Abstract
Inflammasomes are high molecular weight protein complexes that assemble in the cytosol upon pathogen encounter. This results in caspase-1-dependent pro-inflammatory cytokine maturation, as well as a special type of cell death, known as pyroptosis. The Nlrp3 inflammasome plays a pivotal role in pathogen defense, but at the same time, its activity has also been implicated in many common sterile inflammatory conditions. To this effect, several studies have identified Nlrp3 inflammasome engagement in a number of common human diseases such as atherosclerosis, type 2 diabetes, Alzheimer disease, or gout. Although it has been shown that known Nlrp3 stimuli converge on potassium ion efflux upstream of Nlrp3 activation, the exact molecular mechanism of Nlrp3 activation remains elusive. Here, we describe a genome-wide CRISPR/Cas9 screen in immortalized mouse macrophages aiming at the unbiased identification of gene products involved in Nlrp3 inflammasome activation. We employed a FACS-based screen for Nlrp3-dependent cell death, using the ionophoric compound nigericin as a potassium efflux-inducing stimulus. Using a genome-wide guide RNA (gRNA) library, we found that targeting Nek7 rescued macrophages from nigericin-induced lethality. Subsequent studies revealed that murine macrophages deficient in Nek7 displayed a largely blunted Nlrp3 inflammasome response, whereas Aim2-mediated inflammasome activation proved to be fully intact. Although the mechanism of Nek7 functioning upstream of Nlrp3 yet remains elusive, these studies provide a first genetic handle of a component that specifically functions upstream of Nlrp3.
Keywords: caspase 1 (CASP1), cell death, inflammasome, inflammation, NLRP3, NEK7, genetic screen, pyroptosis
Introduction
Employing an evolutionary conserved set of pattern recognition receptors (PRRs),2 the innate immune system senses the presence of microbial pathogens (1). Although PRRs can directly detect microbe-associated molecular patterns, some PRRs also respond to endogenous, host-derived signals that are formed or released upon perturbation or damage that is caused by microbial infections. These signals, which are commonly referred to as damage-associated molecular patterns, can also trigger PRR activation in the context of sterile inflammatory conditions (2).
At the cell-autonomous level, PRR engagement and associated signaling cascades can result in a diverse set of responses, such as the induction of pro-inflammatory gene expression, the control of cytoskeletal rearrangement (e.g. in the context of phagocytosis), as well as the activation of proteolytic cascades, such as the inflammasome pathway. The inflammasome is a cytosolic multiprotein complex that regulates the activation and processing of caspase-1 (3). Inflammasome sensor proteins employ the adapter protein ASC to recruit caspase-1, which in turn results in the proximity-induced autoprocessing and activation of caspase-1. Active caspase-1 cleaves and thereby matures pro-inflammatory cytokines such as IL-1β and IL-18, and at the same time, it triggers an inflammatory type of cell death known as pyroptosis (3). The exact mechanisms of this caspase-1-dependent cell death are currently unknown, but recent evidence suggests that cleavage of the cytosolic protein Gasdermin-D by caspase-1 might in part be responsible for cell death induction (4, 5).
Of all known inflammasome-forming sensors, the Nlrp3 inflammasome plays a central role in antimicrobial defense (6). To this end, a large array of microbial pathogens and microbe-derived molecules have been described to trigger Nlrp3 inflammasome activation. As such, it appears most plausible that Nlrp3 functions as an indirect sensor of cellular perturbation rather than a receptor that would be dedicated to a certain class of exogenous ligands. This notion of Nlrp3 being a general sensor of cellular damage is well in line with its dominant role in sterile inflammatory diseases (7). In fact many sterile inflammatory conditions, in which the cellular integrity of myeloid cells is compromised, display an involvement of Nlrp3-dependent inflammation. Unlike other inflammasome sensors, Nlrp3 activity is subject to a number of regulatory steps that function as additional safeguard mechanisms to prevent unwanted Nlrp3 activation (6). These inputs, commonly known as priming steps (or signal 1), are required to facilitate subsequent Nlrp3 inflammasome activation (or signal 2). On the one hand, given its limited expression under steady state conditions in most cell types, a pro-inflammatory stimulus is required to up-regulate Nlrp3 expression (8). Of note, this requirement for “transcriptional priming” can be overcome by heterologous expression of Nlrp3 (8, 9). On the other hand, pro-inflammatory stimuli can also convey a rapid, “post-translational priming” signal that appears to be required to de-ubiquitinate and thereby to pre-activate Nlrp3 (10, 11). In most studies, LPS is used as a signal 1 stimulus, as it not only functions to prime Nlrp3, but also robustly induces pro-IL-1β expression.
The exact mechanism of Nlrp3 inflammasome activation (signal 2) yet remains to be determined. Nevertheless, a common denominator of its activation seems to be the cytosolic efflux of potassium ions, which appears to be necessary and sufficient to trigger Nlrp3 inflammasome activation (12, 13). Despite enormous research efforts, no molecule has been identified that would specifically and non-redundantly function downstream of potassium efflux, as well as upstream of Nlrp3 inflammasome activation. Driven by the assumption that a genetically encoded factor would indeed serve such a purpose, we carried out a genome-wide, CRISPR/Cas9 loss-of-function screen to identify proteins that would specifically regulate Nlrp3 activation.
Experimental Procedures
gRNA Library Design
A library design tool was written in C++ which for every annotated protein coding gene isoform picks four independent 18-mer gRNA target sites in the first half of the coding region. If more than four target sites were found, target sites fulfilling the scoring criteria detailed in Ref. 14 were prioritized. Duplicate target sites resulting from redundant or similar gene annotations were removed. The resulting library contained 73,177 constructs, which are provided in supplemental Table 1.
gRNA Library Synthesis
Oligonucleotide pools were obtained from CustomArray, and gRNA plasmid libraries were assembled into pLenti-gRNA-GFP using ligation-independent cloning as described (pL-U6-gRNA (14)).
Lentiviral Packaging
For each 15-cm dish of HEK 293T cells grown to a confluence of 70%, 12 μg of pVSV-G, 30 μg of pCMVΔ-8.91, and 40 μg of library plasmid preparation were diluted in 2 ml of 1× HEPES-buffered saline, pH 7.0, and mixed with 100 μl of 2.5 mm CaCl2, vortexed briefly, and incubated for 20 min at room temperature before dropwise addition to the dish. 8 h later, the medium was replaced with DMEM containing 30% FCS. After 48 h, the supernatant was harvested, centrifuged briefly, and filtered through a 0.45-μm filter (GE Healthcare). Virus supernatants were stored at −80 °C in small aliquots.
Cell Culture
Cells were cultured in DMEM supplemented with 10% FCS, 1 mm sodium pyruvate, and 10 μg/ml Ciprofloxacin. Cells were grown at 37 °C and 5% CO2. Wild-type mouse macrophages are immortalized macrophages as described in Ref. 8.
Screening Cell Line
Immortalized murine macrophages stably expressing mNlrp3-FLAG and hASC-CFP (9) were transduced with a lentivirus stably encoding for mCherry-T2A-spCas9 under the CMV promoter. After limiting dilution cloning, a clone with strong and homogenous red fluorescence was selected and expanded (from now on referred to as Nlrp3-Cas9 macrophages throughout this study).
Genome-wide CRISPR/Cas9 Screen
The screen was performed in 12 replicates, and for each replicate, 3 × 106 Nlrp3-Cas9 macrophages were plated in six wells and infected with the library pool at an MOI of 0.05. Starting the next day, cells were trypsinized and expanded every second day. After 1 week, half of the cells were stimulated by adding nigericin to a final concentration of 10 μm, whereas the other half was left unstimulated. Cells were incubated in 15-ml tubes with loose lids at 37 °C 5% CO2 for 6 h. Propidium iodide was added to a final concentration of 5 μg/ml, and cells were incubated on ice for 5 min. Cells were sorted on a BD INFLUX device at 4 °C at a rate of 10,000 cells/s. 2–3 × 107 cells were sorted per stimulated sample, or 5 × 106 cells were sorted per unstimulated sample. Cells were sorted directly into small polypropylene plastic wells pre-filled with 50 μl of a proteinase K-containing lysis buffer (0.2 mg/ml proteinase K, 1 mm CaCl2, 3 mm MgCl2, 1 mm EDTA, 1% Triton X-100, 10 mm Tris, pH 7.5). After sorting, lysates were immediately heated to 65 °C for 10 min and to 95 °C for 15 min and kept at 4 °C until analysis.
Deep Sequencing-based Digital Cell Counting
Each lysate was split into 16 (stimulated samples) or 48 (unstimulated samples) PCR reactions using Phusion HF polymerase (Thermo Fisher) and the primers screen_fwd_2 (stimulated samples, 5′-ACACTCTTTCCCTACACGACGCTCTTCCGATCTCCACTTTTTCAAGTTGATAACGGAC-3′), screen_fwd_1(unstimulated samples, 5′-ACACTCTTTCCCTACACGACGCTCTTCCGATCTNNNNNNNNCTCGGTGCCACTTTTTCAAGTTG-3′), and screen_rev_1 (5′-TGACTGGAGTTCAGACGTGTGCTCTTCCGATCTTACGATACAAGGCTGTTAGAGAG-3′). PCR amplification was performed according to the manufacturer's instructions using an annealing temperature of 65 °C, an elongation time of 30 s, and 19 cycles. From each PCR reaction, 1 μl was transferred to a second PCR reaction using the same cycling conditions, but individual combinations of barcode primers described in Ref. 15. PCR products were pooled, gel-purified, precipitated as described (15) and sequenced using the MiSeq deep sequencing platform. Raw data were evaluated using custom-written software that counts the number of primer barcode combinations with which each individual library gRNA sequence was sequenced, which corresponds to the absolute number of initially sorted cells.
Screening Hit Re-validation
gRNA sequences enriched in nigericin-surviving cells were ordered as single oligonucleotides (supplemental Table 2) and cloned into pLenti-gRNA-GFP as described (14). After lentiviral packaging and infection of Nlrp3-Cas9 macrophages, cells were stimulated in the same way as during the screening and were analyzed on a BD LSR-II flow cytometer. Relative numbers of cells determined to be GFP+ PI− after nigericin stimulation were normalized to the transduction rate in the corresponding unstimulated control sample using the same virus.
KO Clone Generation
Nlrp3-Cas9 macrophages transduced with a gRNA were cloned using limiting dilution. Subsequently, growing cell clones were picked and duplicated. One of the duplicates was lysed for deep sequencing-based genotyping (15, 16). Two all-allelic Nek7 knock-out cell clones as well as two clones generated with a control gRNA transduction were expanded for functional analysis.
Functional Testing of KO Clones
Clones were plated at a density of 50,000 cells per flat-bottom 96-well. On the next day, cells were primed for 3 h using 200 ng/ml LPS (InvivoGen). Cells were stimulated using 6.5 μm nigericin (InvivoGen) for 2–6 h or 200 ng/well poly(dA:dT) (InvivoGen) or plasmid DNA transfected with Lipofectamine 2000 (Thermo Fisher) for 6 h. Unprimed cells were stimulated with 200 ng/well IVT4 5′-triphosphate-dsRNA transfected with Lipofectamine 2000, 1 μg/ml LPS, or 2 μg/ml Pam3CSK4 (InvivoGen) for 16 h. Cytotoxicity was measured by LDH release assay according to the manufacturer's instructions (Pierce), and cytokine release was assessed by ELISA (BD Biosciences).
Immunoblotting
Cells were lysed in 1× Laemmli buffer and heated to 95 °C for 10 min, and supernatants were precipitated as described (17). Blots were incubated in primary antibodies (Nlrp3, Cryo-2, Adipogen International; Nek7, EPR4900, Abcam; α-tubulin, DM1A, New England Biolabs; acetylated α-tubulin, 6-11B-1, Santa Cruz Biotechnology; IL-1β, AF-401-NA, R&D Systems; caspase-1, AG-20B-0042-C100, Adipogen International) overnight in PBS with 0.5% milk. Secondary IgG-HRP conjugates (Santa Cruz Biotechnology) were bound at room temperature for 2 h.
Results and Discussion
To devise a loss-of-function screening strategy aimed at identifying genetic factors functioning upstream of Nlrp3, we made use of a murine macrophage cell line that stably expresses murine Nlrp3. This cell line does not require a priming signal (signal 1) to activate Nlrp3 in response to a potassium efflux-inducing stimulus (signal 2). This poses an important advantage as it focuses a perturbation screen on genuine Nlrp3 activation, rendering the cellular model under study insensitive to the loss of components that are involved in Nlrp3 priming (supplemental Fig. 1A). Employing the ionophoric compound nigericin as a stimulus, pyroptosis as measured by LDH release was readily induced in these cells without additional priming (data not shown). To render these cells amenable to CRISPR-mediated screening, Cas9 was stably transduced into the cells (from now on referred to as Nlrp3-Cas9 macrophages).
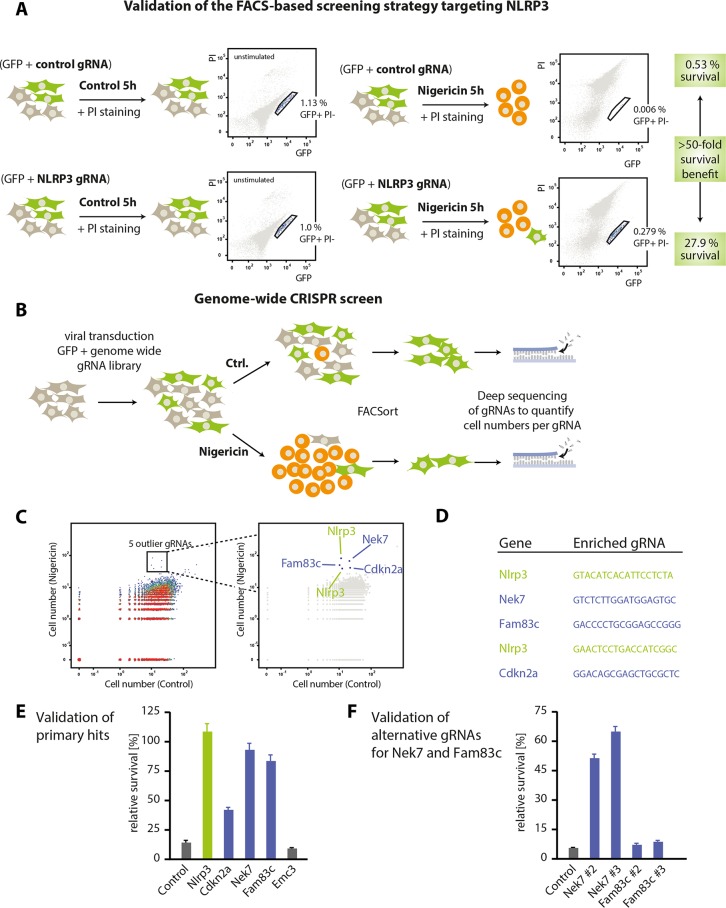
Studying different readouts of Nlrp3 activation at the single cell level, we observed that measuring loss of cytosolic GFP expression with gain in propidium iodide (PI) positivity served as the best signal for Nlrp3-induced cell death. To identify cells resistant to Nlrp3-induced cell death, we defined a stringent gate that contained GFP-positive, yet PI-negative cells (supplemental Fig. 1B). Using these settings, we transduced Nlrp3-Cas9 cells with a gRNA targeting Nlrp3 or a negative control gRNA at an MOI of 0.01. Subsequently, we stimulated cells with nigericin or left them untreated (Fig. 1A). Analyzing these cells via flow cytometry revealed that 27.9% of all cells that had been successfully transduced with a gRNA targeting Nlrp3 still remained in the stringent live gate, whereas only 0.53% of cells transduced with a control gRNA were found in this gate (Fig. 1A). Altogether, these results indicated that targeting Nlrp3 in these cells provided a 52.2-fold survival benefit (27.9%:0.53%) when analyzing cells within this live gate.
FIGURE 1.
A CRISPR loss-of-function screen identifies Nek7 as a component involved in Nlrp3 signaling. A, Nlrp3-Cas9 macrophages were transduced with a control gRNA or a gRNA targeting Nlrp3, which additionally encoded for GFP at an MOI of 0.01. Following stable transduction, macrophages were either stimulated with nigericin for 5 h or left untreated. Subsequently, cells were labeled with PI and subjected to FACS analysis. Depicted are schematic views of the transduction and stimulation modalities as well as FACS plots of a representative result. Highlighted are the frequencies of cells in the stringent live gate that contains GFP-positive and PI-negative cells. Data are representative of three independent experiments with comparable results. B, schematic view of the genome-wide screening approach that was undertaken to identify factors that confer resistance to nigericin-induced cell death. Please see “Experimental Procedures” for details. Ctrl., control. C, dot plot representation of the screening results. Each dot represents the frequency of cells carrying a specific gRNA that were found in the live gate of mock-treated cells (x axis) or of nigericin-treated cells (y axis). Highlighted are five outlier gRNAs targeting Nlrp3 (x2), Nek7, Fam83c, or Cdkn2a. D, depicted are the gRNA sequences found in the outlier population. E, Nlrp3-Cas9 macrophages were transduced with the same gRNAs identified in the screen and a control gRNA targeting Emc3. Subsequently, cells were subjected to nigericin stimulation, and cell survival was analyzed by FACS. The relative survival benefit was calculated by dividing the gated cell fraction from the stimulated condition by that from the unstimulated condition and is depicted as a mean value ± S.E. of three independent experiments. F, two alternative gRNAs targeting Nek7 and Fam83c were used to transduce Nlrp3-Cas9 macrophages. Nigericin stimulation and subsequent cell survival were analyzed as in E.
Based on this technical setup allowing us to efficiently discriminate between Nlrp3-proficient and Nlrp3-defective cells at a single cell level, we conducted a polyclonal genome-wide CRISPR screen by transducing Nlrp3-Cas9 macrophages with a pool of lentiviral particles, each encoding for both GFP and a gene-targeting gRNA (Fig. 1B). After 1 week of expansion, we stimulated half of the cells with nigericin and subsequently enriched the GFP+ PI− population of cells using FACS sorting. As the absolute cell numbers that can be sorted in a reasonable time frame are technically limited, we used a digital PCR barcoding strategy to allow counting single cells that bear an individual gRNA library construct instead of bulk PCR amplification and end point quantification of gRNA sequences as employed in other CRISPR screens (18, 19). From the unstimulated control cells, gRNA sequences targeting 15,600 annotated genes could be retrieved, covering 79.9% of the genes targeted by the library with an average of 1.71 gRNAs per gene. When plotting the numbers of sorted cells bearing each library gRNA with or without nigericin stimulation, five gRNA sequences were readily identified as specifically enriched in the population of cells being resistant to nigericin-induced cell death (Fig. 1C and supplemental Table 3). Of these, two gRNAs targeted Nlrp3 itself, demonstrating the specificity of the screening approach, whereas the three other gRNAs targeted genes that had not been associated with Nlrp3 signaling before (Nek7, Fam83c, and Cdkn2a) (Fig. 1D). We next tested these gRNAs individually for their survival benefit in Nlrp3-Cas9 macrophages using the same FACS readout as during the screening. Here, we found that both the gRNAs targeting Nek7 and Fam83c induced a similarly strong survival benefit as the Nlrp3 control gRNA, whereas a non-targeting control gRNA and a gRNA targeting the unrelated gene Emc3 failed to do so (Fig. 1E). At the same time, the survival benefit of the Cdkn2a-targeting gRNA was only partial, whereas this gRNA induced rapid proliferation in positively transduced cells (data not shown). Therefore, we only validated the target genes Nek7 and Fam83c for their functional involvement in Nlrp3 activation using two alternative gRNA sequences that were not found in the screen (Fig. 1F). Both gRNAs targeting Nek7 could reproduce the survival benefit of the Nek7 gRNA found in the initial screen. However, both alternative gRNAs targeting Fam83c failed to rescue nigericin-induced cell death, despite their target sites being located close to the original hit gRNA within the genetic locus. Altogether, this indicated that the Fam83c gRNA enriched in the screen might be active due to an off-target effect within an unknown second gene, whereas Nek7 was a bona fide hit.
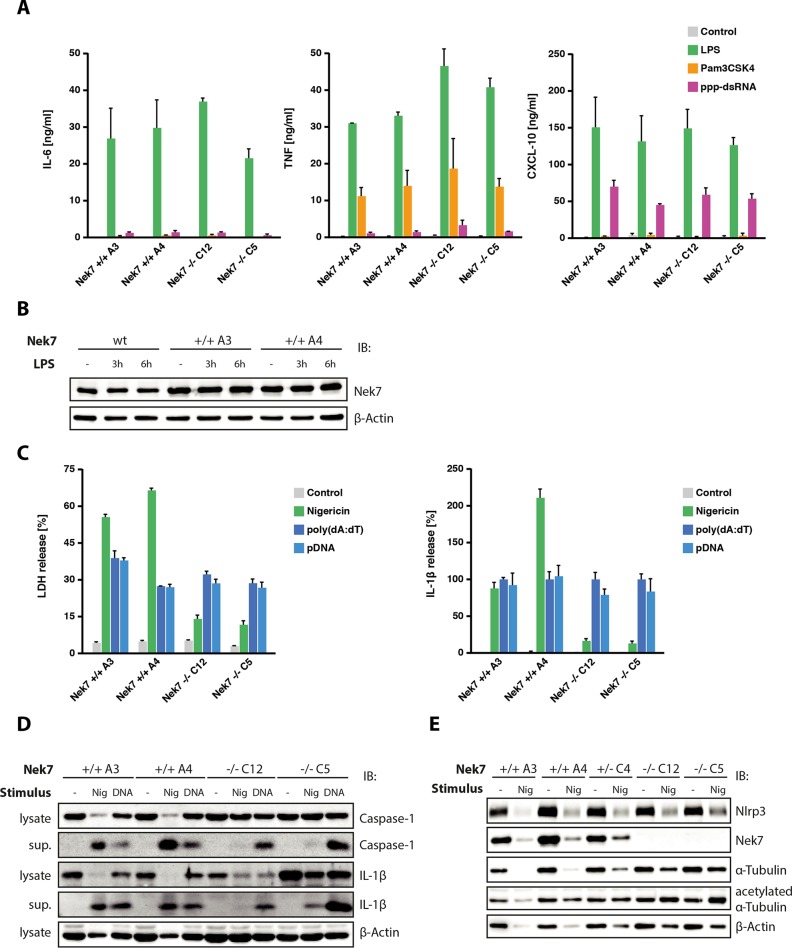
To validate the role of Nek7 in inflammasome signaling and to explore its epistatic role in Nlrp3 activation, we generated single cell clones of Nlrp3-Cas9 macrophages that had been transduced with a gRNA targeting Nek7 (15). Following this targeting approach, we obtained several cell clones bearing all-allelic frameshift mutations within the Nek7 target region. We picked two Nek7 knock-out clones and two clones with a non-targeting control gRNA for further analysis (supplemental Fig. 2). First, we wanted to explore whether Nek7 deficiency had a general impact on pro-inflammatory gene expression. To study this, we stimulated Nek7-deficient and wild-type cells with Pam3CSK4, LPS, and 5′-triphosphate dsRNA to trigger TLR2, TLR4, and RIG-I, respectively. Doing so revealed that Nek7-competent and -deficient cells responded equally well to these stimuli with regard to pro-inflammatory gene expression (Fig. 2A). On the other hand, Nek7-competent macrophages displayed no change in Nek7 protein expression upon TLR4 ligation (Fig. 2B), which is in line with mRNA expression data of primary mouse macrophages stimulated with LPS (20). Studying nigericin- and DNA-mediated inflammasome activation in these cells revealed a different picture. Although Nek7-competent cells readily displayed cell death as well as IL-1β release upon nigericin stimulation, Nek7 knock-out clones showed a greatly blunted cell death as well as IL-1β response following Nlrp3 activation. Of note, Aim2-dependent inflammasome activation was operational in both wild-type cells as well as Nek7-deficient cells. (Fig. 2C). Assessing caspase-1 cleavage as well as IL-1β maturation by immunoblot confirmed the specific role of Nek7 in Nlrp3 inflammasome activation (Fig. 2D). Both caspase-1 cleavage as well as IL-1β cleavage were readily induced by nigericin treatment in Nek7-competent cells, but severely blunted in the absence of Nek7. Again, DNA-mediated inflammasome activation was equally potent in cells of either genotype. Given the fact that Nlrp3-dependent pyroptosome formation was also blunted in the absence of Nek7 (data not shown), these results indicated that Nek7 functioned specifically upstream of Nlrp3 in inflammasome activation.
FIGURE 2.
Nek7 functions specifically upstream of the Nlrp3 inflammasome. A, Nek7-deficient Nlrp3-Cas9 macrophages or wild-type cells were stimulated with Pam3CSK4, LPS, or 5′-triphosphate dsRNA. 16 h later, supernatants were collected and analyzed for TNF, IL-6, and CXCL10 production. Data are presented as mean values ± S.E. of two independent experiments. B, wild-type macrophages or Nlrp3-Cas9 macrophages of the indicated genotype were stimulated with 200 ng/ml LPS for the durations indicated. Nek7 protein expression was analyzed by immunoblot (IB), whereas β-actin served as a loading control. Representative data from one experiment out of two independent experiments are depicted. C, macrophages of the indicated genotypes were primed with LPS and subsequently stimulated with nigericin, poly(dA:dT), or plasmid DNA (pDNA). 6 h after stimulation, supernatants were analyzed for LDH release (left panel) or IL-1β production (right panel). IL-1β data were normalized to the poly(dA:dT) condition. D, lysates or supernatants (sup.) of cells stimulated with nigericin (Nig) for 2 h or poly(dA:dT) for 6 h were analyzed for IL-1β, caspase-1, or β-actin by immunoblot. Representative data from one experiment out of two independent experiments are depicted. E, cells of the indicated genotypes were stimulated with nigericin or left untreated (note that one heterozygote Nek7 cell clone was included in the analysis). 6 h after stimulation, cells were analyzed for Nlrp3, Nek7, α-tubulin, acetylated α-tubulin, and β-actin expression by immunoblot. Representative data from one experiment out of two independent experiments are depicted.
In keeping with the fact that the cells under study were stably expressing Nlrp3, we observed considerable Nlrp3 expression in unprimed macrophages (Fig. 2E). Moreover, there was no alteration in Nlrp3 expression in relation to Nek7 gene targeting.
It has been described that microtubule acetylation triggered by mitochondrial damage is a prerequisite for Nlrp3 inflammasome activation, allowing Nlrp3 and Asc to get into close proximity via active transport in a dynein-dependent mechanism (21). Although Nek7 has been primarily characterized as a factor regulating microtubule network nucleation and spindle formation during mitosis, additional functions within the microtubule network during interphase have also been described. To this effect, it has been shown that Nek7 accelerates microtubule dynamic instability during interphase (22) and that it is able to phosphorylate α- and β-tubulin in vitro (23). Moreover, it has been demonstrated that the related family member Nek3 is able to control microtubule acetylation (24). Consequently, in light of the aforementioned study linking microtubule acetylation and Nlrp3 inflammasome signaling, we wondered whether Nek7 was involved in microtubule acetylation during Nlrp3 inflammasome activation. To test this hypothesis, we blotted α-tubulin and acetylated α-tubulin in wild-type and Nek7-deficient macrophages with or without nigericin stimulation (Fig. 2E). After doing so, we did not detect reduced α-tubulin acetylation in Nek7 knock-out cells; on the contrary, acetylated α-tubulin levels were even increased following nigericin stimulation in the absence of Nek7. Although the latter phenomenon could be due to reduced cell death in response to nigericin, these data clearly indicated that Nek7 was not acting upstream of α-tubulin acetylation in this setting.
Although the mechanism of Nek7 functioning upstream of Nlrp3 currently remains elusive, we consider the unbiased discovery of Nek7 a significant advance in our understanding of Nlrp3 inflammasome biology, as it provides the first “genetic handle” upstream of Nlrp3, yet downstream of potassium efflux. Our screening approach used a stringent setup in which Nlrp3 is steadily expressed to focus only on genetic components relaying signal 2. In light of the fact that Nek7 deficiency did not impact on pro-inflammatory gene expression and that Nek7 expression itself was not regulated as such, we consider it unlikely that Nek7 is involved in providing signal 1. Nevertheless, to fully rule out this possibility, it should be informative to study the role of Nek7 in unmodified macrophages, in which both signal 1 and signal 2 are required. At the same time, it would be interesting to explore the role of Nek7 in vivo, which would, however, require a strategy that circumvents the lethality of Nek7 deficiency in mice (25).
Given its evolutionary conservation (26), its broad expression (20), and its pivotal role in mitosis (27), we consider it unlikely that Nek7 functions as a dedicated and sufficient activator of the Nlrp3 inflammasome. Instead, we favor the hypothesis that Nek7 is involved in the co-regulation of a pathway that impacts on the ability of Nlrp3 to respond to an upstream signal or to relay this signal toward its adapter protein Asc. To this end, Nek7 could be involved in the formation or provision of a common signal that functions upstream of Nlrp3. Although this could be an indirect mechanism, e.g. a phosphorylation event mediated by Nek7, this could also directly involve Nek7 as a protein itself. On the other hand, through its function as a regulator of microtubule dynamics, Nek7 could also be required to facilitate the interaction of Nlrp3 and Asc (21). Without doubt, additional studies are required to obtain insight into the molecular mechanisms of Nek7 facilitating Nlrp3 activation. In this context, it should also be interesting to address the role of its closely related homologue Nek6, as well as its common upstream activator Nek9. Nevertheless, apart from its currently unclear mode of action, the fact that Nek7 is a kinase already makes it a potentially interesting drug target for the treatment of sterile inflammatory conditions known to involve Nlrp3.
Author Contributions
J. L. S. B. and V. H. conceived the project idea and methodology. J. L. S. B. conducted most of the experiments and also analyzed the results. T. S. E., T. S., D. C., and J. R. helped with cell culture experiments, immunoblotting studies and ELISA measurements. E. E. supervised the FACS sorting experiments. J. L. S. B. and V. H. wrote the paper. V. H. supervised the study.
Acknowledgments
We thank Drs. Bernardo Franklin and Eicke Latz (Institute of Innate Immunity, University Hospital, University of Bonn) for providing us with immortalized macrophages expressing Nlrp3.
This work was supported European Research Council Grant 647858–GENESIS and Deutsche Forschungsgemeinschaft Grant EXC1023. The authors declare that they have no conflicts of interest with the contents of this article.

This article contains supplemental Figs. 1 and 2 and supplemental Tables 1–3.
- PRR
- pattern recognition receptor
- gRNA
- guide RNA
- CRISPR
- clustered regularly interspaced short palindromic repeats
- LDH
- lactate dehydrogenase
- PI
- propidium iodide
- MOI
- multiplicity of infection.
References
- 1.Medzhitov R. (2007) Recognition of microorganisms and activation of the immune response. Nature 449, 819–826 [DOI] [PubMed] [Google Scholar]
- 2.Chen G. Y., and Nuñez G. (2010) Sterile inflammation: sensing and reacting to damage. Nat. Rev. Immunol. 10, 826–837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gross O., Thomas C. J., Guarda G., and Tschopp J. (2011) The inflammasome: an integrated view. Immunol. Rev. 243, 136–151 [DOI] [PubMed] [Google Scholar]
- 4.Kayagaki N., Stowe I. B., Lee B. L., O'Rourke K., Anderson K., Warming S., Cuellar T., Haley B., Roose-Girma M., Phung Q. T., Liu P. S., Lill J. R., Li H., Wu J., Kummerfeld S., Zhang J., Lee W. P., Snipas S. J., Salvesen G. S., Morris L. X., Fitzgerald L., Zhang Y., Bertram E. M., Goodnow C. C., and Dixit V. M. (2015) Caspase-11 cleaves gasdermin D for non-canonical inflammasome signaling. Nature 526, 666–671 [DOI] [PubMed] [Google Scholar]
- 5.Shi J., Zhao Y., Wang K., Shi X., Wang Y., Huang H., Zhuang Y., Cai T., Wang F., and Shao F. (2015) Cleavage of GSDMD by inflammatory caspases determines pyroptotic cell death. Nature 526, 660–665 [DOI] [PubMed] [Google Scholar]
- 6.Latz E., Xiao T. S., and Stutz A. (2013) Activation and regulation of the inflammasomes. Nat. Rev. Immunol. 13, 397–411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Masters S. L., Latz E., and O'Neill L. A. (2011) The inflammasome in atherosclerosis and type 2 diabetes. Sci. Transl. Med. 3, 81ps17. [DOI] [PubMed] [Google Scholar]
- 8.Bauernfeind F. G., Horvath G., Stutz A., Alnemri E. S., MacDonald K., Speert D., Fernandes-Alnemri T., Wu J., Monks B. G., Fitzgerald K. A., Hornung V., and Latz E. (2009) Cutting edge: NF-κB activating pattern recognition and cytokine receptors license NLRP3 inflammasome activation by regulating NLRP3 expression. J. Immunol. 183, 787–791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Franklin B. S., Bossaller L., De Nardo D., Ratter J. M., Stutz A., Engels G., Brenker C., Nordhoff M., Mirandola S. R., Al-Amoudi A., Mangan M. S., Zimmer S., Monks B. G., Fricke M., Schmidt R. E., Espevik T., Jones B., Jarnicki A. G., Hansbro P. M., Busto P., Marshak-Rothstein A., Hornemann S., Aguzzi A., Kastenmüller W., and Latz E. (2014) The adaptor ASC has extracellular and 'prionoid' activities that propagate inflammation. Nat. Immunol. 15, 727–737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Juliana C., Fernandes-Alnemri T., Kang S., Farias A., Qin F., and Alnemri E. S. (2012) Non-transcriptional priming and deubiquitination regulate NLRP3 inflammasome activation. J. Biol. Chem. 287, 36617–36622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schroder K., Sagulenko V., Zamoshnikova A., Richards A. A., Cridland J. A., Irvine K. M., Stacey K. J., and Sweet M. J. (2012) Acute lipopolysaccharide priming boosts inflammasome activation independently of inflammasome sensor induction. Immunobiology 217, 1325–1329 [DOI] [PubMed] [Google Scholar]
- 12.Pétrilli V., Papin S., Dostert C., Mayor A., Martinon F., and Tschopp J. (2007) Activation of the NALP3 inflammasome is triggered by low intracellular potassium concentration. Cell Death Differ. 14, 1583–1589 [DOI] [PubMed] [Google Scholar]
- 13.Muñoz-Planillo R., Kuffa P., Martínez-Colón G., Smith B. L., Rajendiran T. M., and Núñez G. (2013) K+ efflux is the common trigger of NLRP3 inflammasome activation by bacterial toxins and particulate matter. Immunity 38, 1142–1153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schmidt T., Schmid-Burgk J. L., and Hornung V. (2015) Synthesis of an arrayed sgRNA library targeting the human genome. Sci. Rep. 5, 14987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schmid-Burgk J. L., Schmidt T., Gaidt M. M., Pelka K., Latz E., Ebert T. S., and Hornung V. (2014) OutKnocker: a web tool for rapid and simple genotyping of designer nuclease edited cell lines. Genome Res. 24, 1719–1723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schmidt T., Schmid-Burgk J. L., Ebert T. S., Gaidt M. M., and Hornung V. (2016) Designer nuclease-mediated generation of knockout THP1 cells. Methods Mol. Biol. 1338, 261–272 [DOI] [PubMed] [Google Scholar]
- 17.Jakobs C., Bartok E., Kubarenko A., Bauernfeind F., and Hornung V. (2013) Immunoblotting for active caspase-1. Methods Mol. Biol. 1040, 103–115 [DOI] [PubMed] [Google Scholar]
- 18.Shalem O., Sanjana N. E., Hartenian E., Shi X., Scott D. A., Mikkelsen T. S., Heckl D., Ebert B. L., Root D. E., Doench J. G., and Zhang F. (2014) Genome-scale CRISPR-Cas9 knockout screening in human cells. Science 343, 84–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang T., Wei J. J., Sabatini D. M., and Lander E. S. (2014) Genetic screens in human cells using the CRISPR-Cas9 system. Science 343, 80–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wu C., Orozco C., Boyer J., Leglise M., Goodale J., Batalov S., Hodge C. L., Haase J., Janes J., Huss J. W. 3rd, and Su A. I. (2009) BioGPS: an extensible and customizable portal for querying and organizing gene annotation resources. Genome Biol. 10, R130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Misawa T., Takahama M., Kozaki T., Lee H., Zou J., Saitoh T., and Akira S. (2013) Microtubule-driven spatial arrangement of mitochondria promotes activation of the NLRP3 inflammasome. Nat. Immunol. 14, 454–460 [DOI] [PubMed] [Google Scholar]
- 22.Cohen S., Aizer A., Shav-Tal Y., Yanai A., and Motro B. (2013) Nek7 kinase accelerates microtubule dynamic instability. Biochim. Biophys. Acta 1833, 1104–1113 [DOI] [PubMed] [Google Scholar]
- 23.O'Regan L., and Fry A. M. (2009) The Nek6 and Nek7 protein kinases are required for robust mitotic spindle formation and cytokinesis. Mol. Cell. Biol. 29, 3975–3990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chang J., Baloh R. H., and Milbrandt J. (2009) The NIMA-family kinase Nek3 regulates microtubule acetylation in neurons. J. Cell Sci. 122, 2274–2282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Salem H., Rachmin I., Yissachar N., Cohen S., Amiel A., Haffner R., Lavi L., and Motro B. (2010) Nek7 kinase targeting leads to early mortality, cytokinesis disturbance and polyploidy. Oncogene 29, 4046–4057 [DOI] [PubMed] [Google Scholar]
- 26.Quarmby L. M., and Mahjoub M. R. (2005) Caught Nek-ing: cilia and centrioles. J. Cell Sci. 118, 5161–5169 [DOI] [PubMed] [Google Scholar]
- 27.Fry A. M., O'Regan L., Sabir S. R., and Bayliss R. (2012) Cell cycle regulation by the NEK family of protein kinases. J. Cell Sci. 125, 4423–4433 [DOI] [PMC free article] [PubMed] [Google Scholar]