Abstract
OBJECTIVES:
Exploring associations between the gut microbiota and colonic inflammation and assessing sequential changes during exclusive enteral nutrition (EEN) may offer clues into the microbial origins of Crohn's disease (CD).
METHODS:
Fecal samples (n=117) were collected from 23 CD and 21 healthy children. From CD children fecal samples were collected before, during EEN, and when patients returned to their habitual diets. Microbiota composition and functional capacity were characterized using sequencing of the 16S rRNA gene and shotgun metagenomics.
RESULTS:
Microbial diversity was lower in CD than controls before EEN (P=0.006); differences were observed in 36 genera, 141 operational taxonomic units (OTUs), and 44 oligotypes. During EEN, the microbial diversity of CD children further decreased, and the community structure became even more dissimilar than that of controls. Every 10 days on EEN, 0.6 genus diversity equivalents were lost; 34 genera decreased and one increased during EEN. Fecal calprotectin correlated with 35 OTUs, 14 of which accounted for 78% of its variation. OTUs that correlated positively or negatively with calprotectin decreased during EEN. The microbiota of CD patients had a broader functional capacity than healthy controls, but diversity decreased with EEN. Genes involved in membrane transport, sulfur reduction, and nutrient biosynthesis differed between patients and controls. The abundance of genes involved in biotin (P=0.005) and thiamine biosynthesis decreased (P=0.017), whereas those involved in spermidine/putrescine biosynthesis (P=0.031), or the shikimate pathway (P=0.058), increased during EEN.
CONCLUSIONS:
Disease improvement following treatment with EEN is associated with extensive modulation of the gut microbiome.
INTRODUCTION
The gut microbiota comprises thousands of bacterial species with a collective gene pool 150 times larger than the human genome with tremendous functional diversity (1). A gut microbiota, characteristic of Crohn's disease (CD), has been repeatedly described (2, 3). Although much progress has been made in understanding interactions between the gut microbiota and intestinal immune system and their involvement CD etiology (4), it remains unclear whether changes in microbiota precede or follow CD onset. Thus far, all studies exploring the colonic microenvironment of CD patients have been of cross-sectional and hence unable to address reverse causality (2, 3). Assessing, during the course of disease modifying treatment, serial changes in the gut microbiota, particularly organisms associated with colonic inflammation, will offer better insights into the role of gut bacteria or their products in the etiology of CD. Such a study has not been performed before.
Exclusive enteral nutrition (EEN) is an established treatment in active pediatric CD (5). In contrast to biologics, the mode of EEN action remains unknown. We have recently shown that EEN treatment in children with active CD induced reduction in numbers of presumptively protective gut bacterial species (e.g., Faecalibacterium prausnitzii) and metabolites (e.g., butyrate) that were paradoxically associated with disease improvement (6). These intriguing observations in conjunction with the simple nutrient and food ingredients of EEN make it plausible to speculate that EEN works by modulating the gut microbiota. Assessing changes in the gut microbiota during a course of EEN can offer clues to which bacteria and/or their metabolites may be implicated in the etiology of CD.
Most previous studies have explored the gut microbiota in CD by targeting certain limited microbial species and bacterial metabolites for which primers/probes were available (6, 7). Recent advances in sequencing technologies allow a hypothesis-free assessment of the entire microbial community dynamics. Using massively parallel ultra-deep sequencing and state-of-the-art computational approaches, we have characterized microbial community structure and genetic functional capacity in fecal samples from children with active CD and healthy controls by both targeting the bacterial 16S rRNA genes and employing shotgun metagenomics. Using the same methodology, we studied, for the first time, the association of the pediatric CD microbiota with intestinal inflammatory markers and changes before, during, and after induction treatment with EEN.
Methods
Subjects
Fecal samples were collected from 23 children (males: n=13; 6.9–14.7 years) with active CD (Montreal classification; Disease location: L2, n=3; L2+L4, n=4; L3, n=3; L3+L4, n=13; Disease behavior: B1, n=20; B2, n=2; B3, n=1) and for 15 of these (11 newly diagnosed), with at least two consecutive samples, during 8 weeks of EEN (Modulen, Nestle, Vevey, Switzerland) as described previously (6). A maximum of five serial samples were collected per patient. The first sample (A) was collected before or within 6 days of EEN initiation (89% collected within 4 days), two during EEN (B:~16 & C:~32 days), and one close to end of treatment (D:~54 days). A final sample (E) was collected when patients returned to habitual diet (E:~63 days after EEN). The number of participants with 1, 2, 3, 4, and 5 samples were 5, 3, 3, 2, and 10, respectively. Two fecal samples were collected, at least 2 months apart, from 21 healthy children (males: n=12; 4.6–16.9 years) with no known family history of inflammatory bowel disease as a control group. The healthy children were recruited from the local community via poster advertisement. No participant had received antibiotics for 3 months prior to the study. Concomitant treatment was prospectively recorded (Supplementary Table S1 online).
Serum albumin, CRP, erythrocyte sedimentation rate, and fecal calprotectin (8) were measured and Paediatric Crohn's Disease Activity Index (PCDAI) (9) calculated at EEN initiation, end of EEN, and when patients returned to habitual diet. All these disease activity indices significantly decreased, and 62% of participants achieved clinical remission (PCDAI <10) at the end of EEN (Supplementary Table S1).
This study received ethics approval by the Yorkhill Research Ethics Committee (05/S0708/66). Carer and patients provided written consent.
16S rRNA gene sequencing
Sequencing of 16S rRNA gene was performed to characterize microbial community composition. Detailed description of the 16S rRNA sequencing is available in Supplementary Methods. Briefly, bacterial DNA was isolated using the chaotropic method (6). 16S rRNA gene sequencing of the V4 region was performed on the MiSeq (Illumina, Essex, UK) platform using 2 × 250 bp paired-end reads. The V4 region was amplified using fusion Golay adapters barcoded on the reverse strand.
Shotgun metagenome sequencing
Shotgun metagenome samples, indicative of microbiota genetic functional capacity, were prepared for 69 of the samples (CD, n=56; Healthy, n=13) with the Nextera XT Prep Kit (Illumina, FC-131–1096, UK) and the Illumina dual-barcoding Nextera XT Index kit (Illumina, FC-131–1002, UK). Detailed description of the shotgun metagenome sequencing is available in Supplementary Methods. Clusters were generated on-board a HiSeq 2500 (Illumina) instrument and sequencing performed using TruSeq Rapid SBS Kit reagents (Illumina, FC-402–4001, FC-402–4002). Sequencing was performed following a paired-end 150 cycle recipe.
Bioinformatics
Detailed description of bioinformatics analysis is available as Supplementary File (Supplementary Methods). In brief, microbiota composition was characterized at multiple levels of resolution using genus, operational taxonomic unit (OTU), and ‘oligotyping' assignments from the 16S rRNA sequencing, the latter two approximating the species level. Shotgun metagenomics reads were used for species taxonomic profiling using MetaPhlAn and assignments to functional modules, orthologs, and pathways through alignment to Kyoto Encyclopedia of Genes and Genomes (KEGG) using HUMAnN (10). The latter are indicative of the genetic functional capacity of the entire gut microbiota.
Statistics
Statistical analysis was performed on log-transformed data, and Benjamini–Hochberg false-discovery rate was used to account for multiple comparisons. We used the Adonis function of the R vegan package to determine the overall amount and significance of differences between controls and CD before EEN. Each marker was tested independently using Kruskal–Wallis. For the 16S rRNA markers (genera, OTU, and oligotypes), only those with mean relative abundance >0.01% were tested. For the metagenome data (MetaPhlAn species and HUManN KEGG modules and orthologs), we used all markers as there were fewer of them. We used a similar procedure as for comparisons between controls and CD children to test for the changes between sample D (end of EEN) and E (habitual diet) for 16S rRNA and metagenome markers.
For CD subjects with two or more samples and who were still on or had just stopped EEN treatment (≤3 days of treatment end), individual marker data regressed against EEN days with a subject-specific intercept using the GLM function. Thus, any confounding effect of different sample collection times, particularly after EEN initiation and completion, was naturally accounted for within the analysis.
We correlated the log-transformed relative abundance of each marker against fecal calprotectin using Kendall's rank correlation. For 16S rRNA markers, we tested only those with mean relative abundance >0.01% but for metagenome markers used all markers. All markers with P-value <0.01 were included in stepwise multivariate regression of calprotectin as a function of all these markers.
RESULTS
We characterized the microbial community using 16S rRNA gene sequencing in 117 samples (n=78 CD participants and n=39 healthy controls) and shotgun metagenomics in 65 (n=53 from CD participants and n=12 controls). Only samples with greater than 5,000 filtered and overlapped 16S rRNA or 100,000 shotgun metagenome reads were included.
Bacterial community structure (16S rRNA gene and shotgun metagenomics)
Controls vs. CD patients before EEN
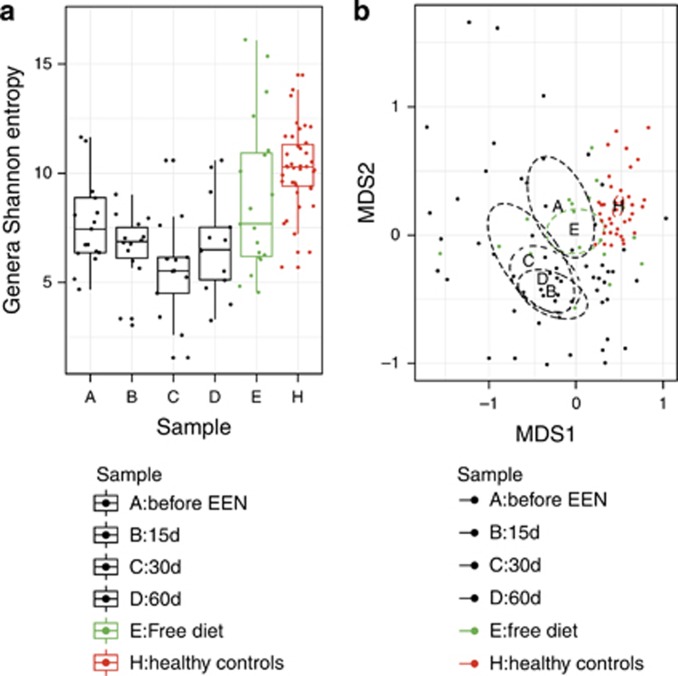
The mean Shannon diversity, a measure of gut microbiota richness and evenness, was higher in controls than CD children prior to EEN (Controls vs. CD: 10.23 vs. 8.00; P=0.006; Figure 1). A higher diversity was observed using OTU than genus analysis. However, the reduction in diversity associated with CD was similar with both approaches (P=4.2 × 10−5, Supplementary Figure S1). In contrast, beta diversity, the variability of the OTU community structure between samples, was higher for CD children than healthy controls. This was apparent from Figure 1 but also using the betadisper function of the R vegan package. The dissimilarity of microbial community structure, among groups, expressed with the average Bray–Curtis distance for samples, was significantly higher in CD children (CD vs. Controls: 0.4913 vs. 0.4089; P=0.048). This effect was more significant using OTU analysis (CD vs. Controls: 0.5848 vs. 0.4868; P<0.001).
Figure 1.
Genus Shannon diversity, in richness equivalents (a), and non-metric multidimensional scaling (NMDS) of operational taxonomic unit (OTU) community structures (b) for each EEN sample time for the Crohn's disease (CD) children and healthy controls. (a) Diversity, in species equivalents, decreased during exclusive enteral nutrition (EEN; P=0.037) and was higher in healthy children vs. CD children at time A (P=0.009). (b) NMDS plot using Bray–Curtis distances of the 3% OTU community structures.
The microbial community structure was also different between controls and CD patients, with 9.9% of the variation in community structure explained by sample groups (P=0.001). An association between age and genus community structure was observed for controls (P=0.033) but not CD children. However, this effect was weak and swamped by the effect that grouping (Controls vs. CD) had on genus community structure. Similarly, no difference was observed (P-value=0.697) in genus community structure between samples of CD patients collected one day before EEN (n=7) compared with samples collected within 6 days of EEN (n=10). The effect of disease affecting the ileum on community diversity and structure was explored and compared with subjects without ileal disease, but the difference did not reach statistical significance (P=0.176).
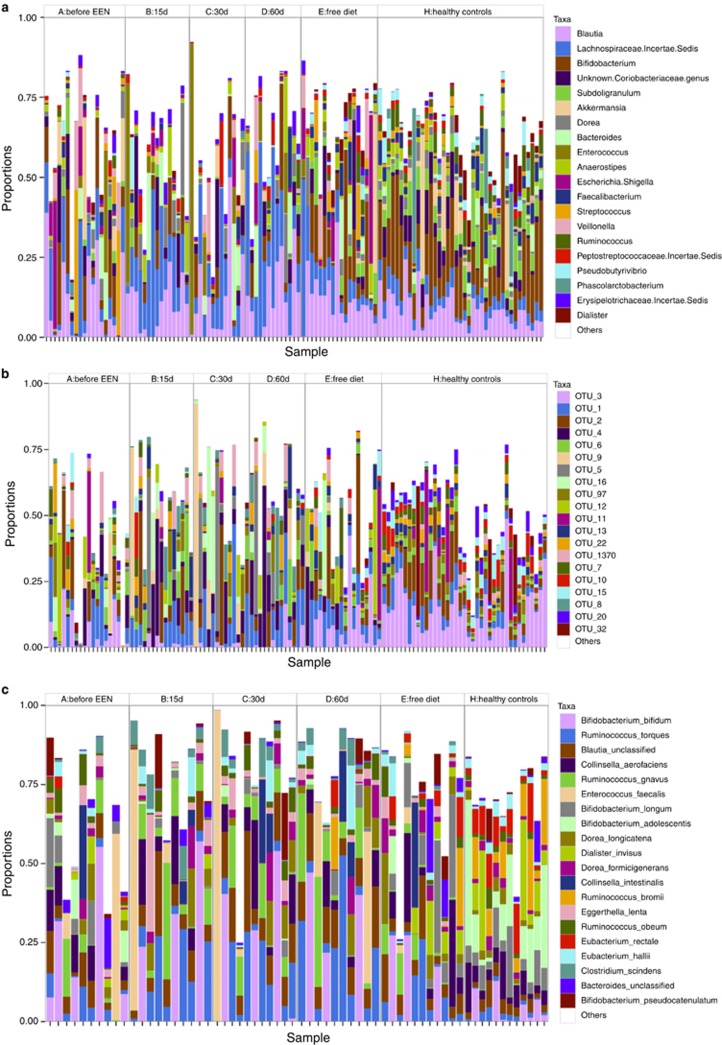
Figure 2 displays the relative abundance of the top 20 genera present in both CD and healthy controls. In total, 36 genera out of 84 (43%) tested differed significantly between CD children and controls (Table 1). Genera belonging to the Lachnospiraceae were more abundant in controls, (e.g., Coprococcus, Pseudobutyrivibrio, and Ruminococcus) as were Subdoligranum and Faecalibacterium from Clostridiales. In contrast, other members of Clostridiales such as Peptostreptococcus were more abundant in CD children (Table 1). A number of genera were also more abundant in CD children from diverse phyla such as the Actinobacteria member Atopobium or the Proteobacteria member Escherichia–Shigella (Table 1). Similar observations resulted from OTU assignments (Figure 2). Overall, 9.6% of the variation (P<0.001) was explained by group (Controls vs. CD). The relative abundance of a total of 141 OTUs out of 351 (42%) tested differed significantly between controls and those with CD. The 20 most significant are presented in Supplementary Table S2. Many OTUs, more abundant in controls, were affiliated with Lachnospiraceae, (e.g., OTU88; OTU10 (Pseudobutyrivibrio); OTU18 (Ruminococcus)), whereas OTU69 (Streptococcus) was more abundant in samples from CD patients (Supplementary Table S2).
Figure 2.
Relative abundance of the 20 most abundant bacterial genera (a), operational taxonomic units (OTUs) (b), and MetaPhlAn species from shotgun metagenome samples (c) for each exclusive enteral nutrition (EEN) sample time for the Crohn's disease (CD) children and healthy controls. OTUs assignments see Supplementary Table S14.
Table 1. Relative abundance of bacterial genera in the healthy control group and children with Crohn's before treatment with exclusive enteral nutrition.
|
Healthy |
Crohn's disease |
|||||
|---|---|---|---|---|---|---|
| Genus | Mean | s.e. | Mean | s.e. | P value | P value adjusted |
| Decreased in CD | ||||||
| Coprococcus | 6.54E−03 | 1.74E−03 | 4.24E−04 | 3.64E−04 | 6.98E−07 | 2.93E−05 |
| Pseudobutyrivibrio | 2.35E−02 | 3.61E−03 | 3.96E−03 | 2.59E−03 | 6.98E−07 | 2.93E−05 |
| Ruminococcus | 3.36E−02 | 4.93E−03 | 5.00E−03 | 2.44E−03 | 1.76E−06 | 4.92E−05 |
| Subdoligranulum | 5.84E−02 | 5.66E−03 | 1.73E−02 | 6.69E−03 | 7.41E−06 | 1.56E−04 |
| Faecalibacterium | 2.96E−02 | 4.23E−03 | 9.27E−03 | 3.34E−03 | 3.14E−04 | 3.30E−03 |
| Lachnospira | 6.57E−04 | 2.14E−04 | 4.13E−05 | 3.21E−05 | 7.48E−04 | 6.28E−03 |
| Clostridium (Clostridiaceae) | 6.65E−03 | 1.14E−03 | 2.48E−03 | 9.89E−04 | 1.35E−03 | 9.22E−03 |
| Alistipes | 4.73E−03 | 1.70E−03 | 1.90E−03 | 1.38E−03 | 1.43E−03 | 9.22E−03 |
| Parabacteroides | 1.44E−03 | 4.56E−04 | 3.78E−04 | 2.57E−04 | 1.89E−03 | 1.12E−02 |
| Atopobium | 1.49E−05 | 5.72E−06 | 1.53E-03 | 1.38E−03 | 2.00E−03 | 1.12E−02 |
| Anaerotruncus | 9.79E−04 | 3.27E−04 | 4.53E−04 | 3.43E−04 | 2.36E-03 | 1.24E−02 |
| Roseburia | 5.66E−03 | 1.05E−03 | 3.51E−03 | 1.62E−03 | 3.10E−03 | 1.53E−02 |
| Turicibacter | 1.69E−03 | 4.75E−04 | 4.90E−04 | 2.31E−04 | 3.84E−03 | 1.79E−02 |
| Anaerostipes | 1.74E−02 | 2.39E−03 | 7.74E−03 | 2.15E−03 | 4.98E−03 | 2.09E−02 |
| Catabacter | 2.61E−04 | 5.04E−05 | 1.24E−04 | 5.34E−05 | 8.64E−03 | 3.30E−02 |
| Bifidobacterium | 1.53E−01 | 1.80E−02 | 7.90E−02 | 2.14E−02 | 9.07E−03 | 3.31E−02 |
| Bilophila | 6.75E−04 | 2.87E−04 | 7.37E−05 | 4.43E−05 | 9.52E−03 | 3.33E−02 |
| Ruminococcaceae Incertae Sedis | 1.14E−02 | 1.25E−03 | 6.66E−03 | 1.78E−03 | 1.05E−02 | 3.38E−02 |
| Unknown Bifidobacteriaceae genus | 1.12E−03 | 1.36E−04 | 6.13E−04 | 1.44E−04 | 1.27E−02 | 3.94E−02 |
| Slackia | 6.81E−04 | 2.38E−04 | 7.76E−06 | 5.68E−06 | 1.67E−02 | 5.00E−02 |
| Unknown Coriobacteriaceae genus | 4.47E−02 | 6.81E−03 | 3.82E−02 | 1.54E−02 | 3.19E−02 | 8.38E−02 |
| Dialister | 1.91E−02 | 5.33E−03 | 4.44E−03 | 2.07E−03 | 3.76E−02 | 9.13E−02 |
| Increased in CD | ||||||
| Unknown Ruminococcaceae genus | 9.81E−04 | 9.83E−05 | 1.05E−02 | 1.02E−02 | 9.35E−06 | 1.57E−04 |
| Parvimonas | 1.37E−05 | 5.01E−06 | 5.08E−04 | 2.25E−04 | 8.92E−05 | 1.25E−03 |
| Actinomyces | 9.59E-04 | 2.11E−04 | 1.09E−02 | 6.15E−03 | 2.43E−04 | 2.91E−03 |
| Anaerococcus | 2.51E−05 | 8.11E−06 | 1.30E−02 | 9.46E−03 | 7.04E−04 | 6.28E−03 |
| Peptostreptococcus | 9.58E−05 | 2.58E−05 | 1.05E−02 | 5.12E−03 | 8.43E-04 | 6.44E−03 |
| Lactococcus | 3.84E−04 | 1.30E−04 | 1.10E−02 | 1.04E−02 | 4.49E−03 | 1.99E−02 |
| Escherichia–Shigella | 1.57E−03 | 4.35E−04 | 4.15E−02 | 2.19E−02 | 7.46E−03 | 2.98E−02 |
| Eggerthella | 1.23E−03 | 5.36E−04 | 6.55E−03 | 2.40E−03 | 9.99E−03 | 3.36E-02 |
| Megasphaera | 5.95E−05 | 3.16E−05 | 3.93E−03 | 3.36E−03 | 2.48E−02 | 7.18E−02 |
| Prevotella | 1.58E−02 | 6.67E−03 | 1.96E−02 | 1.82E-02 | 2.70E−02 | 7.56E−02 |
| Gemella | 5.97E−05 | 1.17E−05 | 1.26E−03 | 6.96E−04 | 2.94E−02 | 7.96E−02 |
| Mogibacterium | 3.40E−05 | 9.51E−06 | 2.59E−04 | 1.48E−04 | 3.61E-02 | 9.13E−02 |
| Peptoniphilus | 5.58E−05 | 1.42E−05 | 3.36E−03 | 2.12E−03 | 3.91E−02 | 9.13E−02 |
| Abiotrophia | 1.29E−06 | 7.94E−07 | 4.27E−04 | 3.70E−04 | 3.91E−02 | 9.13E−02 |
CD, Crohn's disease.
Data are displayed with standard errors and means. P-values for Kruskal–Wallis tests on the log-transformed data using Benjamini–Hochberg false discovery rate to adjust significance values for multiple comparisons. Only genera with a mean abundance >0.01% were tested.
In-depth analysis of the Lachnospiraceae in 1,191,470 reads using oligotyping resulted in 93 high-resolution oligotypes, explaining even more of the variation associated with CD children compared with controls (12.4% P<0.001). Forty-four oligotypes (47%) differed significantly between the two groups. The 20 most significant oligotypes are in Supplementary Table 3. The findings of this high-resolution analysis were consistent with those observed through the genera and OTU assignments. Oligotypes classified as E. rectale and Roseburia inulinivorans were more abundant in controls, whereas R. gnavus and Clostridium clostridioformes were associated with CD.
For classification of shotgun metagenome reads to species level with MetaPhlAn, a tendency for higher mean Shannon diversity was found for controls (CD vs. Controls: 18.49 vs. 14.30; P=0.230, Supplementary Figure S1). The two groups explained 15% of the variation in community structure (P<1.0 × 10−5), but at the whole community level no differences in species proportions were observed (R2=0.0733, P=0.476; Figure S2 online). In total, 36 species had a false discovery rate <10% (Supplementary Table S4). These results were consistent with other computational approaches using 16S rRNA sequencing. Species such as Bifidobacterium adolescentis, Ruminococcus bromii, Eubacterium spp., F. prausnitzii, Coprococcus eutactus, and Subdoligranulum variabile were more abundant in controls, whereas in CD children Streptococcus anginosus, Enterococcus faecalis, and R. gnavus were overrepresented along with other species (Supplementary Table S4).
Impact of EEN on CD
During EEN, the Shannon diversity, at the genus level, decreased (P=0.037; Figure 1). This became apparent even after 15 days on EEN (sample B) with a minimum diversity observed by ~30 days (sample C), a slight recovery toward the end of EEN (sample D) and complete recovery to pre-treatment levels when patients returned to habitual diet (sample E) (Figure 1). When genus diversity was regressed against days on EEN, using subject-dependent linear regression, to overcome any confounding effects of the time of sample collection during EEN, we found (Supplementary Figure S2) that every 10 days on EEN 0.6 genus diversity equivalents are lost (coeff (SE): −0.06 (0.012); P=2.36 × 10−5). Similar results were observed for classification of shotgun metagenome reads to species with MetaPhlAn (P=0.0023) and a trend based on OTU analysis (P=0.140; Supplementary Figure S1).
At the community structure level, EEN made the CD microbial community even more dissimilar to that typical of healthy controls (Figure 1). Sample time explained 6.4% of the variation in the microbial community structure of CD children (P=0.094). At all points during EEN, there was no difference in microbial community structure between patients with and without ileal disease involvement. Changes in the top 20 taxa during EEN are displayed in Figure 2. Microbial community structure at each EEN sample time point was independent of other concomitant treatment or whether this was the first or a subsequent EEN course suggesting that the observed changes were associated with EEN only.
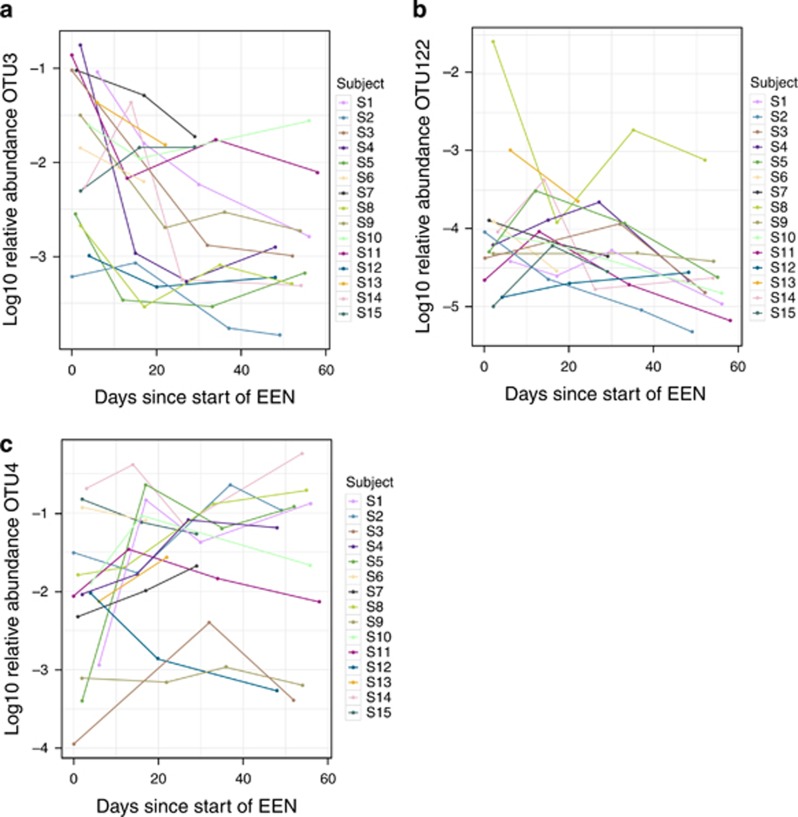
When relative abundance of each bacterial taxon, based on OTU and genus assignments, was regressed against days on EEN, allowing for a subject-dependent intercept, representing different starting abundances in different children, the vast majority of significant changes represented a reduction in relative abundance. Indeed, of 34 genera that significantly changed, only one, Lactococcus, increased with EEN (Table 2). Interestingly, some of the most negatively impacted genera were those whose abundance was already lower in CD children, before EEN initiation, than controls (e.g., Bifidobacterium, Ruminococcus, and Faecalibacterium). A similar impact was observed in OTU analysis (Supplementary Table S5), with a large number of OTUs decreasing during EEN. For example, OTU3, assigned to Bifidobacterium, decreased in some children by two logs of relative abundance (Figure 3), whereas OTU4, assigned to R. gnavus, increased in most subjects during EEN (Figure 3). Oligotyping of Lachnospiraceae confirmed these patterns, with all oligotypes significantly decreasing in abundance during EEN including oligotypes that were significantly higher or lower in CD (Supplementary Table S6).
Table 2. Linear regressions of genus relative abundance against days on EEN.
| Genus | Slope | P value | P value adjusted |
|---|---|---|---|
| Decreased during EEN | |||
| Unknown Bifidobacteriaceae genus | −0.011 | 7.17E−05 | 5.81E−03 |
| Dialister | −0.0212 | 2.05E−04 | 6.12E−03 |
| Bifidobacterium | −0.015 | 2.86E−04 | 6.12E−03 |
| Ruminococcus | −0.0159 | 3.02E−04 | 6.12E−03 |
| Subdoligranulum | −0.0154 | 5.34E−04 | 7.54E−03 |
| Unknown Actinomycetaceae (Actinomycetales) genus | −0.0115 | 5.59E−04 | 7.54E−03 |
| Atopobium | −0.0131 | 6.74E−04 | 7.80E−03 |
| Akkermansia | −0.0155 | 1.02E−03 | 1.03E−02 |
| Solobacterium | −0.0107 | 1.21E−03 | 1.09E−02 |
| Unknown Coriobacteriaceae genus | −0.00919 | 1.64E−03 | 1.33E−02 |
| Megasphaera | −0.0144 | 1.90E−03 | 1.40E−02 |
| Veillonella | −0.0199 | 3.03E−03 | 1.91E−02 |
| Mogibacterium | −0.00849 | 3.75E−03 | 1.91E−02 |
| Anaerococcus | −0.0157 | 3.77E−03 | 1.91E−02 |
| Pseudobutyrivibrio | −0.0103 | 3.90E−03 | 1.91E−02 |
| Peptoniphilus | −0.0137 | 4.04E−03 | 1.91E−02 |
| Actinomyces | −0.0108 | 4.22E−03 | 1.91E−02 |
| Prevotella | −0.0153 | 4.24E−03 | 1.91E−02 |
| Staphylococcus | −0.00978 | 9.30E−03 | 3.82E−02 |
| Faecalibacterium | −0.0144 | 1.09E−02 | 3.82E−02 |
| Gordonibacter | −0.00958 | 1.10E−02 | 3.82E−02 |
| Dorea | −0.0134 | 1.12E−02 | 3.82E−02 |
| Phascolarctobacterium | −0.00807 | 1.12E−02 | 3.82E−02 |
| Unknown Veillonellaceae genus | −0.0103 | 1.13E−02 | 3.82E−02 |
| Varibaculum | −0.0114 | 1.38E−02 | 4.49E−02 |
| Turicibacter | −0.0111 | 1.61E−02 | 5.01E−02 |
| Methanobrevibacter | −0.0118 | 1.69E−02 | 5.08E−02 |
| Unknown Akkermansiaceae genus | −0.00864 | 2.02E−02 | 5.84E−02 |
| Sutterella | −0.00729 | 2.18E−02 | 6.09E−02 |
| Porphyromonas | −0.00881 | 2.58E−02 | 6.96E−02 |
| Unknown Ruminococcaceae genus | −0.00685 | 2.66E−02 | 6.96E−02 |
| Streptococcus | −0.00833 | 2.88E−02 | 7.30E−02 |
| Rothia | −0.00915 | 3.27E−02 | 7.79E−02 |
| Increased during EEN | |||
| Lactococcus | 0.0132 | 3.05E−02 | 7.48E−02 |
EEN, exclusive enteral nutrition.
Linear regression of log genera abundance against days on EEN with subject-dependent intercepts for the children with at least two samples. Benjamini–Hochberg false discovery rate was used to adjust significance values for multiple comparisons.
Figure 3.
Log abundance of OTU3 (Bifidobacterium spp.) (a), OTU122 (Atopobium spp.) (b), and OTU4 (Ruminoccocus gnavus) (c) as a function of days on EEN discriminated by subject. Adjusted P-values: OTU122=0.0338; OTU3=0.00573; OTU4=0.0897. A full color version of this figure is available at the American Journal of Gastroenterology journal online. EEN, exclusive enteral nutrition; OTU, operational taxonomic unit.
Correlations with fecal calprotectin
We analyzed our data set by direct correlation to calprotectin from the same sample to allow proxy analysis of colonic inflammation. Twenty-six genera significantly correlated with calprotectin in CD children (Supplementary Table S7); both strong positive and negative associations were found. Performing stepwise regression analysis, minimizing the overall model Akaike Information Criterion, to determine which associations were most important accounting for correlations between genera, seven of 14 genera were selected (Supplementary Table S8). The relative abundance of three selected genera was negatively associated with calprotectin (Anaerostipes, Coprococcus, and unknown Erysipelotrichaceae). Four were positively associated (Bacteroides, Gemella, Haemophilus, and Atopobium). Overall, the final model explained a remarkable 69% of the variation in fecal calprotectin concentration (Supplementary Table S8). A higher number of 35 OTUs significantly correlated (Supplementary Table S9) with calprotectin, and 14 of these were selected by stepwise regression (Supplementary Table S10). The final model predicted 78% of the variation in calprotectin levels among CD children. Among those OTUs that were most significantly positively associated with fecal calprotectin were OTU122 (Atopobium), OTU1073 (Lachnospiraceae), OTU45 (Streptococcus), OTU1086 (Peptostreptococcus), and OTU41 (Bacteroides). There were a number of OTUs negatively associated with calprotectin including OTU3 (Bifidobacterium), OTU21 (Lachnospiraceae), and OTU106 (Anaerostipes). Using oligotyping, no significant correlations were found between oligotypes and calprotectin, after correcting for multiple comparisons.
Genetic functional capacity using shotgun metagenomics
Healthy children compared with CD patients before EEN
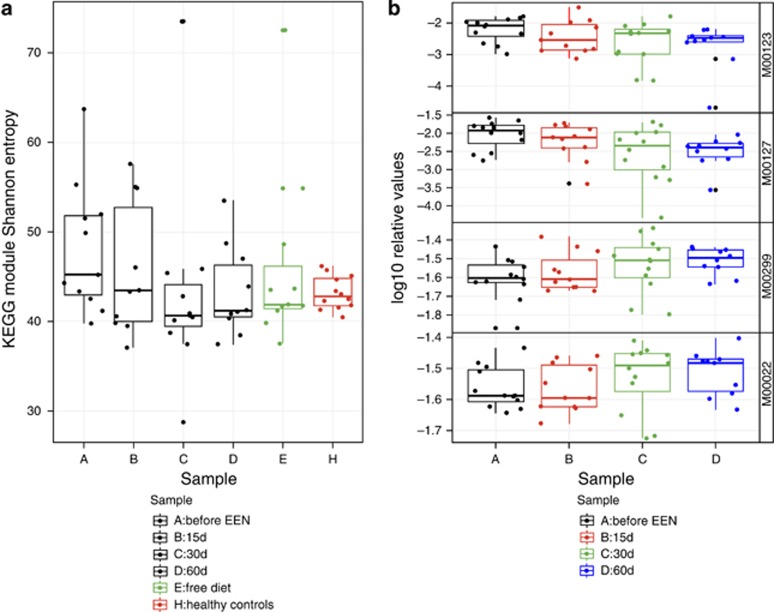
When reads were assigned to metabolic KEGG modules with HUMAnN, a significantly larger mean Shannon diversity, in richness equivalents, was observed in CD children compared with controls (CD vs. Controls: 48.1 vs. 43.2; P=0.05 Figure 4). Healthy and CD children differed at the level of KEGG modules, with marginal significance explaining 9.4% of the variation (P=0.059; Supplementary Figure S3). Several KEGG modules and orthologs (Supplementary Table S11) were different between the two groups, but their significance became marginal (P<0.100) when adjusted for multiple testing (Table 3). Modules more abundant in samples from CD patients included ubiquinone and lipopolysaccharide biosynthesis and the twin-arginine translocation (Tat) system, whereas key processes such as fatty acid biosynthesis, initiation, and sulfur reduction were overrepresented in controls (Table 3).
Figure 4.
KEGG module Shannon diversity for each EEN sample time and participant (a) and log relative abundance of modules that significantly changed during EEN (b). (a) Diversity was not impacted during EEN (P=0.260), but there was a difference between H and CD before EEN (P=0.05). (b): M00123, biotin biosynthesis, P=0.005; M00127, thiamine biosynthesis, P=0.0166; M00299, spermidine/putrescine transport system, P=0.0307; M00022, shikimate pathway, P=0.058. P-values are based on linear regressions of KEGG module log relative abundance against days on EEN with subject-dependent intercepts for 11 children with at least two shotgun metagenome samples. Benjamini–Hochberg false discovery rate was used to adjust significance values for multiple comparisons. All modules with a mean abundance >1.0 × 10−9 were tested; only those with adjusted P-value <0.1 are shown. A full color version of this figure is available at the American Journal of Gastroenterology journal online. CD, Crohn's disease; EEN, exclusive enteral nutrition; KEGG, Kyoto Encyclopedia of Genes and Genomes.
Table 3. Relative abundance of KEGG modules in the healthy control group and children with Crohn's before treatment with exclusive enteral nutrition.
|
Healthy |
Crohn's disease |
||||||
|---|---|---|---|---|---|---|---|
| Module | Name | Mean | s.e. | Mean | s.e. | P value | P value adjusted |
| Decreased in CD | |||||||
| M00312 | 2-oxoisovalerate:ferredoxin oxidoreductase | 9.96E−07 | 6.52E−07 | 3.23E−08 | 3.23E−08 | 1.37E−03 | 6.73E−02 |
| M00082 | Fatty acid biosynthesis, initiation | 1.08E−06 | 8.56E−07 | 2.38E−08 | 1.76E−08 | 3.82E−03 | 6.73E−02 |
| M00029 | Urea cycle | 8.05E−05 | 2.84E−05 | 9.11E−06 | 5.22E−06 | 4.64E−03 | 6.73E−02 |
| M00030 | Lysine biosynthesis | 1.14E−05 | 4.28E−06 | 3.52E−06 | 3.24E−06 | 1.16E−02 | 9.36E−02 |
| M00002 | Glycolysis | 4.07E−02 | 7.60E−04 | 3.68E−02 | 1.32E−03 | 1.64E−02 | 9.50E−02 |
| M00178 | Ribosome, bacteria | 5.09E−02 | 1.10E−03 | 4.52E−02 | 1.97E−03 | 1.64E−02 | 9.50E−02 |
| M00360 | Aminoacyl-tRNA biosynthesis | 4.21E−02 | 1.17E−03 | 3.67E−02 | 1.41E−03 | 1.64E−02 | 9.50E−02 |
| M00361 | Nucleotide sugar biosynthesis, euk. | 8.02E−05 | 4.66E−05 | 6.76E−08 | 3.54E−08 | 1.64E−02 | 9.50E−02 |
| M00362 | Nucleotide sugar biosynthesis, prok. | 8.69E−05 | 4.45E−05 | 1.44E−07 | 7.77E−08 | 1.64E−02 | 9.50E−02 |
| Increased in CD | |||||||
| M00117 | Ubiquinone biosynthesis | 4.81E−05 | 4.81E−05 | 1.35E−03 | 5.24E−04 | 7.12E−04 | 6.73E−02 |
| M00336 | Twin-arginine translocation (Tat) system | 7.03E−05 | 7.03E−05 | 1.22E−03 | 4.68E−04 | 1.70E−03 | 6.73E−02 |
| M00060 | Lipopolysaccharide biosynthesis | 1.83E−04 | 7.82E−05 | 1.78E−03 | 4.94E−04 | 2.09E−03 | 6.73E−02 |
| M00277 | PTS system, N-acetylgalactosamine-specific II component | 2.49E−03 | 6.33E−04 | 6.76E−03 | 1.29E−03 | 3.13E−03 | 6.73E−02 |
| M00019 | Leucine biosynthesis | 1.43E−03 | 6.22E−04 | 9.09E−03 | 2.03E−03 | 3.13E−03 | 6.73E−02 |
| M00324 | Dipeptide transport system | 6.37E−04 | 6.40E−05 | 1.67E−03 | 3.89E−04 | 4.64E−03 | 6.73E−02 |
| M00028 | Ornithine biosynthesis | 7.19E−03 | 9.57E−04 | 1.14E−02 | 1.02E−03 | 4.64E−03 | 6.73E−02 |
| M00231 | Octopine/nopaline transport system | 3.62E−09 | 2.63E−09 | 2.40E−06 | 2.40E−06 | 6.77E−03 | 8.18E−02 |
| M00176 | Sulfur reduction, sulfate => H2S | 6.04E−04 | 1.93E−04 | 1.12E−04 | 4.27E−05 | 6.77E−03 | 8.18E−02 |
| M00276 | PTS system, mannose-specific II component | 6.59E−03 | 8.74E−04 | 1.29E−02 | 2.21E−03 | 1.16E−02 | 9.36E−02 |
| M00335 | Sec (secretion) system | 5.64E−04 | 3.53E−04 | 3.35E−03 | 1.18E−03 | 1.16E−02 | 9.36E−02 |
| M00348 | Glutathione transport system | 1.17E-03 | 9.16E−05 | 2.11E−03 | 3.33E−04 | 1.16E−02 | 9.36E−02 |
| M00136 | GABA biosynthesis | 1.04E−06 | 6.97E−07 | 3.38E−04 | 2.59E−04 | 1.16E−02 | 9.36E−02 |
| M00050 | Guanine nucleotide biosynthesis | 1.62E−02 | 1.09E−03 | 2.26E−02 | 2.13E−03 | 1.16E−02 | 9.36E−02 |
| M00260 | DNA polymerase III complex | 1.71E−04 | 1.18E−04 | 1.60E−03 | 7.21E−04 | 1.64E−02 | 9.50E−02 |
| M00198 | sn-Glycerol 3-phosphate transport system | 2.46E−03 | 2.26E−04 | 3.99E−03 | 6.97E−04 | 1.64E−02 | 9.50E−02 |
CD, Crohn's disease; EEN, exclusive enteral nutrition; GABA, gamma-aminobutyric acid; KEGG, Kyoto Encyclopedia of Genes and Genomes; PTS, phosphotransferase system.
Significance was determined using Kruskal–Wallis tests on the log relative frequencies and using Benjamini–Hochberg false discovery rate to adjust significance values for multiple comparisons; Only 25 modules with adjusted P-value <0.1 are presented.
Impact of EEN on CD and correlations with fecal calprotectin
Four KEGG modules correlated with the number of days on EEN including pathways for biotin and thiamine biosynthesis (negative), the spermidine/putrescine transport system, and the shikimic acid pathway (positive; Figure 4 and Supplementary Table S12). Unlike relationships observed with taxa abundance, there were no significant correlations between calprotectin and relative abundance of KEGG modules.
Comparison between start, end of EEN, and return to habitual diet
In contrast to the impact of EEN on community structure and KEGG modules, there were no significant changes, adjusting for multiple comparisons, between sample A or end of EEN (D) and return to habitual diet (E) for all data types (genera, OTUs, oligotypes, shotgun metagenomics). Although a mean increase in relative abundance of several taxa was observed as CD children returned to habitual diet, these changes were not statistically significant. Those OTUs associated with fecal calprotectin (Supplementary Table S10) showed no significant change, but it is interesting to note that those negatively associated with calprotectin did increase in abundance, whereas those positively associated in general remained low at return to the habitual diet in unadjusted analysis (Supplementary Table S13).
DISCUSSION
In this study, we detailed a comprehensive sequential analysis of the entire fecal microbiota, its taxonomic profile, genetic functional capacity, and associations with fecal calprotectin, during a course of EEN in children with CD. Using a hypothesis-free approach, this study identified various bacterial taxa associated with CD and other that significantly changed during treatment with EEN. This study complements our previous publication of quantitative changes in concentration of selected bacterial metabolites and major bacterial groups previously implicated in the onset of CD.
Several significant differences in taxon community structure and relative abundance between CD children and controls were observed. The gut microbiota of CD children presented lower taxonomic diversity per individual but higher variation between individuals perhaps attributed to variations in disease behavior, location, or inflammation severity. Shotgun metagenome analysis revealed, for the first time, that despite lower taxonomic bacterial diversity a higher level of genetic functional diversity was associated with the CD microbiota prior to EEN treatment. This probably reflects a greater range of functional roles that can be exploited by the gut microbiota in inflamed gut, in active disease. This diverse microbial functional capacity tended to decrease during EEN to levels similar to healthy controls. These data suggest a higher degree of functional redundancy, with multiple species performing similar roles in healthy children, and may be important in maintaining gut health. In active CD, this functional redundancy may be lost and then restored during EEN.
Before treatment, we found bacterial taxa that were both more and less abundant in children with CD. Several of these findings concur with previous work (2, 3, 6), but here the methodology employed is not only hypothesis-free, unselective of certain taxa, but also more comprehensive, detailing bacterial taxonomy down to species, OTU, and oligotype levels. Numerous OTUs and species were less abundant in CD children, including classic commensals such as Faecalibacterium spp. and Bifidobacterium spp., but also a number of organisms that receive less attention, such as Eubacterium rectale and R. obeum. Conversely, some bacteria were more prevalent in CD, including E. coli/Shigella spp., Streptococcus spp., Peptostreptococcus spp., and Atopobium spp. More interestingly, several of these differences were not uniform or of the same direction within a genus, but varied between species and OTUs, which emphasizes the importance and need for an in-depth microbiota analysis when the role of the gut microbiota is explored. For example, R. gnavus was higher, but Ruminococcus obeum was lower in CD.
Although the results agree with previous research (2, 3, 6), possible reverse causality complicates interpretation of their role in the etiology of CD. Their importance and the relevance of these findings can be further unraveled only if these taxa are related to clinical outcomes, colonic inflammation, or changed during the course of disease modifying interventions. In the current study, correlations between fecal calprotectin, global fecal microbial composition, and genetic functional capacity were explored. Several taxa, which were different between CD and controls, did not associate with calprotectin and may have a less protagonist role in the etiology of CD. Instead, we found several other that did associate with calprotectin, negatively and positively. A subset of taxa was identified explaining a sizable amount of the variation in fecal calprotectin. Among them, OTU3 (Bifidobacterium spp.) had the strongest negative and OTU122 (Atopobium spp.) the strongest positive association with calprotectin in multivariate regression analysis.
Treatment with EEN induced a major reduction in the relative abundance of several species, some of which were already at lower abundance compared with controls. This is a paradox, as we would expect EEN treatment to normalize the perceived “dysbiotic” microbiota toward a healthier state. Instead, EEN treatment shifted the microbiota to a more “dysbiotic” state, with a greater NMDS distance from the healthy group compared with samples before EEN (Figure 1). Moreover, EEN caused reduction in the relative abundance of gut bacteria that were negatively associated with calprotectin, as well as those that correlated positively with it. Among these, OTU122 (Atopobium) was strongly negatively associated with the number of EEN days, but the commensal strain Bifidobacterium OTU3 was even more strongly negatively impacted. The particular Atopobium OTU identified, OTU122, has 99% identity in the V4 region with Atopobium parvulum, which is associated with halitosis (11), and has been associated with disease activity in pediatric CD (12) and hence is a good candidate for further research.
In agreement with our previous observations using qPCR (6), we observed a higher abundance of E. coli in CD. The E. coli population is of particular interest, as adherent invasive E. coli are overrepresented in CD patients (13). Moreover, the higher representation of KEGG modules coding for ubiquinone in CD children, before EEN, coincides with this observation of a higher relative abundance of Enterobacteriaceae. Ubiquinone is produced only by organisms capable of aerobic respiration, its main role is maintaining membrane stability by acting as a crucial membrane chain-terminating antioxidant. In nematodes (14), ubiquinone-deficient E. coli strains were more efficiently lysed in the pharynx or the intestine than their wild-type counterparts. Therefore, ubiquinone or bacterial respiration may act as virulence factors, allowing bacteria to colonize and harm the host. Overrepresentation of ubiquinone also fits with previous work (15). Likewise, other KEGG modules that were in higher abundance in CD, such as secretion and (Tat) systems, or module encoding for LPS synthesis, may be linked with overrepresentation of Enterobacteriaceae or other pathobionts in CD.
During EEN treatment, representation of KEGG modules relevant to the biosynthesis of biotin and thiamine, two B-complex vitamins, decreased. This may be indicative of the reduction in bacteria that bear genes encoding for these vitamins during EEN, such as bifidobacteria and E. coli spp., or changes in the synthesis of short or medium-chain fatty acids that requires these vitamins. Alternatively, supply of these vitamins through EEN may induce bacterial redundancy or indolence to produce them. In contrast, genes encoding for aromatic amino acid metabolism and transport of spermidine/putrescine, which have a major role in cell growth, and may indicate increased epithelial cell renewal and tissue healing (16), were overrepresented during EEN. Against expectations, no association between KEGG modules and calprotectin was found in CD. This probably reflects the lack of level in detail of currently available metabolic databases to resolve the key processes by which organisms impact the gut epithelium and that taxonomy serves as a better proxy to functional role.
The observational design of this study impedes us from suggesting a causative association between these findings and their role in CD pathogenesis or indeed in proposing a mechanism of therapeutic action of EEN based on the changes in the gut microbiota. Intervention studies are required with the aim of elucidating whether maintenance of such changes, following a successful course of EEN, can prolong disease remission and whether replication of such changes occur with other types of formula and dietary intervention. Once this is proven, a plausible mechanism of action for EEN, mediated by modulation of the gut microbiota, could be suggested. Conversely, these changes are likely to be direct consequences of a diet low in fiber, rich in protein, fat, simple sugars and micronutrients, modulating various microbial communities, bacterial pathways, and metabolite production. Whether the clinical efficacy of EEN could be enhanced by selectively tailoring its composition to stimulate the growth of beneficial microbes, suppressed during EEN, is also an intriguing concept to explore. Moreover, the very distinct CD microbiome we observed in this study may hold implications for improvement in diagnostic and prognostic classification of the disease, particularly in conjunction with other established disease biomarkers such as fecal calprotectin.
This study has some limitations. We performed analysis in feces, enabling a non-invasive collection of serial samples, but results may differ from mucosal adherent bacteria. Despite correcting for multiple testing, our participant's sample size was modest, and this study may be underpowered to explore secondary outcomes such as associations between disease location and behavior with microbiota characteristics. Also, as a community expresses only a variable subset of its genome, at any time, metatranscriptomic and metabolomic data are required to complement and progress this work further. Some patients were on concomitant treatment, and there was a modest age difference between controls and CD children. However, any effect was minimal with the changes demonstrated driven predominantly by EEN or differences according to grouping (CD vs. Healthy) on community structure. This reconfirms evidence that diet is a strong determinant of the microbial microenvironment (17). Although PCDAI and CRP were highly correlated with fecal calprotectin (r=0.60; P=7.924 × 10−06 and r=0.42; P=4.644 × 10−03), we preferred to use and present data only for the latter, an observer-independent and reliable marker of colonic inflammation, as this study aimed specifically to associate a direct marker of colonic inflammation with microbiota composition and functional capacity measured in the same fecal sample. Moreover, preliminary analysis showed that fecal calprotectin was a better predictor of community structure explaining 10.5% of the variation in genus relative abundances across these samples compared with 9.3% and 3.5% for the PCDAI scores and CRP values, respectively.
Although there is still a degree of reverse causality to address even with the methodological approach applied here, this study successfully removed a great deal of “microbial signal noise”, potentially irrelevant to the microbial origins of CD, and thus focuses the field for more targeted research. Future interventions aiming to induce or maintain EEN-induced changes in the gut microbiota are needed to elucidate the significance of these observed changes in the etiology of CD and mechanism of action of EEN.
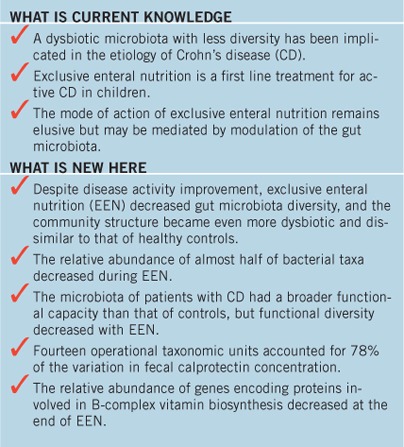
Study Highlights
Acknowledgments
We thank participants and their families, the Department of Paediatric Gastroenterology, Hepatology, and Nutrition. The IBD team at Royal Hospital for Children, Glasgow, is supported by the Catherine McEwan Foundation and the Yorkhill IBD fund. The Open Access publication fee of this article was kindly sponsored by the Crohn's in Childhood Research Association.
Footnotes
SUPPLEMENTARY MATERIAL is linked to the online version of the paper at http://www.nature.com/ajg
Guarantor of the article: Konstantinos Gerasimidis, PhD, MSc, BSc.
Specific author contributions: K.G., C.Q., N.L., R.K.R., C.A.E., and P.M. contributed to study concept and design; K.G., S.J.H., S.T.C., J.Q., J.R., D.S., and M.B. contributed to acquisition of data; all authors contributed to the analysis and interpretation of data, drafting, and critical revision of the manuscript for important intellectual content; A.M.E., C.Q., N.L., and U.Z.I. contributed to bioinformatics and statistical analysis; K.G., A.B., R.K.R., C.A.E., and P.M. obtained funding; K.G., J.R., S.J.H., J.Q., and M.B. contributed to technical aspects of the study; K.G., C.Q., C.A.E., N.L., and R.K.R. contributed in study supervision.
Potential conflicts of interest: R.K. Russell has received speaker's fees, travel support, and participated in medical board meetings with Nestle. P. McGrogan received speaker fees, travel support, and participated in medical board meetings with Nestle. The remaining authors declare no conflict of interest.
Financial support: The study and K. Gerasimidis were funded by the Greek State Scholarship Foundation, the Hellenic Foundation of Gastroenterology, and Nutrition, the Barr Endowment Fund, the Yorkhill Children's Foundation, and the Crohn's in Childhood Research Association. C. Quince is funded by an MRC fellowship as part of the CLIMB consortium Grant Ref: MR/L015080/1. U.Z. Ijaz is funded by NERC IRF NE/L011956/1. R K Russell is supported by an NHS Research Scotland career fellowship award and MRC grant for PICTS (G0800675).
Supplementary Material
References
- 1Qin J, Li R, Raes J et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature 2010;464:59–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2Gevers D, Kugathasan S, Denson LA et al. The treatment-naive microbiome in new-onset Crohn's disease. Cell Host Microbe 2014;15:382–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3Hansen R, Russell RK, Reiff C et al. Microbiota of de-novo pediatric IBD: increased Faecalibacterium prausnitzii and reduced bacterial diversity in Crohn's but not in ulcerative colitis. Am J Gastroenterol 2012;107:1913–1922. [DOI] [PubMed] [Google Scholar]
- 4Haberman Y, Tickle TL, Dexheimer PJ et al. Pediatric Crohn disease patients exhibit specific ileal transcriptome and microbiome signature. J Clin Invest 2014;124:3617–3633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5Cameron FL, Gerasimidis K, Papangelou A et al. Clinical progress in the two years following a course of exclusive enteral nutrition in 109 paediatric patients with Crohn's disease. Aliment Pharmacol Ther 2013;37:622–629. [DOI] [PubMed] [Google Scholar]
- 6Gerasimidis K, Bertz M, Hanske L et al. Decline in presumptively protective gut bacterial species and metabolites are paradoxically associated with disease improvement in pediatric Crohn's disease during enteral nutrition. Inflamm Bowel Dis 2014;20:861–871. [DOI] [PubMed] [Google Scholar]
- 7Sokol H, Seksik P, Furet JP et al. Low counts of Faecalibacterium prausnitzii in colitis microbiota. Inflamm Bowel Dis 2009;15:1183–1189. [DOI] [PubMed] [Google Scholar]
- 8Gerasimidis K, Nikolaou CK, Edwards CA et al. Serial fecal calprotectin changes in children with Crohn's disease on treatment with exclusive enteral nutrition: associations with disease activity, treatment response, and prediction of a clinical relapse. J Clin Gastroenterol 2011;45:234–239. [DOI] [PubMed] [Google Scholar]
- 9Hyams JS, Ferry GD, Mandel FS et al. Development and validation of a pediatric Crohn's disease activity index. J Pediatr Gastroenterol Nutr 1991;12:439–447. [PubMed] [Google Scholar]
- 10Segata N, Waldron L, Ballarini A et al. Metagenomic microbial community profiling using unique clade-specific marker genes. Nat Methods 2012;9:811–814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11Kazor CE, Mitchell PM, Lee AM et al. Diversity of bacterial populations on the tongue dorsa of patients with halitosis and healthy patients. J Clin Microbiol 2003;41:558–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12Muehlbauer M, Mottawea W, Abujamel T et al. Mo1982 atopobium parvulum is a predominant member of the adherent microbiome of pediatric IBD patients and promotes colitis in IL10-/- mice. Gastroenterology 2013;144:S-710. [Google Scholar]
- 13Kotlowski R, Bernstein CN, Sepehri S et al. High prevalence of Escherichia coli belonging to the B2+D phylogenetic group in inflammatory bowel disease. Gut 2007;56:669–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14Gomez F, Monsalve GC, Tse V et al. Delayed accumulation of intestinal coliform bacteria enhances life span and stress resistance in Caenorhabditis elegans fed respiratory deficient E. coli. BMC Microbiol 2012;12:300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15Greenblum S, Turnbaugh PJ, Borenstein E. Metagenomic systems biology of the human gut microbiome reveals topological shifts associated with obesity and inflammatory bowel disease. Proc Natl Acad Sci USA 2012;109:594–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16Slezak K, Hanske L, Loh G et al. Increased bacterial putrescine has no impact on gut morphology and physiology in gnotobiotic adolescent mice. Benef Microbes 2013;4:253–266. [DOI] [PubMed] [Google Scholar]
- 17David LA, Maurice CF, Carmody RN et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature 2014;505:559–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.