Abstract
In order to obtain fine details in 3 dimensions (3D) over time, it is critical for motile biological specimens to be appropriately immobilized. Of the many immobilization options available, the mechanical microcompressor offers many benefits. Our device, previously described, achieves gentle flattening of a cell, allowing us to image finely detailed structures of numerous organelles and physiological processes in living cells. We have imaged protozoa and other small metazoans using differential interference contrast (DIC) microscopy, orientation-independent (OI) DIC, and real-time birefringence imaging using a video-enhanced polychromatic polscope. We also describe an enhancement of our previous design by engineering a new device where the coverslip mount is fashioned onto the top of the base; so the entire apparatus is accessible on top of the stage. The new location allows for easier manipulation of the mount when compressing or releasing a specimen on an inverted microscope. Using this improved design, we imaged immobilized bacteria, yeast, paramecia, and nematode worms and obtained an unprecedented view of cell and specimen details. A variety of microscopic techniques were used to obtain high resolution images of static and dynamic cellular and physiological events.
Keywords: Immobilization, High-resolution imaging, Protozoa, Microbes
1. Introduction
Advancements in the optics of fluorescent and contrast-enhancement microscopy have given rise to a demand for a rapid and delicate means of immobilizing ambulatory organisms. Small, jostling specimens are difficult to image because they move in and out of the focal plane, prohibiting precise or extended imaging. Other, larger organisms can move out of the field of view if they are not held steady. In these larger specimens, it may be critical to immobilize the organisms precisely so that subcellular organelles can be imaged over time. Only when the movement of the whole organism is inhibited can the best information from dynamic molecules, such as fluorescent proteins and RNAs or DNA in live-cells, be obtained. Efforts to reduce the complexity of imaging moving molecules often presents a significant challenge to experimental design so that one can obtain the optimum spatial and temporal resolution.
A myriad of options exist for immobilization of specimens for imagining, each with their own set of advantages and disadvantages [1]. For instance, chemical immobilization utilizes small-molecule anesthetics that induce paralysis. The use of pharmacological drugs can interfere with physiological or biochemical studies, or slowly kill the specimen. Furthermore, these samples are still subject to Brownian motion. Physical inhibition of a specimen includes its placement in a highly viscous fluid, adherence to, or imbedding in, a surface such as agarose, or uncontrolled squeezing of the sample between a coverslip and slide, often without regard for the amount of distortion. Many times these practices do not affect biochemical processes, however recovery of the specimen can be difficult or impossible. Controlled compression of the sample can be obtained with the use of a rotocompressor or a mechanical microcompressor (MMC), a device that allows for manual compression of a specimen between two glass planes. Although the design has evolved over the last 100 years or so, the basic concept has been consistent [2]. Such devices can also be useful for the identification of new species and even novel organelles in living samples [3–5].
Our group has recently developed a more sophisticated MMC that also has the optional feature of microfluidics [6]. The MMC can be used to gently trap and immobilize a wide range of organisms including bacteria, yeast, small protozoa and larger specimens including fly embryos and nematode worms. This device is non-lethal, reusable, and could be used for either long-term or short-term imaging. The addition of the microfluidics to the MMC allowed for long-term growth and imaging of specimens such as the budding of Saccharomyces cerevisiae [6]. The ease by which specimens could be recovered after imaging allowed for future manipulation, analysis, or culture.
Here, we show further uses for the MMC by immobilizing several other specimen types and using an advanced form of DIC optics and polarization microscopy [7–9]. The remarkable optics obtained through the gentle flattening of organism allowed us to obtain unprecedented images of several structures in species of Paramecium and Stentor, as well as several other microbes and pond water specimens. While this new generation of MMCs could be used on inverted microscopes, they were designed mainly for use on an upright microscope. Here we describe a new MMC that takes advantage of several of the updates in the Yan et. al design [6] and is modified specifically to be used on inverted microscopes. The new design allows the user to adjust the degree of compression from above the stage. Additionally, the location of the adjustable components is no longer close to the objectives or inhibited by the size of the objectives. This modification increases the area that may be scanned for imaging. As is the case with the previous MMC, this microcompressor is simple to use, and allows the gentle and reversible trapping of numerous organisms.
2. Materials and methods
2.1. Cell growth and specimen acquisition
Stentor polymorphus, as well as the rotifers, were collected from a small pond in Falmouth, MA. Specimens were mechanically compressed in the pond water at room temperature.
Stentor coeruleus was provided by Wallace Marshall, University of California, San Francisco, and also mounted in Falmouth, MA pond water at room temperature.
The Caenorhabditis elegans was maintained at 18 °C in Nematode Growth Medium(NGM) on plates seeded with OP-50 strain of Escherichia coli, as previously described [10]. The strain LX929 (Punc-17::GFP(vsIs48)) was provided by Dr. Samuel Caito (Albert Einstein College of Medicine, Bronx, NY). After compression, the C. elegans were returned to the NGM plates and grown successfully for 1–6 days, compressed again, and imaged.
Paramecium sonneborni [5] was grown at 17 °C in baked lettuce media inoculated with Klebsiella pneumoniae (ATCC #27889) as food.
Saccharomyces cerevisiae strand CY6002 (MATa his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 SEC61-yEpolylinker-yEGFP::kanMX) [11] was transformed with PhyB-mCherry-NLS, a gift from Chao Tang, (Addgene plasmid # 51571) using the Frozen-EZ Yeast Transformation II Kit (Zymo Research). Transformants were plated on the appropriate synthetic dropout(DO) media (Sigma Aldrich) with kanamycin (50 μg/mL, Sigma-Aldrich). The cells were cultured in the appropriate DO media and grown at 30 °C to mid log phase.
Escherichia coli strain DH5α (Invitrogen) was grown in 2xYT media (Sigma-Aldrich) at 37 °C overnight shaking at 200 rpm.
2.2. Live-cell imaging
For, OI- DIC the light source used was a 100 W halogen lamp. The bandpass interference filter had a central wavelength of 546 nm and 40-nm full width at half maximum, Chroma Technology, Rockingham, Vermont. We selected the green light for illumination. Magnification and image collection used an Olympus BX61 Upright Microscope using either a 10X/0.30NA or 100X/ 1.3NA lens and an Infinity 3M CCD camera (Lumenera, Ottawa, Ontario, Canada).
For the true OI polarized light microscope technique for real-time birefringence imaging, we used a video-enhanced polychromatic polscope. This was performed on an Olympus IX-81 microscope. Low magnification used a 10X/0.30NA lens, while high magnification was with a 40X/0.75NA lens.
L3 and L4 stage C. elegans were picked from NGM plates and washed in a well of NGM media. The worm was then transferred with a pipetter from the well onto the viewing area of the micro-compressor in a small droplet no more than 2 μm in diameter (~1 μL of liquid). C. elegans images were taken on a Marianas Imaging Workstation from Intelligent Imaging and Innovations Inc (3I), Denver, CO, USA. This consists of an inverted Zeiss-Axiovert 200M epifluorescence microscope (Zeiss Inc., Oberkochen, Germany). The lens used was a 40X PlanNeofluar 1.3NA oil immersion objective. A FITC cube (Semrock) was used to excite the green fluorescent protein (GFP) and image acquisition was performed with SlideBook6 software from 3I.
Paramecium were transferred from the lettuce media, stained with DAPI (Life Technologies, 20 ng/mL) and washed with media. One paramecium was placed on the microcompressor in 1 μL media and the top coverslip was lowered until the paramecium was immobilized. It was imaged with the 63X PlanNeofluar 1.4NA oil objective on the Marianas system (3I).
Imaging of S. cerevisiae was performed on a spinning disk (Yokagawa CSU-X1; Andor Technology) confocal microscope using a 100X PlanNeofluar 1.4NA oil immersion objective lens on a TiE microscope equipped with Perfect Focus System (Nikon). A multi-bandpass dichromatic mirror (Semrock) and bandpass filters (Chroma Technology Corp.) in an electronic filter wheel was used for selection of GFP, Texas red and farred emission. The yeast cells were incubated with MitoTracker dye for 30 min (1 μM, Life Technologies) and were washed with the drop-out media and 0.5 μL of mid log phase yeast was trapped for imaging.
E. coli cells were stained with acridine orange (AO) (Fisher Scientific, 40 μM), washed with DO buffer twice and resuspended [12]. 0.25 μL E. coli were imaged with the 63X PlanNeofluar 1.4NA oil objective on the Marianas system (3I).
2.3. Microcompressor
Brass components of the mechanical microcompressors were machined on a computer controlled milling machine. The compressor mount was machined with specifications as previously described. This component, along with the entire MMC, is now available from In Vivo Imaging Solutions, Bel Air, MD [6]. The glass coverslips are all commercially avaliable (Fisher Scientific and Cymbid Scientific, China). To create the base of the MMC, the outer ring (OR) was glued (Eukit, Sigma-Aldrich) to the brass base (BB) and a 30 mm round, glass coverslip (GC) was glued into the recess of the BB.
3. Results
3.1. High resolution imaging of protozoa and small metazoans with MMC
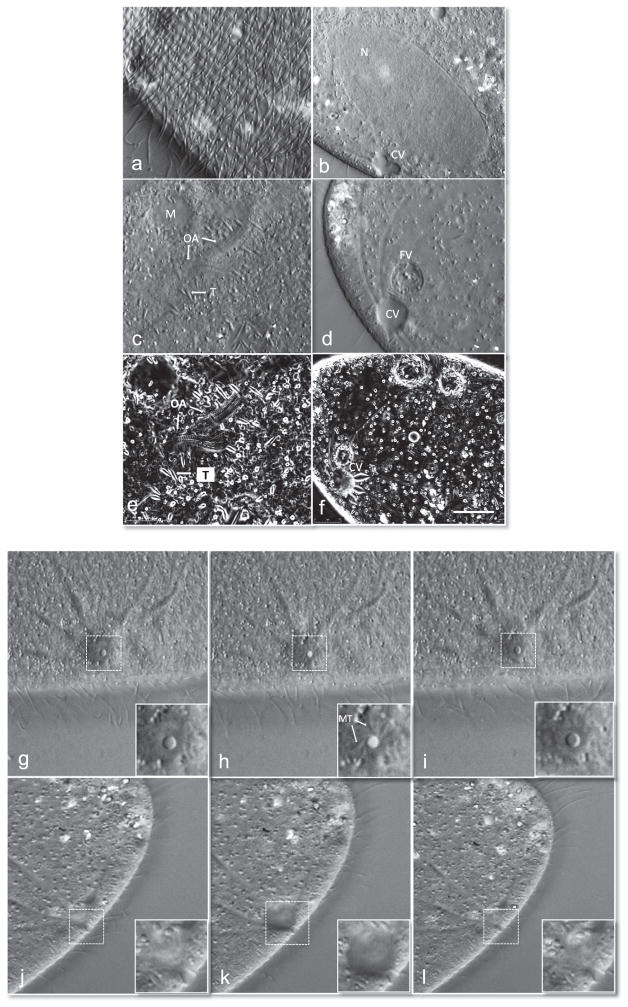
P. sonneborni cells were gently compressed and imaged with differential interference contrast (DIC) optics. Compression occurs by manually rotating the compressor mount in a clockwise direction. This confines the cells between two cover slips (see below for more detailed description). Cells will flatten quite substantially, often times resembling swimming pancakes when reversibly released from mechanical compression. This flattening is valuable, as it allows investigation of the interior of the cell with higher resolution since there is less cytoplasmic –stuff–to cause spherical aberrations. Flattening and immobilization is also useful for watching dynamic events such as the beating of cilia (Fig. 1a). We optically sectioned through 2 cells starting at the cortex. The cortical units that make up the ciliary rows and the underlying kinedesmal fibers and basal bodies were clearly seen. We then scanned along the entire cell (see Video 1). The macronucleus and the contractile vacuole complex are depicted in Fig. 1b. The immobilization of cells allows the investigator to watch very dynamic events over time (Video 1). We have imaged Ciliates for many hours continuously without damage [5,7]. Cells will typically be healthy as long as they are not overly compressed to the point of bursting. Their immobilization normally permits the continuous pumping of the contractile vacuoles and they maintain their osmotic pressure.
Fig. 1.
High resolution images of an immobilized paramecium. Differential interference contrast (DIC) images of paramecium using an Olympus 100X objective (a–d). (a) Paramecium cortex highlighting the ciliary row with basal bodies and kinetodesmal fibers. (b) Parental macronucleus (N) and contractile vacuole (CV) in diastole. (c) Oral apparatus undergoing morphogenesis. The trichocysts (T) and mitochondria (M) are labeled. Also see Video 1. (d) Filled CV prior to discharge and food vacuole (FV) filled with bacteria. Orientation-independent DIC (OI-DIC) images of paramecium using 100X objective (e and f). (e) OI-DIC image of same cells shown in (c) a few seconds later. (f) Filled CV prior to discharge of contents. Image series of cycle of CV opening and closing top down view, inset corresponds to dotted box within image (g–i). (g) Closed pore within CV, (h) opening of the pore within the CV and (i) closing of the pore to complete one cycle of systole. The microtubules (MT) extend from the pore and radiate towards the canals. Image series of side view of CV and pore through cycle of diastole (j–l). The pore gradually fills its contents (j and k) and then releases them (l). Also see Videos 2 and 3. Scale bar is 10 μM.
Supplementary material related to this article can be found online at doi:10.1016/j.yexcr.2015.07.011.
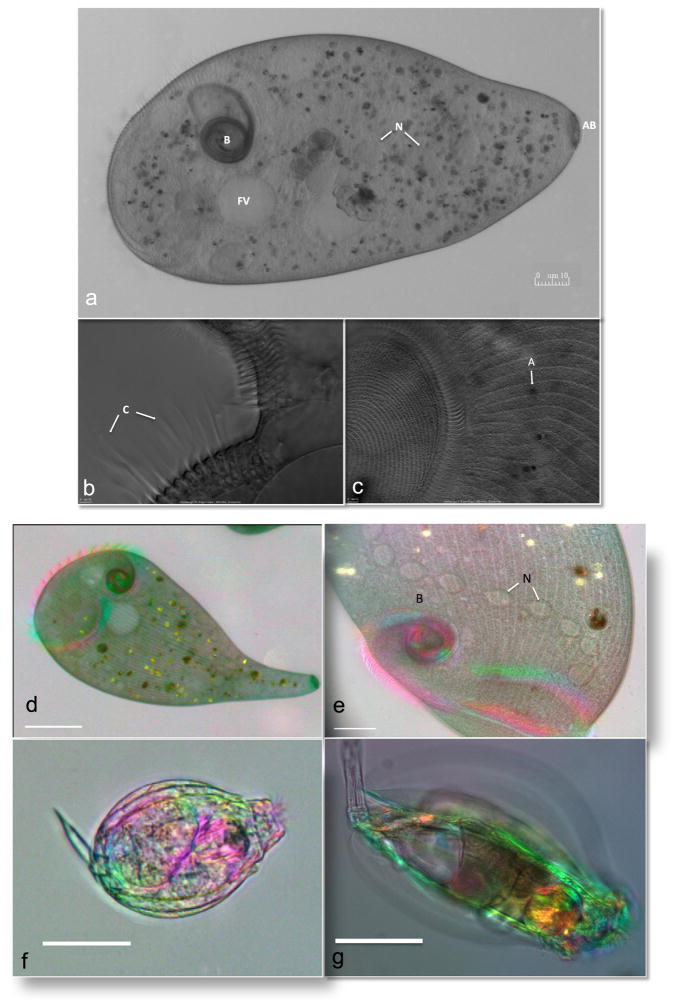
Depicted in Fig. 1c is the morphogenesis of the oral apparatus. Note the elongated structures, the trichocysts, which are scattered throughout the cortex, as well as the more sausage-shaped mitochondria in these high resolution images. Depicted in Fig. 1d is a better view of a fully filled contractile vacuole just prior to discharge with sharply focused radiating canals and an adjacent food vacuole filled with bacteria. Since the cells were completely immobilized, this allowed us to perform OI DIC microscopy where 6 images were rapidly captured. In each case the polarizer is rotated 60° [6, 9]. This allowed us to acquire better-resolved 3D like images of the morphogenesis of the oral apparatus (Fig. 1e) and of a filled contractile vacuole complex just prior to discharge (Fig. 1f). Using DIC optics, we obtained exquisite head on views of the contractile complex undergoing cycles (Fig. 1g–i and Video 2) and from the side (Fig. 1j–l and Video 3). In the first series of images the microtubules that extend from the pore are clearly visible in the inset of Fig. 1h. In the side view, the contractile membrane can be seen lining the pore (inset, Fig. 1k) and disappears as the contractile vacuole undergoes cystole in Fig. 1l. In Fig. 2 are images of the large ciliate Stentor polymorphus, obtained from pond water near Woods Hole, MA. These cells are incredibly difficult to trap. We found that they required considerable compression in order to prevent them from swimming away. Interestingly, they are also quite robust, since they were able to handle these forces quite well. To our knowledge, this is the first time that a device has been used to immobilize this species of ciliate successfully (Fig. 2a). Shown are several views of the membranelles of the oral field. In Fig. 2b, the compound ciliary structures are visible, whereas in Fig. 1c, the oral field is visualized and one can see the lines that make up the contractile myonemes that radiate out to the right of the image. Given the success with S. polymorphus, we also immobilized the larger species Stentor coeruleus. These cells were even more resistant to our attempts to immobilize them. Again, we were able to gently compress them and imaged them on an OI polarized light microscope that allows real-time birefringence imaging using a video-enhanced polychromatic polscope [8]. We obtained remarkable images (Fig. 1d and e) of trapped cells and also obtained cells contracting their myonemes (see Videos 4 and 5, respectively). We also obtained higher magnification images of cells and focused through the cells revealing their inner organelles and very interesting elongated nucleus (Video 6).
Fig. 2.
Imaging of Stentor species and rotifers. DIC imaging of the immobilized Stentor polymorphus with Olympus 20X (a) and 100X objective (b and c). (a) Stentor illustrating the buccal cavity (B), food vacuole (FV), nucleus (N), and attachment base (AB). (b) Close-up of the membranelles with the compound ciliary structures (C). (c) Oral field (left) and algal symbiont (A) are highighted. Birefringence imaging using a video-enhanced polychromatic polscope of Stentor coeruleus with 10X (d) and 40X (e) lens, showing the buccal cavity (B) and the moniliform macronucleus (N) which spans the length of the Stentor. Also see Videos 4, 5, and 6. The immobilized rotifers (f and g) found in pond water were imaged using the 10X lens. Also see Videos 7, 8 and 9. Scale bar is 10 μM.
Supplementary material related to this article can be found online at doi:10.1016/j.yexcr.2015.07.011.
Supplementary material related to this article can be found online at doi:10.1016/j.yexcr.2015.07.011.
Supplementary material related to this article can be found online at doi:10.1016/j.yexcr.2015.07.011.
Supplementary material related to this article can be found online at doi:10.1016/j.yexcr.2015.07.011.
Supplementary material related to this article can be found online at doi:10.1016/j.yexcr.2015.07.011.
Utilizing our access to pond water, we decided to immobilize any organism we observed. We trapped several dozen different single celled and multicellular organisms successfully using the MMC (Fig. 2 and not shown). We demonstrate the usefulness of the compressor for immobilizing and optically sectioning through several immobilized rotifers (Videos 7, 8 and 9).
Supplementary material related to this article can be found online at doi:10.1016/j.yexcr.2015.07.011.
Supplementary material related to this article can be found online at doi:10.1016/j.yexcr.2015.07.011.
Supplementary material related to this article can be found online at doi:10.1016/j.yexcr.2015.07.011.
3.2. A new MMC for inverted microscopes
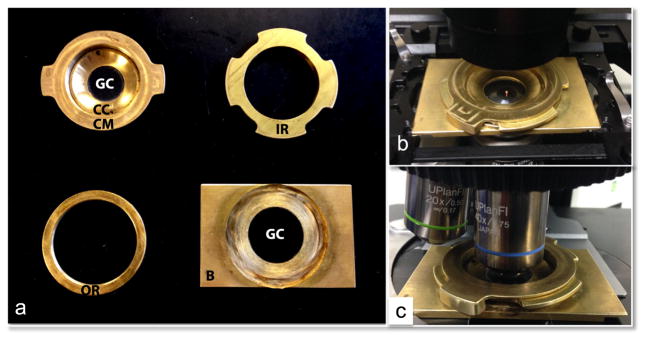
There has been an increase in the use of inverted microscopes over the standard upright microscope design. This has been driven largely by the ability to purchase or make imaging chambers with a coverlip on the bottom so that one can have an open system that allows the addition of chemical or fiuids to the specimen. While the MMC featured above can be used on an inverted microscopes, one has to reach under the stage, to adjust the compression and rotate the compressor mount. This can be cumbersome depending on the system and the amount of objectives in the turret. The general design of the enhanced MMC (eMMC) is similar to that of the MMC except that the base is now machined out of brass (B), with a recess that fits a larger 30 mm round glass coverslip (GC) so that imaging can be performed from the bottom (Fig. 3a). The brass parts were manufactured on computer-controlled milling machine (CCMM). As was the case in the MMC, the glass coverslips are all commercially avaliable. The outer ring (OR) was glued to the brass base. Specimens are placed on the large GC which provides the flat base on which the specimen rests. The inner ring (IR) threads into the OR; the compressor mount (CM) slides and locks into the IR. As with the MMC, the CM holds the coverslip compressor (CC) which positions a 25 mm GC in place. When the CC is tightened into the CM, the GC bows. Bowing of the GC allows the two planes of glass to touch, which can be affirmed by the presence of Newton rings in the center of the device when the two GCs are in close proximity (not shown). The bowed glass keeps the liquid media in the center of the microcompressor by capillary action preventing liquid from dispersing to the outer edges. This bowing action also permits a larger resevoir of liquid than would be held by two parallel flat planes of glass. When compressing a sample in the eMMC, the IR is rotated counter-clockwise reducing the distance between the bowed and flat GCs until the specimen is imobilized. Immobilization can be done at a lower magnification to confirm correct placement of the sample and then small, fine-adjustments can be made at higher magnification for final immobilization.
Fig. 3.
The enhanced mechanical microcompressor (eMMC) device for immobilizing specimens. (a) The main components of the microcompressor consist of a brass base (B), with a commercially available 25 mM coverslip glued in its base and four precision-machined brass pieces. To create the base of the eMMC, the outer ring (OR) was glued to the brass base, and a 30 mm round, glass coverslip (GC) was glued into the recess of B. This GC provides the surface upon which the specimen rests. The inner ring (IR) threads into the OR; the compressor mount (CM) slides and locks into the IR. The CM holds the coverslip compressor (CC) which positions a 25 mm glass coverslip (GC) in place. (b) An assembled unit placed on an inverted Nikon TiE microscope stage. Samples are loaded into the center of the GC when the CM is removed, after which the CM is inserted into IR and rotated until the specimen is immobilized (c) eMMC assembled and positioned on an upright Olympus AX70 microscope.
The eMMC could be fitted with microfluidics for constant flow of fresh media for log-duration time-lapse imaging as was engineered for the MMC [6]. However, if imaging will be for a short duration or does not require a continuous flow of fresh media, a moist atmosphere can be made by just inserting a moistened piece of filter paper, or Kimwipe, or other thin tissue that has been cut into a ring and placed around the periphery of the bottom GC (not shown). The chamber creates a moisture reservoir for long term imaging overnight. Fig. 3b shows the assembled eMMC on an inverted microscope. We also found that this system can be used on an upright microscope (Fig. 3b and not shown).
3.3. Multi-cell, large organism compression
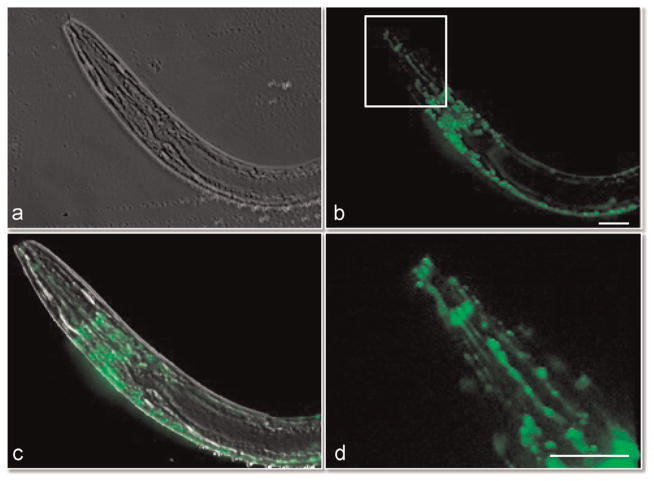
The C. elegans strain described here is under the control of a promoter that expresses GFP in the cholinergic neurons. As was the case for the MMC, the eMMC could be used to immobilize these highly active nematode worms during development and growth. We trapped and imaged worms using phase contrast optics (Fig. 4a). We also used epifluorescence microscopy to visualize the head ganglia, dorsal and ventral cord, and neural system in the pharynx (Fig. 4b, c and d). Imaging of the worm reveals distinct and intricate neurons migrating that without the eMMC would be difficult to observe over time (see Video 10). Worms were trapped and imaged for 3–4 h and then released and collected from the eMMC. The worms were then re-cultured and observing for several days for survival rate. All compressed worms were still alive after 2 days. Two of the worms were monitored for 6 days; both survived.
Fig. 4.
Phase contrast and epifluorescent images of neural network within immobilized Caenorhabditis elegans. The nematode worm was imaged using phase contrast (a) and epifluorescent imaging (b). The neural network within the head and back fluoresce. The overlay of the phase and fluorescent images is shown in (c). (d) Close up view of neural network in head as outlined in white box in (b). Also see Video 10. Scale bar is 10 μM.
Supplementary material related to this article can be found online at doi:10.1016/j.yexcr.2015.07.011.
3.4. Single-cell compression
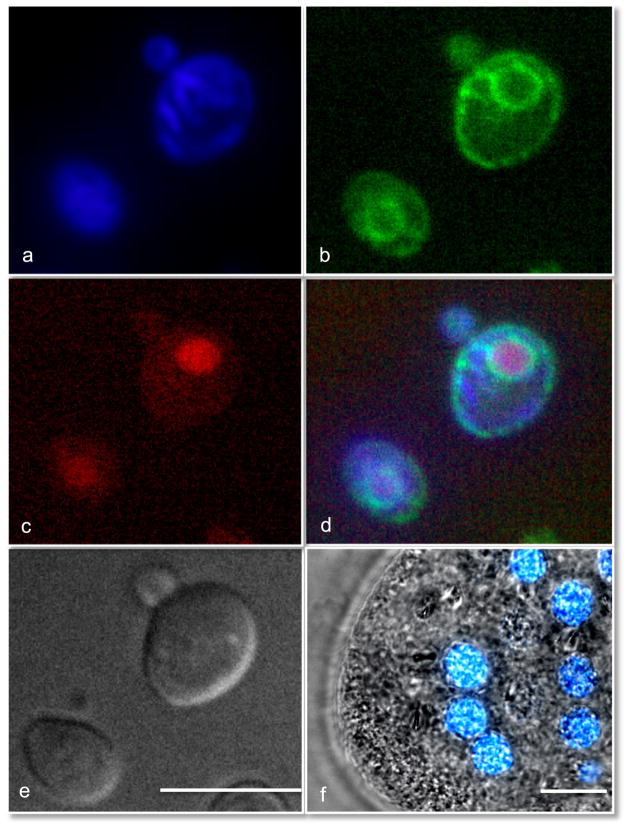
Imaging through the glass cover slips of the eMMC allows for high resolution, detailed images to be taken of small, unicellular organisms. Cells were able to withstand the compression for several hours and were imaged with fluorescence, and phase contrast or DIC. Proper compression of small organisms greatly enhances the quality of images obtained during imaging. The yeast Saccharomyces cerevisiae are small unicellular organisms that are notoriously difficult to immobilize and image. Yeast are commonly immobilized on a bed of agarose, which scatters light, and creates spherical aberrations. We found previously that we could image yeast for many hours in the MMC and watch them grow and bud [6]. The eMMC also could gently trap the yeast and improved the optical performance and resolution. Using the eMMC, we were able to obtain remarkably sharp images of the nucleus, mitochondria and endoplasmic reticulum in living yeast cells (Fig. 5a–d), as well as DIC images (Fig. 5e). To demonstrate the fine trapping ability of this device, we also trapped the bacterium Escherichia coli. These results can also be seen in Videos 11 and 12. Immobilized E. coli in the same focal plane are sharply focused, while the uncompressed cells are blurry and moving within the field.
Fig. 5.
DIC and epifluorescent imaging of immobilized Saccharomyces cerevisiae and Paramecium sonneborni. The organelles labeled by fluorescent protein or dye within yeast are the mitotchondria (a), the endoplasmic reticulum (b), and nucleus (c). The compilation of images a–c is shown in (d), and the DIC is shown in (e). (f) The nuclei of the Paramecium are shown with the blue fluorescent signal overlaid with the brightfield image. See also Paramecium in Video 13. Scale bar is 10 μM.
Supplementary material related to this article can be found online at doi:10.1016/j.yexcr.2015.07.011.
Supplementary material related to this article can be found online at doi:10.1016/j.yexcr.2015.07.011.
To demonstrate the ability of the eMMC to trap larger protozoa, we again turned to P. sonneborni. The paramecia were put into a food-deprivation state to induce autogamy with nuclear reorganization and fragmention of the macronucleus. Shown in Fig. 5f are the nuclear fragments labeled with DAPI overlain with a phase contrast image of the cell. In a lower magnification phase contrast video (Video 13), the cilia of the immobilized paramecia are clearly visible propelling water and the bacterial food source. Additionally, the contractile vacuoles can be clearly seen to going through their systole/diastole cycle.
Supplementary material related to this article can be found online at doi:10.1016/j.yexcr.2015.07.011.
4. Discussion
This work demonstrates the usefulness of immobilizing organisms for high resolution imaging. While there are devices being developed that require complex microfluidics and pumps [13–15], the microcompressor is extremely simple, is cost effective, and uses glass for both interfaces instead of polydimethylsiloxane, making the MMC better for high resolution imaging. The MMC can be used to gently flatten and image a wide variety of organisms ranging from yeast to nematode worms and small embryos [6]. Here, we show the usefulness of this device for obtaining high resolution images of a number of protozoa and metazoan species. The videos of the contractile vacuoles undergoing their systole/ diastole cycles are truly stunning. In addition, we used sophisticated imaging techniques such as OI DIC and real time birefringence imaging [7] to obtain images of highly dynamic events in single cells and small invertebrates. It was demonstrated that this technique could be used to trap two species of Stentor, which until now, have resisted immobilization. We obtained remarkable images of immobilized S. polymorphus and S. coeruleus.
We also demonstrated the usefulness of a new design, the eMMC, which was machined for use on inverted microscopes. The new design is made entirely of brass parts which improves its durability and reduces the risk of breakage. As with the original MMC, all of the glass components are commercially avaliable and are glued into the brass components. The eMMC was fabricated on a CCMM and while not yet commerically available like the MMC, we hope to mass produce multiple devices in the near future for purchasing and testing. The new design fits on to a variety of upright and inverted scope because the base has the same dimensions as a large glass slide. When imaging through the older MMC design, the girth of the objective determined the placement of the CM. A larger diameter objective could only move around the GC until it came in contact with the CC. When imaging through the coverslip in the base of the eMMC, the objective can move freely for a larger field of view because all of the rotating brass parts are on top of base.
We found that the new eMMC is much easier to use on inverted microscopes and permitted trapping of all organisms tested. As is the case with the earlier version, one can smash and damage or kill the specimen, but users can develop a feel for proper compression in just a few attempts. We found that compression does not interfere with the normal life functions or processes while allowing for repeated compression and imaging. The MMC allowed us to take high resolution images of the intricate neural network in the worm–s head; these detailed images could not be taken without complete immobilization of the worm, typically performed by anesthesia, which has been shown to have secondary effects on the worm–s physiology. We also obtained high resolution images of immobilized bacteria and the organelles within yeast and paramecia. This non-toxic means of immobilization allowed for long-term imaging of these organisms without introducing any noticeable artifacts. We have successfully trapped individual nuclei isolated from mammalian cells (not shown) and plan to test the ability to perform high resolution total internal reflection fluorescence to potentially watch transcriptional events or even entrance or exit of mRNAs or proteins through the nuclear pores. The MMC and eMMC devices are extremely useful for identifying microorganisms and observing the physiology of a variety of microbes and small multicellular organisms. In addition to their usefulness as a research device, the microcompressors make an excellent teaching tool for the laboratory classroom.
Supplementary Material
Abbreviations
- 3D
three dimensions
- MMC
mechanical microcompressor
- DIC
differential interference contrast
- OI-DIC
orientation- independent DIC
- ngm
nematode growth media
- DO
drop-out
- GFP
green fluorescent protein
- CV
contractile vacuole
- eMMC
enhanced mechanical microcompressor
- CCMM
computer-controlled milling machine
- OR
outer ring
- BB
brass base
- GC
glass coverslip
- IR
inner ring
- CM
compressor mount
- CC
coverslip compressor
- AO
acridine orange
References
- 1.Aufderheide KJ. An overview of techniques for immobilizing and viewing living cells. Micron. 2008;39:71–76. doi: 10.1016/j.micron.2006.12.002. [DOI] [PubMed] [Google Scholar]
- 2.Spoon DM. A new rotary microcompressor. Trans Am Microsc Soc. 1978;97:412–416. [PubMed] [Google Scholar]
- 3.Aufderheide KJ, Janetopoulos C. Immobilization of living specimens for microscopic observation. In: Méndez-Vilas A, editor. Current Microscopy Contributions to Advances in Science and Technology. Vol. 2. Formatex Research Center; Badajoz, Spain: 2012. pp. 833–838. [Google Scholar]
- 4.Janetopoulos C, Cole E, Smothers JF, Allis CD, Aufderheide KJ. The conjusome: a novel structure in Tetrahymena found only during sexual reorganization. J Cell Sci. 1999;112:1003–1011. doi: 10.1242/jcs.112.7.1003. [DOI] [PubMed] [Google Scholar]
- 5.Aufderheide KJ, Daggett PM, Nerad TA. Paramecium sonneborni, n. sp, a new member of the Paramecium aurelia species-complex. J Protozool. 1983;30:128–131. [Google Scholar]
- 6.Yan Y, Jiang L, Aufderheide KJ, Wright GA, Terekhov A, Costa L, Qin K, McCleery WT, Fellenstein JJ, Ustione A, et al. A microfluidic-enabled mechanical microcompressor for the immobilization of live single- and multi-cellular specimens. Microsc Microanal. 2014;20:141–151. doi: 10.1017/S1431927613014037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shribak M, LaFountain J, Biggs D, Inoue S. Orientation-independent differential interference contrast microscopy and its combination with an orientation-independent polarization system. J Biomed Opt. 2008;13:014011. doi: 10.1117/1.2837406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mehta SB, Shribak M, Oldenbourg R. Polarized light imaging of birefringence and diattenuation at high resolution and high sensitivity. J Opt. 2013:15. doi: 10.1088/2040-8978/15/9/094007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shribak M. Quantitative orientation-independent differential interference contrast microscope with fast switching shear direction and bias modulation. J Opt Soc Am A Opt Image Sci Vis. 2013;30:769–782. doi: 10.1364/JOSAA.30.000769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Young CL, Raden DL, Caplan JL, Czymmek KJ, Robinson AS. Cassette series designed for live-cell imaging of proteins and high-resolution techniques in yeast. Yeast. 2012;29:119–136. doi: 10.1002/yea.2895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mason DJ, Lloyd D. Acridine orange as an indicator of bacterial susceptibility to gentamicin. FEMS Microbiol Lett. 1997;153:199–204. doi: 10.1111/j.1574-6968.1997.tb10482.x. [DOI] [PubMed] [Google Scholar]
- 13.Mondal S, Ahlawat S, Koushika SP. Simple microfluidic devices for in vivo imaging of C. elegans, Drosophila and zebrafish. J Vis Exp. 2012 doi: 10.3791/3780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McCormick KE, Gaertner BE, Sottile M, Phillips PC, Lockery SR. Micro-fluidic devices for analysis of spatial orientation behaviors in semi-restrained Caenorhabditis elegans. PLoS One. 2011;6:e25710. doi: 10.1371/journal.pone.0025710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yanik MF, Rohde CB, Pardo-Martin C. Technologies for micromanipulating, imaging, and phenotyping small invertebrates and vertebrates. Annu Rev Biomed Eng. 2011;13:185–217. doi: 10.1146/annurev-bioeng-071910-124703. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.