Abstract
Conventional estrogen receptor α knockout (neo-ERαKO, neo-ERα−/−) mice contain a truncated and chimeric ERα fusion protein that retains 35% estrogen-dependent transactivation activity, and therefore the in vivo ERα function is difficult to study thoroughly. Furthermore, these neo-ERα−/− mice cannot be used for tissue and temporal specific ERα deletion. Therefore, there is a clear need to establish a floxed ERα mouse line that can knockout ERα specifically and completely in each selected cell type. Here we generated floxed ERα mice using a self-excising ACN (tACE-Cre/Neo) cassette. Mating the floxed ERα mice with ACTB-Cre mice produces a deletion of the floxed allele disrupting the reading frame of the ERα transcript so that no ERα protein is detected in the ACTBCre/ERα−/− mice. Expression of ERα target genes, such as G-6-PD and lactoferrin, is diminished by over 90% in the ACTB-Cre/ERα−/− uterus, but not in the neo-ERα−/− uterus. Furthermore, we also validated that ACTB-Cre/ERα−/− females have a hypoplastic internal genital tract, polycystic ovaries with hemorrhagic follicles, infertility, and higher body weight. Together, our data clearly demonstrate that the newly established floxed ERα mouse is a reliable mouse model for future studies of ERα roles in vivo in the selective estrogen target tissues. The complete knockout of ERα in the ACTB-Cre/ERα−/− mice will also provide an improved mouse model to study the role of ERα in vivo.
Keywords: Estrogen receptor α, Transgenic animal, Female reproduction
Introduction
Estrogens regulate the growth, differentiation, and developmental functions in a broad range of target tissues, such as the reproductive system, bone, and cardiovascular systems [1]. Estrogens actions are mediated by the cell- and tissue-specific transcriptional regulation of estrogen receptors (ERs) target genes via ERs [2]. ERs are members of the nuclear receptor (NR) superfamily and act as ligand-inducible transcription factors [3, 4], which are encoded by two distinct genes, ERα and ERβ. Classically, upon ligand binding, the ERs undergo a conformational change, receptor dimerization, and recruitment of distinct co-regulator complexes for transcriptional control [5]. Although in vitro assays have demonstrated that ERα and ERβ have similar DNA and ligand binding properties [6, 7], ERα and ERβ display a variety of differences in terms of tissue distribution, transcriptional activation, and knockout (KO) phenotypes [1, 6, 8, 9]. Those studies suggest that ERα and ERβ might regulate different target genes’ expression and biological functions [2, 6, 10].
To assay the in vivo ERα functions, Lubahn et al. [11] generated the ERαKO mice (designated as neo-ERα−/−) via the conventional gene KO strategy by insertion of a Neo cassette into exon II of the mouse ERα gene. Neo-ERα−/− mice do not express the full-length ERα, but contain a truncated and chimeric E1 protein, which results from a novel ERα transcript through alternative splicing, has seven amino acids in the N-terminal A/B regions replaced by the neomycin insert, and retains an intact DNA-binding domain (DBD) and ligand-binding domain (LBD). The E1 protein lacks AF-1 transactivation function, but still possesses estrogen-dependent transactivation capability; thus the conventional neo-ERα−/− mice do not represent a complete null ERα mutant [12].
In order to characterize the tissue and temporal-specific ERα function in specific target organs, we have generated the floxed ERα mouse line. By mating floxed ERα mice with ACTB-Cre mice, which ubiquitously express Cre recombinase, we generated ERαKO mice (ACTB-Cre/ERα−/−) to validate KO efficiency and ERα function. Both Dupont et al. [8] and Feng et al. [13] have generated floxed ERα mice to target exon III of the mouse ERα gene. However, it has been reported that variations in the design of the floxed allele in different mouse lines might generate unique or distinct phenotypes. For example, it is expected that the floxed androgen receptor (AR) mouse line should present wild type (Wt) phenotypes with normal hormonal profile and fertility [14]; yet, there is an example of a floxed AR mouse line that has an altered hormonal profile and reduced fertility [15]. Therefore, it is necessary to establish and characterize each independent “floxed” ERα mouse line. The floxed ERα mice developed in our laboratory differ somewhat in the strategy used to “flox” exon III (see Table 1). Importantly, however, we are the first group to characterize whether any residual ERα protein is expressed in ERα−/− mice (Table 1). Using different antibodies recognizing either the N terminus or C terminus of ERα, we demonstrate in this study the generation of a complete null ERα mouse without any remaining ERα protein. We have performed comparisons in expression of ERα, and of ERα transactivation capabilities, between the neo-ERα−/− and the ACTB-Cre/ERα−/− mice.
Table 1.
Comparison of currently available floxed ERα transgenic mice with the conventional neo-ERα−/− mice
| Targeted ERα exon |
Cassette for ES selection |
Neo cassette removal |
Residual ERα protein detected | ERα target gene expression in uterus |
|
|---|---|---|---|---|---|
| Couse et al. [1] | Exon II | PGK-neo | (knock-in) | + E1 protein; + immunoreactive ERα in uterus | Partially reduced |
| Dupont et al. [8] | Exon III | TK-neo | In vivo excision | − C terminus; NR* N terminus | NR* |
| Feng et al. [13] | Exon III | PGK-neo/HSV-TK | In vitro excision | NR* | NR* |
| Chen and Wolfe et al. | Exon III | tACE-Cre/Neo | In vivo excision | − N terminus (H-184); − C terminus (MC-20) | Over 90% diminished |
NR: data not reported
Materials and methods
Injection of targeted embryonic stem (ES) cells into mouse blastocysts and germline transmission of floxed ERα
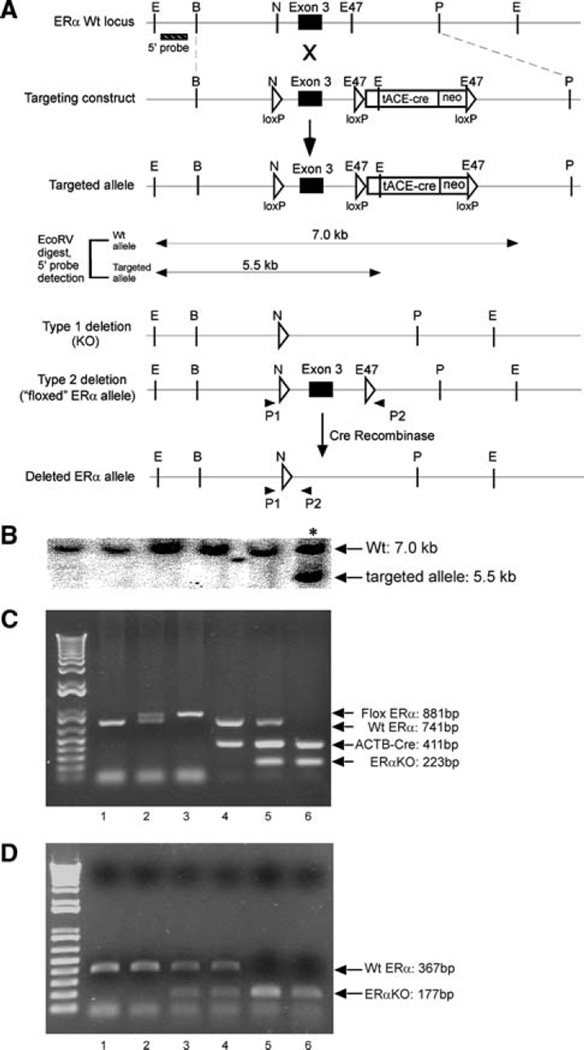
The cloned mouse ERα fragment was obtained from a 129Sv/J BAC clone of the ERα locus from the Microarray and Genomics Facility at Roswell Park Cancer Institute (Buffalo, NY) and used to generate a targeting construct where exon III of the mouse ERα gene was flanked by loxP sites [16]. First, an oligonucleotide containing a 34 bp loxP site was inserted into an NheI site 5′ to exon III. Next, a self-excising ACN (tACE-Cre/Neo) cassette was inserted into an Eco47III site 3′ to the same exon [17]. The self-excising ACN cassette includes a testis-specific promoter tACE (angiotensin-converting enzyme) driving the expression of Cre recombinase. The tACE-Cre fragment is adjacent to a Neo marker gene, and the entire tACE-Cre/Neo cassette is flanked by two loxP sites. The targeting vector was linearized and electroporated into 129 Sv/J mouse ES cells. ES cells containing the targeted ERα allele were identified by Southern Blot using an EcoRV digestion of ES cell genomic DNA. The probe consisted of a PstI/BglII fragment obtained from intron II.
The ES cells heterozygous for the allele containing the integrated homologous recombinant were injected into Wt mouse blastocysts. Chimeric males were mated to Wt C57BL/6 females. Two types of deletions were observed to occur in the testes of chimeric males: a Type I deletion where both the ACN cassette and exon III fragments are deleted and a Type II deletion where the ACN cassette, but not exon III, is deleted. In the Type II deletion, the mutant allele is marked by two loxP sites flanking exon III of the ERα gene and does not contain the ACN cassette.
Southern blot analysis of tail genomic DNA obtained from the Fl progeny was performed to identify Fl mice heterozygous for the Type II deletion (ERαfl/+). These mice were bred to homozygosity to generate homozygote floxed ERα mice (ERαfl/fl) and backcrossed with the pure C57BL/6 strain to generate a more homogenous background for further experimental analyses. All the animals were fed using a standard diet (#5001, LabDiets, Richmond, IN) and all animal procedures were approved by the animal care and use committee of the University of Rochester Medical Center, in accordance with National Institutes of Health guidelines.
Generation and genotyping of ACTB-Cre/ERα−/− mice
In order to generate the ACTB-Cre/ERα−/− mice, the ERαfl/fl female mice were crossed with ACTB-Cre transgenic male mice (Jackson Laboratories, Bar Harbor, ME). These mice ubiquitously express Cre recombinase under the control of the β-actin promoter. After two generations of mating, ACTB-Cre/ERα−/− mice were developed. Genomic DNA was isolated from tail biopsies and used as a template for PCR genotyping as previously described [14]. The primer pairs used were as follows: P1: 5′-AGGC TTTGTCTCGCTTTCC-3′, P2: 5′ -GATCATTCAGAGAG ACAAGAGGAACC-3′ (see Fig. 1a; floxed ER allele). Cre: forward, 5′-AGGTGTAGAGAAGGCACTTAGC-3′; reverse, 5′-CTAATCGCCATCTTCCAGCAGG-3′. The sizes of the P1–P2 fragments from Wt ERα, floxed ERα, and ERα KO allele were 741, 881, and 223 bp, respectively. The size of the Cre fragment was 411 bp. The PCR for the genotyping was performed with the P1, P2, and Cre primer mixture.
Fig. 1.
Targeted disruption of mouse ERα gene using Cre-loxP strategy. a Schematic map of Wt ERα, targeting construct, targeted allele, Type 1 deletion, Type 2 deletion, and deletedERα allele after crerecombination. NheI and Eco47III sites were used to insert the loxP sequence and pACN cassette. The primer set: P1 and P2, was used for genotyping. E (EcoRV), B (BamHI),N(NheI), E47 (Eco47III), P (PstI). b Southern Blot analysis was used to screen ES cell clones using the 1.5 kb 5′ probe. Southern Blot shows the positive clone (*) and five Wt clones. c Genotyping of different ERα mutant mice using primer mixture: P1, P2 and Cre primers. Lane 1, Wt mice (ERα+/+); Lane 2, floxed ERα heterozygote mice (ERαfl/+), the size of floxed ERα is 140 bp more than that of WtERα; Lane 3, floxed ERα homozygote mice (ERαfl/fl); Lane 4, ACTB-Cre transgenic mice (ACTB-cre/ERα+/+), the size of Cre fragment was 411 bp; Lane 5, heterozygote ERαKO mice (ACTB-Cre/ERα+/−), in which the size of floxed ERα allele was reduced to 223 bp after the deletion of intervening DNA via ACTB-cre recombinase; Lane 6, homozygote ERαKOmice (ACTB-Cre/ERα−/−), in which the size of both floxed ERα alleles was reduced to 223 bp by ACTB-cre recombinase and only deleted ERα band was present. d Semi-quantitative RT-PCR using primer sets P3 and P4 to detect the presence of ERα transcripts in the uterine cDNA from Wt, ACTB-Cre/ERα+/−, or ACTB-Cre/ERα−/− mice. Wt: Lanes 1 and 2; ACTB-Cre/ ERα+/−: Lanes 3 and 4; ACTB-Cre/ERα−/−: Lanes 5 and 6
Fertility test
Five 14- to 16-week-old Wt, ACTB-Cre/ERα+/−, and ACTB-Cre/ERα−/− females were mated with known fertile males for 18 weeks. The number of pups and litters was recorded.
RNA isolation, semi-quantitative RT-PCR, and real-time PCR
Total RNA was extracted and purified from the uteri of different female genotypes at 12 weeks of age using Trizol (Invitrogen, Carlsbad, CA), according to the manufacturer’s instructions, then 3 µg RNA was subjected to reverse transcription using Superscript III (Invitrogen, Carlsbad, CA). The resulting first strand cDNA was subjected to semi-quantitative RT-PCR with the following ERα-specific primer pairs: P3, 5′-GGTGCCCTACTACCTGGAG-3′ and P4, 5′-GCCCACTTCGT AACACT TGCG C-3′. P3 and P4 are located in the mouse ERα exon II and exon IV respectively. The sizes of P3–P4 fragments from Wt and ERαKO transcripts were 367 and 177 bp, respectively. The real-time PCR was performed with first strand cDNA, specific gene primers, and SYBR Green PCR Master Mix (Biorad, Hercules, CA). The PCR cycle was performed as follows: 94°C for 3 min, 40 cycles of 94°C for 30 s, 60°C for 30 s, and 72°C for 30 s on an iCycler iQ Multi-color real-time PCR detection system (Biorad, Hercules, CA). Primer sequences were as follows: G-6-PD: sense, 5′-TTAAAGCCACTCCAGAAGAAAGAC-3′; antisense, 5′-TCCACGATGATGCGGTTCC-3′. Lactoferrin: sense, 5′-TAGCAGCAGTTAGAAGAGAAGATG-3′; antisense, 5′-GGGCACAGAGATTGGATTTGG-3′. 18S: sense, 5′-TGCCTTCCTTGGATGTGGTAG-3′; antisense, 5′-CGTCTGCCCTATCAACTTTCG-3′. Each sample was run in triplicate, and data were analyzed using iCycler iQ software (Biorad, Hercules, CA).
Antibodies
Mouse monoclonal anti-GAPDH antibody 6C5, rabbit polyclonal antibody H-184 directed against the 2–185 amino acids (a.a.) of human ERα N terminus, and MC-20 against the last 20 a.a. of the mouse ERα C terminus were obtained from Santa Cruz Biotechnology.
ERαKO N terminus cDNA construct
The putative N-terminal ERαKO cDNAs were generated by PCR from the pcDNA3-flag-mERα plasmid using the forward primer: 5′-CGGGATCCATGACCATGACCCTT CACACCAAAG-3′ and reverse primer: 5′-CGGAATTC TCATTGTGTCCTGTAGAAGGCGGGAG-3′ and subsequently constructed into pcDNA3-flag vector. The correct construction of the plasmid was verified by sequencing and protein expression was confirmed by TNT in vitro expression.
Western blotting
Protein extracts were prepared from the uteri of different mouse genotypes as described previously [18, 19], 80 µg of protein was separated on a 10% gel by SDS-PAGE, and transferred onto a nitrocellulose membrane. ERα protein was detected with the rabbit polyclonal antibodies H-184 and MC-20 (Santa Cruz, CA). The same blot was hybridized with an anti-GAPDH antibody to control for equal loading (Santa Cruz, CA).
Immunohistochemistry (IHC)
The IHC was carried out as described previously [19, 20]. In brief, the paraffin-embedded tissue blocks were cut at 4-µm thickness, dewaxed, and rehydrated. Antigens were retrieved by boiling in 10 mM citrate buffer (pH 7.0) for 10 min. The sections were incubated in 0.5% H2O2 in PBS for 30 min at room temperature to quench endogenous peroxidase. To block nonspecific binding, sections were incubated in 5% normal serum prepared from the host of secondary antibodies for 1 h at 4°C. Sections were incubated with ERα antibodies (MC-20, 1:400) in 3% BSA in PBS overnight at 4°C, and then incubated with 1:200 diluted biotinylated secondary antibody (Vector Laboratories) and ABC solution (Vector Laboratories). The tissues were then stained by AEC followed by Mayer’s hematoxylin counterstaining (DAKO, Carpenteria, CA). Negative controls were incubated without primary antibody.
Results and discussions
Generation of floxed ERα mouse
Exon III of the ERα gene encodes the first zinc finger of the ERα-DBD and plays an important role in ER–ERE binding and transcriptional regulation of ERα target genes [1]. The deletion of exon III results in a translational frameshift in ERα mRNA by the splicing of exon II and IV; thus, we generated a floxed ERα mouse line by targeting exon III. Homologous recombination was used to insert a floxed ERα allele into the ES cell genomic DNA. A self-excising neomycin cassette was used to select for targeting vector incorporation. The strategy is outlined in Fig. 1a. Homologous recombination was determined by Southern Blot of an EcoRV-digested ES cell genomic DNA (Fig. 1b), using the probe consisting of a PstI/BglII fragment obtained from intron II (Probe, Fig. 1a). Targeted ES cells were injected into C57BL/6 blastocysts. The self-excising tACE-Cre/Neo cassette was then removed from the mutant allele in vivo. Retaining a Neo cassette in vivo was shown to have unpredictable consequences on the phenotype of mice, including the generation of a hypomorphic allele with a wide range of phenotypic abnormalities [21, 22], and mis-regulation of adjacent genes [23]. The tACE (angiotensin-converting enzyme) promoter present in the ACN cassette has been shown to specifically activate Cre recombinase activity in the testis sperm cells, but not in other tissues [17]. Thus, self-excision of the ACN cassette in the sperm of chimeric male mice resulted in transmitting the mutant allele that contained the floxed ERα gene, lacking the ACN cassette, to the F1 progeny. F1 mice heterozygous for the Type II deletion were selected (heterozygote floxed ERα mice, ERαfl/+) via Southern blot and PCR analysis of tail DNA and bred to generate homozygous floxed ERα mice (ERαfl/fl). Table 1 includes the targeting strategy used by our laboratory and by two other groups.
Generation of ACTB-Cre/ERα−/− mice
The ACTB-Cre/ERα−/− mice were generated by mating the ERαfl/fl female mice with ACTB-cre transgenic male mice. PCR performed using the Cre primers, and P1 and P2 primers, was able to distinguish the Wt mice (ERα+/+, 741 bp band), heterozygote floxed ERα mice (ERαfl/+; 741 bp and 881 bp band), homozygote floxed ERα mice (ERαfl/fl; only 881 bp band), ACTB-cre transgenic mice (ACTB-Cre/ERα+/+; 411 and 741 bp band), heterozygous ERαKO mice (ACTB-Cre/ERα+/−; 411, 741, and 223 bp band), and homozygous ERαKO mice (ACTB-Cre/ERα−/−; 411 and 223 bp) (Fig. 1c, lanes 1–6, respectively). PCR products were cloned and further sequenced to confirm that the targeted fragment contained the predicted sequence (data not shown). The disruption of ERα gene expression was also confirmed by semi-quantitative RTPCR of messenger RNA from the uterus. Using the primer set of P3 and P4 located in exon II and IV, a PCR product of 177 bp was amplified from reverse transcribed cDNA of ACTB-cre/ERα−/− uterus. Reverse transcribed cDNA of Wt mouse uterus produced a 367 bp product, consistent with the expected size of the Wt locus, while ACTB-cre/ERα+/− heterozygous mice reveal the presence of a Wt (367 bp) and KO (177 bp) transcript (Fig. 1d). DNA sequencing showed that the KO transcripts completely lack exon III (data not shown).
Further confirmation of the complete null ERα mutant in the ACTB-Cre/ERα−/− mice
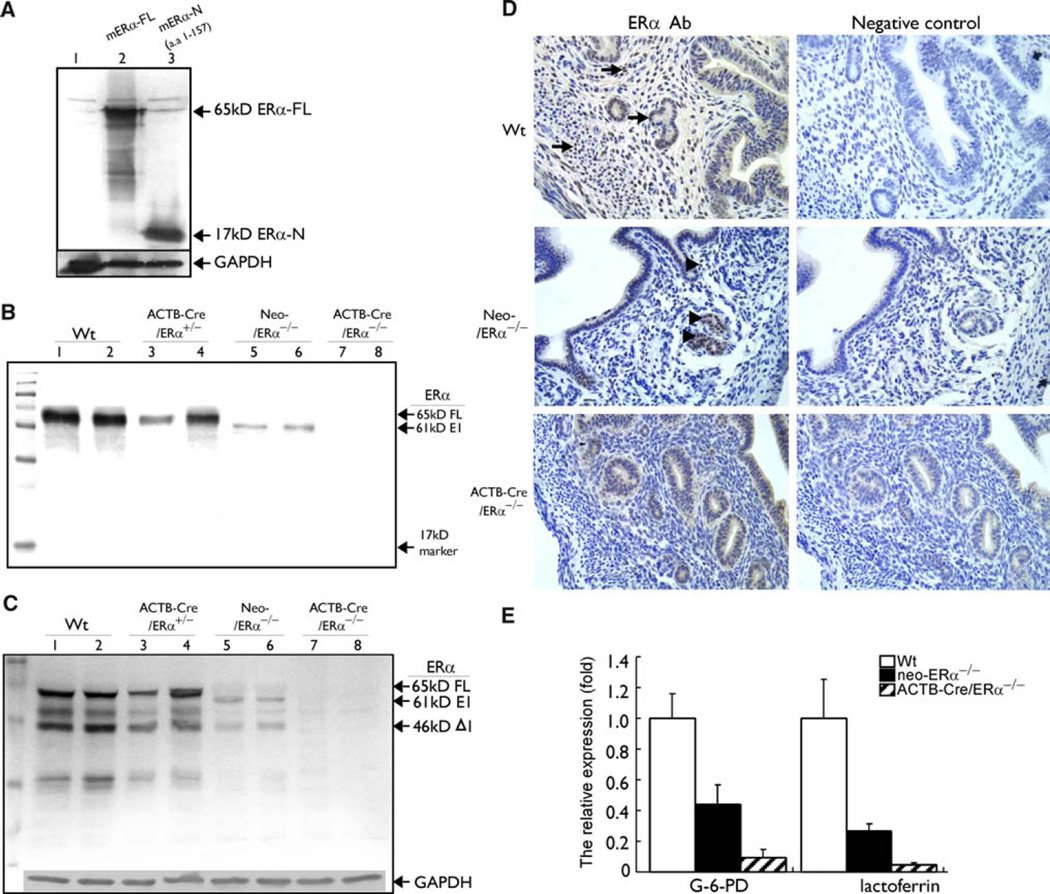
Deletion of exon III by Cre-mediated recombination resulted in a premature stop codon at the new codon 158 position due to the splicing of exon II and IV. Thus, the putative truncated ERα protein from ACTB-Cre/ERα−/− mice would be predicted to be 157 a.a. in length in the absence of a.a. from the ERα DBD and LBD. To examine whether the putative mouse ERα N polypeptide (a.a. 1–157) is expressed in the ACTB-Cre/ERα−/− uterine extracts, we first validated which ERα antibody can recognize mouse ERα N terminus. A rabbit polyclonal antibody, H184, was originally generated to recognize the N terminus of human ERα (a.a. 2–185). Previous studies have shown that this antibody could recognize the mouse ERα [24]; however, the immunoreactive epitopes of H184 are not well characterized. It was unclear whether H184 could recognize the first 157 a.a. of mouse ERα. Therefore, the mouse ERα N terminus (a.a. 1–157) was cloned and expressed in Cos-1 cells. The Cos-1 cell lysates were subjected to western blot analysis using the H184 antibody, and our results indicate that this antibody could detect both mouse ERα full-length and N-terminal polypeptide (a.a. 1–157) (Fig. 2a, lanes 2 and 3).
Fig. 2.
Detection of the expression of ERα protein and estrogen target genes to prove the complete deletion of ERα in the ACTB-Cre/ERα−/− mice. a The recognition of mouse ERα N terminus by the ERα antibody (H-184). The full length and N terminus of mouse ERα were cloned into pCDNA3 plasmids and transfected into Cos-1 cells for the western blot analysis. Our results indicated that both mouse ERα full length and N-terminal (a.a. 1–157) proteins could be recognized by ERα antibody H-184. Note that the H-184 antibody was originally produced against the human ERα. b–c Western blot analysis of ERα protein in the uterus of Wt, neo-ERα−/−, ACTB-Cre/ ERα+/−, and ACTB-Cre/ERα−/− using antibody H-184 against mouse ERα N terminus (b) or antibody MC-20 against the C terminus (c). Note that neo-ERα−/− uteri encode a 61-kD chimeric protein (b and c, Lanes 5 and 6) and a 46-kD ERα Δ1 protein (c, Lanes 5 and 6). No ERα immunoreactivity was detected in ACTB-Cre/ERα−/− uteri (b and c, Lanes 7 and 8). GAPDH was immunoblotted as an indication of equal loading. d IHC analyses of ERα protein in the Wt, neo-ERα−/−, and ACTB-Cre/ERα−/− uterus. MC-20 antibody was used to locate ERα in paraffin-embedded tissue. ERα was mainly expressed in the nuclei of myometrium and glandular epithelial cells (arrow) in the Wt uterus. Note that positive nuclear ERα staining can still be found in the nuclei of some glandular epithelial and stromal cells (arrowhead) in the neo-ERα−/− mice; no positive nuclear ERα staining was detected in our newly generated ACTB-Cre/ERα−/− mice. e The comparison of the expression levels of the estrogen target genes among Wt, neo-ERα−/−, and ACTB-Cre/ERα−/− by real-time PCR. Assays were performed on RNA from the uterus of each individual mouse and then averaged for the gene expression. 18S acted as internal control. Note that the expressions of G-6-PD and lactoferrin were shown more dramatically decreased in the ACTBCre/ERα−/− uterus
We then used western blot analysis to examine whether ERα N terminus was expressed in ACTB-Cre/ERα−/− uterine extracts using the H184 antibody. The Wt and neo-ERα−/− mice revealed a 65-kD ERα protein and a novel 61-kD E1 protein, respectively (Fig. 2b, lanes 1, 2 and 5, 6). In contrast, the disruption of ERα in our ACTB-Cre/ERα−/− mice resulted in the absence of the putative ERα N-terminal 157-amino-acid polypeptide (Fig. 2b, lanes 7 and 8). The deletion of ERα exon III in the ACTB-Cre/ERα−/− mice produces a nonsense mutation that results in a premature termination codon in ERα exon IV. The studies in the eukaryotic cells have shown that the nonsense mutation may lead to mRNA decay and prevent the accumulation of truncated protein [25]. Although it is expected that the premature termination codon in the ERα exon IV will yield a 157-amino-acid N-terminal ERα polypeptide, nonsense-mediated mRNA decay (NMD) could degrade the mutant ERα mRNAs containing the premature termination codon and prevent the synthesis of the 157-amino-acid N-terminal ERα polypeptide in the ACTB-Cre/ERα−/− mice. It is also possible that the ERα N terminus is present, but undetectable, in ACTB-Cre/ERα−/− mice, due to instability of the protein. To our knowledge, this is the first report to definitely show the absence of the ERα N terminus in ERαKO mice.
Moreover, using the MC-20 antibody, recognizing the ERα C terminus, western blotting of Wt uterine extracts revealed not only a 65-kD product, consistent with full-length ERα, but also a 46-kD product ERα Δ1. This is an isoform of ERα splicing of exon II and encodes an in-frame ERα protein with intact DBD and LBD domains by utilizing the start codon in exon III and a favorable Kozak sequence (Fig. 2c, lanes 1 and 2) [26]. The ERα Δ1 protein was found to be expressed in the mouse uterus and human cells [27, 28]. In the previously generated neo-ERα−/− mice, a 65-kD full-length ERα protein was not detectable, but a 61-kD-truncated E1 protein was detected in the uterine extracts of neo-ERα−/− mice, which was consistent with the results detected by ERα antibody H-184 (Fig. 2c, lanes 5 and 6). In addition, a 46-kD ERα Δ1 was detected in the uterine extracts of neo-ERα−/− mice (Fig. 2c, lanes 5 and 6). Our ACTB-Cre/ERα−/− mice did not express ERα Δ1 due to the deletion of exon III. Collectively, the full disruption of ERα in our ACTB-Cre/ERα−/− mice was shown by the absence of any ERα residual protein immunoreactivity with either the ERα-N-terminal antibody (H-184) or ERα-C-terminal antibody (MC-20) in the uterine extracts (Fig. 2b, c).
In addition, we examined ERα expression in the paraffin- embedded uterine tissue by IHC staining. ERα was mainly expressed in the uterine stromal and glandular epithelial cells with little positive nuclear staining in the luminal epithelial cells of 3-month-old Wt mice (Fig. 2d, arrows). The majority of positive ERα staining in the stromal cells was abolished in the neo-ERα−/− uterus, however, the positive nuclear ERα staining can still be found in some glandular epithelial and stromal cells (Fig. 2d, arrowheads). In contrast, there was no positive nuclear ERα staining, which could be detected in the ACTB-Cre/ERα−/− mice, except some diffuse cytoplasmic background staining (Fig. 2d).
Real-time PCR analysis was used to detect the transcription of estrogen target genes in the mouse uterine tissue. Compared to the Wt mice, the expressions of G-6-PD and lactoferrin in neo-ERα−/− uterus were 0.44 ± 0.125 and 0.265 ± 0.057, respectively, whereas, the expressions of G-6-PD and lactoferrin in the ACTB-Cre/ERα−/− uterus were dramatically decreased to 0.093 ± 0.051 and 0.050 ± 0.012, respectively (Fig. 2e). Collectively, these results demonstrate that our newly generated ACTB-Cre/ERα−/− mice are indeed a complete ERα null mutant and provide a mouse model completely lacking in ERα signaling to study the role of ERα in vivo. Furthermore, direct comparisons with the neo-ERα−/− mouse underscore the value of analyzing the ACTB-Cre/ERα−/− mice as a model of ERα deficiency.
Defective reproductive system in ACTB-Cre/ERα−/− females
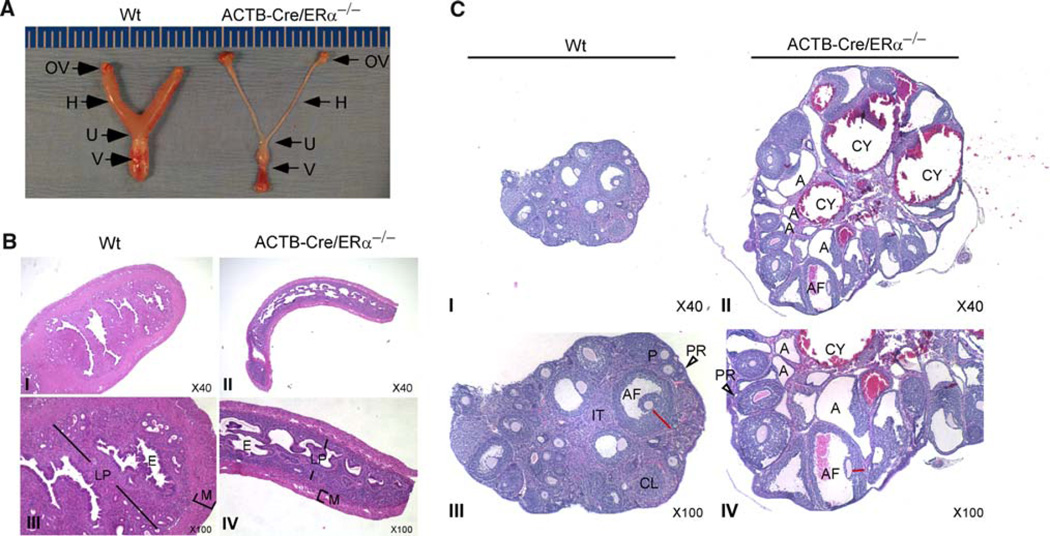
To confirm the phenotypes that have been reported in other ERαK O mouse models, more than five age-matched littermates were used for each experiment. ACTB-Cre/ERα−/− females had similar appearing external genitalia as Wt females; however, the vagina of ACTB-Cre/ERα−/− females did not display any cyclic changes when vaginal smears were performed (data not shown). Gross internal anatomical examination revealed that ACTB-Cre/ERα−/− uterus and vagina were hypoplastic and presented a much more severe uterine phenotype than the hypomorphic phenotype (half the weight of Wt littermates) in the neo-ERα−/− uterus (Fig. 3a) [1]. Histological analyses showed that the ACTB-Cre/ERα−/− uterus contained substantially reduced numbers of stromal cells in both the myometrium and lamina propria, which resulted in significant differences in the uterine diameter between Wt and ACTB-Cre/ERα−/− females, however, the length of uterus had no significant difference between Wt and ACTB-Cre/ERα−/− females (Fig. 3b, I vs. II; III vs. IV), which indicate that the growth of uteri along the body axis is independent of ERα function. This finding agrees with other ERαKO models [8].
Fig. 3.
Comparison of uterus and ovary of 16-week-old Wt and ACTB-Cre/ERα−/−. a Ventral views of the body of the uterus (U), uterine horns (H), ovaries (OV), and vagina (V). b Histological analyses of mouse uterus. Note that ACTB-Cre/ERα−/− uterus was much thinner with a substantial reduction in the total number of stromal cells in the myometrium and lamina propria. M, myometrium; LP, lamina propria, E, epithelia. c Histological analyses of the ovary. Note that ovaries from adult ACTB-Cre/ERα−/− mice (16-week-old) displayed dilated, frequently hemorrhagic cysts (CY); an excess of atretic follicles (A); normal primordial (PR), primary (P) and antral follicles (AF) with few interstitial compartments (IT); and lack of corpus luteum (CL)
As shown in Fig. 3c, in the ovary of Wt mice, primordial follicles (PR, arrowhead), primary follicles (P), antral follicles (AF), and corpus luteum (CL) were all present and well organized, and the interstitial compartment (IT) was abundant and consisted of compact clusters of glandular and fibroblast cells. In contrast, ovaries from adult ACTBCre/ ERα−/− females displayed many large dilated, hemorrhagic cysts (CY). The primordial follicles (PR, arrowhead) and primary follicles were observed in the ACTB-Cre/ERα−/− ovary, but with greatly reduced numbers compared to Wt mice. The antral follicles were also observed in ACTB-Cre/ERα−/− ovaries (AF); however, the thickness of the granulosa cells in the antral follicles of ACTB-Cre/ERα−/− females was significantly reduced (red line, III vs. IV). More prominently, ACTB-Cre/ERα−/− ovaries had few interstitial compartments, but an excess of atretic follicles which were derived from the apoptotic antral follicles due to their inability to develop to the preovulatory stage (A). No corpus luteum was observed in ACTB-Cre/ERα−/− ovaries. Overall, folliculogenesis in ACTB-Cre/ERα−/− females is defective and can only progress into the antral stage. These findings agree with previous studies showing that a functional ERα is required for the functional maturation of preovulatory follicles [8, 29]. Furthermore, our results also suggest that ERα is necessary for interstitial glandular cell development.
Increased body weight and infertility of ACTB-Cre/ERα−/− females
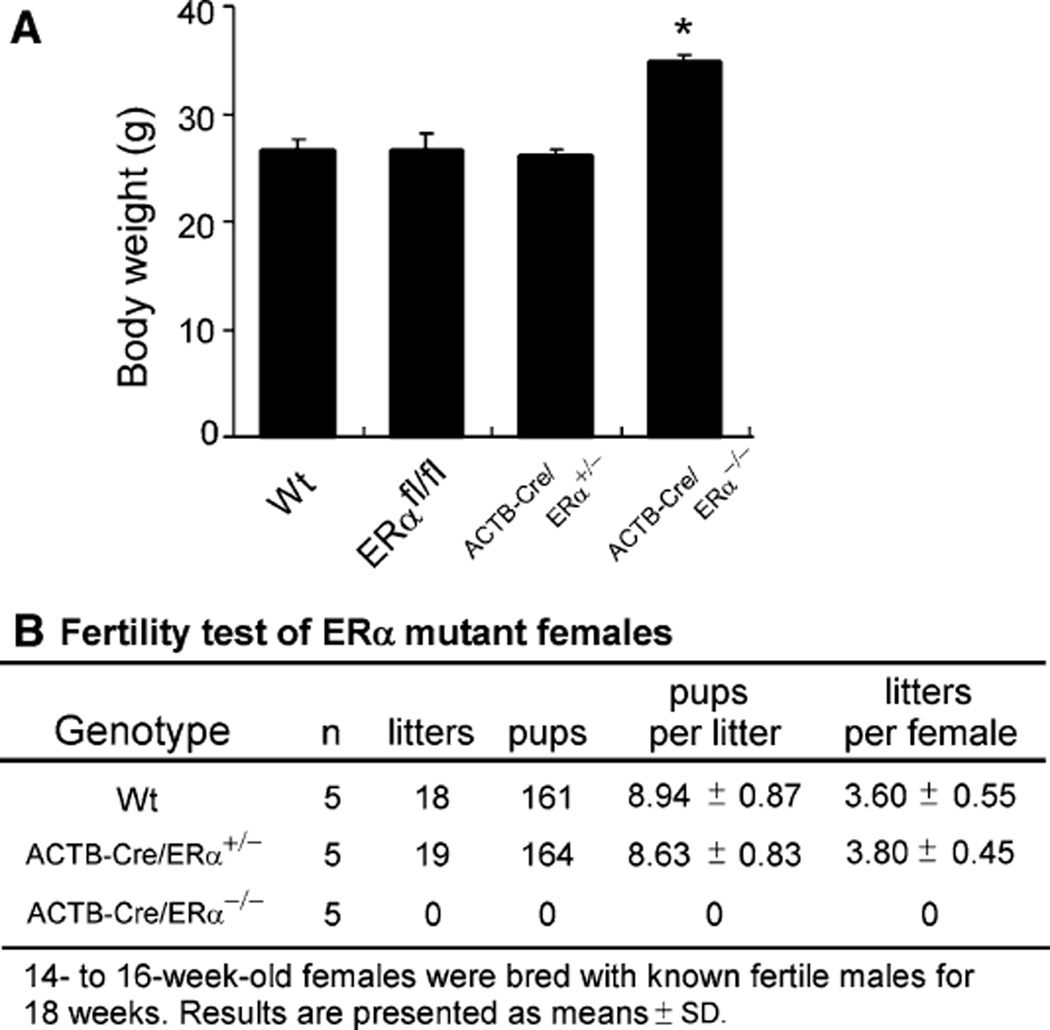
The analyses of ACTB-Cre/ERα−/− females showed that they developed obesity: the body weight of young adult ACTB-Cre/ERα−/− females (12-week-old) was significantly higher than the control mice (P < 0.05, Fig. 4a). ACTB-Cre/ERα−/− females had significant lipid accumulation in the subcutaneous white adipose tissues (WATs) (data not shown). Those results agree with other studies that have examined the increased body weight of ovariectomized mice and neo-ERα−/− females, as well as the role of ERα in suppressing the development of WATs [30, 31]. In agreement with previously published results [8, 11], ACTB-Cre/ERα−/− females were infertile as shown by 18 weeks of continuous mating test (Fig. 4b). Fertility of Wt and ACTB-Cre/ERα+/− females was normal and comparable.
Fig. 4.
Body weight and fertility of ERα mutant females. a Body weights of Wt, ERαfl/fl, ACTB-Cre/ERα+/−, and ACTB-Cre/ERα−/− females. *P < 0.05; ACTB-Cre/ERα−/− versus the other females. Values are presented as mean ± SEM. (n = 8). b ACTB-Cre/ERα−/− females are infertile after 18 weeks of fertility test as compared with the normal fertility of ACTB-Cre/ERα+/− and Wt females
In conclusion, we have generated floxed ERα mice using a self-excising ACN (tACE-Cre/Neo) cassette. Although both Dupont et al. [8] and Feng et al. [13] have generated floxed ERα mice, our floxed ERα mice were developed using a different targeting construct strategy (Table 1). It is true that this mouse model would be one of three reporting a floxed ERα locus, however, ours is the only study that actually compares the effect of an in vivo deletion of exon III with the insertional mutagenesis used to produce the neo- ERα−/− mouse. We show a much more severe uterine phenotype when exon III is deleted in the ACTB-Cre/ERα−/− females than the hypomorphic phenotype in neo-ERα−/− females [1]. Furthermore, we characterized any residual ERα protein being expressed in ERαKO mice (Table 1). We showed in mouse uterine extracts that our ACTB-Cre/ERα−/− mice lack any detectable ERα protein versus a truncated protein in the neo-ERα−/− line using antibodies recognizing either the N-terminus or C-terminus of ERα. To our knowledge, this is the first report to definitely show the absence of the ERα N terminus in ERαKO mice. We also showed that there are almost completely abolished expressions of ERα target genes, G-6-PD and lactoferrin, in the ACTB-Cre/ERα−/− uterus, whereas expression of these genes was still 44% and 27% of Wt values, respectively, in the neo-ERα−/− uterus. Using our ACTB-Cre/ERα−/− mouse model, we have characterized the function of ERα in the female genital tracts. These studies form the basis for the use of the floxed ERα mouse to selectively and completely delete the ERα gene in a tissue- or temporal-specific manner by breeding with transgenic mice harboring promoter specific Cre. The complete knockout of ERα in the ACTB-Cre/ ERα−/− mice also provides an improved mouse model to study the role of ERα in vivo.
Acknowledgments
This work was partly supported by NIH grant DK60912. This research was also supported in part by NICHD/NIH through cooperative agreement (U54 HD 933067, the Baltimore- Chicago Center for Reproductive Research) as part of the Specialized Cooperative Centers Program in Reproduction and Infertility Research (SCCPIR). The technical assistance of Yayoi Shibusawa is gratefully acknowledged. We also thank Karen Wolf and Susan R. Schoen for assisting in manuscript preparation.
Contributor Information
Ming Chen, George Whipple Lab for Cancer Research, Departments of Pathology and Urology, University of Rochester Medical Center, 601 Elmwood Ave, Box 656, Rochester, NY 14642, USA.
Andrew Wolfe, Department of Pediatrics, Johns Hopkins University School of Medicine, 600 North Wolfe Street, CMSC 406, Baltimore, MD 21287, USA.
Xi Wang, George Whipple Lab for Cancer Research, Departments of Pathology and Urology, University of Rochester Medical Center, 601 Elmwood Ave, Box 656, Rochester, NY 14642, USA.
Chawnshang Chang, George Whipple Lab for Cancer Research, Departments of Pathology and Urology, University of Rochester Medical Center, 601 Elmwood Ave, Box 656, Rochester, NY 14642, USA.
Shuyuan Yeh, Email: shuyuan_yeh@urmc.rochester.edu, George Whipple Lab for Cancer Research, Departments of Pathology and Urology, University of Rochester Medical Center, 601 Elmwood Ave, Box 656, Rochester, NY 14642, USA.
Sally Radovick, Email: sradovick@jhmi.edu, Department of Pediatrics, Johns Hopkins University School of Medicine, 600 North Wolfe Street, CMSC 406, Baltimore, MD 21287, USA.
References
- 1.Couse JF, Korach KS. Estrogen receptor null mice: what have we learned and where will they lead us? Endocr Rev. 1999;20:358–417. doi: 10.1210/edrv.20.3.0370. [DOI] [PubMed] [Google Scholar]
- 2.Liu MM, Albanese C, Anderson CM, Hilty K, Webb P, Uht RM, Price RH, Jr, Pestell RG, Kushner PJ. Opposing action of estrogen receptors alpha and beta on cyclin D1 gene expression. J Biol Chem. 2002;277:24353–24360. doi: 10.1074/jbc.M201829200. [DOI] [PubMed] [Google Scholar]
- 3.Kuiper GG, Enmark E, Pelto-Huikko M, Nilsson S, Gustafsson JA. Cloning of a novel receptor expressed in rat prostate and ovary. Proc Natl Acad Sci USA. 1996;93:5925–5930. doi: 10.1073/pnas.93.12.5925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Green S, Walter P, Kumar V, Krust A, Bornert JM, Argos P, Chambon P. Human oestrogen receptor cDNA: sequence, expression and homology to v-erb-A. Nature. 1986;320:134–139. doi: 10.1038/320134a0. [DOI] [PubMed] [Google Scholar]
- 5.Rosenfeld MG, Glass CK. Coregulator codes of transcriptional regulation by nuclear receptors. J Biol Chem. 2001;276:36865–36868. doi: 10.1074/jbc.R100041200. [DOI] [PubMed] [Google Scholar]
- 6.Paech K, Webb P, Kuiper GG, Nilsson S, Gustafsson J, Kushner PJ, Scanlan TS. Differential ligand activation of estrogen receptors ERalpha and ERbeta at AP1 sites. Science. 1997;277:1508–1510. doi: 10.1126/science.277.5331.1508. [DOI] [PubMed] [Google Scholar]
- 7.Pace P, Taylor J, Suntharalingam S, Coombes RC, Ali S. Human estrogen receptor beta binds DNA in a manner similar to and dimerizes with estrogen receptor alpha. J Biol Chem. 1997;272:25832–25838. doi: 10.1074/jbc.272.41.25832. [DOI] [PubMed] [Google Scholar]
- 8.Dupont S, Krust A, Gansmuller A, Dierich A, Chambon P, Mark M. Effect of single and compound knockouts of estrogen receptors alpha (ERalpha) and beta (ERbeta) on mouse reproductive phenotypes. Development. 2000;127:4277–4291. doi: 10.1242/dev.127.19.4277. [DOI] [PubMed] [Google Scholar]
- 9.Kushner PJ, Agard DA, Greene GL, Scanlan TS, Shiau AK, Uht RM, Webb P. Estrogen receptor pathways to AP-1. J Steroid Biochem Mol Biol. 2000;74:311–317. doi: 10.1016/s0960-0760(00)00108-4. [DOI] [PubMed] [Google Scholar]
- 10.Lazennec G, Bresson D, Lucas A, Chauveau C, Vignon F. ER beta inhibits proliferation and invasion of breast cancer cells. Endocrinology. 2001;142:4120–4130. doi: 10.1210/endo.142.9.8395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lubahn DB, Moyer JS, Golding TS, Couse JF, Korach KS, Smithies O. Alteration of reproductive function but not prenatal sexual development after insertional disruption of the mouse estrogen receptor gene. Proc Natl Acad Sci USA. 1993;90:11162–11166. doi: 10.1073/pnas.90.23.11162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Couse JF, Curtis SW, Washburn TF, Lindzey J, Golding TS, Lubahn DB, Smithies O, Korach KS. Analysis of transcription and estrogen insensitivity in the female mouse after targeted disruption of the estrogen receptor gene. Mol Endocrinol. 1995;9:1441–1454. doi: 10.1210/mend.9.11.8584021. [DOI] [PubMed] [Google Scholar]
- 13.Feng Y, Manka D, Wagner KU, Khan SA. Estrogen receptor-alpha expression in the mammary epithelium is required for ductal and alveolar morphogenesis in mice. Proc Natl Acad Sci USA. 2007;104:14718–14723. doi: 10.1073/pnas.0706933104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yeh S, Tsai MY, Xu Q, Mu XM, Lardy H, Huang KE, Lin H, Yeh SD, Altuwaijri S, Zhou X, Xing L, Boyce BF, Hung MC, Zhang S, Gan L, Chang C. Generation and characterization of androgen receptor knockout (ARKO) mice: an in vivo model for the study of androgen functions in selective tissues. Proc Natl Acad Sci USA. 2002;99:13498–13503. doi: 10.1073/pnas.212474399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Maclean HE, Chiu WM, Ma C, McManus JF, Davey RA, Cameron R, Notini AJ, Zajac JD. A floxed allele of the androgen receptor gene causes hyperandrogenization in male mice. Physiol Genomics. 2008;33(1):133–137. doi: 10.1152/physiolgenomics.00260.2007. [DOI] [PubMed] [Google Scholar]
- 16.Lakso M, Sauer B, Mosinger B, Jr, Lee EJ, Manning RW, Yu SH, Mulder KL, Westphal H. Targeted oncogene activation by site-specific recombination in transgenic mice. Proc Natl Acad Sci USA. 1992;89:6232–6236. doi: 10.1073/pnas.89.14.6232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bunting M, Bernstein KE, Greer JM, Capecchi MR, Thomas KR. Targeting genes for self-excision in the germ line. Genes Dev. 1999;13:1524–1528. doi: 10.1101/gad.13.12.1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang M, Altuwaijri S, Yeh S. RRR-alpha-tocopheryl succinate inhibits human prostate cancer cell invasiveness. Oncogene. 2004;23:3080–3088. doi: 10.1038/sj.onc.1207435. [DOI] [PubMed] [Google Scholar]
- 19.Yin Y, Ni J, Chen M, DiMaggio MA, Guo Y, Yeh S. The therapeutic and preventive effect of RRR-alpha-vitamin E succinate on prostate cancer via induction of insulin-like growth factor binding protein-3. Clin Cancer Res. 2007;13:2271–2280. doi: 10.1158/1078-0432.CCR-06-1217. [DOI] [PubMed] [Google Scholar]
- 20.Ni J, Wen X, Yao J, Chang HC, Yin Y, Zhang M, Xie S, Chen M, Simons B, Chang P, di Sant’Agnese A, Messing EM, Yeh S. Tocopherol-associated protein suppresses prostate cancer cell growth by inhibition of the phosphoinositide 3-kinase pathway. Cancer Res. 2005;65:9807–9816. doi: 10.1158/0008-5472.CAN-05-1334. [DOI] [PubMed] [Google Scholar]
- 21.Colledge WH, Abella BS, Southern KW, Ratcliff R, Jiang C, Cheng SH, MacVinish LJ, Anderson JR, Cuthbert AW, Evans MJ. Generation and characterization of a delta F508 cystic fibrosis mouse model. Nat Genet. 1995;10:445–452. doi: 10.1038/ng0895-445. [DOI] [PubMed] [Google Scholar]
- 22.Olson EN, Arnold HH, Rigby PW, Wold BJ. Know your neighbors: three phenotypes in null mutants of the myogenic bHLH gene MRF4. Cell. 1996;85:1–4. doi: 10.1016/s0092-8674(00)81073-9. [DOI] [PubMed] [Google Scholar]
- 23.Meyers EN, Lewandoski M, Martin GR. An Fgf8 mutant allelic series generated by Cre- and Flp-mediated recombination. Nat Genet. 1998;18:136–141. doi: 10.1038/ng0298-136. [DOI] [PubMed] [Google Scholar]
- 24.Kos M, Denger S, Reid G, Korach KS, Gannon F. Down but not out? A novel protein isoform of the estrogen receptor alpha is expressed in the estrogen receptor alpha knockout mouse. J Mol Endocrinol. 2002;29:281–286. doi: 10.1677/jme.0.0290281. [DOI] [PubMed] [Google Scholar]
- 25.Isken O, Maquat LE. Quality control of eukaryotic mRNA: safeguarding cells from abnormal mRNA function. Genes Dev. 2007;21:1833–1856. doi: 10.1101/gad.1566807. [DOI] [PubMed] [Google Scholar]
- 26.Flouriot G, Brand H, Denger S, Metivier R, Kos M, Reid G, Sonntag-Buck V, Gannon F. Identification of a new isoform of the human estrogen receptor-alpha (hER-alpha) that is encoded by distinct transcripts and that is able to repress hER-alpha activation function 1. EMBO J. 2000;19:4688–4700. doi: 10.1093/emboj/19.17.4688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Barraille P, Chinestra P, Bayard F, Faye JC. Alternative initiation of translation accounts for a 67/45 kDa dimorphism of the human estrogen receptor ERalpha. Biochem Biophys Res Commun. 1999;257:84–88. doi: 10.1006/bbrc.1999.0334. [DOI] [PubMed] [Google Scholar]
- 28.Kos M, O’Brien S, Flouriot G, Gannon F. Tissue-specific expression of multiple mRNA variants of the mouse estrogen receptor alpha gene. FEBS Lett. 2000;477:15–20. doi: 10.1016/s0014-5793(00)01750-6. [DOI] [PubMed] [Google Scholar]
- 29.Schomberg DW, Couse JF, Mukherjee A, Lubahn DB, Sar M, Mayo KE, Korach KS. Targeted disruption of the estrogen receptor-alpha gene in female mice: characterization of ovarian responses and phenotype in the adult. Endocrinology. 1999;140:2733–2744. doi: 10.1210/endo.140.6.6823. [DOI] [PubMed] [Google Scholar]
- 30.Wade GN, Gray JM, Bartness TJ. Gonadal influences on adiposity. Int J Obes. 1985;9(Suppl 1):83–92. [PubMed] [Google Scholar]
- 31.Heine PA, Taylor JA, Iwamoto GA, Lubahn DB, Cooke PS. Increased adipose tissue in male and female estrogen receptor-alpha knockout mice. Proc Natl Acad Sci USA. 2000;97:12729–12734. doi: 10.1073/pnas.97.23.12729. [DOI] [PMC free article] [PubMed] [Google Scholar]