Abstract
Appropriate vessel development and its coordinated function is essential for proper embryogenesis and homeostasis in the adult. Defects in vessels cause birth defects and are an important etiology of diseases such as cardiovascular disease, tumor and diabetes retinopathy. The accumulative data indicate that ETV2, an ETS transcription factor, performs a potent and indispensable function in mediating vessel development. This review discusses the recent progress of the study of ETV2 with special focus on its regulatory mechanisms and cell fate determining role in developing mouse embryos as well as somatic cells.
Keywords: cell reprogramming, endothelial cells, ETV2, transcription factors
INTRODUCTION
In consonant with a close proximity in anatomy, coordinated development of the circulatory system including vessels, blood and the heart is prerequisite for securing successful embryogenesis. In developing embryos, the growth of the circulatory system is identifiable first and abnormalities in the establishment of the system frequently cause embryonic lethality. As early as embryonic day (E) 7.5 in mice, the first structure with the signature of endothelial and hematopoietic cells is the blood islands of the extraembryonic yolk sac (Haar and Ackerman, 1971). Shortly after, the blood islands, which have erythrocytes inside the lumen circled by a layer of endothelial cells, fuse together to create primitive forms of vessels (i.e., primary plexus), which then undergo a remodeling process, generating the complex vascular network interwoven by small capillary vessels and large vessels. On the other hand, angioblasts (also known as endothelial precursor cells) initiate the formation of the vascular structures in the embryonic proper such as dorsal aorta, cardinal veins, vitelline vessels (Drake and Fleming, 2000; Flamme et al., 1997; Patan, 2004). The more elaborated vasculatures are further completed through vascular reshaping, recruitment of perivascular cells and deposition of the extracellular matrix (Carmeliet and Jain, 2011; Jain, 2003). While the blood cells that first appear in the yolk sacs are mainly erythrocytes and macrophages in the blood islands (Choi, 2002; Palis et al., 1999), hematopoietic stem cells, which can supply all types of blood cells throughout the adult life, are detected in the specialized region of the dorsal aorta (Bertrand et al., 2010; Boisset et al., 2010; Kissa and Herbomel, 2010; Zovein et al., 2008) and sequentially populate the fetal liver, spleen and bone marrow.
Transcriptional factors have been implicated in a myriad of biological processes including embryogenesis, tumor and cell proliferation. Among are the E26 transforming sequence or E-twenty-six specific sequence (ETS) transcription factors, which are categorized by the presence of the ETS DNA binding domain (Fig. 1A) (Hollenhorst et al., 2004). Extensive studies have revealed important functions of the ETS factors in endothelial and hematopoietic cell development (Bartel et al., 2000; Ciau-Uitz et al., 2013; Dejana et al., 2007; Findlay et al., 2013; Randi et al., 2009). For example, compound knockouts of Ets1 and Ets2 show abnormal endothelial cell branching (Wei et al., 2009). While Fli1 null mouse embryos develop vascular leakage due to enhanced endothelial cell death (Hart et al., 2000; Spyropoulos et al., 2000), the lack of Tel1 in mice leads to defective vascular remodeling in the yolk sac and is accompanied by considerable apoptosis (Wang et al., 1997). However, emergence of the vascular structures is not blocked by the absence of these ETS factors and the inactivation of Ets1 does not cause vascular defects (Barton et al., 1998). This fact suggests the redundant functions of the ETS factors for at least some members in vessel development (Craig et al., 2015; Pham et al., 2007; Wei et al., 2009). In contrast, recent studies have discovered the non-redundant and indispensable role of one of the ETS factors, ETV2 in vessel as well as blood cell development (Ferdous et al., 2009; Kataoka et al., 2011; Lee et al., 2008). In this review, we will discuss the functional significance of ETV2 in embryonic vessel development, postnatal angiogenesis and direct cell reprogramming.
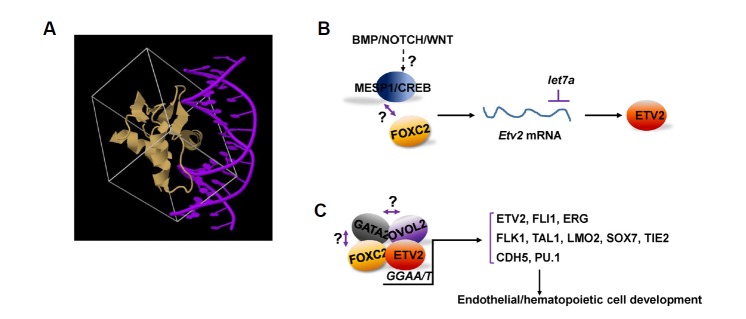
Fig. 1.
Regulation of the expression and function of ETV2. (A) A schematic structural diagram of the complex of the ETS domain of PU.1 in gold and DNA in purple (deposited on The RCSB PDB www.rcsb.org; DOI: 10.2210/pdb1pue/pdb) (Berman et al., 2000; Kodandapani et al., 1996). (B) In early embryos or differentiating mouse ES cells, BMP/NOTCH/WNT pathways act upstream of ETV2 expression. During this process, transcriptional activation of Etv2 is induced by at least MESP1, CREB and FOXC2. let7a functions to inhibit ETV2 protein synthesis. It is of note that the relationship between BMP/NOTCH/WNT pathways to MESP, CREB and FOXC2 is not known. Also, whether the three transcription factors interact each other in regulating Etv2 gene expression remain elucidated. (C) ETV2 can bind and activate promoters/enhancers of genes critical for endothelial and hematopoietic cell development. OVOL2, FOXC2, GATA2 are reported to interact with ETV2 in mediating these regulation. Whether the three transcription factors can form a transcriptionally active complex remains determined.
ETV2 IS ESSENTIAL FOR VASCULAR ENDOTHELIAL AND HEMATOPOIETIC CELL DEVELOPMENT
ETV2 has drawn a great deal of attention as an important regulator for embryonic vessel and blood cell development. Structurally, ETV2 shares a conserved ETS DNA binding domain with other ETS factors but does not exhibit any similarities outside this domain (Brown and McKnight, 1992; De Haro and Janknecht, 2002; 2005). Although identified as a testis specific protein in adults (Brown and McKnight, 1992; De Haro and Janknecht, 2005), accumulative data show that ETV2 is expressed in early mouse embryos. Its expression is first recognizable in mesodermal progenitors, which can generate cardiovascular lineages between E7.0 and E7.5 (Ferdous et al., 2009; Kataoka et al., 2011; Lee et al., 2008; Rasmussen et al., 2011). At later stages, Etv2 is detected in specific vasculatures including the dorsal aorta, endocardium and cardinal vein. From E11.5 and onwards, the Etv2 message becomes extinct (Ferdous et al., 2009; Kataoka et al., 2011; Lee et al., 2008; Rasmussen et al., 2011). The importance of ETV2 in embryogenesis was revealed by a series of seminal findings from three independent groups. In 2005, Lin and fellow colleagues (Sumanas et al., 2005) reported that etsrp, the zebrafish homologue to mammalian ETV2, is one of the significantly downregulated genes in the zebrafish cloche mutant, which displays defects in both vessel and blood cell development (Stainier et al., 1995), compared to controls. Subsequently, the same group demonstrated the vasculogenic function of etsrp in zebrafish (Sumanas and Lin, 2006). In agreement with its endothelial specific expression, etsrp morpholino led to a significant impairment of vessel formation, whereas overexpression of etsrp resulted in an enhanced generation of embryonic vasculature. Rescuing the vascular defects in the cloche mutants upon overexpression of etsrp indicates that etsrp functions downstream of the cloche when generating embryonic vasculatures. In mice, we, for the first time, revealed an indispensable function of ETV2 in the development of vessels and blood cells (Lee et al., 2008). Complete absence of both vascular structures and hematopoietic cells in Etv2 deficient mice embryos leads to embryonic lethality between E9.5 and E10.5. Mechanistically, ETV2 directly binds promoters or enhancers of genes that are critical for endothelial and hematopoietic cell lineages. Similar findings were reported by two groups as the two generated Etv2 gene trap mice and Etv2-gfp knockin mice, respectively (Ferdous et al., 2009; Kataoka et al., 2011). With the addition of the finding that Xenopus er71 has a potent vasculogenic function (Neuhaus et al., 2010), it is clear that ETV2 is indispensable in vessel and blood cell development.
MOLECULAR MECHANISMS OF ETV2 IN REGULATING CARDIOVASCULAR DEVELOPMENT
Figures 1B and 1C summarize the findings regarding the molecular mechanisms of ETV2. The very first clue as to how ETV2 regulates cardiovascular cell lineage development was reported by our group and showed that ETV2 directly binds to the ETS consensus sequence (GGAA/T) (Hollenhorst et al., 2004; 2011) in the promoter of the Flk1 gene, leading to the induction of the gene expression (Lee et al., 2008). In this study, it was also shown that overexpression of ETV2 can generate FLK1+ mesoderm as well as endothelial and hematopoietic cells in differentiating ESCs in a serum-free condition. In subsequent studies, Sox7, Lmo2, Tie2, Nfac1, were identified as direct targets of ETV2 (Behrens et al., 2014; Koyano-Nakagawa et al., 2012; Lee et al., 2011; Palencia-Desai et al., 2011). Rather than performing Chromatin Immunoprecipitation (ChIP) assay on the targeted ones, a recent study (Liu et al., 2015) performed a ChIP-sequencing analysis to reveal the direct downstream target genes of ETV2 at a genome wide analysis level and found that ETV2 can not only bind to promoters or enhancers of already known target genes including Flk1 and Cdh5 but also other genes that perform critical roles in vascular endothelial and hematopoietic cells. Among these genes are Scl, Gata2, Meis1, Dll4, Notch1, Nrp1/2, Flt4, Fli1, RhoJ, Mapk. Thus, these results strongly indicate that ETV2 regulates the endothelial and hematopoietic programs in early stage embryos through direct binding to the ETS elements present in the aforementioned genes (Fig. 1C).
The ETS factors have been shown to interact with other proteins when regulating target genes (Dejana et al., 2007; Sharrocks, 2001; Verger and Duterque-Coquillaud, 2002). Furthermore, several studies showed that ETV2 can form a transcription complex with other proteins (Fig. 1C). In 2008, De Val et al. reported that the interaction of ETV2 and FOXC2 (forkhead transcription factor) synergistically induces the expression of endothelial and hematopoietic genes (De Val et al., 2008). Recently, we demonstrated that OVOL2, a zinc finger transcription factor, directly binds to ETV2 to cooperatively generate FLK1+ mesoderm and vascular endothelial and hematopoietic cell lineages from mouse ESCs (Kim et al., 2014). Interestingly, stability of ETV2 was significantly enhanced upon the overexpression of OVOL2, suggesting a possible mechanism for the cooperative interaction of the two proteins. Additionally, Shi et al. (2014) reported Gata2 as the interacting protein of ETV2. This interaction was cooperative in activating important genes for vascular endothelial and blood cell development. It is of note that all the identified proteins of ETV2 interacting partners have been implicated in embryonic vessel and blood cell development (Kume et al., 2001; Lugus et al., 2007; Seo et al., 2006; Tsai and Orkin, 1997; Tsai et al., 1994; Unezaki et al., 2007). All in all, it is evident that ETV2 can form a multiprotein transcription complex to control the expression of target genes. Thus, revealing more ETV2 interacting proteins in the regulation of endothelial and hematopoietic genes would be an important next step.
One of the major outstanding questions would be the mechanisms, which regulates the expression of ETV2 (Fig. 1B). In the first report conducted by our group, treatment of inhibitors of BMP (noggin), NOTCH (DAPT) and WNT (DKK1) led to a significant decrease in the expression of Etv2 and the generation of FLK1+ mesoderm (Lee et al., 2008). The reduced genesis of the FLK1+ mesoderm in the presence of the inhibitors was reversed upon the overexpression of Etv2, suggesting that ETV2 functions downstream of these signaling pathways. In addition, several studies reported the transcriptional regulation of ETV2. Ferdous et al (2009) reported that the upstream region of the Etv2 promoter can be bound by NKX2-5, a key transcription factor for heart development (Lyons et al., 1995; Tanaka et al., 1999), positing NKX2-5 as an direct upstream factor of Etv2 in the generation of endocardium in the heart (Ferdous et al., 2009). However, no defects were observed in endothelial cell development in Nkx2-5 deficient mouse embryos and the failure of increasing endothelial genes upon overexpression of Nkx2-5 in differentiating mouse ESCs (Caprioli et al., 2011; Lyons et al., 1995; Tanaka et al., 1999). Indeed, overexpression of nkx2-5 in zebrafish significantly decreases the expression of etsrp with concomitant upregulation of cardiac genes (Simoes et al., 2011), suggesting that endothelial expression of ETV2 is NKX2-5 independent. In addition to the role of FOXC2 as an ETV2 interacting protein, it was reported that Fox proteins could function upstream of etsrp when generating endothelial and hematopoietic cells in zebrafish (Veldman and Lin, 2012). In this study, the authors found enhancers that drove the endothelial expression of etsrp. While one of the enhancers of etsrp was occupied by foxc1a/fox1b, the expression of etsrp was significantly reduced by foxc1a/fox1b morpholino. Given the finding that FOXC2 can interact with ETV2 to modulate the expression of vascular endothelial and hematopoietic genes (De Val et al., 2008), these results suggest the critical function of FOXC protein in ETV2-mediated cardiovascular development. Through the use of the mouse ESC differentiation system and mouse genetics, it was shown that Mesoderm posterior 1 (MESP1) and CREB (CRE binding protein) play important roles in Etv2 transactivation (Shi et al., 2015; Yamamizu et al., 2012). Etv2 expression can be activated by cAMP/PKA/CREB signaling in differentiating mouse ESCs (Yamamizu et al., 2012). In this study, the authors showed that CREB can directly bind to the CRE binding elements in Etv2 upstream regions. Blocking ETV2 expression using siRNA leads to the reduction of PKA/CREB-mediated induction of endothelial and hematopoietic cell lineages. In accordance to this, a recent study reported (Shi et al., 2015) that Mesp1 can activate the expression of Etv2 through binding to the CRE element. This transactivation appears to be CREB-dependent as shown by the abrogated Etv2 promoter activity upon co-transfection of MESP1 and dominant negative form of CREB. Interestingly, the study found that the majority of ETV2+ cells originate from MESP1+ cells and that the deficiency of Etv2 in MESP1+ cells causes embryonic lethality that leads to defects in vascular and blood cell generation, which is reminiscent of Etv2 knockout mice. This supports the linear correlation between MESP1 and ETV2 in vascular endothelial and hematopoietic cell development. However, given that the overexpression of Mesp1 can promote the generation of cardiac lineage cells with suppression of hematopoiesis (Bondue et al., 2008; Lindsley et al., 2008), this appears contradictory to the results. Nonetheless, these studies clearly indicate that the transcription factors involved in early mesoderm development as well as cardiovascular generation and maturation play an important role in the regulation of ETV2 expression. Not only mapping sequential order of the transcription factors and identifying additional factors required for ETV2 expression, but it would also be interesting to study how these transcription factors cross talk with the aforesaid signaling pathways in governing ETV2 expression.
INTERPLAY BETWEEN ETV2 AND OTHER ETS FACTORS IN ESTABLISHING CARDIOVASCULAR SYSTEM
As discussed, there are more than 20 ETS factors found in mammals and some of them play significant roles in vascular system development and function (Findlay et al., 2013; Meadows et al., 2011; Randi et al., 2009). Having confirmed the transient but potent vasculogenic activity of ETV2 in early mouse embryos (i.e. its expression is not detectable beyond E11.5) (Ferdous et al., 2009; Kataoka et al., 2011; Lee et al., 2008), many have been puzzled by how the vasculatures develop and are maintained throughout embryogenesis. In this regard, two groups have demonstrated a positive feedback loop mechanism between ETV2 and other ETS factors, at least Fli1 (Abedin et al., 2014; Liu et al., 2015). In 2014, Abedin et al. (2014), showed using Fli1 knockout mice that while Fli1 messages were significantly decreased in Etv2 knockout mouse embryos, other ETS factors such as Ets1/2, Elf1/2 and Etv6 showed comparable levels of expression. The expression of Fli1 is directly upregulated by ETV2 as well as by FLI1 itself through the ETS binding sites on the Fli1 promoter as demonstrated by the ChIP-PCR and luciferase-based promoter assays. Interestingly, that fact that FLI1 binds to its own promoter was observed at E11.5, which is the time in which the ETV2 expression becomes extinct, but not at E9.5, which is when the ETV2 message is still abundant. Also, the ChIP-PCR analysis revealed in vivo occupancy of FLI1 on the promoters of Tie2 and Cdh5 at E11.5, but not at E9.5 embryos. Consistently, several key endothelial genes, such as Tie2 and Cdh5, induced by ETV2 showed reduced levels of expression in Fli1 deficient embryos as well as Fli1 knockdown primary endothelial cells. These results suggest that the function of FLI1 replaces the function of ETV2 at least partly for endothelial cell survival and vascular maintenance at the midgestation stage. Subsequently, Liu et al. (2015) reported a similar feedback regulation mechanism between ETV2 and FLI1. Performing genome wide analysis with ChIP-sequencing comprehensively revealed direct targets of ETV2, which can be classified into VEGF signaling/endothelial lineage specification genes, NOTCH/MAPK signaling and RHO GTPase. The ETS factors such as Fli1, Ets1/2, Erg and notably Etv2 itself were identified as potential targets of ETV2 in differentiating mESCs. In agreement with the results, overexpression of Etv2 led to the immediate induction of Fli1, suggesting that Fli1 is a direct downstream target of ETV2. Indeed, the ChIP-PCR experiment showed in vivo occupancy of ETV2 on Fli1 genomic DNA. The findings that Fli1 null embryos and mouse ESCs showed comparable levels of expression of Etv2 compared to wild type controls but overexpression of Fli1 in mouse ESCs did not induce Etv2 message further support the argument (Liu et al., 2015). Similar to the findings discussed above (Abedin et al., 2014), key endothelial and hematopoietic genes such as Tie2, Cdh5, Lmo2 and Scl, which are the direct targets of ETV2, can also be directly regulated by Fli1 when ETV2 expression is not detected in differentiating ESCs. Given that ETV2 can activate its own promoter, these results suggest the following model; ETV2 triggers a genetic program for endothelial and hematopoietic lineage development through its transcriptional activation function (i.e. positive autoregulation and transactivation of target genes). Once the endothelial cells and hematopoietic cells are generated when the ETV2 expression is silent, other ETS factors, especially FLI1 induced by ETV2 ensure further establishment and maintenance of the vessel and blood systems. Thus, molecular and biochemical studies that uncover the functional significance of other ETS factors in the context of FLI1 in generating vessel and blood cells would be worthy areas to pursue.
CARDIOVASCULAR CELL FATE DETERMINING ROLE OF ETV2
The first emerging FLK1+ mesoderm in developing mouse embryos have the potential to differentiate into vascular endothelial, hematopoietic, muscle cell lineages including cardiomyocyte and smooth muscle cells (Chung et al., 2002; Ema et al., 2003; 2006; Faloon et al., 2000; Motoike et al., 2003; Yamashita et al., 2000), suggesting that FLK1+ mesoderm functions as a multipotent progenitor in cardiovascular cell lineages. A series of studies showed that FLK1+ mesoderm can be subdivided into two distinct cell populations; FLK1+PDGFRα+ (platelet-derived growth factor receptor α) with cardiogenic potential and FLK1+PDGFRα− cells with endothelial and hematopoietic potential (Hirata et al., 2007; Liu et al., 2012; Sakurai et al., 2006). However, mechanisms that determine the cell fate of FLK1+ mesoderm into the cell population remain to be elucidated. Given the role of transcription factors in determining cell identity (Frum and Ralston, 2015; Hatakeyama and Kageyama, 2004; Iwafuchi-Doi and Zaret, 2014; Park et al., 2013; Weintraub et al., 1991), it is plausible that ETV2 regulates the multipotency of the FLK1+ mesoderm. Indeed, the lack of Etv2 in mice and mouse ESCs leads to the failure to generate FLK1+PDGFRα− cells (we refer this hemoangiogenic FLK1+ cells) with concomitant augmentation of FLK1+PDGFRα+ cells (we refer this cardiogenic FLK1+ cells) (Liu et al., 2012; 2015). Also, reduced expression of genes of vascular endothelial and blood cells and augmentation of cardiac genes were observed in Etv2 knockout embryos (Lee et al., 2008). Inversely, the overexpression of Etv2 in differentiating mouse ESCs leads to a significant increment of hemoangiogenic FLK1+ cells at the expense of cardiogenic FLK1+ cells. The capability of ETV2 to induce hemoangiogenic potential is further strengthened when GATA2 and SCL are co-expressed (Liu et al., 2013). Additional supporting results in this field have been reported from studies in zebrafish. In 2007, Schoenebeck et al. (2017), showed that the overexpression of etsrp together with scl leads to the expansion of hematopoietic and endothelial cell area with a reduction in the cardiac field as well as heart size. The observed phenotype was reversed upon injection of etsrp and scl morpholino in zebrafish embryos. A similar finding was reported by the knockdown of etsrp alone in zebrafish embryos (Palencia-Desai et al., 2011). In this study, the authors further showed using etsrp-gfp cells that etsrp+ cells are deficient in etsrp fated to cardiogenic cell lineages. These results suggest that ETV2 functions as an essential cell fate determinant between hemoangiogenic and cardiogenic mesoderms. Likewise, transcriptional regulation of such antagonistic relationships between hemoangiogenic and cardiogenic cell lineage specification is evident in other studies as well. Injection of nkx2-5 into zebrafish embryos reduced the expression of endothelial and blood cell markers such as etsrp, scl and pu.1, but significantly expanded the hand2+ cardiac boundary (Simoes et al., 2011). When Mesp1, another key cardiogenic transcription factor in differentiating mESCs, was overexpressed, the antagonistic developmental outcomes have also been reported (Bondue et al., 2008; Lindsley et al., 2008).
Further insight as to how ETV2 regulates the cell fate of FLK1+ mesoderm was suggested by Liu et al. (2012) as the study reported the first evidence of the role of ETV2 and WNT-β-catenin signaling in this process. The authors found reduced expression of genes involved in WNT-β-CATENINE signaling with decreased cardiomyocyte generation when Etv2 was overexpressed in differentiating ESCs. In sharp contrast, over-expressing β-catenin reversed the ETV2-induced hemoangiogenic cell lineage generation. However, the proposed mechanism was not consonant with the knockout mice study in which deficiency of β-catenin in FLK1+ mesoderm resulted in no obvious phenotypic defects in heart formation (Stenman et al., 2008). Therefore, further investigation to explain the molecular mechanism of ETV2 in determining the cell fate of FLK1+ mesoderm is required.
DIRECT CELL REPROGRAMMING AND ETV2
The monumental findings from the studies conducted by Gurdon (2006), Weintraub et al. (1989) and recently the Yamanaka group (Takahashi and Yamanaka, 2006; Takahashi et al., 2007) have challenged the longstanding consensus that the differentiation process is unidirectional and produces a progressive loss of differentiation potential like a ball rolling from the top of a mountain to the ground and thus generates cells with an irreversibly determined fate (Waddington, 1957). Fueled by the Yamanaka’s finding that four pluripotency factors (OCT4, SOX2, KLF4, and C-MYC) can generate embryonic stem like cells, such as induced pluripotent cells (iPSCs) from differentiated somatic cells (Takahashi and Yamanaka, 2006; Takahashi et al., 2007), considerable efforts have been focused on regenerative medicine, which aims to develop the generation of functional cells or even tissues for autologous cell replacement therapies. However, the clinical applicability of the iPSC-based approaches have been significantly limited due to the inefficient generation of targeted cells and tumorigenic potential (Cohen and Melton, 2011; Knoepfler, 2009), necessitating a novel means to overcome these obstacles. In agreement with the previous studies (Johnson et al., 2008; Kim et al., 2010; Weintraub et al., 1989; Xie et al., 2004), the overexpression of cell type or tissue specific transcription factors is sufficient to directly convert or reprogram somatic cells into targeted cells such as cardiomyocytes, neuron or hepatocytes that bypass the iPSC stages in vivo (Huang et al., 2011; Ieda et al., 2010; Pang et al., 2011; Sekiya and Suzuki, 2011; Song et al., 2012; Vierbuchen et al., 2010).
Considering that cardiovascular diseases (CVDs) are the most serious diseases in both the United States and the world (Mozaffarian et al., 2015), a great number of researchers have been investigating an efficient way to generate autologous functional endothelial cells for cell therapy. Over the past 15 years, interrogating potential of PSCs (i.e., embryonic stem cells and induced pluripotent stem cells) and endothelial progenitor cells that can generate endothelial cells for therapeutic purposes has been an active research area. As discussed, the recent emergence of the direct reprogramming technology has also prompted investigators to seek novel methods to generate functional endothelial cells directly from somatic cells, which can be applied to the treatment of CVD patients. In 2012, Ginsberg et al. showed that ETV2 together with other ETS factors such as FLI1 and ERG can convert human mid gestation c-kit− lineage-committed amniotic cells (ACs) into endothelial cells (Ginsberg et al., 2012). Interestingly, in analogy to embryonic vessel development, the function of ETV2 was only required in the beginning phase of the reprogramming process and the onward steps were completed by FLI1 and ERG in conjunction with the suppression of TGF-β signaling. The reprogrammed endothelial cells were able to evidence neovascularization in vivo. However, the combination of factors was only applied to the ACs, and not to adult somatic cells. In a subsequent study by Han et al. (2014), it was shown that the mixture of transcription factors crucial for vessel development (FOXO1, ETV2, KLF2, TAL1, and LMO2) was able to directly reprogram mouse adult skin fibroblast into endothelial cells. In contrast, we and another group reported direct cell reprogramming of human dermal fibroblasts into endothelial cells by overexpressing ETV2 alone (Lee et al., 2014; Morita et al., 2015). Similarly, skeletal muscles can be converted into endothelial cells in zebrafish. Collectively, these results suggest a potent determinant function of ETV2 in converting non-endothelial cells into endothelial cells and that ETV2 might function as a therapeutic agent for treating CVDs. However, the use of lentiviral or retroviral delivery systems in these studies, which can alter the genomic integrity, is not compatible with clinical use. Therefore, to design novel tools with non-integrating materials such as modified mRNA, small molecules or chemicals is an imperative need in the field of direct reprogramming. Additional efforts should be made to generate specific types of endothelial cells such as arterial, venous and lymphatic ECs.
CONCLUSION
By virtue of extensive studies over the past few years, we have a better understanding on the critical function of ETV2 in the genesis of the vessel, blood and heart in developing mouse embryos. As discussed, one of the salient observations in ETV2 biology is its transient expression in vessel and blood cells (Ferdous et al., 2009; Kataoka et al., 2011; Lee et al., 2008). The intricate interplay between ETV2 and FLI1 is proposed as a means to maintain functional vessels and hematopoietic cells throughout embryogenesis and perhaps in adults (Abedin et al., 2014; Liu et al., 2015). However, the mechanisms, in which the ETV2 expression is off, remain to be determined. The switch-off of the proposed upstream signals as previously discussed could be one possible explanation. Additional means of regulation would be active ways to restrict the expression of ETV2 in a certain narrow window of time to ensure proper development of vessel and hematopoietic cells. Indeed, let7-a miRNA is capable of targeting zebrafish etsrp, resulting in the reduction of expression of both vascular endothelial and hematopoietic markers (Moore et al., 2013). Given the recent report that Kdm1a, histone demethylase, in zebrafish promotes hematopoietic cell development by suppressing etsrp function (Takeuchi et al., 2015), epigenetic modifications of the ETV2 genomic loci would be another possible mechanism. In addition, it was reported that the sustained expression of ETV2 in endothelial and hematopoietic cells caused abnormal development and endothelialization, respectively (Hayashi et al., 2012). Thus, future studies on the safe-guard mechanisms of the ETV2 expression are warranted.
We and others unequivocally proved the potent vasculogenic function of ETV2 in developing mouse embryogenesis. As previously stated, the message becomes extinct once the vessel and hematopoietic cells develop. This raised a question as to the functional significance of ETV2 in post-natal life. In this regard, Lee et al. (2011) found the enriched expression of ETV2 in BM HSCs and reported that Mx1-cre driven deletion of Etv2 led to the decrease in the number and repopulating capacity of BM HSCs. The authors claimed the increased death of BM HSC in the absence of Etv2 as an etiology of the observed phenotypes. Mechanistically, they showed that ETV2 can directly regulate Tie2 expression, but failed to link how the reduced Tie2 expression is related to the death of BM HSCs. Recently, we have found that the endothelial ETV2 acts as a critical regulator in neovascularization in response to injury (Park et al., 2015). The Etv2 expression in endothelial cells is reactivated after injury. Mice deficient in Etv2 in endothelial cells exhibited a significant impairment of new vessel formation upon injury such as wounding, eye injury and hindlimb ischemia. Interestingly, single delivery of lentiviral Etv2 not only promotes the recovery of blood perfusion, but also augments proliferation of endothelial cells as well as smooth muscle cells, leading to neovascularization and tissue repair in the injured hindlimbs. These results suggest that ETV2 in adults plays an important role in vessel and blood construction systems under physiological conditions. Extending from these findings (Park et al., 2015), investigation of the function of ETV2 in pathological settings such as tumor angiogenesis and diabetics related vessel defects would be of significant interest from a therapeutic stand point. Last, but not least, ETV2 alone or together with other endothelial transcription factors were reported to directly convert nonendothelial cells into endothelial cells (Ginsberg et al., 2012; Han et al., 2014; Lee et al., 2014; Morita et al., 2015; Veldman et al., 2013). As reviewed above, the identity of the converted endothelial cells is not clear: are they venous, arterial or lymphatic endothelial cells, functionally and (epi) genetically? What are the mechanism of the direct reprogramming by ETV2? Based upon our results as previously discussed, can ETV2 in clinically compatible forms be developed with high efficiency for clinical use in vivo?
In summary, ETV2 is an indispensable transcription factor and plays a crucial role in vessel development and function. Recent findings have revealed additional capabilities of ETV2 in cell reprogramming. Thus, deciphering the mechanisms by which ETV2 is regulated in governing these processes would provide a novel research venue for the basic and translational aspects of endothelial cell biology.
Acknowledgments
This work was supported by the American Heart Association, 11SDG7390074, National Institutes of Health (NIH) R01 HL119291 and the National Center for Advancing Translational Sciences of the NIH under Award Number UL1TR000454. The content is solely the responsibility of the authors and does not necessarily represents the official views of the NIH.
REFERENCES
- Abedin M.J., Nguyen A., Jiang N., Perry C.E., Shelton J.M., Watson D.K., Ferdous A. Fli1 acts downstream of Etv2 to govern cell survival and vascular homeostasis via positive autoregulation. Circ. Res. 2014;114:1690–1699. doi: 10.1161/CIRCRESAHA.1134303145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel F.O., Higuchi T., Spyropoulos D.D. Mouse models in the study of the Ets family of transcription factors. Oncogene. 2000;19:6443–6454. doi: 10.1038/sj.onc.1204038. [DOI] [PubMed] [Google Scholar]
- Barton K., Muthusamy N., Fischer C., Ting C.N., Walunas T.L., Lanier L.L., Leiden J.M. The Ets-1 transcription factor is required for the development of natural killer cells in mice. Immunity. 1998;9:555–563. doi: 10.1016/s1074-7613(00)80638-x. [DOI] [PubMed] [Google Scholar]
- Behrens A.N., Zierold C., Shi X., Ren Y., Koyano-Nakagawa N., Garry D.J., Martin C.M. Sox7 is regulated by ETV2 during cardiovascular development. Stem Cells Dev. 2014;23:2004–2013. doi: 10.1089/scd.2013.0525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berman H.M., Westbrook J., Feng Z., Gilliland G., Bhat T.N., Weissig H., Shindyalov I.N., Bourne P.E. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertrand J.Y., Chi N.C., Santoso B., Teng S., Stainier D.Y., Traver D. Haematopoietic stem cells derive directly from aortic endothelium during development. Nature. 2010;464:108–111. doi: 10.1038/nature08738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boisset J.C., van Cappellen W., Andrieu-Soler C., Galjart N., Dzierzak E., Robin C. In vivo imaging of haematopoietic cells emerging from the mouse aortic endothelium. Nature. 2010;464:116–120. doi: 10.1038/nature08764. [DOI] [PubMed] [Google Scholar]
- Bondue A., Lapouge G., Paulissen C., Semeraro C., Iacovino M., Kyba M., Blanpain C. Mesp1 acts as a master regulator of multipotent cardiovascular progenitor specification. Cell Stem Cell. 2008;3:69–84. doi: 10.1016/j.stem.2008.06.009. [DOI] [PubMed] [Google Scholar]
- Brown T.A., McKnight S.L. Specificities of protein-protein and protein-DNA interaction of GABP alpha and two newly defined ets-related proteins. Genes Dev. 1992;6:2502–2512. doi: 10.1101/gad.6.12b.2502. [DOI] [PubMed] [Google Scholar]
- Caprioli A., Koyano-Nakagawa N., Iacovino M., Shi X., Ferdous A., Harvey R.P., Olson E.N., Kyba M., Garry D.J. Nkx2-5 represses Gata1 gene expression and modulates the cellular fate of cardiac progenitors during embryogenesis. Circulation. 2011;123:1633–1641. doi: 10.1161/CIRCULATIONAHA.110.008185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmeliet P., Jain R.K. Molecular mechanisms and clinical applications of angiogenesis. Nature. 2011;473:298–307. doi: 10.1038/nature10144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi K. The hemangioblast: a common progenitor of hematopoietic and endothelial cells. J. Hematother. Stem Cell Res. 2002;11:91–101. doi: 10.1089/152581602753448568. [DOI] [PubMed] [Google Scholar]
- Chung Y.S., Zhang W.J., Arentson E., Kingsley P.D., Palis J., Choi K. Lineage analysis of the hemangioblast as defined by FLK1 and SCL expression. Development. 2002;129:5511–5520. doi: 10.1242/dev.00149. [DOI] [PubMed] [Google Scholar]
- Ciau-Uitz A., Wang L., Patient R., Liu F. ETS transcription factors in hematopoietic stem cell development. Blood Cells Mol. Dis. 2013;51:248–255. doi: 10.1016/j.bcmd.2013.07.010. [DOI] [PubMed] [Google Scholar]
- Cohen D.E., Melton D. Turning straw into gold: directing cell fate for regenerative medicine. Nat. Rev. Genet. 2011;12:243–252. doi: 10.1038/nrg2938. [DOI] [PubMed] [Google Scholar]
- Craig M.P., Grajevskaja V., Liao H.K., Balciuniene J., Ekker S.C., Park J.S., Essner J.J., Balciunas D., Sumanas S. Etv2 and fli1b function together as key regulators of vasculogenesis and angiogenesis. Arterioscler. Thromb. Vasc. Biol. 2015;35:865–876. doi: 10.1161/ATVBAHA.114.304768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Haro L., Janknecht R. Functional analysis of the transcription factor ER71 and its activation of the matrix metalloproteinase-1 promoter. Nucleic Acids Res. 2002;30:2972–2979. doi: 10.1093/nar/gkf390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Haro L., Janknecht R. Cloning of the murine ER71 gene (Etsrp71) and initial characterization of its promoter. Genomics. 2005;85:493–502. doi: 10.1016/j.ygeno.2004.12.003. [DOI] [PubMed] [Google Scholar]
- De Val S., Chi N.C., Meadows S.M., Minovitsky S., Anderson J.P., Harris I.S., Ehlers M.L., Agarwal P., Visel A., Xu S.M., et al. Combinatorial regulation of endothelial gene expression by ets and forkhead transcription factors. Cell. 2008;135:1053–1064. doi: 10.1016/j.cell.2008.10.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dejana E., Taddei A., Randi A.M. Foxs and Ets in the transcriptional regulation of endothelial cell differentiation and angiogenesis. Biochim. Biophys. ACTA. 2007;1775:298–312. doi: 10.1016/j.bbcan.2007.05.003. [DOI] [PubMed] [Google Scholar]
- Drake C.J., Fleming P.A. Vasculogenesis in the day 6.5 to 9.5 mouse embryo. Blood. 2000;95:1671–1679. [PubMed] [Google Scholar]
- Ema M., Faloon P., Zhang W.J., Hirashima M., Reid T., Stanford W.L., Orkin S., Choi K., Rossant J. Combinatorial effects of Flk1 and Tal1 on vascular and hematopoietic development in the mouse. Genes Dev. 2003;17:380–393. doi: 10.1101/gad.1049803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ema M., Takahashi S., Rossant J. Deletion of the selection cassette, but not cis-acting elements, in targeted Flk1-lacZ allele reveals Flk1 expression in multipotent mesodermal progenitors. Blood. 2006;107:111–117. doi: 10.1182/blood-2005-05-1970. [DOI] [PubMed] [Google Scholar]
- Faloon P., Arentson E., Kazarov A., Deng C.X., Porcher C., Orkin S., Choi K. Basic fibroblast growth factor positively regulates hematopoietic development. Development. 2000;127:1931–1941. doi: 10.1242/dev.127.9.1931. [DOI] [PubMed] [Google Scholar]
- Ferdous A., Caprioli A., Iacovino M., Martin C.M., Morris J., Richardson J.A., Latif S., Hammer R.E., Harvey R.P., Olson E.N., et al. Nkx2-5 transactivates the Ets-related protein 71 gene and specifies an endothelial/endocardial fate in the developing embryo. Proc. Natl. Acad. Sci. USA. 2009;106:814–819. doi: 10.1073/pnas.0807583106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Findlay V.J., LaRue A.C., Turner D.P., Watson P.M., Watson D.K. Understanding the role of ETS-mediated gene regulation in complex biological processes. Adv. Cancer Res. 2013;119:1–61. doi: 10.1016/B978-0-12-407190-2.00001-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flamme I., Frolich T., Risau W. Molecular mechanisms of vasculogenesis and embryonic angiogenesis. J. Cell. Physiol. 1997;173:206–210. doi: 10.1002/(SICI)1097-4652(199711)173:2<206::AID-JCP22>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- Frum T., Ralston A. Cell signaling and transcription factors regulating cell fate during formation of the mouse blastocyst. Trends Genet. 2015;31:402–410. doi: 10.1016/j.tig.2015.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginsberg M., James D., Ding B.S., Nolan D., Geng F., Butler J.M., Schachterle W., Pulijaal V.R., Mathew S., Chasen S.T., et al. Efficient direct reprogramming of mature amniotic cells into endothelial cells by ETS factors and TGFbeta suppression. Cell. 2012;151:559–575. doi: 10.1016/j.cell.2012.09.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurdon J.B. From nuclear transfer to nuclear reprogramming: the reversal of cell differentiation. Ann. Rev. Cell Devel. Biol. 2006;22:1–22. doi: 10.1146/annurev.cellbio.22.090805.140144. [DOI] [PubMed] [Google Scholar]
- Haar J.L., Ackerman G.A. A phase and electron microscopic study of vasculogenesis and erythropoiesis in the yolk sac of the mouse. The Anatomical Record. 1971;170:199–223. doi: 10.1002/ar.1091700206. [DOI] [PubMed] [Google Scholar]
- Han J.K., Chang S.H., Cho H.J., Choi S.B., Ahn H.S., Lee J., Jeong H., Youn S.W., Lee H.J., Kwon Y.W., et al. Direct conversion of adult skin fibroblasts to endothelial cells by defined factors. Circulation. 2014;130:1168–1178. doi: 10.1161/CIRCULATIONAHA.113.007727. [DOI] [PubMed] [Google Scholar]
- Hart A., Melet F., Grossfeld P., Chien K., Jones C., Tunnacliffe A., Favier R., Bernstein A. Fli-1 is required for murine vascular and megakaryocytic development and is hemizygously deleted in patients with thrombocytopenia. Immunity. 2000;13:167–177. doi: 10.1016/s1074-7613(00)00017-0. [DOI] [PubMed] [Google Scholar]
- Hatakeyama J., Kageyama R. Retinal cell fate determination and bHLH factors. Semin. Cell Devel. Biol. 2004;15:83–89. doi: 10.1016/j.semcdb.2003.09.005. [DOI] [PubMed] [Google Scholar]
- Hayashi M., Pluchinotta M., Momiyama A., Tanaka Y., Nishikawa S., Kataoka H. Endothelialization and altered hematopoiesis by persistent Etv2 expression in mice. Exp. Hematol. 2012;40:738–750.e711. doi: 10.1016/j.exphem.2012.05.012. [DOI] [PubMed] [Google Scholar]
- Hirata H., Kawamata S., Murakami Y., Inoue K., Nagahashi A., Tosaka M., Yoshimura N., Miyamoto Y., Iwasaki H., Asahara T., et al. Coexpression of platelet-derived growth factor receptor alpha and fetal liver kinase 1 enhances cardiogenic potential in embryonic stem cell differentiation in vitro. J. Biosci. Bioeng. 2007;103:412–419. doi: 10.1263/jbb.103.412. [DOI] [PubMed] [Google Scholar]
- Hollenhorst P.C., Jones D.A., Graves B.J. Expression profiles frame the promoter specificity dilemma of the ETS family of transcription factors. Nucleic Acids Res. 2004;32:5693–5702. doi: 10.1093/nar/gkh906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollenhorst P.C., McIntosh L.P., Graves B.J. Genomic and biochemical insights into the specificity of ETS transcription factors. Ann. Rev. Biochem. 2011;80:437–471. doi: 10.1146/annurev.biochem.79.081507.103945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang P., He Z., Ji S., Sun H., Xiang D., Liu C., Hu Y., Wang X., Hui L. Induction of functional hepatocyte-like cells from mouse fibroblasts by defined factors. Nature. 2011;475:386–389. doi: 10.1038/nature10116. [DOI] [PubMed] [Google Scholar]
- Ieda M., Fu J.D., Delgado-Olguin P., Vedantham V., Hayashi Y., Bruneau B.G., Srivastava D. Direct reprogramming of fibroblasts into functional cardiomyocytes by defined factors. Cell. 2010;142:375–386. doi: 10.1016/j.cell.2010.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwafuchi-Doi M., Zaret K.S. Pioneer transcription factors in cell reprogramming. Genes Dev. 2014;28:2679–2692. doi: 10.1101/gad.253443.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain R.K. Molecular regulation of vessel maturation. Nat. Med. 2003;9:685–693. doi: 10.1038/nm0603-685. [DOI] [PubMed] [Google Scholar]
- Johnson N.C., Dillard M.E., Baluk P., McDonald D.M., Harvey N.L., Frase S.L., Oliver G. Lymphatic endothelial cell identity is reversible and its maintenance requires Prox1 activity. Genes Dev. 2008;22:3282–3291. doi: 10.1101/gad.1727208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kataoka H., Hayashi M., Nakagawa R., Tanaka Y., Izumi N., Nishikawa S., Jakt M.L., Tarui H., Nishikawa S. Etv2/ER71 induces vascular mesoderm from Flk1+PDGFRalpha+ primitive mesoderm. Blood. 2011;118:6975–6986. doi: 10.1182/blood-2011-05-352658. [DOI] [PubMed] [Google Scholar]
- Kim H., Nguyen V.P., Petrova T.V., Cruz M., Alitalo K., Dumont D.J. Embryonic vascular endothelial cells are malleable to reprogramming via Prox1 to a lymphatic gene signature. BMC Dev. Biol. 2010;10:72. doi: 10.1186/1471-213X-10-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J.Y., Lee R.H., Kim T.M., Kim D.W., Jeon Y.J., Huh S.H., Oh S.Y., Kyba M., Kataoka H., Choi K., et al. OVOL2 is a critical regulator of ER71/ETV2 in generating FLK1+, hematopoietic, and endothelial cells from embryonic stem cells. Blood. 2014;124:2948–2952. doi: 10.1182/blood-2014-03-556332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kissa K., Herbomel P. Blood stem cells emerge from aortic endothelium by a novel type of cell transition. Nature. 2010;464:112–115. doi: 10.1038/nature08761. [DOI] [PubMed] [Google Scholar]
- Knoepfler P.S. Deconstructing stem cell tumorigenicity: a roadmap to safe regenerative medicine. Stem Cells. 2009;27:1050–1056. doi: 10.1002/stem.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kodandapani R., Pio F., Ni C.Z., Piccialli G., Klemsz M., McKercher S., Maki R.A., Ely K.R. A new pattern for helix-turn-helix recognition revealed by the PU.1 ETS-domain-DNA complex. Nature. 1996;380:456–460. doi: 10.1038/380456a0. [DOI] [PubMed] [Google Scholar]
- Koyano-Nakagawa N., Kweon J., Iacovino M., Shi X., Rasmussen T.L., Borges L., Zirbes K.M., Li T., Perlingeiro R.C., Kyba M., et al. Etv2 is expressed in the yolk sac hematopoietic and endothelial progenitors and regulates Lmo2 gene expression. Stem Cells. 2012;30:1611–1623. doi: 10.1002/stem.1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kume T., Jiang H., Topczewska J.M., Hogan B.L. The murine winged helix transcription factors, Foxc1 and Foxc2, are both required for cardiovascular development and somitogenesis. Genes Dev. 2001;15:2470–2482. doi: 10.1101/gad.907301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee D., Park C., Lee H., Lugus J.J., Kim S.H., Arentson E., Chung Y.S., Gomez G., Kyba M., Lin S., et al. ER71 acts downstream of BMP, Notch, and Wnt signaling in blood and vessel progenitor specification. Cell Stem Cell. 2008;2:497–507. doi: 10.1016/j.stem.2008.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee D., Kim T., Lim D.S. The Er71 is an important regulator of hematopoietic stem cells in adult mice. Stem Cells. 2011;29:539–548. doi: 10.1002/stem.597. [DOI] [PubMed] [Google Scholar]
- Lee S., Park C., Han J.W., Kim J.Y., Cho K., Kim E.J., Kim S., Lee S.-J., An H.J., Sin M.Y., et al. Abstract 18205: Direct Reprogramming of Human Dermal Fibroblasts into Endothelial Cells Using a Single Transcription Factor. Circulation. 2014;130:A18205. [Google Scholar]
- Lindsley R.C., Gill J.G., Murphy T.L., Langer E.M., Cai M., Mashayekhi M., Wang W., Niwa N., Nerbonne J.M., Kyba M., et al. Mesp1 coordinately regulates cardiovascular fate restriction and epithelial-mesenchymal transition in differentiating ESCs. Cell Stem Cell. 2008;3:55–68. doi: 10.1016/j.stem.2008.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F., Kang I., Park C., Chang L.W., Wang W., Lee D., Lim D.S., Vittet D., Nerbonne J.M., Choi K. ER71 specifies Flk-1+ hemangiogenic mesoderm by inhibiting cardiac mesoderm and Wnt signaling. Blood. 2012;119:3295–3305. doi: 10.1182/blood-2012-01-403766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F., Bhang S.H., Arentson E., Sawada A., Kim C.K., Kang I., Yu J., Sakurai N., Kim S.H., Yoo J.J., et al. Enhanced hemangioblast generation and improved vascular repair and regeneration from embryonic stem cells by defined transcription factors. Stem Cell Rep. 2013;1:166–182. doi: 10.1016/j.stemcr.2013.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F., Li D., Yu Y.Y., Kang I., Cha M.J., Kim J.Y., Park C., Watson D.K., Wang T., Choi K. Induction of hematopoietic and endothelial cell program orchestrated by ETS transcription factor ER71/ETV2. EMBO Rep. 2015;16:654–669. doi: 10.15252/embr.201439939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lugus J.J., Chung Y.S., Mills J.C., Kim S.I., Grass J., Kyba M., Doherty J.M., Bresnick E.H., Choi K. GATA2 functions at multiple steps in hemangioblast development and differentiation. Development. 2007;134:393–405. doi: 10.1242/dev.02731. [DOI] [PubMed] [Google Scholar]
- Lyons I., Parsons L.M., Hartley L., Li R., Andrews J.E., Robb L., Harvey R.P. Myogenic and morphogenetic defects in the heart tubes of murine embryos lacking the homeo box gene Nkx2-5. Genes Dev. 1995;9:1654–1666. doi: 10.1101/gad.9.13.1654. [DOI] [PubMed] [Google Scholar]
- Meadows S.M., Myers C.T., Krieg P.A. Regulation of endothelial cell development by ETS transcription factors. Semin. Cell Dev. Biol. 2011;22:976–984. doi: 10.1016/j.semcdb.2011.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore J.C., Sheppard-Tindell S., Shestopalov I.A., Yamazoe S., Chen J.K., Lawson N.D. Post-transcriptional mechanisms contribute to Etv2 repression during vascular development. Dev. Biol. 2013;384:128–140. doi: 10.1016/j.ydbio.2013.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita R., Suzuki M., Kasahara H., Shimizu N., Shichita T., Sekiya T., Kimura A., Sasaki K., Yasukawa H., Yoshimura A. ETS transcription factor ETV2 directly converts human fibroblasts into functional endothelial cells. Proc. Natl. Acad. Sci. USA. 2015;112:160–165. doi: 10.1073/pnas.1413234112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motoike T., Markham D.W., Rossant J., Sato T.N. Evidence for novel fate of Flk1+ progenitor: contribution to muscle lineage. Genesis. 2003;35:153–159. doi: 10.1002/gene.10175. [DOI] [PubMed] [Google Scholar]
- Mozaffarian D., Benjamin E.J., Go A.S., Arnett D.K., Blaha M.J., Cushman M., de Ferranti S., Despres J.P., Fullerton H.J., Howard V.J., et al. Heart disease and stroke statistics--2015 update: a report from the American Heart Association. Circulation. 2015;131:e29–322. doi: 10.1161/CIR.0000000000000152. [DOI] [PubMed] [Google Scholar]
- Neuhaus H., Muller F., Hollemann T. Xenopus er71 is involved in vascular development. Dev. Dyn. 2010;239:3436–3445. doi: 10.1002/dvdy.22487. [DOI] [PubMed] [Google Scholar]
- Palencia-Desai S., Kohli V., Kang J., Chi N.C., Black B.L., Sumanas S. Vascular endothelial and endocardial progenitors differentiate as cardiomyocytes in the absence of Etsrp/Etv2 function. Development. 2011;138:4721–4732. doi: 10.1242/dev.064998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palis J., Robertson S., Kennedy M., Wall C., Keller G. Development of erythroid and myeloid progenitors in the yolk sac and embryo proper of the mouse. Development. 1999;126:5073–5084. doi: 10.1242/dev.126.22.5073. [DOI] [PubMed] [Google Scholar]
- Pang Z.P., Yang N., Vierbuchen T., Ostermeier A., Fuentes D.R., Yang T.Q., Citri A., Sebastiano V., Marro S., Sudhof T.C., et al. Induction of human neuronal cells by defined transcription factors. Nature. 2011;476:220–223. doi: 10.1038/nature10202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park C., Kim T.M., Malik A.B. Transcriptional regulation of endothelial cell and vascular development. Circ. Res. 2013;112:1380–1400. doi: 10.1161/CIRCRESAHA.113.301078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park C., Lee T.J., Bhang S.H., Liu F., Nakamura R., Oladipupo S.S., Pitha-Rowe I., Capoccia B., Choi H.S., Kim T.M., et al. Injury-Mediated Vascular Regeneration Requires Endothelial ER71/ETV2. Arteriosclerosis, thrombosis, and vascular biology. 2015 Nov 19; doi: 10.1161/ATVBAHA.115.306430. pii: ATVBAHA.115.306430. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patan S. Vasculogenesis and angiogenesis. Cancer Treat. Res. 2004;117:3–32. doi: 10.1007/978-1-4419-8871-3_1. [DOI] [PubMed] [Google Scholar]
- Pham V.N., Lawson N.D., Mugford J.W., Dye L., Castranova D., Lo B., Weinstein B.M. Combinatorial function of ETS transcription factors in the developing vasculature. Dev. Biol. 2007;303:772–783. doi: 10.1016/j.ydbio.2006.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randi A.M., Sperone A., Dryden N.H., Birdsey G.M. Regulation of angiogenesis by ETS transcription factors. Biochem. Soc. Trans. 2009;37:1248–1253. doi: 10.1042/BST0371248. [DOI] [PubMed] [Google Scholar]
- Rasmussen T.L., Kweon J., Diekmann M.A., Belema-Bedada F., Song Q., Bowlin K., Shi X., Ferdous A., Li T., Kyba M., et al. ER71 directs mesodermal fate decisions during embryogenesis. Development. 2011;138:4801–4812. doi: 10.1242/dev.070912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakurai H., Era T., Jakt L.M., Okada M., Nakai S., Nishikawa S., Nishikawa S. In vitro modeling of paraxial and lateral mesoderm differentiation reveals early reversibility. Stem Cells. 2006;24:575–586. doi: 10.1634/stemcells.2005-0256. [DOI] [PubMed] [Google Scholar]
- Schoenebeck J.J., Keegan B.R., Yelon D. Vessel and blood specification override cardiac potential in anterior mesoderm. Dev. Cell. 2007;13:254–267. doi: 10.1016/j.devcel.2007.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekiya S., Suzuki A. Direct conversion of mouse fibroblasts to hepatocyte-like cells by defined factors. Nature. 2011;475:390–393. doi: 10.1038/nature10263. [DOI] [PubMed] [Google Scholar]
- Seo S., Fujita H., Nakano A., Kang M., Duarte A., Kume T. The forkhead transcription factors, Foxc1 and Foxc2, are required for arterial specification and lymphatic sprouting during vascular development. Dev. Biol. 2006;294:458–470. doi: 10.1016/j.ydbio.2006.03.035. [DOI] [PubMed] [Google Scholar]
- Sharrocks A.D. The ETS-domain transcription factor family. Nat. Rev. Mol. Cell. Biol. 2001;2:827–837. doi: 10.1038/35099076. [DOI] [PubMed] [Google Scholar]
- Shi X., Richard J., Zirbes K.M., Gong W., Lin G., Kyba M., Thomson J.A., Koyano-Nakagawa N., Garry D.J. Cooperative interaction of Etv2 and Gata2 regulates the development of endothelial and hematopoietic lineages. Dev. Biol. 2014;389:208–218. doi: 10.1016/j.ydbio.2014.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi X., Zirbes K.M., Rasmussen T.L., Ferdous A., Garry M.G., Koyano-Nakagawa N., Garry D.J. The transcription factor Mesp1 interacts with cAMP-responsive element binding protein 1 (Creb1) and coactivates Ets variant 2 (Etv2) gene expression. J. Biol. Chem. 2015;290:9614–9625. doi: 10.1074/jbc.M114.614628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simoes F.C., Peterkin T., Patient R. Fgf differentially controls cross-antagonism between cardiac and haemangioblast regulators. Development. 2011;138:3235–3245. doi: 10.1242/dev.059634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song K., Nam Y.J., Luo X., Qi X., Tan W., Huang G.N., Acharya A., Smith C.L., Tallquist M.D., Neilson E.G., et al. Heart repair by reprogramming non-myocytes with cardiac transcription factors. Nature. 2012;485:599–604. doi: 10.1038/nature11139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spyropoulos D.D., Pharr P.N., Lavenburg K.R., Jackers P., Papas T.S., Ogawa M., Watson D.K. Hemorrhage, impaired hematopoiesis, and lethality in mouse embryos carrying a targeted disruption of the Fli1 transcription factor. Mol. Cell. Biol. 2000;20:5643–5652. doi: 10.1128/mcb.20.15.5643-5652.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stainier D.Y., Weinstein B.M., Detrich H.W., 3rd, Zon L.I., Fishman M.C. Cloche, an early acting zebrafish gene, is required by both the endothelial and hematopoietic lineages. Development. 1995;121:3141–3150. doi: 10.1242/dev.121.10.3141. [DOI] [PubMed] [Google Scholar]
- Stenman J.M., Rajagopal J., Carroll T.J., Ishibashi M., McMahon J., McMahon A.P. Canonical Wnt signaling regulates organ-specific assembly and differentiation of CNS vasculature. Science. 2008;322:1247–1250. doi: 10.1126/science.1164594. [DOI] [PubMed] [Google Scholar]
- Sumanas S., Lin S. Ets1-related protein is a key regulator of vasculogenesis in zebrafish. PLoS Biol. 2006;4:e10. doi: 10.1371/journal.pbio.0040010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumanas S., Jorniak T., Lin S. Identification of novel vascular endothelial-specific genes by the microarray analysis of the zebrafish cloche mutants. Blood. 2005;106:534–541. doi: 10.1182/blood-2004-12-4653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K., Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- Takahashi K., Tanabe K., Ohnuki M., Narita M., Ichisaka T., Tomoda K., Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- Takeuchi M., Fuse Y., Watanabe M., Andrea C.S., Takeuchi M., Nakajima H., Ohashi K., Kaneko H., Kobayashi-Osaki M., Yamamoto M., et al. LSD1/KDM1A promotes hematopoietic commitment of hemangioblasts through downregulation of Etv2. Proc. Natl. Acad. Sci. USA. 2015;112:13922–13927. doi: 10.1073/pnas.1517326112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka M., Chen Z., Bartunkova S., Yamasaki N., Izumo S. The cardiac homeobox gene Csx/Nkx2.5 lies genetically upstream of multiple genes essential for heart development. Development. 1999;126:1269–1280. doi: 10.1242/dev.126.6.1269. [DOI] [PubMed] [Google Scholar]
- Tsai F.Y., Orkin S.H. Transcription factor GATA-2 is required for proliferation/survival of early hematopoietic cells and mast cell formation, but not for erythroid and myeloid terminal differentiation. Blood. 1997;89:3636–3643. [PubMed] [Google Scholar]
- Tsai F.Y., Keller G., Kuo F.C., Weiss M., Chen J., Rosenblatt M., Alt F.W., Orkin S.H. An early haematopoietic defect in mice lacking the transcription factor GATA-2. Nature. 1994;371:221–226. doi: 10.1038/371221a0. [DOI] [PubMed] [Google Scholar]
- Unezaki S., Horai R., Sudo K., Iwakura Y., Ito S. Ovol2/Movo, a homologue of Drosophila ovo, is required for angiogenesis, heart formation and placental development in mice. Genes Cells. 2007;12:773–785. doi: 10.1111/j.1365-2443.2007.01084.x. [DOI] [PubMed] [Google Scholar]
- Veldman M.B., Lin S. Etsrp/Etv2 is directly regulated by Foxc1a/b in the zebrafish angioblast. Circ. Res. 2012;110:220–229. doi: 10.1161/CIRCRESAHA.111.251298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veldman M.B., Zhao C., Gomez G.A., Lindgren A.G., Huang H., Yang H., Yao S., Martin B.L., Kimelman D., Lin S. Transdifferentiation of fast skeletal muscle into functional endothelium in vivo by transcription factor Etv2. PLoS Biol. 2013;11:e1001590. doi: 10.1371/journal.pbio.1001590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verger A., Duterque-Coquillaud M. When Ets transcription factors meet their partners. Bioessays. 2002;24:362–370. doi: 10.1002/bies.10068. [DOI] [PubMed] [Google Scholar]
- Vierbuchen T., Ostermeier A., Pang Z.P., Kokubu Y., Sudhof T.C., Wernig M. Direct conversion of fibroblasts to functional neurons by defined factors. Nature. 2010;463:1035–1041. doi: 10.1038/nature08797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waddington C.H. The strategy of the genes; a discussion of some aspects of theoretical biology. London: Allen & Unwin; 1957. [Google Scholar]
- Wang L.C., Kuo F., Fujiwara Y., Gilliland D.G., Golub T.R., Orkin S.H. Yolk sac angiogenic defect and intraembryonic apoptosis in mice lacking the Ets-related factor TEL. EMBO J. 1997;16:4374–4383. doi: 10.1093/emboj/16.14.4374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei G., Srinivasan R., Cantemir-Stone C.Z., Sharma S.M., Santhanam R., Weinstein M., Muthusamy N., Man A.K., Oshima R.G., Leone G., et al. Ets1 and Ets2 are required for endothelial cell survival during embryonic angiogenesis. Blood. 2009;114:1123–1130. doi: 10.1182/blood-2009-03-211391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weintraub H., Tapscott S.J., Davis R.L., Thayer M.J., Adam M.A., Lassar A.B., Miller A.D. Activation of muscle-specific genes in pigment, nerve, fat, liver, and fibroblast cell lines by forced expression of MyoD. Proc. Natl. Acad. Sci. USA. 1989;86:5434–5438. doi: 10.1073/pnas.86.14.5434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weintraub H., Davis R., Tapscott S., Thayer M., Krause M., Benezra R., Blackwell T.K., Turner D., Rupp R., Hollenberg S., et al. The myoD gene family: nodal point during specification of the muscle cell lineage. Science. 1991;251:761–766. doi: 10.1126/science.1846704. [DOI] [PubMed] [Google Scholar]
- Xie H., Ye M., Feng R., Graf T. Stepwise reprogramming of B cells into macrophages. Cell. 2004;117:663–676. doi: 10.1016/s0092-8674(04)00419-2. [DOI] [PubMed] [Google Scholar]
- Yamamizu K., Matsunaga T., Katayama S., Kataoka H., Takayama N., Eto K., Nishikawa S., Yamashita J.K. PKA/CREB signaling triggers initiation of endothelial and hematopoietic cell differentiation via Etv2 induction. Stem Cells. 2012;30:687–696. doi: 10.1002/stem.1041. [DOI] [PubMed] [Google Scholar]
- Yamashita J., Itoh H., Hirashima M., Ogawa M., Nishikawa S., Yurugi T., Naito M., Nakao K., Nishikawa S. Flk1-positive cells derived from embryonic stem cells serve as vascular progenitors. Nature. 2000;408:92–96. doi: 10.1038/35040568. [DOI] [PubMed] [Google Scholar]
- Zovein A.C., Hofmann J.J., Lynch M., French W.J., Turlo K.A., Yang Y., Becker M.S., Zanetta L., Dejana E., Gasson J.C., et al. Fate tracing reveals the endothelial origin of hematopoietic stem cells. Cell Stem Cell. 2008;3:625–636. doi: 10.1016/j.stem.2008.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]