Abstract
Altered chromatin structures and dynamics are responsible for a range of human malignancies, among which the status of histone lysine methylation remains of paramount importance. Histone lysine methylation is maintained by the relative activities of sequence-specific methyltransferase (KMT) writers and demethylase (KDM) erasers, with aberrant enzymatic activities or expression profiles closely correlated with multiple human diseases. Hence, targeting these epigenetic enzymes should provide a promising avenue for pharmacological intervention of aberrantly marked sites within the epigenome. Here we present an up-to-date critical evaluation on the development and optimization of potent small molecule inhibitors targeted to histone KMTs and KDMs, with the emphasis on contributions of structural biology to development of epigenetic drugs for therapeutic intervention. We anticipate that ongoing advances in the development of epigenetic inhibitors should lead to novel drugs that site-specifically target KMTs and KDMs, key enzymes responsible for maintenance of the lysine methylation landscape in the epigenome.
1. Introduction
Rapid improvements in next-generation sequencing techniques have contributed to the routine application of whole genome and tissue-specific sequencing for the identification of recurrent somatic mutations, many of which are driver mutations of tumorigenesis (Chi et al. 2010; Dawson & Kouzarides, 2012; Shih et al. 2012), thereby providing genomic cues of human disease. Complementary chromatin immunoprecipitation-based sequencing techniques have provided a comprehensive map of the epigenome, in the process revealing that many cancers are accompanied by changes in the epigenome (Dawson & Kouzarides, 2012). Further, accumulating evidence has established that disruption of the epigenome is a fundamental mechanism in cancer, which has prompted the development of therapeutic approaches to target the epigenetic machinery. DNA methyltransferases (DNMTs) and histone deacetylaseses (HDACs) were among the first epigenetic regulatory enzymes to be targeted for drug design (Minucci & Pelicci, 2006; Szyf, 2009), with several of these inhibitors approved by the Food and Drug Administration (FDA) for clinical use in hematological malignancies (Arrowsmith et al. 2012). These advances in turn led to a concerted effort toward identification and characterization of novel inhibitors for other histone-modifying enzymes. Histone methylation has long been considered one of the most prevalent chromatin modifications, one that is written by histone lysine methyltransferases (KMTs) and erased by histone lysine demethylases (KDMs). The establishment and maintenance of an appropriate histone methylation landscape is not only critical for normal cell development, but when altered, closely related to human disease. In addition, an increasing amount of biological data has indicated that aberrant enzymatic activities of a majority of these histone KMTs and KDMs are closely related to human cancer. Within the past decade, tremendous progress has been made toward identification initially of inhibitors that target histone KMTs (Copeland et al. 2009), and soon thereafter, their corresponding KDMs, with potent clinical potential. In this review, we focus on continuing progress toward the design of inhibitors that target human KMTs and KDMs. We start with a short introduction on the targeted enzymes, followed by an emphasis on the contributions of structural biology toward the development of potent inhibitors against specific KMTs and KDMs. We anticipate that a compendium of available successful examples of epigenetic inhibitor discovery against histone KMTs and KDMs will propel the field forward toward the identification and application of new approaches for drug discovery targeted to these epigenetic enzymes in the coming decade.
2. Small molecule inhibitors of histone lysine methyltransferases
Except for DOT1L, all identified histone KMTs are composed of a conserved approx. 130 amino acid (aa)-long SET domain, which adopts a unique topology composed of a series of β-strands folded into three β-sheets that surround a knot-like structure (Cheng et al. 2005). In addition, functional SET domain folds are usually flanked by two closely packed cysteine-rich modules called pre-SET and post-SET domains that are keys to enzymatic activity. Functionally, all histone KMTs catalyze methyl transfer from donor S-adenosyl methionine (SAM) (Fig. 1a) to substrate lysine, resulting in formation of methylated lysine product and S-adenosyl homocysteine (SAH) (Fig. 1b).
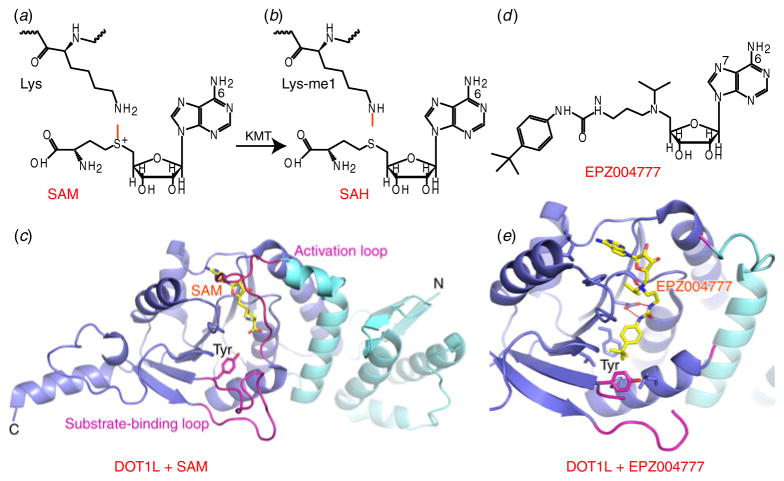
Fig. 1.
Illustration of the methyl-transfer reaction and inhibitors of H3K79 methyltransferase. (a, b) Chemical formula-based illustration of the methyl-transfer reaction. The active methyl group is colored in red. Chemical formulas of SAM and SAH are shown in panels (a) and (b), respectively. (c) Crystal structure of DOT1L with bound SAM, shown in a stick representation (PDB: 1NW3). The activation and substrate-binding loops are colored in purple. The N-terminal domain and the open α/β structure are colored in cyan and blue, respectively. (d) Chemical formula of EPZ004777. (e) Structure of complex of DOT1L with bound EPZ004777 (PDB: 4ER3). Color representations are the same as in panel (c).
2.1 Inhibitors of H3K79 methyltransferases
Methylation of H3K79 was first identified as a bona fide histone modification in the nucleosome core by mass spectroscopy, for which DOT1L was identified as the catalytic enzyme (Feng et al. 2002). Although not a translocation partner of MLL1, DOT1L is recruited by the super elongation complex formed by MLL1 fusion partners (Chi et al. 2010), with aberrant enzymatic activity of DOT1L shown to be a driving force of leukemogenesis (Bernt et al. 2011). Thus, DOT1L has long been considered to be an excellent therapeutic target for MLL1-related leukemia. As the only non-SET domain containing KMT identified in humans, the catalytic core of DOT1L adopts an elongated structure composed of an N-terminal domain and an open α/β-fold for the C-terminal domain (Fig. 1c), a folding topology more similar to histone arginine methyltransferases (RMTs) (Min et al. 2003; Sawada et al. 2004). The bound SAM cofactor exhibited excellent shape complementary within the pocket formed between an activation loop and the open α/β-fold in the structure of human DOT1L, whereas the potential histone substrate is expected to occupy the cleft between the activation loop and the substrate-binding loop (Fig. 1c). Guided by the above structural information, epizyme synthesized a SAM mimic named EPZ004777 (Fig. 1d), which inhibits DOT1L enzymatic activity with extremely high potency (IC50=0.4 nM) (Daigle et al. 2011). The chemical structure of EPZ004777 retains the nucleoside core of the SAM substrate, while the aa moiety was coupled with a bulky para-tert-butylphenyl group through a urea linkage, which should be unable to fit into the SAM-binding pocket. The structure of DOT1L bound to EPZ004777 validated the concept that the deazaa-denosine moiety exhibits similar interactions as SAM with DOT1L, while the tert-butylphenyl end induced a large structural rearrangement within the substrate-binding and activation loops (Yu et al. 2012). Notably, Tyr312, a key residue within the substrate-binding loop known to be important for KMT activity, was rotated away from this cleft (Fig. 1e), thereby indicating that EPZ004777 may also affect substrate binding. Despite the broad utilization of SAM by all histone KMTs, the SAM mimic EPZ004777 displayed remarkable inhibition selectivity toward DOT1L over all other methyltransferases tested in this study. EPZ004777 could selectively inhibit H3K79 methylation and blocked expression of leukemogenic genes in MLL cells, with little effect on non-MLL-translocated cells, indicative of promising potential as a drug candidate. However, poor pharmacokinetic properties of EPZ004777 limited its utility in an in vivo setting (Daigle et al. 2011). Taking advantage of the hydrophobic environment surrounding the adenine ring of SAM in DOT1L, further optimization on EPZ004777 led to the identification of a more potent compound SGC0946, which added a bromine atom at position 7 of the adenine ring (Yu et al. 2012). SGC0946 showed improved potency in in vitro studies (IC50=0.3 nm), as well as in cellular assays. Of interest, just addition of a single bromine atom at the N7 position of SAH (BrSAH) resulted in an 8-fold increase in potency against DOT1L (IC50=77 nM) (Yu et al. 2013), which verified that the N7 position on the adenine ring is available for further optimization. In a parallel study, the Song laboratory showed that addition of a methyl or benzyl group to the 6-NH2 group of SAM resulted in more selective inhibitors toward DOT1L, consistent with DOT1L having a more spacious hydrophobic binding pocket able to accommodate extra groups (Yao et al. 2011). Both examples provide valuable insights into the potential impact of beneficial structure-based modifications positioned either on the adenine ring or the aa moiety toward SAM-based inhibitor design.
2.2 Inhibitors of H3K4 and H3K27 methyltransferases
Histone H3K4 methylation is a hallmark of transcriptional activation, which in humans is established by the Trithorax group of proteins. H3K4 KMTs include SET1A and SET1B, as well as the MLL family proteins MLL1 to MLL4, which exhibit full activity only when complexed with the proteins WDR5, RbBP5, Ash2L and other components (Ruthenburg et al. 2007). Although aberrant expression of these histone KMTs are closely related to human cancer, as shown for MLL1 (Krivtsov & Armstrong, 2007) and MLL2 (Dawson & Kouzarides, 2012), the loss of enzymatic activity is not the causal reason for cancer. In addition, some of these H3K4 methyltransferases exhibit redundant function in cellular processes. In this regard, inhibitors that target the catalytic activity of H3K4 methyltransferases may not be promising candidates for cancer therapy, partially accounting for the slow progress in inhibitor design toward these enzymes.
In strong contrast to H3K4 methylation, H3K27 methylation is a hallmark of gene repression established by the Polycomb group of proteins, which function in opposition to the human Trithorax group of proteins. In practice, both Trithorax and Polycomb group of proteins function together in maintaining ‘ cellular memory ’. The core enzymes that add H3K27 methylation marks are EZH2 and EZH1, which exhibit full activity only when complexed with the proteins EED, Suz12, RbBP4 and other components required to form the PRC2 complex (Margueron & Reinberg, 2011). EZH2 overexpression is implicated in several human cancers including both leukemia and solid tumors (Chi et al. 2010; Shih et al. 2012). Especially, somatic mutations of a specific tyrosine (Tyr641), which result in altered catalytic efficiency, were reported to be associated with follicular lymphoma and diffuse large B-cell lymphoma (Shih et al. 2012), indicating that pharmacological inhibition of EZH2 activity may provide a promising treatment for EZH2-associated cancer. An initial effort aimed toward identification of HDAC inhibitors found that SAH hydrolase inhibitor 3-deazaneplanocin A (DZNep, Fig. 2a) could also function as an EZH2 inhibitor through depletion of the cellular levels of PRC2 components (Tan et al. 2007). Inhibition of SAH hydrolase resulted in the cellular accumulation of SAH, which in turn caused by-product inhibition of SAM-dependent histone KMTs (Chiang & Cantoni, 1979). However, as a SAH hydrolase inhibitor, DZNep could also non-specifically inhibit other KMTs (Miranda et al. 2009), indicating that DZNep was a selective but non-specific EZH2 inhibitor.
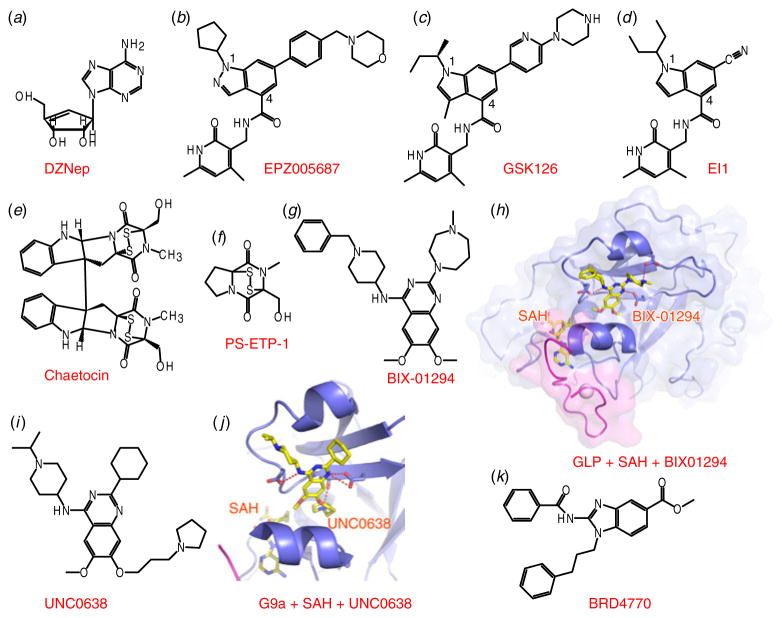
Fig. 2.
Inhibitors of H3K27 and H3K9 methyltransferases. (a, b, c, d) Chemical formulas of inhibitors DZNep (panel a), EPZ005687 (panel b), GSK126 (panel c) and EI1 (panel d) that target EZH2. (e, f, g) Chemical formulas of Chaetocin (panel e), PS-ETP-1 (panel f) and BX-01294 (panel g). (h) Structure of the complex of GLP with bound SAH and inhibitor BIX-01294 (PDB: 3FPD). SAH and BIX-01294 are presented in stick representations. Pre-SET and SET domains are colored in blue, while Post-SET is colored in purple. (i) Chemical formula of UNC0638. (j) Structure of the complex of G9a with bound SAH and inhibitor UNC0638 (PDB: 3RJW). Color representations are the same as in panel (h). (k) Chemical formula of BRD4770.
Recently, three groups reported on the development of a new generation of potent EZH2 specific inhibitors through a combination of high throughput screening and chemistry-based optimization. All three inhibitors exhibited high potency against EZH2-containing PRC2 (EPZ005687 has an IC50 of 54 nM, GSK126 has an IC50 of 9.9 nM and EI1 showed an IC50 of 15 nM; chemical formulas in Figs 2b–2d), while not inhibiting other tested KMTs, indicative of enhanced selectivity (Knutson et al. 2012; McCabe et al. 2012; Qi et al. 2012). Additionally, the inhibition was 50- to 150-fold less potent for closely related EZH1-containing PRC2. The chemical composition of these three compounds are very similar, as reflected in the central indazole group in EPZ005687 and the indole rings in GSK126 and EI1, the invariant pyridone-containing group attached to the 4 position of the indole/indazole rings, as well as the bulky lipophilic groups at the N1 position of the indole/indazole scaffolds. Although structural information is not yet available for the catalytic domain of EZH2, homologous modeling studies indicate that the indole rings could occupy the sugar ring position of SAM, while the pyridone-containing group could occupy the adenine ring position of SAM (McCabe et al. 2012). In addition, it has been proposed that four of the six residue differences between EZH2 and EZH1 within the post-SET domain could account for the observed selectivity, although this claim remains to be validated following eventual structure determination of these key enzymes. All three compounds could decrease global H3K27me3 levels and also effectively inhibit the proliferation of cell lines that harbor EZH2 mutations, indicative of the potential clinical usage of EZH2 inhibitors in cancer therapy. Clearly, the structure of EZH2 and its interacting components would provide a critical platform for future inhibitor design, followed by clinical testing and implementation of epigenetic drugs in cancer treatment.
2.3 Inhibitors of H3K9 methyltransferases
Similar to H3K27 methylation, histone H3K9 methylation is also closely related to repressive chromatin states. The K9me3 and K9me2 marks on H3 serve as docking sites for HP1 (heterochromatin protein 1), a process critical for heterochromatin formation (Grewal & Jia, 2007). Methylation of H3K9 in humans is dependent on the KMTs SUV39H1, SUV39H2, G9a, GLP, SETDB1, SETDB2 and RIZ1 (Krauss, 2008). Most of these H3K9 methyltransferases are reported to be related to various kinds of diseases, with G9a showing increased expression in lung cancer cell lines (Chen et al. 2010), and SUV39H1 associated with colon cancer (Kang et al. 2007). Although it has been shown that the KMT product SAH and the natural product analog Sinefungin could serve as inhibitors through SAM-competitive inhibition (Couture et al. 2006), these natural products lack selectivity. Chaetocin was also shown to efficiently inhibit SUV39H1 (IC50=0.8 μM, Fig. 2e) in a SAM-competitive manner (Greiner et al. 2005). However, as it also inhibited G9a, several other methyltransferases, and some functionally unrelated proteins, further research indicated that Chaetocin may inhibit histone KMTs in a non-specific manner through chemical modification of enzymes mediated by the disulfide groups of this inhibitor (Cherblanc et al. 2013). A simplified Chaetocin derivative PS-ETP-1 (Fig. 2f) was shown to exhibit a comparable potency against G9a (IC50=5.2 μM) with significantly reduced cytotoxicity (Fujishiro et al. 2013), which may help to resolve this controversy. In addition, structural information will be required as part of an effort to resolve this ambiguity.
An extensive screening targeted to G9a led to the discovery of the first potent and selective histone KMT inhibitor, BIX-01294 (Kubicek et al. 2007). The drug BIX-01294 is a diazepin– quinazolin–amine derivative (Fig. 2g), which specifically inhibited G9a (IC50=1.7 μM) and the closely related GLP enzyme. The structure of BIX-01294 bound to the SET domain of GLP established that this inhibitor occupied a region of the histone substrate-binding groove that overlapped with the region bound by the K4 to R8 segment of histone H3 (Fig. 2h) (Chang et al. 2009), indicative of a substrate-competitive inhibition mechanism. However, BIX-01294 exhibited cytotoxicity at concentrations higher than 4.1 μM, which limited its usage in cell-based assays. Subsequent structure-guided chemical optimization resulted in the development of two BIX-01294 derivatives, named E72 (Chang et al. 2010) and UNC0321 (Liu et al. 2010), which exhibited improved inhibitory potency through addition of a 7-alkoxyamine tethered to the quinazoline core so as to mimic the K9 side chain of H3 substrate. Especially, UNC0321 had a Morrision Ki of 63 pM, which was 250-fold more potent than BIX-01294. However, UNC0321 was less potent in cellular assays, possibly due to poor cell membrane permeability. Further optimization aimed at increasing lipophilicity led to the discovery of UNC0638 (Fig. 2i), which was also a substrate-competitive inhibitor, but displayed superior selectivity, as well as improved cellular potency, together with low toxicity (Vedadi et al. 2011). The crystal structure of the G9a-UNC0638-SAH complex established that the core segment of UNC0638 occupied the histone peptide-binding channel, while the 7-(3-pyrrolidin-1-yl)-propoxy side chain was inserted into the lysine-binding pocket (Fig. 2j), thereby explaining the improved potency over BIX-01294 (Vedadi et al. 2011). Recently, a SAM mimic inhibitor BRD4770 (Fig. 2k) was synthesized and tested for efficacy against G9a (Yuan et al. 2012). Although BRD4770 exhibited lower binding affinity (EC50=~5 μM), it displayed no apparent cellular toxicity, indicative of its potential usage to decipher the role of G9a in cancer cell biology.
2.4 Inhibitors of H3K36 methyltransferases
Histone H3K36 methylation is a well-established histone mark primarily associated with transcriptional activation, although it has also been implicated in some instances in transcriptional repression. H3K36 methylation has been identified in processes such as alternative splicing, dosage compensation, as well as DNA repair and recombination (Wagner & Carpenter, 2012). The well-established enzymes that catalyze H3K36 methylation include the NSD family of methyltransferases (NSD1, NSD2 and NSD3), SETD2, ASH1L and SMYD2 (Wagner & Carpenter, 2012). Many of these enzymes are also closely correlated with human cancer, with the SMYD2 gene frequently amplified in esophageal squamous cell carcinoma and other solid tumors (Komatsu et al. 2009; Rooney et al. 1999), whereas defects in SETD2 are correlated with sporadic clear renal cell carcinoma (Wagner & Carpenter, 2012).
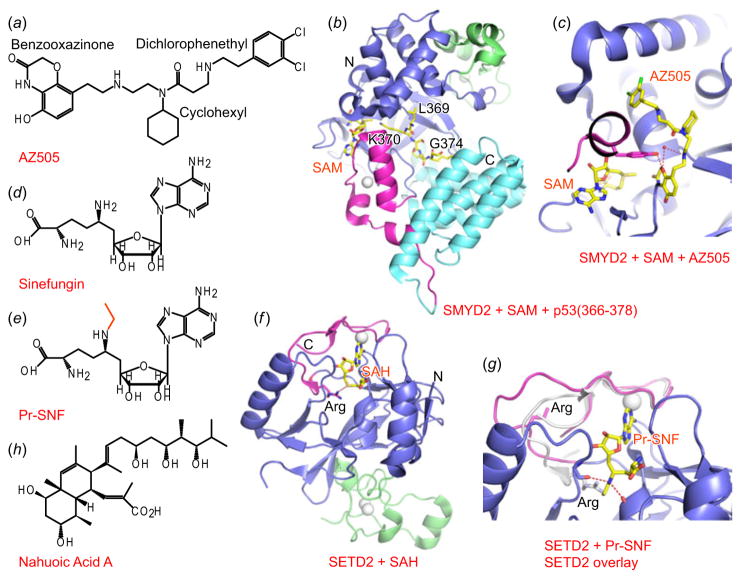
Compared with other members of this family of methyltransferases, SMYD2 may not be a bona fide H3K36 methyltransferase, given that it also targets non-histone substrates, such as p53 and RB (retinoblastoma) proteins. Through an application of AlphaScreen technology (Eglen et al. 2008), Ferguson and colleagues identified a small molecule compound AZ505 (Fig. 3a) as a SMYD2-specific inhibitor (Ferguson et al. 2011). AZ505 has an IC50 of 0.12 μM against SMYD2, while showing much weaker activity against other histone KMTs. Structures of complexes of SMYD2 with bound p53 (366–374) peptide (Fig. 3b) or AZ505 (Fig. 3c) revealed that AZ505 occupied the p53-binding pocket within the concaved cleft of SMYD2, which is formed by the SET domain together with imbedded MYND domain, post-SET and C-terminal lobe of the enzyme (Ferguson et al. 2011), suggestive of AZ505 being a peptide-competitive inhibitor. The benzooxazinone group of AZ505 was positioned within the lysine-binding channel of the substrate, where it formed extensive electrostatic and hydrophobic interactions with the donor methyl group of SAM and residues from SMYD2, consistent with the observation that AZ505 binding was dependent on bound SAM. The cyclohexyl group of AZ505 was positioned in a hydrophobic pocket normally occupied by Leu369 of p53 peptide, whereas the di-chlorophenethyl moiety extended across the peptide-binding groove of SMYD2 to reach another hydrophobic pocket adjacent to the cofactor-binding site, thereby most likely accounting for the observed selectivity. AZ505 binding was mainly mediated by hydrophobic interactions, as shown by limited formation of hydrogen bonds. Given that the inhibitory effect of AZ505 required the presence of SAM, its potency in cellular-based assays needs to be tested.
Fig. 3.
Inhibitors of H3K36 and H4K20 methyltransferase. (a) Chemical formula of AZ505. (b) Structure of complex of SMYD2 with bound SAM and peptide substrate p53 (366–378) (PDB: 3S7F). Pre-SET and SET domain are colored in blue. The imbedded MYDN domain is colored in green, the Post-SET is colored in purple, whereas the C-terminal domain is colored in cyan. Bound SAM and peptide are shown in stick representations. (c) A blow-up view of the structure of the complex of SMYD2 with bound SAM and AZ505 (PDB: 3S7B). Color schemes are the same as in panel (b). (d, e) Chemical formulas of Sinefungin (panel d) and Pr-SNF (panel e). (f) Structure of complex of SETD2 with bound SAH (PDB: 4H12). N-terminal region and the SET domain are colored in blue; the imbedded AWS domain is colored in green, while the Post-SET is colored in purple. Bound zinc ions are shown as white spheres. SAH is shown a stick representation. (g) A blow-up view of the structure of the complex of SETD2 with bound Pr-SNF (PDB: 4FMU), with the overlapped post-SET loop shown in panel f, colored in silver. Note the conformational change of the post-SET loop in the two structures, and the movement of the Arg within the loop.
In an attempt to identify SAM-based inhibitors through modification of the known SAM analog Sinefungin (Fig. 3d), the Luo laboratory synthesized several N-alkyl sinefungin derivatives and identified N-propyl-modified sinefungin (Pr-SNF, Fig. 3e), which displayed 2- to 200-fold preference for SETD2 relative to other KMTs (Zheng et al. 2012). In addition, Pr-SNF also showed more than 10-fold potency against SETD2 compared with unmodified Sinefungin. A structure-activity relationship study revealed that Pr-SNF occupied the same binding pocket as SAH (Fig. 3f), with the propyl group of Pr-SNF partially extending into the lysine-binding pocket to induce a conformational change of the post-SET loop (Fig. 3g), which otherwise adopted an auto-inhibitory conformation with an Arg from the post-SET loop forming a hydrogen bond with the cofactor SAH (Fig. 3f). In this substrate accessible conformation, the secondary amine of Pr-SNF is ideally positioned to form two hydrogen bonds with the backbone carbonyl oxygens from SETD2 (Fig. 3g), thereby potentially accounting for the observed selectivity. This approach has identified a promising route for inhibitor design through optimization of previously identified pan-inhibitors.
2.5 Inhibitors for H4K20 methyltransferases
Histone H4K20 methylation is the only well-known methylation mark on H4, with SETD8, Suv4-20h1 and Suv4-20h2 representing major KMTs responsible for methylation of this site (Beck et al. 2012). H4K20 methylation is related to biological processes ranging from DNA damage response, to mitotic condensation, DNA replication and gene regulation. Generally speaking, H4K20 methylation is considered as a mark for transcriptional repression. Recently, the first selective SAM-competitive inhibitor of SETD8, Nahuoic acid A (Fig. 3h), was identified through a screen using a library of marine organism extracts and pure marine natural products (Williams et al. 2013). Nahuoic acid A specifically inhibited SETD8 (IC50=6.5 μM) but not other tested KMTs. Although cellular based analysis has not yet been reported for Nahuoic acid A, it has been anticipated that this first natural product inhibitor of an H4K20 methyltransfrase would encourage follow-up studies directed toward this family of KMTs.
3. Small molecule inhibitors of histone lysine demethylases
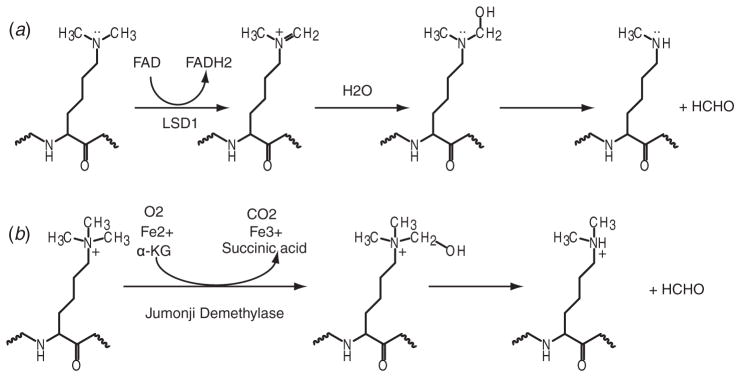
Histone lysine methylation used to be considered as a permanent modification, until the seminal discovery of the first lysine-specific demethylase 1 (LSD1) (Shi et al. 2004). LSD1, and its closely related family member LSD2, are the only KDMs discovered to date that remove methyl groups from methylated-lysines through an flavin adenine dinucleotide (FAD dependent oxidation mechanism. LSD1 and LSD2 cannot remove the methyl group from trimethylated-lysines, since the demethylation mechanism requires a protonated amine (shown schematically in Fig. 4a). In a subsequent groudbreaking discovery, the large Jumonji C-domain-containing family of enzymes were also shown to be histone KDMs (Tsukada et al. 2006), which catalyze the reaction through a hydroxylation pathway using Fe2+ and α-ketoglutarate (α-KG) as cofactors (shown schematically in Fig. 4b). Since they do not require the existence of a lone pair of electrons on the nitrogen atom of the methylated-lysines, the Jumonji family of KDMs can also demethylate trimethylated-lysines.
Fig. 4.
Illustration of two identified mechanisms for lysine demethylation. (a) LSD1-mediated lysine demethylation mechanism. (b) Jumonji demethylase family-mediated lysine demethylation mechanism.
3.1 Inhibitors of H3K4 demethylases
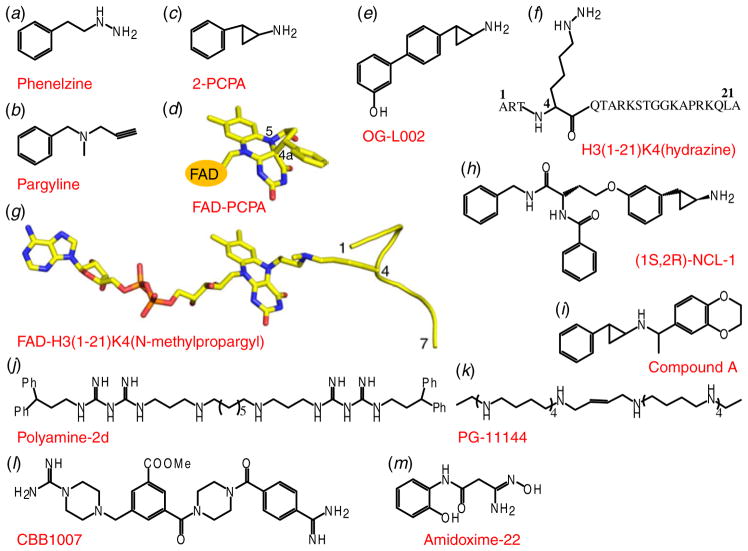
Histone H3K4 demethylases include LSD1, LSD2 and the JARID1 family of proteins, with most of these enzymes impacting on cancer. LSD1 is aberrantly expressed in breast and prostate cancers (Chi et al. 2010), making it an attractive drug target candidate. As the first known histone KDM, extensive efforts have been made using different approaches toward LSD1 inhibitor design (Suzuki & Miyata, 2011). The first generation of LSD1 inhibitors that were discovered was based on its homology with monoamine oxidases (MAOases), which also use FAD as the cofactor. Known MAOase inhibitors such as phenelzine, pargyline and trans-2-phenylcyclopropylamine (PCPA, IC50=243 μM) (chemical formulas in Figs 5a–5c) were tested and found to possess inhibitory activity toward LSD1 (Lee et al. 2006; Metzger et al. 2005; Schmidt & McCafferty, 2007). These inhibitors possibly impact on the demethylation activity by forming a covalent adduct with the FAD, as shown in the structure of LSD1 with bound PCPA (Yang et al. 2007b), wherein the cyclopropyl ring of PCPA adducted with FAD to form a five-membered ring through covalent bonding with the N(5) and C(4a) atoms (Fig. 5d). In turn, structure-guided generation of PCPA derivatives showed improved potency (Benelkebir et al. 2011; Binda et al. 2010; Gooden et al. 2008; Mimasu et al. 2010), but most of them have relatively poor selectivity, as they also inhibit MAOases. Recently, a novel PCPA derivative named OG-L002 (Fig. 5e) has been identified to be a highly specific LSD1 inhibitor (IC50≈0.02 μM), showing a >36-fold selectivity over MAOases (Liang et al. 2013). In search of more selective inhibitors, a series of peptide-based inhibitors, such as compounds with the propargylamine or hydrazine functionality were synthesized in the context of a histone H3(1–21) peptide (Culhane et al. 2006, 2010; Dancy et al. 2012; Szewczuk et al. 2007; Yang et al. 2007a). These peptide analogs are more selective and potent than the MAOase inhibitors, with hydrazine-H3 (Fig. 5f) exhibiting the most potent LSD1 inhibitory activity (Ki=4.35 nM). A structural characterization revealed that the N-methylpropargyl- K4-modified H3 peptide formed a stable covalent complex with the cofactor FAD through the N(5) position (Fig. 5g), which provided a structural basis toward understanding the peptide-based H3K4 demethylation mechanism by LSD1 (Yang et al. 2007a). However, poor membrane permeability limited the usage of these peptide-based inhibitors in cellular assays. In search of a selective LSD1 inhibitor suitable for in vivo assays, a PCPA-lysine hybrid inhibitor NCL-1 was synthesized (Ueda et al. 2009). NCL-1 (IC50=2.5 μM) exhibited selectivity for LSD1 over MAOases of 400 to 11 000 times higher than that of PCPA, with the optically active (1S, 2R)-NCL-1 (Fig. 5h) exhibiting 4-fold more potency than its (1R, 2S)-NCL-1 stereoisomeric counterpart (Ogasawara et al. 2011). Several N-alkylated PCPA analogs, such as compound A (Fig. 5i), which exhibit high selectivity and potency (Ki=5 nM) have been patented (Suzuki & Miyata, 2011), but a detailed description of their mode of action remains to be been disclosed.
Fig. 5.
Inhibitors of LSD1. (a–c) Chemical formulas of inhibitors that target LSD1. Pheneizine is shown in panel (a), Pargyline is shown in panel (b) and 2-PCPA is shown in panel (c). (d) Structural illustration of the FAD–PCPA adduct (PDB: 2UXX). (e, f) Chemical formulas of inhibitors that target LSD1. OG-L002 is shown in panel (e) and H3(1–21)K4(hydrazine) is shown in panel (f). (g) Structural illustration of the FAD-H3(1–21)K4(N-methylpropargyl) adduct (PDB: 2UXN). (h–m) Chemical formulas of inhibitors that target LSD1. (1S, 2R)-NCL-1 is shown in panel (h), Compound A is shown in panel (i), Polyamine 2d is shown in panel (j), PG-11144 is shown in panel (k), CBB1007 is shown in panel (l) and Amidoxine-22 is shown in panel (m).
Besides the above-mentioned family of MAOases, LSD1 was found to be homologous with two polyamine oxidases, SMO (spermine oxidase) and APAO (N1-acetylpolyamine oxidase). Subsequently, polyamine-based inhibitors were designed and proved to be potent and selective LSD1 inhibitors (Huang et al. 2007, 2009; Sharma et al. 2010). The most potent inhibitor, polyamine- 2d (Fig. 5j, IC50≈1 μM), exhibited noncompetitive inhibition kinetics, whereas another compound, PG-11144 (Fig. 5k, IC50=5 μM), functions through a substrate-competitive inhibition mechanism, with both these compounds producing an increase in H3K4me2 levels. Importantly, both compounds when combined with DNMT inhibitor 5-Aza, exhibited marked increase in the inhibition of tumor growth in a human colon cancer tumor model (Huang et al. 2009). These compounds represent a new branch of epigenetic modulators targeted to LSD1 with the potential for usage as antitumor agents.
On the basis of the structural features of the active site of LSD1, several inhibitors with a non-peptide chemical scaffold were also designed, with the most potent compound CBB1007 (Fig. 5l), displaying an IC50 of 5.27 μM (Wang et al. 2011). These compounds selectively inhibited LSD1 but not LSD2 and JARID1A in vitro, and also inhibited the proliferation of pluripotent cancer cells. These compounds may function in a similar manner as the polyamine-derivatives, as they are structurally related to (bis)guanidine and (bis)urea analogs (Huang et al. 2007; Sharma et al. 2010).
A series of small molecule amidoximes were recently reported as LSD1 inhibitors discovered through a virtual screening strategy followed by chemical synthesis and optimization (Hazeldine et al. 2012). The best compound, Amidoxime-22 (Fig. 5m), despite exhibiting moderate LSD1 inhibitory activity in vitro (IC50=16.8 μM), promoted dramatic increases in H3K4me2 levels in Calu-6 lung carcinoma cells. In addition, Amidoxime-22 and its derivatives are more drug-like (Lipinski et al. 2001) and can be easily synthesized, representing a promising scaffold for further optimization.
3.2 Inhibitors of H3K9 demethylases
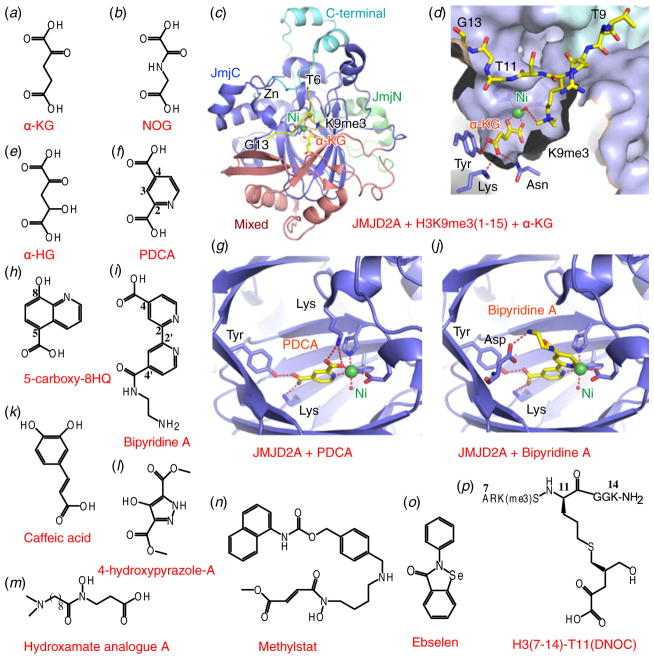
Histone H3K9 demethylases include LSD1, the JMJD2 subfamily of enzymes and JHDM2A (Shi, 2007). Since all members of the Jumonji family of demethylases use Fe2+ and α-KG (Fig. 6a) as cofactors to catalyze the demethylation process, the α-KG analog N-oxalylglycine (NOG, Fig. 6b) has generally been used as an inhibitor for in vitro studies (Cloos et al. 2006). The structure of the catalytic domain of JMJD2A with bound cofactor α-KG (or NOG), in the presence of inactive Ni2+ cation (as a replacement for Fe2+) and substrate H3 (1–15)K9me3 peptide has provided definitive insights into the demethylation mechanism (Couture et al. 2007; Ng et al. 2007). In the complex, the catalytic JmjC domain adopts a conserved β-barrel fold associated with the cupin superfamily, surrounded by the JmjN domain, the connecting mixed domain from the N-terminal segment and a stabilizing C-terminal segment (Fig. 6c). The α-KG interacts with amino acids from the active site from one side and with the Ni2+ cation in a bidentate manner from another side, with the Ni2+ cation hexa-coordinated through additional interactions with the His–His–Glu triad and a water molecule (Fig. 6d).
Fig. 6.
Inhibitors for H3K9 demethylases. (a, b) Chemical formulas of cofactor/inhibitors targeted to H3K9 demethylases. α-KG is shown in panel (a) and NOG is shown in panel (b). (c) Structure of complex of JMJD2A with bound α-KG, Ni2+ cation and its substrate peptide H3(1–15)K9me3 (PDB: 2Q8C). JmjN domain is colored in green, the mixed region is colored in salmon, JmjC domain is colored in blue and the C-terminal region is colored in cyan. Ni2+ cation is colored in green. (d) A blow-up view of the active site shown in panel (c), with the histone peptide shown in a stick representation and JMJD2A shown in a surface view. (e–i) Chemical formulas of cofactor/inhibitors targeted to H3K9 demethylases. α-HG is shown in panel (e), PDCA is shown in panel (f), 5-carboxy-8HQ is shown in panel (h), and Bipyridine A is shown in panle i. (g, j) Blow-up views of the catalytic sites of JMJD2A with bound inhibitor PDCA (panel g) (PDB: 2VD7) and Bipyridine A (panel j) (PDB: 2PDQ). Color schemes are the same as in panel (c). (k–p) Chemical formulas of cofactor/inhibitors targeted to H3K9 demethylases. Caffeic acid is shown in panel (k), 4-hydroxypyrazole-A is shown in panel (l), Hydroxamate analog A is shown in panel (m), Methylstat is shown in panel (n), Ebselen is shown in panel (o) and H3(7–14)-T11(DNOC) is shown in panel (p).
In this regard, α-KG analogs were extensively investigated as cofactor-competitive inhibitors. Besides NOG, another α-KG analog, the oncometabolite α-hydroxyglutarate (α-HG, Fig. 6e) was found to inhibit JMJD2A, JMJD2C and JHDM1A with IC50 values ranging from 24 μM to 106 μM (Chowdhury et al. 2011). To enhance the selectivity, a range of NOG derivatives were synthesized (Hamada et al. 2009; Rose et al. 2008, 2010), most of which showed limited selectivity. A screen using known inhibitors from other Fe2+/α-KG dependent oxygenases identified 2,4-pyridinedicarboxylic acid (PDCA, Fig. 6f), which showed potent inhibitory activity on JMJD2E (IC50=1.4 μM) (Rose et al. 2008). The structure of JMJD2A with bound PDCA verified that PDCA functions in an α-KG competitive manner. PDCA binds the Ni2+ cation in a bidentate manner via its N-atom and 2-carboxylate, whereas the 4-carboxylate mimics α-KG binding by forming two hydrogen bonds with a lysine and a tyrosine in the active site (Fig. 6g). Structural information showed that the C-3 position of PDCA was compatible for further modification, which led to the identification of C-3-substituted PDCA with enhanced selectivity for JMJD2E over PHD2 (Thalhammer et al. 2011). High-throughput screening also identified PDCA-related compound 5-carboxy-8-hydroxyquinoline (5-Carboxy-8HQ, Fig. 6h), which has a greater potency for JMJD2E (IC50=200 nM) (King et al. 2010). Another family of pyridine-based compounds, with a 4-carboxy-2,2′-bipyridyl scaffold (abbreviated here as Bipyridine A, Fig. 6i), have also been reported to show enhanced inhibitory activity toward JMJD2 subfamily of demethylases (Chang et al. 2011). Structural characterization validated that this bipyridine compound also functions through α-KG competition. The carboxylate group of the compound is positioned to interact with the same Tyr and Lys in a manner analogous to that observed for α-KG and PDCA, whereas the active site Ni2+ cation was chelated through both pyridinyl nitrogen atoms (Fig. 6j). This bipyridine scaffold may represent an important element for active site metal chelation, given that it has also been shown to be a key scaffold in the JMJD3 complex with bound inhibitor GSK-J1 (to be discussed below) (Kruidenier et al. 2012).
More recently, screening efforts using a library of pharmacologically active compounds or natural products led to the identification of hydroxamic acids and catechols as JMJD2 demethylase inhibitors (Nielsen et al. 2012; Sakurai et al. 2010). One such compound, caffeic acid (Fig. 6k), could inhibit JMJD2C with an IC50 of 13.7 μM. However, these compounds are more promiscuous as they also inhibit HDACs and DNMTs. A 4-hydroxypyrazole scaffold-containing compound named 4-hydroxypyrazole-A (Fig. 6l) was also reported as an inhibitor of JMJD2C, with the best hit exhibiting an IC50 of 147 μM. Based on the crystal structure of JMJD2A and a homology model of JMJD2C, Hamada and colleagues designed a series of hydroxamate analogs bearing a tertiary amine and identified one such analog, named Hydroxamate analogue A (Fig. 6m), to be a potent and selective JMJD2 inhibitor, showing 500-fold greater JMJD2C inhibitory activity over NOG (Hamada et al. 2010). They also identified prodrugs of this analog, which showed synergistic growth inhibition of cancer cells in combination with a LSD1 inhibitor. With the idea of combining the mimicry of both lysine and α-KG based on available structural information of the active site, Luo et al. synthesized a novel JMJD-selective inhibitor, which could selectively inhibit the JMJD2 family of KDMs (Luo et al. 2011). In addition, its prodrug methylstat (Fig. 6n) selectively inhibited the Jumonji family of demethylases in cells and could inhibit cell growth of the esophageal carcinoma cell line KYSE150 (GI50=5.1 μM). Both methylstat and the hydroxamate analog showed cancer cell inhibitory activity, indicative of clinical promise in anticancer treatment.
In an additional interesting finding, Sekirnik and colleagues showed that Zn-ejecting compounds including disulfiram and ebselen (Fig. 6o, IC50=10.6 μM) could also inhibit JMJD2A by removing the zinc ion from the enzyme (Sekirnik et al. 2009). As zinc finger motifs are a ubiquitous scaffold among many enzymes including several members of the Jumonji KDM family, Zn ejecting compounds should be used with caution for inhibitor design.
Most of above ligand-based inhibitors showed limited selectivity between subfamilies/isoforms of these Jumonji demethylases. To enhance selectivity, a peptidomimetic approach facilitated by structural information of the catalytic site (Fig. 6d) was implemented for inhibitor design (Lohse et al. 2011; Woon et al. 2012), similar to that used for the design of peptide-based LSD1 inhibitors (Szewczuk et al. 2007). Lohse and colleagues combined a uracil-based group for metal chelation and a minimum H3 peptide required for recognition of H3K9 demethylase to obtain a 5-mer inhibitor, which although showing low potency, exhibited a 4-fold selectivity for JMJD2C (Ki=27 μM) over JMJD2A (Lohse et al. 2011). Woon and colleagues examined the structure of JMJD2A with a bound H3K9me3-containing peptide (Fig. 6d), and designed several peptide-based inhibitors by linking the α-KG analog with the peptide through the adjacent position occupied by T11. Subsequently, histone peptide H3(7–14)K9me3 and DNOC (N-oxalyl-D-cysteine), a NOG analog, were linked at peptide position 11 to yield an inhibitor (Fig. 6p) that could potently inhibit both JMJD2E (IC50 of 90 nM) and JMJD2A (IC50 of 270 nM), with little or no inhibition against other Jumonji subfamily members (Woon et al. 2012). They also proved that a change in peptide substrate could enable these inhibitors to discriminate between closely related Jumonji subfamily members. The combination of both cofactor and substrate structural information should provide a promising avenue for future inhibitor design with enhanced selectivity and potency toward this subfamily of Jumonji KDMs.
3.3 Inhibitors of H3K27 demethylases
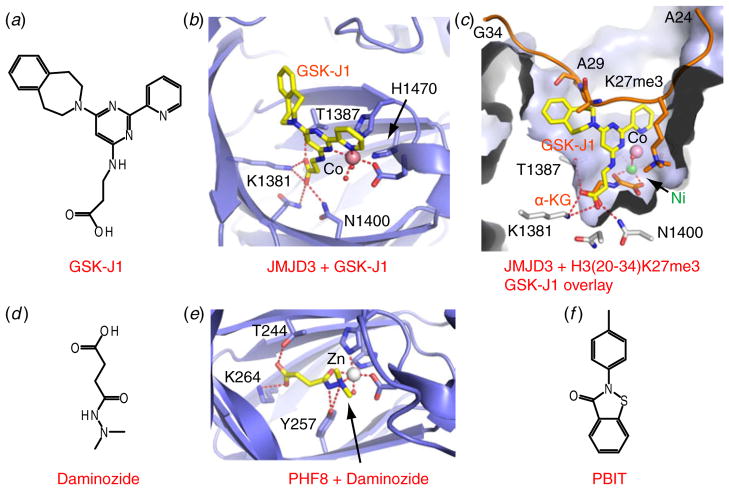
UTX and JMJD3 are H3K27-specific demethylases that function in opposition to EZH2 in regulating H3K27 methylation levels, with both KDMs possessing tumor-suppressive characteristics (Pedersen & Helin, 2010). UTX is a frequent target of somatic mutations in human cancers, whereas JMJD3 has been demonstrated to activate the INK4A-ARF locus (Pedersen & Helin, 2010). An effort championed by the GlaxoSmithKline (GSK) epigenetics group and participation by our group solved crystal structures of JMJD3 with Ni2+ cation, NOG, both in the absence and presence of bound H3K27me3 peptide, as well as potent inhibitors (Kruidenier et al. 2012). An extensive screening approach with subsequent optimization identified the lead compound, GSK-J1 (chemical formula in Fig. 7a), which exhibited an IC50 of 60 nM for JMJD3. In the structure of JMJD3 bound to GSK-J1, the propanoic acid of GSK-J1 mimics α-KG by forming hydrogen bonds with residues within the catalytic site (Figs 7b and 7c), whereas the pyridyl-pyrimidine biaryl ring system makes a bidentate interaction with the catalytic metal (Co2+ was used to mimic Fe2+), which induced a 2.34 Å shift of the cation from its original position. Such cation movement resulted in a disruption of the conserved chelating bond between the cation and conserved His1470 (Fig. 7b). This distinctive dynamic metal shift was unexpected, and introduced a new concept associated with mediating inhibition. Overlaying the structures of inhibitor and peptide bound JMJD3 complexes established that the tetrahydrobenzazepine ring system of GSK-J1 clashes with the Ala29-Pro30 segment of the bound histone peptide (Fig. 7c), indicating that GSK-J1 may also perturb substrate binding. In addition, the prodrug counterpart, GSK-J4, could efficiently reduce lipopolysaccharide induced proinflammatory cytokine production by human primary macrophages (Kruidenier et al. 2012). Such a structure-guided small-molecule and chemoproteomics approach holds promise for the design of inhibitors targeted to other Jumonji family KMTs. Since JMJD3 and UTX function in different pathways, further optimization could facilitate identification of additional inhibitors with the potential for selectively discriminating between these two Jumonji KDM family members.
Fig. 7.
Inhibitors of H3K27, H3K36/H4K20 and H3K4 demethylases. (a) Chemical formulas of inhibitor GSK-J1. (b) A blow-up view of the catalytic site of JMJD3 with bound GSK-J1 (PDB: 4ASK). Co2+ cation is colored in pink and the JmjC domain is colored in blue. Bound GSK-J1 is shown in a stick representation. (c) Structure of complex of JMJD3 with bound H3(20–34)K27me3 substrate (PDB: 4EZH). Bound histone peptide and α-KG are colored in orange and Ni2+ cation is colored in green. Overlayed GSK-J1 and Co2+ are colored as shown in panel (b). (d) Chemical formula of Daminozide. (e) A blow-up view of the catalytic site of PHF8 with bound Daminozide (PDB: 4DO0). Zn2+ is colored in grey. Daminozide is shown in a stick representation. (f) Chemical formula of PBIT.
3.4 Inhibitors of other histone lysine demethylases
To date, there have been no reports of an H3K79 demethylase, and hence the principles underlying removal of this methylation mark remains a mystery. H3K36-specific demethylases include JMJD2 family members JMJD2A, JMJD2B and JMJD2C, which also possess activity for H3K9 demethylation. Their specific inhibitors have been discussed above in the context of H3K9 KDMs. JHDM1A and JHMD1B (FBXL10/KDM2) are also H3K36 histone KDMs. PHF8 (KDM7) and PHF2 have recently been identified as H4K20-specific demethylases, with aberrant PHF8 closely correlated with prostate cancer (Crea et al. 2012).
Recently, plant growth regulator Daminozide (Fig. 7d) was found to be a selective α-KG competitive inhibitor against JHMD1B and PHF8 (IC50=2 μM for both), exhibiting >60-fold selectivity over other families of demethylases tested (Rose et al. 2012). The structure of PHF8 with bound Daminozide revealed the general mode of inhibition, whereby Daminozide occupied the α-KG binding pocket and recognized the catalytic metal through its acylhydrazide carbonyl and dimethylamino groups (Fig. 7e). Importantly, both the nitrogen atoms of Daminozide are within the hydrogen bond distance of the hydroxyl group of Tyr257 within the catalytic site. This tyrosine is not present in the catalytic sites of JMJD2A and JMJD3, which may account for the observed selectivity. Although Daminozide itself is not suitable for medicinal use due to its potential human toxicity, its biological impact on animal and other organisms would be of interest and worth further investigation given the high degree of selectivity.
The JARID1 family of proteins belongs to the Jumonji family of H3K4 demethylases. Progress toward development of potent inhibitors for this family of Jumonji KDMs has lagged behind, most likely due to a lack of structural information. Recently, the Yan laboratory identified several novel selective inhibitors of JARID1B, amongst which N-phenyl-benzisothiazolinone (PBIT, Fig. 7f) exhibited an IC50 of 3 μM (Sayegh et al. 2013). PBIT inhibited several JARID1 subfamily members but not JMJD3 and UTX. Given that its analog ebselen (Fig. 6o), a zinc-rejecting compound (Sekirnik et al. 2009), also inhibited JARID1B (contains several zincfingers) with an IC50 of 6 μM, additional research is needed to elucidate the inhibitory mechanism of PBIT.
4. Inhibitors that target non-catalytic sites
All the inhibitors mentioned above target catalytic active sites of KMTs and KDMs through either cofactor-competitive or substrate-competitive mechanisms or both, while allosteric inhibitors that target non-catalytic sites have not been reported for histone KMTs and KDMs. Allosteric inhibitors could potentially trap alternate states through binding to sites distal from the active site, which could constitute a potentially powerful approach toward regulating these histone-modifying enzymes (Hardy & Wells, 2004). There is however available structure-based information on allosteric inhibition of other epigenetic enzymes that is discussed below.
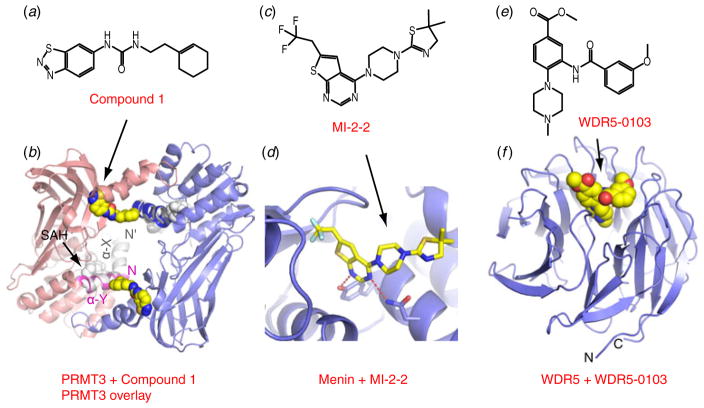
A case in point is the discovery of an allosteric inhibitor for PRMT3 (Siarheyeva et al. 2012), a protein arginine methyltransferase (RMT) that catalyzes arginine methylation using SAM as the cofactor, for both histone and non-histone proteins. A screen of 16 000 drug-like compounds led to the discovery and identification of Compound 1 (Fig. 8a), a potent PRMT3 inhibitor. This small molecule inhibited full-length PRMT3 with an IC50 of 1.6 μM, while showing either weak or no inhibition for several other KMTs and RMTs tested. The structure of PRMT3 bound to Compound 1 revealed that this small molecule bound within a newly created pocket positioned at the dimeric interface that is distal from both the SAM and substrate binding pockets (Fig. 8b). Complex formation induced conformational side-chain rearrangements at the junction of the α-Y and α-X helices of the dimeric pair, which in turn destabilized the α-X segment of the activation helix. The α-X helix, which is critical for the catalytic activity of PRMT3, was disordered in this complex, accounting for the loss of both cofactor binding and activity. This example demonstrates the potential of allosteric inhibitors for targeting epigenetic enzymes, and highlights a potential alternate approach for targeting histone KMTs and KDMs.
Fig. 8.
An allosteric inhibitor of PRMT3 and two inhibitors that target non-catalytic sites of MLL1. (a) Chemical formula of Compound 1. (b) Structure of complex of PRMT3 with bound inhibitor Compound 1 (PDB: 3SMQ). The dimeric pairs are colored in blue and salmon respectively. Bound Compound 1 is shown in a space-filling representation. Overlayed structure of PRMT3 with bound SAH is shown in silver. The related α-X and α-Y helices are labeled, with α-Y, which is close to Compound 1, colored in purple. (c) Chemical formula of MI-2-2. (d) A blow-up view of the structure of the Menin binding pocket with bound MI-2-2 (PDB: 4GQ4). Menin is colored in blue and MI-2-2 is shown in a stick representation. (e) Chemical formula of WDR5-0103. (f) Structure of complex of WDR5 with bound compound WDR5-0103 (PDB: 3UR4). WDR5 is colored in blue. Compound WDR5-0103 is shown in a space-filling representation.
In addition to the promise for targeting allosteric sites within histone KMTs and KDMs, additional potential target sites for inhibitor design include regulatory proteins, as shown by the example of MLL1-binding proteins. MLL1 has been extensively studied due to its close association with human leukemia (Krivtsov & Armstrong, 2007). However, as discussed earlier, the catalytic SET domain of MLL1 does not constitute an optimal drug target. By contrast, the MLL1 N-terminal region, which interacts with Menin, has been shown to be indispensable for leukemogenic transformation (Yokoyama et al. 2005), and could serve as a potential drug target. Systematic screening for inhibitors that target the Menin-MLL1 interaction resulted in the identification of a specific inhibitor named MI-1. Its optimized derivative MI-2-2 (Fig. 8c), displayed excellent potency in disrupting the interaction between MLL1 (4–43) and Menin (IC50=520 nM). High resolution structures of complexes of Menin with bound MI-2-2 or MLL1 (4–15) peptide revealed that both MI-2-2 and MLL1 (4–15) peptide occupied the same binding pocket within Menin (Grembecka et al. 2012; Shi et al. 2012), with MI-2-2 overlapping with the Phe9-Pro13 segment of the MLL1 peptide, indicative a substrate-competitive inhibition mechanism. The bound MI-2-2 is mainly stabilized by hydrophobic interactions, with only a pair of hydrogen bonds formed between N1 and N3 nitrogen atoms of the thienopyrimidine ring and the side chains of a Tyr and an Asn, respectively (Fig. 8d). MI-2-2 displayed specific and pronounced activity on MLL1 leukemia cells, thereby demonstrating proof of concept that chemical inhibitors can be designed to target and inhibit protein-protein interactions.
Similarly, a screen for inhibitors that targeted the interaction between MLL1 WIN-peptide and WDR5 (Patel et al. 2008) identified small molecule WDR5-0101 (Senisterra et al. 2013). Its optimized analog, WDR5-0103 (Fig. 8e), exhibited a peptide-displacement constant Kdis of 3.0 μM. WDR5-0103 bound within the central cavity associated with the arginine-binding pocket of WDR5 mainly through a shape-complementary mechanism (Fig. 8f), thereby displaying direct competition with the peptide substrate. The inhibitory potency of WDR5-0103 on MLL1 core complexes was dependent on substrate concentration, consistent with the stabilization contribution of WDR5 within the MLL1 complex. WDR5-0103 does not inhibit other histone KMTs tested, indicative of good selectivity. However, since these MLL1 binding partners also interact with other regulatory proteins, the side effects of these compounds require further evaluation, due to their potential for competitive targeting of other binding partners. Examples of such side effects include the interaction between Menin and JUND through the same binding pocket within the MLL1 N-terminus (Huang et al. 2012), and the interaction between WDR5 with both histone H3 tails and several other MLL family members through targeting of the same binding pocket within the MLL1 WIN motif (Zhang et al. 2012).
5. Future challenges and prospects
One problem that impedes inhibitor design is the low activity of some of the enzymes, given that quite a few enzymes only show detectable activity in the presence of their associated factors, as shown for MLL1 and EZH2, and reconstitution of the functional complexes would be a critical step for inhibitor screening. Another factor that has not yet been fully appreciated reflects the requirement of the nucleosome as the substrate, as shown for the enzymes DOT1L, EZH2 complex, and NSD2, for which histone peptide-based substrates result in undetectable activity or misleading results. In some cases, modified nucleosomes also exhibit enhanced activity, as in the case of H2B120 ubiqutination, which greatly enhanced DOT1L activity (Nguyen & Zhang, 2011). In such circumstances, designer nucleosomes should be used as substrates (Fierz & Muir, 2012), thereby facilitating the process of inhibitor design at the biologically relevant nucleosomal level.
Recent developments in the successful design of inhibitors that target histone reader modules (Dawson et al. 2012) should draw additional attention to inhibitor design for histone methylated-lysine writers and erasers discussed in this review, given that most histone KMTs and KDMs also possess one or more histone reader modules. Examples include MLL1, which possesses a PHD finger for readout of the H3K4 methylation mark (Wang et al. 2010), and the JMJD2A double Tudor domain, which recognizes both H3K4 and H4K20 methylation marks (Huang et al. 2006; Lee et al. 2008). The histone reader modules of these enzymes are critical for their in vivo enzymatic activity, given that recruitment of these enzymes to the proper loci constitutes an important step toward their in vivo function. Targeting such reader modules within histone KMTs and KDMs offer additional opportunities for inhibitor design.
A major bottleneck in inhibitor design has involved the search for and identification of target-specific hits from within an unlimited choice of candidates. As shown above, the majority of successful studies have made extensive use of structural biology, which although dispensable at the initial hit stage, has been of invaluable assistance in the small molecule optimization stage, and has also been considered as an essential contributor for the fragment-based drug design approach (Murray & Blundell, 2010). Structural biology-based knowledge of enzyme binding pockets has also contributed to in silico approaches to inhibitor design and optimization (Ghosh et al. 2006). We anticipate that the combined improvements in screening technology, medicinal chemistry, structural and computational biology, and cellular-based analysis should greatly expedite the process of pharmacologically potent inhibitor identification and optimization targeted toward these disease-impacted epigenetic modifying enzymes, which in turn should ultimately lead to the application of efficacious and safe drugs beneficial for human health.
Acknowledgments
This research was supported by the Leukemia and Lymphoma Society and the STARR Foundation to D. J. P., as well as the ‘Thousand Young Talents ’ Program of China and start-up funds from Beijing Normal University to Z. W.
Footnotes
7. Competing financial interests
The authors declare no competing financial interests.
References
- Arrowsmith CH, Bountra C, Fish PV, Lee K, Schapira M. Epigenetic protein families : a new frontier for drug discovery. Nature Review Drug Discovery. 2012;11:384–400. doi: 10.1038/nrd3674. [DOI] [PubMed] [Google Scholar]
- Beck DB, Oda H, Shen SS, Reinberg D. PR-Set7 and H4K20me1: at the crossroads of genome integrity, cell cycle, chromosome condensation, and transcription. Genes and Development. 2012;26:325–337. doi: 10.1101/gad.177444.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benelkebir H, Hodgkinson C, Duriez PJ, Hayden AL, Bulleid RA, Crabb SJ, Packham G, Ganesan A. Enantioselective synthesis of tranylcypromine analogues as lysine demethylase (LSD1) inhibitors. Bioorganic and Medical Chemistry. 2011;19:3709–3716. doi: 10.1016/j.bmc.2011.02.017. [DOI] [PubMed] [Google Scholar]
- Bernt KM, Zhu N, Sinha AU, Vempati S, Faber J, Krivtsov AV, Feng Z, Punt N, Daigle A, Bullinger L, Pollock RM, Richon VM, Kung AL, Armstrong SA. MLL-rearranged leukemia is dependent on aberrant H3K79 methylation by DOT1L. Cancer Cell. 2011;20:66–78. doi: 10.1016/j.ccr.2011.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binda C, Valente S, Romanenghi M, Pilotto S, Cirilli R, Karytinos A, Ciossani G, Botrugno OA, Forneris F, Tardugno M, Edmondson DE, Minucci S, Mattevi A, Mai A. Biochemical, structural, and biological evaluation of tranylcypromine derivatives as inhibitors of histone demethylases LSD1 and LSD2. Journal of the American Chemical Society. 2010;132:6827–6833. doi: 10.1021/ja101557k. [DOI] [PubMed] [Google Scholar]
- Chang KH, King ON, Tumber A, Woon EC, Heightman TD, McDonough MA, Schofield CJ, Rose NR. Inhibition of histone demethylases by 4-carboxy-2,2′-bipyridyl compounds. ChemMedChem. 2011;6:759–764. doi: 10.1002/cmdc.201100026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang Y, Ganesh T, Horton JR, Spannhoff A, Liu J, Sun A, Zhang X, Bedford MT, Shinkai Y, Snyder JP, Cheng X. Adding a lysine mimic in the design of potent inhibitors of histone lysine methyltransferases. Journal of Molecular Biology. 2010;400:1–7. doi: 10.1016/j.jmb.2010.04.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang Y, Zhang X, Horton JR, Upadhyay AK, Spannhoff A, Liu J, Snyder JP, Bedford MT, Cheng X. Structural basis for G9a-like protein lysine methyltransferase inhibition by BIX-01294. Nature Structural Molecular Biology. 2009;16:312–317. doi: 10.1038/nsmb.1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen MW, Hua KT, Kao HJ, Chi CC, Wei LH, Johansson G, Shiah SG, Chen PS, Jeng YM, Cheng TY, Lai TC, Chang JS, Jan YH, Chien MH, Yang CJ, Huang MS, Hsiao M, Kuo ML. H3K9 histone methyltransferase G9a promotes lung cancer invasion and metastasis by silencing the cell adhesion molecule Ep-CAM. Cancer Research. 2010;70:7830–7840. doi: 10.1158/0008-5472.CAN-10-0833. [DOI] [PubMed] [Google Scholar]
- Cheng X, Collins RE, Zhang X. Structural and sequence motifs of protein (histone) methylation enzymes. Annual Review of Biophysics and Biomolecular Structure. 2005;34:267–294. doi: 10.1146/annurev.biophys.34.040204.144452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherblanc FL, Chapman KL, Brown R, Fuchter MJ. Chaetocin is a nonspecific inhibitor of histone lysine methyltransferases. Nature Chemical Biology. 2013;9:136–137. doi: 10.1038/nchembio.1187. [DOI] [PubMed] [Google Scholar]
- Chi P, Allis CD, Wang GG. Covalent histone modifications – miswritten, misinterpreted and mis-erased in human cancers. Nature Reviews Cancer. 2010;10:457–469. doi: 10.1038/nrc2876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang PK, Cantoni GL. Perturbation of biochemical transmethylations by 3-deazaadenosine in vivo. Biochemical Pharmacology. 1979;28:1897–1902. doi: 10.1016/0006-2952(79)90642-7. [DOI] [PubMed] [Google Scholar]
- Chowdhury R, Yeoh KK, Tian YM, Hillringhaus L, Bagg EA, Rose NR, Leung IK, Li XS, Woon EC, Yang M, McDonough MA, King ON, Clifton IJ, Klose RJ, Claridge TD, Ratcliffe PJ, Schofield CJ, Kawamura A. The oncometabolite 2-hydroxyglutarate inhibits histone lysine demethylases. EMBO Reports. 2011;12:463–469. doi: 10.1038/embor.2011.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cloos PA, Christensen J, Agger K, Maiolica A, Rappsilber J, Antal T, Hansen KH, Helin K. The putative oncogene GASC1 demethylates tri- and dimethylated lysine 9 on histone H3. Nature. 2006;442:307–311. doi: 10.1038/nature04837. [DOI] [PubMed] [Google Scholar]
- Copeland RA, Solomon ME, Richon VM. Protein methyltransferases as a target class for drug discovery. Nature Reviews Drug Discovery. 2009;8:724–732. doi: 10.1038/nrd2974. [DOI] [PubMed] [Google Scholar]
- Couture JF, Collazo E, Ortiz-Tello PA, Brunzelle JS, Trievel RC. Specificity and mechanism of JMJD2A, a trimethyllysine-specific histone demethylase. Nature Structural and Molecular Biology. 2007;14:689–695. doi: 10.1038/nsmb1273. [DOI] [PubMed] [Google Scholar]
- Couture JF, Hauk G, Thompson MJ, Blackburn GM, Trievel RC. Catalytic roles for carbon- oxygen hydrogen bonding in SET domain lysine methyltransferases. Journal of Biological Chemistry. 2006;281:19280–19287. doi: 10.1074/jbc.M602257200. [DOI] [PubMed] [Google Scholar]
- Crea F, Sun L, Mai A, Chiang YT, Farrar WL, Danesi R, Helgason CD. The emerging role of histone lysine demethylases in prostate cancer. Molecular Cancer. 2012;11:52. doi: 10.1186/1476-4598-11-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Culhane JC, Szewczuk LM, Liu X, Da G, Marmorstein R, Cole PA. A mechanism-based inactivator for histone demethylase LSD1. Journal of the American Chemical Society. 2006;128:4536–4537. doi: 10.1021/ja0602748. [DOI] [PubMed] [Google Scholar]
- Culhane JC, Wang D, Yen PM, Cole PA. Comparative analysis of small molecules and histone substrate analogues as LSD1 lysine demethylase inhibitors. Journal of the American Chemical Society. 2010;132:3164–3176. doi: 10.1021/ja909996p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daigle SR, Olhava EJ, Therkelsen CA, Majer CR, Sneeringer CJ, Song J, Johnston LD, Scott MP, Smith JJ, Xiao Y, Jin L, Kuntz KW, Chesworth R, Moyer MP, Bernt KM, Tseng JC, Kung AL, Armstrong SA, Copeland RA, Richon VM, Pollock RM. Selective killing of mixed lineage leukemia cells by a potent small-molecule DOT1L inhibitor. Cancer Cell. 2011;20:53–65. doi: 10.1016/j.ccr.2011.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dancy BC, Ming SA, Papazyan R, Jelinek CA, Majumdar A, Sun Y, Dancy BM, Drury WJ, 3rd, Cotter RJ, Taverna SD, Cole PA. Azalysine analogues as probes for protein lysine deacetylation and demethylation. Journal of the American Chemical Society. 2012;134:5138–5148. doi: 10.1021/ja209574z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawson MA, Kouzarides T. Cancer epigenetics : from mechanism to therapy. Cell. 2012;150:12–27. doi: 10.1016/j.cell.2012.06.013. [DOI] [PubMed] [Google Scholar]
- Dawson MA, Kouzarides T, Huntly BJ. Targeting epigenetic readers in cancer. New England Journal of Medicine. 2012;367:647–657. doi: 10.1056/NEJMra1112635. [DOI] [PubMed] [Google Scholar]
- Eglen RM, Reisine T, Roby P, Rouleau N, Illy C, Bosse R, Bielefeld M. The use of AlphaScreen technology in HTS: current status. Current Chemical Genomics. 2008;1:2–10. doi: 10.2174/1875397300801010002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Q, Wang H, Ng HH, Erdjument-Bromage H, Tempst P, Struhl K, Zhang Y. Methylation of H3-lysine 79 is mediated by a new family of HMTases without a SET domain. Current Biology. 2002;12:1052–1058. doi: 10.1016/s0960-9822(02)00901-6. [DOI] [PubMed] [Google Scholar]
- Ferguson AD, Larsen NA, Howard T, Pollard H, Green I, Grande C, Cheung T, Garcia-Arenas R, Cowen S, Wu J, Godin R, Chen H, Keen N. Structural basis of substrate methylation and inhibition of SMYD2. Structure. 2011;19:1262–1273. doi: 10.1016/j.str.2011.06.011. [DOI] [PubMed] [Google Scholar]
- Fierz B, Muir TW. Chromatin as an expansive canvas for chemical biology. Nature Chemical Biology. 2012;8:417–427. doi: 10.1038/nchembio.938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujishiro S, Dodo K, Iwasa E, Teng Y, Sohtome Y, Hamashima Y, Ito A, Yoshida M, Sodeoka M. Epidithiodiketopiperazine as a pharmacophore for protein lysine methyltransferase G9a inhibitors : reducing cytotoxicity by structural simplification. Bioorganic and Medical Chemistry Letters. 2013;23:733–736. doi: 10.1016/j.bmcl.2012.11.087. [DOI] [PubMed] [Google Scholar]
- Ghosh S, Nie A, An J, Huang Z. Structure-based virtual screening of chemical libraries for drug discovery. Currunt Opinion in Chemical Biology. 2006;10:194–202. doi: 10.1016/j.cbpa.2006.04.002. [DOI] [PubMed] [Google Scholar]
- Gooden DM, Schmidt DM, Pollock JA, Kabadi AM, McCafferty DG. Facile synthesis of substituted trans-2-arylcyclopropylamine inhibitors of the human histone demethylase LSD1 and monoamine oxidases A and B. Bioorganic and Medical Chemistry Letters. 2008;18:3047–3051. doi: 10.1016/j.bmcl.2008.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greiner D, Bonaldi T, Eskeland R, Roemer E, Imhof A. Identification of a specific inhibitor of the histone methyltransferase SU(VAR)3-9. Nature Chemical Biology. 2005;1:143–145. doi: 10.1038/nchembio721. [DOI] [PubMed] [Google Scholar]
- Grembecka J, He S, Shi A, Purohit T, Muntean AG, Sorenson RJ, Showalter HD, Murai MJ, Belcher AM, Hartley T, Hess JL, Cierpicki T. Menin-MLL inhibitors reverse oncogenic activity of MLL fusion proteins in leukemia. Nature Chemical Biology. 2012;8:277–284. doi: 10.1038/nchembio.773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grewal SI, Jia S. Heterochromatin revisited. Nature Reviews Genetics. 2007;8:35–46. doi: 10.1038/nrg2008. [DOI] [PubMed] [Google Scholar]
- Hamada S, Kim TD, Suzuki T, Itoh Y, Tsumoto H, Nakagawa H, Janknecht R, Miyata N. Synthesis and activity of N-oxalylglycine and its derivatives as Jumonji C-domain-containing histone lysine demethylase inhibitors. Bioorganic and Medical Chemistry Letters. 2009;19:2852–2855. doi: 10.1016/j.bmcl.2009.03.098. [DOI] [PubMed] [Google Scholar]
- Hamada S, Suzuki T, Mino K, Koseki K, Oehme F, Flamme I, Ozasa H, Itoh Y, Ogasawara D, Komaarashi H, Kato A, Tsumoto H, Nakagawa H, Hasegawa M, Sasaki R, Mizukami T, Miyata N. Design, synthesis, enzyme-inhibitory activity, and effect on human cancer cells of a novel series of jumonji domain-containing protein 2 histone demethylase inhibitors. Journal of Medicinal Chemistry. 2010;53:5629–5638. doi: 10.1021/jm1003655. [DOI] [PubMed] [Google Scholar]
- Hardy JA, Wells JA. Searching for new allosteric sites in enzymes. Current Opinion in Structural Biology. 2004;14:706–715. doi: 10.1016/j.sbi.2004.10.009. [DOI] [PubMed] [Google Scholar]
- Hazeldine S, Pachaiyappan B, Steinbergs N, Nowotarski S, Hanson AS, Casero RA, Jr, Woster PM. Low molecular weight amidoximes that act as potent inhibitors of lysine-specific demethylase 1. Journal of Medicinal Chemistry. 2012;55:7378–7391. doi: 10.1021/jm3002845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Gurung B, Wan B, Matkar S, Veniaminova NA, Wan K, Merchant JL, Hua X, Lei M. The same pocket in menin binds both MLL and JUND but has opposite effects on transcription. Nature. 2012;482:542–546. doi: 10.1038/nature10806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Fang J, Bedford MT, Zhang Y, Xu RM. Recognition of histone H3 lysine-4 methylation by the double tudor domain of JMJD2A. Science. 2006;312:748–751. doi: 10.1126/science.1125162. [DOI] [PubMed] [Google Scholar]
- Huang Y, Greene E, Murray Stewart T, Goodwin AC, Baylin SB, Woster PM, Casero RA., Jr Inhibition of lysine-specific demethylase 1 by polyamine analogues results in reexpression of aberrantly silenced genes. Proceedings of the National Academy of Sciences USA. 2007;104:8023–8028. doi: 10.1073/pnas.0700720104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Stewart TM, Wu Y, Baylin SB, Marton LJ, Perkins B, Jones RJ, Woster PM, Casero RA., Jr Novel oligoamine analogues inhibit lysine-specific demethylase 1 and induce reexpression of epigenetically silenced genes. Clinical Cancer Research. 2009;15:7217–7228. doi: 10.1158/1078-0432.CCR-09-1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang MY, Lee BB, Kim YH, Chang DK, Kyu Park S, Chun HK, Song SY, Park J, Kim DH. Association of the SUV39H1 histone methyltransferase with the DNA methyltransferase 1 at mRNA expression level in primary colorectal cancer. International Journal of Cancer. 2007;121:2192–2197. doi: 10.1002/ijc.22953. [DOI] [PubMed] [Google Scholar]
- King ON, Li XS, Sakurai M, Kawamura A, Rose NR, Ng SS, Quinn AM, Rai G, Mott BT, Beswick P, Klose RJ, Oppermann U, Jadhav A, Heightman TD, Maloney DJ, Schofield CJ, Simeonov A. Quantitative high-throughput screening identifies 8-hydroxyquinolines as cell-active histone demethylase inhibitors. PLoS One. 2010;5:e15535. doi: 10.1371/journal.pone.0015535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knutson SK, Wigle TJ, Warholic NM, Sneeringer CJ, Allain CJ, Klaus CR, Sacks JD, Raimondi A, Majer CR, Song J, Scott MP, Jin L, Smith JJ, Olhava EJ, Chesworth R, Moyer MP, Richon VM, Copeland RA, Keilhack H, Pollock RM, Kuntz KW. A selective inhibitor of EZH2 blocks H3K27 methylation and kills mutant lymphoma cells. Nature Chemical Biology. 2012;8:890–896. doi: 10.1038/nchembio.1084. [DOI] [PubMed] [Google Scholar]
- Komatsu S, Imoto I, Tsuda H, Kozaki KI, Muramatsu T, Shimada Y, Aiko S, Yoshizumi Y, Ichikawa D, Otsuji E, Inazawa J. Overexpression of SMYD2 relates to tumor cell proliferation and malignant outcome of esophageal squamous cell carcinoma. Carcinogenesis. 2009;30:1139–1146. doi: 10.1093/carcin/bgp116. [DOI] [PubMed] [Google Scholar]
- Krauss V. Glimpses of evolution: heterochromatic histone H3K9 methyltransferases left its marks behind. Genetica. 2008;133:93–106. doi: 10.1007/s10709-007-9184-z. [DOI] [PubMed] [Google Scholar]
- Krivtsov AV, Armstrong SA. MLL translocations, histone modifications and leukaemia stem-cell development. Nature Reviews Cancer. 2007;7:823–833. doi: 10.1038/nrc2253. [DOI] [PubMed] [Google Scholar]
- Kruidenier L, Chung CW, Cheng Z, Liddle J, Che K, Joberty G, Bantscheff M, Bountra C, Bridges A, Diallo H, Eberhard D, Hutchinson S, Jones E, Katso R, Leveridge M, Mander PK, Mosley J, Ramirez-Molina C, Rowland P, Schofield CJ, Sheppard RJ, Smith JE, Swales C, Tanner R, Thomas P, Tumber A, Drewes G, Oppermann U, Patel DJ, Lee K, Wilson DM. A selective jumonji H3K27 demethylase inhibitor modulates the proinflammatory macrophage response. Nature. 2012;488:404–408. doi: 10.1038/nature11262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubicek S, O’Sullivan RJ, August EM, Hickey ER, Zhang Q, Teodoro ML, Rea S, Mechtler K, Kowalski JA, Homon CA, Kelly TA, Jenuwein T. Reversal of H3K9me2 by a small-molecule inhibitor for the G9a histone methyltransferase. Molecular Cell. 2007;25:473–481. doi: 10.1016/j.molcel.2007.01.017. [DOI] [PubMed] [Google Scholar]
- Lee J, Thompson JR, Botuyan MV, Mer G. Distinct binding modes specify the recognition of methylated histones H3K4 and H4K20 by JMJD2A-tudor. Nature Structural and Molecular Biology. 2008;15:109–111. doi: 10.1038/nsmb1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MG, Wynder C, Schmidt DM, McCafferty DG, Shiekhattar R. Histone H3 lysine 4 demethylation is a target of nonselective antidepressive medications. Chemical Biology. 2006;13:563–567. doi: 10.1016/j.chembiol.2006.05.004. [DOI] [PubMed] [Google Scholar]
- Liang Y, Quenelle D, Vogel JL, Mascaro C, Ortega A, Kristie TM. A Novel Selective LSD1/KDM1A Inhibitor Epigenetically Blocks Herpes Simplex Virus Lytic Replication and Reactivation from Latency. MBio. 2013;4:e00558–12. doi: 10.1128/mBio.00558-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipinski CA, Lombardo F, Dominy BW, Feeney PJ. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Advanced Drug Delivery Reviews. 2001;46:3–26. doi: 10.1016/s0169-409x(00)00129-0. [DOI] [PubMed] [Google Scholar]
- Liu F, Chen X, Allali-Hassani A, Quinn AM, Wigle TJ, Wasney GA, Dong A, Senisterra G, Chau I, Siarheyeva A, Norris JL, Kireev DB, Jadhav A, Herold JM, Janzen WP, Arrowsmith CH, Frye SV, Brown PJ, Simeonov A, Vedadi M, Jin J. Protein lysine methyltransferase G9a inhibitors : design, synthesis, and structure activity relationships of 2,4- diamino-7-aminoalkoxy-quinazolines. Journal of Medicinal Chemistry. 2010;53:5844–5857. doi: 10.1021/jm100478y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohse B, Nielsen AL, Kristensen JB, Helgstrand C, Cloos PA, Olsen L, Gajhede M, Clausen RP, Kristensen JL. Targeting histone lysine demethylases by truncating the histone 3 tail to obtain selective substrate-based inhibitors. Angewandte Chemie International Edition English. 2011;50:9100–9103. doi: 10.1002/anie.201101849. [DOI] [PubMed] [Google Scholar]
- Luo X, Liu Y, Kubicek S, Myllyharju J, Tumber A, Ng S, Che KH, Podoll J, Heightman TD, Oppermann U, Schreiber SL, Wang X. A selective inhibitor and probe of the cellular functions of Jumonji C domain-containing histone demethylases. Journal of the American Chemical Society. 2011;133:9451–9456. doi: 10.1021/ja201597b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margueron R, Reinberg D. The Polycomb complex PRC2 and its mark in life. Nature. 2011;469:343–349. doi: 10.1038/nature09784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCabe MT, Ott HM, Ganji G, Korenchuk S, Thompson C, Van Aller GS, Liu Y, Graves AP, Della Pietra A, 3rd, Diaz E, LaFrance LV, Mellinger M, Duquenne C, Tian X, Kruger RG, McHugh CF, Brandt M, Miller WH, Dhanak D, Verma SK, Tummino PJ, Creasy CL. EZH2 inhibition as a therapeutic strategy for lymphoma with EZH2-activating mutations. Nature. 2012;492:108–112. doi: 10.1038/nature11606. [DOI] [PubMed] [Google Scholar]
- Metzger E, Wissmann M, Yin N, Muller JM, Schneider R, Peters AH, Gunther T, Buettner R, Schule R. LSD1 demethylates repressive histone marks to promote androgen-receptor-dependent transcription. Nature. 2005;437:436–439. doi: 10.1038/nature04020. [DOI] [PubMed] [Google Scholar]
- Mimasu S, Umezawa N, Sato S, Higuchi T, Umehara T, Yokoyama S. Structurally designed trans-2-phenylcyclopropylamine derivatives potently inhibit histone demethylase LSD1/KDM1. Biochemistry. 2010;49:6494–6503. doi: 10.1021/bi100299r. [DOI] [PubMed] [Google Scholar]
- Min J, Feng Q, Li Z, Zhang Y, Xu RM. Structure of the catalytic domain of human DOT1L, a non-SET domain nucleosomal histone methyltransferase. Cell. 2003;112:711–723. doi: 10.1016/s0092-8674(03)00114-4. [DOI] [PubMed] [Google Scholar]
- Minucci S, Pelicci PG. Histone deacetylase inhibitors and the promise of epigenetic (and more) treatments for cancer. Nature Reviews in Cancer. 2006;6:38–51. doi: 10.1038/nrc1779. [DOI] [PubMed] [Google Scholar]
- Miranda TB, Cortez CC, Yoo CB, Liang G, Abe M, Kelly TK, Marquez VE, Jones PA. DZNep is a global histone methylation inhibitor that reactivates developmental genes not silenced by DNA methylation. Molecular Cancer Therapeutics. 2009;8:1579–1588. doi: 10.1158/1535-7163.MCT-09-0013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray CW, Blundell TL. Structural biology in fragment-based drug design. Current Opinion in Structural Biology. 2010;20:497–507. doi: 10.1016/j.sbi.2010.04.003. [DOI] [PubMed] [Google Scholar]
- Ng SS, Kavanagh KL, McDonough MA, Butler D, Pilka ES, Lienard BM, Bray JE, Savitsky P, Gileadi O, von Delft F, Rose NR, Offer J, Scheinost JC, Borowski T, Sundstrom M, Schofield CJ, Oppermann U. Crystal structures of histone demethylase JMJD2A reveal basis for substrate specificity. Nature. 2007;448:87–91. doi: 10.1038/nature05971. [DOI] [PubMed] [Google Scholar]
- Nguyen AT, Zhang Y. The diverse functions of Dot1 and H3K79 methylation. Genes and Development. 2011;25:1345–1358. doi: 10.1101/gad.2057811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen AL, Kristensen LH, Stephansen KB, Kristensen JB, Helgstrand C, Lees M, Cloos P, Helin K, Gajhede M, Olsen L. Identification of catechols as histone-lysine demethylase inhibitors. FEBS Letters. 2012;586:1190–1194. doi: 10.1016/j.febslet.2012.03.001. [DOI] [PubMed] [Google Scholar]
- Ogasawara D, Suzuki T, Mino K, Ueda R, Khan MN, Matsubara T, Koseki K, Hasegawa M, Sasaki R, Nakagawa H, Mizukami T, Miyata N. Synthesis and biological activity of optically active NCL-1, a lysine-specific demethylase 1 selective inhibitor. Bioorganic and Medical Chemistry Letters. 2011;19:3702–3708. doi: 10.1016/j.bmc.2010.12.024. [DOI] [PubMed] [Google Scholar]
- Patel A, Vought VE, Dharmarajan V, Cosgrove MS. A conserved arginine-containing motif crucial for the assembly and enzymatic activity of the mixed lineage leukemia protein-1 core complex. Journal of Biological Chemistry. 2008;283:32162–32175. doi: 10.1074/jbc.M806317200. [DOI] [PubMed] [Google Scholar]
- Pedersen MT, Helin K. Histone demethylases in development and disease. Trends in Cell Biology. 2010;20:662–671. doi: 10.1016/j.tcb.2010.08.011. [DOI] [PubMed] [Google Scholar]
- Qi W, Chan H, Teng L, Li L, Chuai S, Zhang R, Zeng J, Li M, Fan H, Lin Y, Gu J, Ardayfio O, Zhang JH, Yan X, Fang J, Mi Y, Zhang M, Zhou T, Feng G, Chen Z, Li G, Yang T, Zhao K, Liu X, Yu Z, Lu CX, Atadja P, Li E. Selective inhibition of Ezh2 by a small molecule inhibitor blocks tumor cells proliferation. Proceedings of the National Academy of Sciences USA. 2012;109:21360–21365. doi: 10.1073/pnas.1210371110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rooney PH, Murray GI, Stevenson DA, Haites NE, Cassidy J, McLeod HL. Comparative genomic hybridization and chromosomal instability in solid tumours. British Journal of Cancer. 1999;80:862–873. doi: 10.1038/sj.bjc.6690433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose NR, Ng SS, Mecinovic J, Lienard BM, Bello SH, Sun Z, McDonough MA, Oppermann U, Schofield CJ. Inhibitor scaffolds for 2-oxoglutarate-dependent histone lysine demethylases. Journal of Medicinal Chemistry. 2008;51:7053–7056. doi: 10.1021/jm800936s. [DOI] [PubMed] [Google Scholar]
- Rose NR, Woon EC, Kingham GL, King ON, Mecinovic J, Clifton IJ, Ng SS, Talib-Hardy J, Oppermann U, McDonough MA, Schofield CJ. Selective inhibitors of the JMJD2 histone demethylases : combined nondenaturing mass spectrometric screening and crystallographic approaches. Journal of Medicinal Chemistry. 2010;53:1810–1818. doi: 10.1021/jm901680b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose NR, Woon EC, Tumber A, Walport LJ, Chowdhury R, Li XS, King ON, Lejeune C, Ng SS, Krojer T, Chan MC, Rydzik AM, Hopkinson RJ, Che KH, Daniel M, Strain-Damerell C, Gileadi C, Kochan G, Leung IK, Dunford J, Yeoh KK, Ratcliffe PJ, Burgess-Brown N, von Delft F, Muller S, Marsden B, Brennan PE, McDonough MA, Oppermann U, Klose RJ, Schofield CJ, Kawamura A. Plant growth regulator daminozide is a selective inhibitor of human KDM2/7 histone demethylases. Journal of Medicinal Chemistry. 2012;55:6639–6643. doi: 10.1021/jm300677j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruthenburg AJ, Allis CD, Wysocka J. Methylation of lysine 4 on histone H3: intricacy of writing and reading a single epigenetic mark. Molecular Cell. 2007;25:15–30. doi: 10.1016/j.molcel.2006.12.014. [DOI] [PubMed] [Google Scholar]
- Sakurai M, Rose NR, Schultz L, Quinn AM, Jadhav A, Ng SS, Oppermann U, Schofield CJ, Simeonov A. A miniaturized screen for inhibitors of Jumonji histone demethylases. Molecular Biosystems. 2010;6:357–364. doi: 10.1039/b912993f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawada K, Yang Z, Horton JR, Collins RE, Zhang X, Cheng X. Structure of the conserved core of the yeast Dot1p, a nucleosomal histone H3 lysine 79 methyltransferase. Journal of Biological Chemistry. 2004;279:43296–43306. doi: 10.1074/jbc.M405902200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sayegh J, Cao J, Zou MR, Morales A, Blair LP, Norcia M, Hoyer D, Tackett AJ, Merkel JS, Yan Q. Identification of small molecule inhibitors of Jumonji AT-Rich interactive domain 1B (JARID1B) histone demethylase by a sensitive high-throughput screen. Journal of Biological Chemistry. 2013;288:9408–9417. doi: 10.1074/jbc.M112.419861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt DM, McCafferty DG. trans-2-Phenylcyclopropylamine is a mechanism-based inactivator of the histone demethylase LSD1. Biochemistry. 2007;46:4408–4416. doi: 10.1021/bi0618621. [DOI] [PubMed] [Google Scholar]
- Sekirnik R, Rose NR, Thalhammer A, Seden PT, Mecinovic J, Schofield CJ. Inhibition of the histone lysine demethylase JMJD2A by ejection of structural Zn(II) Chemical Communications. 2009:6376–6378. doi: 10.1039/b916357c. [DOI] [PubMed] [Google Scholar]
- Senisterra G, Wu H, Allali-Hassani A, Wasney GA, Barsyte-Lovejoy D, Dombrovski L, Dong A, Nguyen KT, Smil D, Bolshan Y, Hajian T, He H, Seitova A, Chau I, Li F, Poda G, Couture JF, Brown PJ, Al-Awar R, Schapira M, Arrowsmith CH, Vedadi M. Small-molecule inhibition of MLL activity by disruption of its interaction with WDR5. Biochemical Journal. 2013;449:151–159. doi: 10.1042/BJ20121280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma SK, Wu Y, Steinbergs N, Crowley ML, Hanson AS, Casero RA, Woster PM. (Bis)urea and (bis)thiourea inhibitors of lysine-specific demethylase 1 as epigenetic modulators. Journal of Medicinal Chemistry. 2010;53:5197–5212. doi: 10.1021/jm100217a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi A, Murai MJ, He S, Lund G, Hartley T, Purohit T, Reddy G, Chruszcz M, Grembecka J, Cierpicki T. Structural insights into inhibition of the bivalent menin-MLL interaction by small molecules in leukemia. Blood. 2012;120:4461–4469. doi: 10.1182/blood-2012-05-429274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y. Histone lysine demethylases: emerging roles in development, physiology and disease. Nature Reviews Genetics. 2007;8:829–833. doi: 10.1038/nrg2218. [DOI] [PubMed] [Google Scholar]
- Shi Y, Lan F, Matson C, Mulligan P, Whetstine JR, Cole PA, Casero RA. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell. 2004;119:941–953. doi: 10.1016/j.cell.2004.12.012. [DOI] [PubMed] [Google Scholar]
- Shih AH, Abdel-Wahab O, Patel JP, Levine RL. The role of mutations in epigenetic regulators in myeloid malignancies. Nature Reviews Cancer. 2012;12:599–612. doi: 10.1038/nrc3343. [DOI] [PubMed] [Google Scholar]
- Siarheyeva A, Senisterra G, Allali-Hassani A, Dong A, Dobrovetsky E, Wasney GA, Chau I, Marcellus R, Hajian T, Liu F, Korboukh I, Smil D, Bolshan Y, Min J, Wu H, Zeng H, Loppnau P, Poda G, Griffin C, Aman A, Brown PJ, Jin J, Al-Awar R, Arrowsmith CH, Schapira M, Vedadi M. An allosteric inhibitor of protein arginine methyltransferase 3. Structure. 2012;20:1425–1435. doi: 10.1016/j.str.2012.06.001. [DOI] [PubMed] [Google Scholar]
- Suzuki T, Miyata N. Lysine demethylases inhibitors. Journal of Medicinal Chemistry. 2011;54:8236–8250. doi: 10.1021/jm201048w. [DOI] [PubMed] [Google Scholar]
- Szewczuk LM, Culhane JC, Yang M, Majumdar A, Yu H, Cole PA. Mechanistic analysis of a suicide inactivator of histone demethylase LSD1. Biochemistry. 2007;46:6892–6902. doi: 10.1021/bi700414b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szyf M. Epigenetics, DNA methylation, and chromatin modifying drugs. Annual Review of Pharmacology and Toxicology. 2009;49:243–263. doi: 10.1146/annurev-pharmtox-061008-103102. [DOI] [PubMed] [Google Scholar]
- Tan J, Yang X, Zhuang L, Jiang X, Chen W, Lee PL, Karuturi RK, Tan PB, Liu ET, Yu Q. Pharmacologic disruption of Polycomb-repressive complex 2-mediated gene repression selectively induces apoptosis in cancer cells. Genes and Development. 2007;21:1050–1063. doi: 10.1101/gad.1524107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thalhammer A, Mecinovic J, Loenarz C, Tumber A, Rose NR, Heightman TD, Schofield CJ. Inhibition of the histone demethylase JMJD2E by 3-substituted pyridine 2,4-dicarboxylates. Organic and Biomolecular Chemistry. 2011;9:127–135. doi: 10.1039/c0ob00592d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukada Y, Fang J, Erdjument-Bromage H, Warren ME, Borchers CH, Tempst P, Zhang Y. Histone demethylation by a family of JmjC domain-containing proteins. Nature. 2006;439:811–816. doi: 10.1038/nature04433. [DOI] [PubMed] [Google Scholar]
- Ueda R, Suzuki T, Mino K, Tsumoto H, Nakagawa H, Hasegawa M, Sasaki R, Mizukami T, Miyata N. Identification of cell-active lysine specific demethylase 1-selective inhibitors. Journal of the American Chemical Society. 2009;131:17536–17537. doi: 10.1021/ja907055q. [DOI] [PubMed] [Google Scholar]
- Vedadi M, Barsyte-Lovejoy D, Liu F, Rival-Gervier S, Allali-Hassani A, Labrie V, Wigle TJ, Dimaggio PA, Wasney GA, Siarheyeva A, Dong A, Tempel W, Wang SC, Chen X, Chau I, Mangano TJ, Huang XP, Simpson CD, Pattenden SG, Norris JL, Kireev DB, Tripathy A, Edwards A, Roth BL, Janzen WP, Garcia BA, Petronis A, Ellis J, Brown PJ, Frye SV, Arrowsmith CH, Jin J. A chemical probe selectively inhibits G9a and GLP methyltransferase activity in cells. Nature Chemical Biology. 2011;7:566–574. doi: 10.1038/nchembio.599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner EJ, Carpenter PB. Understanding the language of Lys36 methylation at histone H3. Nature Reviews in Molecular Cell Biology. 2012;13:115–126. doi: 10.1038/nrm3274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Lu F, Ren Q, Sun H, Xu Z, Lan R, Liu Y, Ward D, Quan J, Ye T, Zhang H. Novel histone demethylase LSD1 inhibitors selectively target cancer cells with pluripotent stem cell properties. Cancer Research. 2011;71:7238–7249. doi: 10.1158/0008-5472.CAN-11-0896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Song J, Milne TA, Wang GG, Li H, Allis CD, Patel DJ. Pro isomerization in MLL1 PHD3-bromo cassette connects H3K4me readout to CyP33 and HDAC-mediated repression. Cell. 2010;141:1183–1194. doi: 10.1016/j.cell.2010.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams DE, Dalisay DS, Li F, Amphlett J, Maneerat W, Chavez MA, Wang YA, Matainaho T, Yu W, Brown PJ, Arrowsmith CH, Vedadi M, Andersen RJ. Nahuoic acid A produced by a Streptomyces sp. isolated from a marine sediment Is a selective SAM-competitive inhibitor of the histone methyltransferase SETD8. Organic Letters. 2013;15:414–417. doi: 10.1021/ol303416k. [DOI] [PubMed] [Google Scholar]
- Woon EC, Tumber A, Kawamura A, Hillringhaus L, Ge W, Rose NR, Ma JH, Chan MC, Walport LJ, Che KH, Ng SS, Marsden BD, Oppermann U, McDonough MA, Schofield CJ. Linking of 2-oxoglutarate and substrate binding sites enables potent and highly selective inhibition of JmjC histone demethylases. Angewandte Chemie International Edition English. 2012;51:1631–1634. doi: 10.1002/anie.201107833. [DOI] [PubMed] [Google Scholar]
- Yang M, Culhane JC, Szewczuk LM, Gocke CB, Brautigam CA, Tomchick DR, Machius M, Cole PA, Yu H. Structural basis of histone demethylation by LSD1 revealed by suicide inactivation. Nature Structural and Molecular Biology. 2007a;14:535–539. doi: 10.1038/nsmb1255. [DOI] [PubMed] [Google Scholar]
- Yang M, Culhane JC, Szewczuk LM, Jalili P, Ball HL, Machius M, Cole PA, Yu H. Structural basis for the inhibition of the LSD1 histone demethylase by the antidepressant trans- 2-phenylcyclopropylamine. Biochemistry. 2007b;46:8058–8065. doi: 10.1021/bi700664y. [DOI] [PubMed] [Google Scholar]
- Yao Y, Chen P, Diao J, Cheng G, Deng L, Anglin JL, Prasad BV, Song Y. Selective inhibitors of histone methyltransferase DOT1L: design, synthesis, and crystallographic studies. Journal of the American Chemical Society. 2011;133:16746–16749. doi: 10.1021/ja206312b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoyama A, Somervaille TC, Smith KS, Rozenblatt-Rosen O, Meyerson M, Cleary ML. The menin tumor suppressor protein is an essential oncogenic cofactor for MLL-associated leukemogenesis. Cell. 2005;123:207–218. doi: 10.1016/j.cell.2005.09.025. [DOI] [PubMed] [Google Scholar]
- Yu W, Chory EJ, Wernimont AK, Tempel W, Scopton A, Federation A, Marineau JJ, Qi J, Barsyte-Lovejoy D, Yi J, Marcellus R, Iacob RE, Engen JR, Griffin C, Aman A, Wienholds E, Li F, Pineda J, Estiu G, Shatseva T, Hajian T, Al-Awar R, Dick JE, Vedadi M, Brown PJ, Arrowsmith CH, Bradner JE, Schapira M. Catalytic site re-modelling of the DOT1L methyltransferase by selective inhibitors. Nature Communications. 2012;3:1288. doi: 10.1038/ncomms2304. [DOI] [PubMed] [Google Scholar]
- Yu W, Smil D, Li F, Tempel W, Fedorov O, Nguyen KT, Bolshan Y, Al-Awar R, Knapp S, Arrowsmith CH, Vedadi M, Brown PJ, Schapira M. Bromo-deaza-SAH: A potent and selective DOT1L inhibitor. Bioorganic and Medicinal Chemistry. 2013;21:1787–1794. doi: 10.1016/j.bmc.2013.01.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Y, Wang Q, Paulk J, Kubicek S, Kemp MM, Adams DJ, Shamji AF, Wagner BK, Schreiber SL. A small-molecule probe of the histone methyltransferase G9a induces cellular senescence in pancreatic adenocarcinoma. ACS Chemical Biology. 2012;7:1152–1157. doi: 10.1021/cb300139y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P, Lee H, Brunzelle JS, Couture JF. The plasticity of WDR5 peptide-binding cleft enables the binding of the SET1 family of histone methyltransferases. Nucleic Acids Research. 2012;40:4237–4246. doi: 10.1093/nar/gkr1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng W, Ibanez G, Wu H, Blum G, Zeng H, Dong A, Li F, Hajian T, Allali-Hassani A, Amaya MF, Siarheyeva A, Yu W, Brown PJ, Schapira M, Vedadi M, Min J, Luo M. Sinefungin derivatives as inhibitors and structure probes of protein lysine methyltransferase SETD2. Journal of the American Chemical Society. 2012;134:18004–18014. doi: 10.1021/ja307060p. [DOI] [PMC free article] [PubMed] [Google Scholar]