Abstract
Flavivirus methyltransferase (MTase) is essential for viral replication. Here we report the identification of small molecules through virtual screening that putatively bind to the SAM-binding site of flavivirus MTase and inhibit its function. Six of these computationally predicted binders were identified to show significant MTase inhibition with low micromolar inhibitory activity. The most active compounds showed broad-spectrum activity against the MTase proteins of other flaviviruses. Two of these compounds also showed low cytotoxicity and high antiviral efficacy in cell-based assays. Competitive binding analyses indicated that the inhibitors performed their inhibitory function through competitive binding to the SAM cofactor binding site of the MTase. The crystal structure of the MTase–inhibitor complex further supports the mode of action and provides routes for their further optimization as flavivirus MTase inhibitors.
Keywords: flavivirus NS5, RNA cap methylation, West Nile virus, methyltransferase, antiviral development
Viruses of the arthropod-borne Flavivirus genus infect over 200 million people worldwide each year, resulting in a significant disease burden. Infections can result in life-threatening meningitis and encephalitis by West Nile virus (WNV) and Japanese encephalitis virus (JEV) or deadly hemorrhagic fever by Dengue virus (DENV) and yellow fever virus (YFV).1 Dengue has been a worldwide problem and is now endemic in more than 100 countries.2 Although effective vaccines exist for YFV, JEV, and tick-borne encephalitis virus (TBEV),3 there are currently no safe and effective vaccines for WNV or DENV. Furthermore, due to the dangers and difficulties inherent in mass vaccination of large at-risk populations, it is desirable to be able to treat severe flavivirus infections with antiviral therapeutics that could be administered immediately during an outbreak.
The flavivirus methyltransferase (MTase) sequentially methylates the N7 and 2′-O positions of the viral RNA cap (GpppA-RNA→m7GpppA-RNA→ m7GpppAm-RNA), using S-adenosyl-l-methionine (SAM) as a methyl donor and generating S-adenosyl-l-homocysteine (SAH) as a reaction byproduct.4 The flavivirus MTase also methylates the internal adenosine of viral RNA at 2′-O positions.5 It has been demonstrated that defects in N7 methylation are lethal to DENV, WNV, YFV, and Kunjin virus replication, indicating that the first methylation of the viral mRNA cap is an obligate step in the virus life cycle.4,6 Subsequently, the flavivirus MTase became an attractive target for therapeutic inventions.6b,7 Our laboratory recently demonstrated sinefungin (SIN) and several nucleoside analogues display broad-spectrum inhibition of both MTase activity and viral replication of WNV, DENV, and YFV.6b,7c,e We also found an additional flavivirus-conserved pocket next to the pocket holding the adenine base of SAM (or the SIN inhibitor). The conserved pocket has been used to generate flavivirus-specific inhibitors in a recent study.7g However, potential cytotoxicity is a concern with SIN as the lead compound as SIN is too similar to SAM, the methyl donor for essentially all MTases. Many other inhibitors of flavivirus MTases have been identified through the use of a variety of techniques including cell-based assays, virtual screening, fragment-based, and structure-based design.6b,7a,c,h,j–o,8 However, the majority of these compounds have not shown antiviral efficacy, whereas the few that have7c,l,m displayed relatively low potency, high cytotoxicity, and/or low therapeutic index.
To search for novel and potent MTase inhibitors, we performed virtual screening of the Diversity Set II library of 1364 compounds from the National Cancer Institute Developmental Therapeutics Program (NCI DTP). Two compounds, NSC 12155 and NSC 125910, inhibit both the N7 and 2′-O MTase functions of multiple flavivirus MTases. They also inhibit virus growth with low micromolar EC50 values in antiviral analysis. Particularly, compound NSC 12155 displayed a high therapeutic index and broad antiviral spectrum. The crystal structure of the DENV3MTase–12155 complex and competition assay results indicated that NSC 12155 inhibited the MTase function through competitive inhibition of the cofactor SAM binding to the MTase cofactor binding site.
MATERIALS AND METHODS
Compounds
Compounds were obtained from the NCI DTP Open Chemical Repository (http://dtp.nci.nih.gov). [α-32P]-GTP was purchased from MP Biomedicals.
Virtual Screening
Molecular docking was performed using the program Autodock Vina.9 The NCI diversity set II library obtained from http://dtpsearch.ncifcrf.gov/FTP/DIVERSITY in January 2011 was converted to PDB format using the program Babel.10 It was then docked against the WNV MTase with its structure bound to the SAM analogue inhibitor, SIN (PDB ID 3LKZ),7e used as the target structure. To set the input parameters for the Autodock Vina program, SIN was initially docked into its binding site. To reproduce SIN binding, a ligand box extending 30 Å in each direction with its center located at the SAM binding site, and an exhaustiveness parameter of eight was sufficient. These parameters were then used to dock the entire diversity subset library to the WNV MTase. Only compounds that showed binding energies greater than the predicted binding energy for SIN according to the Autodock Vina scoring function (7.2 kcal/mol) were tested experimentally.
Expression and Purification of the NS5MTase from WNV, YFV, DENV2, and DENV3
Recombinant MTases from WNV, YFV, DENV2, and DENV3 containing the N-terminal 300, 266, 265, and 272 amino acids of NS5 protein, respectively, were expressed and purified as described previously.7d The DENV3MTase was also produced through a denaturation and refolding process to remove the copurified SAM as we described previously.11
In Vitro MTase Inhibition Assay
The 5′-end-labeled substrates G*pppA-RNA and m7G*pppA-RNA, representing the first 90 nucleotides of the WNV genome (the asterisk indicates that the following phosphate is 32P labeled), were prepared as described previously.6e,7f The N7 and 2′-O methylation inhibition assays were performed as described previously.4,7f The N7 methylation was measured by conversion of G*pppA-RNA→ m7G*pppA-RNA. The 2′-O methylation was monitored by conversion of m7G*pppA-RNA→ m7G*pppAm-RNA. The methylation reactions were digested with nuclease P1 to release cap moieties (m7G*pppAm, m7G*pppA, and G*pppA). The cap molecules were separated on a thin-layer chromatograph (TLC) and quantified by a PhosphorImager.4,7f All experiments were performed in triplicates unless otherwise stated. The percentage of activity was determined after quantification of m7G*pppA, m7G*pppAm, and G*pppA. The IC50 value, unless specified, was determined by fitting of the dose–response curve of the averaged values using the ORIGIN software package.
Competitive SAM Binding Assay
The DENV3MTase was biotinylated using the EZ-link NHS0biotin kit (Pierce), according to manufactory protocol and as we described previously.7d Competitive SAM binding was performed using the biotinylated DENV3MTase in the presence of [3H]-SAM at a fixed concentration and a concentration series of NSC 12155, using a protocol as we described previously.7d
Cytotoxicity Assay
Cytotoxicity was measured by a MTT cell proliferation assay using the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide method (ATCC), as described previously.7c,d Approximately 2 × 104 BHK-21 or human A549 cells in 100 µL of Eagle’s minimum essential medium (EMEM) containing 2% FBS were seeded into 60 wells of a 96-well plate; the remaining wells held medium. Plates were held at room temperature for 1 h and then incubated for 20–24 h. The medium was removed, and 100 µL of medium containing decreasing concentrations of antiviral compound in 1% DMSO was added to the wells. All determinations were performed in triplicate. After 42 h of incubation at 37 °C, 10 µL of MTT was added to the wells and incubated another 3 h. Detergent (100 µL) was placed in the wells, and the plate was incubated for 3 h at room temperature in the dark. A microtiter plate reader (Ely808, BioTek Instruments, Inc.) with a 570 nm filter was used to record absorbance. All determinations were performed in triplicate. After adjustment of the absorbance for background and comparison to untreated controls, a dose–response curve was plotted using GraphPad Prism software (GraphPad Software, La Jolla, CA, USA). The cytotoxic concentration CC50 (the concentration of inhibitor required to reduce cell viability by 50%) was calculated using nonlinear regression to fit the dose–response curve using the ORIGIN software package.
Antiviral Assay
A viral titer reduction assay was used to determine the compounds’ effect on WNV, DENV-2, SLEV, JEV, and control viruses VSV and WEEV. Approximately 2 × 105 BHK-21 or human A549 cells in 500 µL of the EMEM–2% FBS medium were seeded into each well of a 24-well plate. At 24–30 h after seeding, dilutions at 2 times the desired concentration of the compound were made in 2% DMSO medium, and 250 µL was added to wells in triplicate. Immediately following, 250 µL of medium containing viruses at a concentration to yield a multiplicity of infection (MOI) of 0.1 PFU/cell was added to the wells. After 42 h of incubation at 37 °C, culture medium was collected and stored at −80 °C for later quantification using a plaque assay. For the plaque assay, Vero cell monolayers in 6-well plates were seeded 3–4 days prior to infection to achieve a confluent monolayer. Depending on the virus, three to eight 10-fold serial dilutions of the harvested samples were made, and three to five dilutions of the virus were tested. One hundred microliters of the dilution solutions was inoculated into each of two wells, rocked gently to distribute virus, and incubated for 1 h at 37 °C. Cells were then overlaid with a nutrient medium containing 0.6% oxoid agar. The agar was allowed to solidify, and the plates were then incubated at 37 °C. A second overlay containing 2% neutral red was added after the plaques began to appear on day 2 and then incubated overnight. Plaques were counted daily for 1–3 days until no significant increase was seen. All determinations were performed in triplicate. The effective concentration EC50 (the concentration of inhibitor required to reduce virus growth by 50%) was determined by nonlinear regression fitting of the dose–response curve using the ORIGIN software package.
Crystallization, Data Collection, Structure Determination, and Refinement of the DENV3MTase–12155 Complex
Crystals of apo-form of the refolded DENV3MTase were obtained as described.11 To obtain the MTase–12155 complex, the apo crystals of the refolded DENV3MTase were soaked overnight in the crystallization mother liquor (50 mM sodium citrate, pH 5.6, 24% PEG 4000, 5% saturated ammonium sulfate, 10% glycerol, 20% DMSO, and 5 mM DTT) supplemented with 3 mM NSC 12155 dissolved in DMSO. Co-crystals were harvested in a cryo-solution containing crystallization mother liquor supplemented with 25% glycerol and flash-cooled in liquid nitrogen. Diffraction data were collected to 1.7 Å resolution at 100 K using an MAR325 CCD detector at the BL14-1 beamline of the Stanford Synchrotron Radiation Lightsource. Diffraction data were processed and scaled using the HKL2000 package.12
The structure of the DNV3MTase–12155 complex was determined by the molecular replacement method using the PHENIX program suite,13 with the apo structure of the refolded DNV3MTase (PDB code 4R0511) as a search model. Structural refinement was carried out using PHENIX Refine. Model rebuilding was carried out using Coot.14
RESULTS
Identification of Novel Potent Inhibitors of Flavivirus MTase Using Virtual Screening
The crystal structure for SIN bound to the WNV MTase (PDB ID 3LKZ)7e was chosen as the template to provide a suitable ligand binding pocket for virtual screening. The docking parameters for AutoDock Vina were first optimized by redocking SIN into the SAM-binding site of the MTase. A root-mean-square deviation (rmsd) of only 0.9 Å was obtained upon the best alignment of the redocked and crystallography-determined conformations of SIN (Figure 1A). Repeating the docking procedure 10 times led to rmsd between docked and crystallography-determined conformation of SIN ranging from 0.9 to 1.2 Å, indicating an excellent fidelity of docking. These optimized parameters were applied to dock the NCI diversity set II library (1364 compounds) into the binding sites of the WNV MTase structure, using AutoDock Vina. Previously we also docked the same library using the crystal structure of the DENV2MTase–inhibitor complex (PDB 3P8Z) (data not shown). Therefore, for this study, we selected only 17 compounds that were ranked within the top 40 from the WNV MTase search, but not from the DENV2 search, for further investigation. All of these compounds had scores (>8.9 kcal/mol) better than that of the SIN control (7.2 kcal/mol) (Figure 2). For each of the hit compounds, the top five poses were visually inspected.
Figure 1.
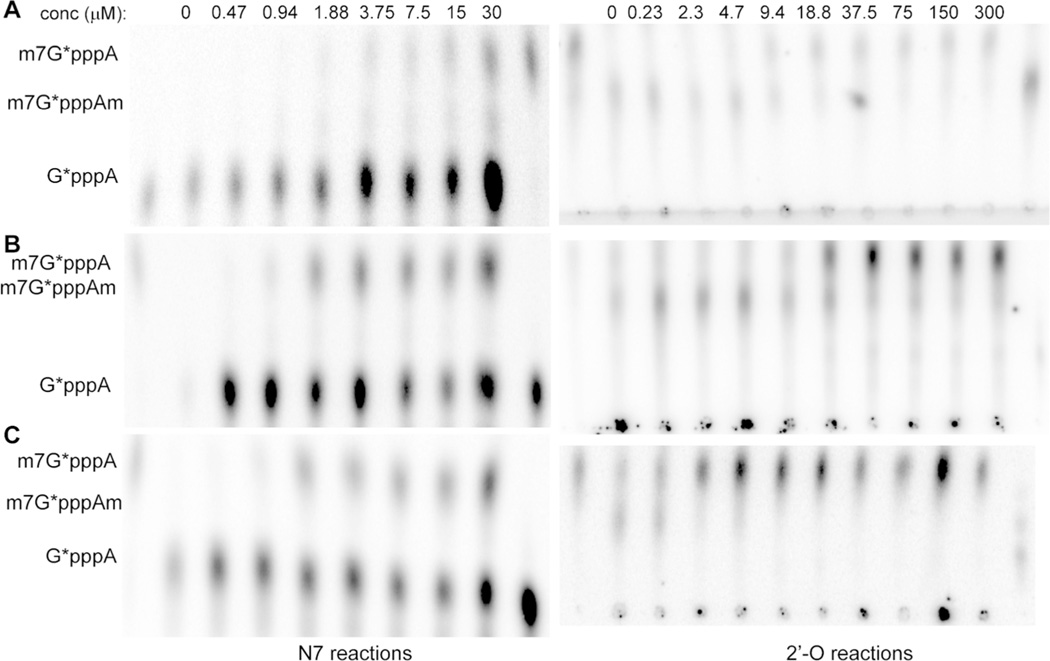
Virtual screening and experimental verification: (A) comparison of experimentally determined and docked conformations of SIN in the SAM-binding pocket of the WNV MTase; (B) inhibition of the N7 and 2′-O methylation activities of the WNV MTase by 17 top ranking compounds at 150 µM concentration, analyzed on TLC plates. The N7 methylation was measured by conversion of G*pppA-RNA→m7G*pppA-RNA; the 2′-O methylation was measured by conversion of m7G*pppA-RNA→m7G*pppAm-RNA (the asterisk indicates that the following phosphate is 32P labeled; the RNA represents the first 90 nucleotides of the WNV genome). The spots representing different cap structures on TLC plates were quantified by a PhosphorImager. The specific activity (%) was defined as intensity (m7G*pppA)/(intensity (G*pppA) + intensity (m7G*pppA)) × 100) for N7 methylation and as intensity (m7G*pppAm)/(intensity (m7G*pppA) + intensity (m7G*pppAm)) × 100 for 2′-O methylation. The methylation activity without compounds was set at 100, and the relative methylation activity with a particular compound was defined as specific activity (compound)/specific activity (no compound) × 100.
Figure 2.
Schematic formulas of selected compounds showing in vitro anti-MTase activities.
Inhibition Assay
Both the N7 and 2′-O MTase activities of the WNV MTase were measured in the presence of the 17 top-ranked compounds at a concentration of 150 µM with SIN as a positive control. As shown in Figure 1B, the N7 activity of the WNV MTase was efficiently inhibited (~80%) by the positive control inhibitor SIN. Six of the 17 compounds showed inhibition of the WNV N7MTase activity by >60% (Figures 1 and 2). Compared to the inhibition of the N7MTase activity, the 2′-O methylation was inhibited more efficiently by these compounds (Figure 1). Similar variations have been observed previously for inhibition of N7 and 2′-O MTase activities.7c,g,8b These are consistent with functional studies, which indicated that the 2′-O MTase activity is more vulnerable to mutagenesis than N7.6b,e Inhibitors were frequently observed to have better inhibition for the 2′-O activity than the N7 activity.7g,8b Because only the N7 methylation activity is essential for the virus life cycle,6e it is believed that the N7 inhibition would be more relevant to the cellular activity. Nevertheless, these six compounds were chosen for further analyses.
Detailed inhibition analyses of these compounds were carried out to determine their IC50 values for both the N7 and 2′-O activities of the WNV MTase (Table 1; Figure 3). In the absence of detergent, the IC50 values for these compounds ranged from 1.4 to 63 µM for the N7 inhibition and from 3.0 to 72 µM for the 2′-O inhibition, respectively. An example dose–response experiment of the best inhibitor, NSC 12155, for both N7 and 2′-O inhibitions is shown in Figure 3. As compounds NSC 371880 and NSC 372295 showed only weak inhibitory activity, they were excluded from further analyses. To rule out nonspecific promiscuous inhibitors,15 detergent CHAPS was used in the N7 inhibition experiment for selected nontoxic compounds (see CC50 below) (Table 1). Addition of CHAPS did not significantly affect the MTase activities (Figure 3D). Actually, CHAPS boosted the MTase activity by about 20%. In contrast, the inhibition activity of the SIN control was not affected at all by CHAPS. The result is consistent with the fact that SIN is a true and specific MTase inhibitor,7e the inhibitory activity of which should not be affected by the addition of detergent. Our results indicated that CHAPS could be used in the MTase assay to distinguish specific and nonspecific inhibitors.
Table 1.
Results of Activity Assays (IC50, CC50, EC50, Therapeutic Index (TI)) for Selected Compounds, with the WNV MTase
| compound ID (NSC) |
IC50 N7 (µM), no CHAPS |
IC50 N7 (µM), with CHAPS |
IC50 2′-O (µM), with CHAPS |
CC50 (µM) |
EC50 (µM) |
TI |
|---|---|---|---|---|---|---|
| 12155 | 1.4 | 0.87 | 3.0 | 49 | 1.0 | 49 |
| 371880 | 63 | nda | 72 | 212 | nd | |
| 372295 | 62 | nd | 57 | 126 | nd | |
| 23217 | 15.7 | >300 | 7.4 | 236 | nd | |
| 125910 | 7.6 | 17.6 | 3.2 | 177 | 2.6 | 68 |
| 293161 | 13.2 | 31 | 54.6 | 25 | nd |
nd, not determined.
Figure 3.
Dose–response inhibition of the N7 and 2′-O methylation activities of the WNV MTase by the most potent compound, NSC 12155. (A) TLC analyses of the N7 and 2′-O inhibition of the WNV MTase by NSC 12155. The migration positions of the G*pppA and m7G*pppA molecules are labeled on the side of the TLC images. (B, C) Curve fitting to determine the IC50 values for each compound on the N7 (B) and 2′-O (C) MTase activities of the WNV MTase. The percentage of activity was determined after quantification of G*pppA and m7G*pppA. The IC50 value was determined by fitting of the dose–response curve as described under Materials and Methods. Each reaction was carried out in triplicate, and the standard deviation is plotted. (D) Analysis of the effects of CHAPS on the N7 methylation activity of the WNV MTase.
Nonetheless, because compound NSC 23217 showed a large difference for IC50 values in the presence and absence of detergent, it was also excluded from further analyses. All other compounds chosen for further analysis, including the most active compound NSC 12155, displayed similar IC50 values with/without CHAPS, indicating that they are likely specific inhibitors.
Broad-Spectrum Anti-MTase Activity
Inhibitors targeted to the SAM-binding site may have broad-spectrum anti-MTase activity as this site is conserved among flavivirus MTases, but not among human MTases.7e,i To identify such broad-spectrum activity, inhibition assays were carried out using the recombinant MTases from DENV2, DENV3, and YFV. As shown in Figure 4, compounds NSC 12155 and NSC 125910 inhibited the N7 activities of MTases from DENV2, DENV3, and YFV in a dose-dependent manner (Figure 4; Table 2; and data not shown). NSC 12155 inhibited these MTase activities with IC50 values in the low micromolar range, which is comparable to those for the WNV MTase. NSC 125910 showed strong inhibition of both the N7 and 2′-O activities of the DENV2 and YFV MTases with IC50 values in the low micromolar range, although it inhibited the DENV3MTase less effectively (Table 2). Overall, our results indicated that NSC 12155 and NSC 125910 are potent broad-spectrum inhibitors for flavivirus MTases.
Figure 4.
Inhibition of the MTase activities by NSC 12155: (A–C) TLC analyses of the dose response of NSC 12155 in inhibition of the N7 and 2′-O activities of the DENV2 (A), DENV3 (B), and YFV (C) MTases.
Table 2.
Inhibition of Flavivirus MTases by Compounds NSC 12155 and NSC 125910
| NSC 12155 | NSC 125910 | |||
|---|---|---|---|---|
| MTase | IC50 N7 (µM) |
IC50 2′-O (µM) |
IC50 N7 (µM) |
IC50 2′-O (µM) |
| DENV2 | 1.2 | 7.6 | 12.6 | 7.4 |
| DENV3 | 3.4 | 7.7 | 74.8 | 20.9 |
| YFV | 2.8 | 0.5 | 7.5 | 0.4 |
Cytotoxicity and Antiviral Analyses
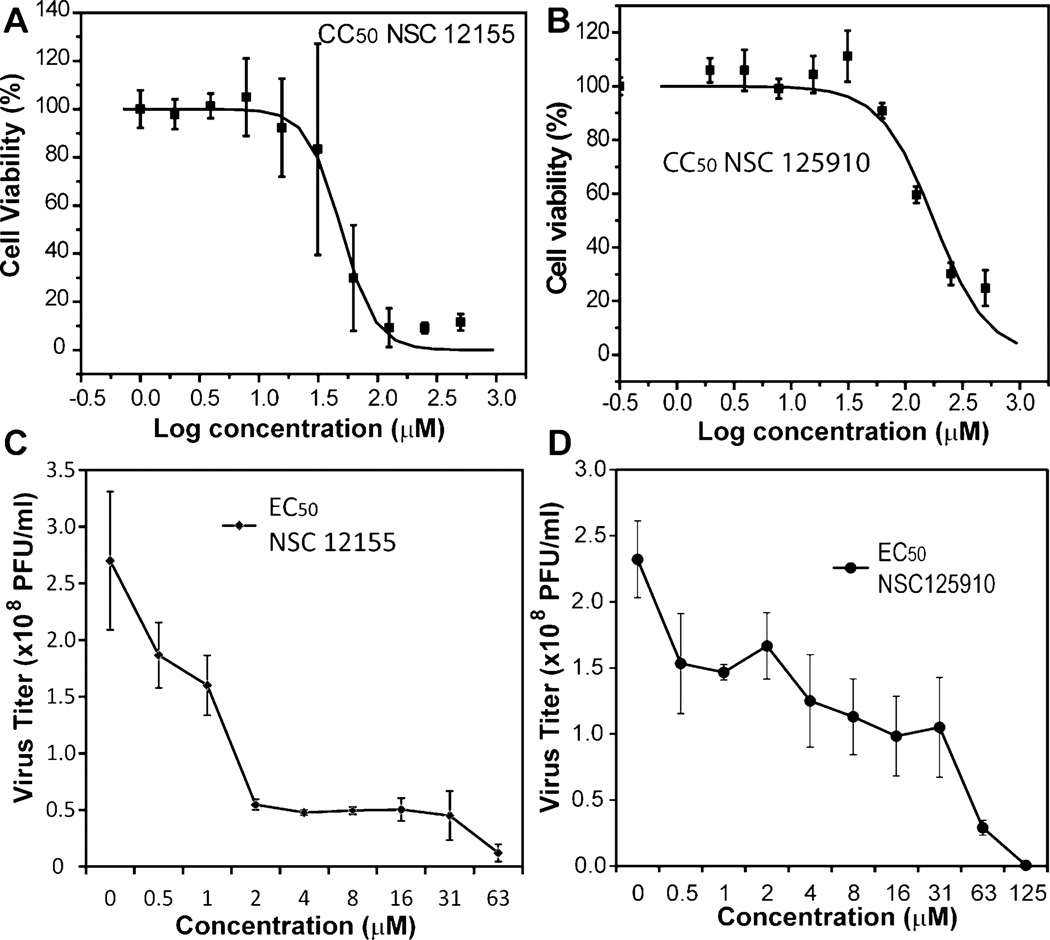
Cell-based assays were used to evaluate the cytotoxicity of the selected compounds. An MTT cell proliferation assay was first used to measure the cytotoxicity to a BHK-21 cell line (Table 1; Figure 5). This eliminated NSC 293161 from further consideration, as it showed a CC50 value similar to its in vitro IC50 value, indicating a potentially poor selectivity. The remaining two compounds, NSC 12155 and NSC 125910, showed much less toxicity. Their CC50 values were 20-fold higher than their IC50 values. Therefore, these two compounds were the only ones chosen for in vitro antiviral efficacy determination.
Figure 5.
Cytotoxicity and antiviral analyses for compounds NSC 12155 and NSC 125910. (A, B) Cytotoxicity of NSC 12155 (A) and 125910 (B). BHK-21 cells were incubated with various concentrations of the compound and then assayed for viability at 42 h post-incubation. (C, D) Inhibition of viral replication by NSC 12155 (C) and NSC 125910 (D). BHK cells were infected with WNV at a multiplicity of infection of 0.1, in the presence or absence of compounds. At 42 h post-infection, viral titers in culture fluids were quantified by plaque assays on Vero cells. Each reaction was carried out in triplicate, and the standard deviation is plotted.
Viral titer reduction assays were performed to evaluate the compounds’ antiviral efficacy (Figure 5; Table 1). The WNV titer was reduced in a dose-dependent manner by compounds NSC 12155 and NSC 125910 (Figure 5). NSC 125910 showed an antiviral efficacy EC50 of 2.6 µM (Figure 5D). NSC 12155 showed an even better antiviral efficacy with an EC50 value of 1.0 µM (Figure 5C).
NSC 12155 Has Broad Antiviral Spectrum
To further investigate the broad-spectrum antiviral activity of these compounds, we carried out viral reduction assays for additional flaviviruses and two control viruses. Because DENV-2, JEV, and St. Louis encephalitis viruses (SLEV) replicate better in human A549 cells than in BHK-21 cells, all of the experiments were performed with A549 cells.
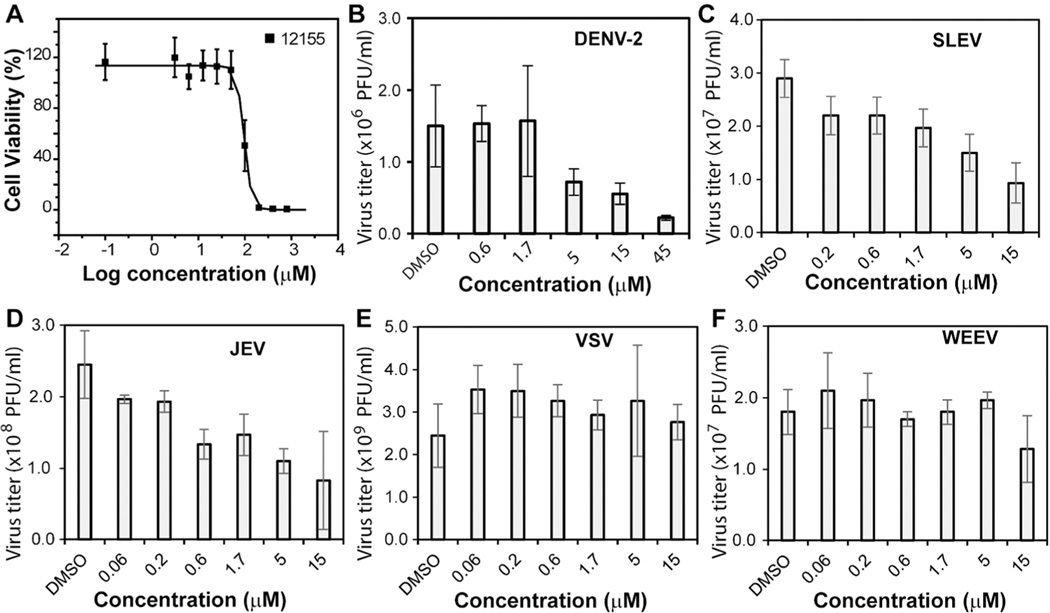
As shown in Figure 6A, NSC 12155 showed a CC50 value of 96 µM on human A549 cells. Virus reduction assays clearly demonstrated that NSC 12155 did not inhibit the growth of negative control viruses VSV and WEEV in the concentration range tested (Figure 6E,F). In contrast, apparent dose-responsive inhibitions of growth of flaviviruses DENV-2, SLEV, and JEV were observed. Although less impressive than that for WNV, the antiviral efficacy of NSC 12155 is also obvious for DENV-2, SLEV, and JEV, with EC50 values of 7.0, 5.1, and 3.1 µM, respectively (Figure 6B–D; Table 3).
Figure 6.
Broad-spectrum antiviral analysis for compound NSC 12155. (A) Cytotoxicity of NSC 12155. Human A549 cells were incubated with various concentrations of the compound and then assayed for viability at 42 h post-incubation. (B–F) Inhibition of viral replication of DENV-2 (B), SLEV (C), JEV (D), and negative control viruses VSV (E) and WEEV (F) by NSC 12155. A549 cells were infected with viruses at a multiplicity of infection of 0.1, in the presence or absence of compounds. At 42 h post-infection, viral titers in culture fluids were quantified by plaque assays on Vero cells. Each reaction was carried out in triplicate, and the standard deviation is plotted.
Table 3.
Inhibition of Flaviviruses by Compound NSC 12155
| DENV2 | SLEV | JEV | |
|---|---|---|---|
| EC50 (µM) | 7.0 | 5.1 | 3.1 |
NSC 12155 Competitively Inhibits the Binding of SAM to the DENV3MTase
To understand the mode of action of the most potent inhibitor, NSC 12155, we performed a competitive SAM-binding assay as we described previously.7d We biotinylated the DENV3MTase, mixed it with streptavidincoated SPA beads (PerkinElmer), and applied a concentration series of NSC 12155 with a fixed amount of [3H]-SAM.
Our data showed that NSC 12155 could competitively inhibit the binding of [3H]-SAM to the DENV3MTase in a dose-dependent manner (Figure 7A). The IC50 value determined using the competition assay is 1.2 µM, which is in a good agreement with the IC50 (0.87 µM) determined using the functional MTase assays. The small discrepancy between IC50 values could be attributed to the use of different assays. Nonetheless, our data demonstrated that NSC 12155 inhibits the MTase activities by direct competition of cofactor SAM binding to the MTase.
Figure 7.
NSC 12155 interactions with the MTase. (A) Dose response of inhibition of the [3H]-SAM–MTase complex formation by NSC 12155. The biotinylated DENV3MTase and 3H-labeled SAM are incubated with or without NSC 12155. A 2-fold dilution series is shown. Streptavidincoated SPA beads are used to mix with the reaction mixtures, and the SAM-binding is quantified using a Microbeta2 scintillation counter. (B) Cartoon representation of the crystal structure of the DENV3MTase–12155 complex. NSC 12155 is in stick representation. Atom colors (unless otherwise specified): carbon, yellow; nitrogen, blue; and oxygen, red. (C) NSC 12155 interaction with the DENV3MTase residues in stick representation, with carbon in cyan color. The omit Fo−Fc electron density map for NSC 12155, contoured at 2δ level, is shown in gray. (D) Superimposition of the structure of the MTase–12155 complex (cyan) with that of SAM-free MTase (gray) (PDB 4R05)11 and that of MTase–SIN complex (pink) (PDB 4R8S).17 Residues interacting with NSC 12155 are shown in stick representation. The carbon atom of SIN is in green.
Crystal Structure of the DENV3MTase–12155 Complex
To further investigate the interaction between NSC 12155 and the flavivirus MTase and its mode of action, we determined the crystal structure of the DENV3MTase in complex with NSC 12155 at 1.7 Å resolution (Figure 7B–D; Table 4). We first refolded the DENV3MTase to remove the bound SAM molecule that was copurified with the MTase,6e using a protocol we described previously.11 Crystals of the refolded DENV3MTase–12155 complex were obtained by soaking the MTase crystals with 3 mM NSC 12155 overnight. The cocrystal structure was determined using the molecular replacement method (Figure 7B; Table 4). During refinement, clear continuous electron density was shown within the SAM-binding pocket of the MTase (Figure 7C). NSC 12155 was therefore modeled into the density map. The complex structure was well refined with statistics as expected at 1.7 Å resolution (Table 4). In the final structure, NSC 12155 displayed an average B factor of 56 Å2, significantly higher than those for the protein and solvent molecules (Table 4). The high B factor indicated that NSC 12155 may only partially occupy the binding pocket. Interestingly, in the crystal structure of NSC 12155 in complex with the anthrax lethal factor, the NSC 12155 molecule also showed high average B factors in the 80s Å2.16 Consistently, the molecule was also considered as highly mobile.16 Nevertheless, although not fully occupied, the consistent continuous electron density map clearly defined the conformation of NSC 12155 in the MTase SAM-binding pocket.
Table 4.
Diffraction Data Collection and Structure Refinement Statistics
| data collection | |
| space group | P21 |
| cell parameters | |
| a = 47.34 Å, b = 99.3 Å, c = 57.85 Å | |
| α = 90°, β = 99.2°, γ = 90° | |
| resolution (Å) | 39−1.7 (1.73−1.7)a |
| no. observed reflections | 171203 |
| no. unique reflections | 54299 |
| redundancy | 2.9 (2.9) |
| completeness (%) | 99.9 (100) |
| average I/σ(I) | 16.9 (1.4) |
| Rsym (%) | 6.5 (82.2) |
| refinement | |
| resolution limits (Å) | 39−1.7 |
| no. of reflections (total) | 54299 |
| no. of reflections (Rfree) | 2003 |
| Rwork (%) | 18.8 |
| Rfree (%) | 22.6 |
| non-H atoms | |
| protein | 4094 |
| ligand | 56 |
| water | 508 |
| average B (Å2) | 18.8 |
| protein | 17 |
| ligand | 56 |
| water | 28.8 |
| geometry | |
| rmsd bond length (Å) | 0.005 |
| rmsd bond angle (deg) | 1.76 |
| Ramachandran plot | |
| favored | 98 |
| allowed | 2 |
| outliers | 0 |
Values in parentheses are those for the highest resolution shell.
The overall structure of the MTase–12155 complex is very similar to that of the SAM-removed MTase (PDB 4R0511) with a Cα rmsd of 0.31 Å (Figure 7D). The rmsd between the 12155–MTase complex and the SIN–MTase complex (PDB 4R8S17) is slightly higher (0.54 Å) (Figure 7D). The slightly higher rmsd between 12155 complex structure and 4R8S could be attributed to the facts that (1) the 4R8S complex was crystallized in a condition different from that for both the 12155 complex and 4R05 and (2) the space groups of 4R8S and the 12155 complex are different, and crystal packing difference could lead to larger structural differences. Nonetheless, the differences are small and well within coordinate error ranges. NSC 12155 binds to the SAM-binding pocket of the DENV3MTase with one quinoline ring of NSC 12155 at a position nearly identical to that of SAM or SIN (Figure 7D). We noted that the crystallography-determined structure of NSC 12155 is slightly different from that predicted by our docking experiment (data not shown), indicating that even though the docking predicted correctly the tight binding energy, crystal structure is required to determine the precise conformation of the compound in contact with the receptor. Regardless, the carboxyl oxygen of 12155 also mimics the oxygen of the ribose group of SAM/SIN, whereas the second quinoline ring of 12155 deviates significantly from the methionine part of SAM (Figure 7D). In the SAM/SIN–MTase complex, the methionine moiety of SAM folds back toward the MTase,6e,7e whereas in the 12155–MTase complex, the second quinolone ring of 12155 folded outward, fitting into a cleft formed by the side chains of H110 and E111 (Figure 7D). NSC 12155 forms numerous contacts with the MTase residues, including G81, T104, K105, H110, E111, K130, D131, V132, F133, D146, and I147. The majority of these contacts are through van de Waals interactions, indicating that hydrophobic interactions play a major role in MTase–12155 recognition.
The majority of flaviviruses are significant human pathogens. The flavivirus MTases have been an attractive target for antiviral development.7i,18 A number of inhibitors have been identified through various approaches.18 Although several inhibitors display subnanomolar inhibition of the MTase in biochemical assay,7g cell permeability issues prohibited them from further improvement.18 In addition, only a limited number of crystal structures of the MTase–inhibitor complexes, particularly for the SAM-binding site inhibitors, are available.6e,7g The successful crystallization of SAM-free MTases may accelerate discovery in this area.11,17
In this study, using a combination of virtual screening, in vitro MTase inhibition assays, cytotoxicity assays, and viral growth inhibition assays, we have identified two promising broad-spectrum antiflaviviral lead compounds with relatively low cytotoxicity. Particularly, compound NSC 12155 not only inhibited the N7 and 2′-O methylation activities with low micromolar IC50 values in in vitro biochemical assays but also inhibited WNV growth with similar low micromolar antiviral efficacy in cell culture (Tables 1–3). Further investigation indicated that it is a broad-spectrum inhibitor, abolishing the activities of MTases from multiple viruses and suppressing the growth of several flaviviruses, whereas the growth of control arboviruses was not affected. By using a competitive SAM-binding assay, we demonstrated that NSC 12155 competitively inhibited the binding of SAM to the MTase cofactor pocket. Finally, we determined the crystal structure of the DENV3MTase–12155 complex.
A PubChem Database search indicated that NSC 12155 is only active in the NCI Yeast Anticancer Drug Screen, indicating that 12155 is less likely a pan-assay interference compound.19 Interestingly, NSC 12155, also named aminoquinuride or surfen, was reported to bind to glycosaminoglycans and prevent cell adhesion by herpes simplex virus (HSV)-1.20 Although it is currently not clear whether DENV and HSV-1 have the same entry mechanism, it is possible that NSC 12155 also inhibited DENV through multiple mechanisms, including inhibitions of binding to both the viral methyltransferase and the host receptors such as glycosaminoglycans.
It was reported that NSC 12155 inhibited the anthrax lethal factor (LF) activity.16 The crystal structure of the LF–12155 complex indicated that NSC 12155 binds to the catalytic site of LF with an extended C-shaped conformation.16 In the cocrystal structure of the DENV3MTase–12155 complex, NSC 12155 displays a curved conformation. One of its quinoline rings binds deeply into the SAM-binding pocket, and the other one is curved from the urea moiety and snugged into a pocket formed between MTase residues H110 and E111 (Figure 7). The cocrystal structures indicated that NSC 12155 is likely a specific inhibitor of the flavivirus MTase. Overall, structural analysis should provide the first structural basis for their further optimization as flavivirus MTase inhibitors and antiflaviviral drugs.
ACKNOWLEDGMENTS
This research was supported by grants (AI09433501 to L.D.K and H.L.) from the National Institute of Health (NIH)). M.B. was partially supported by the NIH Biodefense and Emerging Infectious Disease training grant AI055429. We thank Dr. Vivian Stojanoff at the Brookhaven National Laboratory for helps during X-ray data collection. Use of the Stanford Synchrotron Radiation Lightsource, SLAC National Accelerator Laboratory, is supported by the U.S. Department of Energy, Office of Science, Office of Basic Energy Sciences under Contract No. DE-AC02-76SF00515. The SSRL Structural Molecular Biology Program is supported by the DOE Office of Biological and Environmental Research, and by the National Institutes of Health, National Institute of General Medical Sciences (including P41GM103393). The contents of this publication are solely the responsibility of the authors and do not necessarily represent the official views of NIGMS or NIH. We thank the Wadsworth Center Tissue Culture Core facility for providing cells and media.
Footnotes
ASSOCIATED CONTENT
Accession Codes
The atomic coordinates and structure factors (code 5CUQ) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ, USA (http://www.rcsb.org/).
The authors declare no competing financial interest.
REFERENCES
- 1.(a) Teruel-Lopez E. Dengue. A review. Invest. Clin. 1991;32(4):201–217. [PubMed] [Google Scholar]; (b) Turtle L, Griffiths MJ, Solomon T. Encephalitis caused by flaviviruses. QJM. 2012;105(3):219–223. doi: 10.1093/qjmed/hcs013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.WHO. Dengue factsheet. 2015 http://www.who.int/mediacentre/factsheets/fs117/en/
- 3.Heinz FX, Stiasny K. Flaviviruses and flavivirus vaccines. Vaccine. 2012;30(29):4301–4306. doi: 10.1016/j.vaccine.2011.09.114. [DOI] [PubMed] [Google Scholar]
- 4.Ray D, Shah A, Tilgner M, Guo Y, Zhao Y, Dong H, Deas TS, Zhou Y, Li H, Shi PY. West Nile virus 5′-cap structure is formed by sequential guanine N-7 and ribose 2′-O methylations by nonstructural protein 5. J. Virol. 2006;80(17):8362–8370. doi: 10.1128/JVI.00814-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dong H, Chang DC, Hua MH, Lim SP, Chionh YH, Hia F, Lee YH, Kukkaro P, Lok SM, Dedon PC, Shi PY. 2′-O methylation of internal adenosine by flavivirus NS5 methyltransferase. PLoS Pathog. 2012;8(4):e1002642. doi: 10.1371/journal.ppat.1002642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.(a) Bhattacharya D, Hoover S, Falk SP, Weisblum B, Vestling M, Striker R. Phosphorylation of yellow fever virus NS5 alters methyltransferase activity. Virology. 2008;380(2):276–284. doi: 10.1016/j.virol.2008.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Dong H, Ren S, Li H, Shi PY. Separate molecules of West Nile virus methyltransferase can independently catalyze the N7 and 2′-O methylations of viral RNA cap. Virology. 2008;377(1):1–6. doi: 10.1016/j.virol.2008.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Kroschewski H, Lim SP, Butcher RE, Yap TL, Lescar J, Wright PJ, Vasudevan SG, Davidson AD. Mutagenesis of the dengue virus type 2 NS5 methyltransferase domain. J. Biol. Chem. 2008;283(28):19410–19421. doi: 10.1074/jbc.M800613200. [DOI] [PubMed] [Google Scholar]; (d) Khromykh AA, Kenney MT, Westaway EG. trans-Complementation of flavivirus RNA polymerase gene NS5 by using Kunjin virus replicon-expressing BHK cells. J. Virol. 1998;72(9):7270–7279. doi: 10.1128/jvi.72.9.7270-7279.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]; (e) Zhou Y, Ray D, Zhao Y, Dong H, Ren S, Li Z, Guo Y, Bernard KA, Shi PY, Li H. Structure and function of flavivirus NS5 methyltransferase. J. Virol. 2007;81(8):3891–3903. doi: 10.1128/JVI.02704-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.(a) Benarroch D, Egloff MP, Mulard L, Guerreiro C, Romette JL, Canard B. A structural basis for the inhibition of the NS5 dengue virus mRNA 2′-O-methyltransferase domain by ribavirin 5′-triphosphate. J. Biol. Chem. 2004;279(34):35638–35643. doi: 10.1074/jbc.M400460200. [DOI] [PubMed] [Google Scholar]; (b) Bollati M, Alvarez K, Assenberg R, Baronti C, Canard B, Cook S, Coutard B, Decroly E, de Lamballerie X, Gould EA, Grard G, Grimes JM, Hilgenfeld R, Jansson AM, Malet H, Mancini EJ, Mastrangelo E, Mattevi A, Milani M, Moureau G, Neyts J, Owens RJ, Ren J, Selisko B, Speroni S, Steuber H, Stuart DI, Unge T, Bolognesi M. Structure and functionality in flavivirus NS-proteins: perspectives for drug design. Antiviral Res. 2010;87(2):125–148. doi: 10.1016/j.antiviral.2009.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Chen H, Liu L, Jones SA, Banavali N, Kass J, Li Z, Zhang J, Kramer LD, Ghosh AK, Li H. Selective inhibition of the West Nile virus methyltransferase by nucleoside analogs. Antiviral Res. 2013;97(3):232–239. doi: 10.1016/j.antiviral.2012.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]; (d) Chen H, Zhou B, Brecher M, Banavali N, Jones SA, Li Z, Zhang J, Nag D, Kramer LD, Ghosh AK, Li H. S-adenosyl-homocysteine is a weakly bound inhibitor for a flaviviral methyltransferase. PLoS One. 2013;8(10):e76900. doi: 10.1371/journal.pone.0076900. [DOI] [PMC free article] [PubMed] [Google Scholar]; (e) Dong H, Liu L, Zou G, Zhao Y, Li Z, Lim SP, Shi PY, Li H. Structural and functional analyses of a conserved hydrophobic pocket of flavivirus methyltransferase. J. Biol. Chem. 2010;285(42):32586–32595. doi: 10.1074/jbc.M110.129197. [DOI] [PMC free article] [PubMed] [Google Scholar]; (f) Dong H, Ren S, Zhang B, Zhou Y, Puig-Basagoiti F, Li H, Shi PY. West Nile virus methyltransferase catalyzes two methylations of the viral RNA cap through a substrate-repositioning mechanism. J. Virol. 2008;82(9):4295–4307. doi: 10.1128/JVI.02202-07. [DOI] [PMC free article] [PubMed] [Google Scholar]; (g) Lim SP, Sonntag LS, Noble C, Nilar SH, Ng RH, Zou G, Monaghan P, Chung KY, Dong H, Liu B, Bodenreider C, Lee G, Ding M, Chan WL, Wang G, Jian YL, Chao AT, Lescar J, Yin Z, Vedananda TR, Keller TH, Shi PY. Small molecule inhibitors that selectively block dengue virus methyltransferase. J. Biol. Chem. 2011;286(8):6233–6240. doi: 10.1074/jbc.M110.179184. [DOI] [PMC free article] [PubMed] [Google Scholar]; (h) Lim SP, Wen D, Yap TL, Yan CK, Lescar J, Vasudevan SG. A scintillation proximity assay for dengue virus NS5 2′-O-methyltransferase-kinetic and inhibition analyses. Antiviral Res. 2008;80(3):360–369. doi: 10.1016/j.antiviral.2008.08.005. [DOI] [PubMed] [Google Scholar]; (i) Liu L, Dong H, Chen H, Zhang J, Ling H, Li Z, Shi PY, Li H. Flavivirus RNA cap methyltransferase: structure, function, and inhibition. Front. Biol. 2010;5(4):286–303. doi: 10.1007/s11515-010-0660-y. [DOI] [PMC free article] [PubMed] [Google Scholar]; (j) Luzhkov VB, Selisko B, Nordqvist A, Peyrane F, Decroly E, Alvarez K, Karlen A, Canard B, Qvist J. Virtual screening and bioassay study of novel inhibitors for dengue virus mRNA cap (nucleoside-2′O)-methyltransferase. Bioorg. Med. Chem. 2007;15(24):7795–7802. doi: 10.1016/j.bmc.2007.08.049. [DOI] [PubMed] [Google Scholar]; (k) Milani M, Mastrangelo E, Bollati M, Selisko B, Decroly E, Bouvet M, Canard B, Bolognesi M. Flaviviral methyltransferase/RNA interaction: structural basis for enzyme inhibition. Antiviral Res. 2009;83(1):28–34. doi: 10.1016/j.antiviral.2009.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]; (l) Podvinec M, Lim SP, Schmidt T, Scarsi M, Wen D, Sonntag LS, Sanschagrin P, Shenkin PS, Schwede T. Novel inhibitors of dengue virus methyltransferase: discovery by in vitro-driven virtual screening on a desktop computer grid. J. Med. Chem. 2010;53(4):1483–1495. doi: 10.1021/jm900776m. [DOI] [PubMed] [Google Scholar]; (m) Puig-Basagoiti F, Qing M, Dong H, Zhang B, Zou G, Yuan Z, Shi PY. Identification and characterization of inhibitors of West Nile virus. Antiviral Res. 2009;83(1):71–79. doi: 10.1016/j.antiviral.2009.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]; (n) Sampath A, Padmanabhan R. Molecular targets for flavivirus drug discovery. Antiviral Res. 2009;81(1):6–15. doi: 10.1016/j.antiviral.2008.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]; (o) Selisko B, Peyrane FF, Canard B, Alvarez K, Decroly E. Biochemical characterization of the (nucleoside-2′O)-methyltransferase activity of dengue virus protein NS5 using purified capped RNA oligonucleotides (7Me)GpppAC(n) and GpppAC(n) J. Gen. Virol. 2010;91(1):112–121. doi: 10.1099/vir.0.015511-0. [DOI] [PubMed] [Google Scholar]
- 8.(a) Lim PY, Behr MJ, Chadwick CM, Shi PY, Bernard KA. Keratinocytes are cell targets of West Nile virus in vivo. J. Virol. 2011;85(10):5197–5201. doi: 10.1128/JVI.02692-10. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Coutard B, Decroly E, Li C, Sharff A, Lescar J, Bricogne G, Barral K. Assessment of Dengue virus helicase and methyltransferase as targets for fragment-based drug discovery. Antiviral Res. 2014;106:61–70. doi: 10.1016/j.antiviral.2014.03.013. [DOI] [PubMed] [Google Scholar]
- 9.Trott O, Olson AJ. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010;31(2):455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.O’Boyle NM, Banck M, James CA, Morley C, Vandermeersch T, Hutchison GR. Open Babel: an open chemical toolbox. J. Cheminf. 2011;3:33. doi: 10.1186/1758-2946-3-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brecher MB, Li Z, Zhang J, Chen H, Lin Q, Liu B, Li H. Refolding of a fully functional flavivirus methyltransferase revealed that S-adenosyl methionine but not S-adenosyl homocysteine is copurified with flavivirus methyltransferase. Protein Sci. 2015;24(1):117–128. doi: 10.1002/pro.2594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–325. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 13.Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2010;66(2):213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2010;66(4):486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.(a) Feng BY, Shelat A, Doman TN, Guy RK, Shoichet BK. High-throughput assays for promiscuous inhibitors. Nat. Chem. Biol. 2005;1(3):146–148. doi: 10.1038/nchembio718. [DOI] [PubMed] [Google Scholar]; (b) Feng BY, Shoichet BK. A detergent-based assay for the detection of promiscuous inhibitors. Nat. Protoc. 2006;1(2):550–553. doi: 10.1038/nprot.2006.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Panchal RG, Hermone AR, Nguyen TL, Wong TY, Schwarzenbacher R, Schmidt J, Lane D, McGrath C, Turk BE, Burnett J, Aman MJ, Little S, Sausville EA, Zaharevitz DW, Cantley LC, Liddington RC, Gussio R, Bavari S. Identification of small molecule inhibitors of anthrax lethal factor. Nat. Struct. Mol. Biol. 2004;11(1):67–72. doi: 10.1038/nsmb711. [DOI] [PubMed] [Google Scholar]
- 17.Noble CG, Li SH, Dong H, Chew SH, Shi PY. Crystal structure of dengue virus methyltransferase without Sadenosyl-l-methionine. Antiviral Res. 2014;111:78–81. doi: 10.1016/j.antiviral.2014.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lim SP, Noble CG, Shi PY. The dengue virus NS5 protein as a target for drug discovery. Antiviral Res. 2015;119:57–67. doi: 10.1016/j.antiviral.2015.04.010. [DOI] [PubMed] [Google Scholar]
- 19.Baell JB, Holloway GA. New substructure filters for removal of pan assay interference compounds (PAINS) from screening libraries and for their exclusion in bioassays. J. Med. Chem. 2010;53(7):2719–2740. doi: 10.1021/jm901137j. [DOI] [PubMed] [Google Scholar]
- 20.Schuksz M, Fuster MM, Brown JR, Crawford BE, Ditto DP, Lawrence R, Glass CA, Wang L, Tor Y, Esko JD. Surfen, a small molecule antagonist of heparan sulfate. Proc. Natl. Acad. Sci. U. S. A. 2008;105(35):13075–13080. doi: 10.1073/pnas.0805862105. [DOI] [PMC free article] [PubMed] [Google Scholar]