Background: BMI1 silences the expression of genes located at the facultative heterochromatin.
Results: BMI1 is abundant at repetitive genomic regions, including the pericentromeric heterochromatin (PCH), where it is required for compaction and silencing.
Conclusion: BMI1 is essential for PCH formation.
Significance: BMI1 function at PCH is important to understand how BMI1 regulates genomic stability.
Keywords: BRCA1, heterochromatin, lamin, polycomb, ubiquitylation (ubiquitination), BMI1, histone H2A
Abstract
The polycomb repressive complex 1 (PRC1), containing the core BMI1 and RING1A/B proteins, mono-ubiquitinylates histone H2A (H2Aub) and is associated with silenced developmental genes at facultative heterochromatin. It is, however, assumed that the PRC1 is excluded from constitutive heterochromatin in somatic cells based on work performed on mouse embryonic stem cells and oocytes. We show here that BMI1 is required for constitutive heterochromatin formation and silencing in human and mouse somatic cells. BMI1 was highly enriched at intergenic and pericentric heterochromatin, co-immunoprecipitated with the architectural heterochromatin proteins HP1, DEK1, and ATRx, and was required for their localization. In contrast, BRCA1 localization was BMI1-independent and partially redundant with that of BMI1 for H2Aub deposition, constitutive heterochromatin formation, and silencing. These observations suggest a dynamic and developmentally regulated model of PRC1 occupancy at constitutive heterochromatin, and where BMI1 function in somatic cells is to stabilize the repetitive genome.
Introduction
Chromosomes are structurally organized in distinct subcompartments as determined by the local DNA sequence and chromatin organization. Euchromatin defines “relaxed” chromatin regions containing actively transcribed genes. In contrast, heterochromatin defines “compacted” chromatin regions containing tissue-specific and developmental genes (the facultative heterochromatin) or gene-poor regions (the constitutive heterochromatin) (1). The constitutive heterochromatin is found at the center (centromere) and ends (telomeres) of chromosomes and is mostly constituted of repetitive DNA sequences (1). Numerous (about 10,000) repetitive A/T-rich DNA elements of 231 bp are also found in the pericentromeric heterochromatin (PCH)4 of mouse chromosomes. Because constitutive heterochromatin regions contained repetitive DNA sequences, the maintenance of chromatin compaction is essential to preserve genomic stability (2). During mitosis, repetitive elements can recombine, resulting in non-homologous recombination between different chromosomes or different regions of paired chromosomes and thus chromosomes deletion, translocation, and fusion (3). Repetitive DNA sequences can be also transcribed, resulting in aberrant non-coding RNA. Stabilization of telomeres and centromers is also essential for chromosome end capping and kinetochore attachment during mitosis (4). Finally, about 40% of the mammalian genome is constituted of “parasitic” retro-element located in intergenic regions of chromosomes. Active repression of these elements is important to maintain genomic stability because some of these can self-replicate and randomly integrate the genome (5, 6).
Nucleosomes are the basic building unit of chromatin and are constituted of a 147-bp DNA wrapped against a histone octamer containing two molecules of each of the four histones H2A, H2B, H3, and H4 (the nucleosome core particle) (1). Addition of linker histones, such as histone H1, increases the amount of associated DNA by 20 bp to elicit higher levels of chromatin compaction and high order chromatin structure. The chromatin is also attached at multiple points to the nuclear envelope and spatial organization of the chromatin in the nucleus is important for the regulation of gene transcription (7, 8). Post-translational modifications of histones, such as methylation, acetylation, and ubiquitylation can modify chromatin compaction and stability. For example, silent or compact chromatin is associated with trimethylation of histone H3 at lysine 9 (H3K9me3) or 27 (H3K27me3), whereas open chromatin is associated with histone H3 trimethylation at lysine 4 (H3K4me3) or acetylation at lysine 9 (H3K9ac) (9). Acetylation brings in a negative charge, acting to neutralize the positive charge on histones and decreases the interaction of the N termini of histones with the negatively charged phosphate groups of DNA. In pathological conditions, histones hyper-acetylation can result in chromatin and chromosomes de-condensation (10).
A core of proteins is involved in establishment and maintenance of constitutive heterochromatin. Most of these proteins are conserved in Drosophila and were identified as modifiers of position effect variegation (1). In mammals, the buildup of these proteins to heterochromatic DNA follows a relatively well characterized sequence where zinc finger proteins recognize and bind repetitive DNA sequences. This is followed by enrichment for Histones H1 and H2a/z, accumulation of Hmga1/2, attachment of KAP1/Trim28 (a SUMO E3 ligase), and of the ATP-dependent chromatin remodeler ATRx, deacetylation of histones by HDAC2, and trimethylation of histone H3 at lysine 9 by SUV39h1 and SUV39h2. Association of SUMOylated HP1a and HP1b to a non-coding RNA results in increased binding affinity for H3K9me3 and this is further enhanced and stabilized by the suppressor of variegation protein DEK1 (11, 12). The DEK1-HP1-SUV39 complex then propagates the H3K9me3 marks on the chromatin, resulting in heterochromatin formation (13–17). Interestingly, ATRx localizes at both telomeric and PCH, and germline mutations in ATRx are associated with the α-thalassemia with mental retardation X-linked syndrome (18–20). Surprisingly, it was showed that the BRCA1 protein, which possesses histone H2A monoubiquitin ligase activity when in complex with BARD1, is also enriched at PCH and required for H2A ubiquitinylation, heterochromatin compaction, and silencing (21–23). This novel BRCA1 function was proposed to explain the severe genomic instability phenotype of BRCA1-deficient cells (22).
Polycomb group (PcG) proteins form large multimeric complexes involved in gene silencing through modifications of chromatin organization (24). They are classically subdivided into two groups, namely polycomb repressive complex 1 (PRC1) and PRC2 (25). Histone modifications induced by the PRC2 complex (which includes EZH2, EED, and SUV12) and the PRC1 complex (which includes BMI1, RING1A, and RING1B/RNF2) allows stable silencing of gene expression in euchromatin and facultative heterochromatin (26–28). Notably, previous recruitment models of PcG proteins through sequential histone modifications have been revised following that PRC1-variants could operate independently, and even upstream of PRC2 (29, 30). The PRC2 contains histone H3 trimethylase activity at lysine 27 (H3K27me3), whereas the PRC1 contains histone H2A monoubiquitin ligase activity at lysine 119 (H2Aub) (26–28). A number of observations have implicated these proto-oncogenes in human cancers (31–36). At the opposite, Bmi1-deficient mice display neurological abnormalities, postnatal depletion of stem cells, increased reactive oxygen species, reduced lifespan, and premature aging phenotypes (37–40). Likewise, primary human and mouse cells deficient for BMI1 undergo rapid senescence, in part through activation of the tumor suppressor INK4A locus (39–41). BMI1 was also implicated in DNA damage response and repair, and maintenance of genomic stability (42–45).
Although PRC1 proteins have not been directly implicated in constitutive heterochromatin formation or maintenance, at least some line of evidences support this possibility: 1) immuno-gold localization of BMI1 by electron microscopy in U-2 OS cells revealed high enrichment in electron-dense heterochromatin; 2) BMI1 immunolocalization was found at PCH in transformed human cell lines (46–49). However, based on work performed on mouse embryonic stem cells and oocytes, it is generally assumed that PRC1 proteins are excluded from PCH in normal mammalian somatic cells (30, 50–52). In contrast with this model, we found that BMI1 is abundant at constitutive heterochromatin in mouse and human somatic cells, and required for heterochromatin formation/maintenance and silencing. In Bmi1-null mice, cortical neurons showed loss of heterochromatin compaction and activation of intergenic retro-elements and satellite repeats. Consistently, Bmi1 co-localized with H3K9me3 and was highly enriched at PCH in mouse neurons. BMI1 was also enriched at constitutive heterochromatin, including PCH, in normal human neural precursors. Furthermore, BMI1 co-purified with architectural heterochromatin proteins and with histone H3K9me3. BMI1 localization and H2Aub deposition at constitutive heterochromatin were EZH2 and H3K27me3 independent. In both transformed and normal primary somatic cells, BMI1 inactivation resulted in loss of heterochromatin and alteration in the architecture of the nuclear envelope. Notably, BRCA1 localization was unaffected upon BMI1 deficiency, and both proteins showed partial functional redundancy for H2A ubiquitinylation, heterochromatin formation, and silencing. These findings reveal an essential function for BMI1 in constitutive heterochromatin formation and silencing in mammalian somatic cells.
Experimental Procedures
Animals
Mice were used in accordance with the Animal Care Committee of the Maisonneuve-Rosemont Hospital Research Center (approval ID numbers 2009-40, 2009-42, and 2011-23).
Neuronal Cultures
Embryonic day 18.5 cortices were dissected in oxygenated Hanks' balanced salt solution. Following meninges removal, cortices were cut to ∼1 mm3 pieces, and incubated at 37 °C for 15 min in 2 ml of TrypleEx solution (Invitrogen). Afterward, enzymatic solution was discarded, and cortex pieces dissociated in Hanks' balanced salt solution with a 1-ml tip (10 times up and down). After dissociation, cells were plated at 1.5 × 105 cells/well on poly-l-lysine-coated 6-well plates or 8-well cultures slides (BD Biosciences). Cells were maintained in normal medium composed of Neurobasal-A medium (Invitrogen), Glutamax-I (Gibco), gentamycin (50 μg/ml; Gibco), B27 supplement (Gibco), NGF (50 ng/ml; Invitrogen), and BDNF (0.5 ng/ml; Invitrogen).
Chromatin Immunoprecipitation (ChIP) Assay
ChIP was performed using the ChIP Assay kit (Upstate). Cells were homogenized at room temperature according to the manufacturer's protocol and sonicated on ice for 10 s at 30% amplitude to shear the chromatin (Branson Digital Sonifier 450, Crystal Electronics, On. Canada). Sonicated materials were immunoprecipitated using 2 μg of mouse anti-BMI1, mouse anti-H2AK119ub clone E6C5, mouse anti-RING1B, and mouse anti-HP1 (Millipore), rabbit anti-H3K9me3, and rabbit anti-H3K27me3 (Abcam), rabbit anti-BRCA1 (Santa Cruz Biotechnology), and rabbit anti-mouse IgG (Upstate) antibodies. Fragments were then amplified by real-time PCR in triplicates. Human primers sets used were as described in Ref. 22. ChIP-quantitative PCR (qPCR) data were analyzed according to the Percent Input method. First, the raw Ct of the diluted 1% input fraction is adjusted by subtracting 6.64 cycles (i.e. log2 of the dilution factor 100). Subsequently, the percent input of each immunoprecipitation (IP) fraction is calculated according to this equation: 100 × 2(Adjusted Input Ct−Ct(IP).
Real-time RT-PCR
Mouse cortices or human cells were diced and RNA was isolated using TRIzol reagent (Invitrogen). Reverse transcription (RT) was performed using 1 μg of total RNA and the Moloney murine leukemia virus reverse transcriptase (Invitrogen). Real-time PCR was carried in triplicates using Platinum SYBR Green Supermix (Invitrogen) and Real-time PCR apparatus (ABI Prism 7002).
Micrococcal Nuclease and DNase Assays
One million (106) cells were harvested at the log phase growth and used in either nuclease sensitivity assay. Cells were permeabilized (0.02% l-α-lysolecithin, 150 mm sucrose, 35 mm HEPES, 5 mm KH2PO4, 5 mm MgCl2, 0.5 mm CaCl2) on ice for 90 s, and than washed in ice-cold PBS. The cell pellet was resuspended in nuclease buffer (150 mm sucrose, 50 mm Tris-HCl (pH 7.5), 50 mm NaCl, 2 mm CaCl2) on ice, and nucleases were added. Digestions were performed at 24 °C. Reactions were stopped by adding digestion stop buffer (20 mm Tris-Cl (pH 7.4), 0.2 m NaCl, 10 mm EDTA, 2% SDS) and 0.1 mg/ml of RNase A for 30 min at 37 °C. DNA was extracted by phenol/chloroform and visualized on 0.8% native agarose gel/ethidium bromide.
Plasmid Constructs and Viruses
Sequence-specific oligonucleotides stretch shRNA designed to target the BMI-1 ORF (accession number BC011652) were synthesized. Oligo#1 (nt 1061–1081) 5′-CCTAATACTTTCCAGATTGAT-3′, and oligoScramble (nt 573–591) 5′-GGTACTTCATTGATGCCAC-3′ were used in this study. These sequences are followed by the loop sequence (TTCAAGAGA) and finally the reverse complements of the targeting sequences. The double stranded shRNA sequences were cloned downstream of the H1P promoter of the H1P-UbqC-HygroEGFP plasmid using Age1, SmaI, and XbaI cloning sites. The shRNA-expressing lentiviral plasmids were cotransfected with plasmids pCMVdR8.9 and pHCMV-G into 293FT packaging cells using Lipofectamine (Invitrogen) according to the manufacturer's instructions. Viral containing media were collected, filtered, and concentrated by ultracentrifugation. Viral titers were measured by serial dilution on 293T cells followed by microscopic analysis 48 h later. For viral transduction, lentiviral vectors were added to dissociated cells prior to plating. Hygromycin selection (150 μg/ml) was added 48 h later. shBRCA1 constructs (MISSION shRNA) were from Sigma, and siRING1B (FlexiTube siRNA) from Qiagen. The RNAi-resistant BMI1-Myc construct (BMI1myc−R) was generated by synthesis (GenScript) and where the nucleotide sequence of the human BMI1 cDNA (5′-CCTAATACTTTCCAGATTGAT-3′) was changed to (5-CCCAACACATTTCAAATAGAC-3), thus preserving the original amino acid sequence of BMI1.
Proteomics
293T cells were transfected with the EFv-CMV-GFP (GFP-293T) or EFv-BMI1-Myc-CMV-GFP (Myc-293T) plasmids. Protein extracts were subjected to immunoprecipitation using an anti-Myc antibody. Immunoprecipitates were resolved by SDS-PAGE and LC-MS analysis was performed.
Fixation, Sectioning, and Immunolabeling
Tissues were fixed in 10% buffered formalin and embedded in paraffin according to standard protocols. 5 to 7-μm thick sections were mounted on Super-Frost glass slides (Fisher Scientific) and processed for immunohistochemistry staining. Formalin-fixed paraffin-embedded slices were analyzed by using the Vectastain® ABC kit (Vector) according to the manufacturer's instructions. Peroxidase substrate DAB (brown) was used (Sigma). Observations were made under a fluorescence microscope (Leica DMRE, Leica Microsystems) and images were captured with a digital camera (Retiga EX; QIMAGING; with OpenLab, version 3.1.1 software; Open-Lab, Canada). Antibodies used in this study were mouse anti-BMI1 and anti-HP1 (Millipore), and rabbit anti-H3K9Ac and anti-H3K9me3 (Abcam). Secondary antibodies used were FITC-conjugated donkey anti-mouse and rhodamine-conjugated donkey anti-rabbit (Chemicon).
Immunoprecipitation and Western Blot
For BMI1/Myc immunoprecipitation experiments, 293T cells were transfected with EFv-/CMV-GFP or EFv-BMI1Myc/CMV-GFP plasmids using Lipofectamine according to the manufacturer's instructions. Whole cell extracts were collected in IP buffer (100 mm Tris-HCl, pH 7.5, 150 mm NaCl, 0.1% Tween 20, protease inhibitors Complete (Roche Applied Science)). Following the determination of protein concentration, lysates were subjected to immunoaffinity purification. Briefly, protein extracts (4 mg) were incubated with continuous rotation for 3 h at 4 °C with 50 μl of affinity matrix carrying mouse monoclonal anti-c-Myc IgG (clone 9E10; Covance). The matrix was washed four times with the wash buffer (50 mm Tris-HCl, pH 7.5, 150 mm NaCl, 0.1% Tween 20, protease inhibitors Complete). The bound proteins were eluted by treating the beads twice with 1 bead volume (50 μl) of c-Myc peptide solution (Covance) (400 μg/ml in 20 mm Tris-HCl, pH 7.5, 150 mm NaCl) for 15 min and used in Western blot experiments. Detection and identification of immunoprecipitated proteins were performed by Western blot and LC-MS/MS (liquid chromatography-tandem mass spectrometry). For LC-MS/MS analysis, proteins were revealed in silver-stained gels, according to standard protocol. Protein band cutting, trypsin-based in-gel protein digestions, and subsequent LCMS/MS procedures were performed in the Innovation Centre at Genome Quebec.
Primer Sequences
All primer sequences used in this study are shown in the supplemental Experimental Procedures.
Statistical Analysis
Statistical differences were analyzed using Student's t test for unpaired samples. Two-way analysis of variance test was used for multiple comparisons with one control group. In all cases, the criterion for significance (p value) was set as mentioned in the figures.
Results
Bmi1 Is Required for Constitutive Heterochromatin Formation and Silencing in Mouse Cortical Neurons
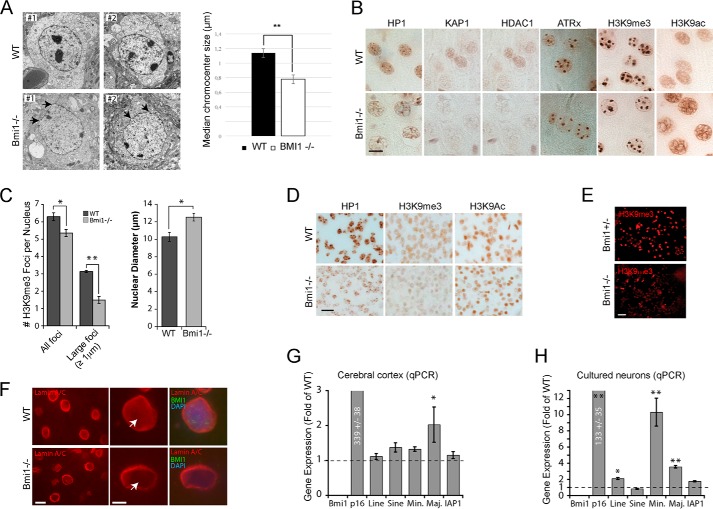
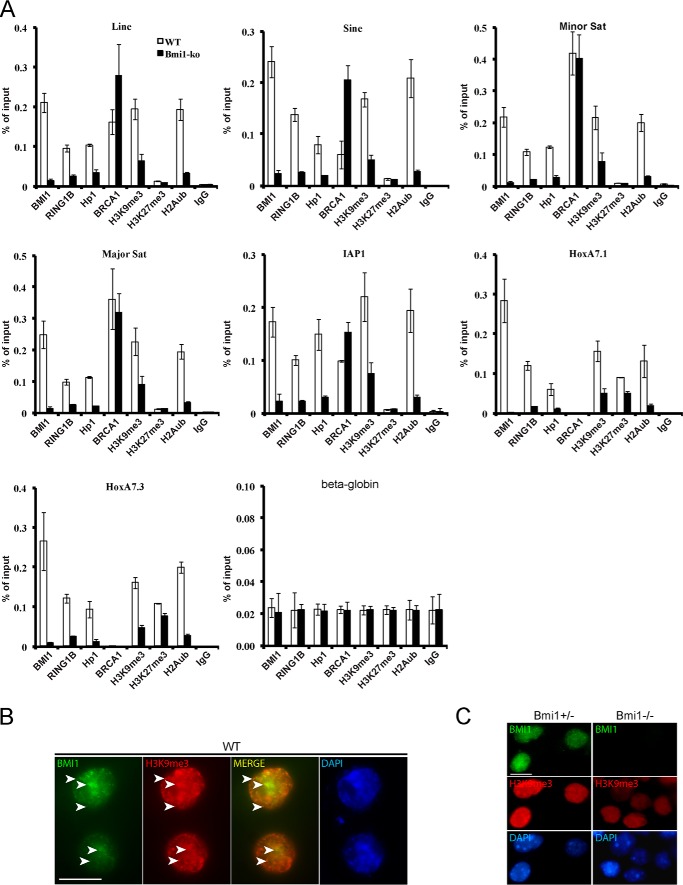
We performed transmission electron microscopy on cortical slices from WT and Bmi1−/− mice at postnatal day 30 (P30). Notably, electron-dense chromocenters were smaller and the nuclear envelope was generally irregular in Bmi1−/− neurons (Fig. 1A). By immunohistochemistry on cortical sections and using antibodies against H3K9me3 and H3K9ac, a mark of open chromatin, we observed reduced H3K9me3 labeling in Bmi1−/− neurons together with increased H3K9ac labeling (Fig. 1B). Immunoreactivity for HP1, KAP1, HDAC1, and ATRx was also reduced in Bmi1−/− neurons, suggesting heterochromatin anomalies (Fig. 1B). Quantitative analysis revealed that the number of H3K9me3-positive chromocenters was reduced in Bmi1−/− neurons, whereas the nuclear diameter of the neuron was increased (Fig. 1C). Because postnatal neurodegeneration may account for the observed chromatin anomalies, we analyzed cortical sections from WT and Bmi1−/− embryos at e18.5. We found that H3K9me3 and HP1 staining were reduced in Bmi1−/− neurons, whereas that of H3K9ac was unaffected, suggesting that histone hyperacetylation is secondary to defective heterochromatinization (Fig. 1D). Likewise, cultured cortical neurons from Bmi1−/− embryos showed reduced H3K9me3 labeling when compared with Bmi1+/− littermates (Fig. 1E). By immunofluorescence (IF) on P30 brain sections, we observed reduced immunolabeling for Lamin A/C at the neuron nuclei center of Bmi1−/− when compared with WT, suggesting anomalies in the nuclear envelope (Fig. 1F). Deficiency in constitutive heterochromatin formation can affect repeat-DNA sequences expression. By qRT-PCR analyses, we found increased expression of Major pericentromeric repeats in Bmi1−/− mouse cortices and of intergenic LINE elements and major and minor pericentromeric repeats in Bmi1−/− cultured e18.5 neurons, when compared with WT (Fig. 1, G and H). To test if Bmi1 was enriched at PCH in mouse neurons, we performed ChIP-qPCR on cultured WT and Bmi1−/− e18.5 neurons after 7 days in vitro. We found that Bmi1 specifically accumulated at all repeat-DNA sequences and at the Bmi1-target gene Hoxa7 (Fig. 2A). Notably, whereas enrichment for H3K27me3 was observed at Hoxa7 in WT neurons (and slightly reduced in Bmi1−/− neurons), H3K27me3 enrichment at repeat-DNA was negligible in both conditions (Fig. 2A). Bmi1 deficiency in mouse neurons also resulted in depletion of RING1B, HP1, H3K9me3, and H2Aub at repeat-DNA sequences, whereas accumulation of BRCA1 was unaffected or increased (Fig. 2A). Bmi1 co-localization with H3K9me3 in mouse cortical neurons was confirmed by IF on brain sections at P30 (Fig. 2B). Bmi1 antibody specificity was further validated by IF on cultured e18.5 cortical neurons (Fig. 2C). These results revealed that Bmi1 is required for heterochromatin formation and repeat-DNA silencing in mouse cortical neurons and enriched at PCH together with RING1B and BRCA1.
FIGURE 1.
Bmi1-deficient mouse cortical neurons present heterochromatin anomalies. A, transmission electron microscopy analysis of cortical neurons in P30 WT and Bmi1−/− mice. Note the reduction in electron dense chromocenters and the anomalies in nuclear membrane architecture in Bmi1−/− neurons (arrows). B, paraffin-embedded brain sections from P30 WT and Bmi1−/− mice were analyzed by immunohistochemistry. Labeled cells are neurons located in the upper cortical layers of the cerebral cortex. Scale bar, 10 μm. C, quantification of the total number of H3K9me3-positive chromocenter and number of large H3K9me3-positive chromocenters. Note that the nuclear diameter of the neuron is increased in Bmi1−/− mice. Where n = 3 brains for each genotype. *, p < 0.05, **, p < 0.01. D, paraffin-embedded brain sections from e18.5 WT and Bmi1−/− embryos were analyzed as in B. E and F, cultured embryonic cortical neurons (E) and P30 cortical sections (F) were analyzed by immunofluorescence, revealing reduced H3K9me3 and Lamin A/C labeling (arrows) in Bmi1−/− neurons. Scale bars: 40 (E), 10 (F), and 5 μm (F′). G, whole cortices or (H) e18.5 neurons from WT and Bmi1−/− mice were analyzed by qPCR for satellite repeats and intergenic retroelements expression. P16Ink4a was used as positive control. Note the up-regulation of minor and major satellite repeats in Bmi1−/− neurons. Where n = 3 independent samples for each genotype. *, p < 0.05; **, p < 0.01.
FIGURE 2.
Bmi1 is required for H2Aub deposition and accumulates at repeat-DNA sequences in mouse cortical neurons. A, WT and Bmi1−/− neurons were analyzed by ChIP for proteins enrichment at satellite repeats, intergenic retroelements, and HoxA7 (positive control). Note the accumulation of Bmi1 and Ring1b at all repeat-DNA sequences, including major and minor satellite repeats. Although HP1 accumulation and H2Aub and H3K9me3 deposition were reduced in Bmi1−/− neurons at all tested loci, BRCA1 accumulation was either unaffected (Minor and Major satellites) or increased (Line, Sine, and IAP). Note the near absence of BRCA1 accumulation at HoxA7.1 and HoxA7.3 in both WT and Bmi1−/− neurons. B, immunofluorescence analysis showing Bmi1 co-localization with H3K9me3 in WT mouse cortical neurons at P30 (arrowheads). Scale bar, 10 μm. C, immunofluorescence analysis showing loss of Bmi1 signal and reduced H3K9me3 labeling in cultured e18.5 Bmi1−/− mouse cortical neurons when compared with Bmi1+/− neurons. Scale bar, 10 μm.
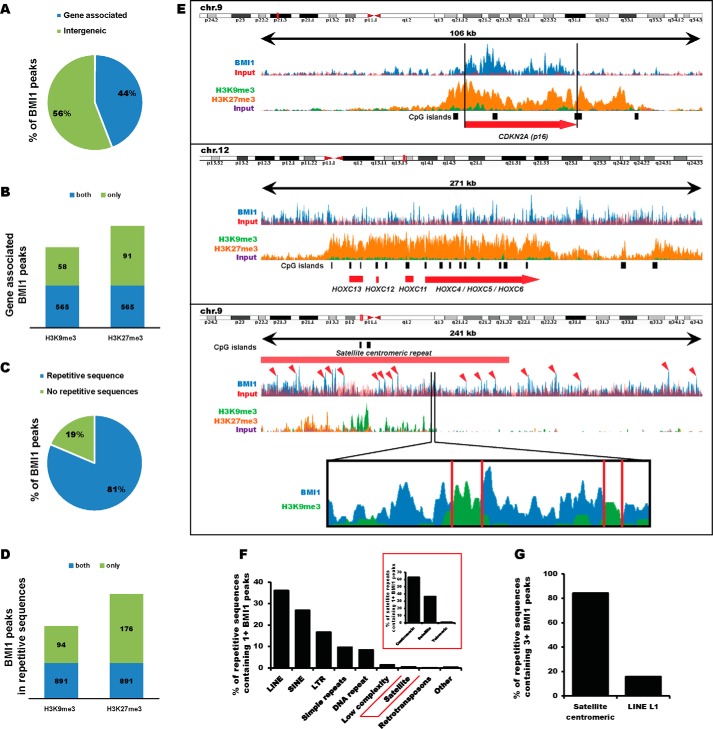
BMI1 Is Highly Enriched at Repetitive Sequences in Human Neural Precursors
To investigate BMI1 distribution on the chromatin genome-wide, we took advantage of publicly available BMI1 ChIP-Seq raw data on normal human neural precursors (53). Using MACS statistical peak calling, we identified 21,525 BMI1 binding sites. The majority of the peaks (56%) were located at intergenic regions, which are highly enriched for constitutive heterochromatin (Fig. 3A). As expected, we observed BMI1 enrichment at the canonical BMI1 targets CDKN2A (p16INK4A) and HOXC locus (Fig. 3E, top). Among 9,471 gene-associated BMI1 peaks, 714 peaks were also enriched by at least 2-fold for either H3K9me3 or H3K27me3. Notably, 565 were marked for both H3K9me3 and H3K27me3 (Fig. 3B). We further annotated BMI1 peaks surrounding DNA repetitive sequences using a repeat masker. Surprisingly, 81% of the total pool of BMI1 was located at repetitive sequences (Fig. 3C) with 985 peaks co-enriched with H3K9me3 and 1,067 peaks co-enriched with H3K27me3 (Fig. 3D). If considering repetitive sequences containing at least one BMI1 peak, LINE, SINE, and LTR were the most represented families of repeats. Among satellite repeats, BMI1 was mainly enriched at PCH regions (Fig. 3F, inset). A closer look at PCH regions on chromosome 9 revealed a “pocket-like” disposition of BMI1 peaks surrounding the H3K9me3 deposition (Fig. 3E). A similar pattern of BMI1 peak distribution was also found at PCH regions in human chromosomes 1–10. Notably, repetitive sequences containing 3 or more BMI1 peaks were largely represented (80%) in centromeric satellite repeats (Fig. 3G). In contrast, although 20% of LINE L1 repeats contained 3 or more BMI1 peaks, no other LINE subfamilies were highly enriched for BMI1 (Fig. 3G). We concluded that in human neural precursors, BMI1 is enriched at constitutive and facultative heterochromatin with prevalence for repetitive sequences.
FIGURE 3.
BMI1 is enriched at repetitive sequences in human neural progenitor cells. A, proportion of BMI1 peaks associated to a gene (surrounding or upstream 2 kb of a gene). Total number of peaks: 21,525. B, proportion of gene-associated BMI1 peaks co-enriched for H3K9me3 or H3K27me3. C, proportion of BMI1 peaks surrounding a repetitive sequence. D, proportion of repeat-associated BMI1 peaks co-enriched for H3K9me3 or H3K27me3. E, example of canonical BMI1 target genes (CDKN2A and HOXC) and the pericentromeric region of human chromosome 9. Red arrowheads indicate BMI1 peaks. Top: physical map on the chromosome. F, families of repeat containing at least one BMI1 peak. G, families of repeat containing three or more BMI1 peak. BMI1 peaks determined by MACS peak calling; p value <0.05. At least 2-fold enrichment for H3K9me3 and H3K27me3 were considered.
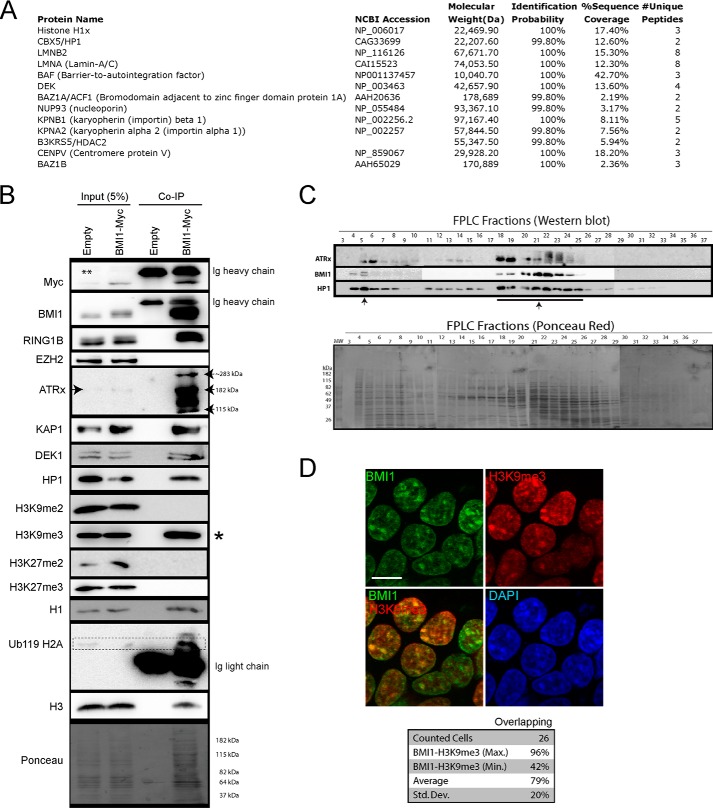
BMI1 Co-purifies with Architectural Heterochromatin Proteins
To identify new BMI1 partner proteins, we infected 293T cells with a lentivirus expressing a Myc-tagged BMI1 fusion protein and GFP (EFv-BMI1Myc/CMV-GFP) or a control virus only expressing GFP. After IP with an anti-Myc antibody, samples were separated on a one-dimensional gel and sequenced by LC-MS (Fig. 4A). We identified several unique peptides in BMI1Myc samples corresponding to proteins involved in heterochromatin organization, including histone H1x, HP1a (also called CBX5), LAMIN A/C and LAMIN B, DEK (also called DEK1), and CENP-V (Fig. 4A) (7, 11, 54, 55). We also identified 2 members of the ISWI family, BAZ1a (also called ACF1) and BAZ1b, which can promote heterochromatin formation and transcription silencing by generating spaced nucleosome arrays (56). To validate some of these findings, we performed IP experiments on control and BMI1Myc virus-infected cells. As expected, we observed that RING1B, but not EZH2, co-precipitated with BMI1 (Fig. 4B). Co-precipitation of ATRx, KAP1, DEK1, and HP1 with BMI1 was also observed, with a notable enrichment of ATRx when compared with input (Fig. 4B), and where the full-length (∼280 kDa) and truncated (∼180 kDa) ATRx isoforms were present, together with a lower molecular mass isoform of ∼115 kDa. Notably, whereas co-precipitation with histones H3K9me3, H3 (total), H1, and H2Aub was robust, co-precipitation was not observed with histones H3K9me2, H3K27me2, and H3K27me3 (Fig. 4B). We use FPLC to separate protein complexes and found that BMI1 was present in one fraction of very large molecular weight and in several other fractions of lower molecular weight all also containing HP1 and ATRx (Fig. 4C). By IF studies, we confirmed that BMI1 largely co-localized with H3K9me3 in interphase nuclei (Fig. 4D).
FIGURE 4.
BMI1 co-purifies with architectural heterochromatin proteins. A and B, 293T cells were infected with EFv/CMV-GFP or EFv-BMI1Myc/CMV-GFP viruses. Protein extracts were subjected to IP using an anti-Myc antibody, and immunoprecipitates were resolved by SDS-PAGE and analyzed either by LC-MS/MS (A) or Western blot (B). A, note the co-purification of BMI1 with several heterochromatin proteins and Lamins. B, note the preferential co-purification of BMI1 with histone H3K9me3 (*) and ATRx. The ** symbol on the panel indicates an artifact coming from partial leakage of the second sample. C, native nuclear extracts were size fractionated by FPLC and analyzed by Western blot (upper panel) and Ponceau Red staining (lower panel). Note BMI1 co-fractionation with ATRx and HP1-containg protein complexes (arrows). D, 293T cells were labeled with BMI1 and H3K9me3 antibodies, counterstained with DAPI, and analyzed by confocal microscopy. Note the co-localization of BMI1 with H3K9me3-positive chromatin domains. Scale bar, 10 μm. Quantitative confocal analysis was used to measure the proportion of overlapping signals.
BMI1 Is Required for Heterochromatin Compaction and Silencing
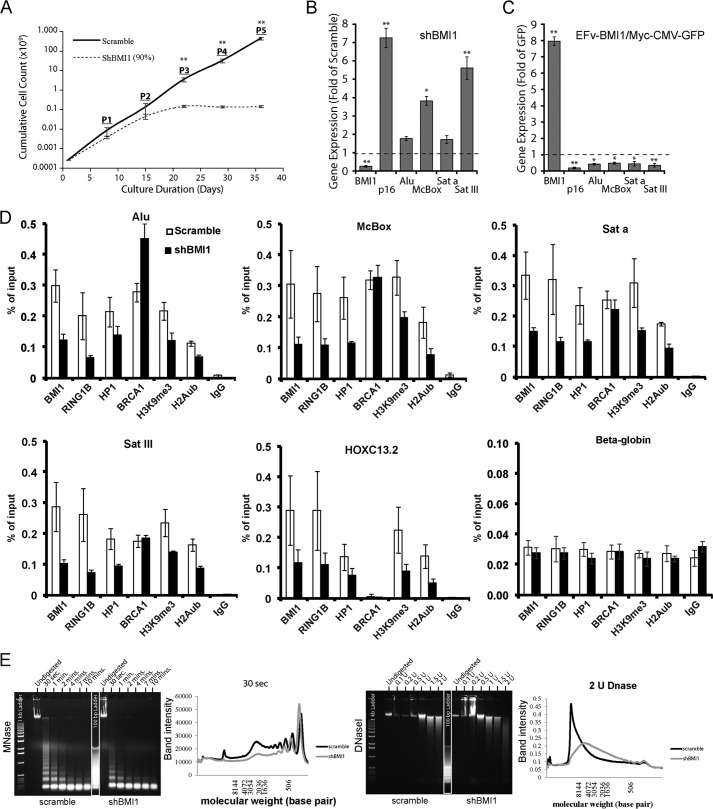
To evaluate BMI1 activity in heterochromatin silencing, we measured gene expression in loss- and gain-of-function experiments. Upon BMI1 deficiency, 293T cells showed reduced proliferation and underwent cell proliferation arrest after 3 passages (Fig. 5A). BMI1 overexpression had, however, no apparent adverse effect on cell proliferation (not shown). In BMI1 knockdown cells, expression of the canonical BMI1 target gene p16Ink4a as well as that of McBox and SATIII was increased (Fig. 5B). Conversely, BMI1 overexpression resulted in transcriptional repression of p16Ink4a and of all tested repeat-DNA sequences (Fig. 5C). By ChIP-qPCR experiments on shScramble and shBMI1-treated cells, we found that BMI1 and RING1B were highly enriched at repeat-DNA sequences and HOXC13 in control cells (Fig. 5D). In shBMI1 cells, BMI1, RING1B, HP1, H3K9me3, and H2Aub were reduced at all chromatin regions tested (Fig. 5D). In contrast, BRCA1 enrichment at repeat-DNA sequences was independent of BMI1 function. Increased BRCA1 enrichment was even observed at ALU sequences upon BMI1 deficiency (Fig. 5D). We tested if RING1B knockdown mimicked the BMI1-deficient phenotype. Although RING1B accumulation at constitutive heterochromatin and HOXC13 was highly reduced in siRING1B-treated cells (clone #4, 80% RING1B knockdown), H2Aub reduction was only detected at HOXC13 (data not shown). Notably, RING1B knockdown had no effect on BMI1, HP1, and H3K9me3 on all tested regions. No significant effect on repeat-DNA sequence expression was observed (data not shown), revealing that RING1B knockdown is not sufficient to reproduce the BMI1-deficient heterochromatin phenotype. BMI1 enrichment at repeat-DNA sequences was also EZH2 and H3K27me3 independent (data not shown). Nuclease hypersensitivity is a common phenotype of cells deficient in heterochromatin condensation (11, 57–59). We used native chromatin extracts isolated from control and shBMI1 293T cells in MNase and DNase I experiments and found that cells knocked down for BMI1 were hypersensitive to both nucleases (Fig. 5E), thus suggesting globally reduced chromatin compaction.
FIGURE 5.
BMI1 is required for heterochromatin compaction and silencing in human cells. A–C, 293T cells were infected with shScramble or shBMI1 viruses (A and B), or with viruses expressing either GFP or the BMI1-myc fusion protein and GFP (C). B and C, gene expression was analyzed by qPCR, and where n = 3 independent cultures. D, 293T knockdown cells for BMI1 were analyzed by ChIP for protein enrichment at satellite repeats, intergenic retro-elements, and HoxC13.2 (positive control). E, 293T cells were infected with shScramble or shBMI1 viruses and treated or not with MNase (0.4 units at 24 °C for different time periods) or DNase I at the indicated concentrations for 20 min at 24 °C. Note the nuclease hypersensitivity phenotype of BMI1 knockdown cells. *, p < 0.05; **, p < 0.01.
Severe Heterochromatin and Nuclear Envelope Alterations in Human Cells Deficient for BMI1
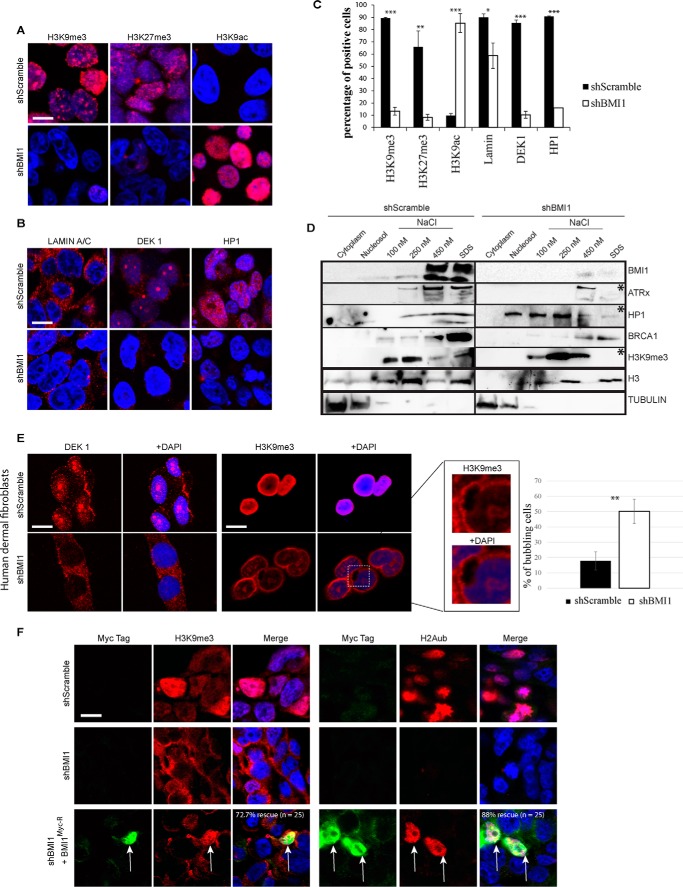
To further characterize the BMI1-deficient phenotype, we analyzed cells by confocal IF using heterochromatin and nuclear envelope markers. We found severe depletion of the H3K9me3 and H3K27me3 histone marks in BMI1-knockdown cells together with dramatic elevation of the H3K9ac mark (Fig. 6, A and C). Likewise, DEK1 and HP1 heterochromatic nuclear foci were lost upon BMI1 knockdown and positive cells for LAMIN A/C were significantly reduced (Fig. 6, B and C), suggesting perturbation of the nuclear envelope architecture.
FIGURE 6.
BMI1 knockdown cells present heterochromatin and nuclear envelope alterations. A–C, formaldehyde-fixed 293FT cells were immunolabeled and counterstained with DAPI. Scale bar, 10 μm. Positive cells were counted on 4 different images for a total of 200 cells per condition, and the percentage of positive cells was calculated accordingly. t test with two tails, where *, p ≤ 0.05; **, ≤0.01; ***, ≤0.001. Note that the apparent localization of Lamin A/C in the cytosol is the result of Triton X-100 treatment. D, 293T cells were infected with shScramble or shBMI1 viruses and the compartments of the cell were fractionated. Note the reduction (*) of ATRx, HP1, and H3K9me3 in SDS fractions of shBMI1-treated cells. E, human dermal fibroblasts were infected with shScramble or shBMI1 viruses, immunolabeled, and counterstained with DAPI. Note the reduced DEK1 and H3K9me3 labeling, and H3K9me3 localization at the nuclear periphery, in BMI1-deficient cells. Bubbling of the nuclear envelope was also observed (inset); **, p ≤ 0.01. Scale bar, 10 μm. F, human dermal fibroblasts were infected with shScramble or shBMI1 viruses, and next transfected with a plasmid encoding an RNAi-resistant BMI1 Myc-tagged construct. Note the rescue of H2Aub and H3K9me3 nuclear labeling in Myc-positive cells knockdown for BMI1 (arrows). Scale bar, 10 μm.
To test whether this correlated with alterations in the subnuclear distribution of heterochromatin proteins, we performed cellular fractionation experiments and where the SDS-soluble fraction is thought to be highly enriched for constitutive heterochromatin proteins (11, 60). In control cells, BMI1 was detected in the 450 nm NaCl and SDS fractions (Fig. 6D). ATRx, HP1, BRCA1, and H3K9me3 were also highly enriched in the 100–450 nm NaCl- and SDS-soluble nuclear fractions. Modest HP1 distribution was also found in the nucleosol fraction (Fig. 6D). In shBMI1 cells, which underwent premature cell proliferation arrest (thus explaining the overall reduced total protein loading), ATRx, HP1, and H3K9me3 were highly reduced in the SDS fraction and displaced in the other fractions. In contrast, the distribution of BRCA1 in chromatin fractions and its overall expression were unaffected upon BMI1 knockdown (Fig. 6D and data not shown). To test BMI1 function in primary human cells, human dermal fibroblasts were infected with the lentiviruses and analyzed by IF. We observed that in contrast to control cells where robust H3K9me3 labeling was widespread throughout the interphase nucleus, H3K9me3 labeling in BMI1 knockdown cells was highly reduced and present at the nuclear periphery where it did not co-localize with DAPI (Fig. 6E). Notably, co-localization of H2Aub and H3K9me3 with DAPI in shBMI1-infected cells could be rescued by an RNAi-resistant BMI1-Myc fusion protein (BMI1myc−R), thus excluding possible off-target effects (Fig. 6F). Bubbling of the nuclear envelope and loss of DEK1 nuclear labeling were also observed (Fig. 6E), revealing BMI1 requirement for constitutive heterochromatin maintenance in both transformed and primary human cells.
BMI1 and BRCA1 Display Partial Functional Redundancy in Heterochromatin Compaction
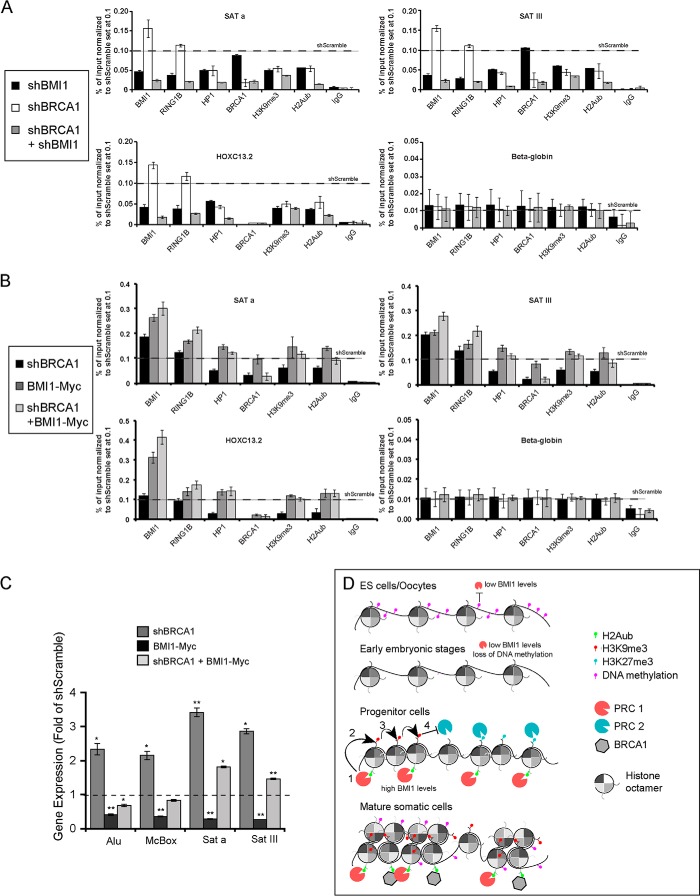
Because BMI1/RING1A/RING1B and BRCA1/BARD1 display H2A monoubiquitinylation activities and that BRCA1 enrichment and distribution at heterochromatin is not affected upon BMI1 depletion, we tested whether BMI1 and BRCA1 displayed functional redundancy. For this, we first inactivated BRCA1 to test the impact on BMI1 localization. In control cells, both BRCA1 and BMI1 were enriched at repeat-DNA sequences (data not shown). In BRCA1 knockdown cells, BRCA1, HP1, H2Aub, and H3K9me3 levels were reduced and transcription of repeat-DNA sequences was increased (data not shown), altogether confirming previous findings (22). Notably, however, BMI1 and RING1B were enriched at all tested regions upon BRCA1 knockdown (data not shown). Next, stably infected shBMI1 cells were transfected with an shBRCA1 plasmid, generating double knockdown (DKN) cells. Although H3K9me3 enrichment was reduced by 55–70% in DKN cells at all tested chromatin regions, enrichment for HP1 and H2Aub was further reduced by 80–90% (Fig. 7A), suggesting additive effects in DKN cells when compared with single BRCA1 or BMI1 knockdown cells. To test whether BMI1 could compensate for BRCA1 deficiency, we overexpressed the BMI1Myc construct. In control cells, BMI1 overexpression could not displace endogenous BRCA1 localization on the chromatin (Fig. 7B). However, chromatin accumulation of both endogenous and ectopic BMI1 proteins was highly increased at all tested regions following BRCA1 knockdown (Fig. 7B). A similar but less dramatic trend was also observed for RING1B. Most notably, whereas BMI1 overexpression could increase H2Aub and H3K9me3 deposition as well as HP1 accumulation at all tested regions in control cells, it could also rescue the corresponding heterochromatin anomalies in shBRCA1 cells (Fig. 7B). BMI1 overexpression in shBRCA1 cells also resulted in normalization of ALU, McBox, Sata, and SatIII expression (Fig. 7C), altogether suggesting functional redundancy in constitutive heterochromatin compaction and silencing between BMI1 and BRCA1.
FIGURE 7.
BRCA1 and BMI1 display redundant activities in constitutive heterochromatin formation and silencing. A, 293T cells were infected with shScramble or shBMI1 viruses. After selection with hygromycin, cells were transfected or not with an shBRCA1-encoding plasmid and analyzed by ChIP. Note the severe reduction for HP1, H3K9me3, and H2Aub at human Satellite repeats in shBMI1/shBRCA1 cells. All data where normalized to shScramble (black horizontal bars). B, 293T cells stably expressing BMI1Myc or not were transfected with shScramble or shBRCA1 plasmids and analyzed by ChIP (B) and qPCR (C). B, endogenous and exogenous BMI1 was enriched in shBRCA1-treated cells at all tested loci. BMI1 overexpression also rescued HP1, H3K9me3, and H2Aub depletion in BRCA1 knockdown cells at human Satellite repeats. All data were normalized to shScramble (black horizontal bars). C, BMI1 overexpression rescues repeat-DNA sequences expression in BRCA1 knockdown cells; *, p ≤ 0.05; **, ≤0.01. D, model depicting the dynamic and developmentally regulated PRC1 (BMI1) occupancy at PCH. In ES cells and oocytes, BMI1 is expressed at low levels and binding to PCH is prevented by DNA methylation. At pre-implantation stages, DNA methylation is erased coincidently with low levels of BMI1. During development, high BMI1 levels in progenitor cells stimulate PCR1-mediated H2Aub (1), which promotes H3K9me3 deposition (2), and propagation (3). In turn, H3K9me3 prevents PRC2-mediated H3K27me3 activity on nucleosomal histones (4). PRC2 occupancy at PCH prior to PRC1 would allow deposition of the H3K27me3 mark (right) and the bivalent histone signature. In mature somatic cells, the PRC1 and BRCA1/BARD complexes are highly enriched at PCH, leading to heterochromatin compaction and silencing. DNA methylation at PCH may occur after spreading the H3K9me3 mark.
Discussion
We showed here that Bmi1-deficient mouse neurons and BMI1 knockdown human cells displayed severe anomalies at the constitutive heterochromatin. These anomalies were accompanied by transcriptional activation of repeat-DNA sequences and correlated with robust accumulation of BMI1 at constitutive heterochromatin. Genome-wide analysis of BMI1 distribution on the chromatin further revealed predominant enrichment at repetitive DNA sequences. BMI1 co-purified with architectural heterochromatin proteins, co-localized with H3K9me3, and was required for HP1, DEK1, and ATRx localization at constitutive heterochromatin. In contrast, BRCA1 localization was BMI1-independent, and both proteins displayed partial functional redundancy for H2Aub deposition, heterochromatin formation, and silencing.
The recruitment mechanisms of PcG proteins are complex and not fully understood. In mouse ES cells and oocytes, evidences suggest that recruitment of PcG proteins at PCH is prevented by a high level of DNA methylation (30, 52). Likewise, observations of Polycomb bodies at PCH in transformed human cells are thought to occur following loss of DNA methylation (30, 47, 52). Based on this, it was proposed that PcG proteins are excluded from PCH in normal somatic cells (52). Using several methods, we demonstrated BMI1 enrichment at PCH and other repetitive elements in mouse neurons, human neural precursors, and immortalized human cells. Our cell fractionation assays further demonstrated that about 50% of the BMI1 pool was bound to the SDS-soluble chromatin fraction, which is enriched for constitutive heterochromatin. Interestingly, we also observed that: 1) BMI1 did not co-purify with EZH2 or H3K27me3; 2) EZH2 and H3K27me3 were not enriched at constitutive heterochromatin; and 3) BMI1 accumulation at constitutive heterochromatin and HOXC13 was EZH2 independent. Although apparently surprising, these results are in agreement with numerous findings showing that PRC1 recruitment can be PRC2 independent, or that the PRC1 can work upstream of the PRC2 (29, 61, 62). It is also notable that RING1B knockdown could not mimic the BMI1-deficient phenotype. More specifically, whereas H2Aub levels were reduced at HOXC13 in RING1B knockdown cells, this was not accompanied by a corresponding reduction in HP1 and H3K9me3 levels, as observed in BMI1 knockdown cells. Furthermore, there was no apparent effect on H2Aub levels at heterochromatin. This finding leaves us open with many explanations, one being functional compensation by RING1A for H2Aub deposition, as shown in other context (50, 63–65). It is also possible that in addition to promote H2Aub at silenced developmental genes and heterochromatin, BMI1 stimulates chromatin compaction and H3K9me3 loading through interactions and activities not shared by RING1A or RING1B (66).
Based on our findings and previously published work, we propose that PRC1 recruitment to PCH is highly dynamic and developmentally regulated (Fig. 7D). The highly variable DNA methylation states between ES cells/oocytes and progenitor cells/somatic cells, in combination with the distinct histone tail modifications and chromatin compactions levels, would explain the re-localization of PRC1 components to PCH in progenitor and somatic cells (67–69). Consistently, BMI1 expression levels are extremely low in human ES cells when compared with human neural progenitors and post-mitotic neurons.5 Although PRC1 proteins are excluded from PCH in ES cells and oocytes, they would start to accumulate at PCH during mid-embryonic development coincidently with H3K9me3 deposition and progressive de novo DNA methylation (69). This is supported by the ChIP-Seq data showing that BMI1 is moderately enriched at PCH when compared with HOX and p16INK4A canonical sites in human neural progenitors (Fig. 3). Interestingly, both H3K9me3 and H3K27me3 marks were also present at PCH but did not clearly overlap (Fig. 3), similarly as reported in DNA methylation-deficient mouse ES cells (30). The histone modification pattern of human neural progenitors at PCH is also distinct from that of mouse neurons where H3K27me3 is excluded (Fig. 2). This could be best explained by the robust accumulation of H3K9me3 at PCH in post-mitotic neurons, because H3K9me3 can prevent PRC2, but not PRC1 recruitment (30). Indeed, Bmi1 is highly enriched in mouse neurons at PCH when compared with canonical sites (Fig. 2). Taken together, these observations suggest a dynamic and developmentally regulated model of PcG occupancy at PCH (Fig. 7D). Because constitutive heterochromatin is intrinsically instable, we further propose that in somatic cells, the main biological function of BMI1 is to stabilize the repetitive genome by promoting chromatin compaction and silencing.
We observed that the Bmi1-null neuronal phenotype was associated with increased nuclear diameter and an irregular nuclear envelope. Human cells knockdown for BMI1 also presented anomalies in nuclear envelope architecture (Fig. 6, B–E). These anomalies are particularly interesting considering that loss of heterochromatin foci can result in disruption of the nuclear lamina (7). Perturbations of the nuclear envelope architecture is also a prominent feature of Hutchinson-Gilford Progeria cells carrying mutations in LAMIN-A and normal aging human cells (70). In most eukaryotes, constitutive heterochromatin perturbations result in genomic instability and premature aging or reduced lifespan (10, 16, 71–74). It is thus notable that Bmi1-deficient mice show reduced lifespan, genomic instability, neurodegeneration, and progeria features (38, 42–45, 75). Similar anomalies were also reported for ATRx-deficient mice (18, 20). Taken together, this raises the possibility that BMI1 requirement for constitutive heterochromatin formation and silencing could underlie the premature aging/senescence and genomic instability phenotypes observed in Bmi1-null mice and cells.
The BRCA1/BARD1 complex is required for heterochromatin formation and silencing through monoubiquitinylation of H2A at PCH, and the genomic instability phenotype of BRCA1−/− cells could be rescued by overexpression of a histone H2A protein fused to an ubiquitin moiety in C terminus (22). How BMI1 or BRCA1-mediated H2Aub deposition at repetitive DNA sequences can translate into H3K9me3 loading and heterochromatin spreading is unknown. One possibility is that H2Aub induces allosteric changes in the histone H3 lysine trimethyltransferases SUV39H1/2 to promote their activity, such as proposed for H2Bub and H3K4 methylation (76, 77). This would be consistent with previous observations that PRC1 components can interact with SUV39H1 (48). Alternatively, BMI1 may directly or indirectly regulate the transcription of H3 lysine methyltransferases or demethylases, thus operating in trans. We showed here that co-inactivation of BMI1 and BRCA1 induces more severe heterochromatin anomalies than individual BMI1 or BRCA1 deficiencies, and that BMI1 overexpression could rescue the BRCA1-deficient heterochromatin phenotype. These results suggest that BMI1 and BRCA1 are at least partially redundant for H2Aub deposition at constitutive heterochromatin, although the BRCA1/BARD1 complex targets H2A at lysines 127–129 (21). This would indicate that the commonly used anti-H2Aub antibody recognizes both the H2AK119ub and H2AK127–129ub motifs. The observation that BMI1 and BRCA1 proteins accumulation is mutually independent and that BMI1 levels are increased in BRCA1-deficient cells (and reciprocally) also suggests that both protein complexes possibly bind to very close substrates to catalyze H2A monoubiquitinylation. Structural analyses have indeed revealed a high degree of conservation between the nucleosome-binding loop of BRCA1 and the corresponding domain of RING1B (78). Notably, the reduced neuronal chromocenter number and size phenotype observed in Bmi1-null neurons is about identical to that reported for mouse cortical neurons conditionally deficient for BRCA1, thus further supporting our findings (22). Taken together, this suggests that although BMI1 and BRCA1 protein complexes target distinct lysine residues on histone H2A, the resulting biological effects on heterochromatin compaction and silencing are highly similar.
In conclusion, we demonstrated for the first time that BMI1 is highly enriched at intergenic repetitive elements and PCH of the mouse and human genomes in normal somatic cells, and required for constitutive heterochromatin formation and silencing. Because BM1 is also present at PCH in cancer cell lines and that several cancer cells were shown to be sensitive to BMI1 inhibition, this raises the possibility that BMI1 may be important to stabilize the transformed heterochromatic genome. BMI1 function at constitutive heterochromatin may be even more critical in BRCA1-deficient tumors, thus opening possibilities for the development of synthetic lethal strategies.
Author Contributions
M. A., R. H., and J. E. H. performed the experiments and analyzed the data. A. F. performed the bio-informatics analysis and wrote the corresponding section. G. B. supervised the experiments and wrote the manuscript. All authors reviewed the results and approved the final version of the manuscript.
Acknowledgments
We are grateful to S. Breault for technical assistance with electron microscopy and Dr. F. Rodier for the dermal fibroblast cell line and critical reading of the manuscript.
This work was supported in part by a grant from the Natural Science and Engineering Research Council of Canada. The authors declare that they have no conflict of interest regarding the content of this manuscript.

This article contains supplemental “Experimental Procedures”.
V. P., A. B., M. Abdouh, A. Flamier, and G. Bernier, unpublished data.
- PCH
- pericentromeric heterochromatin
- PcG
- polycomb group
- PRC
- polycomb repressive complex
- IP
- immunoprecipitation
- IF
- immunofluorescence
- qPCR
- quantitative PCR
- DKN
- double knockdown
- nt
- nucleotide.
References
- 1.Fodor B. D., Shukeir N., Reuter G., and Jenuwein T. (2010) Mammalian Su(var) genes in chromatin control. Annu. Rev. Cell Dev. Biol. 26, 471–501 [DOI] [PubMed] [Google Scholar]
- 2.Kim M., Trinh B. N., Long T. I., Oghamian S., and Laird P. W. (2004) Dnmt1 deficiency leads to enhanced microsatellite instability in mouse embryonic stem cells. Nucleic Acids Res. 32, 5742–5749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kondo Y., Shen L., Ahmed S., Boumber Y., Sekido Y., Haddad B. R., and Issa J. P. (2008) Downregulation of histone H3 lysine 9 methyltransferase G9a induces centrosome disruption and chromosome instability in cancer cells. PLoS ONE 3, e2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Peng J. C., and Karpen G. H. (2008) Epigenetic regulation of heterochromatic DNA stability. Curr. Opin. Genet. Dev. 18, 204–211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Matsui T., Leung D., Miyashita H., Maksakova I. A., Miyachi H., Kimura H., Tachibana M., Lorincz M. C., and Shinkai Y. (2010) Proviral silencing in embryonic stem cells requires the histone methyltransferase ESET. Nature 464, 927–931 [DOI] [PubMed] [Google Scholar]
- 6.Rowe H. M., Jakobsson J., Mesnard D., Rougemont J., Reynard S., Aktas T., Maillard P. V., Layard-Liesching H., Verp S., Marquis J., Spitz F., Constam D. B., and Trono D. (2010) KAP1 controls endogenous retroviruses in embryonic stem cells. Nature 463, 237–240 [DOI] [PubMed] [Google Scholar]
- 7.Pinheiro I., Margueron R., Shukeir N., Eisold M., Fritzsch C., Richter F. M., Mittler G., Genoud C., Goyama S., Kurokawa M., Son J., Reinberg D., Lachner M., and Jenuwein T. (2012) Prdm3 and Prdm16 are H3K9me1 methyltransferases required for mammalian heterochromatin integrity. Cell 150, 948–960 [DOI] [PubMed] [Google Scholar]
- 8.Towbin B. D., González-Aguilera C., Sack R., Gaidatzis D., Kalck V., Meister P., Askjaer P., and Gasser S. M. (2012) Step-wise methylation of histone H3K9 positions heterochromatin at the nuclear periphery. Cell 150, 934–947 [DOI] [PubMed] [Google Scholar]
- 9.Black J. C., and Whetstine J. R. (2011) Chromatin landscape: methylation beyond transcription. Epigenetics 6, 9–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang R. H., Sengupta K., Li C., Kim H. S., Cao L., Xiao C., Kim S., Xu X., Zheng Y., Chilton B., Jia R., Zheng Z. M., Appella E., Wang X. W., Ried T., and Deng C. X. (2008) Impaired DNA damage response, genome instability, and tumorigenesis in SIRT1 mutant mice. Cancer Cell 14, 312–323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kappes F., Waldmann T., Mathew V., Yu J., Zhang L., Khodadoust M. S., Chinnaiyan A. M., Luger K., Erhardt S., Schneider R., and Markovitz D. M. (2011) The DEK oncoprotein is a Su(var) that is essential to heterochromatin integrity. Genes Dev. 25, 673–678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Maison C., Bailly D., Roche D., Montes de Oca R., Probst A. V., Vassias I., Dingli F., Lombard B., Loew D., Quivy J. P., and Almouzni G. (2011) SUMOylation promotes de novo targeting of HP1α to pericentric heterochromatin. Nat. Genet. 43, 220–227 [DOI] [PubMed] [Google Scholar]
- 13.Fan Y., Nikitina T., Zhao J., Fleury T. J., Bhattacharyya R., Bouhassira E. E., Stein A., Woodcock C. L., and Skoultchi A. I. (2005) Histone H1 depletion in mammals alters global chromatin structure but causes specific changes in gene regulation. Cell 123, 1199–1212 [DOI] [PubMed] [Google Scholar]
- 14.Iwase S., Xiang B., Ghosh S., Ren T., Lewis P. W., Cochrane J. C., Allis C. D., Picketts D. J., Patel D. J., Li H., and Shi Y. (2011) ATRX ADD domain links an atypical histone methylation recognition mechanism to human mental-retardation syndrome. Nat. Struct. Mol. Biol. 18, 769–776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Maison C., and Almouzni G. (2004) HP1 and the dynamics of heterochromatin maintenance. Nat. Rev. Mol. Cell Biol. 5, 296–304 [DOI] [PubMed] [Google Scholar]
- 16.Peters A. H., O'Carroll D., Scherthan H., Mechtler K., Sauer S., Schöfer C., Weipoltshammer K., Pagani M., Lachner M., Kohlmaier A., Opravil S., Doyle M., Sibilia M., and Jenuwein T. (2001) Loss of the Suv39h histone methyltransferases impairs mammalian heterochromatin and genome stability. Cell 107, 323–337 [DOI] [PubMed] [Google Scholar]
- 17.Prasanth S. G., Shen Z., Prasanth K. V., and Stillman B. (2010) Human origin recognition complex is essential for HP1 binding to chromatin and heterochromatin organization. Proc. Natl. Acad. Sci. U.S.A. 107, 15093–15098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huh M. S., Price O'Dea T., Ouazia D., McKay B. C., Parise G., Parks R. J., Rudnicki M. A., and Picketts D. J. (2012) Compromised genomic integrity impedes muscle growth after Atrx inactivation. J. Clin. Invest. 122, 4412–4423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Law M. J., Lower K. M., Voon H. P., Hughes J. R., Garrick D., Viprakasit V., Mitson M., De Gobbi M., Marra M., Morris A., Abbott A., Wilder S. P., Taylor S., Santos G. M., Cross J., Ayyub H., Jones S., Ragoussis J., Rhodes D., Dunham I., Higgs D. R., and Gibbons R. J. (2010) ATR-X syndrome protein targets tandem repeats and influences allele-specific expression in a size-dependent manner. Cell 143, 367–378 [DOI] [PubMed] [Google Scholar]
- 20.Watson L. A., Solomon L. A., Li J. R., Jiang Y., Edwards M., Shin-ya K., Beier F., and Bérubé N. G. (2013) Atrx deficiency induces telomere dysfunction, endocrine defects, and reduced life span. J. Clin. Invest. 123, 2049–2063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kalb R., Mallery D. L., Larkin C., Huang J. T., and Hiom K. (2014) BRCA1 Is a histone-H2A-specific ubiquitin ligase. Cell Rep. 8, 999–1005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhu Q., Pao G. M., Huynh A. M., Suh H., Tonnu N., Nederlof P. M., Gage F. H., and Verma I. M. (2011) BRCA1 tumour suppression occurs via heterochromatin-mediated silencing. Nature 477, 179–184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Thakar A., Parvin J., and Zlatanova J. (2010) BRCA1/BARD1 E3 ubiquitin ligase can modify histones H2A and H2B in the nucleosome particle. J. Biomol. Struct. Dyn. 27, 399–406 [DOI] [PubMed] [Google Scholar]
- 24.Sparmann A., and van Lohuizen M. (2006) Polycomb silencers control cell fate, development and cancer. Nat. Rev. Cancer 6, 846–856 [DOI] [PubMed] [Google Scholar]
- 25.Levine S. S., Weiss A., Erdjument-Bromage H., Shao Z., Tempst P., and Kingston R. E. (2002) The core of the polycomb repressive complex is compositionally and functionally conserved in flies and humans. Mol. Cell. Biol. 22, 6070–6078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dellino G. I., Schwartz Y. B., Farkas G., McCabe D., Elgin S. C., and Pirrotta V. (2004) Polycomb silencing blocks transcription initiation. Mol. Cell 13, 887–893 [DOI] [PubMed] [Google Scholar]
- 27.Kuzmichev A., Nishioka K., Erdjument-Bromage H., Tempst P., and Reinberg D. (2002) Histone methyltransferase activity associated with a human multiprotein complex containing the Enhancer of Zeste protein. Genes Dev. 16, 2893–2905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang H., Wang L., Erdjument-Bromage H., Vidal M., Tempst P., Jones R. S., and Zhang Y. (2004) Role of histone H2A ubiquitination in Polycomb silencing. Nature 431, 873–878 [DOI] [PubMed] [Google Scholar]
- 29.Blackledge N. P., Farcas A. M., Kondo T., King H. W., McGouran J. F., Hanssen L. L., Ito S., Cooper S., Kondo K., Koseki Y., Ishikura T., Long H. K., Sheahan T. W., Brockdorff N., Kessler B. M., Koseki H., and Klose R. J. (2014) Variant PRC1 complex-dependent H2A ubiquitylation drives PRC2 recruitment and polycomb domain formation. Cell 157, 1445–1459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cooper S., Dienstbier M., Hassan R., Schermelleh L., Sharif J., Blackledge N. P., De Marco V., Elderkin S., Koseki H., Klose R., Heger A., and Brockdorff N. (2014) Targeting polycomb to pericentric heterochromatin in embryonic stem cells reveals a role for H2AK119u1 in PRC2 recruitment. Cell Rep. 7, 1456–1470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kleer C. G., Cao Q., Varambally S., Shen R., Ota I., Tomlins S. A., Ghosh D., Sewalt R. G., Otte A. P., Hayes D. F., Sabel M. S., Livant D., Weiss S. J., Rubin M. A., and Chinnaiyan A. M. (2003) EZH2 is a marker of aggressive breast cancer and promotes neoplastic transformation of breast epithelial cells. Proc. Natl. Acad. Sci. U.S.A. 100, 11606–11611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lassman A. B., Dai C., Fuller G. N., Vickers A. J., and Holland E. C. (2004) Overexpression of c-MYC promotes an undifferentiated phenotype in cultured astrocytes and allows elevated Ras and Akt signaling to induce gliomas from GFAP-expressing cells in mice. Neuron Glia Biol. 1, 157–163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Orian J. M., Vasilopoulos K., Yoshida S., Kaye A. H., Chow C. W., and Gonzales M. F. (1992) Overexpression of multiple oncogenes related to histological grade of astrocytic glioma. Br. J. Cancer 66, 106–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Valk-Lingbeek M. E., Bruggeman S. W., and van Lohuizen M. (2004) Stem cells and cancer; the polycomb connection. Cell 118, 409–418 [DOI] [PubMed] [Google Scholar]
- 35.Varambally S., Dhanasekaran S. M., Zhou M., Barrette T. R., Kumar-Sinha C., Sanda M. G., Ghosh D., Pienta K. J., Sewalt R. G., Otte A. P., Rubin M. A., and Chinnaiyan A. M. (2002) The polycomb group protein EZH2 is involved in progression of prostate cancer. Nature 419, 624–629 [DOI] [PubMed] [Google Scholar]
- 36.Visser H. P., Gunster M. J., Kluin-Nelemans H. C., Manders E. M., Raaphorst F. M., Meijer C. J., Willemze R., and Otte A. P. (2001) The Polycomb group protein EZH2 is upregulated in proliferating, cultured human mantle cell lymphoma. Br. J. Haematol. 112, 950–958 [DOI] [PubMed] [Google Scholar]
- 37.Bruggeman S. W., Valk-Lingbeek M. E., van der Stoop P. P., Jacobs J. J., Kieboom K., Tanger E., Hulsman D., Leung C., Arsenijevic Y., Marino S., and van Lohuizen M. (2005) Ink4a and Arf differentially affect cell proliferation and neural stem cell self-renewal in Bmi1-deficient mice. Genes Dev. 19, 1438–1443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chatoo W., Abdouh M., David J., Champagne M. P., Ferreira J., Rodier F., and Bernier G. (2009) The polycomb group gene Bmi1 regulates antioxidant defenses in neurons by repressing p53 pro-oxidant activity. J. Neurosci. 29, 529–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jacobs J. J., Kieboom K., Marino S., DePinho R. A., and van Lohuizen M. (1999) The oncogene and Polycomb-group gene bmi-1 regulates cell proliferation and senescence through the ink4a locus. Nature 397, 164–168 [DOI] [PubMed] [Google Scholar]
- 40.Molofsky A. V., Pardal R., Iwashita T., Park I. K., Clarke M. F., and Morrison S. J. (2003) Bmi-1 dependence distinguishes neural stem cell self-renewal from progenitor proliferation. Nature 425, 962–967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sharpless N. E., and DePinho R. A. (1999) The INK4A/ARF locus and its two gene products. Curr. Opin. Genet. Dev. 9, 22–30 [DOI] [PubMed] [Google Scholar]
- 42.Facchino S., Abdouh M., Chatoo W., and Bernier G. (2010) BMI1 confers radioresistance to normal and cancerous neural stem cells through recruitment of the DNA damage response machinery. J. Neurosci. 30, 10096–10111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ismail I. H., Andrin C., McDonald D., and Hendzel M. J. (2010) BMI1-mediated histone ubiquitylation promotes DNA double-strand break repair. J. Cell Biol. 191, 45–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chagraoui J., Hébert J., Girard S., and Sauvageau G. (2011) An anticlastogenic function for the Polycomb Group gene Bmi1. Proc. Natl. Acad. Sci. U.S.A. 108, 5284–5289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liu J., Cao L., Chen J., Song S., Lee I. H., Quijano C., Liu H., Keyvanfar K., Chen H., Cao L. Y., Ahn B. H., Kumar N. G., Rovira I. I., Xu X. L., van Lohuizen M., Motoyama N., Deng C. X., and Finkel T. (2009) Bmi1 regulates mitochondrial function and the DNA damage response pathway. Nature 459, 387–392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hernández-Muñoz I., Taghavi P., Kuijl C., Neefjes J., and van Lohuizen M. (2005) Association of BMI1 with polycomb bodies is dynamic and requires PRC2/EZH2 and the maintenance DNA methyltransferase DNMT1. Mol. Cell. Biol. 25, 11047–11058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Saurin A. J., Shiels C., Williamson J., Satijn D. P., Otte A. P., Sheer D., and Freemont P. S. (1998) The human polycomb group complex associates with pericentromeric heterochromatin to form a novel nuclear domain. J. Cell Biol. 142, 887–898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sewalt R. G., Lachner M., Vargas M., Hamer K. M., den Blaauwen J. L., Hendrix T., Melcher M., Schweizer D., Jenuwein T., and Otte A. P. (2002) Selective interactions between vertebrate polycomb homologs and the SUV39H1 histone lysine methyltransferase suggest that histone H3-K9 methylation contributes to chromosomal targeting of Polycomb group proteins. Mol. Cell. Biol. 22, 5539–5553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Smigová J., Juda P., Cmarko D., and Raška I. (2011) Fine structure of the “PcG body” in human U-2 OS cells established by correlative light-electron microscopy. Nucleus 2, 219–228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Leeb M., Pasini D., Novatchkova M., Jaritz M., Helin K., and Wutz A. (2010) Polycomb complexes act redundantly to repress genomic repeats and genes. Genes Dev. 24, 265–276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Puschendorf M., Terranova R., Boutsma E., Mao X., Isono K., Brykczynska U., Kolb C., Otte A. P., Koseki H., Orkin S. H., van Lohuizen M., and Peters A. H. (2008) PRC1 and Suv39h specify parental asymmetry at constitutive heterochromatin in early mouse embryos. Nat. Genet. 40, 411–420 [DOI] [PubMed] [Google Scholar]
- 52.Saksouk N., Barth T. K., Ziegler-Birling C., Olova N., Nowak A., Rey E., Mateos-Langerak J., Urbach S., Reik W., Torres-Padilla M. E., Imhof A., Déjardin J., and Simboeck E. (2014) Redundant mechanisms to form silent chromatin at pericentromeric regions rely on BEND3 and DNA methylation. Mol. Cell 56, 580–594 [DOI] [PubMed] [Google Scholar]
- 53.Gargiulo G., Cesaroni M., Serresi M., de Vries N., Hulsman D., Bruggeman S. W., Lancini C., and van Lohuizen M. (2013) In vivo RNAi screen for BMI1 targets identifies TGF-beta/BMP-ER stress pathways as key regulators of neural- and malignant glioma-stem cell homeostasis. Cancer Cell 23, 660–676 [DOI] [PubMed] [Google Scholar]
- 54.Tadeu A. M., Ribeiro S., Johnston J., Goldberg I., Gerloff D., and Earnshaw W. C. (2008) CENP-V is required for centromere organization, chromosome alignment and cytokinesis. EMBO J. 27, 2510–2522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shumaker D. K., Dechat T., Kohlmaier A., Adam S. A., Bozovsky M. R., Erdos M. R., Eriksson M., Goldman A. E., Khuon S., Collins F. S., Jenuwein T., and Goldman R. D. (2006) Mutant nuclear lamin A leads to progressive alterations of epigenetic control in premature aging. Proc. Natl. Acad. Sci. U.S.A. 103, 8703–8708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hwang W. L., Deindl S., Harada B. T., and Zhuang X. (2014) Histone H4 tail mediates allosteric regulation of nucleosome remodelling by linker DNA. Nature 512, 213–217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hewish D. R., and Burgoyne L. A. (1973) Chromatin sub-structure. The digestion of chromatin DNA at regularly spaced sites by a nuclear deoxyribonuclease. Biochem. Biophys. Res. Commun. 52, 504–510 [DOI] [PubMed] [Google Scholar]
- 58.Kornberg R. D., LaPointe J. W., and Lorch Y. (1989) Preparation of nucleosomes and chromatin. Methods Enzymol. 170, 3–14 [DOI] [PubMed] [Google Scholar]
- 59.Wu C. (1980) The 5′ ends of Drosophila heat shock genes in chromatin are hypersensitive to DNase I. Nature 286, 854–860 [DOI] [PubMed] [Google Scholar]
- 60.Abdouh M., Facchino S., Chatoo W., Balasingam V., Ferreira J., and Bernier G. (2009) BMI1 sustains human glioblastoma multiforme stem cell renewal. J. Neurosci. 29, 8884–8896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Schoeftner S., Sengupta A. K., Kubicek S., Mechtler K., Spahn L., Koseki H., Jenuwein T., and Wutz A. (2006) Recruitment of PRC1 function at the initiation of X inactivation independent of PRC2 and silencing. EMBO J. 25, 3110–3122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tavares L., Dimitrova E., Oxley D., Webster J., Poot R., Demmers J., Bezstarosti K., Taylor S., Ura H., Koide H., Wutz A., Vidal M., Elderkin S., and Brockdorff N. (2012) RYBP-PRC1 complexes mediate H2A ubiquitylation at polycomb target sites independently of PRC2 and H3K27me3. Cell 148, 664–678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Buchwald G., van der Stoop P., Weichenrieder O., Perrakis A., van Lohuizen M., and Sixma T. K. (2006) Structure and E3-ligase activity of the Ring-Ring complex of polycomb proteins Bmi1 and Ring1b. EMBO J. 25, 2465–2474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.de Napoles M., Mermoud J. E., Wakao R., Tang Y. A., Endoh M., Appanah R., Nesterova T. B., Silva J., Otte A. P., Vidal M., Koseki H., and Brockdorff N. (2004) Polycomb group proteins Ring1A/B link ubiquitylation of histone H2A to heritable gene silencing and X inactivation. Dev. Cell 7, 663–676 [DOI] [PubMed] [Google Scholar]
- 65.Román-Trufero M., Méndez-Gómez H. R., Pérez C., Hijikata A., Fujimura Y., Endo T., Koseki H., Vicario-Abejón C., and Vidal M. (2009) Maintenance of undifferentiated state and self-renewal of embryonic neural stem cells by Polycomb protein Ring1B. Stem Cells 27, 1559–1570 [DOI] [PubMed] [Google Scholar]
- 66.Francis N. J., Kingston R. E., and Woodcock C. L. (2004) Chromatin compaction by a polycomb group protein complex. Science 306, 1574–1577 [DOI] [PubMed] [Google Scholar]
- 67.Meshorer E., Yellajoshula D., George E., Scambler P. J., Brown D. T., and Misteli T. (2006) Hyperdynamic plasticity of chromatin proteins in pluripotent embryonic stem cells. Dev. Cell 10, 105–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Varley K. E., Gertz J., Bowling K. M., Parker S. L., Reddy T. E., Pauli-Behn F., Cross M. K., Williams B. A., Stamatoyannopoulos J. A., Crawford G. E., Absher D. M., Wold B. J., and Myers R. M. (2013) Dynamic DNA methylation across diverse human cell lines and tissues. Genome Res. 23, 555–567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kohli R. M., and Zhang Y. (2013) TET enzymes, TDG and the dynamics of DNA demethylation. Nature 502, 472–479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Scaffidi P., and Misteli T. (2006) Lamin A-dependent nuclear defects in human aging. Science 312, 1059–1063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Oberdoerffer P., Michan S., McVay M., Mostoslavsky R., Vann J., Park S. K., Hartlerode A., Stegmuller J., Hafner A., Loerch P., Wright S. M., Mills K. D., Bonni A., Yankner B. A., Scully R., Prolla T. A., Alt F. W., and Sinclair D. A. (2008) SIRT1 redistribution on chromatin promotes genomic stability but alters gene expression during aging. Cell 135, 907–918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Larson K., Yan S. J., Tsurumi A., Liu J., Zhou J., Gaur K., Guo D., Eickbush T. H., and Li W. X. (2012) Heterochromatin formation promotes longevity and represses ribosomal RNA synthesis. PLoS Genet. 8, e1002473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Pegoraro G., Kubben N., Wickert U., Göhler H., Hoffmann K., and Misteli T. (2009) Ageing-related chromatin defects through loss of the NURD complex. Nat. Cell. Biol. 11, 1261–1267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Peng J. C., and Karpen G. H. (2009) Heterochromatic genome stability requires regulators of histone H3 K9 methylation. PLoS Genet. 5, e1000435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.van der Lugt N. M., Domen J., Linders K., van Roon M., Robanus-Maandag E., te Riele H., van der Valk M., Deschamps J., Sofroniew M., and van Lohuizen M. (1994) Posterior transformation, neurological abnormalities, and severe hematopoietic defects in mice with a targeted deletion of the bmi-1 proto-oncogene. Genes Dev. 8, 757–769 [DOI] [PubMed] [Google Scholar]
- 76.Kim J., Kim J. A., McGinty R. K., Nguyen U. T., Muir T. W., Allis C. D., and Roeder R. G. (2013) The n-SET domain of Set1 regulates H2B ubiquitylation-dependent H3K4 methylation. Mol. Cell 49, 1121–1133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wu L., Lee S. Y., Zhou B., Nguyen U. T., Muir T. W., Tan S., and Dou Y. (2013) ASH2L regulates ubiquitylation signaling to MLL: trans-regulation of H3 K4 methylation in higher eukaryotes. Mol. Cell 49, 1108–1120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.McGinty R. K., Henrici R. C., and Tan S. (2014) Crystal structure of the PRC1 ubiquitylation module bound to the nucleosome. Nature 514, 591–596 [DOI] [PMC free article] [PubMed] [Google Scholar]