Figure 3a-f, •) or H2O (Figure 3a-f, ▼), suggesting that these products were formed directly from CO2 reduction instead of indirectly from CO2-derived CO. GC-MS analysis further confirmed the source of carbon in the C1-C4 hydrocarbons as that from CO2, showing mass shifts of +1, +2, +3 and +4, respectively, of C1, C2, C3 and C4 products upon substitution of 13CO2 for 12CO2 in H2O-based reactions (Figure 4a vs. b). In addition, it demonstrated the source of hydrogen in these products as that from H2O, displaying mass shifts of +4, +6 and +8, respectively, of products containing 4, 6 and 8 hydrogen atoms upon substitution of D2O for H2O in 12CO2-based reactions (Figure 4a vs. c); and additional mass shifts of +2, +3 and +4, respectively, of C2, C3 and C4 products upon further substitution of 13CO2 for 12CO2 in these reactions (Figure 4c vs. d). In the cases of C4H10 and C4D10, the base peaks were monitored to circumvent the problem of detection limit, showing masses of 43.087, 46.064, 50.098 and 58.108, respectively, that represent the predominant fragment ions formed in the cases of 12C4H10, 13C4H10, 12C4D10 and 13C4D10 (Figure 4).
Figure 3.
ATP-independent reduction and coupling of CO2 into C2-C4 products by VFe protein. Comparison of time-dependent formation of a) C2H4, b) C2H6, c) C3H6, d) C3H8, e) C4H8 and f) C4H10 from CO2-reduction in H2O (▽) or D2O (○) with those from CO-reduction in H2O (▼) or D2O (•). The amounts of CO supplied (in a-f) were 136 ppm (▼) and 178 ppm (•), respectively, which simulated the amounts of CO formed in H2O (▽) and D2O (○), respectively, from CO2-reduction by VFe protein at 180 min (see Figure 2a, arrows). Data are presented as mean ± SD (N=3) after background correction.
Figure 4.
GC-MS analyses of hydrocarbons generated by VFe protein in the ATP-independent reaction of CO2-reduction. The products were generated in H2O when a) 12CO2 or b) 13CO2 was supplied as the substrate; or in D2O when c) 12CO2 or d) 13CO2 was supplied as the substrate. The mass-to-charge (m/z) ratios at which the products were traced are indicated. In the cases of C4H10 and C4D10 (a-d), the base peaks (*), or the tallest peaks representing the most common fragment ions of these species, were monitored to overcome the problem of detection limit.
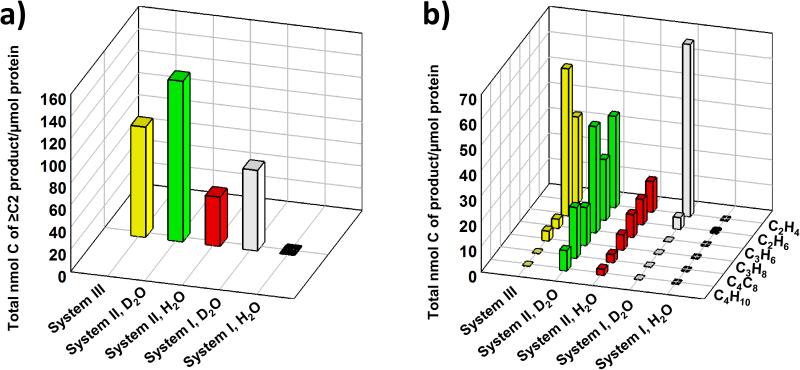
The total activity of C-C coupling (i.e., formation of ≥C2 products) by VFe protein in the ATP-independent, Eu(II) DTPA-driven reaction (designated system II) is higher than that by V-cluster in the ATP-independent, SmI2-driven reaction (designated system III) (Figure 5a, green vs. yellow) and that by V-nitrogenase in the ATP-dependent reaction (designated system I) in D2O (Figure 5a, green vs. grey) or H2O (Figure 5a, red vs. black). Moreover, compared to system I and system III, there is a greater tendency of system II to reduce CO2 to longer carbon chains and generate a broader product profile both in D2O (Figure 5b, green) and in H2O (Figure 5b, red). Given the absence of Fe protein from system II, the active V-cluster site is likely more “open” in this system than that in system I (see Figure 1), which could explain the improved ability of system II to extend the carbon chain and generate larger hydrocarbon products. The protein scaffold that houses the V-cluster in system II, on the other hand, likely provides stability to the cluster while modulating its redox potential in the same time, which may account for a higher activity accomplished by a weaker reductant (i.e., Eu(II) DTPA) in the case of system II as compared to a lower activity accomplished by a stronger reductant (i.e., SmI2) in the case of system III (Figure 5a, green vs. yellow). Apart from the composition-dependent differences, there is a strong deuterium effect on the activity of protein-enabled CO2 reduction in an aqueous buffer, as both system I and system II displayed a dramatic increase of CO2-reducing activity upon substitution of D2O for H2O (Figure 5, green vs. red; grey vs. black) While systematic studies are required to elucidate the mechanism of CO2 reduction by V-nitrogenase, these observations afford the initial insights to the characteristics of this unique reaction and provide a potential template for future design of nitrogenase-based catalysts to recycle the greenhouse gas CO2 into useful hydrocarbon products.
Figure 5.
C-C coupling from CO2 reduction by three V-nitrogenase-based systems. a) Total and b) individual activities of C-C coupling by system I, II and III. Activities were calculated based on the sums of carbons that appeared in all ≥C2 hydrocarbon products (in a) or in each individual ≥C2 hydrocarbon product (in b). For compositions of system I, II and III, please refer to Figure 1 and text. Data of system I and III were taken from Ref. 8 and Ref. 7, respectively.
Supplementary Material
Acknowledgements
This work was supported by National Institutes of Health grant GM 67626 (M.W.R.).
Footnotes
Supporting information for this article is given via a link at the end of the document.
References
- 1.Burgess BK, Lowe DJ. Chem. Rev. 1996;96:2983–3012. doi: 10.1021/cr950055x. [DOI] [PubMed] [Google Scholar]
- 2.Rees DC, Tezcan FA, Haynes CA, Walton MY, Andrade S, Einsle O, Howard JB. Philos. Trans. A. Math. Phys. Eng. Sci. 2005;363:971–984. doi: 10.1098/rsta.2004.1539. [DOI] [PubMed] [Google Scholar]
- 3.Lee CC, Hu Y, Ribbe MW. Science. 2010;329:642. doi: 10.1126/science.1191455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lee CC, Hu Y, Ribbe MW. Angew. Chem. Int. Ed. Engl. 2011;50:5545–5547. doi: 10.1002/anie.201100869. [DOI] [PubMed] [Google Scholar]
- 5.Hu Y, Lee CC, Ribbe MW. Science. 2011;333:753–755. doi: 10.1126/science.1206883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee CC, Hu Y, Ribbe MW. Angew. Chem. Int. Ed. Engl. 2012;51:1947–1949. doi: 10.1002/anie.201108916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rebelein JG, Hu Y, Ribbe MW. Angew. Chem. Int. Ed. Engl. 2014;53:11543–11546. doi: 10.1002/anie.201406863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee CC, Hu Y, Ribbe MW. Angew. Chem. Int. Ed. Engl. 2015;54:1219–1222. doi: 10.1002/anie.201410412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang ZY, Dean DR, Seefeldt LC. J. Biol. Chem. 2011;286:19417–19421. doi: 10.1074/jbc.M111.229344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang ZY, Moure VR, Dean DR, Seefeldt LC. Proc. Natl. Acad. Sci. U. S. A. 2012;109:19644–19648. doi: 10.1073/pnas.1213159109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eady RR. Chem. Rev. 1996;96:3013–3030. doi: 10.1021/cr950057h. [DOI] [PubMed] [Google Scholar]
- 12.Hales BJ. Adv. Inorg. Biochem. 1990;8:165–198. [PubMed] [Google Scholar]
- 13.Lee CC, Hu Y, Ribbe MW. Proc. Natl. Acad. Sci. U. S. A. 2009;106:9209–9214. doi: 10.1073/pnas.0904408106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fay AW, Blank MA, Lee CC, Hu Y, Hodgson KO, Hedman B, Ribbe MW. J. Am. Chem. Soc. 2010;132:12612–12618. doi: 10.1021/ja1019657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vincent KA, Tilley GJ, Quammie NC, Streeter I, Burgess BK, Cheesman MR, Armstrong FA. Chem. Commun. (Camb.) 2003;21:2590–2591. doi: 10.1039/b308188e. [DOI] [PubMed] [Google Scholar]
- 16.Danyal K, Inglet BS, Vincent KA, Barney BM, Hoffman BM, Armstrong FA, Dean DR, Seefeldt LC. J. Am. Chem. Soc. 2010;132:13197–13199. doi: 10.1021/ja1067178. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.