Abstract
The discovery of a gene network regulating lysosomal biogenesis and its transcriptional regulator TFEB revealed that cells monitor lysosomal function and respond to degradation requirements and environmental cues. Here, we report the identification of transcription factor E3 (TFE3) as another regulator of lysosomal homeostasis that induced expression of genes encoding proteins involved in autophagy and lysosomal biogenesis in ARPE-19 cells in response to starvation and lysosomal stress. We found that in nutrient-replete cells, TFE3 was recruited to lysosomes through interaction with active Rag GTPases and exhibited mTORC1-dependent phosphorylation. Phosphorylated TFE3 was retained in the cytosol through its interaction with the cytosolic chaperone 14-3-3. Following starvation, TFE3 rapidly translocated to the nucleus and bound to the CLEAR elements present in the promoter region of many lysosomal genes, thereby inducing lysosomal biogenesis. Depletion of endogenous TFE3 entirely abolished the response of ARPE-19 cells to starvation, suggesting that TFE3 plays a critical role in nutrient sensing and regulation of energy metabolism. Furthermore, overexpression of TFE3 triggered lysosomal exocytosis and resulted in efficient cellular clearance in a cellular model of a lysosomal storage disorder, Pompe disease, thus identifying TFE3 as a potential therapeutic target for the treatment of lysosomal disorders.
Introduction
Lysosomes are the primary degradative organelle in all cells. Lysosomes receive extracellular material destined for degradation through endocytosis, whereas intracellular components reach lysosomes mainly through autophagy1. In addition to their role in biomolecular degradation and recycling, lysosomes are also critical for several cellular and physiological functions including cholesterol homeostasis, downregulation of surface receptors, inactivation of pathogenic organisms, antigen presentation, repair of the plasma membrane, and bone remodeling2.
Lysosomes also function in nutrient sensing and cellular energy homeostasis. This is primarily due to the lysosomal localization of mammalian (or mechanistic) target of rapamycin complex 1 (mTORC1), a protein complex that includes the serine/threonine kinase mTOR and regulates cell growth and division in response to energy levels, growth signals, and nutrients. The activation of mTORC1 by intracellular amino acids is well characterized. In cells in which amino acids are sufficient, mTORC1 is recruited to the lysosomal surface, where it is activated by the guanosine triphosphatase (GTPase) Rheb3,4. The amino acid–dependent translocation of mTOR to the lysosome requires Rag GTPases and Ragulator, a pentameric protein complex that anchors the Rag GTPases to lysosomes5-7. The Rag proteins function as heterodimers in which the active complex consists of GTP-bound RagA or RagB (RagA/B) complexed with GDP-bound RagC or RagD (RagC/D)8,9. The amount of amino acids in the lysosomal lumen signals to the vacuolar-ATPase (v-ATPase)10. When amino acids are abundant, the v-ATPase promotes the guanine exchange factor (GEF) activity of Ragulator, thus triggering the GTP loading and activation of RagA/B proteins5. Active Rags can then bind the mTORC1 component Raptor and recruit mTORC1 to lysosomes. Interestingly, Rheb activity requires growth factors, suggesting that different stimuli (growth factors and amino acids) cooperate to activate mTORC1. Upon activation, mTORC1 promotes cell growth and anabolic processes while simultaneously repressing autophagy.
The Atg family of proteins, such as Atg13 and Atg1 [also known as ULK1 and ULK2 (ULK1/2)], are involved in autophagy induction11,12. Phosphorylation of these proteins by mTORC1 inhibits their activity, thereby repressing autophagy. Indirectly, mTORC1 regulates autophagy by modulating the activity of transcription factor EB (TFEB)13-15. TFEB is a member of the basic helix-loop-helix leucine-zipper family of transcription factors that recognizes a 10 base-pair motif (GTCACGTGAC) enriched in the promoter regions of numerous lysosomal genes16. Activation of TFEB induces expression of many genes associated with lysosomal biogenesis and function. TFEB also stimulates the expression of genes implicated in autophagosome formation, fusion of autophagosomes with lysosomes, and lysosome-mediated degradation of the autophagosomal content17-19. Therefore, TFEB provides coordinated transcriptional regulation of the two main degradative organelles in the cell, autophagosomes and lysosomes.
Under nutrient-rich conditions, active mTORC1 phosphorylates TFEB on several serine and threonine residues, including serine 211 (Ser211)13-15. Phosphorylation of Ser211 creates a binding site for 14-3-3, a cytosolic chaperone that keeps TFEB sequestered in the cytosol. In contrast, under starvation conditions mTORC1 is inactivated, the TFEB and 14-3-3 complex dissociates, and TFEB translocates to the nucleus where it stimulates the expression of hundreds of genes, thus leading to lysosomal biogenesis, increased lysosomal degradation, and autophagy induction13,14. TFEB interacts with active Rag GTPases20. This interaction promotes recruitment of TFEB to lysosomes and facilitates the mTORC1-dependent phosphorylation of TFEB. Inhibition of the interaction between TFEB and Rags results in accumulation of TFEB in the nucleus and constitutive activation of autophagy under nutrient-rich conditions20. Therefore, recruitment of TFEB to lysosomes is critical for the proper negative regulation of this transcription factor.
An important question is whether the regulatory mechanism of TFEB is shared by other transcription factors that belong to the microphthalmia-associated transcription factor (MiTF) and TFE (MiTF/TFE) family, which includes TFEB, MITF, TFEC, and TFE3. MITF1, an isoform of MITF implicated in proliferation and survival of retinal pigment epithelium (RPE) osteoclasts, natural killer cells, and mast cells, interacts with active Rags and translocates to the nucleus upon mTORC1 inactivation20. Here, we assessed the mechanism of TFE3 activation. TFE3 is present in many tissues and plays a major role in activation of the immune system21,22, control of allergic diseases23,24, development of osteoclasts25, and regulation of the expression of critical metabolic regulators26. We found that similarly to TFEB, TFE3 responds to variations in the amount of nutrients and is transported to the nucleus following inactivation of mTORC1 in cells subjected to nutrient starvation. Although TFEB is the only member of the MiTF/TFE family reported to bind to CLEAR motifs and induce multiple genes involved in lysosomal biogenesis16, we found that TFE3 also promoted the expression of genes associated with autophagy and lysosomes and stimulated lysosomal biogenesis. When overexpressed in a model of a lysosomal storage disorder, Pompe disease, TFE3 induced lysosomal exocytosis and cellular clearance. Therefore, our results indicate that cells exhibit distinct reliance on specific members of the MiTF/TFE family to control lysosomal homeostasis with TFE3 serving as a major regulator of this process in some contexts.
Results
mTORC1 promotes retention of TFE3 in the cytosol under nutrient-rich conditions
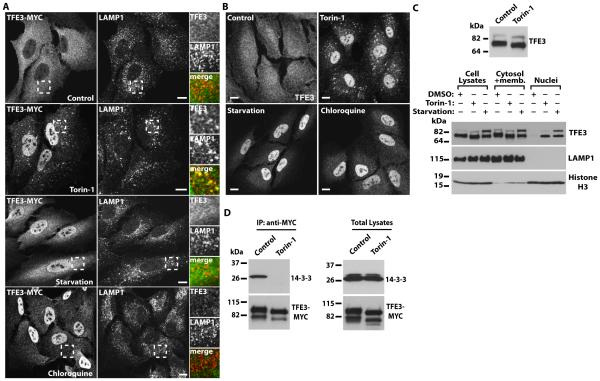
Because mTORC1 phosphorylates and inhibits the transcriptional activity of TFEB and MITF113,20 we tested whether mTORC1 also regulated activation of TFE3. We generated an adenovirus expressing human TFE3 tagged to MYC (TFE3-MYC) and, when expressed in the human retinal epithelial pigment cell line ARPE-19, TFE3-MYC was retained in the cytosol in “fully fed” cells (cultured in normal growth medium) (Fig. 1A). In contrast, incubation with the mTOR inhibitor Torin-1 induced a rapid translocation of some TFE3-MYC from the cytosol to the nucleus; in addition, TFE3-MYC colocalized with LAMP1 (lysosome-associated membrane protein 1; lysosomal marker)-positive vesicles (Fig. 1A). To assess the regulation of TFE3 under a more physiological condition, cells were starved by incubation with amino acid-free medium for 1 hour, a treatment that inactivates mTORC16,7. As expected, starvation caused an accumulation of TFE3-MYC in the nucleus (Fig. 1A). In agreement with previously published data14, we observed that inhibition of lysosomal function with chloroquine, which increases lysosomal pH, also induced transport of TFE3-MYC to the nucleus (Fig. 1A). Therefore, our results indicated that TFE3 was translocated from the cytosol to the nucleus in response to both amino acid starvation and lysosomal stress.
Fig. 1. TFE3 redistributes from the cytosol to the nucleus upon mTORC1 inactivation, nutrient deprivation, or lysosomal stress.
(A) Immunofluorescence confocal microscopy analysis of the subcellular distribution of TFE3-MYC in ARPE-19 cells incubated with Torin-1, starved in serum and amino acid-free medium, or exposed to Chloroquine for 1 hour. Cells were double stained with antibodies against TFE3 (a dilution of 1:7000 was used to detect recombinant TFE3) and LAMP1. Insets show a 2.5-fold magnification of the indicated region. Scale bars, 10 μm. Data are representative of 3 independent experiments and over 90% of cells exhibited the phenotypes shown. (B) Immunofluorescence confocal microscopy analysis of the subcellular distribution of endogenous TFE3 in ARPE-19 cells exposed to the same condition as those indicated in (A). Cells were stained with antibodies against TFE3 (a dilution of 1:200 was used to detect endogenous TFE3). Scale bars, 10 μm. Data are representative of 3 independent experiments and over 90% of cells exhibited the phenotypes shown. (C) Top: Immunoblotting showing changes on TFE3 electrophoretic mobility after 2 h incubation with Torin-1. Bottom: Immunoblotting analysis of the subcellular distribution of endogenous TFE3 in ARPE-19 cells incubated with DMSO or Torin-1 for 2 hours, or starved in a medium without serum and amino acids for 20 hours. The subcellular fractions were probed with antibodies against TFE3, LAMP1 (lysosomal membrane marker), and Histone H3 (nuclear marker). (D) Immunoblotting analysis of coimmunoprecipitated 14-3-3 with TFE3-MYC in ARPE-19 cells treated with DMSO (control) or Torin-1 for 1hour. Protein bands were detected with antibodies against MYC (used to detect TFE3-MYC) and 14-3-3. Data in C and D are representative of 3 independent experiments.
Similar to the effects on exogenously expressed TFE3, inactivation of mTOR by Torin-1, amino acid starvation, or lysosomal stress induced nuclear translocation of endogenous TFE3 in ARPE-19 (Fig. 1B), HeLa, and HepG2 cells (fig. S1A and S1B). Subcellular fractionation experiments also showed that the amount of endogenous TFE3 in the nucleus increased upon treatment with Torin-1 or amino acid starvation (Fig. 1C). TFE3 migrated close to 72 kD and 89 kD (Fig. 1C); the upper 89 kD band may represent a form with uncharacterized posttranslational modifications27. Interestingly, the smaller form of TFE3 detected in the nuclear fractions had a lower molecular weight than that of the smaller form of TFE3 in the cell lysates, suggesting that inactivation of mTORC1 affected TFE3 phosphorylation status. In addition, the amount of the 89-kD form of TFE3 increased after 20h of amino acid starvation, indicating that additional posttranslational modifications may occur upon prolonged TFE3 activation.
To corroborate that mTORC1 regulated the nuclear distribution of TFE3, we genetically inhibited either mTORC1 or mTORC2 by depleting raptor or rictor, respectively, with specific siRNAs. In cells depleted of raptor to inhibit mTORC1, TFE3 primarily accumulated in the nucleus, whereas TFE3 was cytosolic in cells depleted of rictor to inhibit mTORC2 (fig. S1C). In addition, incubation of ARPE-19 cells with rapamycin, a specific inhibitor of mTORC1, promoted redistribution of endogenous TFE3 to the nucleus (fig. S1D). Thus, a clear correlation exists between the activity of mTORC1 and the subcellular distribution of TFE3.
Because phosphorylation by mTORC1 causes 14-3-3-dependent sequestration of TFEB in the cytosol13,14, we tested if TFE3 interacted with 14-3-3 and the effect of mTORC1 activity on this interaction. TFE3-MYC coimmunoprecipitated endogenous 14-3-3 in fully fed cells, whereas this interaction was completely abolished by inactivation of mTORC1 with Torin-1 (Fig. 1D). Protein homology analysis indicated that Ser321 of TFE3 is the equivalent residue to Ser211 of TFEB, which mediates binding between TFEB and 14-3-313,14 (Fig. 2A). Mutation of Ser321 to Ala prevented binding of overexpressed TFE3 to endogenous 14-3-3 in ARPE-19 cells (fig. S2A). TFE3-S321A also accumulated in the nucleus when expressed in ARPE-19 cells (fig. S2B, S2C). As expected, TFE3 was recognized by antibodies against the 14-3-3 binding domain (fig. S2D). Therefore these data suggested that TFE3 is retained in the cytosol through mTORC1-dependent binding to 14-3-3.
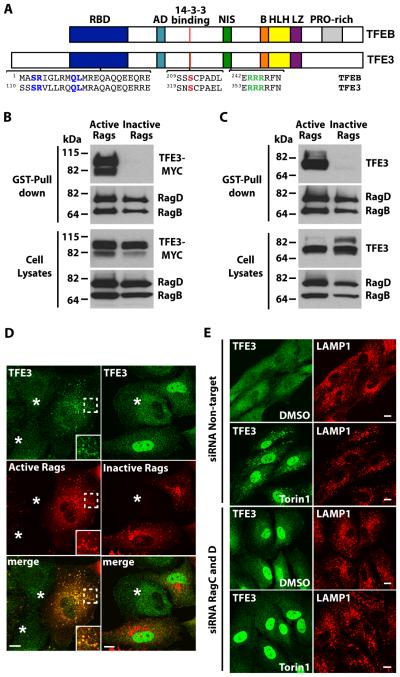
Fig. 2. TFE3 interacts with active Rag heterodimers.
(A) Schematic representation of comparison between TFEB and TFE3. RBD: Rag-binding domain, AD: Activation domain, NIS: Nuclear import signal, B: Basic residues, HLH: Helix-loop-helix, LZ: Leucine-zipper, PRO-rich: Proline rich. Residues proven to be essential in the RBD (S112R113), 14-3-3 binding site (S321), and NIS (R356R357R358) are colored. (B and C) Immunoblotting analysis of TFE3-MYC (B) or endogenous TFE3 (C) affinity purified with glutathione S-transferase (GST) fused to C-terminus of active Rag heterodimers (RagBGTP/RagDGDP) in ARPE-19 cells. The affinity-purified materials were probed with antibodies against GST, MYC, and TFE3 (used to detect Rag proteins, TFE3-MYC, and endogenous TFE3, respectively). Data are representative of 3 independent experiments. (D) Immunofluorescence confocal microscopy analysis of the subcellular distribution of endogenous TFE3 in ARPE-19 cells overexpressing either active or inactive RagB/D heterodimers. Cells were double stained with antibodies against TFE3 and GST (used to detect endogenous TFE3 or Rag proteins, respectively). Asterisks indicate distribution of endogenous TFE3 in non-transfected cells. Scale bars, 10 μm Data are representative of 3 independent experiments and over 90% of cells positive for the RagB/D heterodimers exhibited the phenotypes shown. (E) Immunofluorescence confocal microscopy analysis of the subcellular distribution of endogenous TFE3 in control or Rag-depleted ARPE-19 cells upon incubation with Torin-1 for 1 hour. Cells were double stained with antibodies against TFE3 and LAMP1. Scale bars, 10 μm. Data are representative of 3 independent experiments and over 90% of cells exhibited the phenotypes shown.
Rag GTPases determine the activity and intracellular distribution of TFE3
The first 30 N-terminal residues of TFEB are both necessary and sufficient for interaction with active Rag heterodimers20. Homology analysis revealed that a Rag-binding motif is also present in TFE3, suggesting that TFE3 might interact with Rag GTPases (Fig. 2A). To test this possibility, we co-expressed TFE3 together with different combinations of Rag proteins in ARPE-19 cells. Rags function as heterodimers in which the active complex consists of GTP-bound RagA or B complexed with GDP-bound RagC or D. Therefore, we expressed TFE3-MYC together with Rag mutants predicted to be restricted to the GTP-bound or GDP-bound conformations. TFE3-MYC interacted with Rag heterodimers when they are in an active conformation (RagBGTP/RagDGDP), whereas no binding was observed between TFE3 and inactive Rag heterodimers (RagBGDP/RagDGTP) (Fig. 2B). Similar results were obtained for endogenous TFE3 (Fig. 2C). Furthermore, mutation of conserved residues within the Rag-binding motif dramatically reduced the interaction between TFE3 and active Rags (fig. S2E).
Endogenous TFE3 redistributed from the cytosol to lysosomes in cells overexpressing the active Rag complex and redistributed to the nucleus in cells overexpressing the inactive Rag complex (even in the absence of nutrient starvation) (Fig. 2D). Depletion of endogenous RagC and RagD (see fig. S2F for knockdown efficiency) prevented the redistribution of TFE3 to lysosomes induced by Torin-1 and triggered the accumulation of TFE3 in the nucleus (Fig. 2E), confirming the importance of the Rag complex and mTORC1 in controlling TFE3 subcellular distribution. In addition, mutations in TFE3 in the Rag-binding domain (TFE3-S112A/R113A) that prevented the interaction between TFE3 and Rags (fig. S2E) caused constitutive accumulation of TFE3 in the nucleus of fully fed cells (fig. S2B, S2C).
Together, our data suggested that TFE3 is recruited to lysosomes through direct interaction with active Rags and this recruitment is critical for maintaining TFE3 sequestration in the cytosol under nutrient-rich conditions. Following starvation, Rags and mTORC1 are inactivated, thus allowing transport of TFE3 to the nucleus (see fig. S3 for a model of the mechanism of TFE3 regulation by Rags and mTORC1).
TFE3 and folliculin are involved in a feedback loop stimulated by nutrient depletion
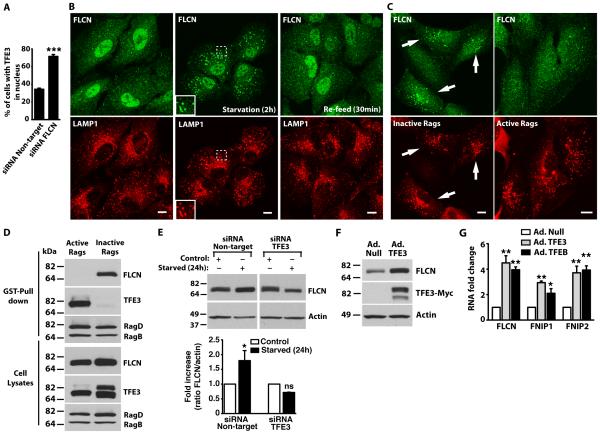
Birt-Hogg-Dube (BHD) syndrome is caused by germline mutations in the tumor suppressor gene folliculin (FLCN) and is characterized by the development of fibrofolliculomas, lung cysts, and renal carcinoma28-30. Mutations in FLCN lead to the dysregulation of TFE3, as indicated by the constitutively nuclear localization of TFE3 in a BHD cancer cell line27. Mutations in FLCN also cause increased TFE3 transcriptional activity in human kidney cells and mouse embryonic fibroblasts27. In agreement with these data, we found that depletion of FLCN by siRNA in ARPE-19 cells increased the proportion of cells with nuclear TFE3, thus confirming the regulatory role for FLCN on TFE3 distribution (Fig. 3A).
Fig. 3. Folliculin is recruited to lysosomes and upregulated by TFE3 during starvation.
(A) ARPE-19 cells were transfected with siRNA duplexes to FLCN or non-target siRNA for 72 hours. Cells were fixed and the nuclear localization was assessed by immunofluorescence microscopy using an antibody that recognizes TFE3. Data are shown as means ± SD and represent 643 cells (siRNA Non-target) and 663 cells (siRNA FLCN) counted in 3 independent experiments. The data were analyzed using paired t test (***, p=0.003). (B) ARPE-19 cells were starved in serum-and amino acid-free medium for 2 hours (Starvation) or starved and re-fed (Re-feed) in complete medium for 30 minutes. Cells were fixed and FLCN subcellular localization was analyzed by immunofluorescence confocal microscopy using antibodies to FLCN and LAMP1. Insets represent a two-fold magnification of the indicated region. Scale bars, 10 μm. Data are representative of 3 independent experiments and over 90% of cells exhibited the phenotypes shown. (C) ARPE-19 cells were transfected with either active or inactive RagB/D heterodimers. After 12 hours, cells were double stained with antibodies against HA (used to detect Rag proteins) and FLCN. Scale bars, 10 μm. Arrows indicate cells transfected with inactive RagB/D heterodimers. Data are representative of 3 independent experiments and over 90% of HA-positive cells exhibited the phenotypes shown. (D) ARPE-19 were nucleofected with the indicated GST-Rag-expressing plasmids. Twenty hours later, cells were lysed, and RagB/D heterodimers were pulled down using glutathione-Sepharose beads. Proteins bound to the beads were analyzed by immunoblotting with antibodies against GST (used to detect Rag proteins), TFE3 and FLCN. Data are representative of 3 independent experiments (E) ARPE-19 cells were transfected with siRNA duplexes to TFE3 or non-target siRNA for 48 hours. Cells were then either kept in complete medium (control) or starved in serum- and amino acid-free medium for 24 hours. A representative blot is shown along with quantification of three independent experiments plotted as the ratio of FLCN to actin is expressed as fold increase relative to the control Non-target siRNA condition. The data were analyzed using one-way ANOVA (*, p ≤ 0.05; and n.s.: not significant starved versus control for each siRNA condition). (F) ARPE-19 cells were infected with either adenovirus expressing Null (Ad. Null) or TFE3-MYC (Ad. TFE3) for 48 hours. Cells were then lysed and the lysates were analyzed by immunoblotting using antibodies to detect FLCN, MYC (TFE3), and actin. Data are representative of 3 independent experiments (G) ARPE-19 cells were infected with either adenovirus expressing Null, TFE3-MYC, or TFEB-FLAG for 48 hours and RNA was extracted. mRNA transcript abundance was assessed by Relative Quantitative Real Time PCR using specific primers for the indicated genes from three independent experiments. The data were analyzed using one-way ANOVA and are shown as means ± SD (*, p ≤ 0.05; **, p ≤ 0.01; versus adenovirus Null-infected cells).
The intracellular localization of endogenous FLCN changed depending on nutrient availability. In fully fed cells, FLCN was mainly cytosolic, although some weak association with lysosomes can be observed (Fig. 3B). Please note that the nuclear staining observed in these cells is probably non-specific as it was also detected in FLCN-depleted cells (fig. S4A) The amount of FLCN bound to lysosomal membranes increased in nutrient-deprived ARPE-19 cells (Fig. 3B). When cells were placed back in a nutrient-rich medium (re-feed), colocalization of FLCN with lysosomes was reduced (Fig. 3B). Relocation of FLCN from lysosomes to the cytosol was observed as early as 10 min after re-feed (fig. S4B). Increased recruitment of endogenous FLCN to lysosomes was also observed upon inactivation of mTORC1 with Torin-1 (fig. S4C).
Next we sought to identify the molecular machinery that regulates the recruitment of FLCN to lysosomes under starvation conditions. Expression of constitutively inactive Rag heterodimers (RagBGDP/RagDGTP) induced the recruitment of endogenous FLCN to lysosomes whereas expression of the active Rag complex did not (Fig. 3C). Consistent with these results, FLCN interacted with inactive but not with active Rag heterodimers (Fig. 3D). We showed that, in the same cells, TFE3 interacted with active (but not inactive) Rags (Fig. 3D). Thus, we propose that under conditions that inactivate the Rag complex, such as starvation, the ability of Rags to interact with mTORC1 and TFE3 would decrease, thereby reducing the lysosomal association and activity of mTORC1 and reducing TFE3 phosphorylation, while the ability to bind FLCN would increase, leading to its recruitment to lysosomes. In this model, FLCN does not regulate TFE3 by direct interaction. In agreement with this, we were unable to detect interaction between both proteins in co-immunoprecipitation assays (Fig. 3D and fig. S4D).
FLCN appears to function as a positive regulator of the mTORC1 pathway, because down-regulation of FLCN inhibits mTORC1 activity both in cultured cells and in mice31,32. Thus, FLCN might be recruited to lysosomes to facilitate rapid or robust re-activation of mTORC1 as cells experience the switch from the nutrient-depleted state to the nutrient-replete state. Because we showed that active mTORC1 was necessary for retention of TFE3 in the cytosol (Fig. 1), we propose that the nuclear accumulation of TFE3 in BHD cells might be a consequence of reduced mTORC1 activity in the absence of FLCN.
We found a TFE3-dependent increase in the amount of FLCN in ARPE-19 cells starved of nutrients for 24 hours (Fig. 3E). Accordingly, we found that overexpression of TFE3 in ARPE-19 cells resulted in an increase in FLCN protein abundance (Fig. 3F), as well as the mRNA abundance of FLCN and two FLCN-interacting proteins, FNIP1 and FNIP2 (Fig. 3G)32. Upregulation of FLCN, FNIP1, and FNIP2 was also observed upon TFEB over-expression (Fig. 3G).
Thus, we propose that by stimulating the production of FLCN and its partners, which stimulate mTORC1, TFE3 may ensure efficient termination of the lysosomal starvation response, including its own activity, once nutrients become available.
TFE3 and MITF1 increase expression of autophagic genes and stimulate autophagy
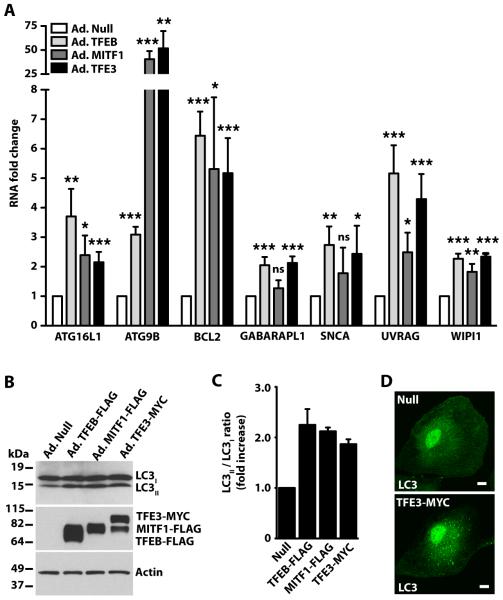
TFEB is a master regulator for the expression of autophagic and lysosomal genes18. Since TFEB, TFE3, and MITF1 share the same mechanism of activation in response to starvation/ mTORC1 inactivation, we asked whether TFE3 and MITF1 might also be implicated in autophagy induction and lysosomal biogenesis. To test the effect of TFE3 and MITF1 overexpression on the transcriptional regulation of autophagy, we used commercially available arrays of primers that allowed us to simultaneously monitor the expression of 83 autophagic genes by relative quantitative real time-PCR (qRT-PCR). We found that TFEB, TFE3, and MITF1 targeted the same autophagic genes (table S1) with 7 of the 83 showing increased expression of at least 2-fold in cells overexpressing any of the three transcription factors (Fig. 4A). The most significantly up-regulated genes encode proteins that play an essential role in formation of autophagosomes (ATG16L1, ATG9B, GABARAPL1, and WIPI1) as well as their degradation (UVRAG). Analysis of the LC3II/LC3I ratio upon TFE3, TFEB, or MITF1 overexpression confirmed autophagy induction (Fig. 4B and 4C). Accordingly, we observed an accumulation of autophagosomes in TFE3-expressing cells (Fig. 4D).
Fig. 4. TFE3 overexpression induces the transcription of autophagy-related genes.
(A) ARPE-19 cells were infected with Null-, MITF1-, TFEB-, or TFE3-expressing adenoviruses for 48 hours and total RNA was extracted. mRNA transcript abundance was assessed using the Human Autophagy RT2 Profiler PCR Array. Represented genes exhibited at least 2-fold increase in cells overexpressing TFE3. Values are means ± SD of 3 independent Profiler experiments. The data were analyzed using one-way ANOVA (*, p ≤ 0.05; **, p ≤ 0.01; ***, p ≤ 0.001; ns: not significant versus Null-infected cells). (B) Immunoblotting analysis of ARPE-19 cells overexpressing adenovirus Null, TFEB-FLAG, MITF1-FLAG or TFE3-MYC. Proteins were detected with antibodies against FLAG (used to detect TFEB-FLAG and MITF-FLAG), MYC (used to detect TFE3-MYC), LC3, and Actin. Data are representative of 3 independent experiments. (C) Quantification of LC3I and LC3II protein bands as shown in B. Data were normalized to actin. Bars represent ratio of LC3II/LC3I expressed as fold increase of the ratio from cells infected with adenovirus Null. Values represent the average ± range of 2 independent experiments. (D) ARPE-19 cells were infected as in A with the indicated adenoviruses, fixed, and then stained with LC3 for detection of autophagosomes. Data are representative of 3 independent experiments and over 80% of cells exhibited the phenotypes shown.
TFE3 binds to CLEAR elements in promoters and stimulates lysosomal biogenesis
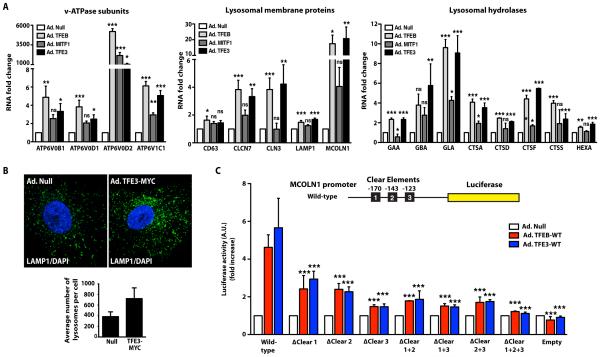
Next, we investigated the role of MITF1 and TFE3 in lysosomal biogenesis. We evaluated the effect of MITF1 or TFE3 overexpression in ARPE-19 cells on several lysosomal genes that are targets of TFEB17. In agreement with previous studies33, MITF1 increased the expression of two of the four genes encoding v-ATPase subunits tested (Fig. 5A). However, MITF1 failed to increase expression of most of the lysosomal genes tested, suggesting that MITF1 does not play a major role in lysosomal biogenesis (Fig. 5A).
Fig. 5. TFE3 overexpression induces transcription of lysosomal genes through the CLEAR element.
(A) ARPE-19 cells were infected with Null-, MITF1-, TFEB-, or TFE3-expressing adenoviruses for 48 hours and RNA was extracted. mRNA transcript abundance was assessed by qRT-PCR using specific primers for the indicated genes. Data are presented as means ± SD of three independent experiments. The data were analyzed using one-way ANOVA(*, p ≤ 0.05; **, p ≤ 0.01; ***, p ≤ 0.001; ns: not significant versus Null-infected cells) (B) Immunofluorescence confocal microscopy analysis of LAMP1 in ARPE-19 cells infected with Null- or TFE3-MYC-expressing adenoviruses. Cells were double stained with antibodies against MYC (to detect recombinant TFE3) and LAMP1. Scale bars, 10 μm. Quantification of LAMP1-positive puncta shown for 25 cells per condition in 3 independent experiments. Data are presented as means ± SD (C) Luciferase activity measured in cells coexpressing the MCOLN1 promoter-luciferase reporter constructs with Null, TFEB-WT or TFE3-WT. Wild-type contains all three CLEAR elements, ΔClear 1 lacks the CLEAR element at position −170, ΔClear 2 lacks the CLEAR element at position −143, ΔClear 3 lacks the CLEAR element at position −123, ΔClear 1+2 lacks the CLEAR elements at position −170 and −123, ΔClear 1+3 lacks the CLEAR elements at position −170 and −123, ΔClear 2+3 lacks the CLEAR elements at position −143 and −123, ΔClear 1+2+3 lacks all three CLEAR elements. Bars represent luciferase activity expressed as fold increase versus cells infected with adenovirus Null. Values are means ± SD of three independent experiments. The data were analyzed using one-way ANOVA (***, p ≤ 0.001 versus Null-infected cells).
Overexpression of TFE3 in ARPE-19 cells increased the mRNA abundance of 16 out of the 17 genes tested, including those encoding several subunits of the v-ATPase (ATP6V0B1, ATP6V0D1, ATP6V0D2, ATP6V1C1) (Fig. 5A), lysosomal transmembrane proteins (CD63, CLCN7, CLCN3, LAMP1, MCOLN1) (Fig. 5A), and lysosomal hydrolases (GAA, GBA, GLA, CTSA, CTSD, CTSF, CTSS, HEXA) (Fig. 5A). Western blotting confirmed the increase in several lysosomal proteins including LAMP1, RagC (encoded by RRAGC), Cathepsin D (encoded by CTSD), and ATP6V1C1 in TFE3-overexpressing cells (fig. S5A).
Consistent with our qRT-PCR data, we found an increased number of LAMP1-positive structures in ARPE-19 cells overexpressing TFE3 when compared with control cells (Fig. 5B). The average number of lysosomes per cell in ARPE-19 cells infected with control (null) adenovirus was 383 ± 90, whereas in TFE3-expressing cells had 722 ± 202, (Fig. 5B). Thus, TFE3 promoted lysosomal biogenesis in ARPE-19 cells.
TFEB regulates lysosomal biogenesis and function through its binding to CLEAR elements (GTCACGTGAC) in the promoters of numerous lysosomal genes16. TFE3 binds the E-box, which is a consensus sequence (CACGTG) that overlaps with the CLEAR sequence (as indicated by the underlining in the CLEAR element), suggesting that TFE3 might recognize CLEAR motifs34,35. MCOLN1 is a lysosomal gene that contains three putative CLEAR sites in its promoter region (at positions −170, −143, and −123)16. To quantitatively assess the ability of TFE3 to bind to CLEAR motifs, we transfected HeLa cells with a luciferase reporter plasmid containing the human MCOLN1 promoter and infected the cells with adenovirus expressing either TFEB or TFE3 or with control (Null) adenovirus (Fig. 5C).
The MCOLN1 promoter had low basal activity in cells infected with control adenovirus. In contrast, expression of either TFEB or TFE3 increased the expression of the MCOLN1 reporter (Fig. 5C). Mutation of the CLEAR1 (ΔCLEAR1) or CLEAR2 (ΔCLEAR2) sites significantly decreased the stimulation by TFEB or TFE3 of reporter activity, and mutation of the CLEAR3 (ΔCLEAR3) site caused a further reduction TFEB-dependent or TFE3-dependent reporter activity (Fig. 5C). The triple-point mutant (ΔCLEAR1+2+3) failed to respond to either TFEB or TFE3. Compared to the wild-type promoter, the ΔCLEAR3 mutant maintained 10% of activity, whereas the triple mutant (ΔCLEAR1+2+3) retained just 2.5% of activity in response to TFE3 expression (Fig. 5C).
Thus, each of the three CLEAR sites in the MCOLN1 promoter contributed to TFEB- or TFE3-mediated stimulation of the expression of the reporter gene. Therefore, we suggest that TFE3 promotes expression of lysosomal genes by binding to CLEAR elements.
TFE3 promotes expression of lysosomal genes independently of TFEB
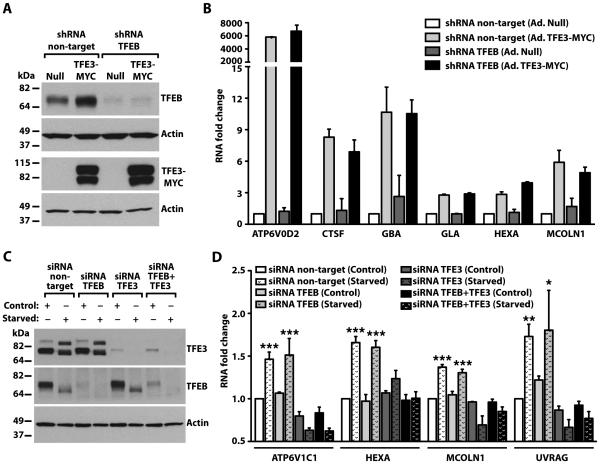
Members of the MiTF/TFE family bind DNA as homo- or heterodimers. Therefore, we tested if the ability of TFE3 to stimulate expression of lysosomal genes was through formation of heterodimers with TFEB. We used lentivirus expressing TFEB shRNA to generate a stable HeLa cell clone with reduced amounts of TFEB (Fig. 6A). Endogenous TFEB increased in the control cells expressing TFE3, suggesting that the gene encoding TFEB may be a target of TFE3 (Fig. 6A). This observation is consistent with a recent report showing the presence of several CLEAR elements in the promoter region of TFEB36. Overexpression of TFE3 in control HeLa cells (shRNA non-target) or in the TFEB-depleted cell line resulted in a dramatic increase in the expression of many lysosomal genes, including ATP6V0D2, CTSF, GBA, GLA, HEXA, and MCOLN1 (Fig. 6B). Because TFE3 did not stimulate TFEB production in the TFEB shRNA stable clone (Fig. 6A), we concluded that TFE3 promotes transcription of lysosomal genes independently of TFEB (Fig. 6B).
Fig. 6. TFE3 promotes expression of lysosomal genes independently of TFEB.
(A) Immunoblotting analysis of control (Null) or TFEB-depleted HeLa cells infected with Null- or TFE3-MYC-expressing adenoviruses for 48 hours. Protein bands were detected with antibodies against TFEB and MYC (used to detect endogenous TFEB and TFE3-MYC, respectively), and Actin. Data are representative of 3 independent experiments. (B) qRT-PCR analysis of lysosomal genes from HeLa cells treated as indicated in (A). Total RNAs were extracted and mRNA transcript abundance was assessed using specific primers for the indicated genes. Data were normalized to Glyceraldehyde 3-phosphate dehydrogenase (GAPDH). Bars represent fold change of the ratio to shRNA non-target cells infected with adenovirus Null. Values are presented as average ± range of 2 independent experiments. (C) Immunoblotting analysis of ARPE-19 cells treated with siRNA to TFEB, TFE3, or TFEB and TFE3 (TFEB+TFE3) under control or starvation conditions. Proteins were detected with antibodies against TFEB, TFE3, and Actin. Note that the immunoblot for endogenous TFEB required a much longer exposure time due to the very low abundance of TFEB in ARPE-19 cells compared to the abundance of endogenous TFE3. Data are representative of 3 independent experiments (D) qRT-PCR analysis of lysosomal- or autophagy-related genes from ARPE-19 cells treated as indicated in (C). Total RNAs were extracted and mRNA transcript abundance was assessed using specific primers for the indicated genes. Bars represent fold change of the ratio to siRNA non-target cells in control condition. Values are means ± SD of 3 independent experiments. The data were analyzed using one-way ANOVA (*, p ≤ 0.05; **, p ≤ 0.01; ***, p ≤ 0.001 versus siRNA non-target control cells).
We determined that different cell lines and tissues exhibited different relative amounts of TFEB and TFE3 (fig. S5B-D). To determine if endogenous TFE3 regulates expression of lysosomal genes in response to starvation, we used ARPE-19 cells, because we found that endogenous TFE3 was more abundant than TFEB in these cells (Fig. 6C, fig. S5B, and S5C). Starvation of ARPE-19 cells for 24h increased the mRNAs of lysosomal and autophagy genes (Fig. 6D). Depletion of TFEB did not prevent the increased expression of selected lysosomal and autophagic genes in response to starvation (Fig. 6D). In contrast, the induction of lysosomal and autophagic genes was entirely abolished in TFE3-depleted cells (Fig. 6D). Similar results were observed upon simultaneous depletion of TFE3 and TFEB (Fig. 6D). Therefore, our data indicated that in some cells, TFE3 plays a critical role in the cellular response to starvation and suggested that whether TFEB or TFE3 serve as the “master” regulator of the lysosomal response may depend on their relative abundance.
Overexpression of TFE3 induces lysosome exocytosis
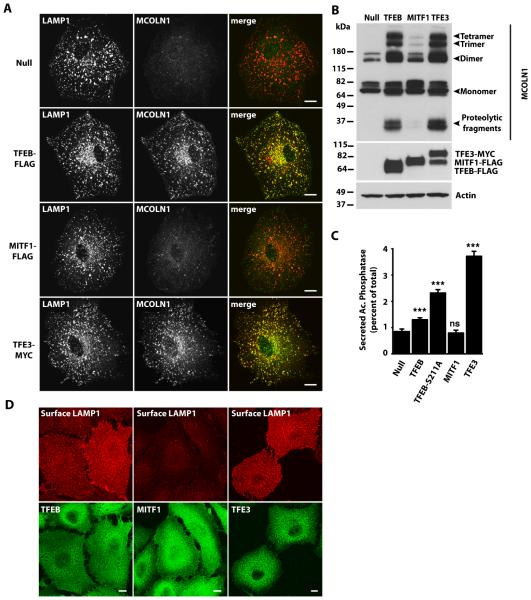
Overexpression of TFEB increases the pool of lysosomes located close to the cell surface and induces fusion of lysosomes with the plasma membrane37. The mechanism by which TFEB promotes lysosomal exocytosis involves increasing intracellular Ca2+ concentrations through the upregulation and activation of the lysosomal Ca2+ release channel MCOLN137. Our luciferase and qRT-PCR data indicated that MCOLN1 mRNA was significantly increased by TFE3 overexpression (Fig. 5A, 5C), suggesting that TFE3 may also induce lysosomal exocytosis. To test this, we used a newly generated affinity purified antibody recognizing MCOLN1 to measure variations in the amount of endogenous MCOLN1 in response to TFE3. The specificity of the MCOLN1 antibody was validated in fibroblasts obtained from Mucolipidosis type IV (MLIV) patients; these cells lack MCOLN138-40 (fig. S6A). Immunofluorescence analysis and Western blot analysis showed that overexpression of either TFEB or TFE3 in ARPE-19 cells increased the amount of endogenous MCOLN1, whereas MITF1 overexpression did not (Fig. 7A, B). Endogenous MCOLN1 colocalized with LAMP1 (Fig. 7A), which is consistent with previous studies describing the lysosomal distribution of overexpressed MCOLN141-43.
Fig. 7. TFE3 overexpression triggers lysosomal exocytosis.
(A) Immunofluorescence confocal microscopy analysis of endogenous MCOLN1 in ARPE-19 cells infected with Null adenovirus or adenovirus expressing TFEB-FLAG, MITF1-FLAG, or TFE3-MYC for 40 hours. Cells were double stained with antibodies against MCOLN1 and LAMP1. Scale bars, 10 μm. Data are representative of 3 independent experiments and over 90% of cells exhibited the phenotypes shown. (B) Immunoblotting analysis of ARPE-19 cells overexpressing adenovirus Null, TFEB-FLAG, MITF1-FLAG, or TFE3-MYC. Proteins were detected with antibodies against MCOLN1, MYC (to detect TFE3), FLAG (to detect TFEB and MITF1), and Actin. Data are representative of 3 independent experiments. (C) Acid phosphatase secretion analysis in HeLa cells infected with adenoviruses expressing the indicated proteins. Bars represent the secreted acid phosphatase activity as a percentage of the total acid phosphatase activity. Values are means ± SD of 3 independent experiments. The data were analyzed using one-way ANOVA (***, p ≤ 0.001; ns: not significant versus Null-infected cells). (D) Immunofluorescence confocal microscopy analysis of LAMP1 present at the cell surface of ARPE-19 cells overexpressing TFEB-FLAG, MITF1-FLAG, or TFE3-MYC. Cells were double stained with antibodies against FLAG and MYC (used to detect TFEB-FLAG, MITF-FLAG, and TFE3-MYC, respectively) and LAMP1. Scale bars, 10 μm. Data are representative of 3 independent experiments and over 90% of cells exhibited the phenotypes shown.
To monitor lysosomal exocytosis from HeLa cells, we measured the activity of lysosomal hydrolases in the extracellular medium. In agreement with previous reports13, we found that overexpression of TFEB caused the release of acid phosphatase into the culture medium (Fig. 7C) and more acid phosphatase activity was present in the culture medium of cells expressing a constitutively active version of TFEB that cannot be retained in the cytosol (TFEB-S211). As expected, secretion of acid phosphatase was not increased when cells were infected with a control (Null) adenovirus or with an adenovirus expressing MITF1 (Fig. 7C). Overexpression of TFE3 caused a robust secretion of lysosomal hydrolases, indicating that TFE3 effectively induces lysosomal exocytosis (Fig. 7C). To ensure that the increased activity of lysosomal enzymes in the medium was not due to cell damage, we measured the amount of cytosolic lactate dehydrogenase (LDH) in the medium, which would indicate cell lysis or leakage. None of overexpressed transcription factors caused increased LDH in the medium (fig. S6B).
To confirm that the presence of acid phosphatase in the medium was due to fusion of lysosomes with the plasma membrane, we monitored accumulation of the lysosomal transmembrane protein LAMP1 at the cell surface by immunofluorescence (Fig. 7D) and flow cytometry (fig. S6C). TFEB and TFE3, but not MITF1, induced the accumulation of LAMP1 at the plasma membrane of ARPE-19 cells. These lysosomal exocytosis data and MCOLN1 abundance data are consistent with a model in which TFE3 promotes lysosomal exocytosis by increasing the abundance of the Ca2+ channel MCOLN1.
TFE3 overexpression promotes cellular clearance in a cell model of Pompe disease
We and others have recently reported that the ability of TFEB to induce lysosomal exocytosis can be exploited to clear cells of the undigested material that accumulates in the lysosomal lumen in numerous Lysosomal Storage Disorders (LSDs)37,44,45. Since TFE3 can also stimulate lysosomal exocytosis, we assessed the ability of TFE3 to promote cellular clearance in Pompe disease (PD; glycogen storage disease type II), a paradigm for LSD. PD is caused by mutations in acid alpha-glucosidase (GAA). The enzyme breaks down glycogen to glucose within the acidic environment of lysosomes46,47. GAA deficiency leads to excessive buildup of glycogen inside lysosomes in many tissues and manifests as severe cardiac and skeletal muscle myopathy. Accumulation of dysfunctional, enlarged lysosomes results in perturbation of many cellular processes, including autophagy48. The autophagic process in the diseased muscle is affected at both the initiation of autophagosomal formation (increase) and at the termination stage (impaired autophagosomal-lysosomal fusion, a condition known as autophagic block).
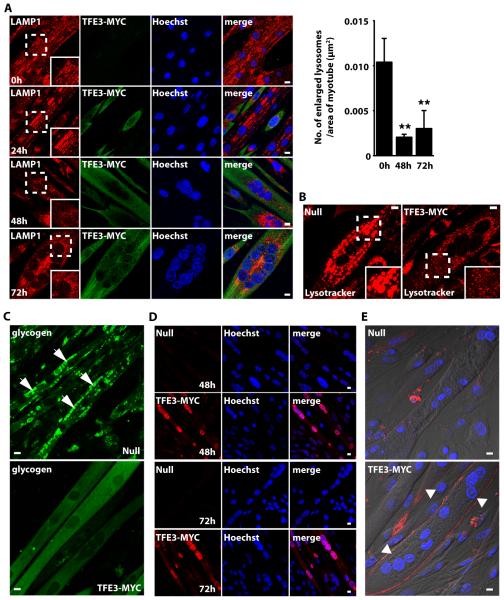
We established a PD conditionally immortalized muscle cell line that replicates the lysosomal pathology (namely, accumulation of enlarged lysosomes and glycogen buildup) characteristic of this disorder45. Infection of PD myotubes with an adenovirus expressing TFE3, but not with a control (Null) adenovirus, significantly reduced the number of enlarged lysosomes, as shown by staining with LAMP-1 and lysotracker (Fig. 8A, 8B). The effect of TFE3 and its nuclear translocation were evident after 48-72h post-infection (Fig. 8A, 8D). Overexpression of TFE3 also reduced the amount of intralysosomal glycogen, as assessed by the incorporation of fluorescent glucose derivative 2-NBDG into glycogen (Fig. 8C). TFE3-mediated lysosomal exocytosis was further corroborated by surface LAMP1 assay. LAMP1 accumulated on the plasma membrane in TFE3-treated myotubes (Fig. 8E lower panel) but not in cells infected with control adenovirus (Fig. 8E upper panel). Therefore, our data indicated that TFE3 promotes cellular clearance in PD myotubes and may be considered an alternative to TFEB for the treatment of different LSDs.
Fig. 8. TFE3 promotes clearance of enlarged lysosomes and reduces glycogen load in PD myotubes.
(A) Confocal microscopy images of PD myotubes (clone 6) infected with adenovirus expressing TFE3-MYC for 24, 48, or 72 hours. Ad. TFE3 was added on day 4, 5, and 6 after the differentiation began (see Materials and Methods). The cells were then fixed on day 7 and stained with LAMP-1 (lysosomes; red) and MYC (TFE3; green) antibodies. Nuclei are stained with Hoechst (blue). Insets show a 1.5 fold magnification of the indicated region. Graphical representation of the quantification of the data in A. The bars represent the number of enlarged lysosomes (>2 μm) per μm2 of the myotube in uninfected cells and cells infected for 48 or 72 hours with adenovirus expressing TFE3-MYC. Values are means ± SD of 3 independent experiments. The data were analyzed using one-way ANOVA (**, p ≤ 0.01). (B) LysoTracker staining of live PD cells infected with the Null or TFE3-MYC-expressing adenovirus. Adenovirus was added to the myotubes for 72 hours on day 7 in differentiation medium. Insets show a 1.7 fold magnification of the indicated region. Data are representative of 3 independent experiments and over 90% of cells exhibited the phenotypes shown. (C) Confocal microscopy images of live noninfected PD myotubes (Top) or PD myotubes infected for 48 hours with adenovirus expressing TFE3-MYC (Bottom). The cells were incubated with the fluorescent glucose (2-NBDG; green), washed, and analyzed by confocal microscopy. Scale bars, 10 μm for all panels. Data are representative of 3 independent experiments and over 90% of cells exhibited the phenotypes shown. (D) Accumulation of TFE3 in the nuclei in myotubes infected with TFE3-MYC for 48 or 72 hours. TFE3 was detected with antibody recognizing MYC (red). (E) Surface LAMP assay of PD myotubes infected with Null- or TFE3-MYC-expressing adenovirus. Confocal microscopy images show LAMP1 staining (red) on plasma membrane in TFE3-treated cells (bottom; arrowheads) but not in noninfected cells (top). Nonpermeabilized cells were incubated with antibody recognizing LAMP1 at 4°C for 40 min, followed by fixation and staining with secondary antibody. Data are representative of 3 independent experiments and over 70% of cells exhibited the phenotypes shown. Bar: 10 μm for all panels.
Discussion
The finding that expression of lysosomal genes is not constitutive but changes in response to nutrient status revealed that cells monitor lysosomal function and respond to degradation requirements and environmental conditions. TFEB was the only previously reported member of the MiTF/TFE family capable of binding to CLEAR motifs and inducing expression of multiple lysosomal genes16. Here, we identified TFE3 as another regulator of lysosomal function and biogenesis in response to starvation. Similar to TFEB, in fully fed cells TFE3 was recruited to lysosomes by active Rag GTPases leading to TFE3 phosphorylation by mTORC1 and retention in the cytosol by 14-3-3. Following starvation or lysosomal stress or other conditions that inhibited mTORC1, TFE3 rapidly translocated to the nucleus and activated genes associated with autophagy and lysosomal biogenesis and function that promote cellular survival during starvation conditions (fig. S3). Our results agree with a previous study showing interaction between 14-3-3 and TFE3 in a large-scale proteomics study of 14-3-3 binding proteins49. However, the present study further expands these observations by showing that the interaction between TFE3 and 14-3-3 is 1) dependent on nutrient levels and mTORC1 activity and 2) critical for retention of TFE3 in the cytosol under nutrient rich conditions.
MITF and TFE3 have previously been implicated in the regulation of certain genes that are important for the biogenesis or function of specialized lysosomal-related organelles, such as secretory lysosomes and melanosomes. For example, MITF and TFE3 stimulate the expression of genes implicated in bone resorption in differentiated osteoclasts50. Furthermore, some MITF targets, including HPS4, PSEN2 and LYST, are important for pigment biogenesis33. However, we found that, despite sharing similar mechanisms of regulation with TFEB and TFE3, MITF1 lacked the ability to induce lysosomal formation or exocytosis. Furthermore, ARPE-19 cells depleted of TFE3 failed to increase expression of lysosomal and autophagic genes despite the presence of MITF1. Overall, our data suggested that MITF1 does not play a major role in mediating cellular adaptation to starvation.
Differences in recognition of response elements in the promoters of the targeted genes may explain why MITF does not induce lysosomal biogenesis, but TFEB and TFE3 do. MITF exhibits a preference for binding to M-boxes35 whereas TFEB binds to the CLEAR elements in the promoter region of many lysosomal genes16. The CLEAR elements are E-box-type DNA sequences that partially overlap with the consensus sequence of other E-boxes recognized by TFE334. Consistent with TFE3 binding to CLEAR elements, we found that TFE3 stimulated expression of a MCOLN1 promoter (containing the CLEAR elements) reporter. Therefore, it is likely that TFE3 stimulates lysosomal genes through interaction with CLEAR elements.
TFE3 also regulates expression of autophagy genes. Of the 83 autophagy genes we tested, less than 10% showed more than a 2-fold increase upon overexpression of TFE3. However, the upregulated genes encode proteins that function as key regulators of the autophagic process, including those involved in the formation of autophagosomes (ATG16L1, ATG9B, GABARAPL1, and WIPI1) and fusion of autophagosomes with lysosomes (UVRAG). An increase of lower than 2-fold for some other autophagy genes was also statistically significant. These results indicated an important role for TFE3 in autophagy induction.
Here, we showed that TFE3 functioned as part of a feedback loop with the positive regulator of the mTORC1 pathway, FLCN. Overexpression of either TFE3 or TFEB increased expression of FLCN, and TFE3 contributed to the increase in FLCN abundance that occurred as part of the cellular response to starvation. Notably, we found that the intracellular distribution of FLCN is dependent on nutrient levels and the activation state of Rag GTPases. The lysosomal recruitment of FLCN under starvation conditions suggested that it may regulate amino acid-dependent re-activation of mTORC1. Further work will help to discern the mechanism by which FLCN regulates mTORC1 activity. One possibility is that FLCN directly facilitates activation of Rag GTPases. Alternatively, FLCN may affect mTORC1 activity through the AMP-activated protein kinase (AMPK)51. Overexpression of the members of the MiTF/TFE family may contribute to different types of cancer52 by altering the regulation of the mTORC1 pathway.
Our results indicate that TFEB and TFE3 are regulated in a similar way, respond to nutrients, and induce expression of many of the same autophagic and lysosomal genes in response to starvation. In addition, previous studies have reported that overexpression of either TFEB or TFE3 regulate expression of metabolic genes and rescue obesity and insulin resistance in vivo26,36. These results raise the question of whether TFEB and TFE3 may be redundant. In Caenhorhabditis elegans the transcription factor HLH-30 has been proposed to be the ortholog of either TFE3 or TFEB36,53,54. Interestingly, HLH-30 regulates expression of autophagic, lysosomal, metabolic, and aging genes and is essential for survival during starvation36,53,54. Moreover, HLH-30 translocates to the nucleus following fasting and inactivation of CeTOR induces HLH-30 transcription54. Thus, the regulation and biological functions of TFE3 and TFEB may be evolutionarily conserved.
It is also possible that TFEB and TFE3 have specific functions in particular cell types or during development. For example, depletion of TFEB in mice is embryonically lethal due to defective vascularization of the placenta55, whereas the TFE3-knockout mouse is viable25. In zebrafish, the expression of TFE3 and TFEB overlaps in many but not all tissues during early embryogenesis56. Finally, in some instances lysosomal biogenesis is required during cell differentiation and occurs not in response to starvation, but rather in response to the presence of specific growth factors or cytokines. For example, TFEB increased the number of lysosomes in osteoclasts in response to RANKL, a process that is critical for bone remodeling57. In this case, activation of TFEB is not dependent on mTORC1 but is mediated by protein kinase C β (PKCβ). Phosphorylation of TFEB by PKCβ on several serine residues located in the C-terminal region increased the stability of TFEB57, thus resulting in an increase in the amount of nuclear TFEB and enhanced transcription of specific lysosomal genes. Likewise, ERK1/2-mediated phosphorylation of TFE3 in osteoclasts promotes the binding to specific co-activators (for example, p300) and TFE3-mediated expression of genes required for osteoclast development and function50. Therefore, in higher organisms TFEB and TFE3 may have evolved to perform specialized functions in specific cell types and their function may not be entirely redundant.
Overexpression of TFEB promotes the elimination of storage material in several animal models and cells from patients with LSDs, including multiple sulfatase deficiency37, mucopolysaccharidosis type IIIA37, Pompe disease45, Gaucher disease44, and Tay-Sachs disease44. TFEB also rescued α–synuclein toxicity in animal models for Parkinson disease58 and enhanced proteostasis in models of Huntington disease59. We presented evidence that TFE3 could be a therapeutic target in LSDs by showing that overexpressed TFE3 increased the abundance of the lysosomal calcium channel MCOLN1, triggered lysosomal exocytosis, and promoted cellular clearance in Pompe myotubes. Ultimately, the goal would be to achieve cellular clearance by activation of endogenous TFEB or TFE3 without the need for overexpression. The success of this strategy will rely heavily on the amount of the endogenous TFEB or TFE3 proteins in a specific tissue or cell type. We found that the amount of endogenous TFEB was low in human brain and muscle, two of the main tissues affected in LSDs. In contrast, TFE3 was moderate to high in these two tissues (fig. S5D). For this reason, the identification of TFE3 as another transcription factor capable of promoting cellular clearance is potentially clinically important.
In summary, our study revealed both similarities and differences in the regulation and functions of members of the MiTF/TFE family. The emerging picture suggests that these members share critical roles in organelle biogenesis, cell survival, and proliferation, as well as tumorigenesis. However, TFEB and TFE3 both function in nutrient sensing, energy metabolism, and cellular clearance, whereas MITF does not. The characterization of TFE3 regulation provides important insights for understanding how cells synchronize environmental signals, such as nutrient availability, with gene expression, energy production, and cellular homeostasis.
Materials and Methods
Cell line cultures and treatment
ARPE-19 is a line of immortalized human retinal pigmented epithelium (RPE) that constitutes an appropriate cellular model for studying regulation of TFE3. They efficiently induce autophagy following starvation and have high levels of endogenous TFE3. In addition, the members of the MiTF/TFE family are known to be critical for the development and survival of RPE60. ARPE-19 cells (CRL-2302, American Type Culture Collection) were grown at 37°C in a 1:1 mixture of DMEM and Ham’s F12 media supplemented with 10% fetal bovine serum (Invitrogen), 2 mM Glutamax™, 100 U/ml penicillin, and 100 μg/ml streptomycin (Gibco) in a humidified 5% CO2 atmosphere. HeLa and Hek293 cells (CCL-2, American Type Culture Collection) and HepG2 cells (HB-8065, American Type Culture Collection) were grown in DMEM and EMEM media, respectively, both supplemented with fetal bovine serum, Glutamax™ and antibiotics as indicated for ARPE-19 cells media. Skin fibroblasts from a MLIV patient (clone WG0909) and unrelated non-diseased skin fibro-blasts (clone MCH065) were obtained from the Repository for Mutant Human Cell Strains of Montreal Children’s Hospital (Montreal, Canada). For infection experiments, cells were infected with adenoviruses according to the manufacturer’s recommendations. Analyses were performed 15-48 h post-infection. For transient expression cells were nucleofected using Cell Line Nucleofector® Kit V (Lonza) following manufacturer’s recommendations. Cells were analyzed 12-24 h post-nucleofection. For drug treatment experiments, cells were incubated for 1-3 h at 37°C in medium containing one of the following reagents: DMSO (Sigma-Aldrich), 250 nM Torin-1 (TOCRIS) or 50 μM Chloroquine (Sigma-Aldrich). For starvation experiments cells were washed three times in Hank’s balances salt solution (Invitrogen) and incubated for 3-24 h at 37°C in Earle’s balanced salt solution (Sigma-Aldrich).
Antibodies
Rabbit polyclonal antibodies used: anti-14-3-3 (1:1,000, 8312S), anti Histone H3 (1:2,000, 9715S), anti-FLCN (1:1000, 3697S), anti-TFEB (1:1000, 4240S), anti-Raptor (1:500; 2280S), anti-Rictor (1:500; 2114S), anti-RagC (1:1000; 9480S) and anti-GST (1:5000, 2625S) were from Cell Signaling Technology, anti-LC3B (1:1000, L7543) and anti-TFE3 (1:2000, HPA023881) were from Sigma-Aldrich. Rabbit polyclonal anti-MCOLN1 NTail) was raised against the N-terminal tail of human MCOLN1 fused to GST. The antibody was then purified from the crude serum by affinity chromatography using a GST-NTail MCOLN1 column. Mouse monoclonal antibodies used: clone Ab5 to actin (1:10,000; 6126570 and clone H4A3 to CD107a-APC (1:50; 560664) were from BD Transduction Laboratories), clone 4C5 to Flag (1:500; TA50011, Origen), clone H4A3 to LAMP1 (1:3,000) and clone 9E10 to MYC (1:5000) were from Developmental Studies Hybridoma Bank and clones M2 (F1804) and M5 (F4240) (1:2,000) to Flag were from Sigma-Aldrich. Alexa Fluor 568-conjugated goat anti-mouse IgG and Alexa Fluor 488-conjugated goat anti-rabbit IgG, were used at a dilution of 1:1,000 (Invitrogen). HRP-conjugated anti-mouse or anti-rabbit IgG were obtained from Cell Signaling Technology and used at a dilution of 1:8,000.
Adenovirus
Adenovirus expressing TFE3-WT-MYC were prepared, amplified and purified by Welgen, Inc. Adenovirus expressing TFEB-WT-FLAG, TFEB-S211A, and MITF1-WT-FLAG have been previously described20.
Recombinant DNA plasmids
TFE3-MYC expression vector was generated by cloning The full-length encoding sequence of human TFE3 obtained by RT-PCR amplification from ARPE-19 cells total RNA followed by in-frame cloning into BamHI-SalI sites of into pCMV-3Tag-4a (Agilent Technologies) with a triple MYC tag fused to the carboxy-termini of TFE3. pLightSwitch-MCOLN1-promoter and pLightSwitch-EMPTY-promoter plasmids were obtained from SwitchGear Genomics. Amino acid substitutions in TFE3 and nucleotide substitutions in the CLEAR elements of MCOLN1-promoter were made using the QuickChange Lightning site-directed mutagenesis kit (Agilent Technologies) according to the manufacturer's instructions. The following constructs were obtained from Addgene: Plasmid 19303: pRK5-HA GST RagBGTP, Plasmid 19302: pRK5-HA GST RagBGDP, Plasmid 19308: pRK5-HA GST RagDGDP, Plasmid 19309: pRK5-HA GST RagDGTP7. Constructs were confirmed by DNA sequencing.
RNA interference (RNAi)
Knockdown of indicated genes was achieved by transfection of siRNA duplexes. In brief, cells grown in six-wells plate were transfected with DharmaFECT transfection reagent and 100 nM of ON-TARGETplus non-targeting pool siRNA duplexes or ON-TARGETplus smart pool siRNA duplexes targeted against Raptor, Rictor, FLCN, TFEB, and TFE3 (Dharmacon-Thermo Scientific), or mission siRNA against RagC and RagD genes (Sigma-Aldrich). Treated cells were analyzed 72 hours after transfection.
Generation of a stable HeLa cell line depleted of TFEB
HeLa cells were infected with lentivirus expressing shRNA non-target or shRNA TFEB (TRCN000013111, Sigma-Aldrich) for 48 hours following manufacturer’s recommendations. Transduced cells were then incubated with medium containing 1.25 μg/ml of Puromycin until resistant colonies were identified. Colonies were then isolated and analyzed to quantify the levels of TFEB transcripts using relative quantitative real time PCR as well as TFEB protein levels by immunoblotting using specific antibodies against endogenous TFEB. Experiments were performed with cells obtained from a puromycin resistant colony with more than 95% depletion of TFEB protein compared to shRNA non-target infected cells.
Human autophagy profiler array
ARPE-19 cells were infected with control (Null) adenovirus or with adenovirus expressing TFEB, TFE3, and MITF1 for 48 hours and total RNAs were extracted using the RNeasy Mini kit (Qiagen). cDNA was synthesized from 500ng of total RNA using the RT2 First Strand Kit (Qiagen) following manufacturer’s recommendations. Relative Quantitative real-time PCR was performed using the Human Autophagy RT2 Profiler PCR Array (PAHS-084ZA, SABiosciences) on an ABI 7900HT real-time PCR system (Applied Biosystems) according to manufacturer instructions. Resultant data were processed and analyzed using the SABiosciences PCR Array Data Analysis Software. Results are expressed as RNA fold change relative to cells expressing Null adenovirus.
Acid Phosphatase and Lactate Dehydrogenase (LDH) activity measurements
ARPE-19 cells were grown to subconfluency in complete medium in 6-wells plate. Cells were washed three times in Hank’s balances salt solution (Invitrogen) and then incubated in 1.1ml of medium without serum for 8 h at 37°C. For acid phosphatase the secreted activity was assayed in 0.45 ml of media by using the acid phosphatase assay kit from Sigma-Aldrich (CS0740) and following manufacturer’s instructions. Intracellular acid phosphatase activity was determined by lysing the cells in 10 mM phosphate buffer, pH 6.0, 0.15 M NaC1, 0.5%Triton X-100 followed by centrifugation at 100,000xg for 15min at 4°C. Fifteen microliters of triton cell extracts were used to measure the activity remaining in the cells. The secreted activity was expressed as percentage of the total (secreted plus intracellular) activity. For lactate dehydrogenase, the release activity was determined by using the LDH kit from Sigma-Aldrich (Tox7-1kt) following manufacturer’s recommendations. The released activity was expressed as percentage of the total (released plus intracellular) activity.
Muscle cell culture
The establishment of immortalized Pompe disease muscle cells has been previously described45. The myoblast cell lines were derived from immortoGAA knockout mice, which express the temperature-sensitive SV40 large T antigen tsA58 under the control of interferon-gamma inducible murine H2Kb promoter. The myoblasts undergo immortalization when grown at 33°C with interferon-γ; the differentiation into multinucleated myotubes proceeds when the oncogene is silenced at 37°C in the absence of interferon-γ. The myogenic cell line clone 6 was used for the experiments. The cells were grown at 33°C in an atmosphere of 5% CO2 in proliferation medium [20% foetal bovine serum, 10% horse serum, 1% chick embryo extract, recombinant IFN- (100 units/ml; Life Technologies), 1× P/S/L-Glutamine in high glucose (4.5 g/L) DMEM). When myoblasts became nearly confluent, the medium was changed to differentiation medium (DMEM containing 2% horse serum, 0.5% chick embryo extract, 1× P/S/L-Glutamine) and the cells were moved to 37°C in an atmosphere of 5% CO2. Myotubes began to form within 2 days.
Infection of myotubes with adenovirus expressing TFE3, immunofluorescence microscopy, and fluorescent glycogen detection
Myotubes were infected with either adenovirus (Ad. Null) or adenovirus expressing TFE3 (Ad. TFE3) for 24, 48 or 72 h. Myotubes were fixed in 2% paraformaldehyde (Electron Microscopy Sciences) for 15 min at room temperature, washed twice in PBS, and permeabilized in 0.2% Triton X-100 (Sigma– Aldrich). Immunostaining with LAMP1 or MYC antibodies was done using M.O.M. kit (Vector Laboratories, Burlingame, CA) as previously described 48. The cell nuclei were stained with 2 μg/ml Hoechst 33342 (Life Technologies) in PBS for 10 min. Alternatively, live cells were used for labeling acidic organelles with LysoTracker® Red DND-99 (Life Technologies) (500 nM). After staining, the cells were imaged on a Carl Zeiss LSM 510 confocal microscope with a 40× or 63× oil immersion objectives. Lysosomal glycogen in live cells was detected by the incorporation of 2-NBDG, a D-glucose fluorescent derivative (2-deoxyglucose), into glycogen as described61.
Immunofluorescence confocal microscopy
Cells grown on glass coverslips were washed with PBS and fixed with 4% formaldehyde at room temperature for 15 min. After fixation cells were washed with PBS and then permeabilized with 0.2% Triton X-100 in PBS at room temperature for 10 min. Cells were then incubated with the indicated primary antibodies in IF buffer (PBS containing 10% Fetal Bovine Serum and 0.1% (wt/v) saponin) for 1h at room temperature. Cells were washed three times with PBS and incubated with the corresponding secondary antibodies conjugated to Alexa Fluor-568 or Alexa Fluor-488 in IF buffer for 30 min at room temperature. PBS washed coverslips were mounted onto glass slides with Fluoromount-G (Southern Biotech). For Lamp1 surface staining analysis, infected ARPE-19 cells grown on coverslips were incubated in ice-cold complete medium containing mouse anti-Lamp1 antibody (1:1000) for 45 minutes at 4°C. Cells were then extensively wash with ice-cold PBS, fixed, permeabilized and incubated with the corresponding primary and secondary antibodies as indicated above. Images were acquired on a Zeiss LSM 510 confocal system equipped with filter sets for FITC and Rhodamine, 488 nm and 543 nm laser excitation, an AxioCam camera, a 63X NA 1.4 oil immersion objective, and LSM 510 operating software (Carl Zeiss). Confocal images taken with the same acquisition parameters were processed with ImageJ software (NIH) and Photoshop CS4 and Adobe Illustrator CS4 software were used to produce the figures.
RNA interference (RNAi)
Cells grown in six-wells plate were transfected with DharmaFECT transfection reagent and 100 nM of ON-TARGETplus non-targeting pool siRNA duplexes or ON-TARGETplus smart pool siRNA duplexes targeted against human TFEB or/and human TFE3 genes (Dharmacon-Thermo Scientific). After 72 hours of transfection cells were either kept in complete medium (Control) or incubated in Earle’s balanced salt solution (Starved) for additional 24 hours before being harvested for analysis.
GST-pull-down, immunoprecipitation, electrophoresis and immunoblotting
Cells washed with ice-cold PBS were lysed in lysis buffer containing 25 mM Hepes-KOH, pH 7.4, 150 mM NaCl, 5mM EDTA and 1% Triton X-100 (wt/v) and supplemented with protease and phosphatase inhibitors cocktail. Cell lysates were incubated on ice for 30 min and then were passed 10 times through a 25-gauge needle. Cell lysates were centrifuged at 16,000 × g for 10 min at 4°C. For immunoprecipitation the soluble fractions were incubated with 2 μl of anti-MYC antibody, and protein G-Sepharose beads (Amersham) for 2 h at 4°C. For GST-pull-down, soluble fractions were incubated with 25 μl of glutathione-sepharose beads for 2 h at 4°C. The immunoprecipitates and pulled-down materials were collected, washed three times with lysis buffer, and proteins were eluted with Laemmli sample buffer. Samples were analyzed by SDS-PAGE (4-20% gradient gels, Invitrogen) under reducing conditions and transferred to nitrocellulose. Membranes were immunoblotted using the indicated antibodies. Horseradish peroxidase-chemiluminiscence was developed by using Western Lightning Chemiluminescence Reagent Plus (PerkinElmer Life Sciences).
Subcellular fractionation
ARPE-19 cells were either starved for 24 hours or treated with DMSO or Torin-1 for 1 hour. Cells were then lysed in lysis buffer containing l0 mM Tris, pH 7.9, 140 mM KCI, 5 mM MgCl2 and 0.5 % NP-40 supplemented with protease and phosphatase inhibitors. Lysed cell were kept on ice for 15 min. The lysates were then centrifugated at 1000 × g for 5 min. The resulting supernatants represent the cytosolic plus the membrane fraction. The corresponding pellets representing the nuclear fractions were washed two times with in NP-40 lysis buffer and sonicated in 0.5% Triton X-100 and 0.5% SDS in 100 mM Tris-HCL buffer pH 7.4.
RNA isolation and relative quantitative real time polymerase chain reaction
RNA from adenovirus-infected cells was isolated by using PureLink RNA Mini Kit (Invitrogen) following manufacturer’s recommendations. Two to four micrograms of RNA were to reverse transcribed in a 20 μl reaction using oligo(dT)20 and SuperScript III First-Strand Synthesis System (Invitrogen). Relative Quantitative Real Time PCR was performed in a total reaction volume of 10 μl, using 2 μl cDNA, 1 μl gene specific primer mix (QuantiTect primer Assays), 5 μl SYBR GreenER qPCR SuperMix (Invitrogen), and 2 μl water. The quantification of gene expression was performed using 7900HT Fast Real-Time PCR System (Applied Biosystems) in triplicate. The thermal profile of the reaction was: 50°C for 2 min, 95°C for 10 min and 35 cycles of 95°C for 15 s followed by 60°C for 1 min. Amplification of the sequence of interest was normalized with a reference endogenous gene Glyceraldehyde 3-phosphate dehydrogenase (GAPDH). The value was expressed as a fold change relative to RNA from cells infected with control adenovirus (Ad. Null). For data analysis the 7900HT Fast Real-Time PCR System Software was used (Applied Biosystems).
Luciferase assay
HeLa cells seeded in 96-wells plate were transfected with 50 ng of pLightSwitch-MCOLN1-promoter or pLightSwitch-Empty-promoter (SwitchGear Genomics) using FuGENE HD (Promega) transfection reagent. After 4 hours of transfection cells were infected with adenovirus Null or expressing TFEB-FLAG or TFE3-MYC and incubated at 37°C. After 48 hours of infection luciferase activity was assayed by adding 100 μl of LightSwitch assay reagent following manufacturer’s instructions. The luciferase activity was expressed as fold increase versus adenovirus Null infected cells.
Flow cytometry analysis
ARPE-19 cells were infected for 48 hour with specific adenoviruses. Cell were harvested in striping solution, washed and resuspended in HBSS supplemented with 0.01% BSA and 0.01% NaN3. Cells were incubated with antibody to Lamp1-APC for 30 minutes on ice and then washed three times in HBSS-BSA buffer. Propidium iodide was added before acquisition to exclude dead cells. Cells were collected on a BD LSR II using FACSDiva software (BD Biosciences). Data were subsequently analyzed using FlowJo (Tree Star).
Statistical Analysis
Obtained data were processed in Excel (Microsoft Corporation) and Prism (GraphPad Software) to generate bar charts and perform statistical analyses. One-way ANOVA and pairwise post-tests were run for each dependent variable. p ≤ 0.05 was considered statistically significant (*) and p ≤ 0.001 extremely significant (***). p > 0.05 was considered not significant (ns).
Supplementary Material
Acknowledgments
Funding: J.M., H.D., S.P., and R.P. were supported by the Intramural Research Program of the National Institutes of Health, National Heart, Lung, and Blood Institute (NHLBI). L.J., L.L., and N.R. were supported in part by the Intramural Research Program of the National Institute of Arthritis and Musculoskeletal and Skin diseases (NIAMS) of the NIH. L.J. and L.L. were supported in part by a CRADA between NIH and Genzyme Corporation.
Footnotes
Author Contribution: J.M. was involved in experimental strategy, performed most of the experiments and analyzed the data; H.D. performed experiments and analyzed the data; S.P. quantified the number of lysosomes in TFE3-expressing cells; L.J., L.L., and N.R. designed, perform, and interpret the experiments in Pompe myotubes; N.R. also edited the manuscript; R.P. designed the research, performed experiments, analyzed the data, supervised the project, and wrote the manuscript. All the authors reviewed the manuscript. Competing interest: The authors declare no competing financial interests.
References
- 1.Luzio JP, Pryor PR, Bright NA. Lysosomes: fusion and function. Nat. Rev. Mol. Cell Biol. 2007;8:622–632. doi: 10.1038/nrm2217. published online EpubAug (10.1038/nrm2217) [DOI] [PubMed] [Google Scholar]
- 2.Saftig P, Klumperman J. Lysosome biogenesis and lysosomal membrane proteins: trafficking meets function. Nat. Rev. Mol. Cell Biol. 2009;10:623–635. doi: 10.1038/nrm2745. published online EpubSep (10.1038/nrm2745) [DOI] [PubMed] [Google Scholar]
- 3.Saucedo LJ, Gao X, Chiarelli DA, Li L, Pan D, Edgar BA. Rheb promotes cell growth as a component of the insulin/TOR signalling network. Nat. Cell Biol. 2003;5:566–571. doi: 10.1038/ncb996. published online EpubJun (10.1038/ncb996) [DOI] [PubMed] [Google Scholar]
- 4.Stocker H, Radimerski T, Schindelholz B, Wittwer F, Belawat P, Daram P, Breuer S, Thomas G, Hafen E. Rheb is an essential regulator of S6K in controlling cell growth in Drosophila. Nat. Cell Biol. 2003;5:559–565. doi: 10.1038/ncb995. published online EpubJun (10.1038/ncb995) [DOI] [PubMed] [Google Scholar]
- 5.Bar-Peled L, Schweitzer LD, Zoncu R, Sabatini DM. Ragulator is a GEF for the rag GTPases that signal amino acid levels to mTORC1. Cell. 2012;150:1196–1208. doi: 10.1016/j.cell.2012.07.032. published online EpubSep 14 (10.1016/j.cell.2012.07.032) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sancak Y, Bar-Peled L, Zoncu R, Markhard AL, Nada S, Sabatini DM. Ragulator-Rag complex targets mTORC1 to the lysosomal surface and is necessary for its activation by amino acids. Cell. 2010;141:290–303. doi: 10.1016/j.cell.2010.02.024. published online EpubApr 16 (10.1016/j.cell.2010.02.024) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sancak Y, Peterson TR, Shaul YD, Lindquist RA, Thoreen CC, Bar-Peled L, Sabatini DM. The Rag GTPases bind raptor and mediate amino acid signaling to mTORC1. Science. 2008;320:1496–1501. doi: 10.1126/science.1157535. published online EpubJun 13 (10.1126/science.1157535) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gao M, Kaiser CA. A conserved GTPase-containing complex is required for intracellular sorting of the general amino-acid permease in yeast. Nat. Cell Biol. 2006;8:657–667. doi: 10.1038/ncb1419. published online EpubJul (10.1038/ncb1419) [DOI] [PubMed] [Google Scholar]
- 9.Sekiguchi T, Hirose E, Nakashima N, Ii M, Nishimoto T. Novel G proteins, Rag C and Rag D, interact with GTP-binding proteins, Rag A and Rag B. J Biol Chem. 2001;276:7246–7257. doi: 10.1074/jbc.M004389200. published online EpubMar 9 (10.1074/jbc.M004389200) [DOI] [PubMed] [Google Scholar]
- 10.Zoncu R, Bar-Peled L, Efeyan A, Wang S, Sancak Y, Sabatini DM. mTORC1 senses lysosomal amino acids through an inside-out mechanism that requires the vacuolar H(+)-ATPase. Science. 2011;334:678–683. doi: 10.1126/science.1207056. published online EpubNov 4 (10.1126/science.1207056) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hosokawa N, Hara T, Kaizuka T, Kishi C, Takamura A, Miura Y, Iemura S, Natsume T, Takehana K, Yamada N, L. Guan J, Oshiro N, Mizushima N. Nutrient-dependent mTORC1 association with the ULK1-Atg13-FIP200 complex required for autophagy. Mol. Biol. Cell. 2009;20:1981–1991. doi: 10.1091/mbc.E08-12-1248. published online EpubApr (10.1091/mbc.E08-12-1248) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hosokawa N, Sasaki T, Iemura S, Natsume T, Hara T, Mizushima N. Atg101, a novel mammalian autophagy protein interacting with Atg13. Autophagy. 2009;5:973–979. doi: 10.4161/auto.5.7.9296. published online EpubOct. [DOI] [PubMed] [Google Scholar]
- 13.Martina JA, Chen Y, Gucek M, Puertollano R. MTORC1 functions as a transcriptional regulator of autophagy by preventing nuclear transport of TFEB. Autophagy. 2012;8 doi: 10.4161/auto.19653. published online EpubJun 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Roczniak-Ferguson A, Petit CS, Froehlich F, Qian S, Ky J, Angarola B, Walther TC, Ferguson SM. The Transcription Factor TFEB Links mTORC1 Signaling to Transcriptional Control of Lysosome Homeostasis. Sci. Signal. 2012;5:ra42. doi: 10.1126/scisignal.2002790. 10.1126/scisignal.2002790) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Settembre C, Zoncu R, Medina DL, Vetrini F, Erdin S, Huynh T, Ferron M, Karsenty G, Vellard MC, Facchinetti V, Sabatini DM, Ballabio A. A lysosome-to-nucleus signalling mechanism senses and regulates the lysosome via mTOR and TFEB. Embo J. 2012;31:1095–1108. doi: 10.1038/emboj.2012.32. published online EpubMar 7 (10.1038/emboj.2012.32) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sardiello M, Palmieri M, di Ronza A, L. Medina D, Valenza M, Gennarino VA, Di Malta C, Donaudy F, Embrione V, Polishchuk RS, Banfi S, Parenti G, Cattaneo E, Ballabio A. A gene network regulating lysosomal biogenesis and function. Science. 2009;325:473–477. doi: 10.1126/science.1174447. published online EpubJul 24 (10.1126/science.1174447) [DOI] [PubMed] [Google Scholar]
- 17.Palmieri M, Impey S, Kang H, di Ronza A, Pelz C, Sardiello M, Ballabio A. Characterization of the CLEAR network reveals an integrated control of cellular clearance pathways. Hum. Mol. Genet. 2011;20:3852–3866. doi: 10.1093/hmg/ddr306. published online EpubOct 1 (10.1093/hmg/ddr306) [DOI] [PubMed] [Google Scholar]
- 18.Settembre C, Di Malta C, Polito VA, Garcia Arencibia M, Vetrini F, Erdin S, Erdin SU, Huynh T, Medina D, Colella P, Sardiello M, Rubinsztein DC, Ballabio A. TFEB links autophagy to lysosomal biogenesis. Science. 2011;332:1429–1433. doi: 10.1126/science.1204592. published online EpubJun 17 (10.1126/science.1204592) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sardiello M, Ballabio A. Lysosomal enhancement: a CLEAR answer to cellular degradative needs. Cell Cycle. 2009;8:4021–4022. doi: 10.4161/cc.8.24.10263. published online EpubDec 15 ( [DOI] [PubMed] [Google Scholar]
- 20.Martina JA, Puertollano R. Rag GTPases mediate amino acid-dependent recruitment of TFEB and MITF to lysosomes. J Cell Biol. 2013;200:475–491. doi: 10.1083/jcb.201209135. published online EpubFeb 18 (10.1083/jcb.201209135) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huan C, Kelly ML, Steele R, Shapira I, Gottesman SR, Roman CA. Transcription factors TFE3 and TFEB are critical for CD40 ligand expression and thymus-dependent humoral immunity. Nat. Immunol. 2006;7:1082–1091. doi: 10.1038/ni1378. published online EpubOct (10.1038/ni1378) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Merrell K, Wells S, Henderson A, Gorman J, Alt F, Stall A, Calame K. The absence of the transcription activator TFE3 impairs activation of B cells in vivo. Mol. Cell. Biol. 1997;17:3335–3344. doi: 10.1128/mcb.17.6.3335. published online EpubJun. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tsicopoulos A, Joseph M. The role of CD23 in allergic disease. Clin Exp Allergy. 2000;30:602–605. doi: 10.1046/j.1365-2222.2000.00871.x. published online EpubMay. [DOI] [PubMed] [Google Scholar]
- 24.Yagil Z, Hadad Erlich T, Ofir-Birin Y, Tshori S, Kay G, Yekhtin Z, Fisher DE, Cheng C, Wong WS, Hartmann K, Razin E, Nechushtan H. Transcription factor E3, a major regulator of mast cell-mediated allergic response. J Allergy Clin Immunol. 2012;129e1355:1357–1366. doi: 10.1016/j.jaci.2011.11.051. published online EpubMay (10.1016/j.jaci.2011.11.051) [DOI] [PubMed] [Google Scholar]
- 25.Steingrimsson E, Tessarollo L, Pathak B, Hou L, Arnheiter H, Copeland NG, Jenkins NA. Mitf and Tfe3, two members of the Mitf-Tfe family of bHLH-Zip transcription factors, have important but functionally redundant roles in osteoclast development. Proc. Natl. Acad. Sci. U.S.A. 2002;99:4477–4482. doi: 10.1073/pnas.072071099. published online EpubApr 2 (10.1073/pnas.072071099) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nakagawa Y, Shimano H, Yoshikawa T, Ide T, Tamura M, Furusawa M, Yamamoto T, Inoue N, Matsuzaka T, Takahashi A, Hasty AH, Suzuki H, Sone H, Toyoshima H, Yahagi N, Yamada N. TFE3 transcriptionally activates hepatic IRS-2, participates in insulin signaling and ameliorates diabetes. Nat. Med. 2006;12:107–113. doi: 10.1038/nm1334. published online EpubJan (10.1038/nm1334) [DOI] [PubMed] [Google Scholar]
- 27.Hong SB, Oh H, Valera VA, Baba M, Schmidt LS, Linehan WM. Inactivation of the FLCN tumor suppressor gene induces TFE3 transcriptional activity by increasing its nuclear localization. PLoS One. 2010;5:e15793. doi: 10.1371/journal.pone.0015793. 10.1371/journal.pone.0015793) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Birt AR, Hogg GR, Dube WJ. Hereditary multiple fibrofolliculomas with trichodiscomas and acrochordons. Arch Dermatol. 1977;113:1674–1677. published online EpubDec. [PubMed] [Google Scholar]
- 29.Nickerson ML, Warren MB, Toro JR, Matrosova V, Glenn G, Turner ML, Duray P, Merino M, Choyke P, Pavlovich CP, Sharma N, Walther M, Munroe D, Hill R, Maher E, Greenberg C, Lerman MI, Linehan WM, Zbar B, Schmidt LS. Mutations in a novel gene lead to kidney tumors, lung wall defects, and benign tumors of the hair follicle in patients with the Birt-Hogg-Dube syndrome. Cancer Cell. 2002;2:157–164. doi: 10.1016/s1535-6108(02)00104-6. published online EpubAug. [DOI] [PubMed] [Google Scholar]
- 30.Schmidt LS, Nickerson ML, Warren MB, Glenn GM, Toro JR, Merino MJ, Turner ML, Choyke PL, Sharma N, Peterson J, Morrison P, Maher ER, Walther MM, Zbar B, Linehan WM. Germline BHD-mutation spectrum and phenotype analysis of a large cohort of families with Birt-Hogg-Dube syndrome. Am J Hum Genet. 2005;76:1023–1033. doi: 10.1086/430842. published online EpubJun (10.1086/430842) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hartman TR, Nicolas E, Klein-Szanto A, Al-Saleem T, Cash TP, Simon MC, Henske EP. The role of the Birt-Hogg-Dube protein in mTOR activation and renal tumorigenesis. Oncogene. 2009;28:1594–1604. doi: 10.1038/onc.2009.14. published online EpubApr 2 (10.1038/onc.2009.14) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Takagi Y, Kobayashi T, Shiono M, Wang L, Piao X, Sun G, Zhang D, Abe M, Hagiwara Y, Takahashi K, Hino O. Interaction of folliculin (Birt-Hogg-Dube gene product) with a novel Fnip1-like (FnipL/Fnip2) protein. Oncogene. 2008;27:5339–5347. doi: 10.1038/onc.2008.261. published online EpubSep 11 (10.1038/onc.2008.261) [DOI] [PubMed] [Google Scholar]
- 33.Hoek KS, C. Schlegel N, Eichhoff OM, Widmer DS, Praetorius C, Einarsson SO, Valgeirsdottir S, Bergsteinsdottir K, Schepsky A, Dummer R, Steingrimsson E. Novel MITF targets identified using a two-step DNA microarray strategy. Pigment Cell Melanoma Res. 2008;21:665–676. doi: 10.1111/j.1755-148X.2008.00505.x. published online EpubDec (10.1111/j.1755-148X.2008.00505.x) [DOI] [PubMed] [Google Scholar]
- 34.Aksan I, Goding CR. Targeting the microphthalmia basic helix-loop-helix-leucine zipper transcription factor to a subset of E-box elements in vitro and in vivo. Mol. Cell. Biol. 1998;18:6930–6938. doi: 10.1128/mcb.18.12.6930. published online EpubDec. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hemesath TJ, Steingrimsson E, McGill G, Hansen MJ, Vaught J, Hodgkinson CA, Arnheiter H, Copeland NG, Jenkins NA, Fisher DE. microphthalmia, a critical factor in melanocyte development, defines a discrete transcription factor family. Genes Dev. 1994;8:2770–2780. doi: 10.1101/gad.8.22.2770. published online EpubNov 15. [DOI] [PubMed] [Google Scholar]
- 36.Settembre C, De Cegli R, Mansueto G, Saha PK, Vetrini F, Visvikis O, Huynh T, Carissimo A, Palmer D, Jurgen Klisch T, Wollenberg AC, Di Bernardo D, Chan L, Irazoqui JE, Ballabio A. TFEB controls cellular lipid metabolism through a starvation-induced autoregulatory loop. Nat. Cell Biol. 2013;15:647–658. doi: 10.1038/ncb2718. published online EpubJun (10.1038/ncb2718) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Medina DL, Fraldi A, Bouche V, Annunziata F, Mansueto G, Spampanato C, Puri C, Pignata A, Martina JA, Sardiello M, Palmieri M, Polishchuk R, Puertollano R, Ballabio A. Transcriptional activation of lysosomal exocytosis promotes cellular clearance. Dev. Cell. 2011;21:421–430. doi: 10.1016/j.devcel.2011.07.016. published online EpubSep 13 (10.1016/j.devcel.2011.07.016) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bargal R, Avidan N, Ben-Asher E, Olender Z, Zeigler M, Frumkin A, Raas-Rothschild A, Glusman G, Lancet D, Bach G. Identification of the gene causing mucolipidosis type IV. Nat Genet. 2000;26:118–123. doi: 10.1038/79095. published online EpubSep (10.1038/79095) [DOI] [PubMed] [Google Scholar]
- 39.Bassi MT, Manzoni M, Monti E, Pizzo MT, Ballabio A, Borsani G. Cloning of the gene encoding a novel integral membrane protein, mucolipidin-and identification of the two major founder mutations causing mucolipidosis type IV. Am J Hum Genet. 2000;67:1110–1120. doi: 10.1016/s0002-9297(07)62941-3. published online EpubNov (10.1016/S0002-9297(07)62941-3) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Slaugenhaupt SA, Acierno JS, Jr., Helbling LA, Bove C, Goldin E, Bach G, Schiffmann R, Gusella JF. Mapping of the mucolipidosis type IV gene to chromosome 19p and definition of founder haplotypes. Am J Hum Genet. 1999;65:773–778. doi: 10.1086/302549. published online EpubSep. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kiselyov K, Chen J, Rbaibi Y, Oberdick D, Tjon-Kon-Sang S, Shcheynikov N, Muallem S, Soyombo A. TRP-ML1 is a lysosomal monovalent cation channel that undergoes proteolytic cleavage. J Biol Chem. 2005;280:43218–43223. doi: 10.1074/jbc.M508210200. published online EpubDec 30 (10.1074/jbc.M508210200) [DOI] [PubMed] [Google Scholar]
- 42.Pryor PR, Reimann F, Gribble FM, Luzio JP. Mucolipin-1 is a lysosomal membrane protein required for intracellular lactosylceramide traffic. Traffic. 2006;7:1388–1398. doi: 10.1111/j.1600-0854.2006.00475.x. published online EpubOct (10.1111/j.1600-0854.2006.00475.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vergarajauregui S, Puertollano R. Two di-leucine motifs regulate trafficking of mucolipin-1 to lysosomes. Traffic. 2006;7:337–353. doi: 10.1111/j.1600-0854.2006.00387.x. published online EpubMar (10.1111/j.1600-0854.2006.00387.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Song W, Wang F, Savini M, Ake A, di Ronza A, Sardiello M, Segatori L. TFEB regulates lysosomal proteostasis. Hum. Mol. Genet. 2013;22:1994–2009. doi: 10.1093/hmg/ddt052. published online EpubMay 15 (10.1093/hmg/ddt052) [DOI] [PubMed] [Google Scholar]
- 45.Spampanato C, Feeney E, Li L, Cardone M, Lim JA, Annunziata F, Zare H, Polishchuk R, Puertollano R, Parenti G, Ballabio A, Raben N. Transcription factor EB (TFEB) is a new therapeutic target for Pompe disease. EMBO Mol. Med. 2013;5:691–706. doi: 10.1002/emmm.201202176. published online EpubMay (10.1002/emmm.201202176) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kroos M, Hoogeveen-Westerveld M, van der Ploeg A, Reuser AJ. The genotype-phenotype correlation in Pompe disease. Am. J. Med. Genet. C Semin. Med. Genet. 2012;160:59–68. doi: 10.1002/ajmg.c.31318. published online EpubFeb 15 (10.1002/ajmg.c.31318) [DOI] [PubMed] [Google Scholar]
- 47.van der Ploeg AT, Reuser AJ. Pompe's disease. Lancet. 2008;372:1342–1353. doi: 10.1016/S0140-6736(08)61555-X. published online EpubOct 11 (10.1016/S0140-6736(08)61555-X) [DOI] [PubMed] [Google Scholar]
- 48.Raben N, Hill V, Shea L, Takikita S, Baum R, Mizushima N, Ralston E, Plotz P. Suppression of autophagy in skeletal muscle uncovers the accumulation of ubiquitinated proteins and their potential role in muscle damage in Pompe disease. Hum. Mol. Genet. 2008;17:3897–3908. doi: 10.1093/hmg/ddn292. published online EpubDec 15 (10.1093/hmg/ddn292) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jin J, Smith FD, Stark C, Wells CD, Fawcett JP, Kulkarni S, Metalnikov P, O'Donnell P, Taylor P, Taylor L, Zougman A, Woodgett JR, Langeberg LK, Scott JD, Pawson T. Proteomic, functional, and domain-based analysis of in vivo 14-3-3 binding proteins involved in cytoskeletal regulation and cellular organization. Current biology : CB. 2004;14:1436–1450. doi: 10.1016/j.cub.2004.07.051. published online EpubAug 24 (10.1016/j.cub.2004.07.051) [DOI] [PubMed] [Google Scholar]
- 50.Hershey CL, Fisher DE. Mitf and Tfe3: members of a b-HLH-ZIP transcription factor family essential for osteoclast development and function. Bone. 2004;34:689–696. doi: 10.1016/j.bone.2003.08.014. published online EpubApr (10.1016/j.bone.2003.08.014) [DOI] [PubMed] [Google Scholar]
- 51.Baba M, Hong SB, Sharma N, Warren MB, Nickerson ML, Iwamatsu A, Esposito D, Gillette WK, Hopkins RF, 3rd, Hartley JL, Furihata M, Oishi S, Zhen W, R. Burke T, Jr., Linehan WM, Schmidt LS, Zbar B. Folliculin encoded by the BHD gene interacts with a binding protein, FNIP1, and AMPK, and is involved in AMPK and mTOR signaling. Proc Natl Acad Sci U S A. 2006;103:15552–15557. doi: 10.1073/pnas.0603781103. published online EpubOct 17 (10.1073/pnas.0603781103) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Linehan WM, Ricketts CJ. The metabolic basis of kidney cancer. Semin Cancer Biol. 2013;23:46–55. doi: 10.1016/j.semcancer.2012.06.002. published online EpubFeb (10.1016/j.semcancer.2012.06.002) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Grove CA, De Masi F, Barrasa MI, Newburger DE, Alkema MJ, Bulyk ML, Walhout AJ. A multiparameter network reveals extensive divergence between C. elegans bHLH transcription factors. Cell. 2009;138:314–327. doi: 10.1016/j.cell.2009.04.058. published online EpubJul 23 (10.1016/j.cell.2009.04.058) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.O'Rourke EJ, Ruvkun G. MXL-3 and HLH-30 transcriptionally link lipolysis and autophagy to nutrient availability. Nat. Cell Biol. 2013;15:668–676. doi: 10.1038/ncb2741. published online EpubJun (10.1038/ncb2741) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Steingrimsson E, Tessarollo L, Reid SW, Jenkins NA, Copeland NG. The bHLH-Zip transcription factor Tfeb is essential for placental vascularization. Development. 1998;125:4607–4616. doi: 10.1242/dev.125.23.4607. published online EpubDec ( [DOI] [PubMed] [Google Scholar]
- 56.Lister JA, Lane BM, Nguyen A, Lunney K. Embryonic expression of zebrafish MiT family genes tfe3b, tfeb, and tfec. Dev. Dyn. 2011;240:2529–2538. doi: 10.1002/dvdy.22743. published online EpubNov (10.1002/dvdy.22743) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ferron M, Settembre C, Shimazu J, Lacombe J, Kato S, Rawlings DJ, Ballabio A, Karsenty G. A RANKL-PKCbeta-TFEB signaling cascade is necessary for lysosomal biogenesis in osteoclasts. Genes Dev. 2013;27:955–969. doi: 10.1101/gad.213827.113. published online EpubApr 15 (10.1101/gad.213827.113) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Decressac M, Mattsson B, Weikop P, Lundblad M, Jakobsson J, Bjorklund A. TFEB-mediated autophagy rescues midbrain dopamine neurons from alpha-synuclein toxicity. Proc. Natl. Acad. Sci. U.S.A. 2013;110:E1817–1826. doi: 10.1073/pnas.1305623110. published online EpubMay 7 (10.1073/pnas.1305623110) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tsunemi T, Ashe TD, Morrison BE, Soriano KR, Au J, Roque RA, Lazarowski ER, Damian VA, Masliah E, La Spada AR. PGC-1alpha rescues Huntington's disease proteotoxicity by preventing oxidative stress and promoting TFEB function. Sci. Transl. Med. 2012;4:142ra197. doi: 10.1126/scitranslmed.3003799. published online EpubJul 11 (10.1126/scitranslmed.3003799) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yasumoto K, Amae S, Udono T, Fuse N, Takeda K, Shibahara S. A big gene linked to small eyes encodes multiple Mitf isoforms: many promoters make light work. Pigment cell research / sponsored by the European Society for Pigment Cell Research and the International Pigment Cell Society. 1998;11:329–336. doi: 10.1111/j.1600-0749.1998.tb00491.x. published online EpubDec ( [DOI] [PubMed] [Google Scholar]
- 61.Louzao MC, Espina B, Vieytes MR, Vega FV, Rubiolo JA, Baba O, Terashima T, Botana LM. "Fluorescent glycogen" formation with sensibility for in vivo and in vitro detection. Glycoconj. J. 2008;25:503–510. doi: 10.1007/s10719-007-9075-7. published online EpubAug (10.1007/s10719-007-9075-7) [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.