Abstract
A new Penicillium species, P. excelsum, is described here using morphological characters, extrolite and partial sequence data from the ITS, β-tubulin and calmodulin genes. It was isolated repeatedly using samples of nut shells and flowers from the brazil nut tree, Bertolletia excelsa, as well as bees and ants from the tree ecosystem in the Amazon rainforest. The species produces andrastin A, curvulic acid, penicillic acid and xanthoepocin, and has unique partial β-tubulin and calmodulin gene sequences. The holotype of P. excelsum is CCT 7772, while ITAL 7572 and IBT 31516 are cultures derived from the holotype.
Introduction
Penicillium species are very important agents in the natural processes of recycling biological matter. Some species cause deterioration of all sorts of man-made goods; some rot fruit or spoil foods; some species secrete secondary metabolites (extrolites) such as mycotoxins (e.g. ochratoxins, patulin, citrinin), while other extrolites are used as pharmaceuticals, including antibiotics such as penicillin and the cholesterol-lowering agent lovastatin [1, 2, 3, 4]. Some species are known for their production of organic acids and diverse enzymes that degrade a wide variety of complex biomolecules [1, 2, 3]. A variety of species are capable of producing or modifying biological chemicals, and this field is set for great expansion. A few species are directly involved in food production: this field is not likely to expand, because many species produce mycotoxins. Penicillium is an ascomycete genus and belongs to the family Aspergillaceae [4]. More than 350 species are currently accepted in this genus [5].
The Amazon rainforest has multiple ecosystems with a huge fungal biodiversity. It has an important role in the global weather balance and is the location of many native people. The equatorial climate is hot and humid, with an average temperature of 26°C and relative humidity 80–95%.
Brazil nuts are one of the most important products taken from the Amazon rainforest region. Brazil nut trees, Bertholletia excelsa Humb. & Bonp., grow wild, take 12 years to bear fruit, may live up to 500 years and reach up to 60 m high. Pollination of the unusual flowers requires wild, large bodied bees, especially from the family Euglossinae [6]. The fungal species most commonly isolated from brazil nuts are Aspergillus flavus, A. nomius, A. pseudonomius, A. niger, A. tamarii, Penicillium glabrum, P. citrinum, Rhizopus spp., Fusarium oxysporum [7, 8, 9, 10, 11, 12] and A. bertholletius, a species described recently [13].
During a study of the mycobiota of the brazil nut tree ecosystem, including flowers, brazil nuts, soil, bees and ants, an undescribed Penicillium species was found. This species is described here as Penicillium excelsum sp. nov.
Materials and Methods
Sample collection, isolation and morphological examination
Samples were collected from the ecosystem of the brazil nut tree, Bertholletia excelsa in the Amazon rainforest in Para and Amazon States, Brazil. Sample collection and methodology have been described previously [13]. Briefly, samples of brazil nut kernels and shells, flowers and leaves, soil from beneath the trees, plus bees and ants. Collecting was carried out in collaboration with the Brazilian Ministry of Agriculture.
For fungal isolation, nuts and shell samples were disinfected in sodium hypochlorite solution, then plated onto dichloran 18% glycerol agar (DG18), according to the methodology of Pitt and Hocking [2]. Soil samples were mixed with sterile water containing peptone (0.1%), then serially diluted and spread plated onto DG18. Flower and leaf samples were surface disinfected as above and plated onto DG18 while bee and ant samples were plated on DG18 without surface disinfection. All plates were incubated at 25°C for 7 days, then all colonies of Penicillium species were transferred onto Czapek yeast extract agar [2] and incubated at 25°C for 7 days for further identification.
The Penicillium isolates were examined on standard identification media for Penicillium species according to Pitt [14] namely: Czapek yeast extract agar (CYA), malt extract agar (MEA, Oxoid), and 25% glycerol nitrate agar (G25N) at 25°C and also on CYA at 37°C and 42°C, plus oatmeal agar (OAT), creatine sucrose agar (CREA) and yeast extract sucrose (YES) agar [3]. The incubation time for all media was 7 days and plates incubated in the dark.
The standard conditions used for the description of Penicillium excelsum are taken from Pitt [14] and Frisvad and Samson [15]. Capitalized colours are from the Methuen Handbook of Colour [16].
DNA extraction, amplification, sequencing and phylogenetic analysis
A standard phenol:chloroform extraction protocol [17] was used for genomic DNA isolation from an extype culture (ITAL 7572). The primer-pairs ITS1-ITS4 [18], Bt2a-Bt2b [19] and cmd5-cmd6 [20] were used to amplify the ITS1-5,8S-ITS2 region (ITS), partial β-tubulin gene (BenA) and partial calmodulin gene (CaM) respectively, adopting a standard amplification cycle, which ran 35 cycles with an annealing temperature of 55°C [5]. Excess primers and dNTPs were removed from the PCR product using the Wizard® SV Gel and PCR Clean-Up System (Promega, Wisconsin, USA). Purified PCR products were sequenced in both directions using a BigDye® Terminator v3.1 Cycle Sequencing kit (Applied Biosystems, California, USA) according to the manufacturer’s instructions. A volume of HiDiformamide (10 μl) was added to the sequencing products, which were processed in an ABI 3500XL Genetic Analyzer (Applied Biosystems). Contigs were assembled using the forward and reverse sequences with the programme SeqMan from the Laser Gene package (DNAStar Inc., Wisconsin, USA). All sequences were subjected to Basic Local Alignment Search Tool (BLAST) against the NCBI database to identify Penicillium species with similar DNA sequences. The ITS and BenA sequences were aligned by ClustalW algorithm using Mega5.1 software (21) with those from Penicillium subgenus Aspergilloides section Lanata-Divaricata type or neotype strains, as recently suggested by Visagie et al [5]. Phylogenetic trees were constructed with Mega5.1 software [21], using the Neighbor-Joining (NJ) and Maximum Likelihood (ML) methods based on the Tamura-Nei model [22]. To determine the support for each clade, a nonparametric bootstrap analysis was performed with 1,000 resamplings.
The ITS, BenA and CaM sequences were deposited in GeneBank under the respective following accession numbers: KR815341, KP691061, KR815342 (strain ITAL 7572); KT749964, KT749957, KT749961 (strain ITAL 7814); KT749965, KT749958, KT749960 (strain 7823); KT749963, KT749959, KT749962 (strain 7804).
Extrolite analysis
Cultures were analysed by High Performance Liquid Chromatography (HPLC) with a diode array detector (HPLC-DAD) as described by Frisvad and Thrane [23] and modified by Houbraken et al. [24], as previously described [13]. Three agar plugs each from CYA and YES medium were pooled and extracted with 0.75 mL of a mixture of ethyl acetate/ dichloromethane/methanol (3:2:1) (v/v/v) with 1% (v/v) formic acid.
Nomenclature
The new name contained in this work has been submitted to MycoBank from where it will be made available to the Global Names Index. The unique MycoBank number can be accessed and the associated information viewed through any standard web browser by appending the MycoBank number contained in this publication to the prefix http://www.mycobank.org/MB811066.
Repository of Penicillium excelsum Taniwaki, Pitt & Frisvad 2015 sp. nov. [urn:lsid:mycobank.org: 811066]
Results and Discussion
Sources of the isolates
In total, 116 isolates of -the new species described here as Penicillium excelsum were found in brazil nut shells and kernels, from soil close to Bertholletia excelsa trees, and from flowers, bees and ants associated with Bertholletia trees. The origins of representative P. excelsum isolates are shown in Table 1. Soil may be the primary habitat of this species, as many species of Penicillium are soil fungi [4, 25]. However, this study shows that P. excelsum also occurs on bees and ants, which may carry spores to the flowers, and other locations by contact or excreta which will all play a role in dispersal of this species.
Table 1. Penicillium excelsum isolates from the Amazon region.
| N° | Substrate | Rainforest | Sample n° |
|---|---|---|---|
| ITAL 2172 | Flowers | Amazon | 85 |
| ITAL 2248 | Flowers | Amazon | 96 |
| ITAL 3000/IBT 30867 | Nut | Pará | 113 |
| ITAL 3030/IBT 30865 | Flowers | Pará | 132 |
| ITAL 3743 | Nut | Pará | 120 |
| ITAL 3904 | Flowers | Pará | 136 |
| ITAL 3931 | Flowers | Pará | 137 |
| ITAL 3985 | Flowers | Pará | 142 |
| ITAL 4005 | Flowers | Pará | 143 |
| ITAL 4067 | Flowers | Pará | 144 |
| ITAL 4419 | Nut | Pará | 152 |
| ITAL 4432 | Nut | Pará | 153 |
| ITAL 4451 | Flowers | Pará | 155 |
| ITAL 4493 | Nut | Pará | 164 |
| ITAL 4545 | Flowers | Pará | 165 |
| ITAL 4570 | Flowers | Pará | 166 |
| ITAL 6705 | Nut | Amazon | 211 |
| ITAL 6706 | Shell | Amazon | 211 |
| ITAL 6722 | Nut | Amazon | 212 |
| ITAL 6752 | Nut | Amazon | 213 |
| ITAL 7014 | Nut | Amazon | 220 |
| ITAL 7035 | Shell | Amazon | 220 |
| ITAL 7572*/CCT 7772/IBT 31516 | Shell | Amazon | 234 |
| ITAL 7613 | Shell | Amazon | 236 |
| ITAL 7732 | Flowers | Amazon | 239 |
| ITAL 7741/CCT 7773/IBT 32953 | Flowers | Amazon | 240 |
| ITAL 7760/CCT 7775/IBT 32732 | Flowers | Amazon | 240 |
| ITAL 7770/CCT 7776 | Flowers | Amazon | 241 |
| ITAL 7788/CCT 7777 | Flowers | Amazon | 242 |
| ITAL 7804/CCT 7778 | Flowers | Amazon | 242 |
| ITAL 7814/CCT 7779 | Bees | Amazon | 244 |
| ITAL 7823/CCT 7780 | Bees | Amazon | 244 |
| ITAL 7875/CCT 7781 | Ants | Amazon | 250 |
| ITAL 8014 | Soil | Amazon | 255 |
* Type culture
Culture collection of: Instituto de Tecnologia de Alimentos (ITAL), Coleção de Cultura Tropical (CCT), Technical University of Denmark (IBT).
Extrolites
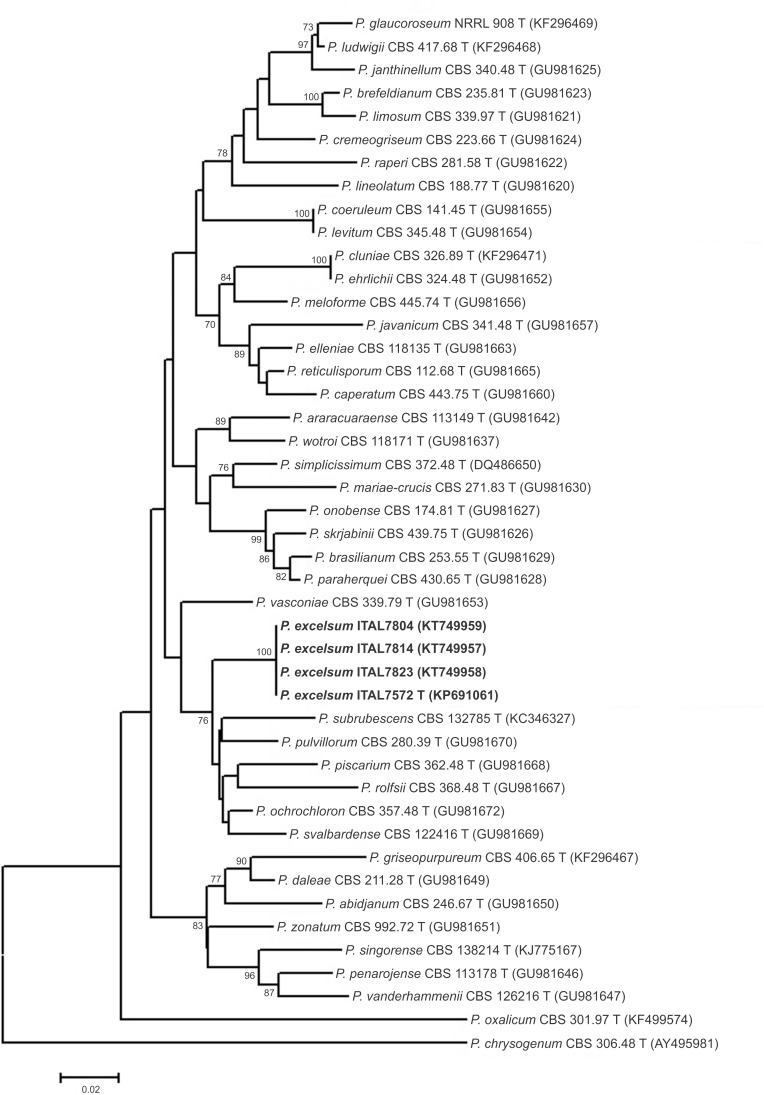
HPLC-DAD analysis of extracts showed that several strains of P. excelsum produce andrastin A, penicillic acid, while some also produce xanthoepocin. Strain ITAL 3000 also produced curvulic acid. Related species also produce penicillic acid, for example P. brasilianum, P. cremeogriseum, P. ochrochloron P. pulvillorum and P. vanderhammenii [24, 26, 27]. P. pulvillorum and P. simplicissimum have also been reported to produce andrastin A, and P. brasilianum, P. ochrochloron, P. pulvillorum, P. rolfsii, P. simplicissimum and P. svalbardense have been reported to produce xanthoepocin [24, 28]. Even though andrastin A, penicilllic acid, and xanthoepocin have been found in species outside section Lanata-Divaricata [15] the particular combination of these extrolites is mostly found in this section. P. excelsum produces a profile of extrolites close to that of P. brasilianum, P. ochrochloron, P. pulvillorum and P. rolfsii and the close relationship is confirmed by sequence and morphological data as shown in Figs 1, 2 and 3.
Fig 1. Neighbour joining tree reconstructed from the partial β-tubulin (BenA) gene sequences aligned with corresponding sequences of Penicillium section Lanata-Divaricata deposited in public databases.
Numbers at branch nodes refer to bootstrap values (1000 replicates), only values of >70% are shown.
Fig 2. Maximum-likelihood tree reconstructed from the partial β-tubulin (BenA) gene sequences aligned with corresponding sequences of Penicillium section Lanata-Divaricata deposited in public databases.
Numbers at branch nodes refer to bootstrap values (1,000 replicates), only values of >70% are shown.
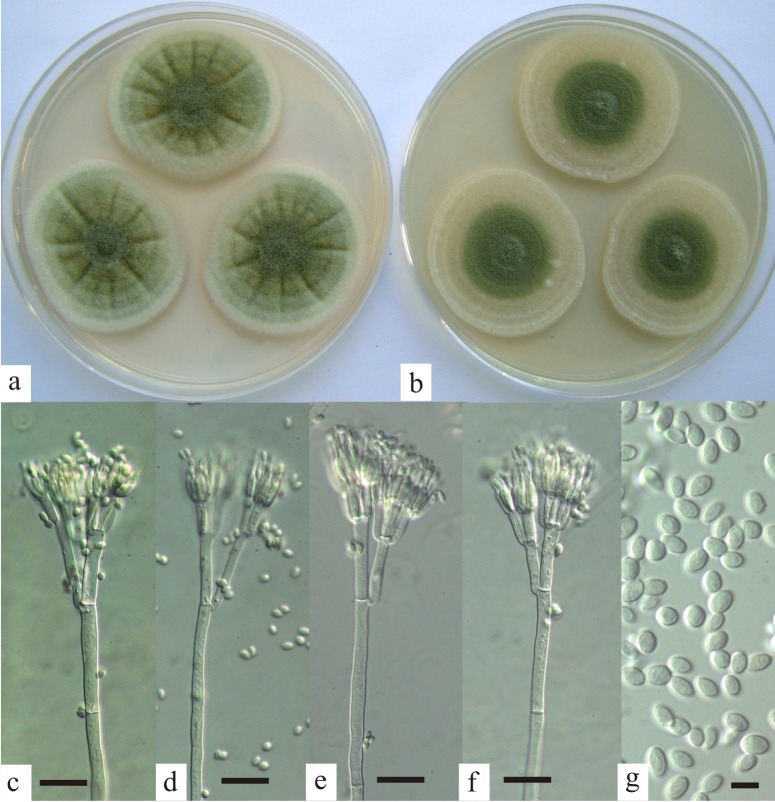
Fig 3. Penicillium excelsum.
Colonies after 7 days at 25°C on (a) Czapek yeast extract agar; (b) malt extract agar; (c–f) penicilli, bar = 20 μm; (g) conidia, bar = 5 μm.
Phylogenetic analyses
P. excelsum ITS, BenA and CaM sequences were found to be different from all other sequences in NCBI (accessed 30 May, 2015). When the BLAST searches were performed using the option “sequences from type material” [29] the sequences harmonized in showing that P. excelsum is most similar to P. ochrochloron neotype strain CBS 357.48 and P. pulvillorum neotype CBS 280.39. Both, P. pulvillorum and P. ochrochloron belong to Penicillium subgenus Aspergilloides section Lanata-Divaricata in the recent phylogenetic reclassification of Penicillium [4].
A more recent study [5] provided GenBank accession numbers to reference sequences for all accepted Penicillium species. Using these reference sequences, ITS-based phylograms (data not shown) generated using Neighbor-Joining and Maximum Likelihood techniques confirmed the placement of P. excelsum in section Lanata-Divaricata. Although the ITS phylograms of P. excelsum clustered and were differentiated from other species of section Lanata-Divaricata, the majority of bootstrap values of branches were low, meaning that the ITS tree was poorly resolved. The ITS region is accepted as the primary fungal barcode [30]; however, it is well known that the ITS region provides only poor resolution of many Penicillium species [5, 31]. In consequence, it has been proposed [5] that β-tubulin (BenA) is an optimal secondary identification marker for Penicillium species. BenA-based phylograms, generated using both Neighbor-Joining and Maximum Likelihood methods, placed P. excelsum on a branch separated from all other species on Penicillium section Lanata-Divaricata (Figs 1 and 2). Neighbor-Joining and Maximum Likelihood based phylograms were consistent and reveled that P. excelsum represent a separated lineage within a clade composed of P. pulvillorum, P. svalbardense, P. piscarium, P. ochrochloron, P. rolfsii, and P. subrubescens.
Taxonomy
Penicillium excelsum sp. nov. Taniwaki, Pitt & Frisvad sp. nov. Mycobank MB 811066 (Fig 3)
On CYA at 7 days, 25°C, colonies 35–50 mm in diameter, dense, lightly sulcate, mycelium white to off-white; lightly sporing, coloured greenish grey (M. 28D2); exudate and soluble pigment absent; reverse pale to slightly brown or pink.
On MEA at 7 days, 25°C, colonies 28–50 mm in diameter, low and sparse, plane, mycelium hyaline to white; lightly sporing, greenish grey; exudate and soluble pigment absent; reverse pale brown.
On G25N at 7 days, 25°C, colonies 10–14 mm in diameter, low and dense, coloured buff with light sporulation; reverse brown to deep brown.
On YES agar at 7 days, 25°C, colonies 34–42 mm in diameter, moderate sporulation and a brown reverse.
On OAT at 7 days, 25°C, colonies 41–48 mm, strong sporulation, reverse brown.
On CREA at 7 days, 25°C, colonies 18–32 mm, weak growth, no sporulation, and no acid production.
At 37°C on CYA, colonies 8–22 mm in diameter, coloured grey to brown; soluble pigment brown, reverse deep brown.
At 42°C on CYA, no growth.
Conidiophores borne from surface hyphae, long and robust, up to 600 x 4.0–5.0 μm, smooth walled, septate, bearing typically biverticillate appressed penicilli, metulae commonly 15–18 x 4–6 μm, but sometimes terverticillate with rami 15–40 μm long; phialides appressed, 10–12 x 3.0–3.5 μm, ampulliform-acerose, bearing ellipsoidal conidia, 4.0–5.0 x 2.0–3.2 μm, smooth walled.
Holotype
CCT 7772, a freeze dried culture in Coleção de Cultura Tropical (Campinas, Brazil) is designated as the holotype of P. excelsum. It was isolated from brazil nut shell, Amazon, Brazil, 2011, by Taniwaki, M.H.
Cultures derived from this type include ITAL 7572 (where ITAL stands for the culture collection of Instituto de Tecnologia de Alimentos, Campinas, Brazil, accredited as Faithful Depositary by the Brazilian Executive Secretary of the Board of the Genetic Heritage Management n° 125/2015) and IBT 31516 (where IBT is the culture collection of the Technical University of Denmark, Lyngby, Denmark).
Etymology: named for the Amazonian brazil nut tree, Bertholletius excelsa, with which this species is associated.
Other isolates examined. ITAL 3000 (= IBT 30867), ITAL 3030 (= IBT 30865), ITAL 7741 (= CCT 7773 = IBT 32953), ITAL 7760 (= CCT 7775 = IBT 32732), ITAL 7770 (= CCT 7776), ITAL 7788 (= CCT 7777), ITAL 7804 (= CCT 7778), ITAL 7814 (= CCT 7779), ITAL 7823 (= CCT 7780) and ITAL 7875 (= CCT 7781).
Distinguishing features
This species is classified in Penicillium subgenus Furcatum section Furcatum in the classification of Pitt [14] and Penicillium subgenus Aspergilloides section Lanata-Divaricata according to Houbraken and Samson [4].
Morphologically, P. excelsum differs from the closely related P. subrubescens, P. pulvillorum, P. piscarium, P. rolfsii, P. ochrochloron and P. svalbardense by having a combination of smooth stipes, the frequent formation of rami, and the production of large, ellipsoidal, smooth walled conidia. P. ochrochloron and P. rolfsii are similar, but have finely roughened conidia. P. subrubescens, P. pulvillorum, P. piscarium and P. svalbardense produce globose to subglobose conidia, and in addition the conidia of P. piscarium are distinctly rough-walled. P. excelsum grows well at 37°C, though not as well as P. rolfsii. Most isolates of P. subrubescens and P. pulvillorum produce a red reverse colour on malt extract agar, whereas the reverse of P. excelsum is pale brown.
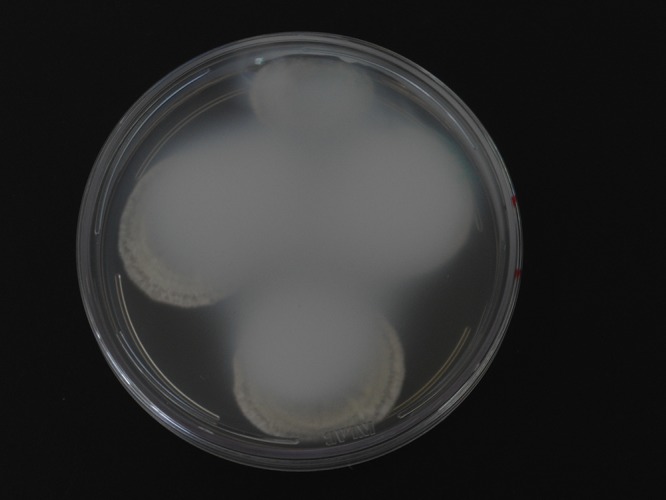
This species is also distinguished by a unique profile of extrolytes and by unique DNA sequences in the ITS, BenA and CaM genes. This species is also notable in that cultures on CYA, MEA and YES agar cause the polystyrene plastic in Petri dishes to become opaque over time (Fig 4). The opaqueness cannot be removed using a scapel, as the chemical reaction with the plastic lids was irreversible. A volatile compound produced by the fungus as it grows must be responsible. A preliminary examination of the volatiles from P. excelsum showed that it produced large amounts of acetic acid. An HPLD-DAD analysis of the opaque layer on the Petri dish lid revealed no detectable extrolites, indicating that the compound responsible for the opaqueness is without a chromophore. This effect has not been reported from any Penicillium or Aspergillus species. Further studies will be carried out in order to determine these compounds.
Fig 4. Plate of Czapek yeast extract agar (CYA) with Penicillium excelsum, opaqueness of petri dish lid after 7 days of incubation at 25°C.
Conclusion
P. excelsum represents a new important phylogenetic species after applying a polyphasic approach using morphological characters, extrolite data, ITS, BenA and CaM partial sequences. P. excelsum is distinguished by a combination of a unique profile of extrolites, DNA sequence, micro-morphological features and the unique capacity to render Petri dish lids irreversible opaque.
Data Availability
The new name contained in this work has been submitted to MycoBank from where it will be made available to the Global Names Index. The unique MycoBank number can be accessed and the associated information viewed by appending the MycoBank number contained in this publication to the prefix http://www.mycobank.org/MB811066.
Funding Statement
This research was supported by Fundação de Amparo à Pesquisa do Estado de São Paulo Process 2011/50136-0, Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq) and Ministério da Agricultura, Pecuária e Abastecimento, (MAPA) Brazil, Process 578485/2008-7. This taxonomic study is supported by the Brazilian Resolution of Genetic Heritage Management Council (MMA/CGEN 21/06). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Geiser DM, Gueidan C, Miadlikowska J, Lutzoni F, Kauff F, Hofstetter V et al. (2006). Eurotiomycetes: Eurotiomycetidae and Chaetothyriomycetidae . Mycologia 2006; 98: 1053–1064. [DOI] [PubMed] [Google Scholar]
- 2. Pitt JI, Hocking AD. Fungi and Food Spoilage 3rd ed. New York: Springer, 2009. [Google Scholar]
- 3. Samson RA, Houbraken J, Thrane U, Frisvad JC, Andersen B. Food and Indoor Fungi, CBS laboratory manual series 2, CBS-Fungal Biodiversity Centre, Utrecht, Netherlands, 2010. [Google Scholar]
- 4. Houbraken J and Samson RA (2011) Phylogeny of Penicillium and the segregation of Trichocomaceae into three families. Stud Mycol. 2011; 70: 1–51. 10.3114/sim.2011.70.01 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Visagie CM, Houbraken J, Frisvad JC, Hong S-B, Klaasen CHW, Perrone G et al. Identification and nomenclature of the genus Penicillium . Stud Mycol. 2014; 78: 343–371. 10.1016/j.simyco.2014.09.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Wadt LHO, Kainer KA, Gomes DAS. Population structure and nut yield of a Bertholletia excelsa stand in southwestern Amazonia. Forest Ecol Manag. 2005; 211: 371–384. [Google Scholar]
- 7. Freire FCO, Kozakiewicz Z, Paterson, RM. Mycoflora and mycotoxins in Brazilian black pepper, white pepper and Brazil nuts. Mycopathologia 2000; 149: 13–19. [DOI] [PubMed] [Google Scholar]
- 8. Bayman P, Baker J, Mahoney N Aspergillus on tree nuts. Mycopathologia 2002; 155: 161–169. [DOI] [PubMed] [Google Scholar]
- 9. Olsen M, Johnsson P, Moller T, Paladino R, Lindblad M. Aspergillus nomius, an important aflatoxin producer in Brazil nuts? World Mycotox J. 2008; 123–126. [Google Scholar]
- 10. Gonçalves JS, Ferracin LM, Vieira MLC, Iamanaka, BT, Taniwaki MH, Fungaro MHP. Molecular analysis of Aspergillus section Flavi isolated from Brazil nuts. World J Microbiol Biotechnol. 2012; 28: 1817–1825. [DOI] [PubMed] [Google Scholar]
- 11. Calderari TO, Iamanaka BT, Frisvad JC, Pitt JI, Sartori D, Pereira JL et al. The biodiversity of Aspergillus section Flavi in brazil nuts: from rainforest to consumer. Int J Food Microbiol. 2013; 160: 267–272. 10.1016/j.ijfoodmicro.2012.10.018 [DOI] [PubMed] [Google Scholar]
- 12. Massi FP, Vieira MLC, Sartori D, Penha RES, Munhoz CF, Ferreira JM, et al. Brazil nuts are subject to infection with B and G aflatoxin-producing fungus, Aspergillus pseudonomius . Int J Food Microbiol. 2014; 186: 14–21 10.1016/j.ijfoodmicro.2014.06.006 [DOI] [PubMed] [Google Scholar]
- 13. Taniwaki MH, Pitt JI, Iamanaka BT, Sartori D, Copetti MV, Balajee A et al. Aspergillus bertholletius sp. nov. from brazil nuts. PLOSONE 2012; 7: e42480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Pitt JI. A Laboratory Guide to Common Penicillium Species. Commonwealth Scientific and Industrial Research Organization, North Ryde, NSW, 2000. [Google Scholar]
- 15. Frisvad JC, Samson RA. Polyphasic taxonomy of Penicillium subgenus Penicillium. A guide to identification of the food and air-borne terverticillate Penicillia and their mycotoxins. Stud Mycol. 2004; 49: 1–173. [Google Scholar]
- 16. Kornerup A, Wanscher JH. Methuen Handbook of Colour Eyre Methuen, London, 1978. [Google Scholar]
- 17. Sambrook J, Russell DW . Molecular Cloning A Laboratory Manual. Cold Spring Harbor Laboratory Press, NY, USA, 2001. [Google Scholar]
- 18. White TJ, Burns T, Lee S, Taylor J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics In: Innis MA, Gelfand DH, Sninsky JJ, White TJ (eds.) PCR Protocols: A guide to methods and applications, Academic Press, San Diego, pp 315–322, 1990. [Google Scholar]
- 19. Glass NL, Donaldson GC. Development of primer sets designed for use with the PCR to amplify conserved genes from filamentous Ascomycetes. Appl Environ Microbiol. 1995; 61: 1323–1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hong SB, Cho HS, Shin HD, Frisvad JC, Samson RA. Novel Neosartorya species isolated from soil in Korea. Int J Syst Evol Microbiol. 2006; 56: 477–486. [DOI] [PubMed] [Google Scholar]
- 21. Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011; 28: 2731–2739. 10.1093/molbev/msr121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Tamura K, Nei M. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol Biol Evol. 1993; 10: 512–526. [DOI] [PubMed] [Google Scholar]
- 23. Frisvad JC, Thrane U. Standardized high performance liquid chromatography of 182 mycotoxins and other fungal metabolites based on alkylphenone retention indices and UV-VIS spectra (diode array detection). J Chromatogr A 1987; 404: 195–214. [DOI] [PubMed] [Google Scholar]
- 24. Houbraken J, López C, Frisvad JC, Boekhout T, Theelen B, Molano AEF. et al. Five new Penicillium species, P. aracuaraense, P. elleniae, P. penarojaense, P. vanderhammenii and P. wotroi, from Colombian leaf litter. Int J Syst Evol Microbiol. 2011; 61: 1462–1475. [DOI] [PubMed] [Google Scholar]
- 25. Pitt JI. The Genus Penicillium and its Teleomorphic States Eupenicillium and Talaromyces Academic Press, London; 1980. [Google Scholar]
- 26.Frisvad JC, Filtenborg O. Revision of Penicillium subgenus Furcatum based on secondary metabolites and conventional characters. In: Samson, R.A. and Pitt, J.I. (eds.): Modern concepts in Penicillium and Aspergillus classification. Plenum Press, New York; 1990. pp. 159-170.
- 27. Mansouri S, Houbraken J, Samson RA, Frisvad JC, Christensen M, Tuthill DE, Koutaniemi S, Hatakka A, Lankinen P. Penicillium subrubescens, a new species efficiently producing inulinase. Antonie van Leeuwenhoek 2013; 103: 1343–1357. 10.1007/s10482-013-9915-3 [DOI] [PubMed] [Google Scholar]
- 28. Sonjak S, Uršic V, Frisvad JC, Gunde-Cimerman N. Penicillium svalbardense, a new species from Arctic glacial ice. Antonie van Leeuwenhoek 2007; 92: 43–51. [DOI] [PubMed] [Google Scholar]
- 29. Schoch CL, Robbertse B, Robert V, Vu D, Cardinali G, Irinyi L. et al. Finding needles in haystacks: linking scientific names, reference specimens and molecular data for Fungi. Database, 2014. Available: 10.1093/database/bau061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Schoch CL, Seifert KA, Huhndorf S, Robert V, Spouge JL, Levesque CA. et al. Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for fungi. Proceedings of the National Academy of Sciences of the United States of America 2012; 109: 6241–6246. 10.1073/pnas.1117018109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Houbraken J, López-Quintero C, Frisvad JC, Boekhout T, Theelen B, Franco-Molano AE. Penicillium araracuarense sp nov., Penicillium elleniae sp. nov., Penicillium penarojense sp. nov., Penicillium vanderhammenii sp. nov. and Penicillium wotroi sp. nov., isolated from leaf litter. Int J Syst Evol Microbiol. 2011; 61: 1462–1475. 10.1099/ijs.0.025098-0 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The new name contained in this work has been submitted to MycoBank from where it will be made available to the Global Names Index. The unique MycoBank number can be accessed and the associated information viewed by appending the MycoBank number contained in this publication to the prefix http://www.mycobank.org/MB811066.