Abstract
This review focuses on a structure-based analysis of histone posttranslational modification (PTM) readout, where the PTMs serve as docking sites for reader modules as part of larger complexes displaying chromatin modifier and remodeling activities, with the capacity to alter chromatin architecture and templated processes. Individual topics addressed include the diversity of reader-binding pocket architectures and common principles underlying readout of methyl-lysine and methyl-arginine marks, their unmodified counterparts, as well as acetyl-lysine and phosphoserine marks. The review also discusses the impact of multivalent readout of combinations of PTMs localized at specific genomic sites by linked binding modules on processes ranging from gene transcription to repair. Additional topics include cross talk between histone PTMs, histone mimics, epigenetic-based diseases, and drug-based therapeutic intervention. The review ends by highlighting new initiatives and advances, as well as future challenges, toward the promise of enhancing our structural and mechanistic understanding of the readout of histone PTMs at the nucleosomal level.
Keywords: histone PTMs, multivalent readout, binding pockets, PTM cross talk, histone mimics, drug discovery
INTRODUCTION
Nucleosomes in cells are packaged into progressively higher-order folds to ultimately form chromosomes. The fundamental unit, the nucleosome core particle, is composed of DNA wrapped around a histone octamer containing histones H2A, H2B, H3, and H4 (reviewed in References 1 and 2), with the histone tails containing dynamic posttranslational modifications (PTMs) ranging from methylation, acetylation, and ubiquitination of lysines to methylation of arginines and phosphorylation of serines, threonines, and tyrosines. These PTMs, which are deposited by writer modules and removed by eraser modules in a histone- and sequence-specific manner, serve as scaffolds for reader modules, which in turn dictate the ordered recruitment of nonhistone proteins and enzymes. Such recruitment of DNA remodeling activities results in dynamic changes in chromatin-templated processes, with associated altered transcription rates reflecting accessibility to particular segments of DNA. The distribution and sequestration of PTMs have now been studied at the genome-wide level using ChIP-chip (chromatin immunoprecipitation with DNA microanalysis) and ChIP-Seq (chromatin immunoprecipitation with next-generation sequence) technologies, thereby permitting the linkage of PTMs with regulatory circuits in defined developmental states in model organisms.
In this review, we first outline structure-function studies on the readout of lysine and arginine methylation, followed by lysine acetylation and serine/threonine/tyrosine phosphorylation. We also focus on multivalent readout by paired reader modules, the cross talk between marks, and their impact on DNA-templated processes ranging from gene transcription to DNA replication, recombination, and repair. The contribution of histone mimics is outlined and so is the topic of dysregulation of PTM readout, which can result in aberrant gene expression patterns and/or genomic alterations, thereby facilitating the onset of disease. This in turn has stimulated research on the development of epigenetic drugs targeted to reader modules, thereby providing an approach to disease-specific therapeutic intervention. The review includes a supplemental Initiatives and Challenges section (at http://www.annualreviews.org) that lists some recent technological developments and outstanding challenges in the field. Several excellent reviews written earlier are available on readout of histone PTMs (3–7).
METHYL-LYSINE AND LYSINE READOUT
There is no net change in charge on lysine methylation, characterized by its relatively stable and multivalent qualities as reflected in monomethyl (Kme1), dimethyl (Kme2), and trimethyl (Kme3) modifications. Methylation of histone H3 lysine 4 (H3K4), H3K36, and H3K79 is linked to transcriptional activation, whereas methylation of H3K9, H3K27, and H4K20 is linked to repression. Of considerable interest is the architecture of methyl-lysine-binding pockets in their respective reader modules, as well as the principles underlying molecular recognition of both the PTM and the flanking sequence, which account for the sequence-specific recognition. Conversely, reader modules have now been identified that recognize unmodified lysines, with their interactions perturbed on methylation. An informative review has been published on the earlier structure-function aspects of the readout of methyl-lysine (Kme) marks (8).
“Royal Family” Modules
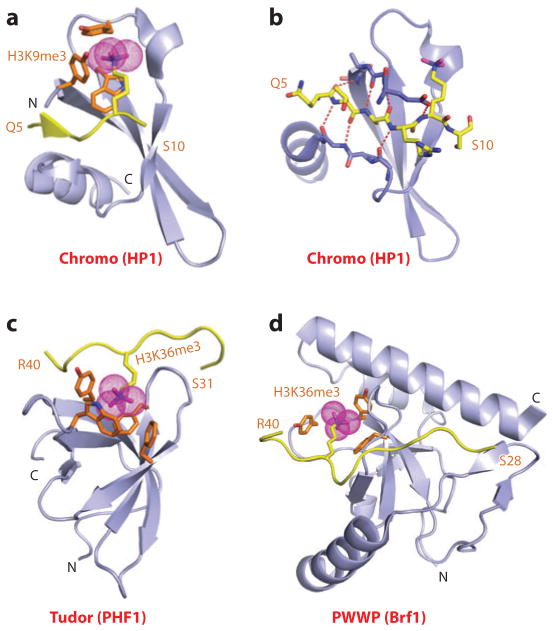
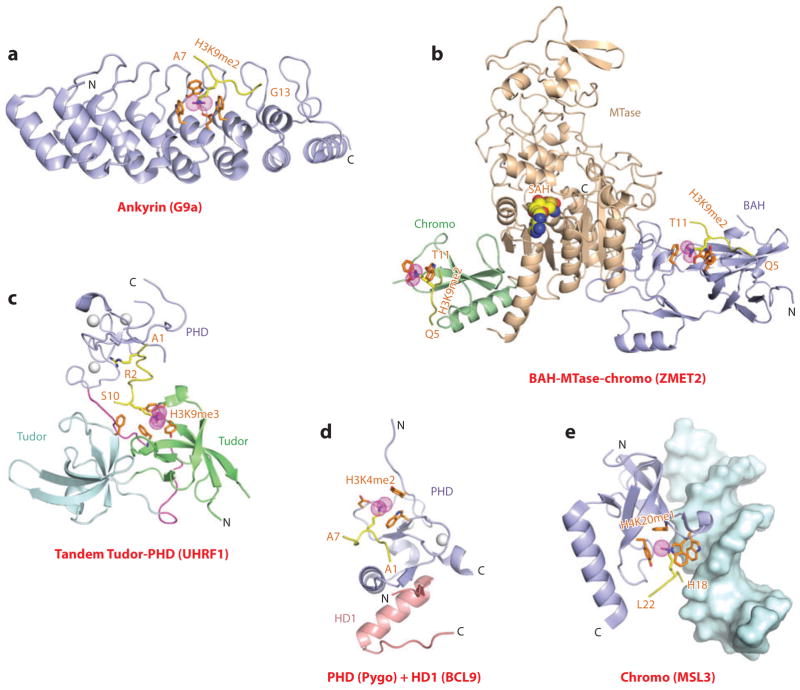
The “Royal family” modules, which include the chromodomain, Tudor, and PWWP domains, provided the first insight into the principles underlying Kme recognition by aromatic cage pockets (reviewed in Reference 9). The initial structural studies focused on the chromo (chromatin organizer modifier) domain of HP1 (10, 11) and on Polycomb (12), known repressors that contribute to epigenetic silencing. Here, we focus on the binding of the H3K9me3 mark to the HP1 chromodomain (Kd = 2.5 μM); the crystal structure of the complex is shown in Figure 1a (10). The peptide binds in an extended β-strand conformation between two β-strands of HP1, thereby completing an antiparallel β-barrel architecture. The directionality of the bound peptide reflects intermolecular contacts involving three amino acids preceding and one amino acid following the K9me3 mark (Figure 1b). The methyl-lysine side chain inserts into a “surface groove” pocket lined by three conserved aromatic amino acids, where it is stabilized by cation-π interactions (Figure 1a) (13). Mutation on any of the cage-forming aromatic amino acids results in a 20-fold loss in binding affinity. Structural studies of additional chromodomains have demonstrated that the sequence context flanking the methyl-lysine PTM determines the sequence specificity of recognition (reviewed in Reference 7).
Figure 1.
Structures of single “Royal family” modules bound to methyl-lysine-containing histone peptides. (a) The 2.4-Å crystal structure of the complex of the HP1 chromodomain bound to the H3(1–15)K9me3 peptide, Protein Data Bank (PDB) reference 1KNE. The bound K9me3-containing H3 peptide can be traced from Gln5 to Ser10 (Q5 to S10). (b) Details of the antiparallel alignment of the β-strand of the bound H3K9me3-containing peptide sandwiched between the β-strands of the HP1 chromodomain, resulting in generation of a three-stranded antiparallel β-pleated sheet on formation of the complex. (c) The 1.85-Å crystal structure of the complex of the PHF1 Tudor domain bound to the H3(31–40)K36me3 peptide complex (PDB: 4HCZ). The bound K36me3-containing H3 peptide can be traced from Ser31 to Arg40 (S31 to R40). (d) The 1.5-Å crystal structure of the complex of the Brf1 PWWP domain bound to the H3(22–42)K36me3 peptide complex (PDB: 2X4W). The bound K36me3-containing H3 peptide can be traced from Ser28 to Arg40 (S28 to R40). Abbreviations: C, C terminus; N, N terminus.
The same aromatic cage capture recognition of methyl-lysine has been observed for H3K36me3 bound to the Tudor domains of PHF1 (Figure 1c) and PHF19 (14–17), thereby facilitating intrusion of the PRC2 (polycomb repressive complex 2) into active chromatin regions to promote gene silencing, as well as H3K36me3 binding by Kme3 cage capture to the PWWP domain of Brf1 (Figure 1d) (18). In each case, the H3K36me3 peptide was bound with directionality (Figure 1c,d), and mutation of the cage-lining amino acids resulted in loss of binding affinity.
PHD Fingers
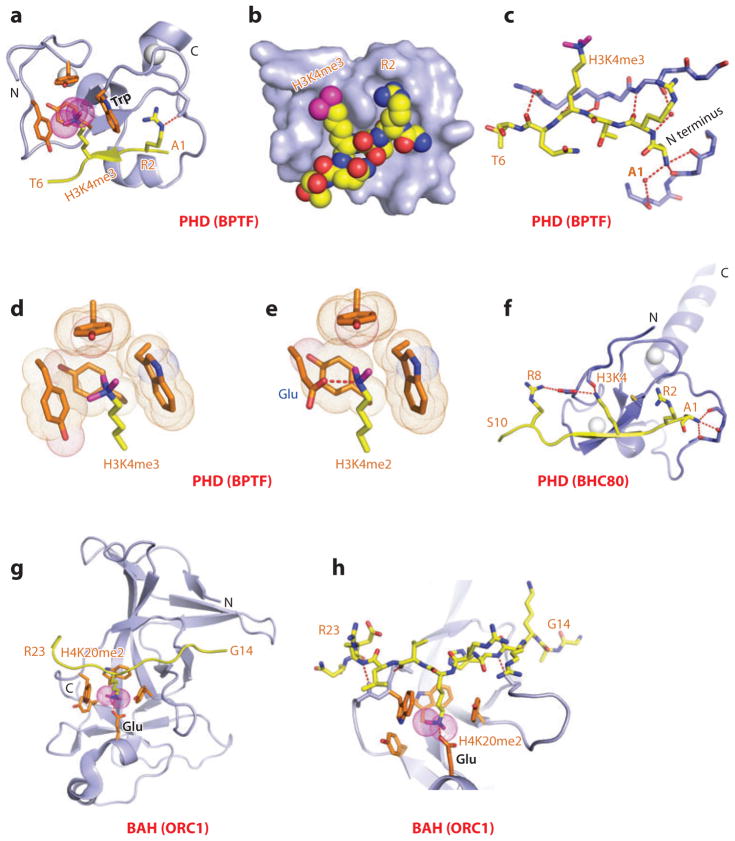
The plant homeodomain (PHD) finger is composed of a two-stranded β-sheet supplemented by one or more helical segments whose cross brace topology is stabilized by a pair of Zn ions coordinated to a Cys4-His-Cys3 segment (19). The function of the PHD finger was poorly understood until structural studies of H3K4me3 recognition by the PHD fingers of BPTF (the largest subunit of the nucleosome-remodeling factor, NURF) (20, 21) and ING2 (inhibitor of growth 2) (22, 23) identified it as a reader of methyl-lysine marks. In this regard, earlier studies had shown that the H3K4me3 mark is associated with nucleosomes near the promoters and the 5′ ends of highly transcribed genes (24, 25).
Here, we focus on readout of the H3K4me3 mark by the PHD finger of BPTF because structure-function studies on the BPTF PHD-bromo cassette have been undertaken at both the peptide and nucleosome level. Indeed, NURF-mediated ATP-dependent chromatin remodeling is directly coupled to trimethylation at H3K4, so as to maintain HOX gene expression patterns during development (21). The BPTF PHD finger bound the H3K4me3 peptide with a Kd equal to 2.7 μM, with a somewhat weaker affinity (Kd = 5.0 μM) for its H3K4me2 peptide counterpart, while discriminating against monomethylated and unmodified counterparts. The crystal structure of the H3K4me3 peptide-BPTF PHD finger complex is shown in Figure 2a,b, with the peptide bound with directionality on the surface of the PHD finger through β-sheet formation (Figure 2c) (20). The K4me3 mark is positioned through a surface groove recognition in a preformed pocket composed of four aromatic residues and stabilized by cation-π interactions. Sequence specificity of recognition is associated with intermolecular recognition of the N terminus and the positioning of Ala1 in a hydrophobic pocket, as well as by insertion of the side chains of R2 and K4me3 in the adjacent surface groove pockets separated by a tryptophan ring that is part of the aromatic cage (Figure 2a,b). Global loss of H3K4me3 resulted in loss of chromatin association with BPTF, and the binding affinity was dramatically reduced on mutation of the cage tryptophan, consistent with developmental defects associated with this mutation (21). The preference of the BPTF PHD finger for the H3K4me3 mark over the H3K4me2 mark could be reversed following replacement of a cage tyrosine by a glutamic acid, thereby facilitating a hydrogen bond between the dimethylammonium proton of Kme2 and the carboxylate of glutamic acid (Figure 2d,e) (26).
Figure 2.
Structures of the PHD finger (BPTF) and BAH domain (mammalian ORC1) bound to methyl-lysine-containing histone peptides. (a) The 2.0-Å crystal structure of the complex of the BPTF PHD finger bound to the H3(1–15)K4me3 peptide, Protein Data Bank (PDB) reference 2F6J. The PHD finger (as part of a PHD-bromo cassette) in a ribbon representation is shown (blue), with two stabilizing bound Zn ions (silver balls). The bound peptide from Ala1 to Thr6 (A1 to T6) is in yellow with the trimethyl group of Kme3 shown as dotted magenta balls. The residues forming the aromatic-lined cage pocket are orange. (b) Positioning of Arg2 (R2) and Lys4me3 (K4me3) side chains in the adjacent open surface pockets (surface groove mode), separated by the indole ring of an invariant Trp in the complex. The PHD finger and peptide in the complex are shown in surface and space-filling representations, respectively. (c) Details showing the antiparallel alignment of the β-strands of the bound H3K4me3-containing peptide and PHD finger, resulting in generation of an antiparallel β-pleated sheet on formation of the complex. Note that the positively charged N terminus is anchored in its own pocket. (d) Positioning of the K4me3 group within the aromatic-lined cage in the complex. (e) Positioning of the K4me2 group into an engineered pocket containing a Glu residue that replaced the Tyr residue in panel d (PDB: 2RIJ). (f) The 1.43-Å crystal structure of the complex of the PHD finger of BHC80 bound to the H3(1–10) peptide (PDB: 2PUY). The bound H3 peptide can be traced from Ala1 to Ser10 (A1 to S10). (g) The 1.95-Å crystal structure of the complex of the mouse ORC1 BAH domain bound to the H4(14–25)K20me2 peptide (PDB: 4DOW). The bound K20me2-containing H4 peptide can be traced from Gly14 to Arg23 (G14 to R23). (h) Details of the alignment of the K20me2-containing H4 peptide from G14 to R23 positioned on the mouse ORC1 BAH domain in the complex. The dimethylammonium group of H4K20 inserts into an aromatic-lined pocket in the BAH domain. Abbreviations: C, C terminus; N, N terminus.
Parallel structure-function studies of H3K4me3 binding to the PHD finger of ING2 have highlighted how, in response to DNA damage, recognition of H3K4me3 by the PHD finger of ING2 stabilizes the mammalian Sin3a transcription regulator–histone deacetylase 1(mSin3a-HDAC1) complex at the promoters of proliferation genes (22, 23). Furthermore, disruption of the binding interactions in the complex impairs the ability of ING2 to induce apoptosis in vivo. Other studies have evaluated the consequences of somatic mutations in ING PHD fingers on solid tumors (reviewed in Reference 27).
The PHD finger of RAG2, an essential component of the RAG V(D)J recombinase that mediates antigen-receptor gene assembly, couples H3K4me3 readout with V(D)J recombination (28). Structure-function studies of H3K4me3 binding to the PHD finger of RAG2 evaluated the effects of methylation of Arg2 (29), whereby both the depletion of the H3K4me3 mark as well as mutations abrogating intermolecular recognition severely impacted V(D)J recombination in vivo. Strikingly, the Trp that is bracketed by the R2 and K4me3 marks in the RAG2 PHD-H3K4me3 complex is mutated in patients with immunodeficiency syndromes, providing the first insight into how disruption of methyl-lysine readout impacts an inherited human disease (28).
Structure-function studies have examined the consequence of fusing the C-terminal H3K4me3-binding PHD finger of JARID1A (a jumonji domain lysine demethylase) to NUP98 (nucleoporin 98), thereby generating potent oncoproteins that arrest hematopoietic differentiation and induce acute myeloid leukemia in murine models (30). The aromatic cage of the JARID1A PHD finger is composed of two orthogonally positioned Trp residues; the mutation of either Trp abrogates H3K4me3 binding and also abolishes leukemic transformation.
Two recent comprehensive reviews have provided further details regarding the recognition of methyl-lysine marks on histone tails by PHD finger reader modules, emphasizing the contribution of plasticity in the recognition process (reviewed in References 31 and 32).
In addition to recognizing methyl-lysine marks, the PHD finger also targets unmodified lysines on histone tails. The unmodified H3(1–10) peptide binds to the PHD finger of BHC80 with a Kd equal to 30 μM (33). The high-resolution structure of the H3 peptide-BHC80 PHD finger complex is shown in Figure 2f. The peptide binds with directionality to the PHD finger with an unmodified K4 hydrogen bonded to a backbone carbonyl group and an acidic side chain, with methylation of K4 abrogating complex formation. Complex formation is stabilized by interdigitation of the side chains involving the H3 peptide (R2 and K4) and the PHD finger (Met and Asp) (Figure 2f). Functionally, RNAi-mediated knockdown of BHC80 results in derepression of the LSD1 target genes, with BHC80 and LSD1 depending reciprocally on each other to associate with chromatin (33).
The same conclusion regarding readout of unmodified lysines by reader modules has also been observed for the ADD domains of DNMT3A (34) and ATRX (35, 36), as well as for additional examples that are discussed below.
BAH Domains
Bromo-adjacent homology (BAH) domains are protein folds associated with gene regulation, but like PHD fingers, their function was poorly understood. Here, we focus on the BAH domain from the mammalian ORC1 (origin of replication protein 1), which has been shown to adopt a conserved β-sheet core from which emerge loop and short helical segments (37). Recently, structure-function studies have established that the BAH domain of mammalian ORC1 is a reader of the H4K20me2 mark (Kd = 5.2 μM) and discriminates against its Kme1 and Kme3 counterparts (38). ORC1, which mediates pre-DNA replication licensing (39), has been shown to contribute both to ORC (a six-subunit origin of replication complex) association and regulation of origin activity. The structure of the H4K20me2 peptide bound to mouse ORC1 is shown in Figure 2g, where the peptide binds with directionality and inserts the K20me2 side chain into an aromatic cage, with the dimethylammonium proton hydrogen bonded to a Glu side chain (Figure 2g,h). Functionally, mutation of the aromatic cage residues of the ORC1 BAH domain impairs ORC1 occupancy at replication origins, ORC chromatin loading, and cell-cycle progression (38). Other mutations in the ORC1 BAH domain have been linked to Meier-Gorlin syndrome, a form of primordial dwarfism (40). Aromatic cage mutants of ORC1, unlike wild-type ORC1, fail to rescue the growth retardation of orc1 morphants, and zebrafish depleted of H4K20me2 marks exhibit diminished body size (38). These studies have identified a direct link between the H4K20me2 mark and the metazoan DNA replication machinery, mediated by ORC1, with a primordial dwarfism syndrome.
In addition to ORC1, BAH domains are observed in a number of proteins associated with epigenetic inheritance processes (reviewed in Reference 41). The BAH domain is often flanked by other reader modules of histone marks, such as PHD and chromodomains, suggesting the potential for a combinatorial readout. In this regard, we discuss below the complex of H3K9me2 peptides bound to both the BAH and chromodomains of the plant CMT3 DNA methyltransferase (42).
MBT Repeats
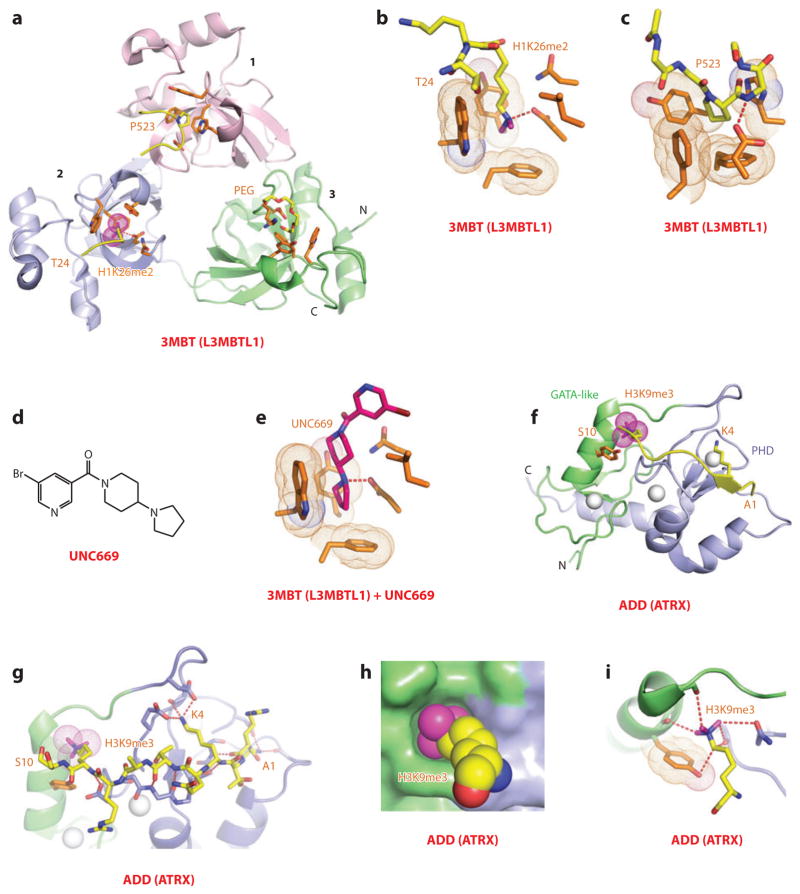
Malignant brain tumor (MBT) repeat–containing proteins impact on diverse processes, ranging from regulation of mitosis and tumor suppression to maintenance of cellular identity and body pattern development (43). The early structure-function studies focused on three MBT repeats containing L3MBTL1 (44) and two MBT repeats containing Drosophila SCML2 (Sex comb on midleg-like 2) (45) proteins. The MBT repeat is composed of a core fold and extended arms with interdigitation between adjacent MBT repeats, resulting in a three-leaved propeller-like architecture for L3MBTL1, with each color-coded MBT repeat containing an aromatic-lined pocket located on the same face of the triangular architecture (Figure 3a). Binding studies established that L3MBTL1 exhibited little sequence specificity for Kme-containing peptides, although it showed a preference for lower-methylation (Kme1 and Kme2 modifications) states (26). Structural studies established that the side chains of Kme1/2 insert deep into aromatic pocket 2 of L3MBTL1 (Figure 3a,b), and a proline ring from a proline-serine segment from the C-terminal tail of an adjacent L3MBTL1 protomer inserts into a shallower aromatic pocket 1 (Figure 3a,c). The methylation state specificity is readily understood because pocket 2 is both deep and narrow, thereby serving as a size-selective filter, with the gating and caging loops restricting access to the larger Kme3 group. Such a “cavity insertion” pocket mode of methyl-lysine recognition is distinct from the surface groove pocket recognition mode outlined in the previous sections. A parallel study of the H4K20me2 peptide bound to L3MBTL1 came to the same conclusions, but the investigators also proposed an unanticipated mode of peptide-mediated dimerization leading to a model of L3MBTL1-mediated chromatin compaction (46). More importantly, structure-function studies have demonstrated that L3MBTL1 compacts nucleosomal arrays containing lower lysine methylation marks at H1K26 and H4K20, such that combinatorial readout of these marks by HP1γ and L3MBTL1 results in chromatin condensation at Rb-regulated genes (47).
Figure 3.
Structures of malignant brain tumor (MBT, i.e., L3MBTL1) and ADD (ATRX) domains bound to methyl-lysine-containing histone peptides. (a) The 1.66-Å crystal structure of the complex of L3MBTL1 bound to the H1(22–26)K26me2 peptide, Protein Data Bank (PDB) reference 2RHI. A C-terminal peptide from an adjacent L3MBTL1 inserts proline 523 (P523) into the aromatic-lined pocket of the MBT domain 1 (pink). The dimethylammonium group of bound K26me2 inserts into the aromatic-lined pocket of the MBT domain 2 (blue), with the K26me2-containing H1 peptide traced from Thr24 to Lys26me2 (T24 to K26me2). A polyethylene glycol (PEG) molecule inserts into the aromatic-lined pocket of the MBT domain 3 (green). (b) Details of insertion (cavity insertion mode) of the dimethylammonium group of bound K26me2 into the aromatic-lined pocket of the MBT domain 2. (c) Details of insertion of the P523 from an adjacent L3MBTL1 into the aromatic-lined pocket of the MBT domain 1. This pocket is shallower than the one shown in panel b. (d) Chemical formula of UNC669, a pyrrolidine-containing small molecule. (e) Details of insertion of UNC669 into the aromatic-lined pocket of MBT domain 2 based on the 2.55-Å crystal structure of L3MBTL1 bound to UNC669 (PDB: 3P8H). (f) The 1.6-Å crystal structure of the complex of the ADD domain of ATRX bound to the H3(1–15)K9me3 peptide (PDB: 3QLA). The GATA-like and PHD fingers are green and blue, respectively. Bound Zn ions are shown (silver balls). The K9me3-containing H3 peptide is traced from A1 to S10. (g) Details of the intermolecular contacts involving the bound K9me3-containing H3 peptide in the complex, with the bound peptide traced from Ala1 to Ser10 (A1 to S10). (h) A surface and space-filling representation of the surface complementarity between K9me3 and the walls of the composite pocket lined by the GATA-like and PHD finger domains in the complex. (i) Ribbon and stick representation of K9me3 positioned to interact with the GATA-like and PHD finger domains in the complex. Abbreviations: C, C terminus; N, N terminus.
ADD Domains
The ATRX-DNMT3A-DNMT3L (ADD) domain is found in the mammalian DNA methyltransferase DNMT3A/DNMT3L and in ATRX, a protein whose mutated form is associated with X-linked mental retardation. The structure of the ADD domain, which contains adjacently positioned zinc-coordinated GATA and PHD fingers, adopts a scaffold where the two domains and a C-terminal α-helix pack against each other to form an overall globular fold (48). Half of the disease-associated missense mutations, many of them related to pancreatic endocrine tumors (49), are clustered within the ADD domain. The structure of the complex between H3K9me3 peptide and the ADD domain (Kd = 0.5 μM) solved at very high resolution (0.9 Å) is shown in Figure 3f, with intermolecular contacts outlined in Figure 3g (35, 36). Molecular recognition involves recognition of the N terminus, unmodified K4, and K9me3 (Figure 3g), readily explaining why binding is promoted by H3K9 trimethylation but inhibited by H3K4 methylation, given that in the latter there is no room to accommodate methylation marks. Unexpectedly, the K9me3 side chain is positioned in an “interfacial composite pocket” involving van der Waals contacts associated with a high degree of surface complementarity (Figure 3h), supplemented by a set of atypical carbon-oxygen hydrogen bonds with the K9me3 group (Figure 3i). Functionally, both the H3K9me3-binding pocket and ATRX syndrome mutants are defective in both H3K9me3 binding and localization at pericentric heterochromatin (36). In addition, the readout of unmodified H3K4 and H3K9me3 marks by the ADD domain of ATRX was shown to be facilitated by recruitment of HP1, a reader module that independently recognizes the H3K9me3 mark, resulting in tripartite recognition with the potential for spanning neighboring nucleosomes (35).
Tandem Repeats
Many reader modules occur as tandem repeats, as discussed above for MBT repeats, with repeat modules showing diverse architecture and principles for recognition of methyl-lysine marks. In this section, we first consider molecular recognition by a tandem chromodomain repeat and then highlight recognition diversity for tandem Tudor repeats.
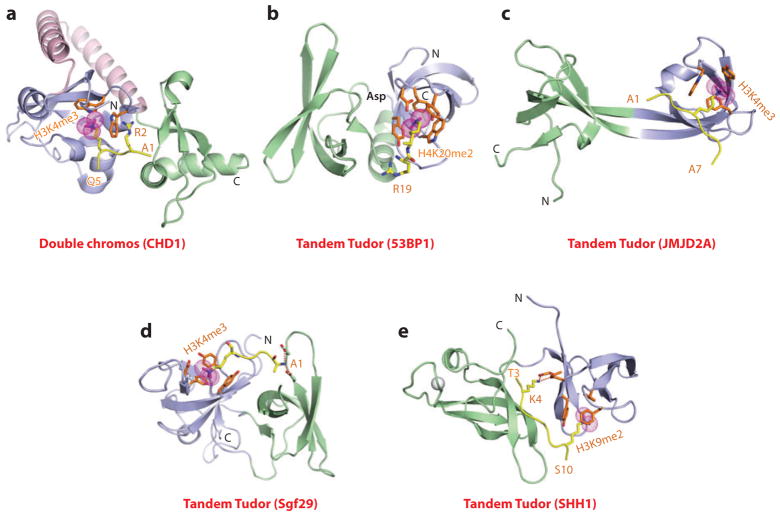
Tandem chromodomains that target the H3K4me3 mark are observed in the CHD (chromo-ATP/helicase-DNA-binding domain) protein, which is involved in regulating ATP-dependent nucleosome assembly and mobilization at sites of transcriptional activity. In the structure of the H3K4me3 peptide bound to CHD1 (Kd = 5 μM), the juxtaposed tandem chromodomains that are linked by a helix-turn-helix motif form a continuous surface, with the peptide positioned between chromodomains (Figure 4a) (50). The K4me3 side chain inserts into a pocket formed by orthogonal alignment of a pair of Trp rings, and the side chain of R2 is stabilized through stacking (cation-π interactions) over one of these Trp rings, with mutation of either Trp impacting binding affinity. It is noteworthy that owing to the unique inserts blocking canonical binding sites, CHD1 uses atypical binding surfaces for peptide recognition.
Figure 4.
Structures of tandem Royal family modules bound to methyl-lysine-containing histone peptides. (a) The 2.4-Å crystal structure of the complex of double chromo domains of human CHD1 proteins bound to the H3(1–19)K4me3 peptide, Protein Data Bank (PDB) reference 2B2W. Tudor domains 1 and 2 are shown (blue and green, respectively) with the connecting helix-turn-helix linker (pink). The bound K4me3-containing H3 peptide can be traced from Ala1 to Gln5 (A1 to Q5). (b) The 1.7-Å crystal structure of the complex of tandem Tudor domains of 53BP1 bound to the H4(15–24)K20me2 peptide (PDB: 2IG0). Tudor domains 1 and 2 are blue and green. The bound K20me2-containing H4 peptide can be traced for the Arg19-Lys20me2 (R19-K20me2) step. (c) The 2.1-Å crystal structure of the complex of tandem Tudor domains of JMJD2A bound to the H3(1–10)K4me3 peptide (PDB: 2GFA). Individual Tudor domains are blue and green. The bound K4me3-containing peptide can be traced from Ala1 to Ala7 (A1 to A7). (d) The 1.26-Å crystal structure of the complex of tandem Tudor domains of Sgf29 bound to the H3(1–11)K4me3 peptide (PDB: 3MEA). Tudor domains 1 and 2 are blue and green. The bound K4me3-containing peptide can be traced from Ala1 to Lys4me3 (A1 to K4me3). (e) The 2.7-Å crystal structure of the complex of tandem Tudor domains of the Arabidopsis thaliana SHH1 protein bound to the H3(1–15)K9me2 peptide (PDB: 4IUT). A bound zinc ion is shown (silver ball). Tudor domains 1 and 2 are blue and green. The bound K9me2-containing H3 peptide can be traced from Thr3 to Ser10 (T3 to S10). Abbreviations: C, C terminus; N, N terminus.
The diversity of methyl-lysine marks recognition by tandem Tudor domains is shown by four examples outlined below. Initial structure-function studies focused on the complex of the H4K20me2 peptide bound to the tandem Tudor domains of 53BP1 (p53-binding protein 1), whereby histone lysine methylation facilitated recruitment of 53BP1 and its relocalization to DNA double-strand breaks upon exposure to DNA-damaging agents. The structure of the H4K20me2 peptide-53BP1 tandem Tudor domain complex (Kd = 19.7 μM) is shown in Figure 4b, with the side chain of K20me2 inserting into an aromatic cage pocket (four aromatic residues and an aspartic acid) of the first Tudor domain, whose dimensions prevent accommodation of a Kme3 modification (51). Mutation of aromatic residues lining the pocket resulted in loss of binding affinity in vitro and impacted on the targeting of 53BP1 to DNA double-strand breaks in vivo.
In contrast to 53BP1, the tandem Tudor domains of JMJD2A (a jumonji histone lysine demethylase) adopt a domain-swapped interdigitated topology with a two-stranded β-sheet separating the two domains, as shown for the structure of the H3K4me3 peptide-JMJD2A tandem Tudor domain complex (Figure 4c) (52). In this hybrid complex, the side chain of K4me3 is inserted in the aromatic cage pocket of one Tudor domain, while intermolecular contacts involve side chains of the other Tudor domain. An extension of these studies has shown that the tandem Tudor domains of JMJD2A bind H3K4me3 (Kd = 0.5 μM) and H4K20me3 (Kd = 0.4 μM) peptides in opposite orientations with the intermolecular contacts in the structures of the two complexes identifying single-point mutations, whereby it was possible to inhibit recognition of H4K20me3 but not H3K4me3, and vice versa (53).
Another example of the diversity of molecular recognition by tandem Tudor domains emerged following structural comparisons of complexes of methyl-lysine peptides bound to Sgf29 on the one hand and the UHRF1 (ubiquitin-like with PHD and Ring finger 1) domain and SHH1 (Sawadee homeodomain homolog 1) on the other. Thus, the tandem Tudor domain of Sgf29, a component of the SAGA (Spt-Ada-Gcn5 acetyltransferase) complex, binds to H3K4me2/3 peptides along one surface of a tightly packed face-to-face dimeric alignment (Figure 4d) (54). Interestingly, the side chain of Ala1 inserts into a pocket of one Tudor domain, and the K4me2/3 side chain inserts into an aromatic cage pocket in the other Tudor domain. Functionally, the tandem Tudor domains of Sgf29 target H3K4me2/3 marks to recruit the SAGA complex to its target sites and mediate acetylation of H3 tails.
By contrast, the H3K9me2 peptide is bound with directionality within a channel between the Tudor domains, thereby interacting with both domains, in complexes with the tandem Tudor domains of mammalian UHRF1 (55) and plant SHH1 (56). The structure of the H3K9me2 peptide-SHH1 tandem Tudor domain complex (Kd = 1.9 μM) is shown in Figure 4e, with sequence specificity attributed to recognition of both unmodified K4 and K9me2 (in an aromatic cage pocket) (56). Functional studies show that the SHH1 protein acts upstream of the RNA-dependent DNA methylation (RdDM) pathway in plants to enable siRNA production from the most active RdDM targets and that SHH1 is required for RNA polymerase IV (Pol IV) occupancy of these same loci. Because key residues within both K4 and K9me2 pockets of SHH1 are required to maintain DNA methylation in vivo, the results provide insights into the mechanism underlying SHH1 recruitment of RNA Pol IV to RdDM targets in plants (56).
Paired Modules
G9a is a euchromatin-associated methyltransferase, which represses transcription following the writing of H3K9me1/2 marks by the C-terminal methyltransferase SET domain and, in addition, reads the same marks using N-terminal ankyrin repeats. The structure of the H3K9me2 peptide bound to the G9a ankyrin repeats (Kd = 6 μM) is shown in Figure 5a. The bound peptide is sandwiched with specificity solely between the β-turns and helices of the fourth and fifth ankyrin repeats, with the K9me2 side chain inserted into an aromatic cage pocket, and the complex is stabilized by intermolecular recognition of H3 residues from positions 9 to 11 (57). Although mutations of the aromatic cage residues perturb complex formation with H3K9me2 peptide, they have no impact on G9a’s SET domain-mediated methyltransferase activity.
Figure 5.
Structures of linked binding and accessory modules involved in multivalent readout of methyl-lysine-containing histone peptides. (a) The 2.99-Å crystal structure of the complex of the ankyrin repeats of G9a bound to the H3(1–15)K9me2 peptide, Protein Data Bank (PDB) reference 3B95. The aromatic-lined binding pocket bound by K9me2 is positioned between the fourth and fifth ankyrin repeats of G9a. The bound K9me2-containing H3 peptide can be traced from Ala7 to Gly13 (A7 to G13). (b) A superposed view of the crystal structures of the complex of the BAH domain–methyltransferase (MTase) domain–chromodomain of maize ZMET2 bound to the H3(1–32)K9me2 peptide (2.7 Å, bound to BAH domain) (PDB: 4FT4) and bound to the H3(1–15)K9me2 peptide (3.2 Å, bound to chromodomain) (PDB: 4FT2). The chromodomain, BAH, and methyltransferase domains are green, blue, and beige, respectively. The bound K9me2-containing H3 peptides can be traced from Gln5 to Thr11 (Q5 to T11) when bound to both the BAH and chromodomains. (c) The 2.9-Å crystal structure of the complex of the tandem Tudor-PHD finger cassette of UHRF1 bound to the H3(1–13)K9me3 peptide (PDB: 3ASK). The tandem Tudor domains are shown in the lower segment (cyan and green), and the PHD finger is shown in the upper segment (blue). The bound H3(1–13)K9me3-containing peptide can be traced from Ala1 to Ser10 (A1 to S10). (d) The 1.7-Å crystal structure of the ternary complex of the Pygopus PHD finger (blue) bound to the H3(1–7)K4me2 peptide in the presence of the homology domain 1 (HD1) domain of BCL9 (pink) (PDB: 2VPE). The bound K4me2-containing H3 peptide can be traced from Ala1 to Ala7 (A1 to A7). (e) The 2.35-Å crystal structure of the complex of MSL3 chromodomain bound to the H4(9–31)K20me1 peptide in the presence of duplex DNA (surface representation) (PDB: 3OA6). The bound K20me1-containing H4 peptide can be traced from His18 to Leu22 (H18 to L22). Abbreviations: C, C terminus; N, N terminus.
A notable example of recognition of the same methyl-lysine marks by two domains in a protein has been highlighted from structure-function studies of plant chromomethylase 3 (CMT3; ZMET2 in maize). CMT3, an H3K9me2-dependent plant-specific DNA methyltransferase responsible for methylation at CpHpG sites, is composed of an N-terminal BAH domain and a DNA methyltransferase domain, within which is embedded a chromo-domain. The structure of the H3K9me2 peptide bound to ZMET2 is shown in Figure 5b, with H3K9me2 peptides bound with directionality to both chromodomain and BAH domain positioned at the corners of a triangular architecture, and complex formation is accompanied by insertion of K9me2 side chains in aromatic-lined pockets in both domains (42). Complementary functional studies demonstrated a perfect correlation between H3K9me3 and CMT3 genome locations and also established that CMT3 is stably associated with H3K9me2-containing nucleosomes. In addition, triple aromatic cage mutants of either the chromodomain or BAH domain disrupted CMT3 binding to nucleosomes and further showed a complete loss of CMT3 activity in vivo (42).
The UHRF1 protein contains an N-terminal tandem Tudor domain followed by a PHD finger. The H3K9me3 peptide binds to the isolated tandem Tudor domain with a Kd equal to 1.75 μM and to the isolated PHD finger with a Kd equal to 1.47 μM. By contrast, the H3K9me3 peptide binds to the UHRF1 tandem Tudor-PHD cassette with a 1:1 stoichiometry and with a Kd equal to 0.37 μM, indicative of multivalent readout of dual marks by linked binding modules. The structure of H3K9me3 peptide bound to the UHRF1 tandem Tudor-PHD cassette is shown in Figure 5c (58, 59). There is minimal contact between the PHD finger and the tandem Tudor domains, with the 17-residue linker packed between the PHD and Tudor domains. The compactly folded bound H3K9me3-containing peptide is positioned within a central hole in the protein scaffold. Key intermolecular contacts involve unmodified H3R2 recognition by the PHD finger through a network of intermolecular hydrogen bonds, with methylation at this position resulting in reduced binding affinity, whereas H3K9me3 recognition involves insertion into an aromatic cage pocket of the first Tudor domain (Figure 5c). Interestingly, phosphorylation of a serine within the UHRF1 linker segment resulted in a 30-fold loss in binding affinity, suggesting that such a modification could act as a switch between the potential regulatory pathways utilized by UHRF1 during maintenance of DNA methylation and transcriptional repression (58).
Accessory Modules
Pygopus, together with its cofactor BCL9, regulates β-catenin-mediated transcription and is critical for Wnt signaling during development. The interacting components of this complex are the PHD finger of Pygopus and homology domain 1 (HD1) of BCL9. Structural studies have been undertaken of the binary complex of Pygopus PHD finger and HD1 (60), as well as of its ternary complex with the added H3K4me2 peptide (60, 61). The H3K4me2 mark and HD1 are positioned on opposite sides of the PHD finger in the ternary complex (Figure 5d), with efficient H3K4me2 mark recognition requiring association of the Pygopus PHD finger with HD1, suggesting that HD1 binding to the PHD finger triggers an allosteric transition, thereby facilitating optimal recognition of the H3K4me2 mark through aromatic cage pocket capture.
MSL3, a subunit of the MSL complex, was shown to be necessary for dosage compensation on Drosophila male X chromosomes (62). Recently, it has been shown that the MSL3 chromodomain targets lower methy-lation states of H4K20 only in the presence of guanine-adenine-rich (GA-rich) DNA (63). A preassembled complex of MSL3 chromodomain and GA-rich DNA that bound H4K20me1 peptide with a Kd equal to 15 μM discriminates against unmodified and trimethylation states. The structure of the ternary complex formed by the MSL3 chromodomain, GA-rich DNA, and H4K20me1 peptide is shown in Figure 5e with the chromodomain packed against the minor groove of the DNA and the K20me1 side chain inserted into an adjacent aromatic amino acid–lined pocket (63). It was speculated that the potential corecognition of the H4K20me1 mark and the DNA of two adjacent nucleosomes could contribute to the in vivo targeting of the MSL complex.
METHYL-ARGININE AND ARGININE READOUT
There is also no net change in charge on arginine methylation. This mark is characterized by its relatively stable and multivalent qualities as reflected in monomethyl (Rme1), symmetrical dimethyl (Rme2s), and asymmetrical dimethyl (Rme2a) modifications, with their major sites at H3R2 and H4R3. Of considerable interest is the architecture of methyl-arginine-binding pockets in their respective reader modules, as well as the principles underlying molecular recognition of both the PTM and the flanking sequences that account for their sequence-specific recognition. Conversely, reader modules have now been identified that recognize unmodified arginines with their interactions perturbed on methylation. An informative and comprehensive review has been published on the structure-function aspects of the readout of methyl-arginine PTMs (64).
Expanded Tudor Domains
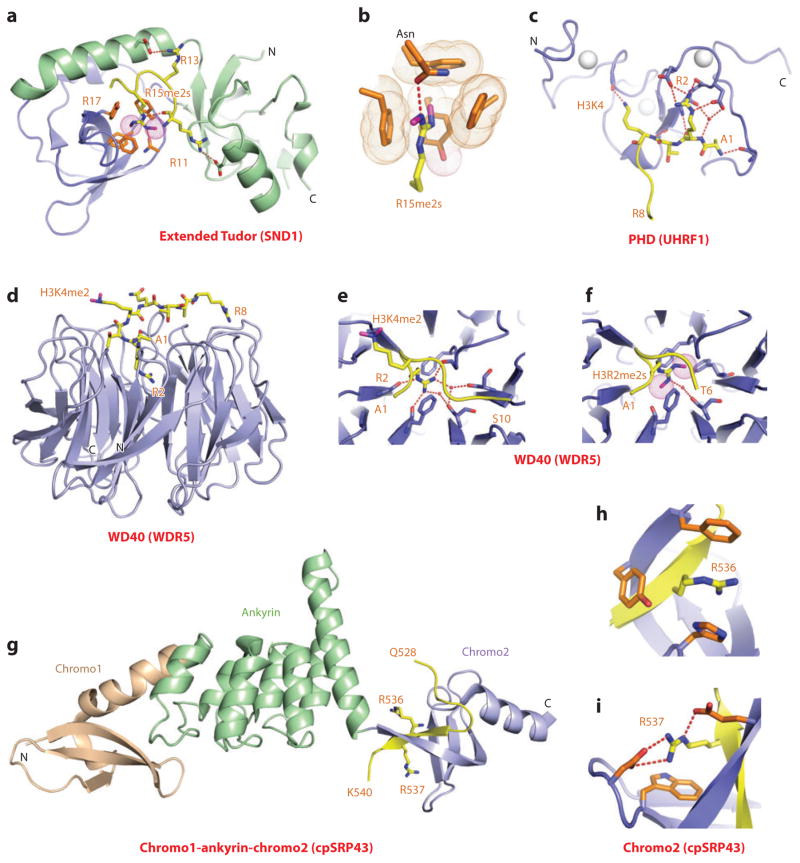
Both canonical (65) and expanded Tudor (66–68) domains are involved in readout of methyl-arginine marks. Because there is no structural study of methyl-arginine-containing histone targets, we discuss, below, examples of methyl-arginine recognition on non-histone targets. Several germ line–expanded Tudor-containing proteins contain repeats of 60–amino acid canonical Tudor domains flanked by N- and C-terminal extension segments. Such expanded Tudor domains target methyl-arginine marks located within the N terminus of PIWI proteins, thereby contributing to the regulation of the PIWI-interacting RNA biogenesis pathway in the germ line and silencing transposable elements in the germ line during early development. The structure of a symmetrically dimethylated R15me2s-containing an N-terminal PIWI peptide bound to the extended Tudor domain of SND1 (staphylococcal nuclease domain-containing 1) is shown in Figure 6a, with the N- and C-terminal extensions combining to adopt an oligonucleotide- and oligosaccharide-binding fold (67). The peptide binds with directionality within a wide and negatively charged groove and in so doing inserts the planar dimethyl-guanidinium group of R15me2s into an aromatic-lined cage pocket (Figure 6b), while making intermolecular contacts with residues flanking the R15me2s mark. Both mutations in the aromatic residues and an asparagine lining the aromatic cage, along with truncations of the canonical Tudor domain and flanking N- and C-terminal extensions, impact on binding affinity. The same structure was observed for the complex of the R4me2s PIWI peptide with the extended Tudor domain of SND1, which had the binding affinity (Kd = 10 μM) dropping by twofold for the R4me1 mark, fourfold for the R4me2a mark, and ninefold for the unmodified R4 mark. Functional studies have identified an intricate interplay between the writers and readers of the Rme2s mark and the mark itself, impacting on the regulation of transposon silencing and germ cell development (reviewed in Reference 64).
Figure 6.
Structures of expanded and paired modules bound to methyl-arginine and unmodified arginine-containing peptides. (a) The 2.8-Å crystal structure of the complex of the extended Tudor module of SND1 bound to R15me2s-containing N-terminal PIWI peptide, Protein Data Bank (PDB) reference 3NTI. The core fold of the Tudor domain is shown in blue, and the extensions are shown in green. The R15me2s-containing N-terminal PIWI peptide can be traced from Arg11 to Arg17 (R11 to R17). (b) Positioning of R15me2s in the aromatic-lined cage pocket of the Tudor domain in the SND1 complex. (c) The 1.8-Å crystal structure of the complex of the PHD finger of UHRF1 bound to the H3(1–9) peptide (PDB: 3SOU). The bound H3 peptide can be traced from Ala1 to Arg8 (A1 to R8). Note the network of hydrogen bonds involving Arg2 (R2) and residues on the PHD finger. (d) The 1.5-Å crystal structure of the complex of the WD40 motif of WDR5 bound to the H3(1–9)K4me2 peptide. The bound K4me2-containing peptide can be traced from Ala1 to Arg8 (A1 to R8) (PDB: 2H6N). (e) Intermolecular hydrogen-bonding interactions stabilizing insertion of Arg2 (R2) into the central channel of the WD40 motif in the H3(1–9)K4me2-WDR5 complex. (f) Insertion of symmetrical Arg2me2 (R2me2s) into the central channel of the WD40 motif in the H3(1–15)R2me2s-WDR5 complex solved at 1.9 Å (PDB: 4A7J). (g) The 3.18-Å crystal structure of the complex of the chromodomains and ankyrin repeats of Arabidopsis thaliana cpSRP43 bound to an Arg-Arg-Lys-Arg (RRKR)-containing peptide (PDB: 3UI2). The side chains of Arg536 (R536) and Arg537 (R537) of the bound RRKR-containing peptide from Gln528 to Lys540 (Q528-K540) are positioned in adjacent pockets at the interface between the fourth ankyrin repeat and the second chromodomain in the complex. (h) Position of Arg536 (R536) of the RRKR-containing peptide within an aromatic-lined cage pocket in the complex. (i) Positioning of Arg537 (R537) of the RRKR-containing peptide in a pocket lined by a Trp and two acidic side chains in the complex. Abbreviation: C, C terminus.
PHD Finger Domains
Similar to recognition of unmodified lysines, reader modules have been identified that recognize unmodified arginines on histone tails. It has been shown recently that the ability of UHRF1 to suppress the expression of target genes is dependent on the PHD finger of UHRF1 binding to unmodified H3R2, thereby linking this recognition event involving UHRF1 to the regulation of euchromatic gene expression (69). The structural basis underlying this functional observation emerged on solving the structure of H3(1–9) peptide bound to the PHD finger of UHRF1 (Kd = 2.1 μM), whereby the guanidinium group of R2 participates in an extensive intermolecular hydrogen-bonding network (Figure 6c), with methylation of H3R2, but not H3K4 or H3K9, disrupting complex formation (69).
WD40 Repeat Domains
The WD40 repeat protein WDR5 with its toroidal β-propeller fold is a common component of the SET1 family of histone methyltransferases, with the latter playing a critical role during proper HOX gene activation and vertebrate gene development. It has been proposed that WDR5 associates with and is essential for H3K4 methylation and vertebrate development (70). To this end, structures have been determined of the complex of H3K4me2 peptide bound to WDR5 (Figure 6d) (71–74). Somewhat unexpectedly, it was the side chain of unmodified R2 rather than K4me2 that inserted into the narrow central channel (cavity insertion mode) of the toroidal β-propeller fold of WDR5, whereby its alignment was stabilized by stacking with staggered phenylalanine side chains and was oriented through direct and water-mediated hydrogen bonds (Figure 6e). More recently, the structure of the complex H3R2me2s peptide bound to WDR5 demonstrated that the dimethyl-guanidinium group of symmetrical R2me2s, but not R2me2a, also inserts into the central channel of WDR5 (Figure 6f) (75). Comparison of the insertion cavities in the H3R2 (Figure 6e) and the H3R2me2s (Figure 6f) complexes indicates that the methyls of the dimethyl-guanidinium group are accommodated following displacement of one of the water molecules together with a protrusion of the dimethylated side chain toward the phenylalanine ring, thereby forming enhanced hydrophobic interactions. Functionally, the H3R2me2s mark retains genes in a poised state in euchromatin for transcriptional activation following transit from the cell cycle and differentiation (75).
Future challenges will be to identify potential readers of Rme2a and Rme1 marks and to understand the principles underlying optimal selectivity as a function of the methylation state of arginine.
Chromodomains
A very interesting mode of unmodified arginine recognition on a nonhistone protein has been recently reported between the cpSRP43(ΔCD3) protein (composed of ankyrin repeats flanked by chromodomains) bound to a peptide containing the RRKR motif from cpsSRP54 associated with the chloroplast signal recognition particle in plants (76). The structure of this complex (Kd = 6.4 μM) is shown in Figure 6g with the peptide binding at the interface between the fourth ankyrin repeat and the second chromodomain, such that the peptide is positioned through β-sheet formation within a hydrophobic binding groove of the second chromodomain. The side chain of unmodified Arg536 is inserted into a three–aromatic residue–lined cage pocket (Figure 6h), and the side chain of the adjacent unmodified Arg537 is anchored through hydrogen bonding in a pocket containing one aromatic and two basic residues (Figure 6i), with replacement of either arginine on the peptide or aromatic amino acids lining the pockets, abolishing both complex formation and functional readout. This insightful contribution introduces the novel concept of readout of adjacent unmodified arginines by a twinned aromatic cage and defines an unexpected nonnuclear function for chromodomains in posttranslational targeting.
ACETYL-LYSINE READOUT
Interest in histone acetylation came to the forefront with the realization that lysine acetylation impacts both the topology of nucleosomes and internucleosome interactions responsible for the higher-order architecture of chromatin. Furthermore, the linkage of histone acetylation and transcription was established early on the basis of the opposing activities of acetyltransferases and deacetylases involved in transcriptional regulation. We outline below the results emerging from structure-function studies on readout of histone acetyl-lysine marks, starting with the initial key discoveries and moving on to more recent contributions, including the exciting development of inhibitors against acetyl-lysine pockets (see Inhibitors of Kac-Binding Bromodomains, below). An overview of acetyl-lysine readout can be found in a comprehensive review on the subject (77).
Bromodomains
As part of an effort toward understanding the molecular principles underlying acetyl-lysine recognition, attention turned to bromodomains (bromo), which are a known component of certain histone acetyltransferases and chromatin remodeling complexes as well as helicases, histone methyltransferases, and transcriptional coactivator complexes. In a pace-setting contribution to the field of epigenetic readout of histone PTMs, NMR studies determined the solution structure of the bromodomain of the histone acetyltransferase coactivator P/CAF (p300/CBP-associated factor) in the free state and bound to an H4 peptide acetylated at K8 (H4K8ac) (78). The P/CAF bromodomain adopted a left-handed antiparallel four-helix bundle with long ZA (connecting helices αZ and αA) and BC (connecting αB and αC) loops packed against each other to form a hydrophobic pocket at one end of the scaffold. The H4K8ac peptide bound the P/CAF bromodomain with modest affinity (Kd = 346 μM), with the observed NMR chemical shift changes characteristic of insertion of the acetylated lysine into the hydrophobic binding pocket formed between the four α-helices and extended by the ZA and BC loops. Mutation of the hydrophobic residues in the pocket, especially aromatic ones, resulted in a loss of binding affinity.
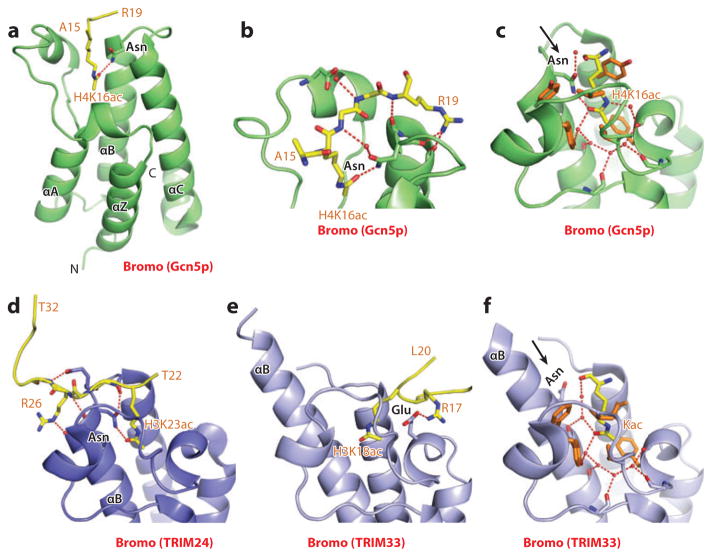
Next, in an equally important contribution, a high-resolution (1.9-Å) crystal structure of the complex of H4K16ac-containing peptide bound to the bromodomain of the histone acetyltransferase Gcn5p identified the role of a key asparagine side chain and bound water molecules in the binding pocket to acetyl-lysine recognition, as well as the contribution of flanking amino acids to sequence-specific recognition (79). The structure of the H4K16ac peptide-Gcn5p bromodomain complex is shown in Figure 7a, with details of intermolecular contacts defining context-dependent readout in Figure 7b and the binding pocket in Figure 7c. The overall structure and positioning of the binding pocket (Figure 7a) are common to both P/CAF and Gcn5p bromodomains. The peptide is positioned with directionality in a shallow depression between the ZA and BC loops, such that the channel is lined with hydrophobic and charged amino acids at opposite ends and with the complex stabilized by intermolecular hydrogen bonds (Figure 7b). The long acetyl-lysine side chain is inserted deep into the central pocket inside the bundle of four helices; the hydrophobic wall is required for stabilization of the aliphatic side chain. Unique interactions in the binding pocket involve a key intermolecular hydrogen bond between the amide nitrogen of a conserved asparagine with the oxygen of the acetyl group of acetyl-lysine (Figure 7c). In addition, intermolecular contacts at the base of the binding pocket are mediated through a network of water molecules hydrogen bonded to the backbone carbonyls lining the walls of the pocket (Figure 7c). The structure elegantly explains why the deep binding cleft targets acetyl-lysine in preference to the unmodified amino acid and also explains the role of peptide sequence in dictating sequence-specific recognition on the basis of the diverse electrostatic surfaces spanning either side of the entrance to the pocket.
Figure 7.
Structures of bromodomains bound to acetyl-lysine-containing histone peptides. (a) The 1.87-Å structure of the bromodomain of Gcn5p bound to the H4(15–29)K16ac peptide, Protein Data Bank (PDB) reference 1E6I. The peptide can be traced from Ala15 to Arg19 (A15 to R19). (b) Details of intermolecular recognition between the peptide backbone and loops that project from the bromodomain scaffold. (c) Details of intermolecular contacts in the binding pocket of the Gcn5p bromodomain-H4(15–29)K16ac complex. The acetyl-lysine side chain inserts into a hydrophobic pocket and is anchored through hydrogen bonding with an asparagine side chain. Also note the extensive use of water-mediated intermolecular recognition in the pocket. (d) The 1.9-Å structure of the bromodomain of TRIM24 bound to the H3(13–32)K23ac peptide (PDB: 3O34). The chain can be traced from Thr22 to Thr32 (T22 to T32), with the side chain of K23ac involved in canonical recognition within the binding pocket. Interactions involving the side chain of Arg26 (R26) of the bound peptide are also show in this panel. (e) The 2.7-Å structure of the bromodomain of TRIM33 bound to the H3(1–22)K9me3K14acK18ac peptide (PDB: 3U5O). The chain can be traced from Ala1 to Leu20 (A1 to L20), with the segment from Arg17 to Leu20 (R17 to L20) shown in this panel. The side chain of K18ac is involved in atypical recognition, in that it is not hydrogen bonded to an asparagine. Interactions involving the side chain of Arg17 (R17) of the bound peptide are also shown in this panel. (f) Details of intermolecular contacts (using the structure at 1.95-Å resolution) in the binding pocket of the TRIM33 bromodomain-H3(1–20)K9me3K14ac complex (PDB: 3U5N). The acetyl-lysine side chain inserts into a hydrophobic pocket and is anchored solely through water-mediated interactions. The extension of the α-helix B (αB) in TRIM33 results in rotation of the asparagine residue away from the pocket such that it is no longer involved in recognition of the Kac side chain. Abbreviations: αA, -C, α-helix A and C; C, C terminus; Kac, acetyl-lysine.
Since the earliest structural studies of bromodomains (78, 79), there has been an outpouring of structures of histone peptide–bromodomain complexes, culminating recently in a large-scale comprehensive structural study of acetyl-lysine recognition by the human bromodomain family (80). The binding affinities of acetyl-lysine (Kac) marks for bromodomains span a range from Kd 3 μM to 200 μM. One of the insights that emerged from this study highlighted the contributions of flanking PTMs for acetyl-lysine recognition.
Tandem Bromodomains
Many bromodomain-containing proteins contain this reader module in tandem repeat with the capacity to bind a pair of acetyl-lysine marks separated by appropriate spacing, thereby increasing the binding affinity as a result of bivalent readout. Examples include the tandem bromodomains of TAF1, the largest subunit of TFIID, where the bromodomains are aligned in an independent side-by-side arrangement, with Kac-binding pockets separated by approximately 25 Å (81). The tandem bromodomains of TAF1 bind di- and tetra-acetylated H4 peptides (acetylated with combinations of K5, K8, K12, and K16) with higher affinity than their monoacetylated counterparts.
By contrast, the tandem bromodomains of yeast Rsc4p interact extensively with each other to form a single structural unit, where the two Kac-binding pockets are oriented on the same face and separated by a shortened (20-Å) separation (82). In this case, the second bromo-domain bound the H3K14ac mark, with binding disrupted on phosphorylation at nearby Ser10 on histone H3 (H3S10). Gcn5p-mediated acetylation of a fused construct linking the H3 tail to the bromodomains of Rsc4p resulted in formation of H3K14ac and Rsc4pK45ac marks. Interestingly, insertion of Rsc4pK45ac mark into the binding pocket of the first bromodomain inhibited insertion of the H3K14ac mark into the binding pocket of the second bromodomain, thereby identifying an autoregulatory mechanism to control Rsc4p activity (82).
At the other extreme, the Polybromo (PB1) protein involved in chromatin remodeling contains six bromodomains in tandem, with individual bromodomains exhibiting distinct preferences for specific acetyl-lysine marks (83).
TRIM Family Bromodomains
Here, we expand on an unanticipated result whereby the PHD-bromo cassette of tripartite motif 24 (TRIM24) reads out H3 peptides containing unmodified K4 and K23ac (84), whereas the PHD-bromo cassette of TRIM33 reads out H3 peptides containing unmodified K4, K9me3, and K18ac (85). Structural studies establish that K23ac recognition by the bromodomain of TRIM24 involves both hydrogen bonding with the conserved asparagine and the network of water-mediated interactions, with sequence specificity defined by the intermolecular hydrogen bonds with the flanking residue Arg26 on the peptide, thereby accounting for its specificity for H3K23ac (Figure 7d) (84). By contrast, in the case of TRIM33, there is an insert in the sequence of the bromodomain compared with TRIM24, which extends the length of α-helix B such that complex formation favors a different sequence context. As a result, K18ac inserts into the binding pocket with an intermolecular hydrogen bond to a flanking residue (Arg17), stabilizing complex formation, and accounting for the specificity of TRIM33 for H3K18ac (Figure 7e) (85). More importantly, extension of the α-helix B rotates the conserved asparagine out of the pocket, so that it no longer participates in hydrogen-bonding recognition of the acetyl-lysine (Figure 7f). Nevertheless, recognition is still mediated by a network of water molecules that now spreads throughout the entire binding pocket (Figure 7f), emphasizing the importance of water-mediated recognition and stabilization to acetyl-lysine readout.
Tandem PHD Fingers
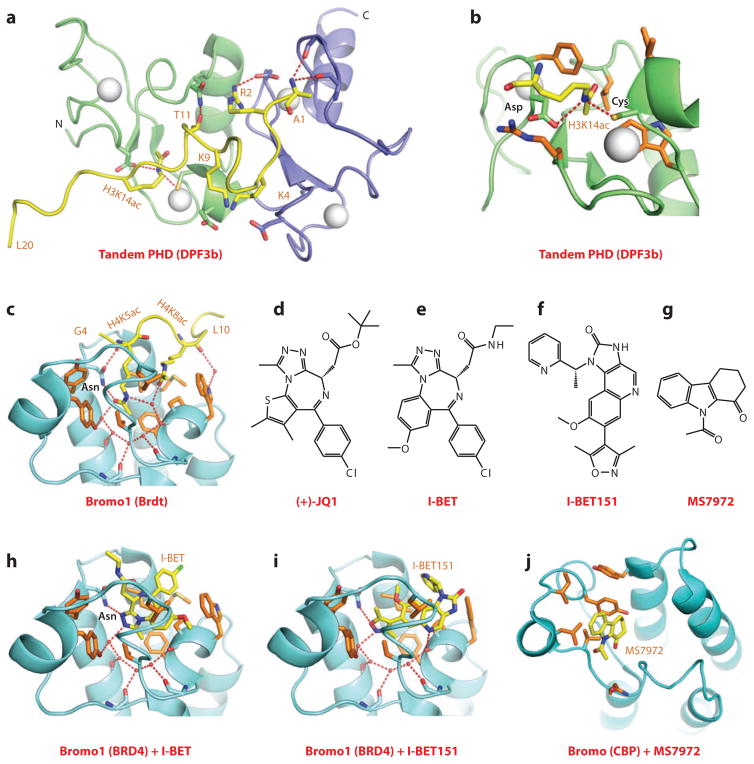
The DPF3 protein, which contains C-terminal tandem PHD fingers, plays a key role in heart and muscle development. DPF3, associated with the BAF chromatin remodeling complex, was shown to bind acetylated histones H3 and H4, with knockdown of DPF3 resulting in cardiac, ventricular, and muscle fiber defects (86). The potential of tandem PHD fingers of DPF3 for targeting acetyl-lysine marks was unanticipated, and the molecular basis underlying recognition emerged from the NMR solution structure of the DPF3 tandem PHD fingers bound to the H3K14ac-containing peptide (Kd = 0.5 μM) (87). The two PHD fingers are packed against each other to form a compact functional unit, thereby generating an extended surface groove shared between the two domains for peptide recognition (Figure 8a). The N terminus of the H3K14ac-containing peptide binds through β-sheet formation to the second PHD finger through recognition of unmodified lysine (K4 and K9) and arginine (R2) residues, while the side chain of K14ac inserts into a hydrophobic pocket in the first PHD finger with the acetylamide positioned within hydrogen bond distance to the side chains of an aspartic acid and a zinc-coordinating cysteine residue (Figure 8b). Complex formation, which is facilitated by acetylation at H3K14, is inhibited by methylation at H3K4. Notably, the H3K14ac-binding pocket on the first PHD finger is on a different surface of the PHD finger from binding pockets that target methyl-lysine marks.
Figure 8.
Structures of tandem PHD fingers (DPF3b) and bromodomains (BET family) bound to acetyl-lysine-containing histone peptides. (a) The NMR-based solution structure of the tandem PHD fingers of DPF3b bound to the H3(1–20)K14ac peptide, Protein Data Bank (PDB) reference 2KWJ. The first and second PHD fingers are shown as green and blue, respectively. The H3 peptide was traced from Ala1 to Leu20 (A1 to L20). Interactions involving the side chains of Arg2 (R2), Lys9 (K9), and Thr11 (T11) of the bound peptide are show in this panel. (b) Details of the alignment and interactions involving the side chain of K14ac with the first PHD finger in the DPF3b tandem PHD finger-H3(1–20)K14ac peptide complex. (c) The 2.37-Å structure of the first bromodomain of Brdt bound to the H4(1–20)K5acK8ac peptide (PDB: 2WP2). The bound peptide can be traced from Gly4 to Leu10 (G4 to L10). The side chain of K5ac inserts into the Kac-binding pocket, whereas the side chain of K8ac is on the outer rim of the pocket and buttresses the insertion of K5ac into the pocket. (d) Chemical formula of (+)-JQ1. (e) Chemical formula of I-BET. (f) Chemical formula of I-BET151. (g) Chemical formula of MS7972. (h) The 1.6-Å structure of the first bromodomain of BRD4 bound to the small-molecule inhibitor I-BET (PDB: 3P5O). I-BET inserts into the Kac-binding pocket and is anchored in place by shape complementarity and direct and water-mediated intermolecular contacts. (i) The 1.5-Å structure of the first bromodomain of BRD4 bound to the small-molecule inhibitor I-BET151 (PDB: 3ZYU). I-BET151 inserts into the Kac-binding pocket and is anchored in place by shape complementarity and direct and water-mediated intermolecular contacts. (j) The NMR-based solution structure of insertion of the small-molecule inhibitor MS7972 into the acetyl-lysine-binding pocket of the bromodomain of CBP. Abbreviation: N, N terminus.
Tandem PH Domains
The H3K56ac mark has been implicated in the regulation of nucleosome assembly during DNA replication and repair, as well as when the nucleosome disassembles during gene transcription. The histone chaperone Rtt106 (regulator of Ty1 transposition 106), which contains tandem plextrin homology (PH) domains toward its C terminus, contributes to the deposition of newly synthesized H3K56ac-labeled H3-H4 tetramer on replicating DNA. A NMR-based chemical shift perturbation study identified the tandem PH domains of Rtt106 as readers of the H3K56ac mark with binding affinity markedly increased on acetylation of K56 (88). In the absence of a structure of the complex, a dimeric model has been proposed, whereby a pair of tandem PH domains within the Rtt106 dimer interacts with each of the two H3K56ac sites in the H3-H4 tetramer. Mutational studies established that the interaction between H3K56ac and Rtt106 tandem PH domains contributes to gene silencing and to the DNA damage response.
BET-Family Bromodomains
Bromodomain and extraterminal (BET) family proteins play a role in transcriptional elongation and cell-cycle progression. One family member, mouse Brdt, contains two bromodomains separated by a long linker and targets multiacetylated histones on H3 and H4. The isolated bromodomain 1 of Brdt bound the H4K5acK8ac peptide with a Kd equal to 21.9 μM and a 1:1 stoichiometry of bromodomain to peptide. The structure of the complex is shown in Figure 8c, with both acetyl-lysines pointing in the direction of an otherwise widened pocket (89). The side chain of K5ac inserts deeply into the pocket where it hydrogen bonds with an asparagine residue adopting the canonical recognition mode, and the side chain of K8ac is positioned near the entrance of the pocket, stabilized through hydrophobic contacts, where it buttresses the inserted K5ac side chain. The structure also explains the requirement for a two-glycine residue separation between K5ac and K8ac within the H4 sequence on the basis of steric principles, as well as why the K8 position has to be acetylated rather than a charged unmodified lysine on the basis of electrostatic principles. Functionally, the H4K5acK8ac ligand-binding capacity of bromodomain 1 of Brdt is required for compaction of hyperacetylated chromatin. This system identifies the Brdt bromodomain 1 as a reader module that mediates positive cooperativity by engaging a pair of Kac marks in a common expanded binding pocket (89).
PHOSPHOSERINE RECOGNITION
Histone phosphorylation at serines, threonines, and tyrosines has been shown to link chromatin function with a diverse range of signal transduction pathways (reviewed in References 90 and 91). Replacement of a hydroxyl by a phosphate introduces a bulky and negatively charged moiety, thereby enhancing the ion-pairing and hydrogen-bonding capabilities following phosphorylation of serine, threonine, and tyrosine residues. Functionally, histone phosphorylation impacts on diverse processes ranging from transcriptional regulation to DNA damage repair and cell-cycle control. Histone phosphorylation sites are located not only on canonical histones but also on histone variants such as H2AX, with phosphorylation at Ser139 and Tyr142 toward the C terminus.
14-3-3 Proteins
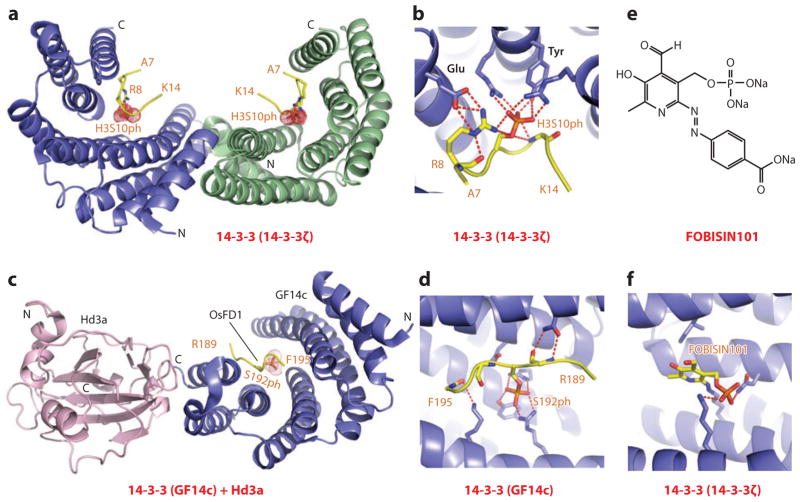
Mammalian 14-3-3 proteins are phosphoserine-binding modules that regulate signal transduction, chromosome condensation, and apoptotic cell death. The 14-3-3ζ isoform targets both H3S10ph (Kd = 78 μM) and H3S28ph (Kd = 23 μM) marks, both of which contain a common Ala-Arg-Lys-Ser sequence motif. In the crystal structure of 14-3-3ζ bound to the H3S10ph-containing peptides (92), the partially extended but kinked H3S10ph-containing peptide was positioned within a V-shaped binding channel of the all α-helical 14-3-3ζ protein scaffold (Figure 9a shows dimeric arrangement of this complex), with the phosphate of S10ph anchored through multiple intermolecular hydrogen bonds, involving the hydroxyl group of a tyrosine and the guanidinium groups of two Arg side chains, thereby facilitating charge neutralization (Figure 9b). In addition, H3R8 is also anchored in the complex through hydrogen bonding to S10ph and a Glu side chain (Figure 9b), thereby accounting for the sequence specificity of the recognition.
Figure 9.
Structures of 14-3-3 proteins bound to phosphoserine marks and inhibitors. (a) The 2.0-Å crystal structure of 14-3-3ζ bound to the H3(7–14)S10ph peptide, Protein Data Bank (PDB) reference 2C1N. The 14-3-3ζ protein in the complex adopts a symmetrical dimeric alignment, with individual monomers shown in blue and green. The bound peptide can be traced from Ala7 to Lys14 (A7 to K14). (b) Details of the alignment and hydrogen-bonding interactions involving the side chain of S10ph of the peptide in the H3(7–14)S10ph-14-3-3ζ complex. Also, note the pair of hydrogen bonds between the guanidinium group of Arg8 (R8) of the peptide and the carboxyl group of a Glu residue of the protein. (c) The 2.4-Å crystal structure of the ternary complex between Hd3a (pink), GF14c (blue), and the bound OsFD1(187–195)S192ph peptide (yellow) (PDB: 3AXY). The side chain can be traced from Arg189 to Phe195 (R189 to F195). (d) Details of the alignment and hydrogen-bonding interactions involving the side chain of phosphorylated Ser192 (S192ph) and the peptide in the ternary complex. (e) The chemical formula of FOBISIN101. (f) Details of interactions of the phosphate group of the cleaved pyridoxal-phosphate moiety of FOBISIN101 with 14-3-3ζ in the FOBISIN101-14-3-3ζ complex. Abbreviations: C, C terminus; N, N terminus.
A fascinating example of phosphoserine recognition in plant biology emerged from studies of 14-3-3 proteins that act as intracellular receptors for rice Hd3a florigen, a protein synthesized in the leaf and transported to the shoot apex, where it induces flowering. Structural studies were undertaken on the florigen activator complex, composed of the rice florigen analog Hd3a, the 14-3-3 protein GF14c, and a phosphoserine peptide from Oryza sativa transcription factor OsFD1, interacting partners identified from yeast two-hybrid studies (93). The structure of one symmetric half of the dimeric topology is shown in Figure 9c, with GF14c forming a stable complex simultaneously with Hd3a and OsFD1, while mediating indirect binding between Hd3a and OsFD1. The S192ph-containing OsFD1 peptide is positioned within the GF14c 14-3-3 domain and anchored in place through hydrogen bonding with arginine and tyrosine side chains (Figure 9d). This study highlights the role of 14-3-3 proteins as intracellular receptors for florigen in shoot apical cells, with new opportunities to manipulate the flowering process.
Tandem BRCT Domains
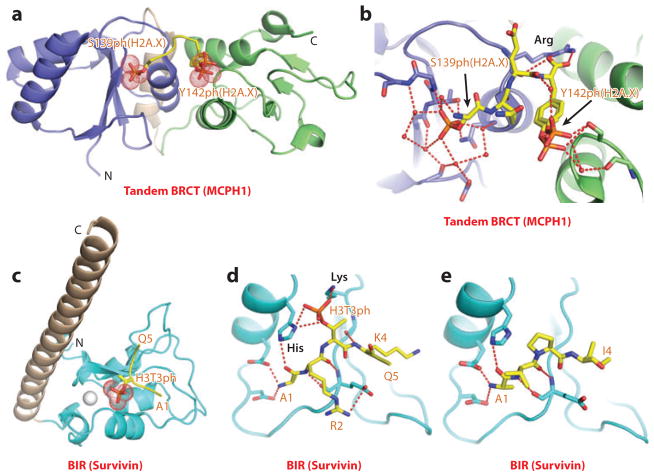
Tandem breast cancer susceptibility (BRCT) domains are readers of phosphorylation marks in response to DNA double-strand breaks as was first demonstrated for recognition of Ser139ph of the histone variant H2A.X by MDC1 (mediator of DNA damage checkpoint protein 1) (94), and more recently by recognition of both Ser139ph along with Ser139ph, Tyr142ph dual mark of H2A.X by MCPH1 (microcephalin 1) (95). Given that the structure of the MDC1 complex was reviewed previously (6), the present review focuses on the structure of the MCPH1 complex.
Under basal cellular conditions, the histone variant H2A.X is phosphorylated at C-terminal Tyr142 by the Williams-Beuren syndrome transcription factor (WSTF), a component of the WSTF-ISWI chromatin remodeling complex (96). Under DNA double-strand break conditions, H2A.X is very rapidly phosphorylated at Ser139 by the ataxia telangiectasia mutated kinase (97), while progressively dephosphorylated at Tyr142 by Eya tyrosine phosphatase (98). This results in a temporal switch from a diphosphorylated (Ser139ph and Tyr142ph; designated di-γH2A.X) to a monophosphorylated (Ser139ph; designated γH2A.X) state of this histone variant. Structural, biochemical, and cellular studies have established that the tandem BRCT domains of MCPH1, an early DNA damage response and maintenance of genome integrity protein, can read both states of H2A.X, thereby serving as a versatile sensor of both γH2A.X and di-γH2A.X phosphorylation marks (95).
The tandem BRCT domains of MCPH1 bound to both Ser139ph-modified γH2A.X (Kd = 0.7 μM) and Ser139ph, Tyr142ph–modified di-γH2A.X (Kd = 4.4 μM) peptides, but not to the Tyr142ph-modified H2A.X peptide. Structures have been determined for the tandem BRCT domains of MCPH1 bound to a Ser139ph-modified γH2A.X peptide and to a Ser139ph,Tyr142ph-modified di-γH2A.X peptide (Figure 10a), thereby defining the intermolecular contacts accounting for the sequence specificity of recognition (95). Specifically, the α/β folds of individual BRCT domains are connected through a helical linker segment, with both γH2A.X and di-γH2A.X (Figure 10b) peptides bound within a cleft at the interface of the tandem BRCT domains and with recognition specificity associated with a tyrosine at position 142 of the bound peptide that is positioned in a hydrophobic pocket. In both complexes, the phosphate group of Ser139ph is recognized by the first BRCT domain (Figure 10b) and hydrogen bonded to acceptor atoms through direct and water-mediated hydrogen bonds, with mutations of phosphate-pocket lining residues resulting in reduced binding affinity. In the di-γH2A.X peptide-MCPH1 complex, the Ser139ph phosphate is recognized by the first BRCT domain, whereas the Tyr142ph phosphate (in two alternate alignments) is recognized by the second BRCT domain (Figure 10b). The phosphate group of Tyr142ph is also hydrogen bonded to acceptor atoms through direct and water-mediated hydrogen bonds, with mutations of phosphate-pocket lining residues resulting in reduced binding affinity. In the structures of both MCPH1 complexes, the C-terminal carboxylate of Tyr142 is anchored by a pair of hydrogen bonds to the side chain of an evolutionarily conserved arginine residue (Figure 10b), whose mutation results in loss of binding.
Figure 10.
Structures of BRCT (MCPH1) and BIR (Survivin) domains bound to phosphoserine/threonine/tyrosine-containing peptides. (a) The 1.5-Å crystal structure of the complex of the tandem BRCT domains of MCPH1 bound to di-γH2A.X(139–142)S139phY142ph peptide, Protein Data Bank (PDB) reference 3U3Z. (b) Details of the alignment and interactions involving the side chain of phosphorylated Ser139 (S139ph) and phosphorylated Tyr142 (Y142ph) (arrows) of the di-γH2A.X(139–142)S139phY142ph peptide with the first (blue) and second (green) BRCT domains of MCPH1. Note that Y142ph adopts two conformations in the structure of the complex. (c) The 2.4-Å crystal structure of the complex of the BIR domain of Survivin bound to the H3(1–15)T3ph peptide (PDB: 3UIG). The peptide chain can be traced from Ala1 to Gln5 (A1 to Q5). (d) Details of the alignment and interactions between the bound H3(1–15)T3ph peptide and the BIR domain of Survivin. Interactions involving the side chains of Arg2 (R2) and Lys4 (K4) are also shown in the panel. (e) Details of the alignment and interactions between the bound Smac/DIABLO(1–15) peptide and the BIR domain of Survivin (PDB: 3UIH). The peptide chain can be traced from Ala1 to Ile4 (A1 to I4). Abbreviations: C, C terminus; MCPH1, microcephalin 1; N, N terminus.
Functionally, MDC1 (and MCPH1) were found to colocalize with γH2AX (or di-γH2A.X) and 53BP1 to DNA damage–induced nuclear foci in a manner dependent on γH2A.X (or di-γH2A.X). Disruption of the MDC1-γH2A.X (or MCPH1-di-γH2A.X) interaction through incorporation of appropriate mutations resulted in a failure to recruit MDC1 (or MCPH1) to DNA damage foci and also prevented the recruitment of downstream DNA damage repair proteins (94, 95).
A recent study determined that the structure of the tandem BRCT repeats of Rtt107, a protein that restores stalled replication forks following DNA damage, bound to phosphorylated H2A (γH2A) (99), thereby reinforcing the recognition principles outlined above for the MDC1 (94) and MCPH1 (95) complexes.
BIR Domains
Survivin is an inhibitor of apoptosis family proteins implicated in mitosis and apoptosis. As such, Survivin is a component of the chromosomal passenger complex, a key player during mitosis. It also recognizes the Smac/DIABLO protein that plays a role in apoptosis. The baculovirus IAP repeat (BIR) domain of Survivin, which adopts an αβ scaffold stabilized by a bound Zn ion (100), recognizes the H3T3ph mark; this interaction is necessary for accumulation of the chromosomal passenger complex at centromeres, an event required for accurate cell division (101–103). Structural studies were undertaken on H3T3ph-containing (104, 105) and Smac/DIABLO N-terminal (104) peptides bound to the BIR domain of Survivin (Figure 10c). The peptides bind in an extended conformation, with the N-terminal Ala residue recognized in both complexes (Figure 10d,e). In addition, the N terminus and Arg2 are hydrogen bonded to acidic side chains, the H3T3ph mark is positioned in a basic pocket, and recognition is associated with intermolecular hydrogen bonding with lysine and histidine side chains (Figure 10d). Survivin binds tighter to the H3T3ph-containing peptide relative to the N-terminal Smac/DIABLO peptide, and this preference can be reversed through structure-guided mutations that increase the hydrophobicity of the phosphate-binding pocket (104). Structural analysis also permitted identification of putative Survivin-binding epitopes in other mitotic proteins, including human Shugoshin 1 (105).
MULTIVALENT READOUT BY LINKED BINDING MODULES
The PHD-bromo cassette involving adjacently positioned PHD finger and bromodomains is the most frequently observed combination of a distinct pair of readers positioned in tandem that impact on epigenetic regulation (reviewed in References 5 and 106). In principle, the PHD-bromo cassette has the potential for readout of different combinations of methyl-lysine (PHD finger) and acetyl-lysine (bromo-domain) marks, and potentially doing so in a combinatorial manner.
BPTF PHD-Bromo Cassette
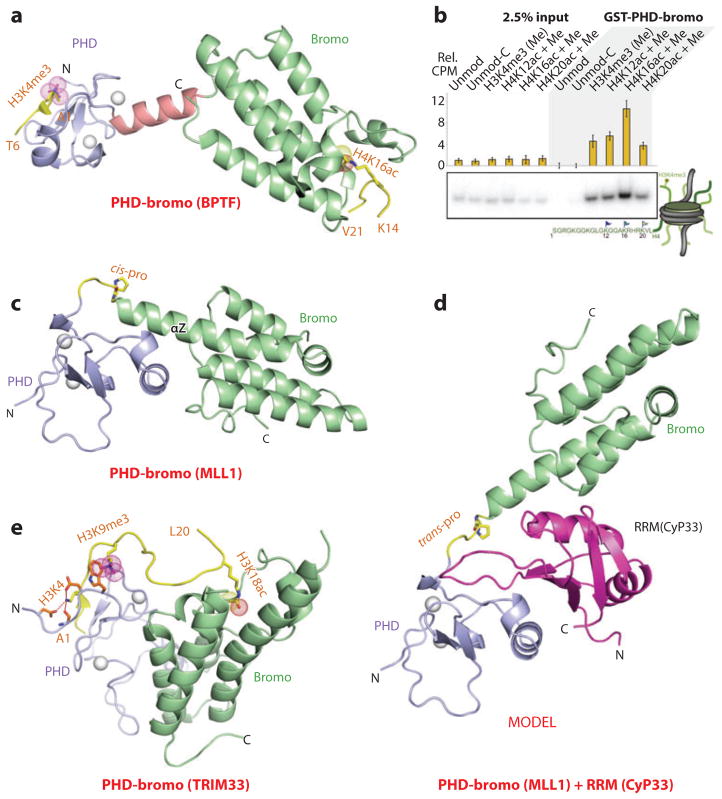
The first insights into structure-function aspects of PHD-bromodomain (PHD-bromo) cassettes emerged from studies of this cassette in BPTF, where structural studies established that the two domains were separated by an α-helical linker, resulting in a defined separation and orientation of the binding pocket domains (Figure 11a) (20). Furthermore, structural and binding studies established that the BPTF PHD finger exhibited selectivity for the H3K4me3 mark (20, 21), whereas the BPTF bromodomain targeted a range of Kac marks, demonstrating a preference for H4K16ac, with cooperative binding observed for simultaneous readout of these two marks on different histone tails (107).
Figure 11.
Structures and in vivo functional studies of PHD-bromo cassettes bound to methyl-lysine- and acetyl-lysine-containing histone peptides. (a) The 2.0-Å crystal structure of the complex of the PHD-bromo cassette of BPTF bound to the H3(1–15)K4me3 peptide, Protein Data Bank (PDB) reference 2F6Z. A separate 1.8-Å crystal structure of the bromodomain of BPTF bound to the H4(12–21)K16ac peptide was also solved (PDB: 3QZS), and that information was superimposed on the structure shown in this panel. The bound H3(1–15)K4me3-containing peptide can be traced from Ala1 to Thr6 (A1 to T6), and the bound H4(12–21)K16ac-containing peptide can be traced from Lys14 to Val21 (K14 to V21) in the complexes. (b) A glutathione S-transferase (GST) pull-down of modified nucleosomes containing semisynthetic histones produced by expressed protein ligation. Nucleosomes containing dual marks involving H4K12ac, H4K16ac, or H4K20ac in combination with H3K4me3 are pulled down with a resin-bound GST-BPTF PHD-bromo cassette and detected by autoradiography after native gel electrophoresis. (c) The 1.72-Å crystal structure of the MLL1 PHD-bromo cassette exhibits a cis linker proline in the free state (PDB: 3LQH). The PHD finger and bromodomains are blue and green, respectively. (d) Model of the MLL1 PHD-bromo cassette with a trans linker proline, with this alignment stabilized by the bound RRM domain of CyP33. (e) The 2.7-Å crystal structure of the complex of the PHD-bromo cassette of TRIM33 bound to the H3(1–22)K9me3K18ac peptide (PDB: 3U5O). The bound H3(1–22)K9me3K18ac peptide can be traced from Ala1 to Leu20 (A1 to L20) in the complex. Abbreviations: C, C terminal; CyP33, cyclophilin 33; MLL1, mixed-lineage leukemia 1 protein; N, N terminal; RRM, RNA recognition motif; Rel CPM, relative counts per minute.
These studies on dual readout by the BPTF PHD-bromo cassette at the histone tail level (20, 21) have been extended to the nucleosomal level (107). Experimentally, despite the BPTF bromodomain displaying limited discrimination among different acetyl-lysines on H4 (K12ac, K16ac, and K20ac) at the histone peptide level, a marked selectivity was observed solely for H4K16ac, in combination with H3K4me3, by the PHD-bromo cassette at the nucleosomal level (Figure 11b) (107). Indeed, a significant pool of doubly modified H3K4me3/H4K16ac nucleosomes was identified in vivo, and additional validation for the multivalent readout emerged on demonstration that the BPTF PHD-bromo cassette colocalizes with the H3K4me3 and H4K16ac marks in the genome (107). Furthermore, structure-based model building suggested that the BPTF PHD-bromo cassette can be docked onto a nucleosome core particle such that the PHD finger has the potential to target the H3K4me3 mark and the bromodomain to target the H4K16ac mark, while positioning the linker in the groove of the DNA (5).
MLL1 PHD-Bromo Cassette
The MLL1 gene is essential for embryonic development and hematopoiesis, and this gene is also a frequent target for recurrent chromosomal translocations, resulting in hematopoietic precursor transformation into leukemia cells. MLL1 is required for maintenance of HOXA9 expression in stem and progenitor cells, as well as for switching expression off during blood cell maturation, with the induction of this epigenetic switch dependent on the PHD finger 3 of MLL1 (reviewed in Reference 108). Structural studies of the MLL1 PHD-bromo cassette have been undertaken in the free and H3K4me3-bound states (109). The individual domains adopt characteristic PHD finger and bromodomain folds with the connecting linker adopting a turn segment that leads into the extended α-helix Z of the bromodomain, such that the two domains interact with each other (Figure 11c). This interaction is manifested in an increase in the binding affinity of the PHD finger for the H3K4me3 mark in the PHD-bromo cassette relative to the isolated PHD finger. Furthermore, MLL1 and H3K4me3 marks colocalize on HOX genes, with these ChIP-based results implying that binding of the H3K4me3 mark by the MLL1 PHD3 contributes to MLL1 localization at target genes (109). By contrast, the MLL1 bromodomain, for as yet to be determined reasons, has lost its ability to bind Kac-containing histone peptides.
Cyclophilin CyP33, a peptidyl-prolyl isomerase that contains an attached RNA recognition motif (RRM) domain, binds to the PHD-bromo cassette of MLL1, wherein it regulates MLL1 function through HDAC recruitment (110, 111). Examination of the structure of the MLL1 PHD-bromo cassette identified a cis proline in the linker segment between domains (Figure 11c), which in the presence of CyP33 undergoes cis-trans proline isomerization. The trans proline isomer, where the two domains no longer interact with each other, is stabilized by binding to the RRM module of CyP33 (model in Figure 11d is based on the structure of CyP33 RRM bound to a fragment of MLL1 PHD3) (109). Thus, disruption of the PHD-bromo interface, as a result of the cis-trans proline transition, facilitates access to an otherwise occluded PHD finger in the cassette to the CyP33 RRM domain. Furthermore, the H3K4me3 and the RRM domain target different surfaces of PHD3, thereby potentially integrating distinct regulatory inputs by coexisting as a ternary complex. Functional experiments demonstrate that the interaction between MLL1 and CyP33 is required for HDAC-mediated repression of HOX target genes in vivo (109). These results collectively establish the role of the MLL1 PHD-bromo cassette both as a regulatory platform and as a switch, whereby cis-trans proline isomerization within the linker segment connects H3K4me3 readout to CyP33-binding and HDAC-mediated repression.
TRIM PHD-Bromo Cassettes
TRIM33, a SMAD (signal transduction)-binding protein, contains a PHD-bromo cassette, implying that it could target histone PTMs. The challenge here focused on understanding how specific chromatin marks keep master regulators of differentiation silent yet poised for activation by cellular signals. Structural studies established that the PHD and bromodomains of the TRIM33 PHD-bromo cassette interact extensively with each other. The TRIM33 PHD-bromo cassette reads out modified H3 tails through multivalent readout by targeting the N terminus, unmodified K4, K9me3 (all by the PHD finger), and K18ac (by the bromodomain) marks (Figure 11e) (85). Binding studies establish that the affinity of TRIM33 for the H3(1–28)K9me3K18ac dual mark peptide (Kd = 0.06 μM) was greater than the sum of binding to individual H3(1–28)K9me3 (Kd = 0.20 μM) and H3(1–28)K18ac (Kd = 0.21 μM) marks, indicative of combinatorial readout of marks at the peptide level. Functionally, nodal (a member of the TGF-β superfamily) induces formation of the TRIM33-Smad2/3 complex, which in turn interacts with the H3K9me3 mark through its PHD-bromo cassette, thereby displacing the chromatin-compacting factor HP1γ and in the process making nodal response elements accessible to the Smad4-Smad2/3 complex for Pol II recruitment. In essence, nodal TGF-β signals use the poised H3K9me3 mark as a platform to switch master regulators of stem cell differentiation from the poised to the active state (85). Related structure-function studies have also been undertaken on the TRIM24 PHD-bromo cassette, where histone PTM recognition involved the H3K23ac mark instead of the H3K18ac mark, with aberrant expression of TRIM24 negatively correlating with survival of breast cancer patients (84).
KAP1 (KRAB-associated protein 1 or TRIM28) is a corepressor containing a PHD-bromo cassette, where the domains also interact extensively with each other but do not bind the anticipated Kme (by the PHD finger) and Kac (by the bromodomain) marks (112, 113). Rather, the PHD finger in the cassette unexpectedly and interestingly functions as an intramolecular E3 SUMO ligase, which facilitates sumoylation of the linked bromodomain. Such PHD-mediated sumoylation facilitates establishment and stabilization of silent chromatin. Interestingly, site-specific phosphorylation of a serine located near the bromodomain of KAP1 appears to inhibit KAP1 sumoylation (114), implying a role for PTM-mediated cross talk in the regulation of KAP1 function.
EPIGENETICS-BASED DRUG DISCOVERY
There is much to be learned from a molecular level understanding of the consequences of genomic alterations and abnormal expression profiles of chromatin regulators on the onset of diseased states, ranging from developmental abnormalities to autoimmune and neurological diseases, and cancer. The development of ongoing chromatin-based therapeutics holds promise given the potential for reversibility of epigenetic-based mutations, as reflected in the recent promising approaches to therapeutic intervention (reviewed in References 27 and 115).
Inhibitors of Kme-Binding MBT Repeats
Some effort has gone into the design of small-molecule inhibitors that target the cavity insertion pockets of L3MBTL1, which are both narrow and deep and function as size-selective filters. A structure-guided approach has identified UNC669 (Figure 3d), a pyrrolidine-containing small molecule, which targets L3MBTL1 with a Kd equal to 5 μM while discriminating against L3MBTL3/4 (116). In the structure of the UNC669 bound to L3MBTL1, the ligand inserts its pyrrolidine ring system into aromatic pocket 2 (Figure 3e), with good shape complementarity between the inserted ligand and the walls of the pocket (116).
A greater challenge, as yet to yield results, would be to target the wider and shallower methyl-lysine-binding surface groove pockets observed in chromodomain, Tudor, PWWP, and PHD finger domains.
Inhibitors of Kac-Binding Bromodomains
Given that bromodomains have both deep and hydrophobic acetyl-lysine-binding pockets, they serve as excellent scaffolds for identification of small-molecule inhibitors of complex formation. The main challenge has been to identify inhibitors that target the bromodomain pockets with nanomolar affinity, exhibit specificity in terms of distinguishing between bromodomain subfamilies, and have favorable clinically relevant disease-related outcomes. The main efforts at inhibitor design have targeted the BET subfamily, which contains a pair of tandem bromodomains. These efforts have resulted in the identification of small-molecule compounds, (+)-JQ1 and I-BET, with promising therapeutic potential (reviewed in References 117 and 118; for more general reviews of epigenetic-based drugs, see reviews in References 115 and 119.
A cell-permeable small-molecule (thienotriazolo-1,4-diazepine-based) inhibitor, named (+)-JQ1 (Figure 8d), was shown to target the acetyl-lysine-binding pocket of bromodomain-containing protein 4 (BRD4) with nanomolar affinity, while competitively displacing bound acetyl-lysine-containing histone peptides (120). The bound (+)-JQ1, but not its (−)-JQ1 stereoisomer, mimics the intermolecular interactions formed by acetyl-lysine, including formation of a hydrogen bond between the triazole ring and the side chain of the conserved asparagine lining the pocket. Intermolecular contacts at the entrance to the pocket explain the specificity of (+)-JQ1 for the BET subfamily of bromodomains. Recurrent translocation of BRD4 with a nuclear protein in testis (NUT) results in formation of the BRD4-NUT oncoprotein, resulting in the generation of an incurable subtype of human squamous carcinoma. (+)-JQ1 competitively displaces the BRD4 fusion oncoprotein from chromatin, facilitating squamous differentiation and specific antiproliferative effects in BRD4-dependent cell lines and patient-derived xenograft models, with the outcome of tumor regression and prolonged survival (120). In addition, (+)-JQ1-based inhibition of BET bromodomains has also been shown to be a therapeutic strategy for targeting c-Myc through generation of a potent antiproliferative effect associated with cell-cycle arrest and cellular senescence (121–123). Equally significant is the recent demonstration of (+)-JQ1-based inhibition of the testis-specific BRT bromodomain, thereby inhibiting bromodomain activity during spermatogenesis, with the potential for male contraception (124).
In a parallel investigation, a high-throughput screen identified a permeable benzodiazepine-based small molecule, I-BET (Figure 8e), that selectively targeted the BET bromodomain subfamily with nanomolar affinity (125). The structure of the complex with I-BET inserted into the binding pocket of bromodomain 1 of BRD4 is shown in Figure 8h. Structural elements within and at the entrance of the pocket, termed the gatekeeper residue, WPF shelf, and ZA channel, account for the spatial complementarity and specificity of recognition. Functionally, I-BET was shown to disrupt chromatin complexes responsible for the expression of key inflammatory genes in activated macrophages, and I-BET also confers protection against lipopolysaccharide-induced endotoxic shock and bacteria-induced sepsis (125). In a follow-up study, a related small molecule, I-BET151 (Figure 8f), was shown to inhibit BET bro-modomain subfamily recruitment (along with a super elongation complex and polymerase-associated factor complex) to chromatin, thereby serving as a potential treatment with survival benefit for patients with MLL fusion leukemia (126). The structural basis for this inhibition can be accessed from the structure of I-BET151 bound within the binding pocket of bromodomain 1 of BRD4 (Figure 8i). Here again, the principle of shape complementarity and distribution of intermolecular contacts account for the specificity of recognition.
Efforts have also been made to identify bromodomain-binding small molecules that inhibit association of human p53 with the transcriptional activator CREB-binding protein (CBP), a process that is essential for p53-induced transcription of the cell-cycle inhibitor p21, in response to DNA damage. This interaction is mediated by the interaction of the p53K382ac mark with the bromo domain of CBP. The structure of one such small molecule, MS7972 (Figure 8g), bound to the bromodomain of CBP is shown in Figure 8j, whereby disruption of the p53-CBP interaction results in inhibition of the transcriptional activity of p53, together with promotion of p53 instability (127). In a related study, a small molecule named ischemin was identified, which when bound to the bromodomain of the coactivator CBP blocked apoptosis in cardiomyocytes (128). One potential limitation to date of the structure-guided approach to identification of small molecules, such as MS7972 (Figure 8g) and ischemin for therapeutic intervention, is that they bind their bromodomain targets with micromolar affinity, in contrast to (+)-JQ1 (Figure 8d) and I-BET (Figure 8e), which bind their bromodomain targets with nanomolar affinity.
Finally, small-molecule bromodomain inhibitors, such as N1-aryl-propane-1,3-diamine compounds, have been identified through structure-based design to target the interaction between HIV trans-activator Tat and the host transcriptional coactivator PCAF, modified by acetylation at K50, as an approach for targeting the host cell PCAF protein in an effort to control transcriptional activation of the integrated HIV-1 provirus (129).
Inhibitors of Sph-Binding 14-3-3 Proteins
14-3-3 Proteins function as adaptor modules that control the function of their target proteins as a consequence of highly regulated protein-protein interactions. 14-3-3 Proteins serve as attractive drug targets given that they are Sph/Tph-binding modules that impact on multiple signaling pathways involved in cell fate and organ development. A chemical screen approach toward identification of small-molecule inhibitors of 14-3-3 proteins yielded FOBISIN101, containing N = N linked pyridoxal-phosphate and benzoate moieties (Figure 9e), which effectively blocked binding of 14-3-3 with both Raf-1 (IC50 = 9.3 μM) and the proline-rich AKT substrate (130). The structure of FOBISIN101 bound to 14-3-3ζ established that X-ray exposure reduced and cleaved the N = N double-bond linkage between the pyridoxal-phosphate and benzoate moieties of FOBISIN101, resulting in a covalent attachment of the pyridoxal-phosphate moiety to Lys120 in the binding groove of 14-3-3ζ (Figure 9f), constituting persistent inactivation of the enzyme (130). In the complex, the phosphate group of FOBISIN101 interacts with lysine and asparagine side chains, and the pyridoxal ring makes van der Waals contacts with the side chain of an isoleucine residue (Figure 9f). These results outline the identification of a novel inhibitor of Sph-binding 14-3-3 proteins that could potentially serve a radiation-triggered therapeutic agent for treatment of 14-3-3-mediated diseases.
SUPPLEMENTAL SECTIONS
The issue of PTM cross talk between adjacent and nearby marks, and the impact of such cross talk on DNA-templated processes, is outlined in the Cross Talk between Histone Marks section in the Supplemental Material: Follow the Supplemental Material link from the Annual Reviews home page at http://www.annualreviews.org. Included are specific examples of pair-wise PTM cross talk between Kme and Rme, Sph, Kac, and Kph marks.
The Supplemental Material also contains a Histone Mimics section on the readout of PTM marks on nonhistone proteins and the extent to which such histone mimics expand our insight into principles of PTM recognition beyond the boundaries of the histone code to regulation of nonhistone proteins.
The Supplemental Material expands on new initiatives and future challenges with the promise for enhancing our current understanding of the readout of histone PTMs at the peptide and nucleosomal level. The topics include additional lysine modifications, technological developments at the genome-wide level, chemical biology approaches to designer nucleosomes, histone mark readout at the nucleosomal level, linking histone and DNA methylation, and the functional roles of noncoding RNAs.
Supplementary Material
PERSPECTIVES.
From a structural perspective, the most important challenges are related to the structural determination of histone PTM readout at the nucleosome level using designer nucleo-somes containing site specifically incorporated PTMs and, in the longer term, to extending these studies to PTM-modified designer nucleosomes bound to ATP-dependent remodeling complexes. Although advances have been made in the area of the function of long-noncoding RNAs (lncRNAs) in epigenetic regulation, the total absence of structural information on lnc-RNAs and their protein complexes opens exciting challenges related to understanding the principles underlying the specificity of complex formation.
SUMMARY POINTS.
The review has outlined structure-based principles underlying posttranslational modification (PTM) readout of methyl-lysine and methyl-arginine marks as a function of a methylation state, as well as their unmodified counterparts, by a range of reader modules.
Structural studies have identified the diversity of recognition scaffolds within a given family of reader modules, reflected in readout of methyl-lysine states by distinct topologies adopted by tandem Tudor domains.
The diversity of methyl-lysine-binding pockets ranging from surface groove and cavity insertion modes of recognition involving cation-π interactions are contrasted with interfacial composite pocket recognition involving surface complementarity and noncanonical hydrogen-bonding interactions.
Related structural studies have defined the principles underlying recognition of acetyl-lysine marks, as well as phosphorylation marks on serine, threonine, or tyrosine by their respective reader modules.
The multivalent readout of two or more marks occurs by linked binding modules, such as PHD-bromo cassettes, with binding affinity enhanced as a result of combinatorial readout.
Structure-function studies outlined in the Supplemental Material evaluate the impact of cross talk between adjacent PTMs on modulation of the readout and its consequences for transcriptional regulation.
The Supplemental Material discusses the feasibility of PTM readout on nonhistone proteins and the degree to which such histone mimics expand the knowledge base beyond boundaries associated with canonical recognition of histone PTMs by reader modules.
The targeting of bromodomain acetyl-lysine-binding pockets by small-molecule inhibitors impacting on tumor suppressor function has been successful, with promising potential against inflammation and as a male contraceptive.
FUTURE ISSUES.
Our current structure-function understanding of readout of methyl-lysine, acetyl-lysine, and phosphoserine marks should be expanded to newly identified PTMs.
Future studies should seek an improved understanding of the distribution, functional readout, and cross talk between PTMs at the genome-wide level.
Developing additional chemical biological approaches directed toward enhancement of biochemical and biophysical studies of PTMs at the nucleosomal level is a necessity.
Focused efforts are required for long-term structural studies of histone PTM readout at the nucleosomal level, along with extension to multivalent PTM readout in a nucleosomal context.
We must gain an improved understanding of larger complexes through structural studies of PTM-modified designer nucleosomes bound to ATP-dependent remodeling complexes.
In the future, studies should address the issue of linkage of histone and DNA methylation marks and the extent to which histone methylation serves as a template for DNA methylation and/or the converse process.
There is a need for an improved understanding of the molecular basis underlying the function of lncRNAs from structural studies of their complexes with their bound protein targets.
Future studies should elucidate the impact of dysfunctional chromatin readers on onset of diseased states and development of chromatin-based therapeutics targeted to reader-binding pockets.
Acknowledgments
This research was supported by funds from the STARR and Maloris foundations and the Abby Rockefeller Mauze Trust to D.J.P.
Glossary
- Posttranslational modification (PTM) cross talk
modulation of the readout of a PTM by the mark status of an adjacent PTM
- ChIP-chip
a technique that combines chromatin immunoprecipitation (ChIP) with microarray technology (chip) to investigate interactions between proteins and DNA in vivo
- ChIP-Seq
a technique that combines chromatin immunoprecipitation (ChIP) with massively parallel DNA sequencing to identify the binding sites of DNA-associated proteins in vivo
- Histone mimics
nonhistone posttranslational modifications (PTMs) that are recognized by reader modules using recognition principles in common with histone PTMs
- Surface groove pocket
a wider and more accessible pocket, with the methyl-lysine side chain positioned along a surface groove with greater tolerance for methylation states
- Cation-π interactions
electrostatic stabilization of the diffuse positive charge of the methylammonium group through stacking with a partial negative charge of the ring of an aromatic residue
- PHD-bromodomain (PHD-bromo) cassette
the formation of a dual reader module by linkage of a PHD finger and a bromodomain capable of simultaneously targeting methyl-lysine and acetyl-lysine marks
- Cavity insertion pocket
a deep and more restricted pocket, with the methyl-lysine inserted into and buried within a deep binding cleft with size-sensitive selection capabilities
- Interfacial composite pocket
a composite binding pocket at the interface of two interacting modules, with methyl-lysine recognition dictated by shape complementarity
- Designer nucleosomes
application of expressed protein ligation technology to couple PTM-modified N-terminal histone peptides with tailless histones, which when combined with 167-bp DNA yield designer nucleosomes
Footnotes
DISCLOSURE STATEMENT
The authors are not aware of any affiliations, membership, funding, or financial holdings that might be perceived as affecting the objectivity of this review.
Contributor Information
Dinshaw J. Patel, Email: pateld@mskcc.org.
Zhanxin Wang, Email: zhanxinw@msn.com.
LITERATURE CITED
- 1.Luger K, Dechassa ML, Tremethick DJ. New insights into nucleosome and chromatin structure: an ordered state or a disordered affair? Nat Rev Mol Cell Biol. 2012;13:436–47. doi: 10.1038/nrm3382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kornberg RD, Lorch Y. Twenty-five years of the nucleosome, fundamental particle of the eukaryote chromosome. Cell. 1999;98:285–94. doi: 10.1016/s0092-8674(00)81958-3. [DOI] [PubMed] [Google Scholar]
- 3.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 4.Musselman CA, Lalonde ME, Cote J, Kutateladze TG. Perceiving the epigenetic landscape through histone readers. Nat Struct Mol Biol. 2012;19:1218–27. doi: 10.1038/nsmb.2436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ruthenburg AJ, Li H, Patel DJ, Allis CD. Multivalent engagement of chromatin modifications by linked binding modules. Nat Rev Mol Cell Biol. 2007;8:983–94. doi: 10.1038/nrm2298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Taverna SD, Li H, Ruthenburg AJ, Allis CD, Patel DJ. How chromatin-binding modules interpret histone modifications: lessons from professional pocket pickers. Nat Struct Mol Biol. 2007;14:1025–40. doi: 10.1038/nsmb1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yap KL, Zhou MM. Keeping it in the family: diverse histone recognition by conserved structural folds. Crit Rev Biochem Mol Biol. 2010;45:488–505. doi: 10.3109/10409238.2010.512001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ruthenburg AJ, Allis CD, Wysocka J. Methylation of lysine 4 on histone H3: intricacy of writing and reading a single epigenetic mark. Mol Cell. 2007;25:15–30. doi: 10.1016/j.molcel.2006.12.014. [DOI] [PubMed] [Google Scholar]
- 9.Maurer-Stroh S, Dickens NJ, Hughes-Davies L, Kouzarides T, Eisenhaber F, Ponting CP. The Tudor domain ‘Royal Family’: Tudor, plant Agenet, Chromo, PWWP and MBT domains. Trends Biochem Sci. 2003;28:69–74. doi: 10.1016/S0968-0004(03)00004-5. [DOI] [PubMed] [Google Scholar]
- 10.Jacobs SA, Khorasanizadeh S. Structure of HP1 chromodomain bound to a lysine 9-methylated histone H3 tail. Science. 2002;295:2080–83. doi: 10.1126/science.1069473. [DOI] [PubMed] [Google Scholar]
- 11.Nielsen PR, Nietlispach D, Mott HR, Callaghan J, Bannister A, et al. Structure of the HP1 chromodomain bound to histone H3 methylated at lysine 9. Nature. 2002;416:103–7. doi: 10.1038/nature722. [DOI] [PubMed] [Google Scholar]
- 12.Min J, Zhang Y, Xu RM. Structural basis for specific binding of Polycomb chromodomain to histone H3 methylated at Lys 27. Genes Dev. 2003;17:1823–28. doi: 10.1101/gad.269603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ma JC, Dougherty DA. The cation-π interaction. Chem Rev. 1997;97:1303–24. doi: 10.1021/cr9603744. [DOI] [PubMed] [Google Scholar]
- 14.Ballare C, Lange M, Lapinaite A, Martin GM, Morey L, et al. Phf19 links methylated Lys36 of histone H3 to regulation of Polycomb activity. Nat Struct Mol Biol. 2012;19:1257–65. doi: 10.1038/nsmb.2434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brien GL, Gambero G, O’Connell DJ, Jerman E, Turner SA, et al. Polycomb PHF19 binds H3K36me3 and recruits PRC2 and demethylase NO66 to embryonic stem cell genes during differentiation. Nat Struct Mol Biol. 2012;19:1273–81. doi: 10.1038/nsmb.2449. [DOI] [PubMed] [Google Scholar]
- 6.Cai L, Rothbart SB, Lu R, Xu B, Chen WY, et al. An H3K36 methylation-engaging Tudor motif of Polycomb-like proteins mediates PRC2 complex targeting. Mol Cell. 2013;49:571–82. doi: 10.1016/j.molcel.2012.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Musselman CA, Avvakumov N, Watanabe R, Abraham CG, Lalonde ME, et al. Molecular basis for H3K36me3 recognition by the Tudor domain of PHF1. Nat Struct Mol Biol. 2012;19:1266–72. doi: 10.1038/nsmb.2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vezzoli A, Bonadies N, Allen MD, Freund SM, Santiveri CM, et al. Molecular basis of histone H3K36me3 recognition by the PWWP domain of Brpf1. Nat Struct Mol Biol. 2010;17:617–19. doi: 10.1038/nsmb.1797. [DOI] [PubMed] [Google Scholar]
- 19.Pascual J, Martinez-Yamout M, Dyson HJ, Wright PE. Structure of the PHD zinc finger from human Williams-Beuren syndrome transcription factor. J Mol Biol. 2000;304:723–29. doi: 10.1006/jmbi.2000.4308. [DOI] [PubMed] [Google Scholar]
- 20.Li H, Ilin S, Wang W, Duncan EM, Wysocka J, et al. Molecular basis for site-specific read-out of histone H3K4me3 by the BPTF PHD finger of NURF. Nature. 2006;442:91–95. doi: 10.1038/nature04802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wysocka J, Swigut T, Xiao H, Milne TA, Kwon SY, et al. A PHD finger of NURF couples histone H3 lysine 4 trimethylation with chromatin remodelling. Nature. 2006;442:86–90. doi: 10.1038/nature04815. [DOI] [PubMed] [Google Scholar]
- 22.Pena PV, Davrazou F, Shi X, Walter KL, Verkhusha VV, et al. Molecular mechanism of histone H3K4me3 recognition by plant homeodomain of ING2. Nature. 2006;442:100–3. doi: 10.1038/nature04814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shi X, Hong T, Walter KL, Ewalt M, Michishita E, et al. ING2 PHD domain links histone H3 lysine 4 methylation to active gene repression. Nature. 2006;442:96–99. doi: 10.1038/nature04835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–26. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- 25.Santos-Rosa H, Schneider R, Bannister AJ, Sherriff J, Bernstein BE, et al. Active genes are tri-methylated at K4 of histone H3. Nature. 2002;419:407–11. doi: 10.1038/nature01080. [DOI] [PubMed] [Google Scholar]
- 26.Li H, Fischle W, Wang W, Duncan EM, Liang L, et al. Structural basis for lower lysine methylation state-specific readout by MBT repeats of L3MBTL1 and an engineered PHD finger. Mol Cell. 2007;28:677–91. doi: 10.1016/j.molcel.2007.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chi P, Allis CD, Wang GG. Covalent histone modifications—miswritten, misinterpreted and mis-erased in human cancers. Nat Rev Cancer. 2010;10:457–69. doi: 10.1038/nrc2876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Matthews AG, Kuo AJ, Ramón-Maiques S, Han S, Champagne KS, et al. RAG2 PHD finger couples histone H3 lysine 4 trimethylation with V(D)J recombination. Nature. 2007;450:1106–10. doi: 10.1038/nature06431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ramón-Maiques S, Kuo AJ, Carney D, Matthews AG, Oettinger MA, et al. The plant homeodomain finger of RAG2 recognizes histone H3 methylated at both lysine-4 and arginine-2. Proc Natl Acad Sci USA. 2007;104:18993–98. doi: 10.1073/pnas.0709170104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang GG, Song J, Wang Z, Dormann HL, Casadio F, et al. Haematopoietic malignancies caused by dysregulation of a chromatin-binding PHD finger. Nature. 2009;459:847–51. doi: 10.1038/nature08036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Musselman CA, Kutateladze TG. Handpicking epigenetic marks with PHD fingers. Nucleic Acids Res. 2011;39:9061–71. doi: 10.1093/nar/gkr613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sanchez R, Zhou MM. The PHD finger: a versatile epigenome reader. Trends Biochem Sci. 2011;36:364–72. doi: 10.1016/j.tibs.2011.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lan F, Collins RE, De Cegli R, Alpatov R, Horton JR, et al. Recognition of unmethylated histone H3 lysine 4 links BHC80 to LSD1-mediated gene repression. Nature. 2007;448:718–22. doi: 10.1038/nature06034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ooi SK, Qiu C, Bernstein E, Li K, Jia D, et al. DNMT3L connects unmethylated lysine 4 of histone H3 to de novo methylation of DNA. Nature. 2007;448:714–17. doi: 10.1038/nature05987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Eustermann S, Yang JC, Law MJ, Amos R, Chapman LM, et al. Combinatorial readout of histone H3 modifications specifies localization of ATRX to heterochromatin. Nat Struct Mol Biol. 2011;18:777–82. doi: 10.1038/nsmb.2070. [DOI] [PubMed] [Google Scholar]
- 36.Iwase S, Xiang B, Ghosh S, Ren T, Lewis PW, et al. ATRX ADD domain links an atypical histone methylation recognition mechanism to human mental-retardation syndrome. Nat Struct Mol Biol. 2011;18:769–76. doi: 10.1038/nsmb.2062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang Z, Hayashi MK, Merkel O, Stillman B, Xu RM. Structure and function of the BAH-containing domain of Orc1p in epigenetic silencing. EMBO J. 2002;21:4600–11. doi: 10.1093/emboj/cdf468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kuo AJ, Song J, Cheung P, Ishibe-Murakami S, Yamazoe S, et al. The BAH domain of ORC1 links H4K20me2 to DNA replication licensing and Meier-Gorlin syndrome. Nature. 2012;484:115–19. doi: 10.1038/nature10956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Duncker BP, Chesnokov IN, McConkey BJ. The origin recognition complex protein family. Genome Biol. 2009;10:214. doi: 10.1186/gb-2009-10-3-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Klingseisen A, Jackson AP. Mechanisms and pathways of growth failure in primordial dwarfism. Genes Dev. 2011;25:2011–24. doi: 10.1101/gad.169037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yang N, Xu RM. Structure and function of the BAH domain in chromatin biology. Crit Rev Biochem Mol Biol. 2013 doi: 10.3109/10409238.2012.742035. In press. [DOI] [PubMed] [Google Scholar]
- 42.Du J, Zhong X, Bernatavichute YV, Stroud H, Feng S, et al. Dual binding of chromomethylase domains to H3K9me2-containing nucleosomes directs DNA methylation in plants. Cell. 2012;151:167–80. doi: 10.1016/j.cell.2012.07.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bonasio R, Lecona E, Reinberg D. MBT domain proteins in development and disease. Semin Cell Dev Biol. 2010;21:221–30. doi: 10.1016/j.semcdb.2009.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang WK, Tereshko V, Boccuni P, MacGrogan D, Nimer SD, Patel DJ. Malignant brain tumor repeats: a three-leaved propeller architecture with ligand/peptide binding pockets. Structure. 2003;11:775–89. doi: 10.1016/s0969-2126(03)00127-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sathyamurthy A, Allen MD, Murzin AG, Bycroft M. Crystal structure of the malignant brain tumor (MBT) repeats in sex comb on midleg-like 2 (SCML2) J Biol Chem. 2003;278:46968–73. doi: 10.1074/jbc.M306469200. [DOI] [PubMed] [Google Scholar]
- 46.Min J, Allali-Hassani A, Nady N, Qi C, Ouyang H, et al. L3MBTL1 recognition of mono- and dimethylated histones. Nat Struct Mol Biol. 2007;14:1229–30. doi: 10.1038/nsmb1340. [DOI] [PubMed] [Google Scholar]
- 47.Trojer P, Li G, Sims RJ, III, Vaquero A, Kalakonda N, et al. L3MBTL1, a histone-methylation-dependent chromatin lock. Cell. 2007;129:915–28. doi: 10.1016/j.cell.2007.03.048. [DOI] [PubMed] [Google Scholar]
- 48.Argentaro A, Yang JC, Chapman L, Kowalczyk MS, Gibbons RJ, et al. Structural consequences of disease-causing mutations in the ATRX-DNMT3-DNMT3L (ADD) domain of the chromatin-associated protein ATRX. Proc Natl Acad Sci USA. 2007;104:11939–44. doi: 10.1073/pnas.0704057104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jiao Y, Shi C, Edil BH, de Wilde RF, Klimstra DS, et al. DAXX/ATRX, MEN1, and mTOR pathway genes are frequently altered in pancreatic neuroendocrine tumors. Science. 2011;331:1199–203. doi: 10.1126/science.1200609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Flanagan JF, Mi LZ, Chruszcz M, Cymborowski M, Clines KL, et al. Double chromodomains cooperate to recognize the methylated histone H3 tail. Nature. 2005;438:1181–85. doi: 10.1038/nature04290. [DOI] [PubMed] [Google Scholar]
- 51.Botuyan MV, Lee J, Ward IM, Kim JE, Thompson JR, et al. Structural basis for the methylation state-specific recognition of histone H4-K20 by 53BP1 and Crb2 in DNA repair. Cell. 2006;127:1361–73. doi: 10.1016/j.cell.2006.10.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Huang Y, Fang J, Bedford MT, Zhang Y, Xu RM. Recognition of histone H3 lysine-4 methylation by the double Tudor domain of JMJD2A. Science. 2006;312:748–51. doi: 10.1126/science.1125162. [DOI] [PubMed] [Google Scholar]
- 53.Lee J, Thompson JR, Botuyan MV, Mer G. Distinct binding modes specify the recognition of methylated histones H3K4 and H4K20 by JMJD2A-tudor. Nat Struct Mol Biol. 2008;15:109–11. doi: 10.1038/nsmb1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bian C, Xu C, Ruan J, Lee KK, Burke TL, et al. Sgf29 binds histone H3K4me2/3 and is required for SAGA complex recruitment and histone H3 acetylation. EMBO J. 2011;30:2829–42. doi: 10.1038/emboj.2011.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nady N, Lemak A, Walker JR, Avvakumov GV, Kareta MS, et al. Recognition of multivalent histone states associated with heterochromatin by UHRF1 protein. J Biol Chem. 2011;286:24300–11. doi: 10.1074/jbc.M111.234104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Law JA, Du J, Hale J, Feng S, Krajewski K, et al. Polymerase-IV occupancy at RNA-directed DNA methylation sites requires SHH1. Nature. 2013 doi: 10.1038/nature12178. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Collins RE, Northrop JP, Horton JR, Lee DY, Zhang X, et al. The ankyrin repeats of G9a and GLP histone methyltransferases are mono- and dimethyllysine binding modules. Nat Struct Mol Biol. 2008;15:245–50. doi: 10.1038/nsmb.1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Arita K, Isogai S, Oda T, Unoki M, Sugita K, et al. Recognition of modification status on a histone H3 tail by linked histone reader modules of the epigenetic regulator UHRF1. Proc Natl Acad Sci USA. 2012;109:12950–55. doi: 10.1073/pnas.1203701109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cheng J, Yang Y, Fang J, Xiao J, Zhu T, et al. Structural insight into coordinated recognition of trimethylated histone H3 lysine 9 (H3K9me3) by the plant homeodomain (PHD) and tandem Tudor domain (TTD) of UHRF1 (ubiquitin-like, containing PHD and RING finger domains, 1) protein. J Biol Chem. 2013;288:1329–39. doi: 10.1074/jbc.M112.415398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Fiedler M, Sánchez-Barrena MJ, Nekrasov M, Mieszczanek J, Rybin V, et al. Decoding of methylated histone H3 tail by the Pygo-BCL9 Wnt signaling complex. Mol Cell. 2008;30:507–18. doi: 10.1016/j.molcel.2008.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Miller TC, Rutherford TJ, Johnson CM, Fiedler M, Bienz M. Allosteric remodelling of the histone H3 binding pocket in the Pygo2 PHD finger triggered by its binding to the B9L/BCL9 co-factor. J Mol Biol. 2010;401:969–84. doi: 10.1016/j.jmb.2010.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Alekseyenko AA, Peng S, Larschan E, Gorchakov AA, Lee OK, et al. A sequence motif within chromatin entry sites directs MSL establishment on the Drosophila X chromosome. Cell. 2008;134:599–609. doi: 10.1016/j.cell.2008.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kim D, Blus BJ, Chandra V, Huang P, Rastinejad F, Khorasanizadeh S. Corecognition of DNA and a methylated histone tail by the MSL3 chromodomain. Nat Struct Mol Biol. 2010;17:1027–29. doi: 10.1038/nsmb.1856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chen C, Nott TJ, Jin J, Pawson T. Deciphering arginine methylation: Tudor tells the tale. Nat Rev Mol Cell Biol. 2011;12:629–42. doi: 10.1038/nrm3185. [DOI] [PubMed] [Google Scholar]
- 65.Tripsianes K, Madl T, Machyna M, Fessas D, Englbrecht C, et al. Structural basis for dimethyl-arginine recognition by the Tudor domains of human SMN and SPF30 proteins. Nat Struct Mol Biol. 2011;18:1414–20. doi: 10.1038/nsmb.2185. [DOI] [PubMed] [Google Scholar]
- 66.Liu H, Wang JY, Huang Y, Li Z, Gong W, et al. Structural basis for methylarginine-dependent recognition of Aubergine by Tudor. Genes Dev. 2010;24:1876–81. doi: 10.1101/gad.1956010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Liu K, Chen C, Guo Y, Lam R, Bian C, et al. Structural basis for recognition of arginine methylated Piwi proteins by the extended Tudor domain. Proc Natl Acad Sci USA. 2010;107:18398–403. doi: 10.1073/pnas.1013106107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mathioudakis N, Palencia A, Kadlec J, Round A, Tripsianes K, et al. The multiple Tudor domain-containing protein TDRD1 is a molecular scaffold for mouse Piwi proteins and piRNA biogenesis factors. RNA. 2012;18:2056–72. doi: 10.1261/rna.034181.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Rajakumara E, Wang Z, Ma H, Hu L, Chen H, et al. PHD finger recognition of unmodified histone H3R2 links UHRF1 to regulation of euchromatic gene expression. Mol Cell. 2011;43:275–84. doi: 10.1016/j.molcel.2011.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wysocka J, Swigut T, Milne TA, Dou Y, Zhang X, et al. WDR5 associates with histone H3 methylated at K4 and is essential for H3 K4 methylation and vertebrate development. Cell. 2005;121:859–72. doi: 10.1016/j.cell.2005.03.036. [DOI] [PubMed] [Google Scholar]
- 71.Couture JF, Collazo E, Trievel RC. Molecular recognition of histone H3 by the WD40 protein WDR5. Nat Struct Mol Biol. 2006;13:698–703. doi: 10.1038/nsmb1116. [DOI] [PubMed] [Google Scholar]
- 72.Han Z, Guo L, Wang H, Shen Y, Deng XW, Chai J. Structural basis for the specific recognition of methylated histone H3 lysine 4 by the WD-40 protein WDR5. Mol Cell. 2006;22:137–44. doi: 10.1016/j.molcel.2006.03.018. [DOI] [PubMed] [Google Scholar]
- 73.Ruthenburg AJ, Wang W, Graybosch DM, Li H, Allis CD, et al. Histone H3 recognition and presentation by the WDR5 module of the MLL1 complex. Nat Struct Mol Biol. 2006;13:704–12. doi: 10.1038/nsmb1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Schuetz A, Allali-Hassani A, Martin F, Loppnau P, Vedadi M, et al. Structural basis for molecular recognition and presentation of histone H3 by WDR5. EMBO J. 2006;25:4245–52. doi: 10.1038/sj.emboj.7601316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Migliori V, Muller J, Phalke S, Low D, Bezzi M, et al. Symmetric dimethylation of H3R2 is a newly identified histone mark that supports euchromatin maintenance. Nat Struct Mol Biol. 2012;19:136–44. doi: 10.1038/nsmb.2209. [DOI] [PubMed] [Google Scholar]
- 76.Holdermann I, Meyer NH, Round A, Wild K, Sattler M, Sinning I. Chromodomains read the arginine code of post-translational targeting. Nat Struct Mol Biol. 2012;19:260–63. doi: 10.1038/nsmb.2196. [DOI] [PubMed] [Google Scholar]
- 77.Sanchez R, Zhou MM. The role of human bromodomains in chromatin biology and gene transcription. Curr Opin Drug Discov Dev. 2009;12:659–65. [PMC free article] [PubMed] [Google Scholar]
- 78.Dhalluin C, Carlson JE, Zeng L, He C, Aggarwal AK, Zhou MM. Structure and ligand of a histone acetyltransferase bromodomain. Nature. 1999;399:491–96. doi: 10.1038/20974. [DOI] [PubMed] [Google Scholar]
- 79.Owen DJ, Ornaghi P, Yang JC, Lowe N, Evans PR, et al. The structural basis for the recognition of acetylated histone H4 by the bromodomain of histone acetyltransferase Gcn5p. EMBO J. 2000;19:6141–49. doi: 10.1093/emboj/19.22.6141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Filippakopoulos P, Picaud S, Mangos M, Keates T, Lambert JP, et al. Histone recognition and large-scale structural analysis of the human bromodomain family. Cell. 2012;149:214–31. doi: 10.1016/j.cell.2012.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Jacobson RH, Ladurner AG, King DS, Tjian R. Structure and function of a human TAFII250 double bromodomain module. Science. 2000;288:1422–25. doi: 10.1126/science.288.5470.1422. [DOI] [PubMed] [Google Scholar]
- 82.VanDemark AP, Kasten MM, Ferris E, Heroux A, Hill CP, Cairns BR. Autoregulation of the Rsc4 tandem bromodomain by Gcn5 acetylation. Mol Cell. 2007;27:817–28. doi: 10.1016/j.molcel.2007.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Charlop-Powers Z, Zeng L, Zhang Q, Zhou MM. Structural insights into selective histone H3 recognition by the human Polybromo bromodomain 2. Cell Res. 2010;20:529–38. doi: 10.1038/cr.2010.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Tsai WW, Wang Z, Yiu TT, Akdemir KC, Xia W, et al. TRIM24 links a non-canonical histone signature to breast cancer. Nature. 2010;468:927–32. doi: 10.1038/nature09542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Xi Q, Wang Z, Zaromytidou AI, Zhang XH, Chow-Tsang LF, et al. A poised chromatin platform for TGF-β access to master regulators. Cell. 2011;147:1511–24. doi: 10.1016/j.cell.2011.11.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Lange M, Kaynak B, Forster UB, Tonjes M, Fischer JJ, et al. Regulation of muscle development by DPF3, a novel histone acetylation and methylation reader of the BAF chromatin remodeling complex. Genes Dev. 2008;22:2370–84. doi: 10.1101/gad.471408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Zeng L, Zhang Q, Li S, Plotnikov AN, Walsh MJ, Zhou MM. Mechanism and regulation of acetylated histone binding by the tandem PHD finger of DPF3b. Nature. 2010;466:258–62. doi: 10.1038/nature09139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Su D, Hu Q, Li Q, Thompson JR, Cui G, et al. Structural basis for recognition of H3K56-acetylated histone H3-H4 by the chaperone Rtt106. Nature. 2012;483:104–7. doi: 10.1038/nature10861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Moriniere J, Rousseaux S, Steuerwald U, Soler-Lopez M, Curtet S, et al. Cooperative binding of two acetylation marks on a histone tail by a single bromodomain. Nature. 2009;461:664–68. doi: 10.1038/nature08397. [DOI] [PubMed] [Google Scholar]
- 90.Baek SH. When signaling kinases meet histones and histone modifiers in the nucleus. Mol Cell. 2011;42:274–84. doi: 10.1016/j.molcel.2011.03.022. [DOI] [PubMed] [Google Scholar]
- 91.Seet BT, Dikic I, Zhou MM, Pawson T. Reading protein modifications with interaction domains. Nat Rev Mol Cell Biol. 2006;7:473–83. doi: 10.1038/nrm1960. [DOI] [PubMed] [Google Scholar]
- 92.Macdonald N, Welburn JP, Noble ME, Nguyen A, Yaffe MB, et al. Molecular basis for the recognition of phosphorylated and phosphoacetylated histone H3 by 14-3-3. Mol Cell. 2005;20:199–211. doi: 10.1016/j.molcel.2005.08.032. [DOI] [PubMed] [Google Scholar]
- 93.Taoka K, Ohki I, Tsuji H, Furuita K, Hayashi K, et al. 14-3-3 Proteins act as intracellular receptors for rice Hd3a florigen. Nature. 2011;476:332–35. doi: 10.1038/nature10272. [DOI] [PubMed] [Google Scholar]
- 94.Stucki M, Clapperton JA, Mohammad D, Yaffe MB, Smerdon SJ, Jackson SP. MDC1 directly binds phosphorylated histone H2AX to regulate cellular responses to DNA double-strand breaks. Cell. 2005;123:1213–26. doi: 10.1016/j.cell.2005.09.038. [DOI] [PubMed] [Google Scholar]
- 95.Singh N, Basnet H, Wiltshire TD, Mohammad DH, Thompson JR, et al. Dual recognition of phosphoserine and phosphotyrosine in histone variant H2A.X by DNA damage response protein MCPH1. Proc Natl Acad Sci USA. 2012;109:14381–86. doi: 10.1073/pnas.1212366109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Xiao A, Li H, Shechter D, Ahn SH, Fabrizio LA, et al. WSTF regulates the H2A.X DNA damage response via a novel tyrosine kinase activity. Nature. 2009;457:57–62. doi: 10.1038/nature07668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Burma S, Chen BP, Murphy M, Kurimasa A, Chen DJ. ATM phosphorylates histone H2AX in response to DNA double-strand breaks. J Biol Chem. 2001;276:42462–67. doi: 10.1074/jbc.C100466200. [DOI] [PubMed] [Google Scholar]
- 98.Cook PJ, Ju BG, Telese F, Wang X, Glass CK, Rosenfeld MG. Tyrosine dephosphorylation of H2AX modulates apoptosis and survival decisions. Nature. 2009;458:591–96. doi: 10.1038/nature07849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Li X, Liu K, Li F, Wang J, Huang H, et al. Structure of C-terminal tandem BRCT repeats of Rtt107 protein reveals critical role in interaction with phosphorylated histone H2A during DNA damage repair. J Biol Chem. 2012;287:9137–46. doi: 10.1074/jbc.M111.311860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Jeyaprakash AA, Klein UR, Lindner D, Ebert J, Nigg EA, Conti E. Structure of a Survivin-Borealin-INCENP core complex reveals how chromosomal passengers travel together. Cell. 2007;131:271–85. doi: 10.1016/j.cell.2007.07.045. [DOI] [PubMed] [Google Scholar]
- 101.Kelly AE, Ghenoiu C, Xue JZ, Zierhut C, Kimura H, Funabiki H. Survivin reads phosphorylated histone H3 threonine 3 to activate the mitotic kinase Aurora B. Science. 2010;330:235–39. doi: 10.1126/science.1189505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Wang F, Dai J, Daum JR, Niedzialkowska E, Banerjee B, et al. Histone H3 Thr-3 phosphorylation by Haspin positions Aurora B at centromeres in mitosis. Science. 2010;330:231–35. doi: 10.1126/science.1189435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Yamagishi Y, Honda T, Tanno Y, Watanabe Y. Two histone marks establish the inner centromere and chromosome bi-orientation. Science. 2010;330:239–43. doi: 10.1126/science.1194498. [DOI] [PubMed] [Google Scholar]
- 104.Du J, Kelly AE, Funabiki H, Patel DJ. Structural basis for recognition of H3T3ph and Smac/DIABLO N-terminal peptides by human Survivin. Structure. 2012;20:185–95. doi: 10.1016/j.str.2011.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Jeyaprakash AA, Basquin C, Jayachandran U, Conti E. Structural basis for the recognition of phosphorylated histone H3 by the Survivin subunit of the chromosomal passenger complex. Structure. 2011;19:1625–34. doi: 10.1016/j.str.2011.09.002. [DOI] [PubMed] [Google Scholar]
- 106.Wang Z, Patel DJ. Combinatorial readout of dual histone modifications by paired chromatin-associated modules. J Biol Chem. 2011;286:18363–68. doi: 10.1074/jbc.R111.219139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Ruthenburg AJ, Li H, Milne TA, Dewell S, McGinty RK, et al. Recognition of a mononucleosomal histone modification pattern by BPTF via multivalent interactions. Cell. 2011;145:692–706. doi: 10.1016/j.cell.2011.03.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Grow EJ, Wysocka J. Flipping MLL1’s switch one proline at a time. Cell. 2010;141:1108–10. doi: 10.1016/j.cell.2010.06.013. [DOI] [PubMed] [Google Scholar]
- 109.Wang Z, Song J, Milne TA, Wang GG, Li H, et al. Pro isomerization in MLL1 PHD3-bromo cassette connects H3K4me readout to CyP33 and HDAC-mediated repression. Cell. 2010;141:1183–94. doi: 10.1016/j.cell.2010.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Fair K, Anderson M, Bulanova E, Mi H, Tropschug M, Diaz MO. Protein interactions of the MLL PHD fingers modulate MLL target gene regulation in human cells. Mol Cell Biol. 2001;21:3589–97. doi: 10.1128/MCB.21.10.3589-3597.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Xia ZB, Anderson M, Diaz MO, Zeleznik-Le NJ. MLL repression domain interacts with histone deacetylases, the polycomb group proteins HPC2 and BMI-1, and the corepressor C-terminal-binding protein. Proc Natl Acad Sci USA. 2003;100:8342–47. doi: 10.1073/pnas.1436338100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Ivanov AV, Peng H, Yurchenko V, Yap KL, Negorev DG, et al. PHD domain-mediated E3 ligase activity directs intramolecular sumoylation of an adjacent bromodomain required for gene silencing. Mol Cell. 2007;28:823–37. doi: 10.1016/j.molcel.2007.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Zeng L, Yap KL, Ivanov AV, Wang X, Mujtaba S, et al. Structural insights into human KAP1 PHD finger-bromodomain and its role in gene silencing. Nat Struct Mol Biol. 2008;15:626–33. doi: 10.1038/nsmb.1416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Li X, Lee YK, Jeng JC, Yen Y, Schultz DC, et al. Role for KAP1 serine 824 phosphorylation and sumoylation/desumoylation switch in regulating KAP1-mediated transcriptional repression. J Biol Chem. 2007;282:36177–89. doi: 10.1074/jbc.M706912200. [DOI] [PubMed] [Google Scholar]
- 115.Dawson MA, Kouzarides T. Cancer epigenetics: from mechanism to therapy. Cell. 2012;150:12–27. doi: 10.1016/j.cell.2012.06.013. [DOI] [PubMed] [Google Scholar]
- 116.Herold JM, Wigle TJ, Norris JL, Lam R, Korboukh VK, et al. Small-molecule ligands of methyl-lysine binding proteins. J Med Chem. 2011;54:2504–11. doi: 10.1021/jm200045v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Oliver SS, Denu JM. Disrupting the reader of histone language. Angew Chem Int Ed Engl. 2011;50:5801–3. doi: 10.1002/anie.201101414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Prinjha RK, Witherington J, Lee K. Place your BETs: the therapeutic potential of bromodomains. Trends Pharmacol Sci. 2012;33:146–53. doi: 10.1016/j.tips.2011.12.002. [DOI] [PubMed] [Google Scholar]
- 119.Arrowsmith CH, Bountra C, Fish PV, Lee K, Schapira M. Epigenetic protein families: a new frontier for drug discovery. Nat Rev Drug Discov. 2012;11:384–400. doi: 10.1038/nrd3674. [DOI] [PubMed] [Google Scholar]
- 120.Filippakopoulos P, Qi J, Picaud S, Shen Y, Smith WB, et al. Selective inhibition of BET bromo-domains. Nature. 2010;468:1067–73. doi: 10.1038/nature09504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Delmore JE, Issa GC, Lemieux ME, Rahl PB, Shi J, et al. BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Cell. 2011;146:904–17. doi: 10.1016/j.cell.2011.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Mertz JA, Conery AR, Bryant BM, Sandy P, Balasubramanian S, et al. Targeting MYC dependence in cancer by inhibiting BET bromodomains. Proc Natl Acad Sci USA. 2011;108:16669–74. doi: 10.1073/pnas.1108190108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Zuber J, Shi J, Wang E, Rappaport AR, Herrmann H, et al. RNAi screen identifies Brd4 as a therapeutic target in acute myeloid leukaemia. Nature. 2011;478:524–28. doi: 10.1038/nature10334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Matzuk MM, McKeown MR, Filippakopoulos P, Li Q, Ma L, et al. Small-molecule inhibition of BRDT for male contraception. Cell. 2012;150:673–84. doi: 10.1016/j.cell.2012.06.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Nicodeme E, Jeffrey KL, Schaefer U, Beinke S, Dewell S, et al. Suppression of inflammation by a synthetic histone mimic. Nature. 2010;468:1119–23. doi: 10.1038/nature09589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Dawson MA, Prinjha RK, Dittmann A, Giotopoulos G, Bantscheff M, et al. Inhibition of BET recruitment to chromatin as an effective treatment for MLL-fusion leukaemia. Nature. 2011;478:529–33. doi: 10.1038/nature10509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Sachchidanand, Resnick-Silverman L, Yan S, Mutjaba S, Liu WJ, et al. Target structure-based discovery of small molecules that block human p53 and CREB binding protein association. Chem Biol. 2006;13:81–90. doi: 10.1016/j.chembiol.2005.10.014. [DOI] [PubMed] [Google Scholar]
- 128.Borah JC, Mujtaba S, Karakikes I, Zeng L, Muller M, et al. A small molecule binding to the coactivator CREB-binding protein blocks apoptosis in cardiomyocytes. Chem Biol. 2011;18:531–41. doi: 10.1016/j.chembiol.2010.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Pan C, Mezei M, Mujtaba S, Muller M, Zeng L, et al. Structure-guided optimization of small molecules inhibiting human immunodeficiency virus 1 Tat association with the human coactivator p300/CREB binding protein–associated factor. J Med Chem. 2007;50:2285–88. doi: 10.1021/jm070014g. [DOI] [PubMed] [Google Scholar]
- 130.Zhao J, Du Y, Horton JR, Upadhyay AK, Lou B, et al. Discovery and structural characterization of a small molecule 14-3-3 protein-protein interaction inhibitor. Proc Natl Acad Sci USA. 2011;108:16212–16. doi: 10.1073/pnas.1100012108. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.