Abstract
Studying the genetic diversity and natural polymorphisms of HIV-1 would benefit our understanding of HIV drug resistance (HIVDR) development and predict treatment outcomes. In this study, we have characterized the HIV-1 genetic diversity and natural polymorphisms at the 5′ region of the pol gene encompassing the protease (PR) and reverse transcriptase (RT) from 271 plasma specimens collected in 2008 from HIV-1-infected patients who were eligible for initiating antiretroviral therapy in Abuja (Nigeria). The analysis indicated that the predominant subtype was subtype G (31.0%), followed by CRF02-AG (19.2 %), CRF43-02G (18.5%), and A/CRF36-cpx (11.4%); the remaining (19.9%) were other subtypes and circulating (CRF) and unique (URF) recombinant forms. Recombinant viruses (68.6%) were the major viral strains in the region. Eighty-four subtype G sequences were further mainly classified into two major and two minor clusters; sequences in the two major clusters were closely related to the HIV-1 strains in two of the three major subtype G clusters detected worldwide. Those in the two minor clusters appear to be new subtype G strains circulating only in Abuja. The pretreatment DR prevalence was < 3%; however, numerous natural polymorphisms were present. Eleven polymorphic mutations (G16E, K20I, L23P, E35D, M36I, N37D/S/T, R57K, L63P, and V82I) were detected in the PR that were subtype or CRF specific while only three mutations (D123N, I135T, and I135V) were identified in the RT. Overall, this study indicates an evolving HIV-1 epidemic in Abuja with recombinant viruses becoming the dominant strains and the emergence of new subtype G strains; pretreatment HIVDR was low and the occurrence of natural polymorphism in the PR region was subtype or CRF dependent.
Introduction
ONE OF THE MAJOR CHALLENGES in controlling the HIV/AIDS pandemic is the genetic variability of HIV and its consequences for the development of antiretroviral (ARV) drugs and vaccine. HIV vaccine development has been hindered by its extensive genetic heterogeneity.1,2 Currently, the genetic diversity of HIV-1 in the worldwide epidemic is characterized by four groups, M, N, O, and P.3 The group M is the leading cause of the global epidemics and is composed of nine subtypes (A,B, C, D, F, G, H, J, and K),3 more than 49 circulating recombinant forms (CRFs), and 100 unique recombinant forms (URFs).4–6 While subtype B is the predominant strain in the developed countries, the non-B subtypes as well as CRFs and URFs are the major epidemic strains characterized in the African region.7–30 In sub-Saharan Africa, multiple HIV-1 subtypes are found along with various CRFs such as CRF01-AE in Central Africa and CRF02-AG in West Africa.7,9,15,16,31–37
In Nigeria, studies have shown a diversified HIV-1 epidemic with the viral subtype G, CRF06-cpx, CRF02-AG, sub-subtype A3, and other recombinants cocirculating.16,18,34,38–40 In a study published in 2000, subtype A was predominant (about 70%) in the southwest-Lagos state and subtype G was predominant in the northwest-Kano state (about 58%), while both subtypes A (49%) and G (47%) were observed to be equally distributed in the northeast (Maiduguri).18 In 2006, a study in Oyo state (southeast) showed the predominance of CRF02-AG (57%), subtype G (26%), and CRF06-cpx (11%),16 and similar results with 39–45% for CRF02-AG and 38% for subtype G were reported in 200941 and 2012.39
Characterization of the polymorphisms within the protease (PR) and reverse transcriptase (RT) genes have been conducted mostly for subtype B viruses; few studies have been conducted for non-B subtype viruses, and their impact on highly active antiretroviral therapy (HAART) is undetermined.9,29,42–46 Indeed, it has been shown that differences in codon sequences at positions associated with drug resistance mutations (DRMs) might predispose viral isolates of different subtypes to encode different amino acid substitutions that can affect the rate of emergence of resistance, cross-resistance to same-class drugs, and potentially drug susceptibility and clinical outcomes.8,47 Data from virological and biochemical analysis revealed that natural variations in amino acids can affect the degree of drug resistance (DR) conferred by some mutations.48 It has been shown that HIV-2 and group O HIV-1 viruses are naturally resistant to nonnucleoside RT inhibitors (NNRTIs) due to mutations present in their RT gene.49,50 Moreover, differences in nucleotide and mutational motifs (these are transitions and transversions needed to develop DR to different antivirals) between subtypes can affect the genetic barrier for resistance.51,52
One good example of this is the V106M polymorphism in the RT of subtype C viruses inducing resistance to NNRTIs.53 However, study of the influence of genetic variability and polymorphisms on HIV-1 DR development in places where diverse HIV-1 non-B subtypes, CRFs, and URFs are co-circulating is limited. We undertook this study in Abuja, Nigeria’s capital city, using specimens collected from HIV-1-infected patients who were eligible for initiation of ART at two treatment sites. The aims of this analysis were to (1) determine the HIV-1 subtype distribution in the cohort; (2) identify and characterize baseline polymorphisms and DRMs at pretreatment and the association of any specific mutational pattern with HIV-1 subtypes or CRFs; and (3) evaluate the potential impact of these polymorphisms on DR development.
Materials and Methods
Specimens
Patients were recruited for a prospective cohort study to monitor HIVDR development. The median age was 34 years [interquartile range (IQR), 28–40 years]; 38% were male, while 62% were female at the time of ART initiation. The median CD4 count was 149 cells/μl (IQR, 73–205 cells/μl) and 72% of participants had a CD4 count of ≤200 cells/μl. Detailed demographic and clinical data of this cohort were described previously.54 The specimens were collected from HIV-infected patients initiating ART from two sites in Abuja, between January and July 2008. Samples were systematically collected from all the patients who were eligible for ART to limit the introduction of bias in the nature and quantity of polymorphisms and/or DRMs detected.
RT-PCR and DNA sequencing
Sequences were generated from these plasma specimens using the broadly sensitive HIV-1 genotyping assay that amplifies the 5′ region of the HIV-1 pol gene including DR mutation sites in the PR and RT regions.55 Sequences were edited with the ReCall program56 and the consensus sequences were made for further analysis.
HIV-1 subtype determination and characterization
HIV-1 subtypes and CRFs for the newly obtained 271 sequences were initially determined by phylogenetic analysis using MEGA 557 along with 109 reference sequences including subtypes A to K and all available CRFs, downloaded from the Los Alamos HIV sequence database (2010 version, www.hiv.lanl.gov/). Neighbor-joining and maximum likelihood with algorithms of Tamura-Nei were used for the analysis with 1,000 bootstrap replicates. The bootstrap values above 70% were considered significant.58 For the sequences whose HIV-1 subtypes or CRFs could not be determined by the initial phylogenetic analysis, pairwise genetic distances (MEGA57), similarity, and bootscan analysis (SimPlot, v.3.5159) were performed for gene structure (recombination) analysis. If a segment of the query sequences with a length of < 200 nucleotides had a similarity of < 95% to the reference sequences, a U was assigned to indicate an unclassifiable segment within a sequence; the relatedness of these unclassifiable sequences to any sequence in the public database was then determined by using a standard GenBank Blast search. If the similarity scores were < 95%, the sequences were considered not closely related.
The phylogeny of 84 subtype G sequences identified from the initial subtyping was further assessed by using the MrBayes tool (Geneious, v.6.1.8, Biomatters Ltd., San Francisco, CA) with the references of subtype G (n = 36) and other major HIV-1 subtypes (n = 21) downloaded from the Los Alamos HIV sequence database (www.hiv.lanl.gov/). The posterior probabilities of trees were estimated using Markov chain Monte Carlo (MCMC) approach with a chain length of 1,100,000 and an HKY85 substitution model.60 Only probability values > 70% at the nodes of major clusters were presented in the tree.
Identification of major drug resistance-associated mutations and natural polymorphisms
Major HIVDR mutations of the 271 quality-assured sequences were identified by the Calibrated Population Resistance Tool (CPR), version 6.0, deployed at the Stanford HIV drug resistance website (http://cpr.stanford.edu/cpr.cgi). Sequences were aligned using Clustal W (BioEdit version 7.0.9.1, Carlsbad, CA) and were visually compared across subtypes to identify both synonymous and nonsynonymous substitutions at polymorphic and/or DR sites in both RT and PR genes, which also allowed us to determine any subtype-specific mutational pattern or motif. The mutational motif is defined as the number of transitions or transversions needed to develop resistance to different classes of ARVs.51,52 The specific mutational pattern analysis was focused on the sites that are polymorphic and/or involved in HIVDR development to any drug class of ARVs.
Results
HIV-1 subtype distribution
Phylogenetic analyses of the 271 newly obtained nucleotide sequences along with 109 HIV-1 subtype and CRF reference sequences revealed that 93.7% (254/271) of the sequences could be subtyped with our sequence analysis procedures while 6.3% (17/271) could not be assigned a standard subtype or CRF, which were denoted as unclassifiable sequences (Table 1). A total of 16 possible types of HIV-1 strains were identified. The predominant strains were subtype G (31.0%), CRF02-AG (19.2%), and CRF43-02G (18.5%), accounting for 68.7% of the total sequences analyzed. The remaining strains (31.3%) were A/CRF36-cpx (11.4%), CRF06-cpx (5.5%), CRF25-cpx (3.3%), CRF36-cpx (2.2%), A/CRF02-AG (1.5%), CRF15-01B/G (0.4%), CRF43-02G/G (0.4%), and unclassifiable (6.3%). Overall, the recombinant viral strains, including CRFs and URFs, accounted for 68.6%, while pure subtypes (G and C) were only 31.4%.
TABLE 1.
HIV-1 SUBTYPE DISTRIBUTION AMONG 271 SEQUENCES OBTAINED FROM NIGERIAN HIV-1-INFECTED PATIENTS ELIGIBLE FOR INITIATION OF ANTIRETROVIRAL THERAPY IN 2008
| Subtype/CRF | No. | % |
|---|---|---|
| C | 1 | 0.4 |
| G | 84 | 31.0 |
| CRF02-AG | 52 | 19.2 |
| CRF06-cpx | 15 | 5.5 |
| CRF25-cpx | 9 | 3.3 |
| CRF36-cpx | 6 | 2.2 |
| CRF43-02G | 50 | 18.5 |
| A/CRF02-AG | 4 | 1.5 |
| A/CRF36-cpx | 31 | 11.4 |
| CRF15-01B/G | 1 | 0.4 |
| CRF43-02G/G | 1 | 0.4 |
| Unclassifiable (U) | 17 | 6.3 |
| Pure subtypes | 85 | 31.4 |
| CRFs | 186 | 68.6 |
CRF, circulating recombinant form.
The figures in bold are subtype or CRFs having higher prevalence.
It is interesting to note that the subtype G sequences identified in this study (n = 84) exhibited intrasubtype diversity by segregating into two large distinct clusters in the initial phylogenetic analysis (data not shown). To further understand the diversity of these G sequences, we analyzed them with all available subtype G reference sequences from the Los Alamos database. Results indicated that the G sequences mainly formed three major and two minor clusters (Fig. 1). For description purpose, we named them G1-3 and Gm1-2, respectively. Most of the G sequences from this study (n = 64) fell into two of the three known major subtype G clusters, G1 (n = 19) and G3 (n = 45), and only one clustered in the G2 along with reference sequences from other countries. Interestingly, the remaining study sequences formed two independent minor clusters of Gm1 and Gm2 without clustering with any sequence from other countries with an exception of two sequences that didn’t group into clusters.
FIG. 1.
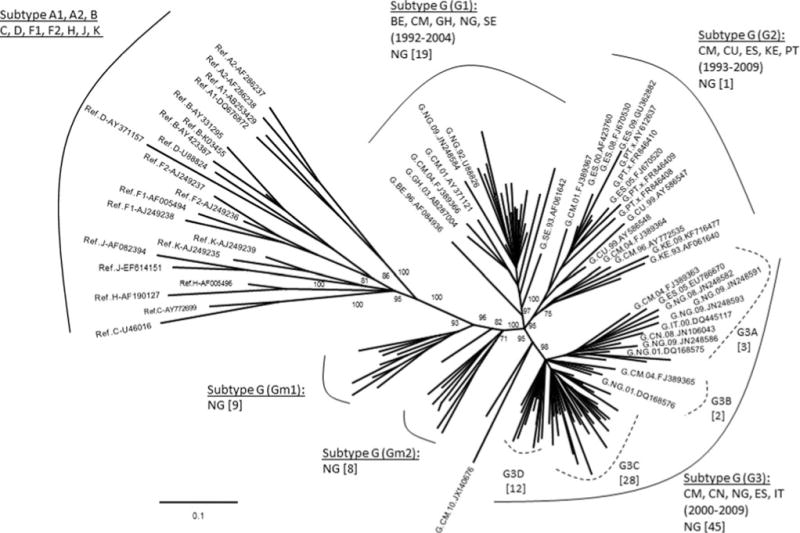
Phylogenetic analysis of subtype G sequences with HIV-1 reference sequences. The tree was constructed with 84 newly characterized Nigerian subtype G sequences and 57 reference sequences downloaded from the Los Alamos HIV database (www.hiv.lanl.gov/) representing major subtypes (n = 21) of A, B, C, D, F, H, K, and G (n = 36). Only the names of reference sequences were given in the tree. The number in the bracket gives the year spanning the reference sequences in the cluster or branch; the number in the square bracket indicates the number of Nigerian sequences in the cluster or branch. The numbers labeled at the nodes were bootstrapping values. Country code: BE (Belgium), CM (Cameroon), CN (China), CU (Cuba), ES (Spain), IT (Italy), GH (Ghana), KE (Kenya), NG (Nigeria), PT (Portugal), and SE (Sweden).
For those 17 sequences that a subtype/CRF could not be assigned, 13 representatives were selected for further analyses. Genetic distance analysis showed that they were not closely related to any subtype reference sequences, but had approximately equal distances to two or multiple subtype sequences (Table 2). Sequence similarity analysis indicated that they had complicated gene structures with multiple subtypes involved (Fig. 2a). Bootscan analysis further revealed that eight of them had strong evidence of recombination between multiple subtypes (Fig. 2b). Blast search for closely related sequences in the GenBank database showed the highest similarity hits were 93–95% to the GenBank sequences (data not shown), indicating they might be new recombinant viruses.
TABLE 2.
CHARACTERIZATION OF 13 UNCLASSIFIABLE (U) NIGERIAN HIV-1 SEQUENCES
| Sequence ID |
Method used for subtypinga
|
Possible subtype | ||
|---|---|---|---|---|
| Phylogenetic tree | Genetic distance | SimPlot | ||
| 5186 | CRF43-02G/G | 0.0449/0.0593 | CRF43-02G/G | CRF43-02G/G |
| 5287 | CRF02-AG/CRF36-cpx | 0.049/0.049 | CRF36/A/CRF36 | U/CRF36-cpx |
| 5290 | CRF02-AG/CRF36-cpx | 0.0605/0.0521 | CRF36/CRF06/CRF36 | U/CRF36-cpx |
| 5303 | CRF02-AG/CRF36-cpx | 0.0581/0.0572 | A/C/CRF36 | U/CRF36-cpx |
| 5298 | CRF02-AG/CRF36-cpx | 0.0419/0.0431 | A/CRF36-cpx | A/CRF36-cpx |
| 5261 | CRF02-AG/CRF36-cpx | 0.0498/0.0554 | A/CRF02-cpx | A/CRF36-cpx |
| 5102 | CRF02-AG/CRF45-cpx | 0.0644/0.0675 | A/K | A/U |
| 5305 | CRF15-01B/CRF01-AE | 0.0663/0.0764 | A/CRF15-01B | A/U |
| 5089 | CRF23-BG/CRF19-cpx | 0.0669/0.0679 | CRF36-cpx/B | U/B |
| 5343 | CRF21-A2D/D/CRF19-cpx | 0.0621/0.0525/0.0611 | U/D | U/D |
| 5202 | CRF37-cpx/CRF02-AG/G | 0.741/0.0699/0.699 | CRF15-01B/G | CRF15-01B/G |
| 5078 | CRF36-cpx/CRF43-02G/G | 0.0552/0.0598/0.0628 | CRF43/CRF36/G | U |
| 5299 | CRF36-cpx/CRF43-02G | 0.0687/0.0691 | CRF43/CRF36/G | U |
Details of methodology were described in Materials and Methods.
FIG. 2.
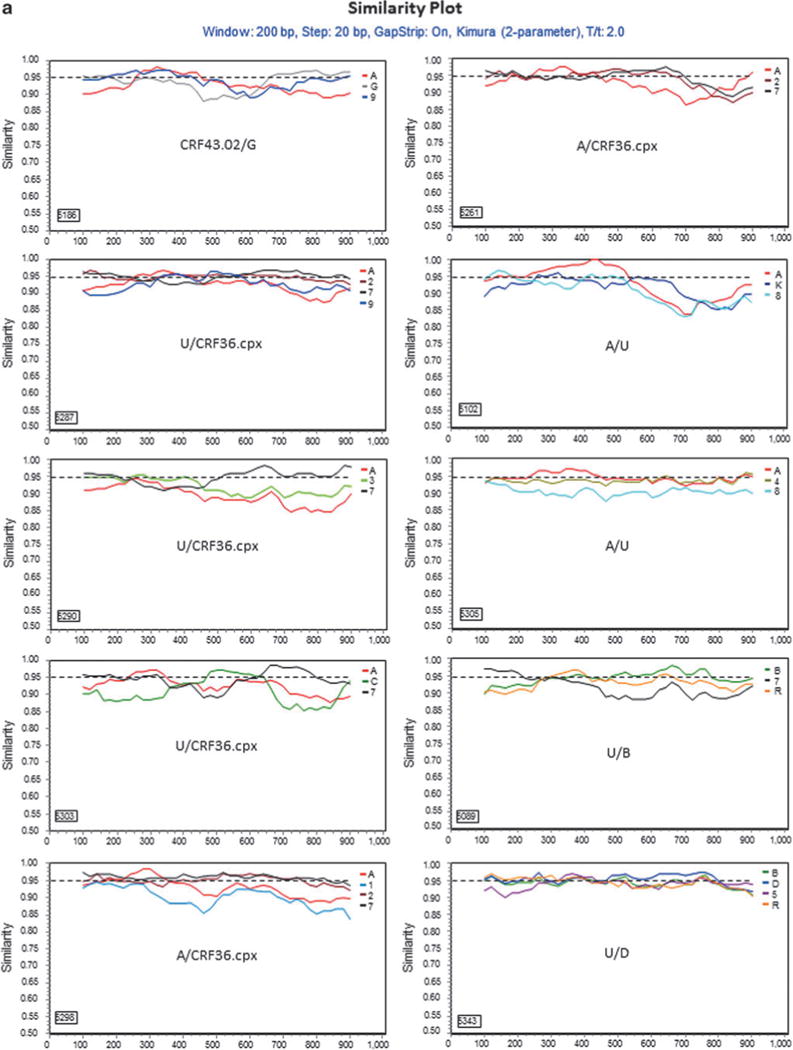
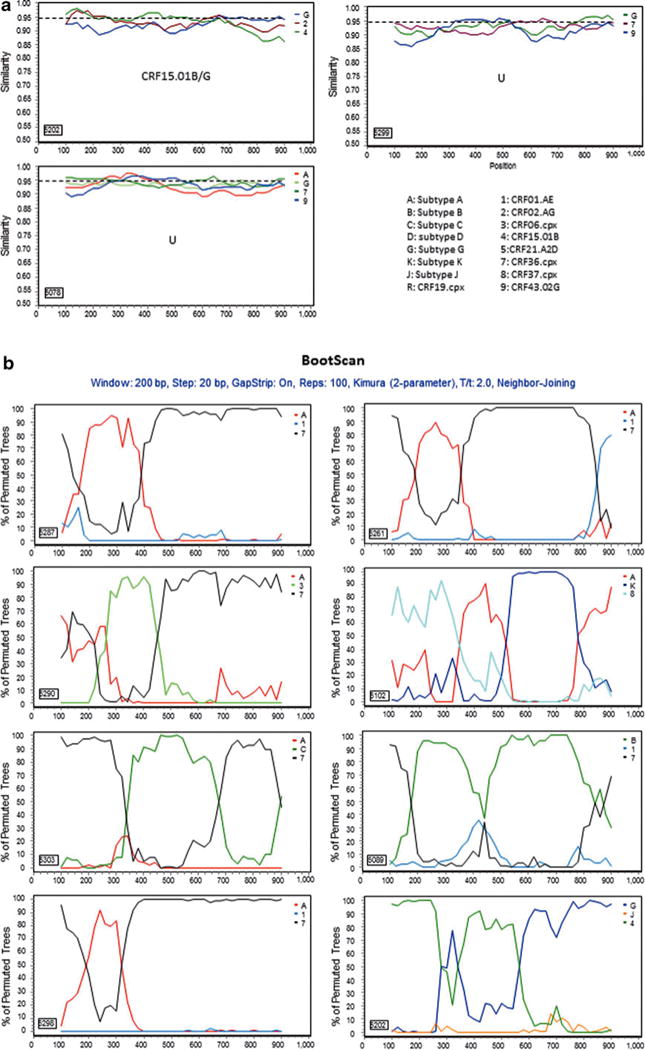
Further characterization of phylogenetically unresolved Nigerian sequences. The 13 representative Nigerian unclassifiable sequences (Table 1) were further analyzed individually using SimPlot59 for genetic similarity (200-bp window, 20-bp step, and two-parameter Kimura algorithm). The selection of HIV-1 reference subtypes for analysis was based on the genetic distance analysis (Table 2). Only three to four relevant reference subtypes were chosen to generate the graph to show the gene structure of query sequences (a). The dashed line was used to determine the relatedness of the query sequence to the selected reference subtypes at the 95% similarity threshold across the gene. The segment of the query sequence with a length of less than 200 nucleotides and less than 95% similarity to the references was assigned a “U” indicating an unresolved subtyping for the sequence portion. The chimeric subtyping of the query sequence was given in each of the SimPlot diagrams (a) and populated in Table 2. To explore the recombinant events, 8 among the 13 unclassifiable (U) sequences were again analyzed using the BootScan59 method, demonstrating a clear recombination event (b).
HIV-1 drug resistance-associated mutations
Among the 271 sequences analyzed, 28% (76/271) had polymorphisms and/or primary and/or accessory DRMs in the PR and RT region (Table 3). The CPR analysis identified the PI mutations M46L, I85V, and F53Y as well as the NRTI mutations M41L and V75M, and the NNRTI mutations K101E, K103N, and G190A as Surveillance Drug Resistance Mutations (SDRM).61,62 The total number of detected mutations in the PR was 11, distributed at 9 sites among 11 samples. In the RT, 81 mutations were identified and distributed at 15 sites among 69 samples. Seven specimens had PI mutations but no NRTI or NNRTI mutations, while the remaining 69 specimens with resistance-associated mutations had no PI mutations.
TABLE 3.
SUMMARY OF SPECIMENS WITH POLYMORPHISMS AND OR DRUG RESISTANCE MUTATIONS AT BASELINE
| IDs | Subtype |
Scored resistance mutations
|
Resistance levela
|
||||
|---|---|---|---|---|---|---|---|
| PR | RT | Sensitiveb | Low | Intermediate | High | ||
| 5149 | A/CRF02-AG | None | V179I | ||||
| 5292 | A/CRF02-AG | None | V179I | ||||
| 5152 | A/CRF15-01B | None | V179I | ||||
| 5165 | A/CRF15-01B | None | V179I | ||||
| 5313 | A/CRF15-01B | None | V179IV | ||||
| 5123 | A/CRF15-01B | None | V179I | ||||
| 5305 | A/CRF15-01B | None | V90I | ||||
| 5168 | A/CRF36-cpx | None | T69N, V106I | ||||
| 5213 | A/CRF36-cpx | T74S | T69NT | All remaining PI | NFV | ||
| 5214 | A/CRF36-cpx | None | G190AG | ETR, RPV | EFV | NVP | |
| 5225 | A/CRF36-cpx | None | T69N, K101E, V108IV, G190A | ETR | RPV | EFV, NVP | |
| 5251 | A/CRF36-cpx | None | V179AT | ||||
| 5253 | A/CRF36-cpx | None | E138A | ||||
| 5312 | A/CRF36-cpx | I85V | T69NT | ||||
| 5330 | A/CRF36-cpx | None | E138A | ||||
| 5345 | A/CRF36-cpx | F53FY | V118I | ||||
| 5217 | C | T74S | None | All remaining PIs | NFV | ||
| 5074 | CRF02-AG | None | V90I | ||||
| 5093 | CRF02-AG | T74S | None | All remaining PIs | NFV | ||
| 5169 | CRF02-AG | None | T69ST | ||||
| 5188 | CRF02-AG | None | E138A | ||||
| 5228 | CRF02-AG | None | K103N | ETR, RPV | EFV, NVP | ||
| 5289 | CRF02-AG | None | V118I | ||||
| 5293 | CRF02-AG | None | T69ST | ||||
| 5304 | CRF02-AG | None | E138A | ||||
| 5306 | CRF02-AG | None | E138AE | ||||
| 5183 | CRF06-cpx | None | M41L, E44DE, V179E | 3TC, ABC, ddI, FTC, TDF | AZT, d4T | ||
| 5208 | CRF06-cpx | None | V90I | ||||
| 5241 | CRF06-cpx | None | V179E | ||||
| 5250 | CRF06-cpx | None | V179E | ||||
| 5279 | CRF06-cpx | None | V179E | ||||
| 5294 | CRF06-cpx | None | V179E | ||||
| 5328 | CRF06-cpx | None | V179E | ||||
| 5206 | CRF25-cpx | A71V | None | ||||
| 5248 | CRF25-cpx | None | V179I | ||||
| 5318 | CRF25-cpx | None | V179I | ||||
| 5100 | CRF36-cpx | None | V179I | ||||
| 5326 | CRF36-cpx | None | V118I | ||||
| 5089 | CRF36-cpx/B | None | V179I | ||||
| 5223 | CRF36-cpx/B | None | V179I | ||||
| 5079 | CRF43-02G | None | E138A | ||||
| 5084 | CRF43-02G | None | V179EV | ||||
| 5087 | CRF43-02G | T74S | None | All remaining PIs | NFV | ||
| 5106 | CRF43-02G | None | V179E | ||||
| 5174 | CRF43-02G | None | K101Q | ||||
| 5184 | CRF43-02G | None | V118I, E138A | ||||
| 5195 | CRF43-02G | None | V179E | ||||
| 5211 | CRF43-02G | T74S | T69A | All remaining PIs | NFV | ||
| 5238 | CRF43-02G | None | T69S | ||||
| 5315 | CRF43-02G | None | V118IV | ||||
| 5116 | G | None | V90I, V179IV | ||||
| 5119 | G | None | V118I | ||||
| 5142 | G | None | V106I | ||||
| 5161 | G | None | V179IMV | ||||
| 5194 | G | M46L | None | All remaining PIs | NFV | ||
| 5201 | G | None | E138A | ||||
| 5205 | G | None | A98G | EFV, ETR, RPV | NVP | ||
| 5220 | G | None | V75MV, K103R | ||||
| 5222 | G | None | V118IV | ||||
| 5226 | G | None | V179E | ||||
| 5233 | G | None | E138A | ||||
| 5237 | G | None | E138AE | ||||
| 5242 | G | None | V179E | ||||
| 5247 | G | None | V118I | ||||
| 5256 | G | None | V118I | ||||
| 5278 | G | None | K101Q, K103R | ||||
| 5296 | G | None | T69ST | ||||
| 5300 | G | None | V106IV, V118IV | ||||
| 5308 | G | None | V106I | ||||
| 5314 | G | None | M41L, V90I | 3TC, ABC, ddI, FTC, TDF | AZT, d4T | ||
| 5316 | G | T74S | None | All remaining PIs | NFV | ||
| 5336 | G | K43KT | None | ||||
| 5338 | G | None | V106I, V118IV | ||||
| 5320 | G | None | E138A | ||||
| 5325 | G | None | E138A | ||||
| 5299 | U | None | K101EK, E138A | DLV, ETR | EFV | NVP, RPV | |
Sensitivity column is filled only when drugs are resistant.
Potential low level of resistance is classified as sensitive as per WHO interpretation.
NFV, nelfinavir; EFV, efavirenz; NVP, nevirapine; ETR, etravirine; RPV, rilpivirine; AZT, zidovudine; d4T, stavudine; ABC, abacavir; 3TC, lamivudine; ddI, didanosine; FTC, emtricitabine; TDF, tenofovir.
In the PR region, one sample displayed the K43T mutation; another one had the M46L, while a third specimen had K53Y, A71V, and I85V. The T74S mutation was found in the PR of six specimens.
We also detected NRTI selected mutations M41L, E44D, T69ANS, V75M, and V118I, and NNRTI mutations V90I, A98G, K101EQ, K103NR, V106I, V108I, E138A, V179EI, and G190A (Table 3). Due to the M41L mutation detected in one specimen, the virus would have reduced susceptibility to zidovudine (AZT) and stavudine (d4T). Five specimens had A98G, K101EK, V108IV, K103N, E138A, and G190A occurring alone or in combination with each other, which could cause intermediate and/or high-level resistance to delavirdine (DLV), efavirenz (EFV), etravirine (ETR), and nevirapine (NVP). The mutation E138A associated with a decreased response to ETR, the second generation NNRTI, was found in 16% (12/76) of the specimens displaying DR mutations. There were only three specimens that had DR mutations associated with two combined drug classes (NRTIs plus NNRTIs). The overall resistance rate within the cohort was < 3% and the affected drugs were one PI [nelfinavir, (NFV)], two NRTIs (AZT and d4T), and five NNRTIs [EFV, DLV, ETR, rilpivirine (RPV), and NVP].
Among all the mutations detected, T74S (54.5%) in PR and E138A (17%), V179I (16%), followed by V118I and V179E (14% each) in RT were the most prevalent, while T69NS, V90I, and V106I were present at only 5% each. Within the cohort, the rate of these mutations was 2.2% for T74S, 4% for V118I and V179E each, 4.5% for V179I, and 5% for E138A. The prevalence for T69NS, V90I, and V106I was 1.5%, respectively. L89T/I, the nonpolymorphic PI-selected mutation of uncertain phenotypic and clinical significance was also detected.
Natural polymorphisms
Of the 271 sequences analyzed, 11 polymorphic mutations (G16E, K20I, L23P, E35D, M36I, N37D/S/T, R57K, L63P, and V82I) in the PR and 3 (D123N and I135T/V) in the RT were identified. According to the Stanford DR algorithm, these substitutions occurred in the wild type (WT) of non-B subtype viruses and have no effect on DR for PIs or RTIs; however, when combined with other mutations, they increase the resistance to PIs or RTIs in subtype B viruses.
Uncommon mutations
Numerous uncommon mutations with unknown impact and/or function within the PR and the RT were identified. In PR, 58 uncommon mutations were detected at 28 positions, while in RT the number was 96 at 58 positions. Some of these mutations occurred at a very high rate and were subtype specific in the PR gene. For example, the E35Q mutation occurred in 67% of all subtype G sequences and in only 5% of CRF02-AG sequences, but was absent in all other subtypes and CRFs. E35K occurred in 5% of CRF02-AG but was absent in the rest of the other subtypes. However, in the RT gene, we did not detect a high prevalence of an uncommon mutation or subtype-specific mutational pattern. The random mutation rates ranged from 0.5% to 15%.
Synonymy and subtype-specific codon mutational pattern
We next performed a codon-by-codon comparison of the nucleotide sequences of subtype B (HXB2) to the sequences in the cohort to detect any subtype-specific mutational patterns (Table 4). We found that at position 179 of the RT gene, all sequences of CRF06-cpx (6/6), subtype G (2/2), and CRF43-02G (2/2) harbored the V179E mutation, while all sequences of CRF36-cpx (1/1), A/CRF02-AG (2/2), CRF36-cpx/B (2/2), A/CRF15-01B (4/4), and CRF25-cpx (2/2) had the V179I mutation. The WT CRF06-cpx and subtype G mainly had a codon GTG at position 179, while the WT of the other subtypes and CRFs possessed the GTT at this position except for CRF36-cpx, which had a codon ATA.
TABLE 4.
CHANGES OF CODON PATTERN AT POSITION 179 OF THE REVERSE TRANSCRIPTASE
| Position 179 nucleotides | IDs—amino acid | Subtypes |
|---|---|---|
| GTT | HXB2-V | B |
| GTG/GTA | G-V | G |
| CRF02-AG-V | CRF02-AG | |
| CRF06-cpx-V | CRF06-cpx | |
| GAG | 5106-E | CRF43-02G |
| GAA | 5195-E | CRF43-02G |
| GAG | 5242-E | G |
| GAG | 5226-E | G |
| GAG | 5183-E | CRF06-cpx |
| GAG | 5241-E | CRF06-cpx |
| GAG | 5250-E | CRF06-cpx |
| GAG | 5328-E | CRF06-cpx |
| GAG | 5294-E | CRF06-cpx |
| GAG | 5279-E | CRF06-cpx |
| ATA | 5100-I | CRF36-cpx |
| ATT | 5149-I | A/CRF02-AG |
| ATT | 5292-I | A/CRF02-AG |
| ATT | 5089-I | CRF36-cpx/B |
| ATT | 5223-I | CRF36-cpx/B |
| ATT | 5152-I | A/CRF15-01B |
| ATT | 5165-I | A/CRF15-01B |
| ATT | 5313-I | A/CRF15-01B |
| ATT | 5123-I | A/CRF15-01B |
| ATT | 5248-I | CRF25-cpx |
| ATT | 5318-I | CRF25-cpx |
Discussion
We have analyzed the PR and RT regions of the pol gene of 271 sequences generated from plasma specimens collected from ART-naive HIV-1-infected patients initiating ART in Abuja, Nigeria in 2008. Phylogenetic analysis revealed that recombinant viruses have become the dominant strains over pure subtypes. The subtype G viruses have further diversified into three major subtype G clusters using worldwide sequences, and most Nigerian sequences from this study were closely related to two of the three clusters (Fig. 1, G1 and G3), indicating a close epidemiological relationship of HIV epidemics between Nigeria and countries in Europe, West Africa, and Asia during the periods of 1992–2004 (G1) and 2000–2009 (G3), respectively. We also noticed that the HIV-1 strains in the G2 cluster formed by sequences obtained from countries in Europe, East and West Africa, and South America from 1993 to 2009 had a minimal impact to the HIV epidemic in Nigeria, and strains in the two minor clusters, Gm1 and Gm2, appear to be indigenous viruses that had no relationship to the HIV subtype G strains circulating in other countries.
The three major subtype G clusters were reported previously as GWA-I, GWA-II, and GCA6,22,63 However, within a G cluster, different strains may be present. For example, in the G3 cluster, there are possibly four different strains, denoted as G3A–D (Fig. 1), while strain G3C and G3D seem to circulate only in Nigeria. Studies have suggested that subtype G emerged in Central Africa in 1968 (1956–1976), and between the middle and late 1970s two subtype G strains were probably introduced into Nigeria.63 Since then, they were disseminated locally and to neighboring countries, leading to the origin of two major western African clades (GWA-I and GWA-II). For the strains in the two minor clusters of Gm1 and Gm2, we have not identified any phylogenetic relationship of these strains to those from other countries (> 95% similarity) or to those detected in other regions of the country (data not shown), indicating that they might be newly emerging and locally circulating only in the Abuja area.
We also observed an evolved subtype distribution in the country. Prior to 2000, subtypes A (61%) and G (31%) were dominant in the south and north, respectively, and CRF02-AG was not detected in the country.18 However, after 2000, studies revealed that the prevalence of subtype A was greatly reduced with a slight reduction of subtype G and a remarkable increase of CRF02-AG (39–57%).16,34,38–41 In this study, we did not detect pure subtype A and had rates for subtype G (31.0%) and CRF02-AG (19.2%) detected in the capital area of Abuja similar to those detected in other areas, which is in agreement with the subtype distribution patterns reported in the country.16,34,38,39 Our data and other studies suggest that among the two previously dominant subtypes, one (subtype A) is disappearing and the other (subtype G) is still playing an important role but is circulating at different rates in the country. Meanwhile the progeny (CRF02-AG) of subtypes A and G are emerging quickly in the HIV epidemic in the country. In addition, CRF43-02G is becoming one of the dominant subtypes in Abuja, which was not reported in other areas of the country. Nevertheless, the majority of the current circulating HIV strains were derived from the subtypes or recombinant forms between subtypes A and G, again indicating localized transmission and circulation.
The overall pretreatment HIVDR rate in the cohort was less than 3%, which is expected due to the short time of ART implementation in the country at the time of the study. This rate is in agreement with the rates reported in Jos, in the Plateau State,40 and the North Central region of Nigeria. In the protease region, one sample displayed the K43T mutation, which was associated with a decreased virological response to tipranavir boosted with ritonavir (TPV/r) in the RESIST trials.64 One sample harbored the M46L mutation, which is a primary mutation inducing an intermediate level of resistance to NFV. A rare treatment-associated mutation K53Y, the polymorphic mutation A71V, as well as the PI-selected mutation I85V were also found in one sample each. Six specimens had the T74S mutation that induces a potential low level of resistance [classified as sensitive per the World Health Organization (WHO) interpretation]65 to PIs.
We also identified subtype/CRF-specific mutation patterns in the PR and RT gene regions. For instance, K20I in the PR region was present in all subtypes and CRFs except for eight sequences that were all subtype A-containing recombinants.34 The polymorphic mutations M36I, L63P, V82I, and M89I were proportionately present in the sequences of CRF02-AG, CRF43-02G, and CRF06-cpx and subtype G. A study has shown that the polymorphic mutation M36I in the PR region was associated with a higher rate of PI treatment failure.66 Furthermore, biochemical studies of this mutation with subtypes A and C recombinant I36 PR revealed that the PR enzymes of non-subtype B viruses have a higher inhibition constant (Ki) and a greater catalytic efficiency than subtype B viruses67 and that introduction of I36 into subtype B HIV-1 resulted in a higher virus replication capacity in both the absence and presence of PIs.7 In addition, we found that mutations E35D, N37DST, R57K, and K70R in the PR and D123N and I135TV in the RT were present at high rates in this cohort. These mutations have been shown to be associated with increased or decreased DR to some ARVs.68–70 Several other mutations observed in this study were not classically associated with resistance and their biological functionality remains to be elucidated. Since some of the uncommon mutations occurred at a very high frequency in a subtype/CRF-specific manner, they may warrant further investigation to determine their biological functions within the virus genome.
The analysis of the mutational pattern suggests that the V179I mutation in the RT is not only the preferred mutation in subtype B viruses, but also occurs in non-B subtypes and CRFs. Subtype G, CRF43-02G, and CRF06-cpx viruses utilized the codon GTG/GTA to encode valine and more likely to develop V179E mutations,16,34,38,39,71 whereas other CRF viruses are biased toward V179I and V179D. This pattern of amino acid substitution by a single nucleotide change is called quasisynonymy; it may explain the high rate of the V179E found in subtype G and CRF06-cpx in this cohort and is concordant with published data and statistics predicting the occurrence of this mutation at RT position 179 of these viruses.16,71,72
In summary, this study has shown an evolving HIV-1 epidemic in Abuja, with recombinant viruses becoming the prevailing strains and locally circulating strains emerging. This evolving trend in the HIV-1 epidemic deserves closer monitoring since a recent study has shown that a recombinant HIV-1 sub-subtype A3 and CRF02-AG virus was more virulent than the original viral strains.73 In addition, it has also confirmed that the occurrence of DR may be influenced by quasisynonymy and the genetic cost of mutations.
Acknowledgments
This research was supported by the President’s Emergency Plan for AIDS Relief (PEPFAR) through the Centers for Disease Control and Prevention.
The findings and conclusions in this report are those of the authors and do not necessarily represent the official position of the Centers for Disease Control and Prevention/the Agency for Toxic Substances and Disease Registry.
Footnotes
Sequence Data
The GenBank accession numbers for sequences analyzed in this study are JX083986–JX084256.
Author Disclosure Statement
No competing financial interests exist.
References
- 1.Chen J, Powell D, Hu W. High frequency of genetic recombination is a common feature of primate lentivirus replication. J Virol. 2006;80(19):9651–9658. doi: 10.1128/JVI.00936-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rhodes T, Wargo H, Hu WS. High rates of human immunodeficiency virus type 1 recombination: Near-random segregation of markers one kilobase apart in one round of viral replication. J Virol. 2003;77(20):11193–11200. doi: 10.1128/JVI.77.20.11193-11200.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Robertson DL, Anderson JP, Bradac JA, et al. HIV-1 nomenclature proposal. Science. 2000;288(5463):55–56. doi: 10.1126/science.288.5463.55d. [DOI] [PubMed] [Google Scholar]
- 4.Zhang M, Foley B, Schultz AK, et al. The role of recombination in the emergence of a complex and dynamic HIV epidemic. Retrovirology. 2010;7:25. doi: 10.1186/1742-4690-7-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hemelaar J, Gouws E, Ghys PD, Osmanov S. Global and regional distribution of HIV-1 genetic subtypes and recombinants in 2004. AIDS. 2006;20(16):W13–23. doi: 10.1097/01.aids.0000247564.73009.bc. [DOI] [PubMed] [Google Scholar]
- 6.Hemelaar J, Gouws E, Ghys PD, Osmanov S. Global trends in molecular epidemiology of HIV-1 during 2000– 2007. AIDS. 2011;25(5):679–689. doi: 10.1097/QAD.0b013e328342ff93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Holguin A, Sune C, Hamy F, et al. Natural polymorphisms in the protease gene modulate the replicative capacity of non-B HIV-1 variants in the absence of drug pressure. J Clin Virol. 2006;36(4):264–271. doi: 10.1016/j.jcv.2006.05.001. [DOI] [PubMed] [Google Scholar]
- 8.Kantor R. Impact of HIV-1 pol diversity on drug resistance and its clinical implications. Curr Opin Infect Dis. 2006;19(6):594–606. doi: 10.1097/QCO.0b013e3280109122. [DOI] [PubMed] [Google Scholar]
- 9.Kantor R, Katzenstein D. Polymorphism in HIV-1 non-subtype B protease and reverse transcriptase and its potential impact on drug susceptibility and drug resistance evolution. AIDS Rev. 2003;5(1):25–35. [PubMed] [Google Scholar]
- 10.Kantor R, Katzenstein DA, Efron B, et al. Impact of HIV-1 subtype and antiretroviral therapy on protease and reverse transcriptase genotype: Results of a global collaboration. PLoS Med. 2005;2(4):e112. doi: 10.1371/journal.pmed.0020112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kosakovsky Pond SL, Smith DM. Are all subtypes created equal? The effectiveness of antiretroviral therapy against non-subtype B HIV-1. Clin Infect Dis. 2009;48(9):1306–1309. doi: 10.1086/598503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Martinez-Cajas JL, Pai NP, Klein MB, Wainberg MA. Differences in resistance mutations among HIV-1 non-subtype B infections: A systematic review of evidence (1996–2008) J Int AIDS Soc. 2009;12(1):11. doi: 10.1186/1758-2652-12-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McCutchan FE. Understanding the genetic diversity of HIV-1. AIDS. 2000;14(Suppl 3):S31–44. [PubMed] [Google Scholar]
- 14.McCutchan FE, Carr JK, Bajani M, et al. Subtype G and multiple forms of A/G intersubtype recombinant human immunodeficiency virus type 1 in Nigeria. Virology. 1999;254(2):226–234. doi: 10.1006/viro.1998.9505. [DOI] [PubMed] [Google Scholar]
- 15.Ojesina AI, Chaplin B, Sankale JL, et al. Interplay of reverse transcriptase inhibitor therapy and gag p6 diversity in HIV type 1 subtype G and CRF02_AG. AIDS Res Hum Retroviruses. 2008;24(9):1167–1174. doi: 10.1089/aid.2007.0308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ojesina AI, Sankale JL, Odaibo G, et al. Subtype-specific patterns in HIV type 1 reverse transcriptase and protease in Oyo State, Nigeria: Implications for drug resistance and host response. AIDS Res Hum Retroviruses. 2006;22(8):770–779. doi: 10.1089/aid.2006.22.770. [DOI] [PubMed] [Google Scholar]
- 17.Paredes R, Lalama CM, Ribaudo HJ, et al. Pre-existing minority drug-resistant HIV-1 variants, adherence, and risk of antiretroviral treatment failure. J Infect Dis. 2010;201(5):662–671. doi: 10.1086/650543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Peeters M, Esu-Williams E, Vergne L, et al. Predominance of subtype A and G HIV type 1 in Nigeria, with geographical differences in their distribution. AIDS Res Hum Retroviruses. 2000;16(4):315–325. doi: 10.1089/088922200309197. [DOI] [PubMed] [Google Scholar]
- 19.Richman DD. Antiviral drug resistance. Antiviral Res. 2006;71(2–3):117–121. doi: 10.1016/j.antiviral.2006.03.004. [DOI] [PubMed] [Google Scholar]
- 20.Robertson DL, Sharp PM, McCutchan FE, Hahn BH. Recombination in HIV-1. Nature. 1995;374(6518):124–126. doi: 10.1038/374124b0. [DOI] [PubMed] [Google Scholar]
- 21.Sagay AS, Kapiga SH, Imade GE, et al. HIV infection among pregnant women in Nigeria. Int J Gynaecol Obstet. 2005;90(1):61–67. doi: 10.1016/j.ijgo.2005.03.030. [DOI] [PubMed] [Google Scholar]
- 22.Sankale JL, Langevin S, Odaibo G, et al. The complexity of circulating HIV type 1 strains in Oyo state, Nigeria. AIDS Res Hum Retroviruses. 2007;23(8):1020–1025. doi: 10.1089/aid.2006.0304. [DOI] [PubMed] [Google Scholar]
- 23.Soares EA, Makamche MF, Siqueira JD, et al. Molecular diversity and polymerase gene genotypes of HIV-1 among treatment-naive Cameroonian subjects with advanced disease. J Clin Virol. 2010;48(3):173–179. doi: 10.1016/j.jcv.2010.04.008. [DOI] [PubMed] [Google Scholar]
- 24.Sungkanuparph S, Oyomopito R, Sirivichayakul S, et al. HIV-1 drug resistance mutations among antiretroviral-naive HIV-1-infected patients in Asia: Results from the TREAT Asia Studies to Evaluate Resistance-Monitoring Study. Clin Infect Dis. 2011;52(8):1053–1057. doi: 10.1093/cid/cir107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tebit DM, Sangare L, Tiba F, et al. Analysis of the diversity of the HIV-1 pol gene and drug resistance associated changes among drug-naive patients in Burkina Faso. J Med Virol. 2009;81(10):1691–1701. doi: 10.1002/jmv.21600. [DOI] [PubMed] [Google Scholar]
- 26.Tong CY, Mullen J, Kulasegaram R, et al. Genotyping of B and non-B subtypes of human immunodeficiency virus type 1. J Clin Microbiol. 2005;43(9):4623–4627. doi: 10.1128/JCM.43.9.4623-4627.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ugbena RE, Wurie I. Emerging HIV-1 reverse transcriptase mutation patterns amongst antiretroviral therapy (ART)-naive patients in Abuja, federal capital of Nigeria—potential impact on treatment efficacy. Antivir Ther. 2009;14(Suppl 1):A196. [Google Scholar]
- 28.van de Vijver DA, Wensing AM, Angarano G, et al. The calculated genetic barrier for antiretroviral drug resistance substitutions is largely similar for different HIV-1 subtypes. J Acquir Immune Defic Syndr. 2006;41(3):352–360. doi: 10.1097/01.qai.0000209899.05126.e4. [DOI] [PubMed] [Google Scholar]
- 29.Vergne L, Peeters M, Mpoudi-Ngole E, et al. Genetic diversity of protease and reverse transcriptase sequences in non-subtype-B human immunodeficiency virus type 1 strains: Evidence of many minor drug resistance mutations in treatment-naive patients. J Clin Microbiol. 2000;38(11):3919–3925. doi: 10.1128/jcm.38.11.3919-3925.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vicente AC, Agwale SM, Otsuki K, et al. Genetic variability of HIV-1 protease from Nigeria and correlation with protease inhibitors drug resistance. Virus Genes. 2001;22(2):181–186. doi: 10.1023/a:1008123508416. [DOI] [PubMed] [Google Scholar]
- 31.Agwale SM, Zeh C, Robbins KE, et al. Molecular surveillance of HIV-1 field strains in Nigeria in preparation for vaccine trials. Vaccine. 2002;20(16):2131–2139. doi: 10.1016/s0264-410x(02)00059-2. [DOI] [PubMed] [Google Scholar]
- 32.Burda ST, Konings FA, Williams CA, et al. HIV-1 CRF09_cpx circulates in the North West Province of Cameroon where CRF02_AG infections predominate and recombinant strains are common. AIDS Res Hum Retroviruses. 2004;20(12):1358–1363. doi: 10.1089/aid.2004.20.1358. [DOI] [PubMed] [Google Scholar]
- 33.Butler IF, Pandrea I, Marx PA, Apetrei C. HIV genetic diversity: Biological and public health consequences. Curr HIV Res. 2007;5(1):23–45. doi: 10.2174/157016207779316297. [DOI] [PubMed] [Google Scholar]
- 34.Chaplin B, Eisen G, Idoko J, et al. Impact of HIV type 1 subtype on drug resistance mutations in Nigerian patients failing first-line therapy. AIDS Res Hum Retroviruses. 2011;27(1):71–80. doi: 10.1089/aid.2010.0050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ellenberger DL, Pieniazek D, Nkengasong J et al. Genetic analysis of human immunodeficiency virus in Abidjan, Ivory Coast reveals predominance of HIV type 1 subtype A and introduction of subtype G. AIDS Res Hum Retroviruses. 1999;15(1):3–9. doi: 10.1089/088922299311655. [DOI] [PubMed] [Google Scholar]
- 36.Esbjornsson J, Mild M, Mansson F, et al. HIV-1 molecular epidemiology in Guinea-Bissau, West Africa: Origin, demography and migrations. PLoS One. 2011;6(2):e17025. doi: 10.1371/journal.pone.0017025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Keele BF, Giorgi EE, Salazar-Gonzalez JF, et al. Identification and characterization of transmitted and early founder virus envelopes in primary HIV-1 infection. Proc Natl Acad Sci USA. 2008;105(21):7552–7557. doi: 10.1073/pnas.0802203105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ajoge HO, Gordon ML, de Oliveira T, et al. Genetic characteristics, coreceptor usage potential and evolution of Nigerian HIV-1 subtype G and CRF02_AG isolates. PLoS One. 2011;6(3):e17865. doi: 10.1371/journal.pone.0017865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ajoge HO, Gordon ML, Ibrahim S, et al. Drug resistance pattern of HIV type 1 isolates sampled in 2007 from therapy-naive pregnant women in North-Central Nigeria. AIDS Res Hum Retroviruses. 2012;28:115–118. doi: 10.1089/aid.2011.0115. [DOI] [PubMed] [Google Scholar]
- 40.Imade GE, Sagay AS, Chaplin B, et al. Transmitted HIV drug resistance in antiretroviral-naive pregnant women in north central Nigeria. AIDS Res Hum Retroviruses. 2014;30(2):127–133. doi: 10.1089/aid.2013.0074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hawkins CA, Chaplin B, Idoko J, et al. Clinical and genotypic findings in HIV-infected patients with the K65R mutation failing first-line antiretroviral therapy in Nigeria. J Acquir Immune Defic Syndr. 2009;52(2):228–234. doi: 10.1097/QAI.0b013e3181b06125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bessong PO. Polymorphisms in HIV-1 subtype C proteases and the potential impact on protease inhibitors. Trop Med Int Health. 2008;13(2):144–151. doi: 10.1111/j.1365-3156.2007.01984.x. [DOI] [PubMed] [Google Scholar]
- 43.Champenois K, Bocket L, Deuffic-Burban S, et al. Expected response to protease inhibitors of HIV-1 non-B subtype viruses according to resistance algorithms. AIDS. 2008;22(9):1087–1089. doi: 10.1097/QAD.0b013e3282ff629b. [DOI] [PubMed] [Google Scholar]
- 44.Champenois K, Deuffic-Burban S, Cotte L, et al. Natural polymorphisms in HIV-1 protease: Impact on effectiveness of a first-line lopinavir-containing antiretroviral therapy regimen. J Med Virol. 2008;80(11):1871–1879. doi: 10.1002/jmv.21315. [DOI] [PubMed] [Google Scholar]
- 45.Lawrence P, Lutz MF, Saoudin H, et al. Analysis of polymorphism in the protease and reverse transcriptase genes of HIV type 1 CRF02-AG subtypes from drug-naive patients from Saint-Etienne, France. J Acquir Immune Defic Syndr. 2006;42(4):396–404. doi: 10.1097/01.qai.0000221675.83950.4a. [DOI] [PubMed] [Google Scholar]
- 46.Sagoe KW, Dwidar M, Lartey M, et al. Variability of the human immunodeficiency virus type 1 polymerase gene from treatment naive patients in Accra, Ghana. J Clin Virol. 2007;40(2):163–167. doi: 10.1016/j.jcv.2007.07.016. [DOI] [PubMed] [Google Scholar]
- 47.Brenner BG. Resistance and viral subtypes: How important are the differences and why do they occur? Curr Opin HIV AIDS. 2007;2(2):94–102. doi: 10.1097/COH.0b013e32801682e2. [DOI] [PubMed] [Google Scholar]
- 48.Martinez-Cajas JL, Pant-Pai N, Klein MB, Wainberg MA. Role of genetic diversity amongst HIV-1 non-B subtypes in drug resistance: A systematic review of virologic and biochemical evidence. AIDS Rev. 2008;10(4):212–223. [PubMed] [Google Scholar]
- 49.Descamps D, Collin G, Letourneur F, et al. Susceptibility of human immunodeficiency virus type 1 group O isolates to antiretroviral agents: In vitro phenotypic and genotypic analyses. J Virol. 1997;71(11):8893–8898. doi: 10.1128/jvi.71.11.8893-8898.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tuaillon E, Gueudin M, Lemee V, et al. Phenotypic susceptibility to nonnucleoside inhibitors of virion-associated reverse transcriptase from different HIV types and groups. J Acquir Immune Defic Syndr. 2004;37(5):1543–1549. doi: 10.1097/00126334-200412150-00001. [DOI] [PubMed] [Google Scholar]
- 51.Brenner BG, Oliveira M, Doualla-Bell F, et al. HIV-1 subtype C viruses rapidly develop K65R resistance to tenofovir in cell culture. AIDS. 2006;20(9):F9–13. doi: 10.1097/01.aids.0000232228.88511.0b. [DOI] [PubMed] [Google Scholar]
- 52.Maiga AI, Malet I, Soulie C, et al. Genetic barriers for integrase inhibitor drug resistance in HIV type-1 B and CRF02_AG subtypes. Antivir Ther. 2009;14(1):123–129. [PubMed] [Google Scholar]
- 53.Loemba H, Brenner B, Parniak MA, et al. Genetic divergence of human immunodeficiency virus type 1 Ethiopian clade C reverse transcriptase (RT) and rapid development of resistance against nonnucleoside inhibitors of RT. Antimicrob Agents Chemother. 2002;46(7):2087–2094. doi: 10.1128/AAC.46.7.2087-2094.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ugbena R, Diallo K, Bassey O, et al. Virological response and HIV-1 drug resistance development profile among patients treated with the first-line antiretroviral regimens in Nigeria. Clin Infect Dis. 2012;54(Suppl 11):S375–S380. doi: 10.1093/cid/cir1064. [DOI] [PubMed] [Google Scholar]
- 55.Yang C, McNulty A, Diallo K, et al. Development and application of a broadly sensitive dried-blood-spot-based genotyping assay for global surveillance of HIV-1 drug resistance. J Clin Microbiol. 2010;48(9):3158–3164. doi: 10.1128/JCM.00564-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Woods CK, Brumme CJ, Liu TF, et al. Automating HIV drug resistance genotyping with RECall, a freely accessible sequence analysis tool. J Clin Microbiol. 2012;50(6):1936–1942. doi: 10.1128/JCM.06689-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tamura K, Dudley J, Nei M, Kumar SL. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24(8):1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- 58.Hills DM, Bull JJ. An empirical test of bootstrapping as a method for assessing confidence in phylogenetic analysis. System Biol. 1993;42:182–192. [Google Scholar]
- 59.Lole KS, Bollinger RC, Paranjape RS, et al. Full-length human immunodeficiency virus type 1 genomes from subtype C-infected seroconverters in India, with evidence of intersubtype recombination. J Virol. 1999;73(1):152–160. doi: 10.1128/jvi.73.1.152-160.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Huelsenbeck JP, Ronquist F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics. 2001;17(8):754–755. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
- 61.Bennett DE, Camacho RJ, Otelea D, et al. Drug resistance mutations for surveillance of transmitted HIV-1 drug-resistance: 2009 update. PLoS One. 2009;4(3):e4724. doi: 10.1371/journal.pone.0004724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gifford RJ, Liu TF, Rhee SY, et al. The calibrated population resistance tool: Standardized genotypic estimation of transmitted HIV-1 drug resistance. Bioinformatics. 2009;25(9):1197–1198. doi: 10.1093/bioinformatics/btp134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Delatorre E, Mir D, Bello G. Spatiotemporal dynamics of the HIV-1 subtype G epidemic in West and Central Africa. PLoS One. 2014;9(2):e98908. doi: 10.1371/journal.pone.0098908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rhee SY, Gonzales MJ, Kantor R, et al. Human immunodeficiency virus reverse transcriptase and protease sequence database. Nucleic Acids Res. 2003;31(1):298–303. doi: 10.1093/nar/gkg100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Shafer RW, Rhee SY, Bennett DE. Consensus drug resistance mutations for epidemiological surveillance: Basic principles and potential controversies. Antiviral Ther. 2008;13(Suppl 2):59–68. [PMC free article] [PubMed] [Google Scholar]
- 66.Perno CF, Cozzi-Lepri A, Forbici F, et al. Minor mutations in HIV protease at baseline and appearance of primary mutation 90M in patients for whom their first protease-inhibitor antiretroviral regimens failed. J Infect Dis. 2004;189(11):1983–1987. doi: 10.1086/386307. [DOI] [PubMed] [Google Scholar]
- 67.Velazquez-Campoy A, Todd MJ, Vega S, Freire E. Catalytic efficiency and vitality of HIV-1 proteases from African viral subtypes. Proc Natl Acad Sci USA. 2001;98(11):6062–6067. doi: 10.1073/pnas.111152698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Abecasis AB, Deforche K, Bacheler LT, et al. Investigation of baseline susceptibility to protease inhibitors in HIV-1 subtypes C, F, G and CRF02_AG. Antivir Ther. 2006;11(5):581–589. [PubMed] [Google Scholar]
- 69.Abecasis AB, Deforche K, Snoeck J, et al. Protease mutation M89I/V is linked to therapy failure in patients infected with the HIV-1 non-B subtypes C, F or G. AIDS. 2005;19(16):1799–1806. doi: 10.1097/01.aids.0000188422.95162.b7. [DOI] [PubMed] [Google Scholar]
- 70.Masquelier B, Droz C, Dary M, et al. R57K polymorphism in the human immunodeficiency virus type 1 protease as predictor of early virological failure in a cohort of antiretroviral-naive patients treated mostly with a nelfinavir-containing regimen. Antimicrob Agents Chemother. 2003;47(11):3623–3626. doi: 10.1128/AAC.47.11.3623-3626.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Dumans AT, Soares MA, Machado ES, et al. Synonymous genetic polymorphisms within Brazilian human immunodeficiency virus type 1 subtypes may influence mutational routes to drug resistance. J Infect Dis. 2004;189(7):1232–1238. doi: 10.1086/382483. [DOI] [PubMed] [Google Scholar]
- 72.Kijak GH, Currier JR, Tovanabutra S, et al. Lost in translation: implications of HIV-1 codon usage for immune escape and drug resistance. AIDS Rev. 2004;6(1):54–60. [PubMed] [Google Scholar]
- 73.Palm AA, Esbjornsson J, Mansson F, et al. Faster progression to AIDS and AIDS-related death among seroincident individuals infected with recombinant HIV-1 A3/CRF02_AG compared with sub-subtype A3. J Infect Dis. 2014;209(5):721–728. doi: 10.1093/infdis/jit416. [DOI] [PubMed] [Google Scholar]


