Abstract
The regulatory protein collybistin (CB) recruits the receptor-scaffolding protein gephyrin to mammalian inhibitory glycinergic and GABAergic postsynaptic membranes in nerve cells. CB is tethered to the membrane via phosphoinositides. We developed an in vitro assay based on solid-supported 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine membranes doped with different phosphoinositides on silicon/silicon dioxide substrates to quantify the binding of various CB2 constructs using reflectometric interference spectroscopy. Based on adsorption isotherms, we obtained dissociation constants and binding capacities of the membranes. Our results show that full-length CB2 harboring the N-terminal Src homology 3 (SH3) domain (CB2SH3+) adopts a closed and autoinhibited conformation that largely prevents membrane binding. This autoinhibition is relieved upon introduction of the W24A/E262A mutation, which conformationally “opens” CB2SH3+ and allows the pleckstrin homology domain to properly bind lipids depending on the phosphoinositide species with a preference for phosphatidylinositol 3-monophosphate and phosphatidylinositol 4-monophosphate. This type of membrane tethering under the control of the release of the SH3 domain of CB is essential for regulating gephyrin clustering.
Keywords: GABA receptor, inositol phospholipid, lipid bilayer, lipid-protein interaction, membrane function
Introduction
The function of neuronal synapses and the dynamic regulation of their efficacy depend on the assembly of the postsynaptic neurotransmitter receptor apparatus. The main scaffolding protein of inhibitory glycinergic and GABAergic postsynapses in mammals is gephyrin (1, 2), whose recruitment to the postsynaptic membrane is controlled by the adaptor protein collybistin (CB)2 (3). Loss of CB results in a strong reduction of gephyrin and GABAA receptor clusters in several regions of the forebrain, which demonstrates the essential role of CB in the assembly and maintenance of GABAergic postsynaptic structures (4).
CB belongs to the Dbl family of guanine nucleotide exchange factors. In mouse, four differently spliced CB mRNAs are present (CB1SH3+, CB2SH3−, CB2SH3+, and CB3SH3+). All four mRNAs encode a Dbl homology (DH) and a pleckstrin homology (PH) domain. The three major variants (CB1SH3+, CB2SH3+, and CB3SH3+) encode CBs with an additional N-terminal Src homology 3 (SH3) domain but differ in their C termini. A fourth variant (CB2SH3−) encodes a CB2 isoform lacking the SH3 domain (Fig. 1) but is very rare (5) as its protein product is not detectable in mouse brain (6). Importantly, the PH domain of the different CBs is required for proper function as indicated by the fact that its deletion abolishes the plasma membrane targeting of gephyrin-CB complexes when cotransfected in HEK293 cells and causes a loss of dendritic gephyrin clusters in dissociated rat cortical neurons (5).
FIGURE 1.
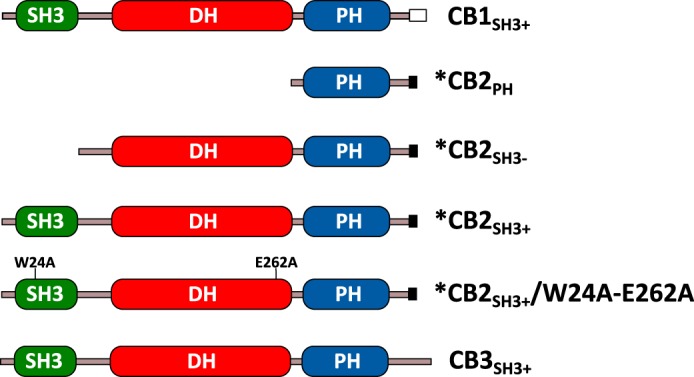
Domain architectures of the CB variants. Proteins that were used in the present study are marked with an asterisk (*).
In vitro binding studies utilizing a variety of inositol headgroups, soluble phosphoinositide analogs, and liposomes containing phosphoinositides showed that PH domains bind phosphoinositides with a broad range of selectivity and affinity (7–10). An early membrane activation model suggested that the PH domain of CB binds to phosphatidylinositol 3,4,5-trisphosphate (PI(3,4,5)P3) (1). In contrast, experiments with immobilized phosphoinositides and purified glutathione S-transferase (GST)-tagged CB variants in overlay assays indicated that the PH domain of CB specifically binds phosphatidylinositol 3-monophosphate (PI(3)P) (11, 12), and subsequent studies verified the binding of CB to PI(3)P (6, 13). However, most of the relevant experiments were conducted with lipids spotted on blotting membranes, which have been shown to be less reliable than other techniques (14). Hence, the question arises as to whether the phosphoinositide specificity of CB observed with overlay assays properly reflects the lipid binding specificity of CB in intact phospholipid bilayers. That this is a critical issue is also illustrated by the fact that PI(3)P, the best characterized CB ligand so far, is mainly concentrated in early endosomes (Fig. 2) (15) and is present at the plasma membrane where CB is ultimately required for GABAergic synapse formation only under specific stimulatory conditions (16). Instead, PI(3,4)P2, PI(4,5)P2, and PI(3,4,5)P3 are primarily localized to the plasma membrane; PI(4)P is enriched in the Golgi complex and present at the plasma membrane; and PI(3,5)P2 is found in compartments of the late endosomal pathway (Fig. 2) (17, 18).
FIGURE 2.
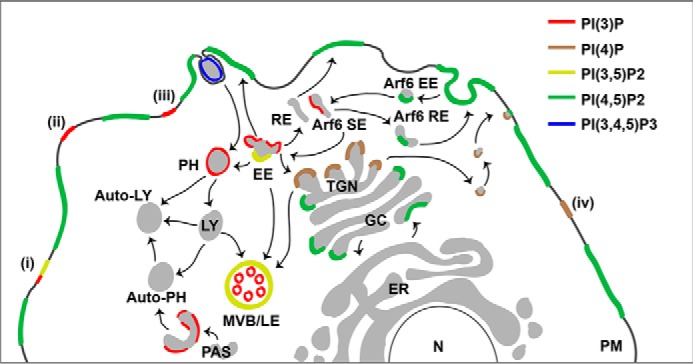
Subcellular distribution of PIPs. Shown is the distribution of the predominant PIP species in different cell compartments according to Vicinanza et al. (18) and modified according to Kong et al. (49) (i), Maffucci et al. (50) (ii), Falasca and Maffucci (51) (iii), and Nakatsu et al. (52) (iv). PIP-metabolizing enzymes and other proteins involved in the synthesis, degradation, and trafficking of PIPs (indicated by arrows) are not shown for simplicity. Of note, many of the PIP-metabolizing enzymes are present in more than one cellular compartment, and their overall distribution does not completely fit to the PIP distribution. PM, plasma membrane; EE, early endosome; RE, recycling endosome; LY, lysosome; MVB/LE, multivesicular body/late endosome; PAS, preautophagosomal structure; PH, phagosome; TGN, trans-Golgi network; GC, Golgi complex; ER, endoplasmic reticulum; N, nucleus; SE, sorting endosome.
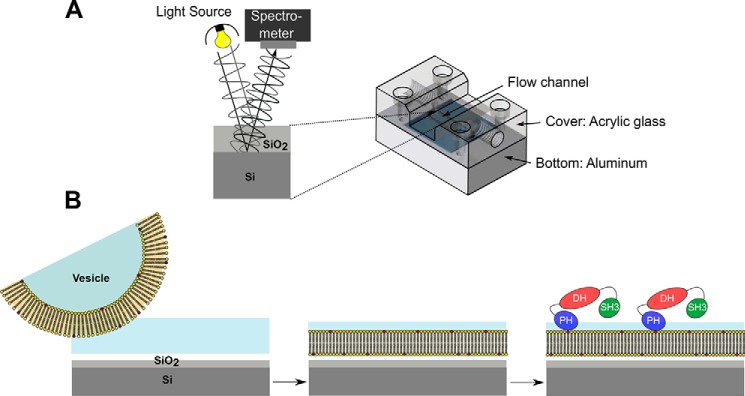
In the present study, we assessed the lipid binding specificity of CB in a more natural phospholipid bilayer environment that reflects the situation in live cells more closely. For that purpose, we used fluid planar lipid bilayers composed of 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC) doped with different phosphoinositides. Immobilized on a silicon/silicon dioxide substrate, these membranes enabled us to monitor the specific interaction of CB variants with receptor lipids in a time-resolved and label-free manner by means of reflectometric interference spectroscopy (RIfS) (Fig. 3A) (19–21). RIfS is a well established technique to quantitatively monitor protein-receptor interactions at solid-supported membranes (22–25).
FIGURE 3.
Setup of the RIfS experiments. A, schematic drawing of the RIfS setup. B, scheme of membrane preparation and protein binding on the silicon dioxide surface.
Experimental Procedures
Materials
Phosphoinositides (PI(3)P, PI(4)P, PI(3,4)P2, PI(3,5)P2, PI(4,5)P2, and PI(3,4,5)P3) were obtained as C16 derivatives from Echelon Biosciences (Salt Lake City, UT). POPC, 1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-l-serine (POPS), l-α-phosphatidylinositol (PI) from soy and 1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-(1′-racemic glycerol) (POPG) were purchased from Avanti Polar Lipids (Alabaster, AL). BL21(DE3) competent Escherichia coli cells were from Invitrogen. Chitin resin was obtained from New England Biolabs (Ipswich, MA). Silicon wafers were purchased from Silicon Materials (Kaufering, Germany).
Protein Purification
The CB2 variants CB2PH, CB2SH3−, CB2SH3+, and CB2SH3+/W24A/E262A were obtained by recombinant expression as described previously (6). Briefly, plasmids based on the IMPACT system vector pTXB1 (CB2SH3+ and CB2SH3+/W24A/E262A) or pTYB12 (CB2PH and CB2SH3−) encoding CB2 isoforms with an N-/C-terminal intein tag were transformed into E. coli BL21(DE3) cells. Cells were grown to an A600 of 0.8–1.0 at 37 °C in LB-Miller medium. Protein expression was induced by addition of 0.5 mm isopropyl β-d-thiogalactopyranoside. After overnight growth at 15 °C, cells were harvested by centrifugation (4,500 × g, 20 min, 4 °C). Cells were resuspended in lysis buffer (250 mm NaCl, 20 mm HEPES, 2 mm EDTA, 10% (v/v) glycerol, pH 8.0), and cell lysis was performed using a microfluidizer (1 kbar, three cycles, ice-cooled; LM 10 processor, Microfluidics, Westwood, MA). After centrifugation (70,000 × g, 30 min, 4 °C), the supernatant was applied to the equilibrated chitin resin for 1 h. The resin was rinsed with >1 liter of washing buffer (1 m NaCl, 20 mm HEPES, 2 mm EDTA, pH 8.0), and protein cleavage was induced by incubation with 50 mm dithiothreitol (DTT) in buffer G (250 mm NaCl, 20 mm HEPES, 2 mm EDTA, pH 8.0) for >40 h. After elution with buffer G containing 5 mm DTT, the protein solution was diluted to 50 mm NaCl. The protein was purified using a Mono Q 10/100GL column (GE Healthcare) with a linear NaCl gradient from 50 mm to 1 m NaCl in buffer (20 mm HEPES, 2 mm EDTA, 2 mm β-mercaptoethanol, pH 8.0). Proteins were stored at 4 °C. In the case of CB2SH3−, 20 mm Tris instead of HEPES was used in all buffers. Concentration and dialysis to buffer A (100 mm NaCl, 25 mm HEPES, 0.5 mm DTT, 0.5 mm EDTA, pH 7.2) was performed by ultrafiltration using spin concentrators (Sartorius, Göttingen, Germany). Protein concentrations were determined by UV/visible spectroscopy using extinction coefficients of ϵ280 = 37,930 m−1 cm−1 (CB2PH), ϵ280 = 70,000 m−1 cm−1 (CB2SH3−), ϵ280 = 98,945 m−1 cm−1 (CB2SH3+), or ϵ280 = 93,445 m−1 cm−1 (CB2SH3+/W24A/E262A).
Vesicle Preparation
Stock solutions of POPC, POPS, POPG, and PI were prepared in chloroform, whereas the phosphoinositides were dissolved in a mixture of chloroform/methanol/water according to the manufacturer's instructions. Lipid stock solutions were added to chloroform to obtain the desired lipid molar ratios, and the organic solvents were evaporated with a gentle nitrogen stream at 30 °C. Lipid films were further dried under vacuum for at least 3 h at 32 °C. For the preparation of small unilamellar vesicles (SUVs), the lipid film was rehydrated by incubation with 500 μl of buffer for 20 min followed by vortexing for 30 s (three cycles, every 5 min). The vesicle suspension was then sonicated using an ultrasonic homogenizer (30 min, 65%, four cycles; Sonopuls HD2070, Bandelin, Berlin, Germany) to generate SUVs.
Reflectometric Interference Spectroscopy
RIfS was used to monitor the formation of supported lipid bilayers and subsequent protein binding in a time-resolved manner (26). The experimental setup was described in detail previously (Fig. 3A) (22). Reflectometric interference spectra were monitored using a NanoCalc-2000 visible/near-IR spectrometer (Ocean Optics, Dunedin, FL). Spectra were recorded every 2 s and were analyzed using a MATLAB routine. To provide a substrate for interference fringes, silicon wafers with a 5-μm-thick SiO2 layer were used. Silicon substrates (1 × 2 cm2) were rinsed with ethanol and water. To hydrophilize the silicon substrates, they were incubated in an aqueous solution of NH3 and H2O2 (H2O/NH3/H2O2, 5:1:1) for 20 min at 70 °C followed by treatment with oxygen plasma (30 s, 50% power). Afterward, the hydrophilic substrate was rinsed with buffer, and SUVs were added. After the spreading process of the SUVs was finished, as indicated by a constant optical thickness, the remaining lipid material was removed by rinsing with buffer A. To prevent nonspecific protein binding, the measuring chamber was flushed with a bovine serum albumin-containing buffer solution (1 mg/ml in buffer A) and rinsed again with buffer A.
CD Spectroscopy
CD spectra were used to determine the secondary structure elements of the isolated proteins and to compare them with the crystal structure of CB2SH3+ (Protein Data Bank code 4MT6) (6). CD spectra (λ = 190–260 nm) were recorded at 20 °C with a J-1500 spectrometer (Jasco, Gross-Umstadt, Germany) equipped with a cuvette (d = 0.1 mm). Five spectra were averaged, background was subtracted (buffer solution), and the mean residue ellipticity was calculated. Secondary structure analysis was performed with the program DichroWeb using the CDSSTR method and reference data set 4 (27). Table 1 summarizes the fractions of the secondary structure elements as obtained from data evaluation using DichroWeb.
TABLE 1.
Secondary structure elements of the different CB variants used in this study in comparison with the fractions calculated (Calc.) from the crystal structure of CB2SH3+ (Protein Data Bank code 4MT6) (6)
| CB2PH | Calc. | CB2SH3− | Calc. | CB2SH3+ | CB2SH3+/W24A/E262A | Calc. | |
|---|---|---|---|---|---|---|---|
| α-Helix | 0.14 | 0.18 | 0.54 | 0.44 | 0.51 | 0.53 | 0.39 |
| β-Strand | 0.39 | 0.31 | 0.09 | 0.14 | 0.23 | 0.11 | 0.15 |
| β-Turn | 0.17 | 0.06 | 0.13 | 0.07 | 0.07 | 0.14 | 0.06 |
| Random coil | 0.29 | 0.45 | 0.23 | 0.35 | 0.18 | 0.22 | 0.40 |
Results
Formation of Supported Lipid Bilayers
To obtain planar supported lipid bilayers, SUVs were spread onto the silicon/silicon dioxide surface (Fig. 3B). To obtain membranes with high surface coverage, optimum spreading conditions of the vesicles to form planar bilayers are required. In a previous study, we showed that SUVs composed of POPC doped with PI(4,5)P2 do not spread to a continuous bilayer on silicon/silicon dioxide substrates at pH 7.4 if the PI(4,5)P2 concentration is larger than 4 mol % (23). However, if the pH is reduced to 4.8, the net negative charge of PI(4,5)P2 and the negative surface charge of the silicon substrate are reduced (28, 29). In the present study, we doped POPC membranes with 10 mol % of the different phosphatidylinositol phosphates (PIPs), namely PI(3)P, PI(4)P, PI(3,4)P2, PI(3,5)P2, PI(4,5)P2, and PI(3,4,5)P3. 10 mol % of the potential receptor lipid was chosen to ensure a good signal-to-noise ratio. As the PIPs vary in their phosphorylation pattern and thus in their overall net negative charge, the pH value of the buffer was adjusted accordingly. For SUVs containing 10 mol % PI(3)P and PI(4)P, a buffer at pH 6.4 was used (20 mm citrate, 50 mm KCl, 0.1 mm EDTA, 0.1 mm NaN3, pH 6.4), whereas for SUVs composed of POPC and 10 mol % PI(3,4)P2, PI(3,5)P2, PI(4,5)P2, or PI(3,4,5)P3, a citrate buffer at pH 4.8 (20 mm citrate, 50 mm KCl, 0.1 mm EDTA, 0.1 mm NaN3, pH 4.8) was chosen. SUVs containing POPC only, POPC/POPS (8:2), POPC/POPG (9:1 and 8:2), or POPC/PI (9:1) were prepared and spread in buffer A, which was also used for protein binding studies.
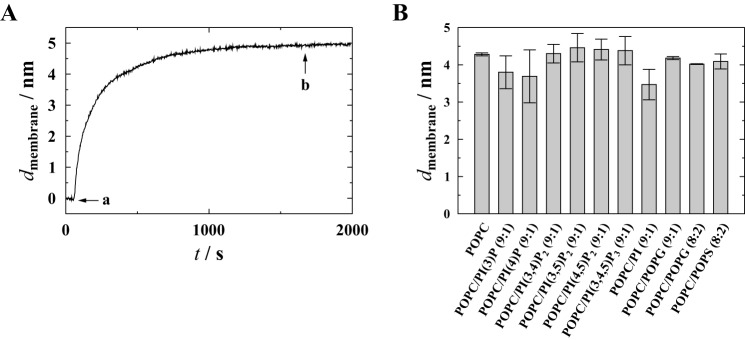
The spreading process of the SUVs was monitored in a time-resolved manner by means of RIfS. A characteristic time trace is depicted in Fig. 4A. From the maximum change in optical thickness (ΔOT), the quality of the resulting bilayer can be assessed. After the spreading process was completed, the system was rinsed with buffer A. No changes in bilayer thickness were observed, which is indicative of a stable planar solid-supported bilayer (Fig. 4A).
FIGURE 4.
Membrane preparations analyzed by RIfS. A, time-resolved change in membrane thickness dmembrane during the spreading of SUVs composed of POPC/PI(3,4,5)P3 (9:1) on a silicon wafer. a, addition of SUVs (0.4 mg/ml) in 20 mm citrate buffer, pH 4.8; b, rinsing with buffer. B, membrane thicknesses dmembrane for different lipid compositions. Error bars show the S.D. of the mean obtained from at least nine independent membrane preparations.
From the ΔOT values at saturation, the physical thickness of the bilayer (dmembrane) was calculated using ΔOT = nlipiddmembrane with a refractive index of nlipid = 1.5. For POPC membranes, a bilayer thickness of dmembrane = 4.28 ± 0.04 nm was obtained (Fig. 4B). This value is in good agreement with measurements using small angle neutron and x-ray scattering (dmembrane = 3.98 ± 0.08 nm) (30). For POPC membranes containing POPG (9:1 and 8:2), membrane thicknesses of 4.18 ± 0.04 and 4.02 ± 0.01 nm were obtained, whereas for POPC/POPS (8:2) membranes, a thickness of 4.1 ± 0.2 nm was measured; these values are in good agreement with the thickness of a pure POPC bilayer and thus indicate rather defect-free bilayers. For membranes containing PI, PI(3)P, and PI(4)P, slightly thinner bilayers were obtained (3.8 ± 0.4 nm for PI(3)P, 3.7 ± 0.7 nm for PI(4)P, and 3.5 ± 0.4 nm for PI-containing bilayers; Fig. 4B), which indicates that some defects are present in these bilayers. Compared with pure POPC bilayers, a membrane surface coverage of at least 90% was still obtained. To prevent nonspecific protein binding to these defects, the membranes were incubated with a bovine serum albumin (BSA) solution prior to the addition of the CB variant as described under “Experimental Procedures.” The slightly lower membrane surface coverage is probably a result of the repulsion between the negatively charged vesicles and the negatively charged silicon dioxide surface at the given pH (23) but is still fully sufficient for the experiments given the fact that the free surface area is blocked with BSA. Membranes doped with PI(3,4)P2, PI(3,5)P2, PI(4,5)P2, or PI(3,4,5)P3 show slightly larger membrane thicknesses as compared with POPC-only membranes (Fig. 4B), which might be explained by the strongly phosphorylated inositol headgroups that protrude about 0.6 nm from the membrane surface (31).
Secondary Structure Analysis of CB Variants
Circular dichroism spectra were taken and analyzed for all CB variants. The fraction of the secondary structure elements was compared with the crystal structure to provide evidence for proper protein folding under the chosen buffer conditions (Table 1). The crystal structure of CB2PH shows nine antiparallel β-strands and a C-terminal α-helix, which is reflected in 39% β-strands and 14% α-helices. CB2SH3− consists of a DH/PH tandem domain with a DH domain formed only by α-helices. This is reproduced in the large increase (54%) in α-helical content of CB2SH3−. Full-length CB2SH3+ is composed of the DH/PH tandem domain and the N-terminal SH3 domain. The SH3 domain consists of five short β-strands connected by unordered loop regions, leading to an increase in the β-strand fraction (23%) as compared with CB2SH3−. In contrast to the crystal structure, we found a larger α-helical and larger β-strand fraction at the expense of random coil. This can be explained by the fact that the linker between the SH3 and DH domain is missing in the crystal structure and hence does not contribute to our calculations.
Interaction of CB2PH with Different Phosphoinositides
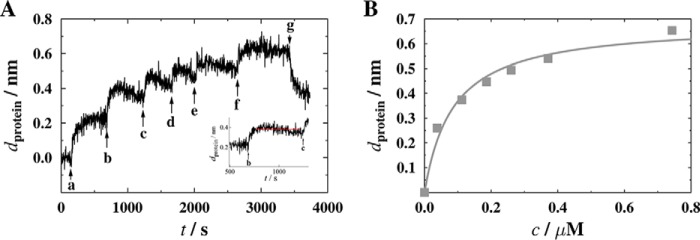
The planar continuous lipid bilayers doped with different PIPs and prepared as described above are a prerequisite for the analysis of the specificity of CB binding. As binding to phosphoinositides is known to be mediated by a region of positively charged amino acids in the PH domain of CB (32), we started out by studying the isolated PH domain of CB (CB2PH) and its specificity for certain PIPs. Protein concentration-dependent binding studies were performed by means of the RIfS setup. Fig. 5A shows a typical result of a RIfS experiment. Adding CB2PH to a membrane composed of POPC doped with 10 mol % of PI(3)P results in a change in thickness due to interactions between the protein and the membrane-embedded lipids. With increasing protein concentration, the thickness of the protein layer increases until the maximum protein layer thickness (dprotein-max)is reached. Rinsing with buffer A leads to a partial decrease in thickness, indicating that about 50% of the protein is reversibly bound and desorbs upon rinsing. It is a general phenomenon that proteins bound via a receptor lipid to a planar membrane only partially desorb from the membrane upon rinsing (23, 33). This can be explained as follows. (i) Proteins that bind in a one-to-one fashion to the receptor lipid desorb from the membrane upon rinsing with the corresponding rate constant of desorption, koff. However, part of the proteins might bind more than one receptor lipid or bind additionally to other lipid headgroups so that they do not desorb upon rinsing. (ii) If the membrane-bound proteins laterally interact with each other and form protein clusters, this second interaction on the membrane surface prevents the proteins from desorption upon rinsing.
FIGURE 5.
Adsorption of CB2PH to PI(3)P. A, characteristic time trace of a RIfS experiment showing the specific binding of CB2PH to a PI(3)P-containing membrane. Different concentrations of CB2PH were added at time points as indicated by arrows (a, 0.038 μm; b, 0.11 μm; c, 0.19 μm; d, 0.26 μm; e, 0.37 μm; f, 0.74 μm). After rinsing the system with buffer A (g), a fraction of the bound protein desorbs from the membrane, indicating reversible binding. The inset shows the determination of the saturation value dprotein. B, adsorption isotherm obtained from the data shown in A. The solid gray line is the result of fitting a Langmuir isotherm (Equation 1) to the data with dprotein-max = 0.69 ± 0.04 nm and KD = 0.09 ± 0.02 μm.
From the saturation values reached after each addition of the respective protein, dprotein was extracted (see Fig. 5A, inset) and was plotted versus the bulk protein concentration. Fig. 5B shows the resulting adsorption isotherm. The dissociation constant KD as well as dprotein-max, which reflects the binding capacity of the membrane, were determined by fitting a simple Langmuir adsorption isotherm (Equation 1) to the data.
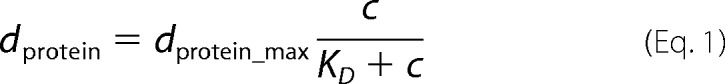 |
We are aware of the fact that the Langmuir model is only valid in the case of fully reversible protein binding, and more elaborate models are required to account for the irreversibility (33–35). However, as the origin of the irreversibility is as yet not known, we refrained from a more sophisticated model and used the obtained KD values to compare the affinities among the different PIPs. Control experiments with POPC-only membranes demonstrated that binding of CB2PH is specific; i.e. no interaction of the protein with POPC membranes lacking phosphoinositides was observed (data not shown). Also no protein binding was observed on membranes composed of POPC/POPS (8:2), indicating that a mere negative surface charge density is not sufficient for CB2PH to bind.
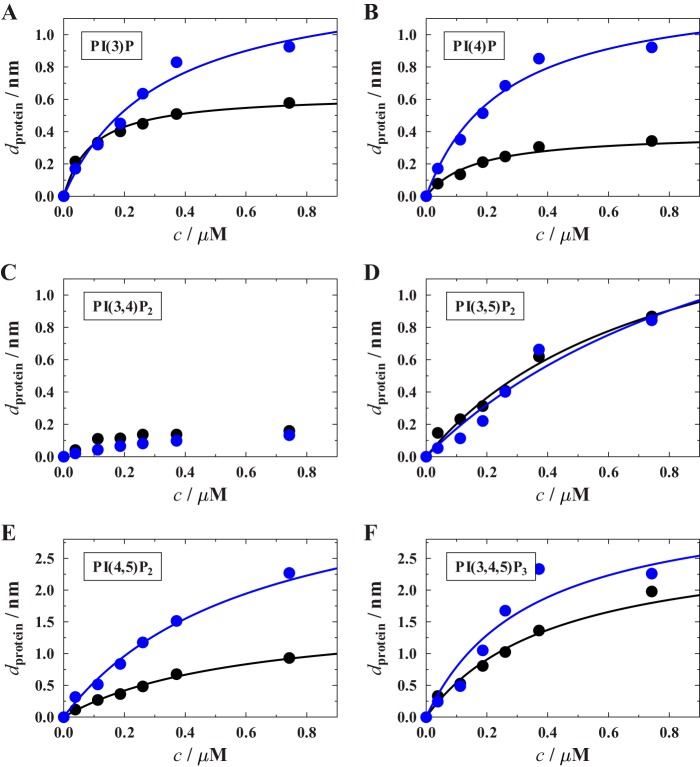
We obtained adsorption isotherms for different PIPs (Fig. 6) from which the dissociation constants and protein layer thicknesses were determined (Table 2). The isotherms indicate that CB2PH does not significantly bind to PI(3,4)P2 as indicated by the maximum change in protein layer thickness of only 0.2–0.3 nm. These values are too low to be attributed to specific binding. Thus, all measurements with such low protein surface coverage were not subjected to a determination of dissociation constants.
FIGURE 6.
Binding isotherms of CB2PH (black circles) and CB2SH3− (blue circles) obtained for different lipid compositions. A, POPC/PI(3)P (9:1); B, POPC/PI(4)P (9:1); C, POPC/PI(3,4)P2 (9:1); D, POPC/PI(3,5)P2 (9:1); E, POPC/PI(4,5)P2 (9:1); F, POPC/PI(3,4,5)P3. For each isotherm, at least three independent RIfS experiments were performed, and the data represent the mean values. The solid lines are results of fitting Equation 1 to the data. The obtained KD values are summarized in Table 2.
TABLE 2.
Dissociation constants (KD) for the binding of CB2PH, CB2SH3−, and CB2SH3+/W24A/E262A to PIP-containing solid-supported membranes
|
KD |
||||||
|---|---|---|---|---|---|---|
| PI(3)P | PI(4)P | PI(3,4)P2 | PI(3,5)P2 | PI(4,5)P2 | PI(3,4,5)P3 | |
| μm | ||||||
| CB2PH | 0.10 ± 0.01 | 0.16 ± 0.03 | 0.8 ± 0.2 | 0.60 ± 0.06 | 0.4 ± 0.1 | |
| CB2SH3− | 0.32 ± 0.08 | 0.26 ± 0.06 | 1.2 ± 0.5 | 0.63 ± 0.09 | 0.3 ± 0.1 | |
| CB2SH3+/mutant | 0.27 ± 0.02 | 0.25 ± 0.04 | 0.9 ± 0.1 | 1.9 ± 0.2 | ||
In contrast to this low nonspecific binding, CB2PH binds to all other PIPs with a dissociation constant in the 0.1 μm range with a slight preference for monophosphorylated PIPs. The dissociation constants for the diphosphorylated PI derivatives PI(3,5)P2 and PI(4,5)P2 and the triphosphorylated PI(3,4,5)P3 are about 4–8 times larger. The largest protein layer thickness was found for PI(3,4,5)P3-doped membranes with dprotein-max = 2.8 ± 0.3 nm. Assuming a maximum protein surface coverage of 56% according to the scaled particle theory (25) and a diameter of the PH domain of 4.2 nm as obtained from the crystal structure (12), one would expect a protein layer thickness of 2.4 nm. Comparing the theoretical and experimental values, we conclude that full protein coverage is reached in the case of PI(3,4,5)P3, whereas for the other PIPs, a lower surface coverage is found.
Interaction of CB2SH3− with Different Phosphoinositides
It has been proposed that CB exists in a closed and autoinhibited conformation in which the SH3 domain interacts with the DH and PH domains, thus preventing its membrane recruitment (6). Hence, for CB2 lacking the SH3 domain, it is expected that it preferentially binds to phosphoinositides, whereas the full-length protein interacts only weakly. In light of this hypothesis, we first studied the binding of CB2SH3−, which harbors the DH domain and the PH domain but lacks the SH3 domain, to PIP-containing membranes. Again, adsorption isotherms were monitored for different PIPs, and the dissociation constants as well as the maximum protein layer thicknesses were determined by fitting a Langmuir adsorption isotherm to the data (Fig. 6). As expected, CB2SH3− binds with submicromolar affinity to all PIPs except for PI(3,4)P2, demonstrating that the PH domain of CB is accessible for PIP binding. The same trend was observed for the di- and triphosphorylated PIPs with slightly larger KD values (Fig. 6 and Table 2). Due to the size of the additional DH domain, the maximum protein layer thickness on the membrane increased as expected except for PI(3,5)P2, which also shows the smallest binding affinity among the different PIPs (Fig. 6). The largest maximum protein layer thicknesses of 3.5–4 nm were determined for PI(4,5)P2 and PI(3,4,5)P3, similar to the results obtained for CB2PH. Based on the protein dimensions obtained from the crystal structure of CB2SH3−, a theoretical protein layer thickness of 4.4 nm was estimated.
We next tested the impact of the chemical nature of the receptor lipid on the binding affinity of CB2SH3−. In accordance with the results obtained for CB2PH, no binding to negatively charged POPC/POPS (8:2) membranes was found, supporting the notion that a mere electrostatic interaction of the protein with the membrane is not responsible for CB binding. However, CB2SH3− binds to lipid membranes composed of POPC/PI (9:1) with a dissociation constant (KD = 0.14 ± 0.01 μm) similar to those of the phosphorylated PI derivatives but with a smaller protein layer thickness of dprotein-max = 0.8 ± 0.2 nm. This observation is compatible with the idea that the OH groups of the inositol headgroup together with the phosphate group at the glycerol backbone might be involved in the binding specificity of the protein.
Binding of CB2SH3+
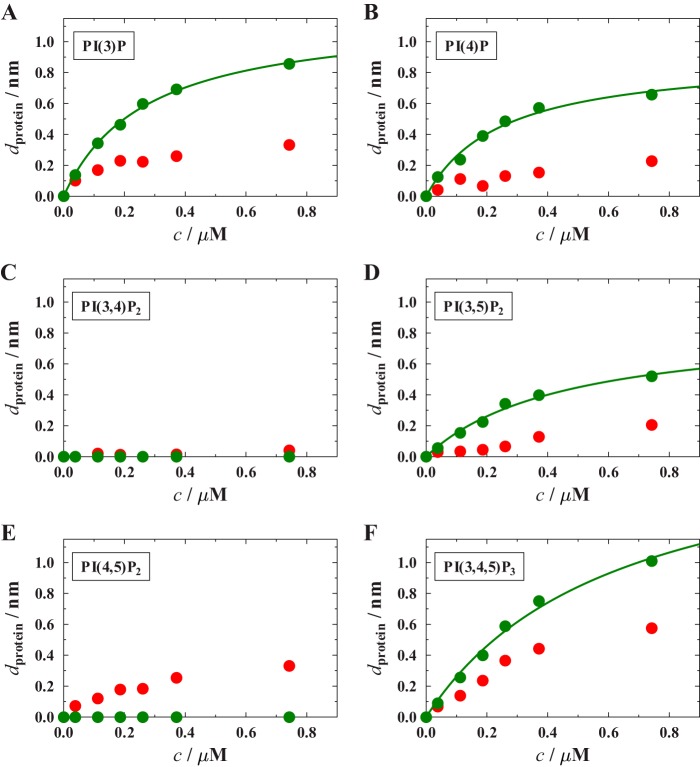
In view of the notion that CB harboring an SH3 domain forms a closed and autoinhibited conformation where the SH3 domain interacts with the DH and PH domains, we further analyzed the influence of the SH3 domain on the binding behavior of CB by studying CB2SH3+ binding to PIP-doped membranes. If CB2SH3+ bound to PIP-doped membranes, we expected to observe a larger protein layer thickness than for CB2SH3− as CB2SH3+ is larger in size than CB2SH3−. However, in contrast to this, the observed protein layer thicknesses were greatly diminished by at least a factor of 5 (Fig. 7) independently of the chosen PIP. This finding indicates that only a minor fraction of CB2SH3+ is still capable of binding to the membrane. The majority remains in an inactive state not capable of membrane binding. This is in line with the proposed “closed” and thus autoinhibited conformation of CB2SH3+ (6).
FIGURE 7.
Binding isotherms of CB2SH3+ (red circles) and CB2SH3+/W24A/E262A (green circles) obtained for different lipid compositions. A, POPC/PI(3)P (9:1); B, POPC/PI(4)P (9:1); C, POPC/PI(3,4)P2 (9:1); D, POPC/PI(3,5)P2 (9:1); E, POPC/PI(4,5)P2 (9:1); F, POPC/PI(3,4,5)P3. For each isotherm, at least three independent RIfS experiments were performed, and the data represent the mean values. The solid lines are results of fitting Equation 1 to the data. The obtained KD values are summarized in Table 2.
Partial Activation of CB2SH3+
Point mutations in CB2SH3+ (e.g. CB2SH3+/W24A/E262A) that perturb the intramolecular interactions within CB2SH3+ relieve autoinhibition, leading to an increase in CB-triggered gephyrin clustering in COS7 cells, CB−/− neurons, and neuroligin-2−/− neurons, indicating that the corresponding mutant adopts an open conformation with an increased capacity to bind to phosphoinositide-containing membranes (6). We tested whether CB2SH3+/W24A/E262A is also capable of binding to PIP-containing membranes in vitro (Fig. 7). Indeed, a significant increase in protein layer thickness for the mutant CB2SH3+/W24A/E262A compared with wild-type CB2SH3+ was observed for the monophosphorylated PIPs and for PI(3,5)P2 and PI(3,4,5)P3. These results indicate that a larger fraction of CB2SH3+/W24A/E262A is in an open conformation and capable of binding to phosphoinositides with submicromolar to micromolar affinities as compared with wild-type CB2SH3+. Consistent with our previous results, no binding to PI(3,4)P2 was found. However, for PI(4,5)P2 (Fig. 7E), also no binding of CB2SH3+/W24A/E262A was observed. Importantly, the CB2SH3+/W24A/E262A variant is partially selective for certain phosphoinositides with a preference for monophosphorylated phosphoinositols (PI(3)P and PI(4)P) and for PI(3,5)P2.
Discussion
Based on an in vitro phospholipid bilayer membrane system, the present study provides important new insights into the specificity of CB2 binding to phosphoinositides and elucidates the mechanism of autoinhibition of CB2 at the molecular level. We made use of solid-supported planar bilayers doped with different phosphoinositides to provide a membrane system that partially resembles, but at the same time simplifies, the natural situation at cellular membranes. It allowed for a quantitative analysis of the binding properties of CB2 in a label-free manner by means of reflectometric interference spectroscopy. Previous studies on the specificity of CB used pure phosphoinositides spotted on synthetic membranes (protein-lipid overlay assays) (6, 12, 13). In these assays, phosphoinositides are not embedded in a lipid membrane, and thus the headgroup positions of the phosphorylated inositols are not defined and aligned, which can alter the binding specificity (14).
Here we first tested the specificity of binding of the PH domain of CB2 to different phosphoinositides, which is of key relevance as the PH domain of CB2 is functionally essential (5, 12, 36). In general, PH domains are best known for their ability to bind phosphoinositides and to be targeted to cellular membranes (10, 37). Most lipid-binding PH domains show a preference for one or several phosphoinositides, such as the PH domain of phospholipase C-δ1, which binds specifically to PI(4,5)P2 (38). For the isolated PH domain of CB2, the dissociation constants for the phosphoinositides tested were in the 0.1–0.8 μm range with an up to 8 times higher affinity for monophosphorylated PI(3)P and PI(4)P. In the literature, quite a large range of KD values can be found for PH domain binding to phosphoinositides, ranging from about 30 μm in the case of the interaction of the pleckstrin PH domain with PI(4,5)P2 (7) to 10 nm values for the PH domains of phospholipase C-δ1 and Grp1-PH (39, 40). Another important aspect in this regard is the binding capacity of the membrane as a function of different phosphoinositides. We used 10 mol % of the corresponding receptor lipid, which is roughly 10 times higher than the concentrations of phosphoinositides found in native cellular membranes and was chosen to optimize the signal-to-noise ratio in our assay. If the receptor lipids were homogeneously distributed in the POPC membranes and assuming a 1:1 stoichiometry of PIP-CB2 binding as has been shown for other PH domain-containing proteins (37, 41, 42), a lower phosphoinositide concentration should have been sufficient. However, we found that a larger PIP concentration was required to obtain a good signal-to-noise ratio as the overall protein surface coverage was, depending on the phosphoinositide, rather low. Assuming that at 10 mol % the maximum protein surface is reached (43), the maximum protein layer thickness obtained by RIfS experiments is a measure of the binding capacity of the membrane. Interestingly, a large protein layer thickness for CB2PH was obtained for phosphoinositides with a phosphate group at the 5-position with the largest dprotein-max found for the most strongly phosphorylated form, PI(3,4,5)P3. The monophosphorylated phosphoinositols showed a lower binding capacity. This might be explained by a non-homogeneous, clustered distribution of the receptor lipids prior to and after protein binding (17), respectively, resulting in more than one phosphoinositide bound to CB and would require further investigations.
The submicromolar affinity of the isolated PH domain of CB2 was also seen with the DH/PH tandem domain of CB2. The slightly lower binding affinity of CB2SH3− for monophosphorylated phosphoinositols as compared with the isolated PH domain of CB2 is likely due to the multiple interactions between the PH and DH domains, which influence the interactions between the DH/PH domains and membrane lipids (13, 32). The maximum protein layer thickness was largest for PI(4,5)P2 and PI(3,4,5)P3, which is similar to the results obtained for CB2PH and fits to the dimensions estimated from the crystal structure of CB2SH3− (long axis) (6). Based on the results obtained with POPC membranes containing either phosphatidylinositol or phosphatidylserine, we conclude that the molecular recognition of the OH groups of the inositol headgroup and the specific phosphorylation state are more relevant for the specific recognition of CB than an interaction driven purely by electrostatics.
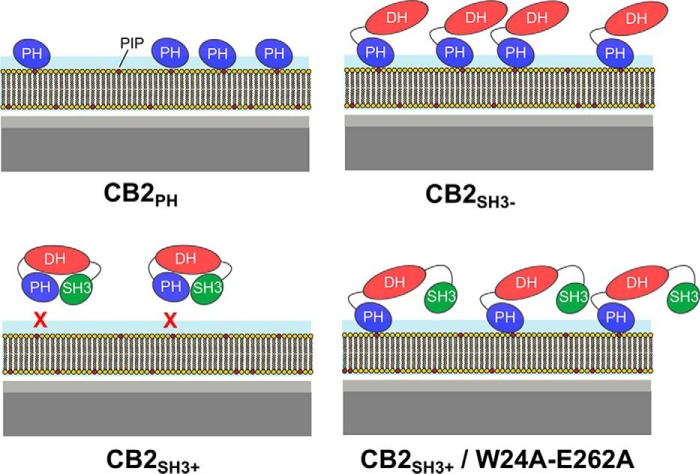
In mouse brain, CB2SH3− exists only in trace amounts, and the majority of CB isoforms contain an additional SH3 domain (6). This SH3 domain causes CB to adopt a closed and autoinhibited conformation in which the SH3 domain interacts with the DH/PH tandem domain (6), thus preventing its membrane recruitment (Fig. 8). To analyze the phosphoinositide binding characteristics of the biologically relevant CB variants, i.e. the ones with an N-terminal SH3 domain, we studied CB2SH3+ and a constitutively activated variant in which the autoinhibitory effect of the SH3 domain is eliminated (CB2SH3+/W24A/E262A). With CB2SH3+, the amount of bound protein is greatly diminished as compared with CB2PH and CB2SH3−, independently of the phosphoinositide used. Taking into account the more compact structure of CB2SH3+, which has a smaller radius of gyration (Rg = 26.3 Å) than CB2SH3− (Rg = 28.3 Å) (6), the protein size does not explain this significant decrease in protein layer thickness. Instead, the results indicate that a large fraction of CB2SH3+ is not capable of binding to the PIP-containing membranes. This provides key support for the notion that CB2SH3+ adopts an autoinhibited conformation that is stabilized by contacts between the SH3 domain and the DH/PH tandem domain (Fig. 8). Furthermore, our data show that a main consequence of the SH3 domain-mediated autoinhibition of CB is an inhibition of phosphoinositide binding and membrane tethering of CB (6), whereas the guanine nucleotide exchange factor activity does not seem to be affected (6, 32, 44, 45). This is different in Asef, the closest CB homologue, where the SH3 domain-mediated autoinhibition of Asef affects the enzymatic guanine nucleotide exchange factor activity of the DH domain (46).
FIGURE 8.
Schematic drawing of the proposed modes of protein binding. Proteins bind to solid-supported POPC membranes doped with 10 mol % of the respective PIP.
To further assess the autoinhibitory influence of the SH3 domain on phosphoinositide binding of CB2, we made use of the CB2SH3+/W24A/E262A variant in which two amino acids in the SH3-DH/PH interface are exchanged. The corresponding mutation leads to an open conformation exhibiting a larger flexibility and a more elongated protein shape in solution. In lipid overlay assays, the mutant protein binds more strongly to PI(3)P as compared with wild-type CB2SH3+ albeit not as well as CB2SH3− (6). A similar behavior of CB2SH3+/W24A/E262A was observed with solid-supported POPC membranes containing PI(3)P. Although the binding capacity of CB2SH3+ was estimated to be about 3 times lower than that of CB2SH3− (Fig. 7A), it was regained to about 90% in the case of the CB2SH3+/W24A/E262A mutant. Interestingly, this regain in activity is a function of the phosphoinositide. Although the binding capacity of the mutant for the monophosphorylated phosphoinositols and for PI(3,5)P2 is large, it remains partially diminished for the other PIPs, and the mutant protein does not significantly bind to phosphoinositides with two juxtaposed phosphate groups, i.e. PI(3,4)P2 and PI(4,5)P2.
The present results provide the first insights into the phosphoinositide binding specificity of CB in a phospholipid bilayer context. Of most biological relevance are the data on CB2SH3+ and its open and disinhibited CB2SH3+/W24A/E262A variant because essentially all CB isoforms expressed in murine brain carry an N-terminal SH3 domain (6). Our corresponding data nicely corroborate the notion that CB2SH3+ is autoinhibited with respect to phosphoinositide binding and that this autoinhibition is relieved upon introduction of the W24A/E262A mutation, which “opens” the CB2SH3+ conformation and exposes the PH domain for proper lipid binding. In neurons expressing wild-type CB, this conformational activation of phosphoinositide binding of CB is mediated by neuroligin-2 (47), TC10 (44), and neuroligin-4 (48), likely along with other interactors of the SH3 and PH domains of CB, to promote CB-dependent gephyrin clustering at nascent GABAergic synapses. Furthermore, our data show that CB2SH3+ can bind to several phosphoinositide variants but show a significant preference for PI(3)P and PI(4)P in bilayer membranes. This partial selectivity, which has been observed with other assay systems as well (6, 12, 13, 36), is also seen with the isolated PH domain of CB (CB2PH) but only to a smaller degree with the N-terminally truncated CB2SH3− variant, which lacks the SH3 domain. In view of these findings and given that neurons almost exclusively express SH3 domain-containing CB isoforms, future studies on the regulation of CB function will have to focus on PI(3)P and PI(4)P signaling toward CB.
Author Contributions
M. L. and J. S. isolated the proteins and performed the RIfS experiments. D. S. and H. S. designed the protein constructs and helped with protein purification. T. S. and T. P. performed lipid overlay assays and critically reviewed the manuscript. C. S. and N. B. designed the experiments and wrote the manuscript. All authors reviewed the results and approved the final version of the manuscript.
This work was supported by the Max Planck Society (to N. B.), the German Research Foundation (a Center of Nanoscale Microscopy and Molecular Physiology of the Brain grant (to N. B.) and Grants PA 2087/1-1 (to T. P.) and SCHI 425/8-1 (to H. S.)) as well as funding through the Rudolf Virchow Center for Experimental Biomedicine (to H. S.), and European Commission Innovative Medicines Initiative FP7-115300 (to N. B.). The authors declare that they have no conflicts of interest with the contents of this article.
- CB
- collybistin
- DH
- Dbl homology
- OT
- optical thickness
- PH
- pleckstrin homology
- PIP
- phosphatidylinositol phosphate
- POPC
- 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine
- POPG
- 1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-(1′-racemic glycerol)
- POPS
- 1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-l-serine
- RIfS
- reflectometric interference spectroscopy
- SH3
- Src homology 3
- PI(3,4,5)P3
- phosphatidylinositol 3,4,5-trisphosphate
- PI(3)P
- phosphatidylinositol 3-monophosphate
- PI(3,4)P2
- phosphatidylinositol 3,4-bisphosphate
- PI(4,5)P2
- phosphatidylinositol 4,5-bisphosphate
- PI(4)P
- phosphatidylinositol 4-monophosphate
- PI(3,5)P2
- phosphatidylinositol 3,5-bisphosphate
- SUV
- small unilamellar vesicle.
References
- 1.Kneussel M., and Betz H. (2000) Clustering of inhibitory neurotransmitter receptors at developing postsynaptic sites: the membrane activation mode. Trends Neurosci. 23, 429–435 [DOI] [PubMed] [Google Scholar]
- 2.Kneussel M., and Betz H. (2000) Receptors, gephyrin and gephyrin-associated proteins: novel insights into the assembly of inhibitory postsynaptic membrane specializations. J. Physiol. 525, 1–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Papadopoulos T., and Soykan T. (2011) The role of collybistin in gephyrin clustering at inhibitory synapses: facts and open questions. Front. Cell. Neurosci. 5, 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Papadopoulos T., Korte M., Eulenburg V., Kubota H., Retiounskaia M., Harvey R. J., Harvey K., O'Sullivan G. A., Laube B., Hülsmann S., Geiger J. R., and Betz H. (2007) Impaired GABAergic transmission and altered hippocampal synaptic plasticity in collybistin-deficient mice. EMBO J. 26, 3888–3899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Harvey K., Duguid I. C., Alldred M. J., Beatty S. E., Ward H., Keep N. H., Lingenfelter S. E., Pearce B. R., Lundgren J., Owen M. J., Smart T. G., Lüscher B., Rees M. I., and Harvey R. J. (2004) The GDP-GTP exchange factor collybistin: an essential determinant of neuronal gephyrin clustering. J. Neurosci. 24, 5816–5826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Soykan T., Schneeberger D., Tria G., Buechner C., Bader N., Svergun D., Tessmer I., Poulopoulos A., Papadopoulos T., Varoqueaux F., Schindelin H., and Brose N. (2014) A conformational switch in collybistin determines the differentiation of inhibitory postsynapses. EMBO J. 33, 2113–2133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kavran J. M., Klein D. E., Lee A., Falasca M., Isakoff S. J., Skolnik E. Y., and Lemmon M. A. (1998) Specificity and promiscuity in phosphoinositide binding by pleckstrin homology domains. J. Biol. Chem. 273, 30497–30508 [DOI] [PubMed] [Google Scholar]
- 8.Ferguson K. M., Kavran J. M., Sankaran V. G., Fournier E., Isakoff S. J., Skolnik E. Y., and Lemmon M. A. (2000) Structural basis for discrimination of 3-phosphoinositides by pleckstrin homology domains. Mol. Cell 6, 373–384 [DOI] [PubMed] [Google Scholar]
- 9.Lietzke S. E., Bose S., Cronin T., Klarlund J., Chawla A., Czech M. P., and Lambright D. G. (2000) Structural basis of 3-phosphoinositide recognition by pleckstrin homology domains. Mol. Cell 6, 385–394 [DOI] [PubMed] [Google Scholar]
- 10.Lemmon M. A. (2007) Pleckstrin homology (PH) domains and phosphoinositides. Biochem. Soc. Symp. 74, 81–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dowler S., Currie R. A., Campbell D. G., Deak M., Kular G., Downes C. P., and Alessi D. R. (2000) Identification of pleckstrin-homology-domain-containing proteins with novel phosphoinositide-binding specificity. Biochem. J. 351, 19–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kalscheuer V. M., Musante L., Fang C., Hoffmann K., Fuchs C., Carta E., Deas E., Venkateswarlu K., Menzel C., Ullmann R., Tommerup N., Dalprà L., Tzschach A., Selicorni A., Lüscher B., Ropers H. H., Harvey K., and Harvey R. J. (2009) A balanced chromosomal translocation disrupting ARHGEF9 is associated with epilepsy, anxiety, aggression, and mental retardation. Hum. Mutat. 30, 61–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Papadopoulos T., Schemm R., Grubmüller H., and Brose N. (2015) Lipid binding defects and perturbed synaptogenic activity of a collybistin R290H mutant that causes epilepsy and intellectual disability. J. Biol. Chem. 290, 8256–8270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Busse R. A., Scacioc A., Hernandez J. M., Krick R., Stephan M., Janshoff A., Thumm M., and Kühnel K. (2013) Qualitative and quantitative characterization of protein-phosphoinositide interactions with liposome-based methods. Autophagy 9, 770–777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Di Paolo G., and De Camilli P. (2006) Phosphoinositides in cell regulation and membrane dynamics. Nature 443, 651–657 [DOI] [PubMed] [Google Scholar]
- 16.Lodhi I. J., Bridges D., Chiang S. H., Zhang Y., Cheng A., Geletka L. M., Weisman L. S., and Saltiel A. R. (2008) Insulin stimulates phosphatidylinositol 3-phosphate production via the activation of Rab5. Mol. Biol. Cell 19, 2718–2728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Saarikangas J., Zhao H., and Lappalainen P. (2010) Regulation of the actin cytoskeleton-plasma membrane interplay by phosphoinositides. Physiol. Rev. 90, 259–289 [DOI] [PubMed] [Google Scholar]
- 18.Vicinanza M., D'Angelo G., Di Campli A., and De Matteis M. A. (2008) Function and dysfunction of the PI system in membrane trafficking. EMBO J. 27, 2457–2470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gauglitz G., Brecht A., Kraus G., and Nahm W. (1993) Chemical and biochemical sensors based on interferometry at thin (multi-)layers. Sens. Actuators B Chem. 11, 21–27 [Google Scholar]
- 20.Roth G., Freund S., Möhrle B., Wöllner K., Brünjes J., Gauglitz G., Wiesmüller K. H., and Jung G. (2007) Ubiquitin binds to a short peptide segment of hydrolase UCH-L3: a study by FCS, RIfS, ITC and NMR. Chembiochem 8, 323–331 [DOI] [PubMed] [Google Scholar]
- 21.Stephan M., Kramer C., Steinem C., and Janshoff A. (2014) Binding assay for low molecular weight analytes based on reflectometry of absorbing molecules in porous substrates. Analyst 139, 1987–1992 [DOI] [PubMed] [Google Scholar]
- 22.Krick R., Busse R. A., Scacioc A., Stephan M., Janshoff A., Thumm M., and Kühnel K. (2012) Structural and functional characterization of the two phosphoinositide binding sites of PROPPINs, a β-propeller protein family. Proc. Natl. Acad. Sci. U.S.A. 109, E2042–E2029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Braunger J. A., Kramer C., Morick D., and Steinem C. (2013) Solid supported membranes doped with PIP2: influence of ionic strength and pH on bilayer formation and membrane organization. Langmuir 29, 14204–14213 [DOI] [PubMed] [Google Scholar]
- 24.Vilardi F., Stephan M., Clancy A., Janshoff A., and Schwappach B. (2014) WRB and CAML are necessary and sufficient to mediate tail-anchored protein targeting to the ER membrane. PLoS One 9, e85033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schütte O. M., Patalag L. J., Weber L. M., Ries A., Römer W., Werz D. B., and Steinem C. (2015) 2-Hydroxy fatty acid enantiomers of Gb3 impact Shiga toxin binding and membrane organization. Biophys. J. 108, 2775–2778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Proll G., Markovic G., Steinle L., and Gauglitz G. (2009) in Biosensors and Biodetection (Rasooly A., and Herold K., eds) pp. 167–178, Humana Press, New York [Google Scholar]
- 27.Sreerama N., and Woody R. W. (2000) Estimation of protein secondary structure from circular dichroism spectra: comparison of CONTIN, SELCON, and CDSSTR methods with an expanded reference set. Anal. Biochem. 287, 252–260 [DOI] [PubMed] [Google Scholar]
- 28.Fernandes F., Loura L. M., Fedorov A., and Prieto M. (2006) Absence of clustering of phosphatidylinositol-(4,5)-bisphosphate in fluid phosphatidylcholine. J. Lipid Res. 47, 1521–1525 [DOI] [PubMed] [Google Scholar]
- 29.Larson I., and Attard P. (2000) Surface charge of silver iodide and several metal oxides. Are all surfaces Nernstian? J. Colloid Interface Sci. 227, 152–163 [DOI] [PubMed] [Google Scholar]
- 30.Kučerka N., Nieh M.-P., and Katsaras J. (2011) Fluid phase lipid areas and bilayer thicknesses of commonly used phosphatidylcholines as a function of temperature. Biochim. Biophys. Acta 1808, 2761–2771 [DOI] [PubMed] [Google Scholar]
- 31.Lupyan D., Mezei M., Logothetis D. E., and Osman R. (2010) A molecular dynamics investigation of lipid bilayer perturbation by PIP2. Biophys. J. 98, 240–247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xiang S., Kim E. Y., Connelly J. J., Nassar N., Kirsch J., Winking J., Schwarz G., and Schindelin H. (2006) The crystal structure of Cdc42 in complex with collybistin II, a gephyrin-interacting guanine nucleotide exchange factor. J. Mol. Biol. 359, 35–46 [DOI] [PubMed] [Google Scholar]
- 33.Herrig A., Janke M., Austermann J., Gerke V., Janshoff A., and Steinem C. (2006) Cooperative adsorption of ezrin on PIP2-containing membranes. Biochemistry 45, 13025–13034 [DOI] [PubMed] [Google Scholar]
- 34.Latour R. A. (2015) The Langmuir isotherm: a commonly applied but misleading approach for the analysis of protein adsorption behavior. J. Biomed. Mater. Res. A 103, 949–958 [DOI] [PubMed] [Google Scholar]
- 35.Faiss S., Kastl K., Janshoff A., and Steinem C. (2008) Formation of irreversibly bound annexin A1 protein domains on POPC/POPS solid supported membranes. Biochim. Biophys. Acta 1778, 1601–1610 [DOI] [PubMed] [Google Scholar]
- 36.Reddy-Alla S., Schmitt B., Birkenfeld J., Eulenburg V., Dutertre S., Böhringer C., Götz M., Betz H., and Papadopoulos T. (2010) PH-domain-driven targeting of collybistin but not Cdc42 activation is required for synaptic gephyrin clustering. Eur. J. Neurosci. 31, 1173–1184 [DOI] [PubMed] [Google Scholar]
- 37.Hyvönen M., Macias M. J., Nilges M., Oschkinat H., Saraste M., and Wilmanns M. (1995) Structure of the binding site for inositol phosphates in a PH domain. EMBO J. 14, 4676–4685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lemmon M. A., Ferguson K. M., O'Brien R., Sigler P. B., and Schlessinger J. (1995) Specific and high-affinity binding of inositol phosphates to an isolated pleckstrin homology domain. Proc. Natl. Acad. Sci. U.S.A. 92, 10472–10476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ferguson C. G., James R. D., Bigman C. S., Shepard D. A., Abdiche Y., Katsamba P. S., Myszka D. G., and Prestwich G. D. (2005) Phosphoinositide-containing polymerized liposomes: stable membrane-mimetic vesicles for protein-lipid binding analysis. Bioconjug. Chem. 16, 1475–1483 [DOI] [PubMed] [Google Scholar]
- 40.Knight J. D., and Falke J. J. (2009) Single-molecule fluorescence studies of a PH domain: new insights into the membrane docking reaction. Biophys. J. 96, 566–582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Garcia P., Gupta R., Shah S., Morris A. J., Rudge S. A., Scarlata S., Petrova V., McLaughlin S., and Rebecchi M. J. (1995) The pleckstrin homology domain of phospholipase C-δ1 binds with high affinity to phosphatidylinositol 4,5-bisphosphate in bilayer membranes. Biochemistry 34, 16228–16234 [DOI] [PubMed] [Google Scholar]
- 42.Rameh L. E., Arvidsson A.-k., Carraway K. L. 3rd, Couvillon A. D., Rathbun G., Crompton A., VanRenterghem B., Czech M. P., Ravichandran K. S., Burakoff S. J., Wang D. S., Chen C. S., and Cantley L. C. (1997) A comparative analysis of the phosphoinositide binding specificity of pleckstrin homology domains. J. Biol. Chem. 272, 22059–22066 [DOI] [PubMed] [Google Scholar]
- 43.Janke M., Herrig A., Austermann J., Gerke V., Steinem C., and Janshoff A. (2008) Actin binding of ezrin is activated by specific recognition of PIP2-functionalized lipid bilayers. Biochemistry 47, 3762–3769 [DOI] [PubMed] [Google Scholar]
- 44.Mayer S., Kumar R., Jaiswal M., Soykan T., Ahmadian M. R., Brose N., Betz H., Rhee J. S., and Papadopoulos T. (2013) Collybistin activation by GTP-TC10 enhances postsynaptic gephyrin clustering and hippocampal GABAergic neurotransmission. Proc. Natl. Acad. Sci. U.S.A. 110, 20795–20800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tyagarajan S. K., Ghosh H., Harvey K., and Fritschy J. M. (2011) Collybistin splice variants differentially interact with gephyrin and Cdc42 to regulate gephyrin clustering at GABAergic synapses. J. Cell Sci. 124, 2786–2796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mitin N., Betts L., Yohe M. E., Der C. J., Sondek J., and Rossman K. L. (2007) Release of autoinhibition of ASEF by APC leads to CDC42 activation and tumor suppression. Nat. Struct. Mol. Biol. 14, 814–823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Poulopoulos A., Aramuni G., Meyer G., Soykan T., Hoon M., Papadopoulos T., Zhang M., Paarmann I., Fuchs C., Harvey K., Jedlicka P., Schwarzacher S. W., Betz H., Harvey R. J., Brose N., Zhang W., and Varoqueaux F. (2009) Neuroligin 2 drives postsynaptic assembly at perisomatic inhibitory synapses through gephyrin and collybistin. Neuron 63, 628–642 [DOI] [PubMed] [Google Scholar]
- 48.Hoon M., Soykan T., Falkenburger B., Hammer M., Patrizi A., Schmidt K. F., Sassoè-Pognetto M., Löwel S., Moser T., Taschenberger H., Brose N., and Varoqueaux F. (2011) Neuroligin-4 is localized to glycinergic postsynapses and regulates inhibition in the retina. Proc. Natl. Acad. Sci. U.S.A. 108, 3053–3058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kong A. M., Horan K. A., Sriratana A., Bailey C. G., Collyer L. J., Nandurkar H. H., Shisheva A., Layton M. J., Rasko J. E., Rowe T., and Mitchell C. A. (2006) Phosphatidylinositol 3-phosphate [PtdIns3P] is generated at the plasma membrane by an inositol polyphosphate 5-phosphatase: endogenous PtdIns3P can promote GLUT4 translocation to the plasma membrane. Mol. Cell. Biol. 26, 6065–6081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Maffucci T., Brancaccio A., Piccolo E., Stein R. C., and Falasca M. (2003) Insulin induces phosphatidylinositol-3-phosphate formation through TC10 activation. EMBO J. 22, 4178–4189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Falasca M., and Maffucci T. (2007) Role of class II phosphoinositide 3-kinase in cell signalling. Biochem. Soc. Trans. 35, 211–214 [DOI] [PubMed] [Google Scholar]
- 52.Nakatsu F., Baskin J. M., Chung J., Tanner L. B., Shui G., Lee S. Y., Pirruccello M., Hao M., Ingolia N. T., Wenk M. R., and De Camilli P. (2012) PtdIns4P synthesis by PI4KIIIα at the plasma membrane and its impact on plasma membrane identity. J. Cell Biol. 199, 1003–1016 [DOI] [PMC free article] [PubMed] [Google Scholar]