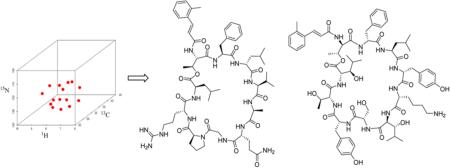
Abstract
Despite the advances in NMR, structure determination is often slow and constitutes a bottleneck in natural products discovery. Removal of this bottleneck would greatly improve the throughput for antibiotic discovery as well as other therapeutic areas. Overall, faster structure methods for structure determination will serve the natural products community in a broad manner. This report describes the first application of 3D NMR for elucidation of two microbially produced peptide natural products with novel structures. The methods are cost-effective and greatly improve the confidence in a proposed structure.
INTRODUCTION
Drug resistant infectious diseases, in particular Gram-negative infections, continue to threaten global health as well as contemporary medical practices.1 For example, as pathogens become increasingly resistant, semiroutine procedures such as organ transplants and joint replacements will become increasingly dangerous. Combined with the emergence of pan-resistant pathogens, new antibiotics are sorely needed. Although natural products have provided the antibiotic basis upon which many medical practices were built, discovering novel antibiotics has become more challenging. Nonetheless, advances in sequencing and genomics has not only indicated that there is more potential than previously thought, but also have provided tools for nearly routine analyses of the genetic potential of bacteria to produce small molecules, many of which would have antibiotic potential.2,3 In parallel, analytical strategies that take advantage of increasing sensitivity of mass spectrometers as well as the speed and resolving power of UPLC systems have enabled rapid identification of novel antibiotics.3–12 Combining these new tools with the bacterial diversity found in underexplored ecological niches such as insect symbionts,13 thermal vents,14 caves,15 coal mines,16 and marine ecosystems17 has demonstrated great promise for discovering the next generation of antibiotic therapies.
To address the challenges of resistant infectious disease, the potential of these genetic and analytical methods must be fully realized. Our lab has focused on LCMS-based metabolomics as a discovery platform to discover novel natural products from marine invertebrate-associated bacteria. This has led to a high discovery rate of novel natural products.8–11 As these methods pave the way for rapid discovery, structure determination becomes a limiting factor for providing valuable lead structures. Therefore, we have developed methods such as isotopic labeling followed by 13C–13C gCOSY11,18,19 for rapid assembly of carbon backbones in polyketides and 13C–15N NMR11 to assist with thiazolines and other heterocyclic systems. While our previous studies demonstrated a high level of 13C enrichment could be achieved for molecules of acetate origin, questions remained surrounding isotopically labeling peptides. For example, typical growth media used were rich in natural abundance precursors such as amino acids from sources like yeast extract and peptone. On the other hand, if uniform labeling could be achieved for both 13C and 15N, structure determination of peptide-based natural products could be achieved using biomolecular NMR strategies such as 3D NMR techniques. These approaches would also provide a gateway to automated backbone assignment of structures containing typical amino acids. Therefore, we evaluated isotopic labeling in a medium that contained typical quantities of nonenriched components. To demonstrate proof of concept for both isotopic labeling and 3D NMR strategies, we tested the methods using two novel antibiotic peptides, one of which was prioritized as part of an antibiotic drug discovery program. In particular, peptide 2 showed broad spectrum activity including against Gram-negative pathogens. Although peptide 2 did not show high potency, the difference between the MIC (22.7 μM) and MBC (45.5 μM) was small indicating potential for Gram-negative infections. The overall goal of this study was to provide a basis for reducing structure determination times from months to hours or days.
RESULTS AND DISCUSSION
Uniform Labeling Peptides with 13C and 15N
Our previous studies showed that fermentation of actinobacteria upon addition of uniformly 13C-labeled glucose and subsequent purification yielded 13C-labeled natural products.19 However, peptides could be more challenging since in most cases U–13C glucose was utilized for 13C enrichment in natural products which have been biosynthesized via a polyketide pathway.11,20 For the 15N incorporation, 15NH4Cl was evaluated as the nitrogen source. As mentioned, a potential issue was 15N incorporation with peptides in rich media. To evaluate the 13C and 15N incorporation, a marine invertebrate-derived Streptomyces sp. (strain WMMB 705) that produced two novel peptides that we named eudistamides A (1) and B (2) was grown in multiple media and extracts were analyzed by LC–MS data (Supporting Information). Although the medium containing 15NH4Cl as the only nitrogen source (M3, Supporting Information) yielded the highest 15N abundance of the peptides, taking into consideration the various factors such as the price of the medium, the yield, and the 13C and 15N abundance, the labeled medium containing peptone and yeast extract (20 g soluble starch, 10 g [U–13C] glucose, 2.5 g peptone, 2.5 g yeast extract, 5 g 15NH4Cl, 5 g CaCO3/L of artificial seawater) was used for producing 13C- and 15N-labeled peptides.
Strategies and Experiments for the Resonance Assignment of Uniformly 13C, 15N-Labeled Peptides
Backbone Assignment Strategy
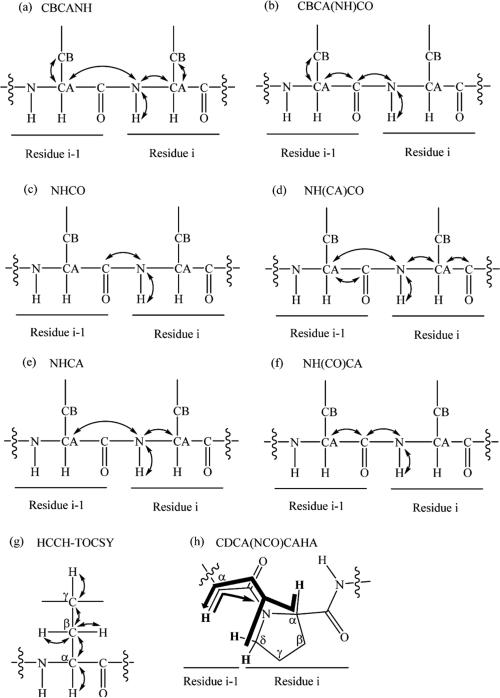
A typical triple resonance 3D NMR spectrum observes correlations of three resonances, a proton, a carbon, and a nitrogen. Many 3D NMR experiments have been designed for assigning the backbone of 15N/13C isotopically labeled protein, such as HNCO,21 NH(CA)CO,22 HNCA,21 CBCANH,23 CBCA(CO)NH,24 HN(CO)CA,25 HCACO,21 HCA(CO)N,21 H(CCO)NH,26 C(CO)NH,26 and HBHA(CBCACO)NH.27 Ideally, standard triple resonance backbone assignment of a peptide has relied on a combination of the CBCANH, CBCA(CO)NH, and NHCO spectra (Figure 1a–c). The general idea was that the CBCANH correlated each NH group with the Cα and Cβ chemical shifts of its own residue (strongly) and of the preceding residue (weakly), whereas the CBCA(CO)NH only correlates the NH group to the preceding Cα and Cβ chemical shifts. The NHCO experiment provides the connectives between each NH group with the carbonyl carbon of the preceding residue, thus establishing the backbone assignment. In our experience, the quality of the CBCANH and CBCA(CO)NH spectra was sometimes not sufficient in terms of S/N. The Cα or Cβ resonances were, for example, not visible above the noise level. In this case, the combination of HNCA and HN(CO)CA experiments (Figure 1e–f) which were more sensitive provided the same information as the CBCANH and CBCA(CO)NH spectra, except without the Cβ resonances. The HN(CA)CO experiment (Figure 1d) was powerful for linking each NH group with the carbonyl carbon of the same residue, particularly helpful to the assignment of depsipeptides. A depsipeptide is a peptide in which one or more of its –C(O)NHR groups are replaced by the corresponding ester, –C(O)OR. Taking eudistamides A (1) and B (2) for example, no correlation for C-1 could be seen in NHCO spectrum, whereas HN(CA)CO experiment could correlate NH-7 with the carbonyl carbon of the ester (C-1). Therefore, our backbone assignment strategy used three 3D NMR experiments, HNCO, CBCANH, and CBCA(CO)NH, to establish the connectivity of the backbone as the first step. The other 3D NMR experiments, NHCA, HN(CO)CA, NH(CA)CO, were utilized to make further confirmation regarding to some complicated cases, such as overlapping carbonyl carbon signals and any crosspeak that was very weak or missing in either CBCANH or CBCA(CO)NH spectrum. We were thereby able to make unambiguous backbone assignment of the eudistamides A and B and provided a general strategy for non-Pro containing peptide natural products. However, due to the absence of an amide proton in proline, these 3D NMR experiments cannot be used to sequentially connect proline residue to the preceding residue. The CDCA(NCO)CAHA (Figure 1h) was designed for the sequential assignment of proline residues,28 and the other option is to rely on the observation of NOEs to neighboring residues.
Figure 1.
Connectives observed in seven different types of 3D NMR experiment. (a) The CBCANH experiment correlates the chemical shift of amide resonance with the Cα and Cβ of both inter- and intraresidue. (b) The CBCA(CO)NH experiment only correlates the amide resonance with Cα and Cβ of the preceding residue. (c) The NHCO experiment provides the connectives between the amide of a residue (NHi) with the carbonyl carbon of the preceding residue (COi–1). (d) The NH(CA)CO experiment shows the amide resonance (NHi) is correlated with the carbonyl carbon of the same residue (COi), as well as that of the preceding residue (COi–1). (e) The NHCA experiment correlates the amide resonance (NHi) with the Cα of the same residue and preceding residue. (f) The NH(CO)CA experiment only correlates the resonances of the amide with the Cα of the preceding residue. (g) The HCCH-TOCSY correlates a 1H/13C pair to all other protons in the same aliphatic side chain. (h) The thick arrow represents pathways utilizing the 1J (Cα/δ, N) coupling, whereas the thin arrow represents the less efficient pathway via 2J (Cα, N). Protons directly involved in the transfer pathways are shown in bold font.
Side Chain Assignment Strategy
Depending on the structure, side chains can be assigned using TOCSY and HSQC. However, the most useful NMR experiment for side chain assignment is an HCCH-TOCSY spectrum (Figure 1g).29 The general principle behind using the HCCH-TOCSY spectrum is as follows: Using the known Cα and Cβ chemical shifts from the backbone assignment, the side chain chemical shifts would be assigned by viewing 2D slices at each carbon shift, typically at the Cα and Cβ chemical shifts. Each “slice” contains resonances for the protons attached to carbons in that 13C–13C spin system. The HCCH-TOCSY experiment combined with 13C chemical shift ranges of the common amino acids provides a very powerful means for the assignment of the aliphatic side chain. Some of the proteinogenic amino acids, such as alanine, serine, threonine and glycine, were easily identified since alanine, serine, and threonine's Cβ chemical shifts were unique compared to those of the other amino acids and glycine has no Cβ. Valine, isoleucine, and proline could also be easily identified because they have lower than normal Cα chemical shifts. For the aromatic side chain assignment, such as tyrosine, phenylalanine, tryptophan, and histidine, additional 2D NMR data are needed to complete the whole side chain assignment.
Structure Elucidation of Eudistamides A (1) and B (2)
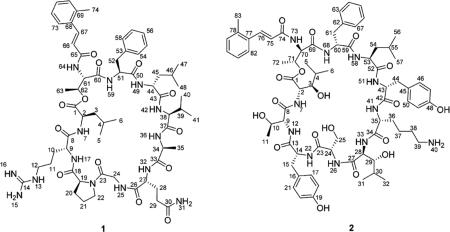
Eudistamide A (1) was obtained as a white powder with a molecular formula of C61H90N14O13, determined by HRESIMS. Interpretation of HCCH-TOCSY data (Table S1; Supporting Information) of the 13C- and 15N-doubly labeled 1 established the amino acid residues leucine (Leu; 2×), arginine (Arg), proline (Pro), glycine (Gly), glutamine (Gln), alanine (Ala), valine (Val), and phenylalanine (Phe), which were further confirmed by natural abundance experiments, 1H–1H COSY, HSQC and HMBC data (Table S3; Supporting Information). The Pro residue was assigned as cis based on the 13C NMR chemical shifts of the β- and γ-carbon atoms (Δδβγ). Δδβγ for trans-Pro is regularly less than 5 ppm, while Δδβγ for cis-Pro is regularly between 5 and 10 ppm.30 Δδβγ for the proline residue in 1 was 5.8 ppm, supporting the assignment as cis. The 3-(2-methylphenyl)acrylic acid (Me-acyl) moiety was established by HMBC, whereas geometry of the double bond (C66 and C67) was assigned as E on the basis of large coupling constant (15.5 Hz) through the double bond.
The connectivity of the partial structures Thr-Phe-Leu2-Val-Ala-Gln-Gly and Pro-Arg-Leu1 was established by using 3D NMR data which were discussed for the backbone assignment (CBCANH, CBCACONH, NHCO, HNCA, NH(CO)CA, NH(CA)CO; Table S1; Supporting Information) of the 13C- and 15N-doubly labeled 1. The HMBC correlation from H-62 to C-1 and the ROESY correlation between H3-63 and H-2 connected Leu1 to Thr via an ester on the basis of the chemical shift of C-62 (δc 68.5). Moreover, for the uncommon amino acid residue, the HMBC correlations from H-66 and H-67 to C-65 supported the linkage of Me-acyl moiety to Thr. The ROESY correlations between H-24a and H2-22 secure the ring closure by connecting the Gly unit to Pro, which satisfied the 24 degrees of unsaturation deduced from the molecular formula.
The advanced Marfey's method31 was applied to assign the absolute configurations of the amino acid residues from acid hydrolysis of 1. The 1-fluoro-2,4-dinitrophenyl-5-leucine-amide (FDLA) derivatives of the hydrolysate of 1 and authentic d- and l-amino acids were subjected to LC–MS analysis. The absolute configuration of all amino acids was established by comparison of their HPLC retention time and molecular weights with those of corresponding authentic d- and l-standards. Upon analysis, the amino acid residues Arg, Pro, Ala, Val, allo-Thr, and Phe were determined to have the l-configuration, whereas the Gln and Leu were deduced to have the d-configuration (Table S5; Supporting Information).
Eudistamide B (2) was isolated a white amorphous powder with a molecular formula of C72H99N11O18 as determined by HRESIMS. A detailed analysis of HCCH-TOCSY (Table S2; Supporting Information) of the 13C- and 15N-doubly labeled 2 indicated an amino acid content as follows: Thr (×2), Tyr (×2), Ser, Lys, 3-OH-Leu (×2), Leu, and Phe. CBCANH, CBCA(CO)NH NHCO, HNCA, NH(CO)CA, and NH(CA)-CO spectra (Table S2; Supporting Information) revealed the amino acids sequence as 3-OH-Leu1-Thr1-Tyr1-Ser-3-OH-Leu2-Lys-Tyr2-Leu-Phe-Thr2, which was further confirmed by ESIMS/MS fragmentation analysis (Figure S22; Supporting Information). The linkage of Me-acyl moiety to Thr2 was supported by the HMBC correlation from H-76 to C-74. The ROESY correlation between H3-72 and H-2 and consideration of unsaturation deduced from the molecular formula connected 3-OH-Leu1 to Thr2 via an ester on the basis of the chemical shift of C-71 (δc 68.5).
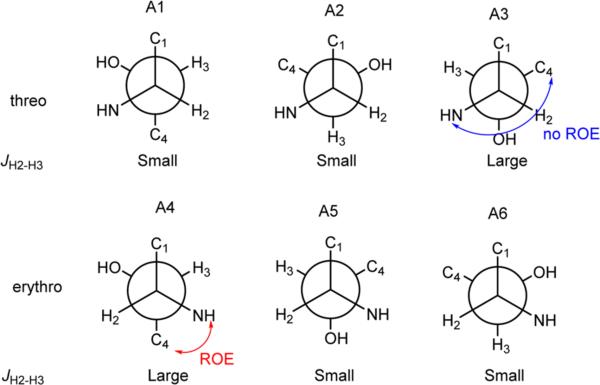
The absolute configurations of the amino acids were determined using acid hydrolysis followed by the advanced Marfey's method. The chromatographic comparison between Marfey's derivatives of the hydrolysate of 2 and appropriate amino acid standards assigned the l-configurations for Ser and Leu and d-configurations for Lys, Phe, allo-Thr, and Tyr (Table S4; Supporting Information). The (2R, 3S) and (2R, 3R)-3-hydroxyleucine were synthesized following Bonnard's procedure32 and derivatized with l- and l/d-FDLA respectively. Since enantiomers would exhibit identical retention behavior under nonchiral HPLC conditions, the (2R, 3S)-d-FDLA derivative showed the same retention time as (2S, 3R)-l-FDLA, whereas the (2R, 3R)-d-FDLA derivative showed the same retention time as (2S, 3S)-l-FDLA. By comparing the above the four retention times of the 3-OH-Leu standards with the l-FDLA derivatives of the hydrolysate of 2, both the 3-OH-Leu in 2 were assigned as 2S, 3R (Table S6; Supporting Information). The 2S, 3R configuration of the two 3-hydroxyleucines in 2 were also confirmed by J-based analysis and NOESY data. The large coupling constants observed from DQF-COSY in MeOD between H-2 and H-3 (11.5 Hz), and between H-28 and H-29 (11.0 Hz) established their anti-relationship. The ROESY correlation between H-4 and NH-7, as well as the ROESY correlation between H-30 and NH-33, established the rotamer (A4) in Figure 2.
Figure 2.
J-based configuration analysis of C-2 and C-3 of compound 2.
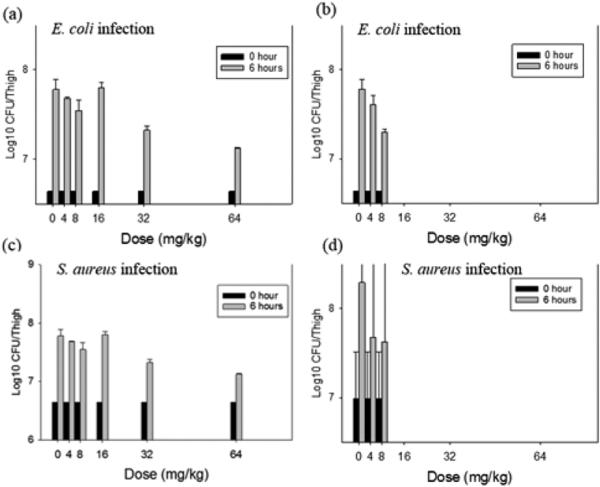
Compounds 1 and 2 were tested for antibacterial activity against Pseudomonas aeruginosa, Methicillin-resistant Staphylococcus aureus (MRSA), Escherichia coli, and Bacillus subtilis. Compound 2 showed antibacterial activity against MRSA, E. coli and B. subtilis, with the MIC values of 22.7, 22.7, and 2.8 μM, respectively, while compound 1 did not show any detectable antibacterial activities at 52.2 μM. Compound 2 was also tested using a murine thigh model infected with either E. coli ATCC 25922 or S. aureus ATCC 25923 (Figure 3). Over the 16-fold intraperitoneal dose range, no toxicity associated with compound administration was observed (Figure 3a,c). Conversely, intravenous dose levels above 8 mg/kg resulted in animal death (Figure 3b,d). Modest, but broad spectrum (both Gram-positive and Gram-negative bacteria) efficacy was observed with two highest tolerated dose levels given by both administration routes. Compared to untreated control animals, compound 2 reduced the organism burden in the thighs of immunocompromised mice by more than 0.5 log10 cfu/thigh.
Figure 3.
In vivo time kill experiment with compound 2 using a neutropenic mouse thigh model. Each symbol represents the mean and standard deviation from four thighs of two mice infected with either E. coli or S. aureus. The error bars represent the standard deviation. Five single doses (4, 8, 16, 32, and 64 mg/kg) of compound 2 were administered to mice. (a) E. coli infected murine thighs treated with five single doses of 2 by intraperitoneal route. (b) E. coli infected murine thighs treated with five single doses of 2 by intravenous route. (c) S. aureus infected murine thighs treated with five single doses of 2 by intraperitoneal route. (d) S. aureus infected murine thighs treated with five single doses of 2 by intravenous route. The (b) and (d) regimens do not include the 16, 32, and 64 mg/kg since the mice died following administration.
Although the cyclopetides and cyclodepsipeptides have been encountered frequently as bacterial secondary metabolites, eudistamides A (1) and B (2) differ markedly from the known peptides by virtue of the presence of a unique 3-(2-methylphenyl)acrylic acid moiety, which has not been previous reported in a peptide. Only some analogues of 3-(2-methylphenyl)acrylic acid residue were rarely descried in peptides, such as WS9326s,33 pepticinnamins,34 and mohangamides35 incorporating a 2-pentenyl cinnamic acid moiety, skyllamycins A and B36,37 incorporating a 2-propenyl]-cinnamic acid moiety, and corprisamides A and B bearing 2-heptatrienyl cinnamic acid moiety.38 Moreover, another remarkable feature of compound 2 is the high content of the β-hydroxy-α-amino acids, including genetically coded proteinogenic amino acids Ser and Thr, and nonstandard amino residue 3-OH-Leu. Members of the β-hydroxy-α-amino acid class occur as constituents of many important antibiotics, such as vancomycin39 and lysobacin.40 Furthermore, compounds 1 and 2 could be produced in sufficient quantities by scale-up microbial fermentation, which would greatly facilitate follow-up mechanistic and preclinical studies.
CONCLUSION
The introduction of 3D NMR has dramatically improved the speed and reliability of the protein assignment process.22,41 However, we expanded the scope of 3D NMR experiments from protein applications to a strategy that is universally applicable for labeled peptides. We have demonstrated a rapid and efficient structure elucidation protocol for complex peptides containing typical amino acids. The protocol we propose consists of a typical set of 3D NMR experiments for backbone and side chain assignment of uniformly labeled peptides with 13C and 15N. While 3D NMR experiments may not be optimal for natural product peptides, this proof of concept provides a basis for using triple resonance strategies for rapidly establishing the backbone of peptide natural products. Development of specific NMR experiments would be advantageous. The triple resonance experiments used in this study were highly complementary to more traditional methods such as HMBC and ROESY. This report describes the first application of 3D NMR to two microbially produced peptide natural products with novel structures. The methods are cost-effective and greatly improve the confidence in a proposed structure.
EXPERIMENTAL SECTION
General Experimental Procedures
Optical rotations were measured on a Polarimeter. UV spectra were recorded on a UV–vis Spectrophotometer. IR spectra were measured with a FT–IR Spectrophotometer. 1D and 2D NMR spectra were obtained in DMSO with a NMR spectrometer equipped with a 13C/15N{1H} cryoprobe. 3D NMR spectra were obtained in DMSO with a NMR spectrometer with a (1H{13C,15N}) cold probe. HRMS data were acquired with a QTOF mass spectrometer (Ionization method: Spray Voltage (+) 4000; Spray Voltage (−) 3700; Capillary temperature 320; full MS scan 200–2000). MSMS data were acquired with a hybrid quadrupole-Orbitrap mass spectrometer. RP HPLC was performed using a HPLC system and a Phenomenex Luna C18 column (250 × 10 mm, 5 μm), as well as a preparative HPLC and Phenomenex Gemini C18 column (250 × 30 mm, 5 μm). The Advanced Marfey's method utilized a HPLC coupled with a mass spectrometer.
Biological Material
Ascidian specimens were collected in September 2011 from the Florida Keys (24°33.416′, 81°21.611′). Identification was confirmed by Shirley Parker-Nance. A voucher specimen for Eudistoma olivaceum (Van Name, 1902)42 is housed at the University of Wisconsin—Madison. For cultivation, a sample of ascidian (1 cm3) was rinsed with sterile seawater and macerated using a sterile pestle in a microcentrifuge tube, and dilutions were made in sterile seawater, with vortexing between steps to separate bacteria from heavier tissues. Dilutions were separately plated on three media: ISP2 supplemented with artificial seawater,43 R2A,44 and M4.45 Each medium was supplemented with 50 μg/mL cycloheximide and 25 μg/mL nalidixic acid. Plates were incubated at 28 °C for at least 28 days, and strain WMMB 705 was purified from an ISP2 isolation plate.
Sequencing
Genomic DNA was extracted using the UltraClean Microbial DNA Isolation kit (Mo Bio Laboratories, Inc.). 16S rDNA genes were amplified using 100–200 ng genomic DNA template with the primers 8–27F (5′ to 3′ GAGTTTGATCCTGGCTCAG) and 1492R (5′ to 3′ GGTTACCTTGTTACGACTT). The following PCR conditions were used: 94 °C for 5 min, followed by 30 cycles of 94 °C for 30 s, 55 °C for 1 min, 72 °C for 1.5 min, with a final step of 72 °C for 5 min. The PCR bands were excised from the gel and purified using the QIAquick Gel Extraction kit (QIAGEN). One microliter of purified product was sequenced. Sequencing reactions were performed by the UW Biotechnology Center and reactions were sequenced with an ABI 3730xl DNA Analyzer. WMMB 705 were identified as Streptomyces sp. by 16S sequencing, and demonstrated 99% sequence similarity to Streptomyces sp. FXJ6.141 (accession number GU 339421).
Fermentation, Extraction, and Isolation
Two 10 mL of seed cultures (25 × 150 mm tubes) in medium ASW-A (20 g soluble starch, 10 g glucose, 5 g peptone, 5 g yeast extract, 5 g CaCO3 per liter of artificial seawater) were inoculated with strain WMMB 705 and shaken (200 rpm, 28 °C) for 4 days. Two-liter flasks (2 × 500 mL) containing ASW-A medium with Diaion HP20 (7% by weight) were inoculated with 8 mL from the culture tube and shaken at 200 rpm and 28 °C for 7 days. Filtered HP20 was washed with water and extracted with acetone. The acetone extract was chromatographed on Diaion HP20ss, eluting sequentially with methanol/water (10:90 and 95:5). The 95% methanol eluate was evaporated to dryness to afford a yellow oil (460 mg), and then it was subjected to RP HPLC (30%/70% to 60%/40% ACN/H2O with H2O containing 0.1% acetic acid over 30 min, 25 mL/min, followed by 90/10% to 100% of the same solvents for 3 min, 25 mL/min, and a hold at 100%/0% of the same solvents) using a Phenomenex Gemini C18 column (250 × 30 mm, 5 μm), yielding a fraction containing 1 (50 mg, tR 11.5 min), and compound 2 (21 mg, tR 12.5 min). The fraction containing 1 was further subjected to RP HPLC (55–65% MeOH–H2O with H2O containing 0.1% acetic acid over 25 min, 4.0 mg/mL) using a Phenomenex Luna C18 column (250 × 10 mm, 5 μm), yielding 1 (22 mg, tR 19 min). For 13C and 15N incorporation, the same procedure was used (1 × 250 mL) with labeled medium ASW-A (20 g soluble starch, 10 g U13C-glucose, 2.5 g peptone, 2.5 g yeast extract, 5 g 15NH4Cl, 5 g CaCO3 per liter of artificial seawater).
Antibacterial Assay
Eudistamides A (1) and B (2) were tested for antibacterial activity against MRSA (ATCC #33591), E. coli (ATCC #25922), B. subtilis, and P. aeruginosa (ATCC #27853), and MICs were determined using a dilution antimicrobial susceptibility test for aerobic bacteria.46 Eudistamides A (1) and B (2) were dissolved in DMSO, serially diluted to 10 concentrations (0.125–64 μg/mL), and tested in a 96-well plate. Vancomycin was used as a control and exhibited an MIC of 0.69 μmol against MRSA and 0.69 μmol against B. Subtilis. Gentamicin was used as a control and exhibited an MIC of 8.4 μmol against E. coli and 8.4 μmol against P. aeruginosa. Peptides 1 and 2 were tested in triplicate, and vancomycin and gentamicin were also tested in triplicate. Six untreated media controls were included on each plate. The plates were incubated at 35 °C for 18 h. The MIC was determined as the lowest concentration that inhibited visible growth of bacteria.
Eudistamide A (1)
White solid, [α]25d −51 (c 0.1, MeOH); UV (MeOH) λ (log ε) 209 (3.55), 224 (3.17), 280 (3.23); IR (ATR) vmax 3300, 2953, 1648, 1535, 1519, 1450, 1320, 1242, 1072, 760, 677 cm−1; 1H and 13C NMR (see Table S3; Supporting Information); HRMS [M + H]+ m/z 1227.6882 (calcd for C61H91N14O13, 1227.6890).
Eudistamide B (2)
White solid, [α]25d + 37 (c 0.1, MeOH); UV (MeOH) λ (log ε) 207 (3.32), 223 (3.17), 279 (2.82); IR (ATR) vmax 3290, 2932, 1738, 1647, 1532, 1517, 1455, 1314, 1244, 1072, 761, 669 cm−1; 1H and 13C NMR (see Table S4; Supporting Information); HRMS [M + H]+ m/z 1406.7253 (calcd for C72H100N11O18, 1406.7248).
Acid Hydrolysis of Eudistamides A (1) and B (2)
Separate solutions of compounds 1 and 2 (0.5 mg each) in 6 N HCl (1 mL) were hydrolyzed at 110 °C for 4 h and dried under vacuum.
Determination of Proteinogenic Amino Acid Configurations
l- and l/d-FDLA were synthesized as previously reported.47 The hydrolysate was mixed with 1 N NaHCO3 (40 μL), and 35 μL of l-FDLA (10 mg/mL in acetone). Each solution was stirred at 45 °C for 1 h, cooled to room temperature, quenched with 1 N HCl (40 μL), and dried under vacuum. Similarly, the standard l- and d-amino acid were derivatized separately. The derivatives of the hydrolysate of compound 1 and the standard amino acids were subjected to LC–MS analysis with a Phenomenex Kinetex C18 reversed-phase column (2.6 μm, 100 × 4.6 mm) at a flow rate of 0.5 mL/min and with a linear gradient of H2O (containing 0.1% formic acid) and MeOH (90:10 to 0:100 over 15 min, and a hold at 100% MeOH for 5 min). The absolute configurations of the amino acids were determined by comparing the retention times of the l- and d-amino acids derivatives, which were identified by MS. The absolute configurations of the amino acids in 1 were assigned based on a comparison of retention time of amino acid standards derivatized with l-FDLA. In a similar manner, the derivatives of the hydrolysate of compound 2 and the standard amino acids were subjected to LC–MS analysis with a Phenomenex Luna C18 reversed-phase column (5.0 μm, 250 × 4.6 mm) at a flow rate of 1.0 mL/min and with a linear gradient of H2O (containing 0.1% formic acid) and ACN (90:10 to 0:100 over 30 min, and a hold at 100% ACN for 5 min). The retention times and ESIMS data for l-FDLA derivatives of the hydrolysates and the standard amino acids are summarized in Tables S5 and S6 (Supporting Information).
Synthesis of (2R,3S)-3-Hydroxyleucine and (2R,3R)-3-Hydroxyleucine and Advanced Marfey's Analysis of 3-Hydroxyleucines
(2R,3S)-3-Hydroxyleucine and (2R,3R)-3-hydroxyleucine were synthesized using the procedures by Bonnard et al.32 We obtained 20.0 mg of major precursor (S)-1-((2R,5S)-5-isopropyl-3,6-dimethoxy-2,5-dihydropyrazin-2-yl)-2-methylpropan-1-ol and 2.5 mg of minor precursor (R)-1-((2R,5S)-5-isopropyl-3,6-dimethoxy-2,5-dihydropyrazin-2-yl)-2-methylpropan-1-ol. The hydroxyleucine precursors were hydrolyzed following the general peptide acid hydrolysis process. The major precursor gave a mixture of (2R,3S)-3-hydroxyleucine and d-valine, and the minor precursor gave a mixture of (2R,3R)-3-hydroxyleucine and d-valine. These two hydrolysate mixtures were derivatized with l-FDLA and l/d-FDLA, respectively, resulting in four products: (2R, 3S)-3-hydroxyleucine-l-FDLA, (2R, 3S)-3-hydroxyleucine-d-FDLA, (2R, 3R)-3-hydroxyleucine-l-FDLA, and (2R, 3R)-3-hydroxyleucine-d-FDLA. Among these four products, (2R, 3S)-3-hydroxyleucine-d-FDLA will show the same retention time as (2S, 3R)-3-hydroxyleucine-L-FDLA, and (2R, 3R)-3-hydroxyleucine-d-FDLA will show the same retention time as (2S, 3S)-3-hydroxyleucine-l-FDLA, under the nonchiral HPLC condition we used in advanced Marfey's analysis. The resulting four derivatization mixtures were subjected to LC–MS analysis with a Phenomenex Luna C18 reversed-phase column (5.0 μm, 250 × 4.6 mm) at a flow rate of 1.0 mL/min and with a linear gradient of H2O (containing 0.1% formic acid) and ACN (25:75 to 65:35 over 50 min). When these four retention times were compared with the retention time of hydrolysate-l-FDLA derivative of compound 2, the two 3-hydroxyleucines in the peptide were assigned as 2S, 3R. Detailed retention times of four isomers and hydrolysate could be found in Table S6 (Supporting Information). 1H NMR data of (S)-1-((2R,5S)-5-isopropyl-3,6-dimethoxy-2,5-dihydropyrazin-2-yl)-2-methylpropan-1-ol (CDCl3, 500 MHz): δ 4.09 (1H, m), 3.99 (1H, m), 3.73 (3H, s), 3.69 (3H, s), 3.63 (1H, br), 2.25 (1H, m), 2.01, (1H, m), 1.64 (1H, br), 1.04 (3H, d, J = 7.0 Hz), 1.03 (3H, d, J = 7.0 Hz), 0.99 (3H, d, J = 7.0 Hz), 0.71 (3H, d, J = 7.0 Hz). 1H NMR data of (R)-1-((2R,5S)-5-isopropyl-3,6-dimethoxy-2,5-dihydropyrazin-2-yl)-2-methylpropan-1-ol (CDCl3, 500 MHz): δ 4.10 (1H, t, J = 3.5 Hz), 3.92 (1H, t, J = 3.5 Hz), 3.65 (3H, s), 3.64 (3H, s), 3.59 (1H, br), 2.20 (1H, m), 1.76, (1H, m), 1.51 (1H, br), 0.98 (3H, d, J = 7.0 Hz), 0.85 (3H, d, J = 7.0 Hz), 0.84 (3H, d, J = 7.0 Hz), 0.63 (3H, d, J = 6.9 Hz).
Organisms, Media, and Antibiotic of in Vivo Studies
Eudistamide B (2) was tested for antibacterial activity in vivo studies. One isolate of E. coli ATCC 25922 and S. aureus ATCC 25923, separeatly, was used for these studies. Organisms were grown, subcultured, and quantified using Mueller-Hinton broth (MHB) and agar (Difco Laboratories, Detroit, MI).
Animals
Six week-old, specific pathogen-free, female ICR/Swiss mice weighing 24–27 g were used for all studies (Harlan Sprague–Dawley, Indianapolis, IN). Animals were maintained in accordance with the American Association for Accreditation of Laboratory Animal Care (AAALAC) criteria. All animal studies were approved by the Animal Research Committees of the William S. Middleton Memorial VA Hospital and the University of Wisconsin.
Murine Thigh Infection Model
Mice were rendered neutropenic (neutrophils <100/mm3) by injecting cyclophosphamide (Mead Johnson Pharmaceuticals, Evansville, IN) intraperitoneally 4 days (150 mg/kg) and 1 day (100 mg/kg) before thigh infection. Previous studies have shown that this regimen produces neutropenia in this model for 5 days. Broth cultures of freshly plated bacteria were grown to logarithmic phase overnight to an absorbance of 0.3 at 580 nm (Spectronic 88; Bausch and Lomb, Rochester, NY). After a 1:10 dilution into fresh MHB, bacterial counts of the inoculum ranged from 106.3 to 6.9 cfu/mL. Thigh infections with each of the isolates were produced by injection of 0.1 mL of inoculum into the thighs of isoflorane-anesthetized mice 2 h before therapy.
In Vivo Time Kill and PAE
Two hours after thigh infection with either single doses of compound 2 (4, 8, 16, 32, and 64 mg/kg) by the intraperitoneal or intravenous route, groups of two treated and untreated mice were sacrificed at each treatment end point (0 h at the start of therapy and 6 h after treatment). The thighs (four per treatment group) were immediately removed upon euthanasia and processed for cfu determination. The burden of organisms in the thigh was measured by viable plate counts of tissue homogenates. The impact of each dose on the burden of organisms was measure over time of the study period.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by funding from the University of Wisconsin—Madison School of Pharmacy. This work was also funded in part by the U19 AI109673, NIH, and NIGMS Grant R01 GM107557. We thank the Analytical Instrumentation Center (AIC) at the University of Wisconsin—Madison School of Pharmacy for the facilities to acquire spectroscopic data, especially MS data. This study made use of the National Magnetic Resonance Facility at Madison (NMRFAM), which is supported by NIH grants P41RR02301 (BRTP/NCRR) and P41GM66326 (NIGMS).
Footnotes
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.joc.5b01486.
NMR data, MS data, and Marfey's analysis for compounds 1 and 2. (PDF)
The authors declare no competing financial interest.
REFERENCES
- 1.Livermore DM. Clin. Microbiol. Infect. 2004;10:1–9. doi: 10.1111/j.1465-0691.2004.1004.x. [DOI] [PubMed] [Google Scholar]
- 2.a Lautru S, Deeth RJ, Bailey LM, Challis GL. Nat. Chem. Biol. 2005;1:265–269. doi: 10.1038/nchembio731. [DOI] [PubMed] [Google Scholar]; b Bentley SD, et al. Nature. 2002;417:141–147. doi: 10.1038/417141a. [DOI] [PubMed] [Google Scholar]
- 3.Doroghazi JR, Albright JC, Goering AW, Ju K, Haines RR, Tchalukov KA, Labeda DP, Kelleher NL, Metcalf WM. Nat. Chem. Biol. 2014;10:963–968. doi: 10.1038/nchembio.1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Forner D, Berrué F, Correa H, Duncan K, Kerr RG. Anal. Chim. Acta. 2013;805:70–79. doi: 10.1016/j.aca.2013.10.029. [DOI] [PubMed] [Google Scholar]
- 5.Gill KA, Berrué F, Arens JC, Kerr RG. J. Nat. Prod. 2014;77:1372–1376. doi: 10.1021/np500122s. [DOI] [PubMed] [Google Scholar]
- 6.Hoffmann T, Krug D, Hüttel S, Müller R. Anal. Chem. 2014;86:10780–10788. doi: 10.1021/ac502805w. [DOI] [PubMed] [Google Scholar]
- 7.Nguyen DD, Wu C, Moree WJ, Lamsa AL, Medema MH, Zhao X, Gavilan RG, Aparicio M, Atencio L, Jackson C, Ballesteros J, Sanchez J, Watrous JD, Phelan VV, van de Wiel C, Kersten RD, Mehnaz S, Mot RD, Shank EA, Charusanti P, Nagarajan H, Duggan BM, Moore BS, Bandeira N, Palsson BØ, Pogliano K, Gutiérrez M, Dorrestein PC. Proc. Natl. Acad. Sci. U. S. A. 2013;110:E2611–E2620. doi: 10.1073/pnas.1303471110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hou Y, Braun DR, Michel CR, Klassen JL, Adnani N, Wyche TP, Bugni TS. Anal. Chem. 2012;84:4277–4283. doi: 10.1021/ac202623g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wyche TP, Hou Y, Vazquez-Rivera E, Braun D, Bugni TS. J. Nat. Prod. 2012;75:735–740. doi: 10.1021/np300016r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hou Y, Tianero DB, Kwan JC, Wyche TP, Michel CR, Ellis GA, Vazquez-Rivera E, Braun DR, Rose WE, Schmidt EW, Bugni TS. Org. Lett. 2012;14:5050–5053. doi: 10.1021/ol3022758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wyche TP, Piotrowski JS, Hou Y, Braun D, Deshpande R, McIlwain S, Ong IM, Myers CL, Guzei IA, Westler WM, Andes DR, Bugni TS. Angew. Chem., Int. Ed. 2014;53:11583–11586. doi: 10.1002/anie.201405990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Samat N, Tan PJ, Shaari K, Abas F, Lee HB. Anal. Chem. 2014;86:1324–1331. doi: 10.1021/ac403709a. [DOI] [PubMed] [Google Scholar]
- 13.a Kikuchi Y. Microbes Environ. 2009;24:195–204. doi: 10.1264/jsme2.me09140s. [DOI] [PubMed] [Google Scholar]; b Poulsen M, Oh D, Clardy J, Currie CR. PLoS One. 2011;6:e16763. doi: 10.1371/journal.pone.0016763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.a Sogin ML, Morrison HG, Huber JA, Welch DM, Huse SM, Neal PR, Arrieta JM, Herndl GJ. Proc. Natl. Acad. Sci. U. S. A. 2006;103:12115–12120. doi: 10.1073/pnas.0605127103. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Andrianasolo EH, Haramaty L, Rosario-Passapera R, Bidle K, White E, Vetriani C, Falkowski P, Lutz R. J. Nat. Prod. 2009;72:1216–1219. doi: 10.1021/np800726d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.a Schabereiter-Gurtner C, Saiz-Jimenez C, Pinar G, Lubitz W, Rölleke S. FEMS Microbiol. Ecol. 2003;1606:1–13. doi: 10.1016/S0168-6496(03)00280-0. [DOI] [PubMed] [Google Scholar]; b Engel AS, Porter ML, Stern LA, Quinlan S, Bennett PC. FEMS Microbiol. Ecol. 2004;51:31–53. doi: 10.1016/j.femsec.2004.07.004. [DOI] [PubMed] [Google Scholar]
- 16.a Wang X, Shaaban KA, Elshahawi SI, Ponomareva LV, Sunkara M, Zhang Y, Copley GC, Hower JC, Morris AJ, Kharel MK, Thorson JS. J. Nat. Prod. 2013;76:1441–1447. doi: 10.1021/np400231r. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Shaaban KA, Wang X, Elshahawi SI, Ponomareva LV, Sunkara M, Copley GC, Hower JC, Morris AJ, Kharel MK, Thorson JS. J. Nat. Prod. 2013;76:1619–1626. doi: 10.1021/np400308w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.a Lam KS. Curr. Opin. Microbiol. 2006;9:245–251. doi: 10.1016/j.mib.2006.03.004. [DOI] [PubMed] [Google Scholar]; b Abdelmohsen UR, Bayer K, Hentschel U. Nat. Prod. Rep. 2014;31:381–399. doi: 10.1039/c3np70111e. [DOI] [PubMed] [Google Scholar]
- 18.Reibarkh M, Wyche TP, Bugni TS, Martin GE, Williamson RT. Structure elucidation of uniformly 13C labeled small molecule natural products. Magn. Reson. Chem. 2015 doi: 10.1002/mrc.4384. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ellis GA, Wyche TP, Fry CG, Braun DR, Bugni TS. Mar. Drugs. 2014;12:1013–1022. doi: 10.3390/md12021013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.a Fellermeier M, Eisenreich W, Bacher A, Zenk MH. Eur. J. Biochem. 2001;268:1596–1604. doi: 10.1046/j.1432-1033.2001.02030.x. [DOI] [PubMed] [Google Scholar]; b Watanabe H, Tetsuo T, Oikawa H. Tetrahedron Lett. 2006;47:1399–1402. [Google Scholar]; c Kwon Y, Park S, Shin J. Arch. Pharmacal Res. 2014;37:967–971. doi: 10.1007/s12272-013-0254-8. [DOI] [PubMed] [Google Scholar]
- 21.Kay LE, Ikura M, Tschudin R, Bax AJ. Magn. Reson. 1990;89:496–514. doi: 10.1016/j.jmr.2011.09.004. [DOI] [PubMed] [Google Scholar]
- 22.Clubb RT, Thanabal V, Wagner GJ. Magn. Reson. 1992;97:213–217. [Google Scholar]
- 23.Grzesiek S, Bax AJ. Magn. Reson. 1992;99:201–207. [Google Scholar]
- 24.Grzesiek S, Bax AJ. Am. Chem. Soc. 1992;114:6291–6293. [Google Scholar]
- 25.Bax A, Ikura MJ. Biomol. NMR. 1991;1:99–104. doi: 10.1007/BF01874573. [DOI] [PubMed] [Google Scholar]
- 26.Grzesiek S, Anglister J, Bax AJ. Magn. Reson., Ser. B. 1993;101:114–119. [Google Scholar]
- 27.Grzesiek SJ, Bax AJ. Biomol. NMR. 1993;3:185–204. doi: 10.1007/BF00178261. [DOI] [PubMed] [Google Scholar]
- 28.Bottomley MJ, Macias MJ, Liu Z, Sattler MJ. Biomol. NMR. 1999;13:381–385. doi: 10.1023/a:1008393903034. [DOI] [PubMed] [Google Scholar]
- 29.Bax A, Clore M, Gronenborn AM. J. Magn. Reson. 1990;88:425–431. [Google Scholar]
- 30.Siemion IZ, Wieland T, Pook KH. Angew. Chem., Int. Ed. Engl. 1975;14:702–703. doi: 10.1002/anie.197507021. [DOI] [PubMed] [Google Scholar]
- 31.a Harada K, Fujii K, Mayumi T, Hibino Y, Suzuki M. Tetrahedron Lett. 1995;36:1515–1518. [Google Scholar]; b Harada K, Fujii K, Hayashi K, Suzuki MM. Tetrahedron Lett. 1996;37:3001–3004. [Google Scholar]; c Fujii K, Ikai Y, Oka H, Suzuki M, Harada K. Anal. Chem. 1997;69:5146–5151. [Google Scholar]
- 32.Bonnard I, Rolland M, Salmon J, Debiton E, Bartheomeuf C, Banaigs BJ. Med. Chem. 2007;50:1266–1279. doi: 10.1021/jm061307x. [DOI] [PubMed] [Google Scholar]
- 33.a Hayashi K, Hashimoto M, Shigematsu N, Nishikawa M, Ezaki M, Yamashita M, Kiyoto S, Okuhara M, Kohsaka M, Imanaka H. J. Antibiot. 1992;45:1055–1063. doi: 10.7164/antibiotics.45.1055. [DOI] [PubMed] [Google Scholar]; b Yu Z, Vodanovic-Jankovic S, Kron M, Shen B. Org. Lett. 2012;14:4946–4949. doi: 10.1021/ol302298k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shiomi K, Yang H, Inokoshi J, Van der Pyl D, Nakagawa A, Takeshima H, Omura S. J. Antibiot. 1993;46:229–234. doi: 10.7164/antibiotics.46.229. [DOI] [PubMed] [Google Scholar]
- 35.Bae M, Kim H, Moon K, Nam S, Shin J, Oh K, Oh D. Org. Lett. 2015;17:712–715. doi: 10.1021/ol5037248. [DOI] [PubMed] [Google Scholar]
- 36.Toki S, Agatsuma T, Ochiai K, Saitoh Y, Ando K, Nakanishi S, Lokker NA, Giese N, Matsuda YJ. Antibiot. 2001;54:405–414. doi: 10.7164/antibiotics.54.405. [DOI] [PubMed] [Google Scholar]
- 37.Pohle S, Appelt C, Roux M, Fieldler H, Süssmuth RD. J. Am. Chem. Soc. 2011;133:6194–6205. doi: 10.1021/ja108971p. [DOI] [PubMed] [Google Scholar]
- 38.Um S, Park SH, Kim J, Park HJ, Ko K, Bang H, Lee SK, Shin J, Oh D. Org. Lett. 2015;17:1272–1275. doi: 10.1021/acs.orglett.5b00249. [DOI] [PubMed] [Google Scholar]
- 39.a Wohlleben W, Stegmann E, Süssmuth RD. Methods Enzymol. 2009;458:459–486. doi: 10.1016/S0076-6879(09)04818-6. [DOI] [PubMed] [Google Scholar]; b Gunasekera SP, Ritson-Williams R, Paul VJ. J. Nat. Prod. 2008;71:2060–2063. doi: 10.1021/np800453t. [DOI] [PubMed] [Google Scholar]; c Cai G, Napolitano JG, McAlpine JB, Wang Y, Jaki BU, Suh J, Yang SH, Lee I, Franzblau SG, Pauli GF, Cho S. J. Nat. Prod. 2013;76:2009–2018. doi: 10.1021/np400145u. [DOI] [PubMed] [Google Scholar]; d Hou Y, Tianero DB, Kwan JC, Wyche TP, Michel CR, Ellis GA, Vazquez-Rivera E, Braun DR, Rose WE, Schmidt EW, Bugni TS. Org. Lett. 2012;14:5050–5053. doi: 10.1021/ol3022758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.O'Sullivan J, McCullough JE, Tymiak AA, Kirsch DR, Trejo WH, Principe PA. J. Antibiot. 1988;41:1740–1744. doi: 10.7164/antibiotics.41.1740. [DOI] [PubMed] [Google Scholar]
- 41.a Salzmann M, Pervushin K, Wider G, Senn H, Wüthrich K. J. Am. Chem. Soc. 2000;122:7543–7548. [Google Scholar]; b Liang B, Tamm LK. Proc. Natl. Acad. Sci. U. S. A. 2007;104:16140–16145. doi: 10.1073/pnas.0705466104. [DOI] [PMC free article] [PubMed] [Google Scholar]; c Sakakibara D, Sasaki A, Ikeya T, Hamatsu J, Hanashima T, Mishima M, Yoshimasu M, Hayashi N, Mikawa T, Wälchli M, Smith BO, Shirakawa M, Güntert P, Ito Y. Nature. 2009;458:102–106. doi: 10.1038/nature07814. [DOI] [PubMed] [Google Scholar]
- 42.Van Name WGZ. Kristallogr. - Cryst. Mater. 1902;11:325–412. [Google Scholar]
- 43.Harrison PJ, Waters RE, Taylor FJR. J. Phycol. 1980;16:28–35. [Google Scholar]
- 44.Reasoner DJ, Geldreich EE. Appl. Environ. Microbiol. 1985;49:1–7. doi: 10.1128/aem.49.1.1-7.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Maldonado LA, Fragoso-Yańez D, Pérez-García A, Rosellón-Druker J, Quintana ET. Antonie van Leeuwenhoek. 2009;95:111–120. doi: 10.1007/s10482-008-9294-3. [DOI] [PubMed] [Google Scholar]
- 46.National Committee for Clinical Laboratory Standards . Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria that Grow Aerobically. 7th ed. NCCLS; Villanova, PA: 2006. pp. M7–A7. Approved standard. [Google Scholar]
- 47.Marfey P. Carlsberg Res. Commun. 1984;49:591–596. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.