Abstract
Rheumatoid arthritis (RA) is a chronic systemic inflammatory disease characterized by synovial inflammation and joint disability. Curcumin is known to be effective in ameliorating joint inflammation in RA. To obtain new insights into the effect of curcumin on primary fibroblast-like synoviocytes (FLS, N = 3), which are key effector cells in RA, we employed gas chromatography/time-of-flight mass spectrometry (GC/TOF-MS)-based metabolomics. Metabolomic profiling of tumor necrosis factor (TNF)-α-stimulated and curcumin-treated FLS was performed using GC/TOF-MS in conjunction with univariate and multivariate statistical analyses. A total of 119 metabolites were identified. Metabolomic analysis revealed that metabolite profiles were clearly distinct between TNF-α-stimulated vs. the control group (not stimulated by TNF-α or curcumin). Treatment of FLS with curcumin showed that the metabolic perturbation by TNF-α could be reversed to that of the control group to a considerable extent. Curcumin-treated FLS had higher restoration of amino acid and fatty acid metabolism, as indicated by the prominent metabolic restoration of intermediates of amino acid and fatty acid metabolism, compared with that observed in TNF-α-stimulated FLS. In particular, the abundance of glycine, citrulline, arachidonic acid, and saturated fatty acids in TNF-α-stimulated FLS was restored to the control level after treatment with curcumin, suggesting that the effect of curcumin on preventing joint inflammation may be elucidated with the levels of these metabolites. Our results suggest that GC/TOF-MS-based metabolomic investigation using FLS has the potential for discovering the mechanism of action of curcumin and new targets for therapeutic drugs in RA.
Introduction
Rheumatoid arthritis (RA) is a chronic systemic inflammatory disease characterized by synovial inflammation and hyperplasia, and concomitant destruction of the cartilage and bone. Proinflammatory transcription factors such as NF-κB and proinflammatory cytokines such as tumor necrosis factor (TNF)-α, are closely associated with the pathological process of RA [1].
Curcumin (diferuloylmethane), a polyphenol derived from the rhizomes of the plant Curcuma longa (turmeric), has been used as a traditional medicine to treat many inflammatory disorders [2,3]. Many researchers have shown the potent anti-inflammatory, anti-carcinogenic, and antioxidant action of curcumin against cancer and inflammatory diseases [3–7]. Human clinical trials have also shown beneficial effects against cancer and inflammatory diseases, such as inflammatory bowel disease, uveitis, and orbital pseudotumor [3,8]. Despite the beneficial effects of curcumin in cancer and inflammatory diseases, it has not yet been approved to treat chronic inflammatory arthritis such as RA.
Curcumin is also effective in reducing joint inflammation, based on studies conducted in fibroblast-like synoviocytes (FLS) and animal models in RA [9–12]. Although the exact mechanism underlying the effect in inflammatory diseases remains to be elucidated, the anti-inflammatory activity of curcumin appears to be closely related to the suppression of proinflammatory cytokines such as TNF-α, interleukin (IL)-1β and the down-regulation of cyclooxygenases (COX)-2, nitric oxide synthase, mitogen-activated kinases, and NF-κB [3,8,13,14].
In this study, we applied metabolomics to investigate the beneficial effect of curcumin on FLS in RA. Metabolomics is a tool for comprehensively analyzing all small-molecule metabolites generated in a given biological system, and has been widely used in several biomedical areas, such as cellular responses to drugs or nutrients and new drug development [15]. For example, metabolomics has provided insight into the mechanism of action underlying curcumin in breast cancer cell lines [16,17].
The important pathologic feature of RA FLS is the characteristic ability to express inflammatory cytokines, chemokines, adhesion molecules, and matrix-degrading enzymes. FLS also increase in number and become a prominent component of the destructive pannus in RA [18,19]. Although the clinical activity of curcumin is well known, the mechanism of action of curcumin remains to be elucidated at the cellular level [16,20]. In this study, through metabolomic analysis using FLS with the same genetic background and treated in a uniform manner, a deep and new understanding of the therapeutic effects of curcumin in RA was targeted.
Materials and Methods
Preparation of Curcumin
Curcumin was purchased from Sigma-Aldrich (St. Louis, MO, USA). Curcumin 10mg was dissolved in dimethyl sulfoxide (DMSO) 1mL, and then further diluted in phosphate-buffered saline (PBS).
Isolation and culture of RA FLS
Because immortalized mammalian FLS cell lines in RA are not yet available for scientific research, primary FLS cultures were employed. Synovial tissues were obtained from 3 RA patients undergoing arthroscopic wrist synovectomy or total knee joint replacement. The clinical characteristics of the patients are depicted in Table A in S1 File. The experimental protocols used in this study were approved by the Samsung Medical Center (#2006-08-117) institutional review board and written informed consent was obtained from each patient included in this study. This study was conducted in accordance with the principles expressed in the Helsinki Declaration.
Patients with RA were diagnosed according to the standard criteria reported by the American College of Rheumatology [21]. Synovial tissue was minced, and digested overnight with 5 mg/mL type IV collagenase (Sigma, Poole, UK) and 150 μg/mL type I DNase (Sigma, Poole, UK), and separated from undigested tissue using unit gravity sedimentation. After collecting the suspended cells into fresh tubes, the cells were harvested by centrifugation at 500 g (relative centrifuge force) for 10 min. The pellet was washed twice with Dulbecco’s Modified Eagle’s Media (DMEM; Life Technologies, Grand Island, NY, USA) containing 10% fetal bovine serum (FBS). The resuspended cells were plated at a concentration of 2 x 106/mL in a total volume of 1 mL/200 mm2 into T-25 culture flasks. After overnight incubation, the non-adherent cells were removed by replacing the medium with fresh culture medium. The attached cells were cultured in DMEM with 10% FBS and 50 units/mL penicillin, 50 mg/mL streptomycin, and 0.025 mg/mL amphotericin B, until 90% confluent growth was achieved. Primary cultured cells were passaged three to five times over several weeks for subsequent experiments. Between the third and fourth passages, cultures were a homogeneous population of fibroblastic cells.
To investigate the metabolic perturbation and anti-inflammatory effects of curcumin on RA FLS, FLS were divided into three groups as follows: 1) RA FLS pretreated with curcumin (40 μM) for 1hr before stimulation with TNF-α (100 ng/ml) for 24 hour; 2) RA FLS only stimulated with TNF-α (100 ng/ml) for 24 hours; 3) control RA FLS not treated with curcumin or TNF-α at all.
Treatment of RA FLS with curcumin and TNF-α
To study the effects of curcumin on IL-6, IL-8, and matrix metalloproteinase (MMP) production, RA FLS (N = 3) were incubated with DMSO-containing vehicle or curcumin (40 μM) for 1 h, followed by stimulation with TNF-α (100 ng/mL) for 24 h. Then, the culture supernatant was collected and centrifuged at 10,000 g for 5 min at 4°C to remove particulate matter, and was stored at -20°C in fresh tubes. Using commercially available enzyme immunoassay kits (ELISA; R&D Systems, Minneapolis, MN, USA), the culture supernatants were used to determine the quantities of IL-6, IL-8, MMP-1, and MMP-3.
Sample preparation for metabolomics
To obtain the metabolome sample from RA FLS (N = 3), we used fast filtration methods with a slight modification of the methods used in previous studies [22]. Briefly, RA FLS were detached in PBS buffer using a rubber-tipped cell scraper. The 2 mL of FLS were vacuum-filtered using a nylon membrane filter (0.45-μm pore size, 30 mm diameter; Whatman, Piscataway, NJ, USA) and washed with 2 mL PBS buffer. The cells with membrane filter were mixed with a water:2-propanol:methanol mixture (5:2:2, v/v/v) at -20°C, and then, the extraction mixture was directly immersed in liquid nitrogen. The extraction mixture was thawed on ice and vortexed for 3 min. After centrifugation at 13,000 g for 5 min at 4°C, the supernatant was collected and completely vacuum-dried using a vacuum concentrator (Labconco, Kansas City, MO, USA). The dried samples were derivatized with 5 μL of methoxyamine hydrochloride in pyridine (40 mg/mL; Sigma-Aldrich, St. Louis, MO, USA) at 30°C for 90 min and 45 μL of N-methyl-N-(trimethylsilyl) trifluoroacetamide (Fluka; Buchs, Switzerland) at 37°C for 30 min. A mixture of fatty acid methyl esters (C08-C30) as retention index markers was added to the derivatized extract.
Metabolite analysis using gas chromatography with time-of-flight mass spectrometry (GC/TOF-MS)
We used an Agilent 7890B GC (Agilent Technologies, Wilmington, DE, USA) equipped with Pegasus HT TOF-MS (LECO, St. Joseph, MI, USA) for sample analysis. A 1 μL sample was injected into an RTX-5Sil MS column (30 m × 0.25 mm, 0.25-μm film thickness; Restek, Bellefonte, PA, USA) and an integrated guard column (10 m × 0.25 mm, 0.25-μm film thickness; Restek, Bellefonte, PA, USA) on splitless mode for separation of metabolome in samples. The oven temperature was set for 5 min at 50°C, and was then increased to 330°C at the rate of 20°C/min, where it was then held for 5 min. The mass spectra were collected at the mass range of 85 to 500 m/z at an acquisition rate of 17 spectra/s. The temperatures of ion source and transfer line were 250°C and 280°C, respectively, and the impact of electron ionization was at 70 eV.
Statistical analysis
The data obtained from GC/TOF-MS was preprocessed by using Chroma TOF software (v. 4.50; LECO, St. Joseph, MI, USA). After that, we used BinBase, an in-house database for further processing of the preprocessed data [23]. The processed data were then normalized by abundance of the sum of identified metabolites. The median of coefficient of variation of the identified metabolites in control group, TNF-α-stimulated, and curcumin-treated RA FLS were 0.21, 0.10, and 0.09, respectively. Therefore, all identified metabolites were used for statistical analyses in this study. We conducted univariate and multivariate statistical analyses of normalized data using Statistica (v. 7.1; StatSoft, Tulsa, OK, USA) [24]. After transforming the data with unit variance scaling, hierarchical clustering analysis (HCA) based on the Euclidean distance coefficient and average linkage method was performed using MultiExperiment Viewer (MeV; Dana-Farber Cancer Institute, Boston, MA, USA) to cluster both conditions and the identified metabolites [25]. The pathway analysis was conducted using the web-based MetaboAnalyst (http://www.metaboanalyst.ca) [26]. The measured levels of IL-6, IL-8, and MMP were expressed as mean ± standard deviation for the number of experiments. Statistical significance was determined by comparisons among each group using the Mann-Whitney test or the Kruskal-Wallis test. Values with a P < 0.05 were considered significant.
Results
Overview of metabolite profiles among untreated, TNF-α-stimulated, and curcumin-treated RA FLS
The metabolites obtained from the control group, TNF-α-stimulated, and curcumin-treated RA FLS were analyzed using GC/TOF–MS, and over 1,000 unique m/z values with retention time were detected. After deconvolution and alignment using ChromaTOF software and BinBase, a total of 119 metabolites were identified (Table 1). These metabolites are classified into various chemical classes, including amino acids (19.3% of identified metabolites), organic acids (19.3%), sugars and sugar alcohols (18.5%), fatty acids (17.6%), and amines (12.6%). These metabolites are major intermediates of metabolic pathways, such as glycolysis, amino acid and fatty acid metabolisms, as well as those of the urea and tricarboxylic acid cycles.
Table 1. Classification of 119 metabolites identified according to metabolic pathway and comparison of relative levels of metabolites between three groups.
| Metabolite | Control | TNF-α-stimulated | Curcumin + TNF-α-stimulated |
|---|---|---|---|
| Amine | |||
| 5ʹ-deoxy-5ʹ-methylthioadenosine | 1 ± 0.228 | 0.345 ± 0.042* | 0.973 ± 0.340 |
| adenosine | 1 ± 0.283 | 0.433 ± 0.053# | 1.227 ± 0.239* |
| guanine | 1 ± 0.059 | 0.492 ± 0.165* | 0.909 ± 0.107# |
| inosine | 1 ± 0.499 | 0.516 ± 0.316 | 1.004 ± 0.097 |
| methylamine NIST a | 1 ± 0.513 | 0.333 ± 0.102 | 0.683 ± 0.162# |
| O-phosphorylethanolamine | 1 ± 0.172 | 0.736 ± 0.029 | 1.171 ± 0.210# |
| spermidine | 1 ± 0.319* | 0.788 ± 0.559 | 2.321 ± 0.165# |
| thymine | 1 ± 0.367 | 0.662 ± 0.493 | 0.579 ± 0.155 |
| uracil | 1 ± 0.239 | 0.999 ± 0.107 | 0.814 ± 0.091 |
| xanthine | 1 ± 0.013 | 0.745 ± 0.037* | 0.682 ± 0.234 |
| citrulline | 1 ± 0.384 | 3.365 ± 0.263* | 0.883 ± 0.085* |
| cysteine | 1 ± 0.200* | 1.020 ± 0.210 | 2.333 ± 0.082* |
| hypoxanthine | 1 ± 0.691 | 1.917 ± 0.167 | 1.260 ± 0.097* |
| nicotinamide | 1 ± 0.111 | 1.513 ± 0.014# | 1.076 ± 0.151# |
| putrescine | 1 ± 0.219 | 1.644 ± 0.119# | 0.996 ± 0.093* |
| Amino acid | |||
| asparagine | 1 ± 0.148 | 0.604 ± 0.317 | 0.978 ± 0.127 |
| asparagine dehydrated | 1 ± 0.275 | 0.597 ± 0.136 | 0.750 ± 0.717 |
| aspartate | 1 ± 0.400# | 0.427 ± 0.044 | 2.715 ± 0.031* |
| β-alanine | 1 ± 0.166 | 0.649 ± 0.141# | 1.140 ± 0.178# |
| alanine | 1 ± 0.312 | 1.185 ± 0.329 | 1.046 ± 0.218 |
| glutamate | 1 ± 0.054* | 1.003 ± 0.024 | 2.381 ± 0.032* |
| glutamine | 1 ± 0.177 | 1.630 ± 0.099* | 1.183 ± 0.018# |
| glycine | 1 ± 0.162 | 1.354 ± 0.003# | 1.038 ± 0.051* |
| histidine | 1 ± 0.440 | 15.36 ± 2.694* | 3.308 ± 2.543* |
| isoleucine | 1 ± 0.178 | 2.155 ± 0.040* | 1.285 ± 0.016* |
| leucine | 1 ± 0.258 | 2.420 ± 0.177* | 1.526 ± 0.059* |
| lysine | 1 ± 0.147 | 1.814 ± 0.101* | 0.940 ± 0.027* |
| methionine | 1 ± 0.262 | 1.617 ± 0.351 | 0.798 ± 0.169# |
| N-methylalanine | 1 ± 0.206 | 1.212 ± 0.235 | 1.025 ± 0.269 |
| ornithine | 1 ± 0.309 | 1.928 ± 0.016# | 1.034 ± 0.100* |
| oxoproline | 1 ± 0.030 | 1.607 ± 0.005* | 1.046 ± 0.011* |
| phenylalanine | 1 ± 0.012* | 1.612 ± 0.078* | 0.942 ± 0.011* |
| proline | 1 ± 0.391# | 1.397 ± 0.140 | 1.997 ± 0.129* |
| serine | 1 ± 0.044* | 1.523 ± 0.018* | 1.125 ± 0.010* |
| threonine | 1 ± 0.031* | 1.691 ± 0.038* | 1.147 ± 0.018* |
| tryptophan | 1 ± 0.097 | 1.862 ± 0.057* | 1.049 ± 0.004* |
| tyrosine | 1 ± 0.064 | 2.096 ± 0.032* | 1.030 ± 0.017* |
| valine | 1 ± 0.146# | 2.201 ± 0.092* | 1.349 ± 0.066* |
| Fatty acid | |||
| 1-monopalmitin | 1 ± 0.340 | 0.193 ± 0.018# | 1.206 ± 0.041* |
| 2-ketoisocaproic acid | 1 ± 0.439 | 0.841 ± 0.195 | 0.801 ± 0.156 |
| arachidic acid | 1 ± 0.181 | 0.457 ± 0.076* | 0.847 ± 0.101* |
| arachidonic acid | 1 ± 0.190# | 0.347 ± 0.109* | 0.644 ± 0.057# |
| behenic acid | 1 ± 0.744 | 0.525 ± 0.051 | 0.386 ± 0.414 |
| capric acid | 1 ± 0.083* | 0.783 ± 0.386 | 1.277 ± 0.043 |
| fructose-6-phosphate | 1 ± 0.211 | 0.483 ±0.083# | 1.984 ±0.826# |
| heptadecanoic acid | 1 ± 0.258 | 0.466 ± 0.028 | 1.052 ± 0.088* |
| lauric acid | 1 ± 0.176 | 0.614 ± 0.027 | 0.760 ± 0.013* |
| lignoceric acid | 1 ± 0.232# | 0.478 ± 0.103# | 0.605 ± 0.033 |
| linoleic acid | 1 ± 0.273 | 0.629 ± 0.054 | 0.488 ± 0.039# |
| linolenic acid | 1 ± 0.402 | 0.476 ± 0.059 | 0.746 ± 0.067* |
| myristic acid | 1 ± 0.141 | 0.657 ± 0.011 | 1.058 ± 0.022* |
| octadecanol | 1 ± 0.035 | 0.523 ± 0.116* | 1.153 ± 0.152* |
| oleic acid | 1 ± 0.224 | 0.410 ± 0.037# | 1.023 ± 0.119* |
| palmitic acid | 1 ± 0.076 | 0.534 ± 0.045* | 0.949 ± 0.113* |
| palmitoleic acid | 1 ± 0.212 | 0.531 ± 0.302 | 0.839 ± 0.190 |
| pelargonic acid | 1 ± 0.150 | 0.595 ± 0.144# | 1.121 ± 0.152# |
| pentadecanoic acid | 1 ± 0.739 | 3.336 ± 0.242* | 1.703 ± 0.753# |
| stearic acid | 1 ± 0.085 | 0.533 ± 0.034* | 0.922 ± 0.100* |
| Organic acid | |||
| 2-hydroxyvalerate | 1 ± 0.252 | 0.577 ± 0.221 | 0.917 ± 0.112 |
| 2-ketoadipate | 1 ± 0.456 | 0.322 ± 0.152 | 0.619 ± 0.148 |
| 3-phenyllactate | 1 ± 0.178 | 0.357 ± 0.032* | 0.693 ± 0.169# |
| adipate | 1 ± 0.357 | 0.319 ± 0.055 | 0.870 ± 0.174* |
| benzoate | 1 ± 0.380 | 0.404 ± 0.246 | 0.762 ± 0.010 |
| β-hydroxybutyrate | 1 ± 0.264 | 0.935 ± 0.169 | 1.177 ± 0.486 |
| fumarate | 1 ± 0.371* | 0.962 ± 0.028 | 2.254 ± 0.132* |
| galactonate | 1 ± 0.099# | 0.951 ± 0.637 | 1.483 ± 0.214* |
| gluconate | 1 ± 0.095* | 0.238 ± 0.400 | 0.016 ± 0.001 |
| glycerate | 1 ± 0.067 | 0.815 ± 0.142 | 0.900 ± 0.068 |
| indole-3-lactate | 1 ± 0.068# | 0.572 ± 0.052* | 0.746 ± 0.111 |
| lactate | 1 ± 0.048 | 0.884 ± 0.052# | 0.924 ± 0.023 |
| malate | 1 ± 0.282# | 0.656 ± 0.102 | 1.511 ± 0.111* |
| malonate | 1±0.027 | 0.473 ± 0.051* | 1.135 ± 0.119* |
| oxalate | 1 ± 0.211 | 0.512 ± 0.044 | 0.929 ± 0.007* |
| phenylacetate | 1 ± 0.367 | 0.612 ± 0.196* | 0.939 ± 0.081 |
| phosphogluconate | 1 ± 0.392 | 0.699 ± 0.122 | 1.815 ± 0.251* |
| pyrrole-2-carboxylate | 1 ± 0.177 | 0.630 ± 0.110# | 1.171 ± 0.154* |
| pyruvate | 1 ± 0.181 | 0.496 ± 0.045* | 0.639 ± 0.137 |
| terephthalate | 1 ± 0.343 | 0.894 ± 0.011 | 1.070 ± 0.044* |
| aminomalonate | 1 ± 0.219# | 1.159 ± 0.048 | 1.947 ± 0.012* |
| citramalate | 1 ± 0.255 | 1.116 ± 0.044 | 0.801 ± 0.049* |
| succinate | 1 ± 0.181 | 1.453 ± 0.002# | 1.061 ± 0.009* |
| Phosphate | |||
| adenosine-5-monophosphate | 1 ± 0.287 | 0.256 ± 0.055# | 1.052 ± 0.030* |
| β-glycerolphosphate | 1 ± 0.342 | 0.970 ± 0.049 | 1.018 ± 0.363 |
| mannose-6-phosphate NIST a | 1 ± 0.272 | 0.621 ± 0.261 | 0.906 ± 0.355 |
| phosphate | 1 ± 0.015* | 0.624 ± 0.014* | 0.941 ± 0.009* |
| pyrophosphate | 1 ± 0.344* | 0.292 ± 0.022 | 3.546 ± 0.213* |
| glucose-6-phosphate | 1 ± 0.127 | 1.290 ± 0.916 | 1.334 ± 0.228 |
| Sugar and sugar alcohols | |||
| 3,6-anhydro-D-galactose | 1 ± 0.065 | 0.327 ± 0.022* | 0.944 ± 0.247# |
| cellobiose | 1 ± 0.625 | 0.540 ± 0.121 | 1.419 ± 0.054* |
| citrate | 1 ± 0.182* | 0.940 ± 0.027 | 1.747 ± 0.029* |
| glycerol | 1 ± 0.521 | 0.659 ± 0.108 | 1.235 ± 0.101* |
| glycolate | 1 ± 0.073# | 0.714 ± 0.147# | 1.183 ± 0.017# |
| lactulose | 1 ± 0.142# | 0.579 ± 0.303 | 0.467 ± 0.190 |
| maltotriose | 1 ± 0.461 | 0.397 ± 0.072 | 0.828 ± 0.171# |
| mannose | 1 ± 0.089 | 0.654 ± 0.007* | 0.965 ± 0.020* |
| palatinitol | 1 ± 0.286 | 0.443 ± 0.148# | 0.958 ± 0.089* |
| phytol | 1 ± 0.271 | 0.449 ± 0.051# | 0.835 ± 0.071* |
| threose | 1 ± 0.260 | 0.593 ± 0.050* | 2.274 ± 0.142* |
| xylose | 1 ± 0.098# | 0.786 ± 0.846 | 1.448 ± 0.141 |
| lyxose | 1 ± 0.752 | 1.465 ± 1.221 | 0.893 ± 0.690 |
| mannitol | 1 ± 0.488 | 1.406 ± 0.087 | 0.475 ± 0.005* |
| melibiose | 1 ± 0.361 | 1.105 ± 0.189 | 1.147 ± 0.240 |
| myo-inositol | 1 ± 0.126* | 1.099 ± 0.018 | 1.839 ± 0.021* |
| arabitol | 1 ± 0.237 | 1.067 ± 0.191 | 0.942 ± 0.073 |
| fructose | 1 ± 0.064# | 1.740 ± 0.033* | 1.137 ±0.040* |
| galactose | 1 ± 0.123 | 1.892 ± 0.073* | 1.063 ± 0.019 |
| glucose | 1 ± 0.113 | 1.672 ± 0.043* | 1.036 ± 0.056* |
| tagatose | 1 ± 0.126 | 1.835 ± 0.044* | 1.127 ± 0.029* |
| threitol | 1 ± 0.013 | 1.501 ± 0.011* | 0.991 ± 0.017* |
| Others | |||
| tyrosol | 1 ± 0.091 | 0.462 ± 0.082* | 1.080 ± 0.149* |
| benzamide | 1 ± 0.177 | 0.413 ± 0.286# | 0.942 ± 0.051 |
| 2,3-butanediol NIST a | 1 ± 0.256 | 0.538 ± 0.076# | 0.784 ± 0.069# |
| carnitine | 1 ± 0.204 | 0.511 ± 0.491 | 1.104 ± 0.179 |
| dihydroxyacetone | 1 ± 0.322 | 0.317 ± 0.017# | 0.886 ± 0.078* |
| salicylaldehyde | 1 ± 0.455 | 0.329 ± 0.086 | 0.589 ± 0.023# |
| squalene | 1 ± 0.028 | 0.405 ± 0.038* | 0.834 ± 0.216# |
| taurine | 1 ± 0.293 | 0.649 ± 0.106 | 1.899 ± 0.697# |
| urea | 1 ± 0.243* | 0.001 ± 0.000* | 0.002 ± 0.000* |
Mean control value is set to 1 while dispersion of control data is maintained. Unless otherwise indicated, values are mean ± standard deviation.
In comparison between Control vs. TNF-α or TNF-α vs. Curcumin pretreatment, or Curcumin pretreatment vs. Control, significant difference is observed in the TNF-α-stimulated, curcumin treatment, and control groups, respectively.
#, P < 0.05
*, P < 0.01
aThe metabolite identified by National Institute of Standards and Technology (NIST) mass spectral library, but not verified by its authentic standard chemical.
Principal component analysis (PCA) and hierarchical cluster analysis (HCA) of metabolomic data
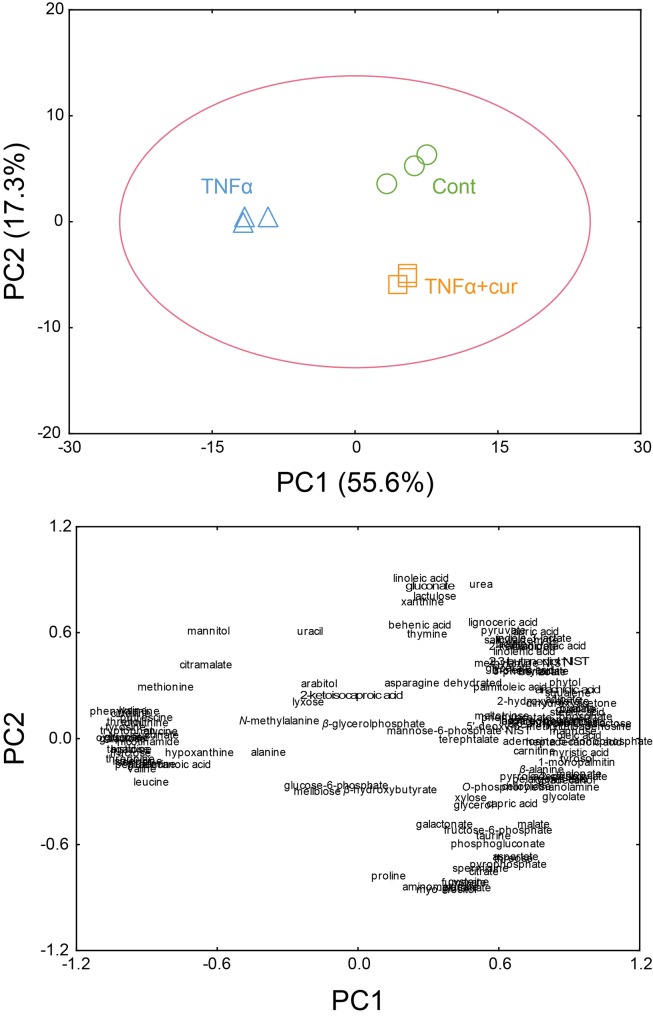
After the normalization by the sum of peak intensity of identified metabolites, multivariate statistical modeling using PCA was performed on mass spectral data. The PCA model showed a variation value (R 2 X) of 0.73 and predictive capability (Q 2) of 0.81, indicating the excellence of the PCA model. From the PCA score plots, the metabolite profiles were clearly separated between the three groups (i.e., control, TNF-α-stimulated, and curcumin-treated FLS) (Fig 1A). The principal component (PC) 1 had the largest possible variation (55.6%) between these three groups, explaining the significant separation of metabolite profiles between TNF-α-stimulated group and the control and curcumin-treated group. These results indicated that the metabolite profiles of RA FLS were altered by TNF-α and the altered metabolic pattern of TNF-α-stimulated FLS was recovered to that of control group by curcumin treatment.
Fig 1. PCA score (A) and loading plots (B) of RA fibroblast-like synoviocytes (FLS), which were not stimulated (Control), stimulated with TNF-α (TNF), and treated with curcumin (Curcumin).
(A) Principal component (PC)1 explained the significant separation of metabolite profiles between the TNF-α-stimulated group on the negative region of the PC1, and the control and curcumin-treated groups on the positive region of the PC1. Further, the control group was clearly separated from the curcumin-treated group on PC2. (B) PC1 was explained by 84 metabolites that correlated positively with the axis, and 35 metabolites that correlated negatively.
A total of 84 metabolites, including fatty acid metabolism derivatives (i.e., myristic acid, oleic acid, 1-monopalmitin, and stearic acid), phosphate, adenosine-5-monophosphate, and malonate positively contributed to PC 1 (Fig 1B). However, 35 metabolites, including amino acids (tyrosine, phenylalanine, and oxoproline), tagatose, threitol, glucose, and galactose negatively contributed to PC1. A total of 63 metabolites, such as linoleic acid, urea, and gluconate positively contributed to PC2. The other 56 metabolites, including myo-inositol, glutamate, aminomalonate, and fumarate negatively contributed to PC2.
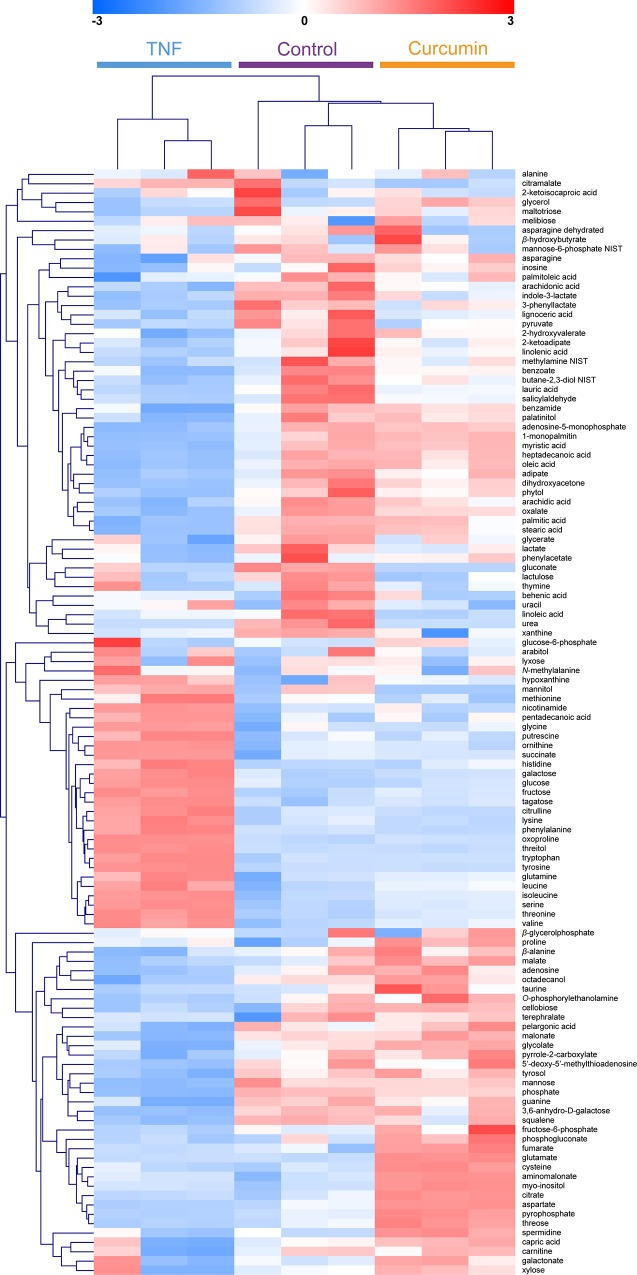
A total of 119 identified metabolites were clustered and visualized by the HCA using the Euclidean distance and average linkage method to determine possible variations in the metabolite profiling of the three groups (Fig 2). The clustering of the metabolites led to good separation among the three groups. Similar metabolic patterns were shown between the control and curcumin-treated group. On the other hand, a clear separation of the metabolic profile of the TNF-α-stimulated group from the control and curcumin-treated groups was shown. This indicated that the metabolic pattern of the TNF-α-stimulated group was completely different from those of the control and curcumin-treated groups, similar to the PCA result.
Fig 2. Hierarchical clustering analysis of 119 identified metabolites from RA FLS.
The results of heat mapping generated through metabolomic analysis and the relevant changes discovered. A heat map showed that the metabolite profiles of controls were similar to those of the curcumin-treated group. Red color reflects an increase, and blue color a decrease.
Significant metabolites for elucidating the effects of TNF-α and curcumin
The metabolomic analysis revealed that many metabolites were severely perturbed when RA FLS were exposed to TNF-α. To compare changes in metabolites between the control group and TNF-α-stimulated FLS, a Student’s t-test at the 5% significance level was conducted. The levels of 41 metabolites, including xanthine, palmitic acid, stearic acid, pyruvate, and myristic acid were lower in the TNF-α-stimulated group than those in the control group. Twenty four metabolites, such as oxoproline, tyrosine, threonine, serine, phenylalanine, tryptophan, and valine were higher in the TNF-α-stimulated group than in the control group. Subsequently, the metabolic pattern of the curcumin-treated RA FLS before TNF-α stimulation became similar to that in the control group. Among 65 metabolites altered by TNF-α, the levels of 54 metabolites reached levels similar to those in the control group after curcumin treatment (Table 1).
Effects of curcumin on TNF-α-induced IL-6, IL-8, MMP-1, and MMP-3 production in RA FLS
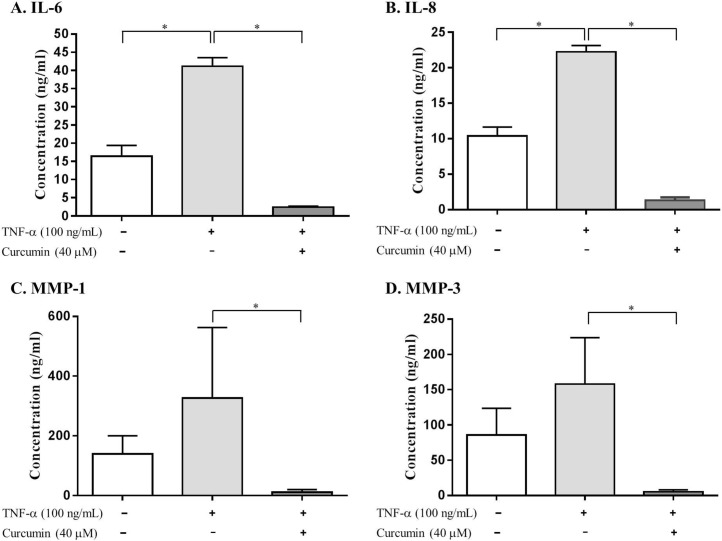
Stimulation of RA FLS with TNF-α (100 ng/mL) for 24 h resulted in a 2.5-, 2-, 2.3-, and 1.8-fold induction of IL-6, IL-8, MMP-1, and MMP-3 production, respectively, compared to that in the control group, when measured in culture media using ELISA (Fig 3A–3D). Treatment with curcumin significantly inhibited TNF-α-induced IL-6, IL-8, MMP-1, and MMP-3 production. Interestingly, curcumin resulted in almost 94% inhibition in IL-6 and IL-8 production, and blocked MMP-1 and MMP-3 production by 96%, compared to that in the TNF-α-stimulated FLS.
Fig 3. Inhibition of TNF-α-induced production of (A) IL-6, (B) IL-8, (C) MMP-1, and (D) MMP-3 by curcumin in RA FLS.
RA FLS (N = 3) were incubated with DMSO-containing vehicle or curcumin for 1 h, followed by stimulation with TNF-α (100 ng/mL) for 24 h. Production of IL-6, IL-8, MMP-1, and MMP-3 in the culture supernatants was measured using a commercially available ELISA kit. Values are mean ± standard deviation. * = P < 0.05, by the Kruskal-Wallis test.
Discussion
Our metabolomic analysis using RA FLS reaffirmed that curcumin has a protective effect against the inflammatory response of TNF-α. Exposure to TNF-α caused prominent changes in the intracellular metabolites of RA FLS that was reversed by curcumin treatment. In addition, TNF-α stimulation and curcumin treatment led to perturb metabolite profiling of amino acids and fatty acids. It is suggested that curcumin could weaken obvious metabolic effects of TNF-α exposure in RA FLS and prominent metabolic target of TNF-α and curcumin is amino acid and fatty acid metabolism.
Recently, metabolomic analysis has been used to monitor the activity of tumor cells, the effects of anticancer drugs in tumor cells, and to detect specific genetic alterations in tumor cells [27–29]. RA FLS are key effector cells in the invasion of the synovium and have a role in the initiation and perpetuation of destructive joint inflammation [19]. In this regard, we used the metabolomic approach with GC/TOF-MS to research the role of FLS in RA. This study demonstrated clear differentiation of the metabolic patterns of TNF-α-stimulated FLS from those of the control or curcumin-treated FLS. One of the most important findings is that these differences were identified using an unsupervised analysis, with no information about the samples. Since all three samples were cultured under the same conditions, the observed discrimination demonstrates that the changes in metabolic profile strongly reflect the effects of TNF-α and curcumin. From the PCA plots, TNF-α-stimulated FLS moved from the left to the right after treatment with curcumin, suggesting that metabolic perturbation by TNF-α, which has a role in the pathogenesis of RA, was restored to the metabolic profile of the control group after curcumin treatment. Also, little differences were shown in metabolic profiles between the control and curcumin-pretreated groups by PC2. From the clinical view points, clinicians frequently have experienced that patients with RA does not make a full recovery but have recovered considerably by treating RA with disease modifying drugs. We think that these findings in clinical practice are consistent with making a difference between control group and curcumin-pretreated group by PC2. On the other hand, in Fig 2 and Table 1, there are some metabolites showing significant differences between the control and crucumin-pretreated groups. Further study is required to determine the exact role of each metabolite including anti-inflammatory effects.
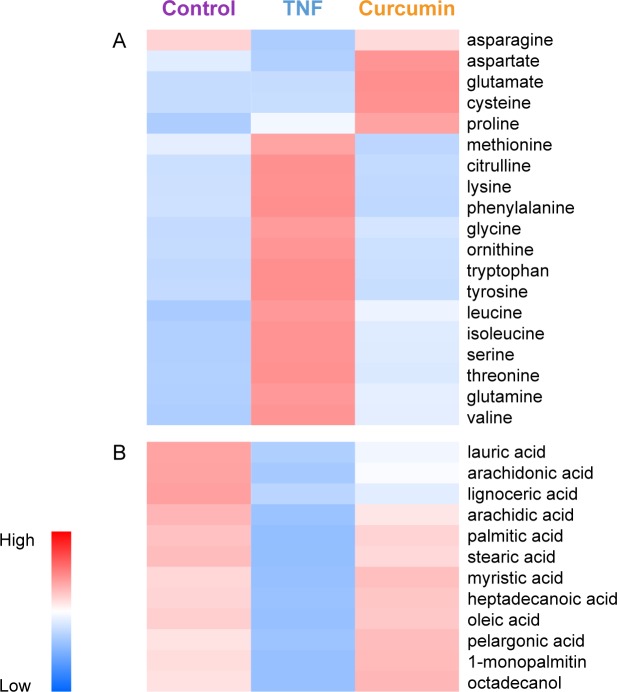
The pathway analysis conducted using MetaboAnalyst based on enrichment analysis and pathway topology analysis to reveal the change of metabolisms by TNF-α and curcumin. These results showed that metabolite levels, which are involved in amino acid biosynthesis, purine metabolism, glycolysis, and fatty acid metabolism, were affected by TNF-α in RA FLS (Table 2), but returned to their original levels after curcumin treatment (Table 3). In our study, most of the glucogenic and ketogenic amino acids levels increased more in the TNF-α-stimulated FLS group than the control group (Fig 4A). The metabolic profiling of RA FLS after TNF-α stimulation was in accordance with a previous study in which RA synovial fluid was used [23]. Treatment with curcumin reversed these TNF-α-induced perturbations of amino acids metabolites to normal. The level of glycine in FLS increased after TNF-α treatment and decreased after curcumin treatment. Glycine has been reported to stimulate prostaglandin E2 and COX-2 in IL-1β-stimulated human gingival fibroblasts [30]. In addition, the levels of citrulline were significantly higher in the TNF-α-stimulated group than in the control group (3.6-fold increase). Similarly, synovial tissue and FLS from RA patients had higher levels of citrullinated protein than those from osteoarthritis patients [31]. Citrulline is an essential constituent of antigenic determinants recognized by RA-specific autoantibodies. Citrullinated proteins may be detected as antigens by the immune system, thereby generating an immune response. http://en.wikipedia.org/wiki/Anti%E2%80%93citrullinated_protein_antibody-cite_note-pmid17558653-3 The high levels of citrulline in TNF-α-stimulated FLS reduced to the level of the control group after curcumin treatment. Collectively, this suggested that the effect of curcumin on preventing joint inflammation may be associated with the restoration of amino acid metabolite levels, such as citrulline or glycine. In addition to the known mechanism of action of curcumin, our metabolomic study revealed the new mechanism of action of curcumin for reducing synovial inflammation.
Table 2. The major metabolic pathways changed by TNF-α stimulation compared with the control group.
| Pathway | -log (P)a | Impactb |
|---|---|---|
| Purine metabolism | 12.544 | 0.075 |
| Histidine metabolism | 11.959 | 0.140 |
| Pyrimidine metabolism | 11.868 | 0.114 |
| Phenylalanine, tyrosine and tryptophan biosynthesis | 10.836 | 0.008 |
| Nitrogen metabolism | 10.302 | 0.008 |
| Ubiquinone and other terpenoid-quinone biosynthesis | 10.053 | <0.001 |
| Arginine and proline metabolism | 9.776 | 0.359 |
| Sulfur metabolism | 9.304 | 0.000 |
| Phenylalanine metabolism | 9.188 | 0.173 |
| β-Alanine metabolism | 9.184 | 0.334 |
| Aminoacyl-tRNA biosynthesis | 9.023 | 0.169 |
| Glycine, serine and threonine metabolism | 8.906 | 0.421 |
| Methane metabolism | 8.858 | 0.034 |
| Glycolysis or Gluconeogenesis | 8.797 | 0.095 |
| Glyoxylate and dicarboxylate metabolism | 8.238 | 0.172 |
| Glycerolipid metabolism | 7.824 | 0.230 |
| Alanine, aspartate and glutamate metabolism | 7.786 | 0.520 |
| Propanoate metabolism | 7.681 | 0.086 |
| Caffeine metabolism | 7.420 | 0.031 |
| Fatty acid metabolism | 7.223 | 0.030 |
P a value and Impactb were calculated from enrichment and pathway topology analysis.
Table 3. The major metabolic pathways recovered by curcumin-pretreatment compared with TNF-α stimulation group.
| Pathway | -log (P)a | Impactb |
|---|---|---|
| Valine, leucine and isoleucine biosynthesis | 4.213 | 0.074 |
| Glycerolipid metabolism | 4.049 | 0.230 |
| Aminoacyl-tRNA biosynthesis | 4.014 | 0.169 |
| Nitrogen metabolism | 3.974 | 0.008 |
| Purine metabolism | 3.949 | 0.075 |
| Butanoate metabolism | 3.926 | 0.125 |
| Galactose metabolism | 3.915 | 0.109 |
| β-Alanine metabolism | 3.861 | 0.334 |
| Pantothenate and CoA biosynthesis | 3.846 | 0.073 |
| Propanoate metabolism | 3.836 | 0.086 |
| Glycine, serine and threonine metabolism | 3.824 | 0.421 |
| Cyanoamino acid metabolism | 3.819 | <0.001 |
| Arginine and proline metabolism | 3.799 | 0.359 |
| Glyoxylate and dicarboxylate metabolism | 3.791 | 0.172 |
| Lysine biosynthesis | 3.788 | 0.159 |
| Fatty acid biosynthesis | 3.777 | <0.001 |
| Alanine, aspartate and glutamate metabolism | 3.763 | 0.520 |
| Nicotinate and nicotinamide metabolism | 3.739 | 0.038 |
| Citrate cycle (TCA cycle) | 3.738 | 0.185 |
| Phenylalanine metabolism | 3.716 | 0.173 |
P a value and Impactb were calculated from enrichment and pathway topology analysis.
Fig 4. The intensities of metabolites related to amino acid metabolism (A) and fatty acid metabolism (B).
Accumulated data have revealed that TNF-α perturbs the normal regulation of lipid metabolism [32]. According to our results, total fatty acid levels, including saturated fatty acids (SFAs: arachidic acid, lignoceric acid, lauric acid, myristic acid, palmitic acid, and stearic acid), polyunsaturated fatty acids (PUFAs: arachidonic acid and oleic acid), octadecanol, pelargonic acid, and heptadecanoic acid were significantly lower in the TNF-α-stimulated FLS group than in the control group. On the contrary, the lower levels of SFAs and PUFAs in the TNF-α-stimulated FLS restored to the levels of the control group after curcumin treatment (Fig 4B). SFAs were reported to generate reactive oxygen species (ROS) [33]. Curcumin is a potent in vitro antioxidant owing to its ability to scavenge ROS [34]. These findings suggest that the effect of curcumin as an antioxidant may be affected by the change of SFA abundance. In addition, the levels of arachidonic acid were significantly different between the TNF-α-stimulated and control groups. The levels of arachidonic acid were significantly decreased in the TNF-α-stimulated FLS group compared to that in the control group, while its levels increased in curcumin-treated group. Arachidonic acid mediates inflammation and can be metabolized by COX or lipoxygenase. TNF-α has no apparent effect on the expression of COX-1, but induces the expression of COX-2 [35]. Curcumin inhibited the inflammatory response through the suppression of COX-2 in FLS, followed by the inhibition of prostaglandin E2 synthesis [3,8]. The findings in this study are thought to reaffirm the effect of TNF-α and curcumin on COX-2 with respect to metabolomics. In our study, treatment with curcumin also inhibited TNF-α-induced IL-6, IL-8, MMP-1, and MMP-3 production and restored altered fatty acids levels by TNF-α. Recently, it was reported that free fatty acids, such as palmitic acid, stearic acid, or oleic acid, induce IL-6, IL-8, pro-MMP-1, and MMP-3 in RA FLS [36]. In this context, perturbation of fatty acids levels by TNF-α and curcumin may cause changes in inflammatory cytokines and MMP production. Thus, a reasonable conclusion is that free fatty acids are one of the major components in the mechanism of action of curcumin.
In conclusion, metabolomic data and multivariate statistical analysis clearly revealed strong TNF-α- and curcumin-induced effects on the central metabolism of RA FLS. A prominent metabolic target of TNF-α and curcumin is amino acid and fatty acid metabolism, as revealed by perturbation of metabolite profiles. This is the first report of GC/TOF-MS-based metabolomic application to evaluate the effect of curcumin on FLS which are key effector cells in RA. These results also suggest that metabolomic investigation of FLS has the potential for discovering new biologic targets for therapeutic drugs in RA.
Supporting Information
(DOCX)
Acknowledgments
We thank Eun-Kyung Bae for her excellent technical assistance and help.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported by the Advanced Biomass R&D Center of Korea (http://www.biomass.re.kr/en, 2011-0031353) to KHK, the National Research Foundation of Korea funded by the Ministry of Education (http://www.nrf.re.kr/nrf_eng_cms/, NRF-2013R1A1A2059103) to HSC, and a grant of the Korea Health Technology R&D Project through the Korea Health Industry Development Institute, funded by the Ministry of Health & Welfare (http://www.khidi.or.kr/kps, HI14C2285) to HSC, which are funded by the Korean Government. Experiments were performed using facilities of the Institute of Biomedical Science and Food Safety at the Korea University Food Safety Hall.
References
- 1. McInnes IB, Schett G (2011) The pathogenesis of rheumatoid arthritis. N Engl J Med 365: 2205–2219. 10.1056/NEJMra1004965 [DOI] [PubMed] [Google Scholar]
- 2. Ammon HP, Wahl MA (1991) Pharmacology of Curcuma longa. Planta Med 57: 1–7. [DOI] [PubMed] [Google Scholar]
- 3. Gupta SC, Tyagi AK, Deshmukh-Taskar P, Hinojosa M, Prasad S, Aggarwal BB (2014) Downregulation of tumor necrosis factor and other proinflammatory biomarkers by polyphenols. Arch Biochem Biophys 559: 91–99. 10.1016/j.abb.2014.06.006 [DOI] [PubMed] [Google Scholar]
- 4. Iqbal M, Okazaki Y, Okada S (2009) Curcumin attenuates oxidative damage in animals treated with a renal carcinogen, ferric nitrilotriacetate (Fe-NTA): implications for cancer prevention. Mol Cell Biochem 324: 157–164. 10.1007/s11010-008-9994-z [DOI] [PubMed] [Google Scholar]
- 5. Surh YJ, Chun KS (2007) Cancer chemopreventive effects of curcumin. Adv Exp Med Biol 595: 149–172. [DOI] [PubMed] [Google Scholar]
- 6. Kim HY, Park EJ, Joe EH, Jou I (2003) Curcumin suppresses Janus kinase-STAT inflammatory signaling through activation of Src homology 2 domain-containing tyrosine phosphatase 2 in brain microglia. J Immunol 171: 6072–6079. [DOI] [PubMed] [Google Scholar]
- 7. Punithavathi D, Venkatesan N, Babu M (2000) Curcumin inhibition of bleomycin-induced pulmonary fibrosis in rats. Br J Pharmacol 131: 169–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Shehzad A, Rehman G, Lee YS (2013) Curcumin in inflammatory diseases. Biofactors 39: 69–77. 10.1002/biof.1066 [DOI] [PubMed] [Google Scholar]
- 9. Mun SH, Kim HS, Kim JW, Ko NY, Kim do K, Lee BY, et al. (2009) Oral administration of curcumin suppresses production of matrix metalloproteinase (MMP)-1 and MMP-3 to ameliorate collagen-induced arthritis: inhibition of the PKCdelta/JNK/c-Jun pathway. J Pharmacol Sci 111: 13–21. [DOI] [PubMed] [Google Scholar]
- 10. Funk JL, Oyarzo JN, Frye JB, Chen G, Lantz RC, Jolad SD, et al. (2006) Turmeric extracts containing curcuminoids prevent experimental rheumatoid arthritis. J Nat Prod 69: 351–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kloesch B, Becker T, Dietersdorfer E, Kiener H, Steiner G (2013) Anti-inflammatory and apoptotic effects of the polyphenol curcumin on human fibroblast-like synoviocytes. Int Immunopharmacol 15: 400–405. 10.1016/j.intimp.2013.01.003 [DOI] [PubMed] [Google Scholar]
- 12. Moon DO, Kim MO, Choi YH, Park YM, Kim GY (2010) Curcumin attenuates inflammatory response in IL-1beta-induced human synovial fibroblasts and collagen-induced arthritis in mouse model. Int Immunopharmacol 10: 605–610. 10.1016/j.intimp.2010.02.011 [DOI] [PubMed] [Google Scholar]
- 13. Zhou H, Beevers CS, Huang S (2011) The targets of curcumin. Curr Drug Targets 12: 332–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Jurenka JS (2009) Anti-inflammatory properties of curcumin, a major constituent of Curcuma longa: a review of preclinical and clinical research. Altern Med Rev 14: 141–153. [PubMed] [Google Scholar]
- 15. Cuperlovic-Culf M, Barnett DA, Culf AS, Chute I (2010) Cell culture metabolomics: applications and future directions. Drug Discov Today 15: 610–621. 10.1016/j.drudis.2010.06.012 [DOI] [PubMed] [Google Scholar]
- 16. Bayet-Robert M, Morvan D (2013) Metabolomics reveals metabolic targets and biphasic responses in breast cancer cells treated by curcumin alone and in association with docetaxel. PLoS One 8: e57971 10.1371/journal.pone.0057971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Cao H, Zhang A, Zhang H, Sun H, Wang X (2014) The application of metabolomics in traditional Chinese medicine opens up a dialogue between Chinese and Western medicine. Phytother Res. [DOI] [PubMed] [Google Scholar]
- 18. Bartok B, Firestein GS (2010) Fibroblast-like synoviocytes: key effector cells in rheumatoid arthritis. Immunol Rev 233: 233–255. 10.1111/j.0105-2896.2009.00859.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Bottini N, Firestein GS (2013) Duality of fibroblast-like synoviocytes in RA: passive responders and imprinted aggressors. Nat Rev Rheumatol 9: 24–33. 10.1038/nrrheum.2012.190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Sandur SK, Ichikawa H, Pandey MK, Kunnumakkara AB, Sung B, Sethi G, et al. (2007) Role of pro-oxidants and antioxidants in the anti-inflammatory and apoptotic effects of curcumin (diferuloylmethane). Free Radic Biol Med 43: 568–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Arnett FC, Edworthy SM, Bloch DA, McShane DJ, Fries JF, Cooper NS, et al. (1988) The American Rheumatism Association 1987 revised criteria for the classification of rheumatoid arthritis. Arthritis Rheum 31: 315–324. [DOI] [PubMed] [Google Scholar]
- 22. Kim S, Lee DY, Wohlgemuth G, Park HS, Fiehn O, Kim KH (2013) Evaluation and optimization of metabolome sample preparation methods for Saccharomyces cerevisiae. Analytical Chemistry 85: 2169–2176. 10.1021/ac302881e [DOI] [PubMed] [Google Scholar]
- 23. Kim S, Hwang J, Xuan J, Jung YH, Cha HS, Kim KH (2014) Global metabolite profiling of synovial fluid for the specific diagnosis of rheumatoid arthritis from other inflammatory arthritis. PLoS One 9: e97501 10.1371/journal.pone.0097501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lee DY, Fiehn O (2008) High quality metabolomic data for Chlamydomonas reinhardtii. Plant Methods 4: 7 10.1186/1746-4811-4-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Saeed AI, Bhagabati NK, Braisted JC, Liang W, Sharov V, Howe EA, et al. (2006) TM4 microarray software suite. Methods Enzymol 411: 134–193. [DOI] [PubMed] [Google Scholar]
- 26. Xia J, Psychogios N, Young N, Wishart DS (2009) MetaboAnalyst: a web server for metabolomic data analysis and interpretation. Nucleic Acids Res 37: W652–660. 10.1093/nar/gkp356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Bullinger D, Neubauer H, Fehm T, Laufer S, Gleiter CH, Kammerer B (2007) Metabolic signature of breast cancer cell line MCF-7: profiling of modified nucleosides via LC-IT MS coupling. BMC Biochem 8: 25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Griffin JL, Shockcor JP (2004) Metabolic profiles of cancer cells. Nat Rev Cancer 4: 551–561. [DOI] [PubMed] [Google Scholar]
- 29. D'Alessandro A, Zolla L (2013) Proteomics and metabolomics in cancer drug development. Expert Rev Proteomics 10: 473–488. 10.1586/14789450.2013.840440 [DOI] [PubMed] [Google Scholar]
- 30. Rausch-Fan X, Ulm C, Jensen-Jarolim E, Schedle A, Boltz-Nitulescu G, Rausch WD, et al. (2005) Interleukin-1beta-induced prostaglandin E2 production by human gingival fibroblasts is upregulated by glycine. J Periodontol 76: 1182–1188. [DOI] [PubMed] [Google Scholar]
- 31. Ling S, Cline EN, Haug TS, Fox DA, Holoshitz J (2013) Citrullinated calreticulin potentiates rheumatoid arthritis shared epitope signaling. Arthritis Rheum 65: 618–626. 10.1002/art.37814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Chen X, Xun K, Chen L, Wang Y (2009) TNF-alpha, a potent lipid metabolism regulator. Cell Biochem Funct 27: 407–416. 10.1002/cbf.1596 [DOI] [PubMed] [Google Scholar]
- 33. Noguchi Y, Young JD, Aleman JO, Hansen ME, Kelleher JK, Stephanopoulos G (2011) Tracking cellular metabolomics in lipoapoptosis- and steatosis-developing liver cells. Mol Biosyst 7: 1409–1419. 10.1039/c0mb00309c [DOI] [PubMed] [Google Scholar]
- 34. Visioli F, De La Lastra CA, Andres-Lacueva C, Aviram M, Calhau C, Cassano A, et al. (2011) Polyphenols and human health: a prospectus. Crit Rev Food Sci Nutr 51: 524–546. 10.1080/10408391003698677 [DOI] [PubMed] [Google Scholar]
- 35. Gallois C, Habib A, Tao J, Moulin S, Maclouf J, Mallat A, et al. (1998) Role of NF-kappaB in the antiproliferative effect of endothelin-1 and tumor necrosis factor-alpha in human hepatic stellate cells. Involvement of cyclooxygenase-2. J Biol Chem 273: 23183–23190. [DOI] [PubMed] [Google Scholar]
- 36. Frommer KW, Schaffler A, Rehart S, Lehr A, Muller-Ladner U, Neumann E (2015) Free fatty acids: potential proinflammatory mediators in rheumatic diseases. Ann Rheum Dis 74: 303–310. 10.1136/annrheumdis-2013-203755 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.