Abstract
Pro-angiogenic drugs hold great potential to promote reperfusion of ischemic tissues and in tissue engineering applications, but efficacy is limited by poor targeting and short half-lives. Methods to control release duration or provide enzymatically-responsive drug delivery have independently improved drug efficacy. However, no material has been developed to temporally control the rate of enzymatically-responsive drug release. To address this void, hydrogels were developed to provide sustained, tunable release of Qk, a pro-angiogenic peptide mimic of vascular endothelial growth factor, via tissue-specific enzymatic activity. After confirmation that sustained delivery of Qk is necessary for pro-angiogenic effects, a variety of previously-identified matrix metalloproteinase (MMP)-degradable linkers were used to tether Qk to hydrogels. Of these, three (IPES↓LRAG, GPQG↓IWGQ, and VPLS↓LYSG) showed MMP-responsive peptide release. These linkers provided tunable Qk release kinetics, with rates ranging from 1.64 to 19.9 × 10−3 hours−1 in vitro and 4.82 to 8.94 × 10−3 hours−1 in vivo. While Qk was confirmed to be bioactive as released, hydrogels releasing Qk failed to induce significant vascularization in vivo after one week, likely due to non-enzymatically degradable hydrogels employed. While Qk was the focus of this study, the approach could easily be adapted to control the delivery of a variety of therapeutic molecules.
Keywords: drug delivery, hydrogel, stimuli-responsive materials, therapeutic peptides, temporal control
1. Introduction
Pro-angiogenic therapies hold great potential for treatment of ischemic tissue disorders[1] and in tissue engineering applications, where tissues larger than 100–200 µm in any dimension require supporting vasculature.[2] Delivery of pro-angiogenic proteins[3] and peptides[4] have shown promising therapeutic results. Peptides offer advantages over proteins as they do not require complex tertiary structure for bioactivity, and due to small molecular weights, can be produced synthetically and delivered at higher concentrations to target tissues.[4a] While peptides do not always fully recapitulate the bioactivity of the proteins they mimic, some, such as the vascular endothelial growth factor (VEGF) mimic Qk, show comparable bioactivity to protein counterparts.[4] However, when simply injected in vivo, both proteins and peptides suffer from poor targeting and short half-lives due to rapid clearance and degradation, necessitating the use of controlled release systems.[5]
VEGF is one of the most common pro-angiogenic proteins exploited for therapeutic angiogenesis.[3, 6] However, tight spatio-temporal control over VEGF presentation is required to exploit its full potential and prevent the formation of immature, leaky vasculature.[3, 6e] Poly(lactic–co-glycolic acid)[6c, 6d] and alginate[3] materials have successfully extended the duration of encapsulated VEGF delivery through hindered diffusion or upon hydrolytic degradation, increasing tissue vascularization. However, these approaches deliver VEGF over pre-dictated, and not necessarily therapeutically-relevant, time frames. Alternately, enzymatically-responsive hydrogels that release VEGF in response to local matrix metalloproteinase (MMP) expression have been developed.[6a, 6b] These materials are based on the theory that micro-environmental concentrations of VEGF are critical for angiogenesis,[6e] and cell-dictated release would result in ideal VEGF delivery profiles.[6a, 6b] While the use of MMP-degradable tethers provides cell-dictated drug release, release rates were not manipulated due to use of a single, common MMP-cleavable substrate, precluding temporal control over enzymatically-responsive VEGF delivery. Therefore, these previously developed materials are unable to investigate the role of temporally controlled enzymatically-responsive growth factor delivery in angiogenesis.
In this work, the first materials approach to temporally control enzymatically-responsive drug delivery was developed. Poly(ethylene glycol) (PEG)-based hydrogels were modified to provide enzymatically-responsive release of the VEGF mimic Qk,[4] a potent pro-angiogenic peptide that mimics the α-helix region of VEGF (Figure 1). Hydrogels were formed with Qk tethered to the gel via MMP-degradable peptide linkers (Table 1) utilizing five distinct linkers with a variety of reported kcatKM−1 values to temporally control release (Table S1). The ability of Qk to retain bioactivity as released (“tail” form; Table 1) was assessed. Hydrogel swelling ratio, peptide incorporation efficiency, and enzymatically-responsive peptide release behavior were also characterized. Hydrogels that demonstrated MMP-responsive release were implanted subcutaneously in vivo to track the in vivo release of fluorescently labeled peptides. Hydrogels vascularization in vivo was also assessed by measuring hemoglobin (Hb) content and imaged using multiphoton fluorescent microscopy.
Figure 1.
Temporal control over enzymatically-responsive peptide delivery. 8-arm 20 kDa norbornene functionalized poly(ethylene glycol) (PEGN) is crosslinked with enzymatically stable peptide crosslinkers using a 2:3 thiol:ene molar ratio. The remaining norbornene groups are linked to the therapeutic peptide Qk via enzymatically-degradable peptide tethers. By changing the enzymatically degradable tether and associated kcat Km−1 values, temporal control over enzymatically-responsive peptide release was achieved. Not to scale.
Table 1.
Peptide sequences utilized.
| Type | Full name | Abbreviation | Sequence |
|---|---|---|---|
| “Linker” forms (for hydrogel tethering) | Qk with fast linker | Qk(FL) | "Qk"-PES↓LRAG-C-G (no extra C-terminal I) |
| Qk with moderate linker | Qk(ML) | "Qk"-GPQG↓IWGQ-C-G | |
| Qk with slow linker | Qk(SL) | "Qk"-VPLS↓LYSG-C-G | |
| Qk with non-releasing linker 1 | Qk(NRL1) | "Qk"-SGESPAY↓YTA-C-G | |
| Qk with non-releasing linker 2 | Qk(NRL2) | "Qk"-PVS↓LRSG-C-G (no extra C-terminal I) | |
| Scrambled with fast linker | Scrambled(FL) | "Scrambled"-IPES↓LRAG-C-G | |
| “Tail” forms (predicted release forms) | Qk with fast linker tail | Qk(FT) | "Qk"-PES-G (no extra C-terminal I) |
| Qk with moderate linker tail | Qk(MT) | "Qk"-GPQG-G | |
| Qk with slow linker tail | Qk(ST) | "Qk"-VPLS-G | |
| Qk with non-releasing linker 1 tail | Qk(NRT1) | "Qk"-SGESPAY-G | |
| Qk with non-releasing linker 2 tail | Qk(NRT2) | "Qk"-PVS-G (no extra C-terminal I) | |
| “Native” form (drug alone) | Native Qk | Qk(N) | "Qk"-G |
| Scrambled control peptide | Scrambled | "Scrambled" (no extra C-terminal G) | |
| Miscellaneous | Non-enzymatically degradable crosslinker | NDL | CLGKGKGKGLCG |
| Cell adhesion peptide | RGD | CG-RGDS-G | |
Standard amino acid abbreviations are used. ↓ indicates MMP cleavage site. "Qk" is the VEGF peptide mimic KLTWQELYQLKYKGI;[4b] "Scrambled" is the scrambled control peptide GLKEQSPRKHRLG. While all linkers used here were reported in literature to be susceptible to cleavage by MMP2,[7] the observed Qk release behaviors were not consistent with literature predictions. Therefore, peptide linkers are named based on their observed Qk release profiles as used in this application, and not based on their published linker-alone cleavage kinetics.
2. Results and Discussion
2.1. Pro-angiogenic potential of Qk
2.1.1. Qk benefits from sustained release
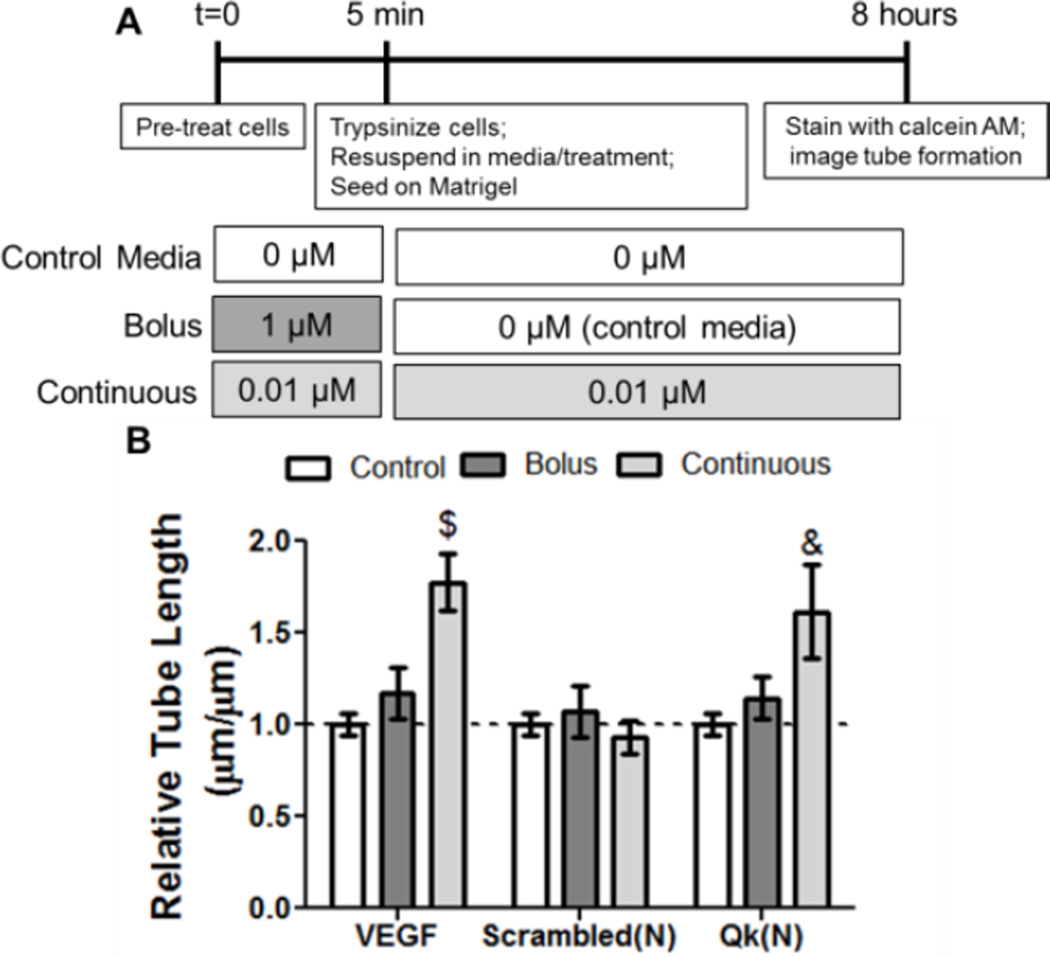
The pro-angiogenic effect of Qk presented to cells transiently and continuously was first analyzed. A modified human umbilical vein endothelial cell (HUVEC) tube formation assay was developed that emulates bolus and sustained treatment (Figure 2A) with Qk or full-length VEGF. Bolus treatment (high dose for 5 minutes, followed by control media for 8 hours) with VEGF did not induce significant tube networks (1.2-fold over control media); however, sustained treatment (low dose for 5 minutes, followed by low dose for 8 hours) resulted in a significant 1.8-fold increase in tube length (Figure 2B). This is unsurprising, as activation of receptor tyrosine kinase signaling due to interactions of VEGF with VEGFR1 and VEGFR2 has been shown to be both time- and dose-dependent.[8] Additionally, our assay results are consistent with in vivo data showing VEGF benefits from continuous, long-term delivery.[3, 6c, 6d] Similar to VEGF, only when continuously presented to cells did Qk significantly affect tube network formation (1.6-fold over control media). The control scrambled peptide did not significantly affect tube formation upon bolus or continuous treatment. To alleviate concerns about trypsin affecting the effects of the bolus treatment, a modified assay was performed when cells were treated in suspension with washes and media changes performed by centrifugation. VEGF and scrambled peptide showed similar trends regardless of trypsin use (Figure 2 and S1). Similar results between Qk and VEGF was expected, as Qk mimics the α-helix region of VEGF, and has been shown to bind both VEGFR1 and VEGFR2, resulting in similar levels of phosphorylation and ERK1/2 and Akt signaling.[4a, 4c] Overall, these data suggest that the sustained availability of Qk is critical for its bioactivity, and sustained Qk release is likely to result in more robust pro-angiogenic effects. This is also consistent with previous in vivo reports, exploiting methods to provide sustained Qk delivery to ensure bioactivity, including osmotic pumps, Matrigel, and diffusive release from hydrogels.[4a, 4b]
Figure 2.
In vitro analysis of the effect of bolus versus continuous delivery of Qk. A) Scheme of experimental setup, and B) results of tube formation data. A) To simulate bolus treatment, cells were pre-treated with a high dose for 5 minutes then given control media alone for the remainder of the 8 hour experiment. To simulate continuous delivery, cells received a low dose for the pre-treatment phase as well as the remainder of the 8 hours. Peptide doses were selected to have approximately the same dose-duration (0.01 µM × 8 hours ≈ 1.0 µM × 5 min). VEGF doses were 10 ng mL−1 (bolus) and 1 ng mL−1 (continuous). & p<0.01, $p<0.001 vs. control media. n=9, error bars represent SEM.
2.1.2. Qk bioactivity is unaffected by amino acids left after MMP-degradable sequence cleavage
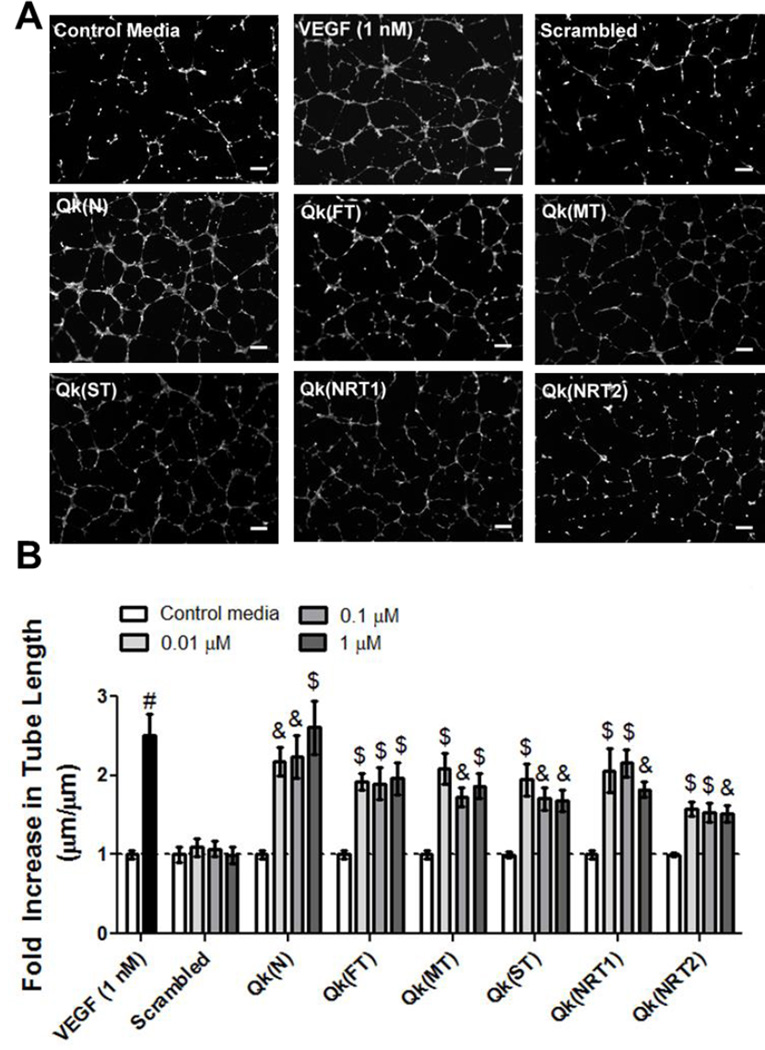
MMP-degradable linkers described in literature and adapted for this work are cleaved in the middle of the sequence, and therefore leave residual MMP substrates, or “tails”, on Qk when it is released (i.e., Qk “fast linker”, or Qk(FL) is KLTWQELYQLKYKGI-PES↓LRAG-C-G, and upon MMP cleavage releases Qk “fast linker tail”, or Qk(FT) KLTWQELYQLKYKGI-PES; Table 1). To determine if the residual peptide substrates affect bioactivity, Qk was tested in the HUVEC tube formation assay in both native “N” and as released “tail” forms (Figure 3). While the presence of some “tails” decreased the extent of tube formation induced by Qk (i.e. at 1 µM Qk(NRT2) increased tube length ~ 1.5-fold, while Qk(N) increased tube formation ~ 2.6-fold control media), all “tail” forms of Qk significantly increased tube length over the same 100-fold concentration range as Qk(N). This confirms that Qk remains bioactive in its predicted released form for all five linkers, and all linkers warrant further investigation of temporally controlled enzymatically-responsive Qk release.
Figure 3.
In vitro screening of the impact of peptide linker “tail” on Qk bioactivity. A) Representative images of HUVEC tube formation at 0.1 µm, and B) fold increase in average tube length over within-plate control media. & p<0.01, $ p<0.001, # p<0.0001 vs. control media. Scale bars = 250 µm, n=9, error bars represent SEM.
2.2. Hydrogel formation and characterization
To facilitate hydrogel incorporation, Qk was synthesized in linker form, with an MMP-degradable linker and cysteine to allow for hydrogel incorporation on the C-terminus (Qk-linker-C-G; Table 1). Hydrogels were formed using a 2:3 crosslinking thiol:ene molar ratio, with the remaining 1:3 norbornene groups used for drug tethering (Figure 1).
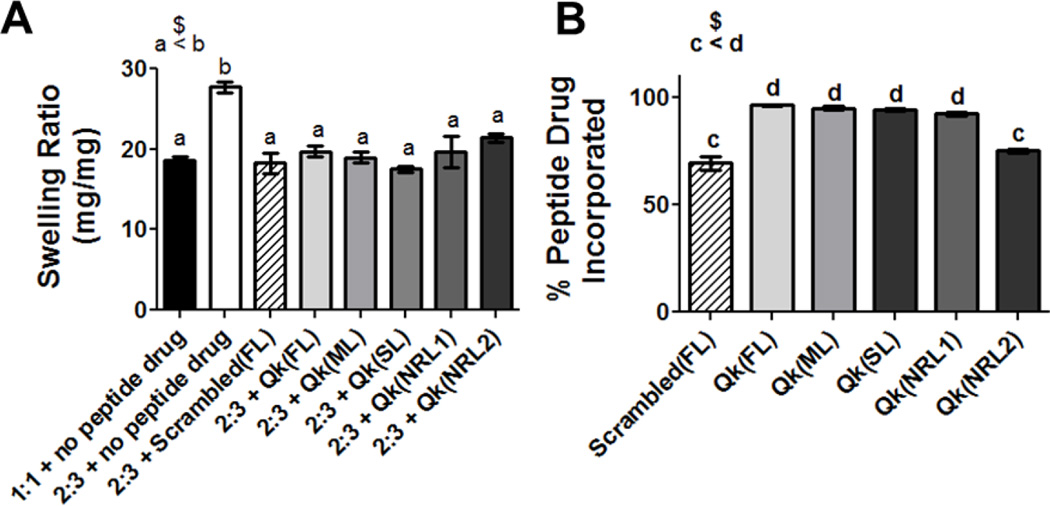
2.2.1. All linkers result in Qk hydrogel incorporation and altered hydrogel swelling ratio
In comparison to fully crosslinked (1:1) hydrogels, the use of a 2:3 crosslinking thiol:ene molar ratio increased hydrogel swelling ratio from 19 to 28 mg mg−1 (Figure 4A). This is consistent with previous work showing swelling ratio increases as network crosslinking decreases.[8] Interestingly, including tethered peptides reduced swelling ratio to levels similar to the fully crosslinked network, ranging from 18 to 21 mg mg−1, but the swelling ratio was not affected by the specific peptide incorporated. This decrease in swelling ratio is likely due to intermolecular peptide interactions, similar to our previous observations.[9] All peptide drug/linker combinations were successfully incorporated into hydrogels, with incorporation efficiencies varying from 75 and 69% for Qk(NRL2) and Scrambled(FL) to above 90% for Qk(FL), Qk(ML), Qk(SL), and Qk(NRL1) (Figure 4B).
Figure 4.
Characterization of enzymatically-responsive hydrogels in terms of A) swelling ratio and B) % peptide incorporated. n.s. p>0.05, $ p<0.001, a–d indicate statistically equivalent groups. n=9, error bars represent SEM.
2.2.2. Temporal control over enzymatically-responsive peptide release in vitro was achieved
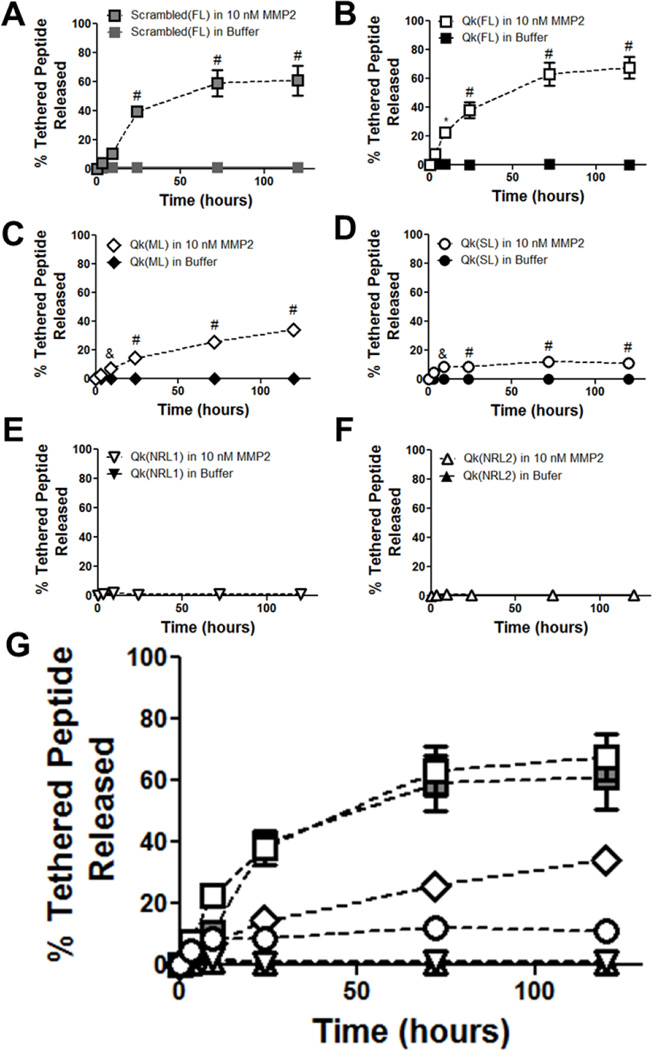
Hydrogels were formed with peptides tethered via MMP-responsive linkers and swollen overnight. Gels were incubated with 10 nM MMP2, and peptide release was analyzed and compared with control gels incubated in buffer alone. Of the five linkers investigated, all which have been previously used as crosslinks in enzymatically-degradable PEG hydrogels,[7] only three resulted in enzymatically-responsive Qk release: FL, ML, and SL (Figure 5B–D). The other two linkers, NRL1 and NRL2 did not result in detectable Qk release (Figure 5E–F). FL was also exploited to provide enzymatically-responsive release of the negative control scrambled peptide (Figure 5A). As seen in Figure 5G, varying the enzymatically-responsive linker used to tether Qk to the hydrogel successfully achieved temporal control over enzymatically-responsive Qk release. Mass spectrometry on released peptides (Figure S2) confirmed release in predicted “tail” forms used in bioactivity testing (Figure 3). Hydrogel mass over the time course of peptide release remained stable, confirming that peptide release was successfully uncoupled from hydrogel degradation (Figure S3).
Figure 5.
Peptide release from enzymatically-responsive hydrogels. Release of tethered peptide in the expected “tail” form from A) Scrambled(FL), B) Qk(FL), C) Qk(ML), D) Qk(SL), E) Qk(NRL1), and F) Qk(NRL2) hydrogels, assessed using HPLC. G) shows all MMP-treated gels on the same plot, for comparison purposes (significances not indicated for clarity). * p<0.05, & p<0.01, # p<0.0001 vs. buffer alone at same time point. n=6–12, error bars represent SEM; some obscured by symbols.
Interestingly, there was no clear relationship between reported kcat/KM values for the linker and MMP2 and release of Qk by that linker (Figure 5, Table S1). For example, NRL1 had the highest reported kcat KM−1 of any linker investigated but resulted in no detectable Qk release, while ML had the smallest kcat KM−1 investigated and resulted in moderate rates of enzymatically-responsive Qk release. Changes in degradable linker peptide structure upon inclusion of the Qk peptide was examined using PEP-FOLD prediction software, shown to deviate from NMR structures by less than 3 Å.[10] However, investigations into the length, number, and location of amorphous regions, α-helix, and β-sheets were uninformative in predicting release profiles (Figure S4). Further investigation as to the role of linker size, hydrophobicity, and amino acid composition, as well as reported kcat values for the linker and time to degradation of hydrogels formed using the peptide as a crosslinker[7] were unable to satisfactorily predict Qk release rates (Figure S5 and S6). This makes design of similar systems challenging, as the literature values for sequence cleavage in solution cannot be used to predict linker release profiles. These results are not surprising, as linker sequence, MMP, linker-adjacent peptide modifications, and hydrogel incorporation have all been shown to affect cleavage kinetics.[7, 9, 11] For example, the same modification to GPLGLWAQ increases its MMP3 cleavage rate 3-fold, but does not affect cleavage by MMP1.[11] Modification of the linker IPES↓LRAG (FL) with different adjacent peptides affects MMP2 cleavage rate.[9] Hydrogels crosslinked using MMP-responsive sequences SGESPAY↓YTA (NRL1) and GPQG↓IWGQ (ML) produces gels that degraded in ~ 1 and ~ 4 days in MMP2, while RPFS↓MIMG-crosslinked gels are stable for 10 days, despite contradictory reported kcat KM−1 values for MMP2 (440000, 555, and 4600 M−1s−1 for NRL1, ML, and RPFS↓MIMG, respectively).[7] As additional materials are developed utilizing MMP-degradable sequences for controlled, enzymatically-responsive drug release, the relationship between drug, degradable sequence, and linker cleavage kinetics will likely be further illuminated, facilitating predictive drug release profiles.
2.3. Temporal control over enzymatically-responsive Qk release was achieved in vivo, but did not affect hydrogel vascularization
Hydrogels were formed using fluorescently labeled peptides and implanted subcutaneously in mice after in vitro testing confirmed that live animal fluorescent imaging (IVIS) is an accurate and non-invasive method to track peptide release (Figure S7). Subcutaneous implantation was selected as it is a commonly used method that allows for rapid assessment of vascularization (e.g., 1 week after implantation).[12] Hydrogels were formed within non-degradable silicone reactors to aid in localization of the target tissue for collection and imaging (Figure S8). Soluble controls were similarly injected into reactors after placement in the subcutaneous pocket. Specifically, hydrogels with fluorescently-labeled Qk(FL), Qk(ML), Qk(SL), and Scrambled(FL) were studied.
2.3.1. Temporal control over enzymatically-responsive peptide release in vivo was achieved
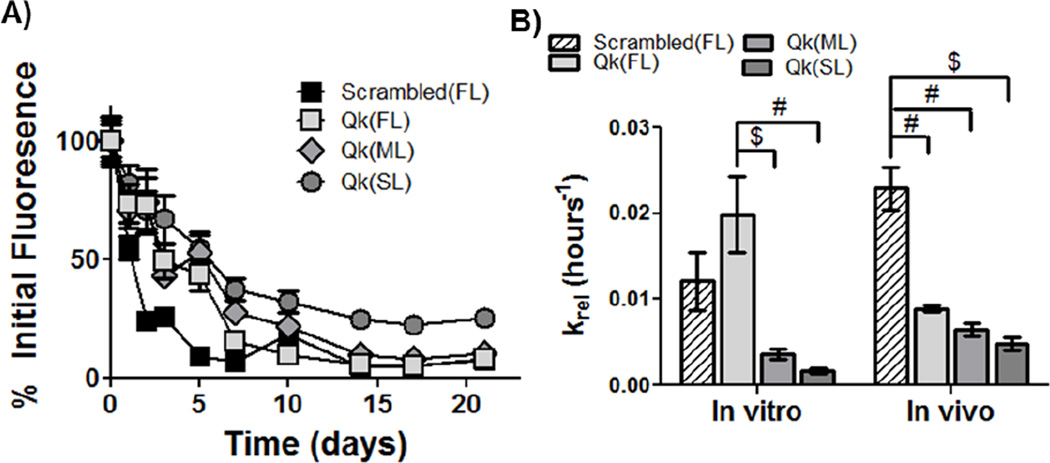
Upon subcutaneous implantation, temporal control over enzymatically-responsive peptide released was demonstrated, with Scrambled(FL), Qk(FL), Qk(ML) and Qk(SL) releasing ~ 50% of the tethered peptide after ~ 1, 3, 5, and 7 days, respectively (Figure 6A). Peptide release was significantly affected by time and hydrogel type, with a significant interaction between the two factors (p<0.0001). Undetectable amounts of peptide remained in gels after ~ 1 to ~ 2.5 weeks, depending on the linker utilized and the drug delivered. To facilitate quantitative comparisons of release profiles, in vitro and in vivo release data was fit to a pseudo-first order release model, and the release coefficients calculated (Figure 6B). In vitro, the release coefficient for Qk(FL) was the highest, followed by Scrambled(FL), Qk(ML), and Qk(SL) (19.9, 12.1, 3.59, and 1.64 × 10−3 hours−1, respectively). In vivo, the release coefficient for Scrambled(FL) was the highest, followed by Qk(FL), Qk(ML), and Qk(SL) (22.9, 8.9, 6.4, and 4.8 × 10−3 hours−1, respectively).
Figure 6.
Release of fluorescently labeled peptides in vivo, and comparison to in vitro release. A) Quantification of fluorescence over 3 weeks in vivo, normalized to average within-group day 0 fluorescence. Peptide release in vivo is significantly affected by both gel type and time (p<0.0001), and there was a significant interaction between the factors (p<0.0001). B) Pseudo-first order release coefficients (krel) were calculated for both in vitro and in vivo peptide release, to facilitate comparison of release kinetics. $ p<0.001, # p<0.0001. n=12 for A, 6–12 for B; error bars represent SEM.
Interestingly, and despite releasing peptide with approximately the same profile in vitro (Figure 5G), the Scrambled(FL) hydrogels released peptide more quickly than Qk(FL) gels when implanted in vivo (Figure 6). This is likely due to unexpected complete degradation of Scrambled(DL) gels. While all Qk-releasing gels were intact when collected for vascularization measurements at 1 week post-implantation, all Scrambled(FL) gels had degraded, contributing to the decrease in gel fluorescence observed and confounding the krel calculations. Due to the presence of the ester bond between the PEG and norbornene groups,[13] all hydrogels used here are expected to be hydrolytically degradable over 1–2 months based on in vitro studies[13b], with PEG hydrogel degradation in vitro correlating well with in vivo behavior.[14] However, exclusive degradation in vivo of the scrambled-peptide releasing gels was unexpected, as they were formed with the same enzymatically stable peptide crosslinker, had similar swelling ratios (Figure 4), and were stable over the same time frames in vitro as the Qk-releasing gels (Figure S3). While the crosslinking peptide was shown to be stable against MMP2 (Figure S3), it is possible that non-specific enzymatic degradation of the crosslinking peptide occurred. It is possible that degradation specifically of the Scrambled(FL) gels is due to inflammatory macrophage recruitment. The RGD peptide present in these hydrogels intended to facilitate vascular infiltration has also been shown to mediate macrophage adhesion and foreign body giant cell formation involved in the inflammatory response after implantation.[15] Macrophages have been shown to degrade biomaterials by releasing hydrolytic enzymes, esterases, hydroxyl radicals, and many other reactive species that could contribute to accelerated cleavage of ester, ether, or amide bonds within hydrogel crosslinks.[16] Our results indicate delivery of the VEGF mimic Qk may have decreased the activity or number of macrophages present at the implant site, either by directly affecting macrophages or by initiating a feedback loop between tissue and immune cells.[15] VEGF has been shown to shift macrophages from an inflammatory (M1) to a regenerative (M2) phenotype.[17] As the VEGF mimic Qk shows similar receptor binding and biological effects to the full-length protein,[4] it could be polarizing macrophages to an M2 rather than M1 phenotype, delaying macrophage-mediated degradation of Qk-releasing gels. However, additional investigations are necessary to support these theories.
All Qk-releasing gels remained stable in vitro and in vivo, allowing comparison of the release coefficients. Kinetics of drug release followed similar trends in vitro and in vivo, with Qk(FL) exhibiting the fastest release kinetics, followed by Qk(ML) and Qk(SL), although the krel values were not significantly different in vivo. Qk(ML) and Qk(SL) had statistically equivalent release rates in vitro and in vivo, but Qk(FL) released more rapidly in vitro. These results show that by altering the specific MMP-responsive linkage used to tether the drug to the hydrogel, cell-dictated peptide release can be temporally manipulated. This provided a method for exquisite control over the in vitro and in vivo presentation of Qk to target cells/tissues. While the relative release rates were consistent between in vitro and in vivo conditions, the relative differences in release were not. There was a 12-fold difference in krel between Qk(FL) and Qk(SL) in vitro, but only a 1.9-fold difference in vivo. This provides useful information for design of similar systems; differences in release profiles are dampened when exposed to the more complex in vivo environment. Based on these results, degradable linkers should be selected that provide ~ 5–10-fold larger differences in release rates in vitro than are desired for in vivo release (i.e., if a 3-fold difference in release rate is desired in vivo, linkers should be identified that produce 15 to 30-fold differences in in vitro rates).
Differences in release rates between in vitro and in vivo conditions were not unexpected, and are consistent with other studies.[6a] Numerous MMPs are expressed in vivo, with differential expression profiles based on the tissue and extent of injury or disease state.[18] Additionally, tissue inhibitors of metalloproteinases (TIMPs) present in vivo could affect MMP activation levels.[18b] As the linkers investigated here are reported to be susceptible to multiple MMPs (Table S1), it is possible that MMPs other than the MMP2 used in vitro are contributing to peptide release in vivo. Due to differential MMP expression that occurs based on tissue type and injury,[18] it is likely that the release profiles achieved in this study would not directly correlate with release in a different tissue (i.e., ischemic hindlimb or cardiac tissue). However, the material developed here would likely still provide differential enzymatically-responsive release profiles, and alternate MMP-degradable linkages[7, 19] could be investigated if the desired temporal release profiles were not achieved.
2.3.2. Non-enzymatically degradable Qk-releasing hydrogels do not induce vascularization after one week
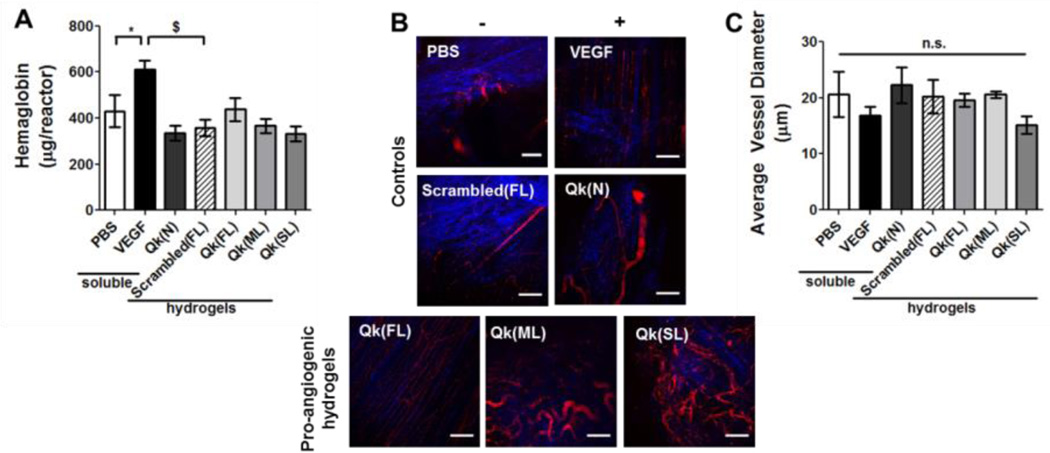
While the main goal of this work was to develop a method to temporally control enzymatically-responsive peptide delivery, a secondary goal was to study the effects of temporal control over enzymatically-responsive Qk release on angiogenesis in vivo. Therefore, vascularization resulting from Qk-releasing hydrogels was assessed. One week after implantation, hydrogel-containing reactors were explanted and vascular ingrowth was quantified by measuring hemoglobin (Hb). Multiphoton fluorescent microscopy was also used to qualitatively assess vasculature. Neither the Qk-releasing hydrogels, nor soluble Qk significantly affected Hb relative to PBS or scrambled-peptide releasing (Figure 7A). Similarly, while variations in average vessel size between treatment groups was observed (from 15 to 22 µm for Qk(SL) and Qk(N), respectively), there was no significant difference between treatment groups (Figure 7C). However, preliminary studies examining vascularization of hydrogels four weeks after implantation shows significantly greater Hb resulting from Qk(ML) and Qk(SL) vs. Qk(FL) gels (Figure S9), suggesting the therapeutic potential of these hydrogels and the influence of temporally-controlled enzymatically-responsive peptide release.
Figure 7.
Enzymatically-responsive Qk-releasing hydrogels were formed in non-degradable reactors and implanted subcutaneously in mice. One week after implantation, vascularization into the reactor was assessed via A) hemoglobin content and B–C) by multiphoton fluorescent imaging. B) Representative images of collagen (SHG, blue) and vasculature (FITC-dextran, red) formed within reactors. C) Quantification of average vessel diameter. n.s. p>0.05, * p<0.05, $ p<0.001. n=14–16 for A, 6–8 for C; error bars represent SEM. Scale bar = 100 µm.
While it did not significantly induce vascularization as a soluble, injected drug here, Qk(N) has been shown to be pro-angiogenic in vivo when delivered using alternate approaches. For example, when Qk is encapsulated within Matrigel and implanted subcutaneously, vascularization is significantly increased compared to controls.[4b] Matrigel provides a degree of sustained delivery and a physical matrix facilitating cellular adhesion and infiltration not provided by the soluble injection control here.[20] This is likely the reason the Qk(N) peptide did not induce a significant angiogenic response in this study. Lackluster vascularization one week after implanting hydrogels providing enzymatically-responsive Qk release was observed. One week is a commonly used assessment point for vascularization of subcutaneous implantats,[12] and the positive control VEGF significantly increased vascularization, supporting the validity of the in vivo model. The temporal release profiles obtained here (~ 1 to 2.5 weeks in vivo) are likely not the cause for the lackluster vascularization, as they are within the range of release profile ranges previously shown to improved vascularization via VEGF delivery (~ 2–3 weeks).[3a, 6a, 6d] However, some materials have shown improved vascularization when release occurred over ~ 3–4 weeks,[21] suggesting it could be advantageous to identify MMP-degradable tethers that further prolongs Qk delivery. It is possible that released Qk lacks bioactivity. However, this is unlikely, as Qk was confirmed to be bioactive in the form it is released from the gels (Figure 3 and S2). It is more likely that minimal vascularization observed with Qk-releasing hydrogels is due to the non-enzymatically degradable nature of the hydrogels employed. In this initial study, hydrogels were designed to be stable over the time course investigated, to prevent confounding Qk release with hydrogel degradation (as was observed with the Scrambled(FL) gels). However, this causes hydrogels to present physical resistance to infiltrating vasculature due to the small mesh sizes of the gels (~ 16–17 nm),[13b] regardless of the pro-angiogenic signals delivered. It is possible that released peptides diffused into nearby tissue and induced vascularization near the hydrogel implant. However, vasculature in nearby tissue was not assessed due to difficulty discriminating between pre-existing host vasculature and vasculature that developed in response to hydrogels. Preliminary studies looking at vascularization of these hydrogel after four weeks, e.g., once gels were degraded, showed greater vascularization than at the one-week time point used here (Figure 7 and S9), further supporting this conclusion. Use of degradable hydrogels either via enzymatic or temporally-relevant hydrolytic mechanisms has been shown to greatly improve cell invasion in vitro[7] and in vivo.[6a] Future studies could employ an enzymatically-degradable crosslinking peptide for hydrogel formation,[7, 22] facilitating simultaneous cell-dictated hydrogel degradation and drug release, likely increasing the pro-angiogenic efficacy of the hydrogels.
While this material was designed to achieve temporal control over enzymatically-responsive release of Qk, the hydrogel system could be easily translated to deliver any number of therapeutic peptides with a range of functionality (e.g., pro-angiogenic,[23] anti-inflammatory,[24] or chemotherapeutic).[25] The material could also be used for controlled delivery of protein drugs, with the inclusion of degradable linkages at either the C- or N-termini. It could also be used to deliver non-peptide therapeutics that contain complementary chemical functionalities to allow for tethering to MMP-degradable linkers.[26] However, as linker-adjacent modification has been shown to affect cleavage kinetics, delivery of different therapeutic agents would likely require different MMP-degradable linkers to achieve similar temporal release profiles.[9, 11] It is possible that inclusion of a spacer such as linear PEG or poly(glycine) between the MMP-degradable linkage and the therapeutic sequence could decrease the influence of the therapeutic on linker cleavage kinetics (e.g., by reducing steric hindrance),[27] and result in drug release profiles that more closely emulate well-established soluble linker cleavage kinetics.
3. Conclusions
In conclusion, we developed a PEG hydrogel system providing temporally controlled enzymatically-responsive release of the pro-angiogenic peptide Qk, for which sustained release was confirmed necessary for bioactivity. Qk was successfully incorporated into PEG hydrogels using five MMP-degradable linkers. Three of the linkers (IPES↓LRAG (FL), GPQG↓IWGQ (ML) and VPLS↓LYSG (SL)) provided a range of MMP-responsive peptide release rates both in vitro and in vivo. While unable to promote significant vascularization in vivo one week after implantation, likely due to the slow degradation of the hydrogels, temporally controlled enzymatically-responsive peptide release was attained. This approach can be readily adapted for controlled delivery of a number of therapeutic peptides, proteins, and small molecule drugs, and provides a means to study how temporal control over cell-dictated release affects drug efficacy.
4. Experimental Section
Materials and Methods
Unless otherwise noted, all chemicals were obtained from Sigma-Aldrich, all amino acids from AAPPTec, and all cell culture materials obtained from Lonza.
Peptide synthesis
Peptides were synthesized as previously described (Table 1).[9] O-benzotriazole-N,N,N’,N’-tetramethyl-uronium-hexafluoro-phosphate (0.5 M, AnaSpec) in dimethylformamide (DMF) and diisopropylethylamine (2 M, DIEA, Alfa Aesar) in N-methylpyrrolidone was used as the activator and activator base for RGD, NDL, and “N” and “T” peptides, and diisopropylcarbodiimide (0.5 M, Chem-Impex International) and hydroxybenzotriazole (1 M, Advanced ChemTech) in DMF as activator and activator base for all “L” peptides. Peptides were cleaved in trifluoroacetic acid (TFA, Alfa Aesar) containing 3,6-dioxa-1,8-octanedithiol (Alfa Aesar), triisopropylsilane (Alfa Aesar), and distilled, deionized water (ddH2O) (2.5 vol% each), with thioanisole (2.5 vol%, Alfa Aesar) if the sequence included arginine. After 3–6 hours, peptides were precipitated and washed in ether.
Peptide purification and assessment
Synthesis was confirmed using Matrix Assisted Laser Desorption Ionization-Time of Flight (MALDI-ToF) mass spectrometry (Bruker). Peptides were dialyzed in ddH2O overnight using 1–500 or 1,000 MWCO tubing (Spectrum Laboratories). After collection by lyophilization, percent peptide was determined by measuring absorbance at 205 nm (Evolution 300 UV/Vis detector, Thermo Scientific).[28] Peptide purity was determined to be ~ 90% as measured by High Performance Liquid Chromatography (HPLC, Shimadzu Prominence) on a Kromasil Eternity C18 column (4.6 × 50 mm) running a gradient from 5 to 95% acetonitrile in water (both containing 1% TFA).
Fluorescent peptide labeling
Qk(FL), Qk(ML), Qk(SL), and Scrambled(FL) were selectively labeled at the N-terminal primary amine by reacting on-resin peptides with Texas Red succinimidyl ester (Life Technologies). Texas Red in DMF (2 mg, 10 mg mL−1) was added drop-wise to peptide in DMF (0.25 mM in 3 mL) containing 3 drops DIEA. The mixture was protected from light and stirred overnight at room temperature. The peptide was cleaved and precipitated as above. The labeled peptide was purified by dialysis in ddH2O until the water remained clear for at least 6 hours, and labeled peptide collected by lyophilization.
HUVEC cell culture
Human umbilical vein endothelial cells (HUVECs), originally from Lonza, were a gift from Dr. Stephen Dewhurst. After thawing from cryostorage, cells were maintained in Endothelial Growth Media 2 (Endothelial Basal Media-2 (EBM-2) containing EGM-2 SingleQuots; EGM-2), at 37 °C with 5% CO2. Cells were used at least two passages after thawing, but prior to passage 10. Control media consisted of EBM-2 containing fetal bovine serum (2.5 vol%, Atlanta Biologicals), penicillin (100 U mL−1), streptomycin (100 U mL−1), and amphotericin B (250 ng mL−1, Thermo Scientific).
HUVEC tube formation assay
Tube formation was assessed as previously described,[9] with minor modifications to simulate bolus versus continuous treatment. HUVECs were grown to 60–70% confluency, then washed and treated for 5 minutes. Cells were then washed, trypsinized, resuspended in the continuous treatment, and seeded on Matrigel (2.4×104 cells well−1; Figure 2A). For the dose response testing, HUVECs were suspended directly in the treatment solution and seeded on Matrigel, with no pre-treatment step. All cells were fluorescently imaged (0.5 µL mL−1 calcein AM, Invitrogen) 8 hours after seeding on Matrigel, using a Nikon Eclipse Ti 2000 inverted light microscope with temperature/humidity control (Pathology Devices).[29] Tube length was quantified using the image processing program Angioquant.[30]
Poly(ethylene glycol) norbornene synthesis
Poly(ethylene glycol) norbornene (PEGN) was synthesized from 8-arm 20 kDa PEG (JenKem Technologies USA) as previously described.[31] Functionalization was determined to be >95% by 1H-NMR (400 MHz, CDCl3, δ): 6.25 – 5.8 (m, 8H, norbornene vinyl protons), 4.35 – 4.05 (d, 8H, -COOCH2), 3.9 – 3.35 (m, 892H, -CH2CH2O-)). The final product was dialyzed overnight in ddH2O using 1000 MWCO dialysis tubing, collected by lyophilization, and stored at −20 °C.
Hydrogel formation
Hydrogels were formed as previously described,[9] with minor modifications. All gels contained 10 wt% PEGN with a 2:3 crosslinking thiol:ene ratio using NDL as the crosslinking peptide, with the remaining 1:3 thiol:ene containing tethered drug, unless otherwise specified. Lithium phenyl-2,4,6-trimethylbenzoylphosphinate (0.05 wt%, LAP, synthesized as described[32]) was used as photoinitiator, and acetonitrile/water (25/75 vol/vol) used as the solvent to improve Qk solubility.[9] 40 µL of hydrogel precursor solution was injected into a custom cylindrical Teflon mold (~ 5 mm in diameter, ~2 mm in height) and exposed to 10 min 365 nm UV light (~ 2.5 mW cm−2) to induce polymerization. All hydrogels were swollen overnight in buffer (10 mM CaCl2, 50 mM NaCl, 50 µM ZnCl2 (Alfa Aesar), 50 mM Tricine (Acros Organics), and 0.05 wt% Brij35 (Alfa Aesar) in ddH2O, pH 7.4) at 37 °C prior to use. For in vivo studies, gels also contained 2.8 mM RGD to facilitate cell adhesion, and fluorescently labeled peptides were used to facilitate tracking. Gels were polymerized within reactors (OD=7.94 mm, ID=4.76 mm, h=2.25 mm) cut from pharmaceutical grade silicone tubing (Thermo Scientific), to facilitate tissue identification for collection. These reactors are similar to the Angioreactors used in the directed in vivo angiogenesis assay (DIVAA) which have been shown to induce only mild inflammatory infiltration.[33] However, the increased exposed surface area of the reactors used in this study allows for a more accurate simulation of reactor-free implantation. Reactors were placed in a custom Teflon holder temporarily capping the bottom end. Precursor solution (40 µL) was then pipetted into the reactor and gels were polymerized in situ. Gels in reactors were then transferred to a 24-well plate, where they were incubated in buffer overnight before subcutaneous implantation (Figure S8).
Hydrogel characterization
Hydrogel swelling ratio was determined by measuring gel mass prior to and after lyophilization (Q=Mwet/Mdry; mg mg−1). Peptide incorporation efficiency was assessed by forming gels with known amounts of drug in the precursor solution, incubating gels overnight in buffer, collecting buffer, and assessing the amount of peptide released using HPLC as described for peptide purification. Peak areas (absorbance at 214 nm for Scrambled peptides and 278 nm for Qk peptides) were compared to a standard curve to determine the amount of peptide released into buffer. The incorporation efficiency was then back-calculated based on the amount used to form the gel and the amount of peptide released (not incorporated into the gel) (%Drug Incorporated = (Mprecursor solution – Mreleased in buffer)/Mprecursor solution*100%).
Characterizing peptide release
After overnight swelling, gels were transferred to fresh buffer or buffer with 10 nM recombinant human MMP2 (PeproTech). Solutions were collected and replaced periodically, at a minimum once every 48 hours as MMP2 inactivates fairly rapidly.[7] Solutions were stored at −20 °C until peptide release was quantified using HPLC, as before. Peptide release data was fit to a pseudo-first order release equation (Equation 1):
| (1) |
where Dt is the drug remaining in the gel at time t, D0 is the initial drug loaded in the gel, and krel is the pseudo-first order release rate.[34]
In vivo testing
All animal procedures were approved by the University Committee of Animal Resources (UCAR) at the University of Rochester. 6–8 week old female BALB/c mice (Taconic) were used for all studies. Buprenorphine (3.25 mg kg−1) was given immediately prior to surgery and every 12 hours thereafter as necessary for pain. Mice were randomly assigned to treatment groups, making sure each treatment group spanned at least two surgery days.
Subcutaneous implantation model
Mice were anesthetized (60 mg kg−1ketamine, 4 mg kg−1 xylazine). Fur was removed using clippers followed by Nair. A subcutaneous pocket was formed on each side of the mouse dorsal flank, and a hydrogel within a reactor (formed as described above) placed in each pocket. For soluble treatment (PBS, 11.7 mM Qk(N), and 80 µg mL−1 VEGF), empty reactors were placed in the pocket and 40 µL of solution injected into the reactor in situ. Incisions were closed with skin glue.
Peptide release tracking
Peptide release was tracked using the IVIS live animal imaging system (IVIS) as previously described,[14] using λex/em = 570/620 nm (Xenogen IVIS-200 Optical In Vivo Imaging System, Caliper Life Sciences Inc.). Mice were imaged immediately following surgery and isoflurane inhalation used as anesthesia for subsequent imaging sessions. Mice were re-depilated as necessary to reduce background signal from fur. Fluorescence (total radiant efficiency, [(p s−1) (µW cm−2) −1]) was measured using a ROI of constant area, and normalized by within group average day 0 fluorescence. Release data was fit to a pseudo-first order model, using a modified version of Equation 1 where Ft/F0 (gel fluorescence at time t over initial gel fluorescence) was used in place of Dt/D0.
Quantification of vascular ingrowth
7 days after implantation, vascular ingrowth was assessed by measuring hemoglobin within the reactor, which has been shown to correlate well with other measures of angiogenesis.[35] Reactors were explanted, and residual hydrogel and ingrown material removed and placed in 1 mL Drabkin’s reagent (RICAA chemical). Samples were homogenized via sonication, centrifuged for 20 minutes at 14,000 g, and then filtered through 0.45 µm polyvinylidene (PVDF) filters (PerkinElmer). Absorbance at 540 nm was determined relative to the reference wavelength of 650 nm, and a hemoglobin standard curve used to determine the hemoglobin concentration.[35c]
Multiphoton imaging
Mice were anesthetized by ketamine/xylazine injection, and 2M MW fluorescein isothiocynate-dextran (10 mg mL−1, FITC, LifeTechnologies) in PBS (50 µL) injected retro-orbitally. After one hour, mice were sacrificed and the dorsal flank opened to expose the hydrogel reactor for in situ imaging. Excitation light generated using a MaiTai Ti:Sapphire laser was directed towards the sample through a BX61WI upright microscope (Olympus). An Olympus Fluoview FV300 scanning system further controlled beam scanning and image acquisition. 350 mW of 810 nm light, with 100 fs pulses at 80 MHz was directed into the scan box to image all samples. Excitation light was focused and the backscattered fluorescent and second harmonic generation (SHG, collagen) signal was collected using an Olympus UMPLFL20×W water immersion lens (20×, 0.95 N.A.). Signals were separated from the excitation beam first using a 670 nm short pass dichroic mirror, and then separated from each other using a 475nm long pass filter. 535 nm band-pass (HQ535/40m-2P, Chroma) and 405 nm band-pass filters (HQ405/30m-2P, Chroma) were used to filter the FITC and SHG emission signals, respectively, with hamamatsu HC125-02 photomultiplier tubes used to collected both channels. Images were collected from the same five locations in each reactor. 21 images taken every 5 µm were taken at each location and then maximum intensity projected to form a 100 µm stack. Vessel diameter was measured manually in ImageJ, with 5 of the largest vessels in each image quantified, and the average diameter for the gel reported. Images were collected and vessel measurements made by a user blinded to treatment groups.
Statistical analysis
Data assembly and calculations were performed using Microsoft Excel (2010 v14.0). Figure preparation and statistical analysis were performed using Graphpad Prism (5.04). All data is shown as mean ± standard error of the mean (SEM). Data presented in Figure 2, 5, and 6 was analyzed using a two-way ANOVA; data in Figure 3, 4, and 7 was analyzed using a one-way ANOVA, with post-hoc testing (Bonferroni, Tukey, or Dunn) as necessary. All experiments were performed in at least duplicate to reach n=6–16 as indicated in legends. p<α=0.05 was considered significant for all analyses.
Supplementary Material
Acknowledgements
The authors would like to thank the Howard Hughes Medical Institute Med-into-Grad fellowship (AVH), the National Institute of Health (R01 AR064200 (DB), F31CA183351 (KB), and T32AI007285 (KB)), and the Department of Defense (W81XWH-09-1-0405 (EB)) for funding. The authors would also like to thank Dr. Alex Shestopalov and Dr. James McGrath for the use of their equipment, and Dr. Stephen Dewhurst for providing HUVECs.
Footnotes
Supporting Information is available from the Wiley Online Library or from the author.
Contributor Information
Amy H. Van Hove, Department of Biomedical Engineering, 207 Robert B. Goergen Hall, University of Rochester, Rochester, NY 14627, USA
Erin Antonienko, Department of Biomedical Engineering, 207 Robert B. Goergen Hall, University of Rochester, Rochester, NY 14627, USA.
Kathleen Burke, Department of Biomedical Engineering, 207 Robert B. Goergen Hall, University of Rochester, Rochester, NY 14627, USA.
Edward Brown, III, Department of Biomedical Engineering, 207 Robert B. Goergen Hall, University of Rochester, Rochester, NY 14627, USA; Department of Neurobiology and Anatomy, University of Rochester, 601 Elmwood Ave, Rochester, NY, 14642, USA.
Danielle S.W. Benoit, Email: benoit@bme.rochester.edu, Department of Biomedical Engineering, 207 Robert B. Goergen Hall, University of Rochester, Rochester, NY 14627, USA; Department of Biomedical Genetics, 601 Elmwood Ave, University of Rochester, Rochester, NY 14642, USA; Department of Chemical Engineering, 206 Gavett Hall, University of Rochester, Rochester, NY 14627 USA; Center for Musculoskeletal Research, 601 Elmwood Ave, University of Rochester Medical Center, Rochester, NY 14642, USA.
References
- 1.Zachary I, Morgan RD. Heart. 2011;97:181. doi: 10.1136/hrt.2009.180414. [DOI] [PubMed] [Google Scholar]
- 2.Lovett M, Lee K, Edwards A, Kaplan DL. Tissue Eng Part B-Re. 2009;15:353. doi: 10.1089/ten.teb.2009.0085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.a) Silva EA, Mooney DJ. Biomaterials. 2010;31:1235. doi: 10.1016/j.biomaterials.2009.10.052. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Silva EA, Mooney DJ. J Thromb Haemost. 2007;5:590. doi: 10.1111/j.1538-7836.2007.02386.x. [DOI] [PubMed] [Google Scholar]
- 4.a) Finetti F, Basile A, Capasso D, Di Gaetano S, Di Stasi R, Pascale M, Turco CM, Ziche M, Morbidelli L, D'Andrea LD. Biochem Pharm. 2012;84:303. doi: 10.1016/j.bcp.2012.04.011. [DOI] [PubMed] [Google Scholar]; b) Santulli G, Ciccarelli M, Palumbo G, Campanile A, Galasso G, Ziaco B, Altobelli GG, Cimini V, Piscione F, D'Andrea LD, Pedone C, Trimarco B, Iaccarino G. J of Trans Med. 2009;7:41. doi: 10.1186/1479-5876-7-41. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) D'Andrea LD, Iaccarino G, Fattorusso R, Sorriento D, Carannante C, Capasso D, Trimarco B, Pedone C. P Natl Acad Sci USA. 2005;102:14215. doi: 10.1073/pnas.0505047102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.a) Craik DJ, Fairlie DP, Liras S, Price D. Chem Biol Drug Des. 2013;81:136. doi: 10.1111/cbdd.12055. [DOI] [PubMed] [Google Scholar]; b) Laham RJ, Rezaee M, Post M, Sellke FW, Braeckman RA, Hung D, Simons M. Drug Metab Dispos. 1999;27:821. [PubMed] [Google Scholar]
- 6.a) Phelps EA, Landazuri N, Thule PM, Taylor WR, Garcia AJ. P Natl Acad Sci USA. 2010;107:3323. doi: 10.1073/pnas.0905447107. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Zisch AH, Lutolf MP, Ehrbar M, Raeber GP, Rizzi SC, Davies N, Schmokel H, Bezuidenhout D, Zilla DVP, Hubbell JA. Faseb J. 2003;17:2260. doi: 10.1096/fj.02-1041fje. [DOI] [PubMed] [Google Scholar]; c) Formiga FR, Pelacho B, Garbayo E, Abizanda G, Gavira JJ, Simon-Yarza T, Mazo M, Tamayo E, Jauquicoa C, Ortiz-de-Solorzano C, Prosper F, Blanco-Prieto MJ. J Control Release. 2010;147:30. doi: 10.1016/j.jconrel.2010.07.097. [DOI] [PubMed] [Google Scholar]; d) Sun QH, Chen RR, Shen YC, Mooney DJ, Rajagopalan S, Grossman PM. Pharm Res. 2005;22:1110. doi: 10.1007/s11095-005-5644-2. [DOI] [PubMed] [Google Scholar]; e) Ozawa CR, Banfi A, Glazer NL, Thurston G, Springer ML, Kraft PE, McDonald DM, Blau HM. J Clin Invest. 2004;113:516. doi: 10.1172/JCI18420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Patterson J, Hubbell JA. Biomaterials. 2010;31:7836. doi: 10.1016/j.biomaterials.2010.06.061. [DOI] [PubMed] [Google Scholar]
- 8.a) Gupta K, Kshirsagar S, Li W, Gui LZ, Ramakrishnan S, Gupta P, Law PY, Hebbel RP. Exp Cell Res. 1999;247:495. doi: 10.1006/excr.1998.4359. [DOI] [PubMed] [Google Scholar]; b) Marshall CJ. Cell. 1995;80:179. doi: 10.1016/0092-8674(95)90401-8. [DOI] [PubMed] [Google Scholar]; c) Zhu WH, MacIntyre A, Nicosia RF. Am J Pathol. 2002;161:823. doi: 10.1016/S0002-9440(10)64242-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Van Hove AH, Beltejar MJ, Benoit DS. Biomaterials. 2014;35:9719. doi: 10.1016/j.biomaterials.2014.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thevenet P, Shen YM, Maupetit J, Guyon F, Derreumaux P, Tuffery P. Nucleic Acids Res. 2012;40:W288. doi: 10.1093/nar/gks419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vessillier S, Adams G, Chernajovsky Y. Protein Eng Des Sel. 2004;17:829. doi: 10.1093/protein/gzh097. [DOI] [PubMed] [Google Scholar]
- 12.a) Tabata Y, Miyao M, Ozeki M, Ikada Y. J Biomat Sci-Polym E. 2000;11:915. doi: 10.1163/156856200744101. [DOI] [PubMed] [Google Scholar]; b) Sun G, Shen YI, Kusuma S, Fox-Talbot K, Steenbergen CJ, Gerecht S. Biomaterials. 2011;32:95. doi: 10.1016/j.biomaterials.2010.08.091. [DOI] [PubMed] [Google Scholar]
- 13.a) Roberts JJ, Bryant SJ. Biomaterials. 2013;34:9969. doi: 10.1016/j.biomaterials.2013.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Shih H, Lin CC. Biomacromolecules. 2012;13:2003. doi: 10.1021/bm300752j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hoffman MD, Van Hove AH, Benoit DS. Acta Biomater. 2014;10:3431. doi: 10.1016/j.actbio.2014.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Franz S, Rammelt S, Scharnweber D, Simon JC. Biomaterials. 2011;32:6692. doi: 10.1016/j.biomaterials.2011.05.078. [DOI] [PubMed] [Google Scholar]
- 16.Xia Z, Triffitt JT. Biomed Mater. 2006;1:R1. doi: 10.1088/1748-6041/1/1/R01. [DOI] [PubMed] [Google Scholar]
- 17.a) Eirin A, Zhu XY, Li ZL, Ebrahimi B, Zhang X, Tang H, Korsmo MJ, Chade AR, Grande JP, Ward CJ, Simari RD, Lerman A, Textor SC, Lerman LO. Arterioscl Throm Vas. 2013;33:1006. doi: 10.1161/ATVBAHA.113.301164. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Jay SM, Shepherd BR, Andrejecsk JW, Kyriakides TR, Pober JS, Saltzman WM. Biomaterials. 2010;31:3054. doi: 10.1016/j.biomaterials.2010.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.a) Muhs BE, Plitas G, Delgado Y, Ianus I, Shaw JP, Adelman MA, Lamparello P, Shamamian P, Gagne P. J Surg Res. 2003;111:8. doi: 10.1016/s0022-4804(02)00034-3. [DOI] [PubMed] [Google Scholar]; b) Phatharajaree W, Phrommintikul A, Chattipakorn N. Can J Cardiol. 2007;23:727. doi: 10.1016/s0828-282x(07)70818-8. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Luttikhuizen DT, van Amerongen MJ, de Feijter PC, Petersen AH, Harmsen MC, van Luyn MJ. Biomaterials. 2006;27:5763. doi: 10.1016/j.biomaterials.2006.07.004. [DOI] [PubMed] [Google Scholar]
- 19.Turk BE, Huang LL, Piro ET, Cantley LC. Nat Biotechnol. 2001;19:661. doi: 10.1038/90273. [DOI] [PubMed] [Google Scholar]
- 20.Kleinman HK, Martin GR. Semin Cancer Bio. 2005;15:378. doi: 10.1016/j.semcancer.2005.05.004. [DOI] [PubMed] [Google Scholar]
- 21.a) Ennett AB, Kaigler D, Mooney DJ. J Biomed Mater Res A. 2006;79A:176. doi: 10.1002/jbm.a.30771. [DOI] [PubMed] [Google Scholar]; b) Murphy WL, Peters MC, Kohn DH, Mooney DJ. Biomaterials. 2000;21:2521. doi: 10.1016/s0142-9612(00)00120-4. [DOI] [PubMed] [Google Scholar]
- 22.Seliktar D, Zisch AH, Lutolf MP, Wrana JL, Hubbell JA. J Biomed Mater Res A. 2004;68A:704. doi: 10.1002/jbm.a.20091. [DOI] [PubMed] [Google Scholar]
- 23.Pickart L. J Biomat Sci-Polym E. 2008;19:969. doi: 10.1163/156856208784909435. [DOI] [PubMed] [Google Scholar]
- 24.Akeson AL, Woods CW, Hsieh LC, Bohnke RA, Ackermann BL, Chan KY, Robinson JL, Yanofsky SD, Jacobs JW, Barrett RW, Bowlin TL. J Biol Chem. 1996;271:30517. doi: 10.1074/jbc.271.48.30517. [DOI] [PubMed] [Google Scholar]
- 25.Yang LL, Mashima T, Sato S, Mochizuki M, Sakamoto H, Yamori T, Oh-hara T, Tsuruo T. Cancer Res. 2003;63:831. [PubMed] [Google Scholar]
- 26.Chen XY, Plasencia C, Hou YP, Neamati N. J Med Chem. 2005;48:5874. doi: 10.1021/jm049165z. [DOI] [PubMed] [Google Scholar]
- 27.a) Hern DL, Hubbell JA. J Biomed Mater Res. 1998;39:266. doi: 10.1002/(sici)1097-4636(199802)39:2<266::aid-jbm14>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]; b) Salinas CN, Anseth KS. J Tissue Eng Regen Med. 2008;2:296. doi: 10.1002/term.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Anthis NJ, Clore GM. Protein Sci. 2013;22:851. doi: 10.1002/pro.2253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Arnaoutova I, Kleinman HK. Nat Protoc. 2010;5:628. doi: 10.1038/nprot.2010.6. [DOI] [PubMed] [Google Scholar]
- 30.Niemisto A, Dunmire V, Yli-Harja O, Zhang W, Shmulevich I. Ieee T Med Imaging. 2005;24:549. doi: 10.1109/tmi.2004.837339. [DOI] [PubMed] [Google Scholar]
- 31.Fairbanks BD, Schwartz MP, Halevi AE, Nuttelman CR, Bowman CN, Anseth KS. Adv Mater. 2009;21:5005. doi: 10.1002/adma.200901808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fairbanks BD, Schwartz MP, Bowman CN, Anseth KS. Biomaterials. 2009;30:6702. doi: 10.1016/j.biomaterials.2009.08.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.a) Napoli C, Giordano A, Casamassimi A, Pentimalli F, Ignarro LJ, De Nigris F. Int J Cancer. 2011;128:1505. doi: 10.1002/ijc.25743. [DOI] [PubMed] [Google Scholar]; b) Guedez L, Rivera AM, Salloum R, Miller ML, Diegmueller JJ, Bungay PM, Stetler-Stevenson WG. Am J Pathol. 2003;162:1431. doi: 10.1016/S0002-9440(10)64276-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ehrbar M, Metters A, Zammaretti P, Hubbell JA, Zisch AH. J Control Release. 2005;101:93. doi: 10.1016/j.jconrel.2004.07.018. [DOI] [PubMed] [Google Scholar]
- 35.a) Belo AV, Ferreira MAND, Bosco AA, Machado RDP, Andrade SP. Inflammation. 2001;25:91. doi: 10.1023/a:1007114404953. [DOI] [PubMed] [Google Scholar]; b) Andrade SP, Machado RDP, Teixeira AS, Belo AV, Tarso AM, Beraldo WT. Microvasc Res. 1997;54:253. doi: 10.1006/mvre.1997.2047. [DOI] [PubMed] [Google Scholar]; c) Barcelos LS, Talvani A, Teixeira AS, Cassali GD, Andrade SP, Teixeira MM. Inflamm Res. 2004;53:576. doi: 10.1007/s00011-004-1299-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.