Abstract
Infection with mosquito-borne West Nile virus (WNV) is usually asymptomatic but can lead to severe WNV encephalitis. The innate cytokine, macrophage migration inhibitory factor (MIF), is elevated in patients with WNV encephalitis and promotes viral neuroinvasion and mortality in animal models. In a case-control study, we examined functional polymorphisms in the MIF locus in a cohort of 454 North American patients with neuroinvasive WNV disease and found patients homozygous for high-expression MIF alleles to be >20-fold (p=0.008) more likely to have WNV encephalitis. These data indicate that MIF is an important determinant of severity of WNV neuropathogenesis and may be a therapeutic target.
Keywords: West Nile virus, mosquito vector, immune response, macrophage, neurologic disease
1. Introduction
West Nile virus (WNV) is a mosquito-borne enveloped positive-strand RNA virus that has caused the largest arboviral neuroinvasive disease outbreaks in the US and is now the country’s leading cause of arthropod-borne encephalitis (1). An estimated 3 million people in the USA may have been infected since 1999, resulting in more than 1,700 fatalities (2). While WNV infections are predominantly subclinical, clinical manifestations may range from fever and myalgias to encephalitis and death. Less than one percent of infected individuals develop neuroinvasive complications and the likelihood of adverse outcomes (death, admission to long-term care facilities) has been related to developing encephalitis, advanced age, immunosuppression, presence of comorbid medical conditions, and perhaps individual differences in elements of the immune response (3, 4). Investigating WNV infection and its clinical complications is important because there are no human vaccines available, and the only treatment for patients is supportive care (3).
Host factors mediate neuroinvasive WNV disease and genetic predisposition for contracting neuroinvasive WNV has been considered to play a role in disease outcome after infection. Recent studies show that permeability of the blood brain barrier, which is enhanced by pro-inflammatory responses, appear to be critical to susceptibility of neuroinvasive WNV infection (3). The use of experimental animal models, especially genetically modified mice with defects in specific immune system molecules, has provided important insight into the immunopathogenesis of WNV. Macrophage Migration Inhibitory Factor (MIF) is an upstream mediator of innate immunity that plays a key role in the balance between a protective immune response and inflammatory pathology. MIF expression is increased in WNV-infected mice and genetic deletion, immuoneutralization, or small molecule antagonism of MIF reduces viral load and inflammatory responses in the brain, and renders mice resistant to WNV lethality (5).
There are functional polymorphisms in the human MIF gene that separate the population into high and low MIF producers, and these polymorphisms correlate with disease outcome in patients suffering from the inflammatory sequelae of different autoimmune or infectious conditions (6). The best characterized polymorphism is a tetranucleotide repeat (CATT5–8) at −794 position in the MIF promoter, where the 5-CATT variant is a low expression allele and the >5-CATT variants are higher expression alleles. A single nucleotide polymorphism (SNP) at −173(G/C) of the MIF promoter also has been the focus of some studies, with the C allele considered high expression and found in linkage disequilibrium with the high expression, −794 7-CATT allele (7). Increased MIF expression and corresponding high-expression MIF alleles are well known to have a deleterious effect for patients suffering from autoimmune conditions such as systemic lupus erythematosus and rheumatoid arthritis as well as those with bacterial sepsis (6). We previously reported that patients suffering from acute WNV infection presented with increased levels of MIF in serum and in cerebrospinal fluid (5). In this work we explore genetic polymorphisms in MIF in patients with different manifestations of neuroinvasive WNV disease.
2. Material and Methods
Human Cohorts
The cohort of patients with WNV infection from 6 sites in North America was enrolled previously over a 5 year period (2004–2009), with approved consent as described (8). Study subjects were >age 18, Caucasian, and had a history of infection with WNV based on laboratory diagnostic criteria from the Centers for Disease Control (8). Subjects were separated into cases meeting the criteria for neuroinvasive disease (including meningitis, encephalitis, or acute flaccid paralysis) and controls (WNV infection without neuroinvasive disease). Details of demographic criteria (age, gender) and clinical symptoms were reported previously for both the primary and replication sample cohorts (8). For the current study, DNA was available from a subset of subjects, from 518 cases (85%) and 514 controls (52%).
MIF genotyping
DNA was isolated from peripheral blood samples using a commercially-available kit (Invitrogen). Analysis of the MIF promoter −794 CATT5–8 microsatellite [rs5844572] and −173 G/C SNP [rs755622] was performed as previously described (9) with 909 successfully genotyped from 1032 samples attempted (88% of subjects in both groups).
MIF Promoter Experiments
A Dual-luciferase reporter assay system was employed (Promega) where 106 human THP-1 monocytes (ATCC) were cultured in RPMI1640/10% fetal bovine serum (FBS) and co-transfected with 1 μg of individual plasmids containing −794 5–8-CATT (or 0-CATT) MIF promoter sequences ligated to a luciferase gene together with a β-actin promoter luciferase control using the Amaxa nucleofactor platform (Lonza) (6). The SNP status of these MIF constructs, in accordance with the most commonly occurring genotypes, were: 5G, 6G, 7C, and 8G. The cells were incubated at 37° C for 24 hrs, infected with virulent WNV strain CT-2741 at a multiplicity of infection (MOI) of 1 (10), and luciferase activity quantified after 48 hrs. The luciferase activity was measured by following the protocol “Dual-luciferase Report Assay System” (Promega).
Statistics
Demographic differences in the enrolled WNV cohort between neuroinvasive disease and controls without neuroinvasive disease were analyzed using Student’s t-test. The proportion of MIF genotypic low-expressers in the cases and controls and among cases, those with encephalitis and those without, were compared by chi square analysis. MIF expression and stimulated MIF responses were compared by Student’s t-test with p<0.05 considered significant.
3. Results
MIF polymorphisms in WNV patients
We used a case-control study design to investigate the frequency of MIF promoter polymorphisms in a cohort of WNV patients recruited from the US and Canada and divided into cases, patients with neuroinvasive WNV disease (N = 518), and WNV-infected controls without neuroinvasion (N = 514) (8). The distribution of clinical findings indicated that case patients were more likely to be male (cases: 50.8% male vs. controls: 44.5% male, p=0.04) and older (cases: mean age 60.9 ± 15.0 vs. controls: 53.6 ± 13.6, p<0.001)] when compared to controls. Among patients with neuroinvasive WNV disease in whom clinical classification was available, severity was distributed across a spectrum that included meningitis (M; n=83, 16.3%), meningoencephalitis (ME; n=138, 27.1%), encephalitis (E; n=100, 19.6%), and acute flaccid paralysis (AFP; n=188, 36.9%). There were no differences in distribution of gender among patients with different neuroinvasive manifestations of WNV disease, but patients with encephalitis were older than their counterparts with other types of neuroinvasive disease (encephalitis: 65.5 ± 14.8 vs. other disease: 60.0 ± 14.9, p=0.0026).
MIF alleles were analyzed in genomic DNA samples from the cases and controls to ascertain the −794 CATT repeat number and the −173 G/C SNP. Since MIF alleles are ethnically stratified, this study was limited to Caucasian subjects. No data was available on relatedness of study subjects, but as the cohort was collected over a wide geographic area, relatedness was considered to be unlikely. Genotype data was available from 454 case patients and 455 controls (88% of subjects in both groups). Neither group deviated from Hardy-Weinberg Equilibrium. There were no differences between the overall −794 CATT and −173 G/C distributions between cases and controls (case: 5-CATT – 24.6%, 6-CATT - 63.2%, 7-CATT - 12.3% and control: 5-CATT – 23.3%, 6-CATT – 62%, 7-CATT – 14.6%, p=0.4) and (case: G – 81.8%, C – 18.2% and control: G – 82.3%, C – 18.3%, p=0.5).
We next investigated the relationship between these MIF polymorphisms and manifestations of neuroinvasive disease. In comparing the frequency distribution of the possible −794 CATT alleles among the clinically defined neuroinvasive manifestations in the cases, we found the low-expresser 5-CATT allele to be less common among encephalitis cases (Table 1). This difference was significant when comparing encephalitis (E) cases with all other neuroinvasive manifestations (19.9% vs. 25.5%) as well as with meningitis and meningoencephalitis (M + ME) cases (19.9% vs. 25.3%). Patients with encephalitis also were more likely to carry a high expression MIF 7-CATT allele compared to those with all other neuroinvasive disease as well as with M+ME [18% vs. 11%, 0.037, OR 1.73CI (1.07–2.79), and 18% vs. 10% 0.027, OR 1.89 (1.10–3.21)]. Finally, the homozygous 7/7-CATT MIF high-expresser genotype was 12-fold more common among encephalitis (E) cases when compared with meningitis and meningoencephalitis (M + ME) cases (p=0.0174), and approached but did not reach statistical significance when compared to all other disease (p=0.08). This difference also was significant in multivariate analysis accounting for age and gender (p=0.0084, OR: 21.54 [2.20–210.90]). Although the −173 C SNP is in linkage disequilibrium with the 7-CATT allele, no association between the SNP and neuroinvasive disease were observed. This is the first report of an association between the MIF −794 CATT polymorphisms and WNV encephalitis.
Table 1.
The MIF -794 CATT5–8 promoter variants correlate with severity of WNV disease.
| E (%) n=73 |
All Other Disease (%) n=381 |
p-value, OR (CI) | M+ME (%) n=204 |
p-value, OR (CI) | |
|---|---|---|---|---|---|
| Alleles | |||||
| 5-CATT | 29 (19.86%) | 194 (25.46%) | 0.049 | 103 (25.25%) | 0.042 |
| 6-CATT | 91 (62.33%) | 483 (63.39%) | 263 (64.46%) | ||
| 7-CATT | 26 (17.81%) | 85 (11.5%) | 42 (10.29%) | ||
| Groups | |||||
| 7-CATT | 26 (17.81%) | 85 (11.15%) |
0.037, OR 1.73 CI (1.07–2.79) |
42 (10.29%) | 0.027, OR 1.89 (1.10–3.21) |
| All Others | 120 (82.19%) | 677 (88.85%) | 366 (89.71%) | ||
| Genotype | |||||
| 77-CATT | 4 (5.5%) | 7 (1.83%) | 0.0833 | 1 (0.5%) | 0.0174, 11.94 (1.32–108.70) |
| All other genotypes | 69 (94.5%) | 374 (98.2%) | 203 (99.5%) | ||
| Adjusted for Age/ Gender | |||||
| 0.0084, 21.54 (2.20, 210.90) |
In comparing the distribution of different CATT alleles among the different disease manifestations in the cases, the low-expresser 5-CATT allele was less common among encephalitis (E) cases, and the high-expresser 7-CATT allele was more common. This difference was significant comparing encephalitis (E) with all other neuroinvasive disease as well as with meningitis and meningoenceophaltitis alone (M + ME).
WNV induces allele-dependent MIF expression in human monocytes
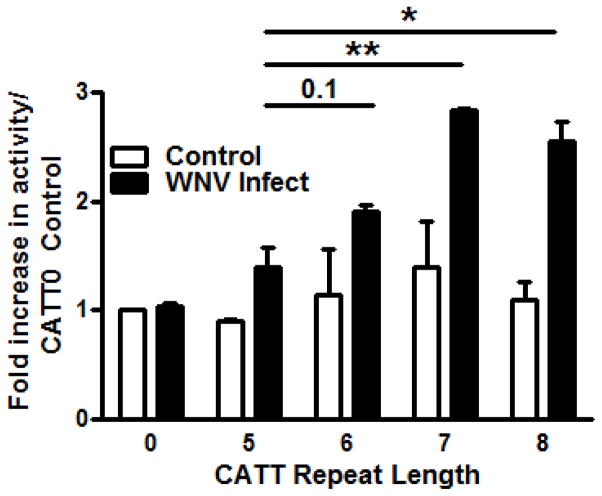
Experimental WNV infection induces MIF expression in mice and genetic Mif deletion, anti-MIF, or pharmacologic MIF antagonism protects mice from lethal neuroinvasive disease (5). Whether WNV directly induces MIF expression or if the increased MIF observed during infection is simply a consequence of downstream inflammatory end-organ damage is unknown. We examined the ability of WNV to directly induce MIF expression by measuring the impact of initial infection on the transcriptional activity of human monocytes transfected with individual −794 CATT5–8 MIF promoter luciferase fusion plasmids. Infection with WNV led to increased expression of MIF in a CATT length-dependent manner (Figure 1). The differences between the 6-, 7-, or 8-CATT promoter variants were statistically significant when compared to the lowest expression 5-CATT promoter allele. Notably, maximal transcriptional induction was observed with the 7-CATT MIF allele.
Figure 1. WNV induces MIF expression in a.
−794 CATT5–8 length-dependent manner in human monocytes.
Cultured human THP-1 monocytes were transfected with MIF promoter/luciferase reporter fusion plasmids bearing 0, 5, 6, 7, and 8 CATT repeats and infected with WNV in vitro for 48 hrs prior to assessment of luciferase activity. Methods follow ref; (10). Data are means ± SD for 4 determination. * p< 0.05, ** p< 0.01 for comparisons to 5-CATT.
4. Discussion
Genome-wide association studies of single nucleotide polymorphisms (SNP) have identified key regulators of immune function – including interferon pathway elements such as IRF3, OAS-1, and MX-1 – to be associated with an increased risk for initial infection with WNV and severe neurological disease (>750 subjects) (11, 12). A dominant negative splice variant of RNaseL, which functions in the anti-proliferative roles of interferon, was detected more often in WNV cases than control patients (13).
In addition to interferon pathway elements, other immune regulators have been identified that influence the severity of infection with WNV. In particular, HLA types have been associated either with risk of more severe outcome (HLA-A*68 and C*08) or better resistance (B*40 and C*03) (14). A study reported differential frequency of Natural Killer cell receptors (KIRs) between WNV patients and a healthy cohort in Macedonia (patients n=4 and healthy controls n=214) (15). Homozygous deletion in the CCR5 gene (CCR5Δ32), which confers protection to HIV infection, also is a risk factor for severe WNV disease (16). In addition, SNPs in the RFC1, SCN1A and ANPEP genes have been identified as risk candidates but the immunological mechanisms linking these loci with disease outcome are not known (8).
While we did not find the distribution of MIF polymorphisms to be different among patients with WNV non-neuroinvasive infection vs. those with neuroinvasive disease, we did find the high MIF expresser 7-CATT allele to be significantly correlated with the severe WNV clinical manifestations. Carriage of the homozygous 7/7-CATT MIF genotype conferred a 20-fold increase in risk for encephalitis compared to meningitis and meningoencephalitis. Together, these results suggest that the propensity to express high levels of MIF does not predispose a person to develop neuroinvasive sequealae but that once these sequelae develop, it is associated with progression to the more severe and lethal complications of the infection. Higher levels of MIF can cause a patient to experience a more severe clinical manifestation of neuroinvasive WNV, such that the high expressers are more likely to experience encephalitis after neuroinvasion than low expressers. The role of variant MIF alleles in the clinical outcome of viral infection has not been shown previously; however, these results are in accord with published observations in meningococcal and pneumococcal meningitis, where high-expression MIF alleles were associated with the development of invasive disease (17, 18). The lack of association between the −173 G/C SNP and neuroinvasive WNV disease may be explained by the relatively small size of the cohorts. Additional evaluation of both the MIF −794 CATT and the −173 SNP in larger cohorts of subjects with neuroinvasive WNV disease is necessary to replicate these findings.
The present findings support a concordance in human infection in the relationship between WNV infection and MIF expression reported previously in mouse models (5). MIF may be of special importance in the context of neurotrophic viruses, inflammation, and blood brain barrier permeability and a critical effector cytokine because of its role in sustaining inflammatory responses at sites of tissue invasion (17, 18). The functional MIF polymorphisms studied herein occur commonly in the population and are established risk factors for inflammatory end-organ damage in autoimmunity as well as with the development of gram-negative and mycobacterial bacteremia (7). Additionally, pharmacologic MIF inhibition has been shown to attenuate morbidity and mortality due to overwhelming inflammation in pneumococcal pneumonia (19). Although WNV disease morbidity and mortality occurs in only a small proportion of those infected with virus, there are currently no effective vaccines or specific antiviral therapies. The present findings indicate that MIF genotype determination in WNV disease may provide useful prognostic information and that MIF inhibition, which is currently also in clinical testing, may be of therapeutic value in patients at risk for encephalitis.
Highlights.
West Nile virus infection is usually asymptomatic but may cause severe encephalitis
The cytokine macrophage migration inhibitory factor is elevated in WNV encephalitis
Patients with high-expression MIF alleles are more likely to have WNV encephalitis
MIF genotype in WNV patients may be useful for prognostic or therapeutic targets
Acknowledgments
This work was supported in part by awards from the National Institutes of Health (N01-HHSN272201100019C, RO1AI042310, F32 AI085712, U19 AI089992 and K08 AI097223). Conflicts that the editors consider relevant to the content of the manuscript have been disclosed. None of the authors have any commercial or other association that may pose a conflict of interest for this work. An earlier version of this work was presented in poster form at the meeting of the Human Immunology Project Consortium (HIPC). The authors are grateful to members of their laboratories for helpful discussions and to the study subjects who provided samples for analysis.
Abbreviations
- MIF
Macrophage Migration Inhibitory Factor
- WNV
West Nile virus
- SNP
single nucleotide polymorphism
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Literature Cited
- 1.Lindsey NP, Lehman JA, Staples JE, Fischer M. West Nile Virus and Other Arboviral Diseases — United States, 2013. MMWR Morb Mortal Wkly Rep. 2014;63:521–526. [PMC free article] [PubMed] [Google Scholar]
- 2.Petersen LR, Carson PJ, Biggerstaff BJ, Custer B, Borchardt SM, Busch MP. Estimated cumulative incidence of West Nile virus infection in US adults, 1999–2010. Epidemiol Infect. 2013;141:591–595. doi: 10.1017/S0950268812001070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cho H, Diamond MS. Immune responses to West Nile virus infection in the central nervous system. Viruses. 2012;4:3812–3830. doi: 10.3390/v4123812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tyler KL, Pape J, Goody RJ, Corkill M, Kleinschmidt-DeMasters BK. CSF findings in 250 patients with serologically confirmed West Nile virus meningitis and encephalitis. Neurology. 2006;66:361–365. doi: 10.1212/01.wnl.0000195890.70898.1f. [DOI] [PubMed] [Google Scholar]
- 5.Arjona A, Foellmer H, Town T, Leng L, McDonald C, Wong S, Montgomery RR, Fikrig E, Bucala R. Abrogation of Macrophage Migration Inhibitory Factor Decreases West Nile Virus Lethality by Limiting Viral Neuroinvasion. J Clin Invest. 2007;117:3059–3066. doi: 10.1172/JCI32218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Das R, Subrahmanyan L, Yang I, van Duin D, Levy R, Piecychna M, Leng L, Montgomery RR, Shaw AC, Schwartz D, Bucala R. Functional Macrophage Migration Inhibitory Factor (MIF) polymorphisms are associated with gram-negative bacteremia in older adults. J Infect Dis. 2014;209:764–768. doi: 10.1093/infdis/jit571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Baugh JA, Chitnis S, Donnelly SC, Monteiro J, Lin X, Plant BJ, Wolfe F, Gregersen PK, Bucala R. A functional promoter polymorphism in the macrophage migration inhibitory factor (MIF) gene associated with disease severity in rheumatoid arthritis. Genes Immun. 2002;3:170–176. doi: 10.1038/sj.gene.6363867. [DOI] [PubMed] [Google Scholar]
- 8.Loeb M, Eskandarian S, Rupp M. Genetic variants and susceptibility to neurological complications following West Nile virus infection. J Infect Dis. 2011;204:1031–1037. doi: 10.1093/infdis/jir493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhong XB, Leng L, Beitin A, Chen R, McDonald C, Hsiao B, Jenison RD, Kang I, Park SH, Lee A, Gregersen P, Thuma P, Bray-Ward P, Ward DC, Bucala R. Simultaneous detection of microsatellite repeats and SNPs in the macrophage migration inhibitory factor (MIF) gene by thin-film biosensor chips and application to rural field studies. Nucleic Acids Res. 2005;33:e121. doi: 10.1093/nar/gni123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Qian F, Goel G, Meng H, Wang X, You F, Devine L, Raddassi K, Garcia MN, Murray KO, Bolen CR, Gaujoux R, Shen-Orr SS, Hafler D, Fikrig E, Xavier RJ, Kleinstein SH, Montgomery RR. Systems Immunology reveals markers of susceptibility to West Nile virus infection. Clin Vacc Immunol. 2015;22:6–16. doi: 10.1128/CVI.00508-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lim JK, Lisco A, McDermott DH, Huynh L, Ward JM, Johnson B, Johnson H, Pape J, Foster GA, Krysztof D, Follmann D, Stramer SL, Margolis LB, Murphy PM. Genetic variation in OAS1 is a risk factor for initial infection with West Nile virus in man. PLoS Pathog. 2009;5:e1000321. doi: 10.1371/journal.ppat.1000321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bigham AW, Buckingham KJ, Husain S, Emond MJ, Bofferding KM, Gildersleeve H, Rutherford A, Astakhova NM, Perelygin AA, Busch MP, Murray KO, Sejvar JJ, Green S, Kriesel J, Brinton MA, Bamshad M. Host genetic risk factors for West Nile virus infection and disease progression. PLoS One. 2011;6:e24745. doi: 10.1371/journal.pone.0024745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yakub I, Lillibridge KM, Moran A, Gonzalez OY, Belmont J, Gibbs RA, Tweardy DJ. Single nucleotide polymorphisms in genes for 2′-5′-oligoadenylate synthetase and RNase L inpatients hospitalized with West Nile virus infection. J Infect Dis. 2005;192:1741–1748. doi: 10.1086/497340. [DOI] [PubMed] [Google Scholar]
- 14.Lanteri MC, Kaidarova Z, Peterson T, Cate S, Custer B, Wu S, Agapova M, Law JP, Bielawny T, Plummer F, Tobler LH, Loeb M, Busch MP, Bramson J, Luo M, Norris PJ. Association between HLA class I and class II alleles and the outcome of West Nile virus infection: an exploratory study. PLoS One. 2011;6:e22948. doi: 10.1371/journal.pone.0022948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Spiroski M, Milenkovic Z, Petlichkovski A, Ivanovski L, Topuzovska IK, Djulejic E. Killer cell immunoglobulin-like receptor genes in four human West Nile virus infections reported 2011 in the Republic of Macedonia. Hum Immunol. 2013;74:389–394. doi: 10.1016/j.humimm.2012.11.015. [DOI] [PubMed] [Google Scholar]
- 16.Glass WG, McDermott DH, Lim JK, Lekhong S, Yu SF, Frank WA, Pape J, Cheshier RC, Murphy PM. CCR5 deficiency increases risk of symptomatic West Nile virus infection. J Exp Med. 2006;203:35–40. doi: 10.1084/jem.20051970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Doernberg S, Schaaf B, Dalhoff K, Leng L, Beitin A, Quagliarello V, Bucala R. Association of macrophage migration inhibitory factor (MIF) polymorphisms with risk of meningitis from Streptococcus pneumoniae. Cytokine. 2011;53:292–294. doi: 10.1016/j.cyto.2010.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Renner P, Roger T, Bochud PY, Sprong T, Sweep FC, Bochud M, Faust SN, Haralambous E, Betts H, Chanson AL, Reymond MK, Mermel E, Erard V, van Deuren M, Read RC, Levin M, Calandra T. A functional microsatellite of the macrophage migration inhibitory factor gene associated with meningococcal disease. FASEB J. 2012;26:907–916. doi: 10.1096/fj.11-195065. [DOI] [PubMed] [Google Scholar]
- 19.Weiser JN, Roche AM, Hergott CB, LaRose MI, Connolly T, Jorgensen WL, Leng L, Bucala R, Das R. Macrophage Migration Inhibitory Factor Is Detrimental in Pneumococcal Pneumonia and a Target for Therapeutic Immunomodulation. J Infect Dis. 2015 doi: 10.1093/infdis/jiv262. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]