Abstract
Calcium flux through store-operated calcium entry is a central regulator of intracellular calcium signaling. The two key components of the store-operated calcium release-activated calcium channel are the Ca2+-sensing protein stromal interaction molecule 1 (STIM1) and the channel pore-forming protein Orai1. During store-operated calcium entry activation, calcium depletion from the endoplasmic reticulum triggers a series of conformational changes in STIM1 that unmask a minimal Orai1-activating domain (CRAC activation region (CAD)). To gate Orai1 channels, the exposed STIM1-activating domain binds to two sites in Orai1, one in the N terminus and one in the C terminus. Whether the two sites operate as distinct binding domains or cooperate in CAD binding is unknown. In this study, we show that the N and C-terminal domains of Orai1 synergistically contribute to the interaction with STIM1 and couple STIM1 binding with channel gating and modulation of ion selectivity.
Keywords: calcium channel, calcium release-activated calcium channel protein 1 (ORAI1), gating, ion channel, stromal interaction molecule 1 (STIM1)
Introduction
Store-operated calcium entry represents a key mechanism by which cells generate Ca2+ signals and maintain Ca2+ homeostasis by replacing Ca2+ lost from the endoplasmic reticulum (ER)2 with Ca2+ that enters the cytoplasm through plasma membrane channels. The Ca2+ release-activated Ca2+ (CRAC) channel is a prototypical store-operated calcium entry channel whose essential components are STIM1, the Ca2+ sensor of the ER, and Orai1, the CRAC channel pore-forming subunit. STIM1 is a single-pass ER membrane protein with several functional domains, including three coiled-coil domains (CC1, CC2, and CC3) facing the cytosol, with CC2 and CC3 forming part of a minimal CRAC channel activation domain called CAD (1) or STIM-Orai activating region (SOAR) (2) (spanning residues 339–448 in hSTIM1, referred to hereafter only as CAD). Each Orai1 subunit has four transmembrane segments, with the N and C termini of the protein facing the cytosol, and Orai1 channels are assembled from hexamers of Orai1 subunits. The activation of CRAC channels starts when depletion of calcium from the ER causes rearrangement of STIM1 and unmasking of CAD (3–8). When exposed, CAD binds to a site in the Orai1 N terminus (N-terminal binding domain (NBD), residues 74–87) and to a second site in the Orai1 C terminus (C-terminal binding domain (CBD), residues 267–292) (1, 9). This interaction results in clustering of STIM-Orai1 at ER-PM (plasma membrane) junctions and in pore opening of Orai1 channels, which mediate calcium influx into cells (10–17). We and others have shown recently that STIM1 binding to both the Orai1 NBD and CBD is critical for channel activation and that binding of STIM1 at these N- and C-terminal domains of Orai1 likely induces rearrangements in proximal membrane segments to open the channel (18–20). Whether the Orai1 NBD and CBD contribute to STIM1 binding independently or through a more complex manner is unknown.
In this study, we determine the relative contribution of the Orai1 NBD and CBD to interaction with STIM1 by studying the effect of cycling mutations between the NBD and CBD on STIM1-Orai1 physical interaction and on channel gating. Our results reveal that STIM1 binding to the NBD and CBD of Orai1 occurs in a cooperative manner to control the gating and ion selectivity of CRAC channels.
Materials and Methods
Cell Culture and Transfection
HEK293 cells were cultured in DMEM as described previously (21). Plasmid transfection of cells was performed using Lipofectamine 2000 (Invitrogen) according to the protocol of the manufacturer. For electrophysiological experiments, 6–8 h after plasmid transfection and 12–15 h before the start of experiments, cells were plated onto 18-mm coverglass coated with l-polylysine. For Ca2+ imaging, co-localization, and FRET experiments, cells were plated onto 18-mm coverglass coated with l-polylysine, and, 6–8 h after plasmid transfection, culture medium was replaced to wash off the transfection reagent. To avoid a constitutive calcium rise in cells expressing all forms of Orai1-SS or V102A mutants, cells were cultured in high-glucose, Ca2+-free DMEM supplemented with 50 μm La3+.
Electrophysiological Recordings
Membrane currents were recorded under voltage clamp conditions using the whole-cell patch clamp configuration on an Axopatch 200B amplifier (Axon Instruments). Patch pipettes were fabricated from borosilicate glass capillaries (5–8 MΩ). Signals were analog-filtered using a 2-kHz low-pass Bessel filter. Data acquisition and analysis were performed using pCLAMP 10 software (Axon Instruments). Voltage protocols consisted of a 100-ms ramp from −100 to +100 mV delivered every 1 or 4 s from a holding potential of 0 mV. Current densities were calculated by normalizing currents measured at −90 mV to cell capacitance. The internal solution contained 150 mm cesium aspartate, 8 mm MgCl2, 8 mm BAPTA, and 10 mm HEPES (pH 7.2 with CsOH). External Ringer's solution contained 145 mm NaCl, 2.8 mm KCl, 10 mm HEPES, and 10 mm glucose (pH 7.4 with NaOH), and either 10 mm CaCl or 10 mm MgCl2 was added to the external solution for high-Ca2+ or Ca-free solution, respectively. STIM1 activation was induced by passive store depletion, and currents measured from cells co-expressing STIM1 and Orai1 normally reached maximum within 2 min from cell break-in. Whole-cell recordings in cells co-expressing STIM1 with mutant Orai1 channels were therefore performed for a period of 4–10 min to ensure sufficient time for STIM-Orai1 activation. All data were leak-corrected using the current elicited in high Ca2+ Ringer's solution supplemented with 10–100 μm La3+ or in Ca2+ free Ringer's solution, as appropriate.
Quantitation of Expression Levels for the Orai1 Constructs Used in This Work
For quantitation of relative expression of Orai1 constructs, we relied on fluorescence emitted by a tagged fluorescent protein (EGFP) as a quantitative reporter of cDNA expression (22). Images of cells expressing the indicated Orai1-SS constructs were collected with identical confocal settings. Images from untransfected cells (control) were used to assess background fluorescence values subsequently used to establish a threshold. All fluorescent pixels with more than five times the threshold value were recorded and normalized to the pixel area in the analyzed field.
Measurements of Intracellular Calcium Levels
Transfected cells grown on glass coverslips were incubated for 30 min with 5 μm Fura-2/AM (Molecular Probes, Life Technologies) in standard Ringer's solution (140 mm NaCl, 10 mm HEPES, 10 mm glucose, 0.8 mm MgCl, 2.8 mm KCl, and 2 mm CaCl (pH 7.4)) and allowed to equilibrate for an additional 5–10 min without dye. Following dye loading, coverslips were mounted in a chamber, and emission (510-nm-long pass) images were collected during alternate excitation at 350 ± 5 nm and 380 ± 5 nm using a filter wheel (Lambda-10, Sutter Instruments). The imaging system consisted of a Nikon Diaphot inverted microscope equipped with a ×20 objective and charge-coupled device camera (SenSys, Photometrics). Axon Imaging Workbench 4.0 (Axon Instruments) controlled both filters and collection of data.
Co-localization Analysis
Cells co-expressing mCherry-Orai1 or various mCherry Orai1 mutants together with EGFP-S1C (an hSTIM1 fragment corresponding to residues 343–465) were plated and cultured as described above. Midplane sections of transfected cells were captured using an LSM 780 confocal microscope (Zeiss) controlled by Zen imaging software (Zeiss). EGFP was excited at 488 nm, and emission was collected from 493–540 nm, and mCherry was excited at 561 nm, and emission was collected from 583–685 nm. The parameters used for image acquisition were kept constant across all sets of experiments, and analysis was restricted to cells with similar EGFP and mCherry fluorescence to ensure similar ratios of S1C/Orai1 expression. Line scans of regions spanning the extracellular, plasma membrane and cytosol regions were analyzed from individual images using ImageJ. Each line had a total scan width of 36 pixels, corresponding to a length of 5 μm. The fluorescence intensity along each individual scan was normalized to the peak value. Finally, normalized line scans of both mCherry-Orai1 and EGFP-S1C from individual cells were aligned according to the peak maximum of mCherry-Orai1 to produce cellular intensity plots.
Acceptor Photobleaching FRET
Cells co-expressing mCherry-Orai1 or various mCherry-Orai1 mutants, together with EGFP-S1C, were plated and cultured as described above. Cells were fixed in 4% fresh paraformaldehyde for 30 min and washed three times for 10 min with PBS. Cells were subsequently imaged in PBS on a LSM 780 confocal microscope (Zeiss). The donor (EGFP) was excited at 488 nm, and emission was collected from 493–524 nm, and the acceptor (mCherry) was excited at 561 nm, and emission was collected from 583–685 nm. EGFP-CAD donor fluorescence was imaged before and after bleaching a region of interest of mCherry-Orai1. Following bleaching, mCherry-Orai1 fluorescence was decreased to <10% of its initial intensity. FRET energy transfer (FRETefficiency) was calculated as % (FRETefficiency) = 100 × (Dpost − Dpre)/Dpost, where Dpre and Dpost are the donor (EGFP) intensities before and after acceptor (mCherry) bleaching, respectively.
Generation of Plasmids
The STIM1-GFP construct has been described previously (21), and S1C (the fragment corresponding to residues 342–465 of hSTIM1) cDNA was PCR-amplified from this construct and inserted into p3XFLAG 7.1 in-frame with an existing cDNA coding for mCherry to create mCherry-S1C. EGFP-S1C was created by insertion of the S1C fragment from mCherry-S1C into EGFP-C1 using BsrGI-BamHI sites. The mCherry-hOrai1 construct was obtained from the laboratory of Prof. Shmuel Muallem (National Institutes of Health). The Orai1-SS-EGFP and Orai1-S constructs were obtained from the laboratory of Prof. Tao Xu (Chinese Academy of Sciences, Beijing, China). All point mutations were created by standard long-range PCR reaction, and all plasmids were sent to the DNA Sequencing Facility at University of California Berkeley for verification of the modified plasmid regions.
Statistical Analysis
Statistical significance of data was calculated using one-way analysis of variance with Bonferroni correction (Kaleidagraph, Synergy Software) when comparing three or more dataset groups and unpaired two-tailed Student's t test when comparing two dataset groups (Microsoft Excel 2010). Predicted parameters for linear additive effects of double mutants were calculated by first scaling the measured parameters of each single mutant to the corresponding control. The resultant single mutation parameters were then multiplied pairwise to generate values for the relevant combinations of single mutations. To calculate the standard deviation of these predicted parameters, we employed standard propagation of error analysis using the standard deviation values derived from each single mutant dataset. The MSD (Mean, Size, Standard Deviation) format of the Student's t test analysis (SigmaPlot 11, Systat) was used to evaluate significance between measured and predicted parameters for double mutants. All data are reported as mean ± S.E.
Results
Cooperativeness of the N- and C-terminal Domains of Orai1 Determines Channel Gating by S1C
In vitro binding assays have shown that C-terminal fragments of STIM1, harboring the CAD domain, can bind to synthetic fragments of either the NBD or CBD of Orai1 (1, 9, 23–25). Cell-based binding assays, however, showed that, although the NBD contributes to CAD-Orai1 interaction, it does not interact with CAD when key residues in the CBD are deleted (19). This has been interpreted to indicate that the NBD-CAD interaction is of such low affinity that it cannot be detected. We wondered whether this observation could, instead, reflect a more complex interaction of CAD with the NBD and CBD in which it is more than a sum of NBD and CBD interactions.
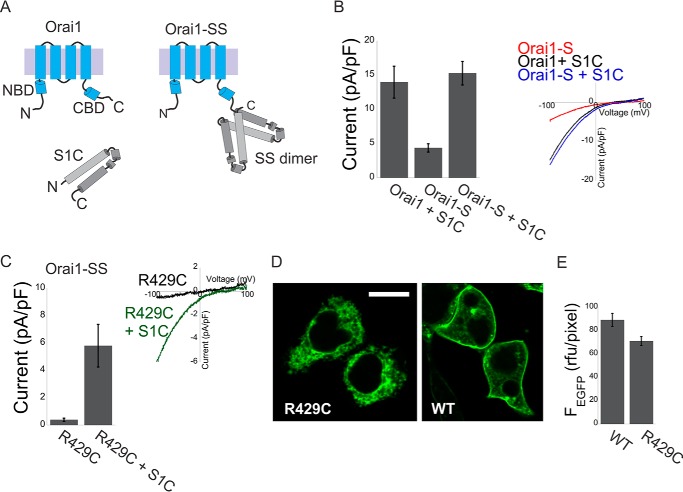
To investigate the functional interaction between the NBD and CBD, we introduced point mutations into the NBD, the CBD, or both and analyzed the effect on channel activation. Because several mutations in the Orai1 CBD abolish STIM1 binding, we forced a high local concentration of CAD near the channel by employing an earlier approach in which dimers of STIM1 C-terminal fragments (residues 340–485, S domain) are directly linked to Orai1 (Fig. 1A and Refs. 18, 19, 27, 28). Recent studies have suggested that pairs of Orai1 CBD from neighboring subunits interact in a conformation that is optimal for STIM1 binding. This raises the concern that the Orai1 CBD may be distorted simply by the covalent attachment of the STIM1 fragments. To verify that the Orai1-SS constructs represent a reliable framework to study the nature of interactions between STIM1 and Orai1, we asked whether tethering the STIM1 fragments to the Orai1 C terminus would prevent binding of untethered STIM1 fragments. Fig. 1 shows that tethering of the STIM1 fragment to the Orai1 C terminus does not prevent functional interaction between Orai1 and untethered STIM1 fragments. Despite its poor membrane expression, Orai1-SS R429C, which carries the loss-of-function mutation R429C in both of the tethered STIM1 fragments, is activated when co-expressed with a soluble STIM1 C-terminal fragment that is similar to CAD (S1C, residues 342–465 in hSTIM1; Fig. 1, C–E). Similarly, the Orai1 S channel, which displays submaximal channel activation when expressed alone, is maximally activated when co-expressed with the soluble S1C fragment (Fig. 1B). These results suggest that the CBD of Orai1-SS retains the ability to bind to and be activated by STIM1.
FIGURE 1.
Untethered STIM1 fragments interact with Orai1-S or with the R429C Orai1-SS mutant. A, schematic of Orai1, the SOAR/CAD-like S1C fragment, and the Orai1-SS channels used in this work. B, summary of current densities (n = 5–8 cells, left panel) and representative plots of the current-voltage relationship (right panel) of currents recorded from cells expressing Orai1-S alone or from cells co-expressing S1C together with Orai1 or Orai1-S. pF, picofarad. C, summary of current densities (n = 5 cells, left panel) and representative plots of the current-voltage relationship (right panel) recorded from cells expressing Orai1-SS with the loss-of-function mutation R429C alone or together with WT S1C (left panel). D and E, representative fluorescence images (D) and summary of fluorescence densities (E) measured from cells expressing the WT or the R429C Orai1-SS-EGFP mutant (n = 6 regions in each). rfu, relative fluorescence units. Scale bar = 10 μm.
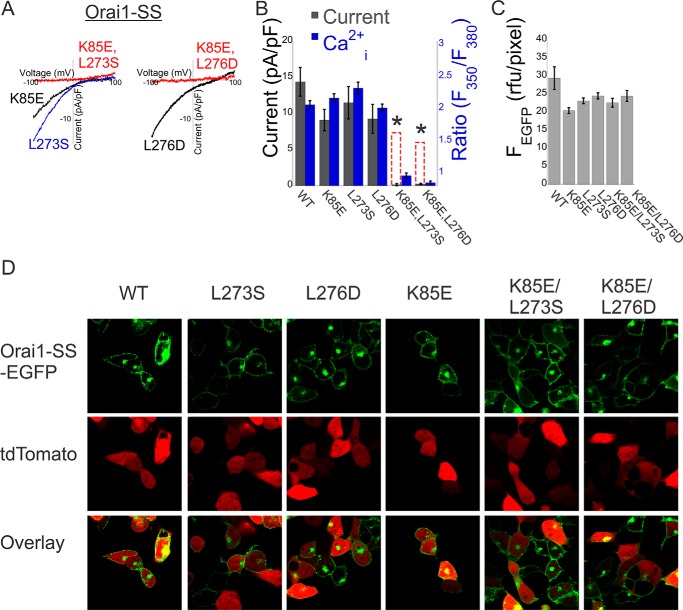
Having seen that STIM1 binding and activation are retained in the STIM1-tethered Orai1, we proceeded to ask how mutations in Orai1 that compromise activation by STIM1 interact in combination. We analyzed three mutants of Orai1 that are not activated by soluble STIM1 but retain activation by tethered STIM1, indicating that they weaken interaction with STIM1: K85E in the NBD and L276D and L273S in the CBD (data not shown and Refs. 18, 19, 27). We reasoned that if each interaction contributed separately to STIM1 binding, then the effect of NBD/CBD double mutants would reflect the sum of the effects of the single mutations (Fig. 2B, dashed red columns) but that a cooperative process would result in a deviation from linearity. We found that even though the single mutants had normal or almost normal channel activation in Orai1-SS, as assessed either by the density of membrane current or by intracellular Ca2+ levels, channel activation was abolished in both double NBD-CBD mutants (K85E/L273S or K85E/L276D) (Figs. 2B). Expression profiles of the single and double mutants were comparable with that of WT channels (Fig. 2, C and D), and the conductance of the double mutant channels could be restored by the constitutively activating V102A mutation (Fig. 4), indicating that the effect of the mutations was on their ability to be activated by CAD and not on their expression levels or their ability to conduct large currents when the gate is constitutively open. These supralinear effects therefore suggest that the Orai1 NBD and CBD operate synergistically during channel activation by CAD.
FIGURE 2.
Cooperative interaction between STIM1 fragments and the N and C termini of Orai1 controls channel activation. A, representative plots of the current-voltage relationship of currents recorded from cells expressing the indicated single or double Orai1-SS mutants. pF, picofarad. B, summary of current densities (n = 5–8 cells) or basal Ca2+ levels (n = 25–35 cells) recorded from cells expressing Orai1-SS with the indicated single or double mutations. Dashed red columns show the predicted additive effect for the double mutants. C and D, quantitation of expression levels for the Orai1 constructs used in this work. C, summary of fluorescence densities measured for the indicated Orai1-SS-EGFP mutants (n = 6 regions in each). D, representative fluorescence images of cells co-expressing the indicated Orai1-SS-EGFP mutant together with td-Tomato (as a marker for the cytosol and nucleus regions). *, p < 0.05.
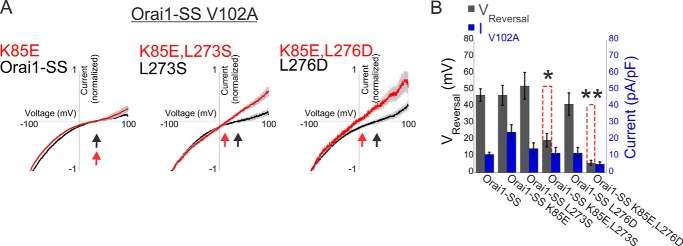
FIGURE 4.
Cooperative interaction between STIM1 fragments and the N and C termini of Orai1 controls regulation of ion selectivity. A, average of normalized plots of the current-voltage relationship recorded in 10 mm Ca2+ Ringer's solution from cells expressing the indicated double or triple Orai1-SS mutants. Arrows indicate the reversal potential in each instance. B, summary of reversal potentials and current densities recorded from cells expressing the indicated Orai1-SS mutants (n = 5–6 cells). Dashed red columns show the predicted additive effect for the double mutants. *, p < 0.05; **, p < 0.01. pF, picofarad.
Cooperativeness of the N- and C-terminal Domains of Orai1 Determines Channel Gating by Full-length STIM1
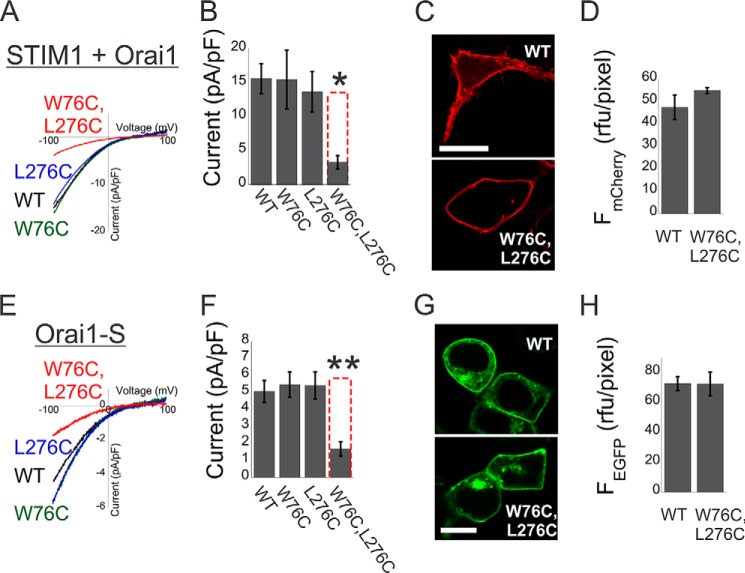
Having observed non-linearity in the effect of mutations on the activation of Orai1 by CADs that are directly fused to the channel, we next searched for mutations that would enable us to ask whether normal coupling to full-length STIM1 has the same properties. Previous studies (18, 19, 24, 29) have shown that NBD or CBD point or deletion mutations at positions Trp-76 or Leu-276, respectively, introduce mild to strong effects on channel activation and physical interaction between Orai1 and full-length unlinked STIM1. We therefore examined the effect of combining mutations in these positions on activation of Orai1 by STIM1 or by tethered STIM1 fragment (Orai1-S). We recorded similar current densities from cells that co-expressed STIM1 together with WT Orai1 or with the individual Orai1 mutants W76C or L276C (Fig. 3, A and B). Similarly, cells that expressed WT Orai1-S or Orai1-S carrying the W76C or L276C mutations displayed similar current densities. Although all WT and mutant Orai1 or Orai1-S displayed similar expression levels and plasma membrane localization (Figs. 3, C, D, G, and H, and 4A), a significant decrease in current density was measured for the double NBD/CBD mutant Orai1-S W76C/L276C or when STIM1 was co-expressed with the Orai1 W76C/L276C (Fig. 3, A and B). Therefore, non-linear interaction with the Orai1 NBD and CBD is not only a characteristic of activation by tethered CAD fragments but also of normal activation by full-length STIM1.
FIGURE 3.
Cooperative interaction between STIM1 and the N and C termini of Orai1 controls channel activation. A, representative plots of the current-voltage relationship of currents recorded from cells expressing STIM1 with mCherry-Orai1 or with the indicated mCherry-Orai1 mutants. pF, picofarad. B, summary of current densities recorded from cells expressing STIM1 with the indicated single or double mCherry-Orai1 mutations (n = 10–15 cells). C and D, representative fluorescence images (C) and summary of fluorescence densities (D) measured from cells expressing the WT or the W76C/L276C Orai1 double mutant (n = 6 regions in each). E, representative plots of the current-voltage relationship of currents recorded from cells expressing WT Orai1-S or Orai1-S with the indicated Orai1 mutation. F, summary of current densities recorded from cells expressing Orai1-S with the indicated single or double mutations (n = 6–9 cells). G and H, representative fluorescence images (G) and summary of fluorescence densities (H) measured from cells expressing the WT Orai1-S or the W76C/L276C Orai1-S double mutant (n = 6 regions in each). Dashed red columns show the predicted additive effect for the double mutants. Scale bars = 10 μm. *, p < 0.05; **, p < 0.01.
Cooperativeness of the N- and C-terminal Domains of Orai1 Determines the Regulation of Ion Selectivity in V102A Orai1-SS Channels
Activation of Orai1 channels by STIM1 has been shown earlier to occur in a stepwise manner that involves a non-linear dependence on the number of STIM1s that bind to the channel (27, 30), leading to the proposal that Orai1 subunits operate cooperatively (30). Having observed a non-linear interaction in channel activation by STIM1 or its C-terminal fragment S1C with the NBD and CBD of Orai1, we wondered whether cooperativity was restricted to the activation process. We therefore asked whether the regulation of ion selectivity in constitutively active channels follows the same rules. We analyzed the ion selectivity of V102A Orai1-SS mutants that contain the NBD point mutation K85E, the CBD point mutations L273S or L276D, or the double NBD/CBD mutations K85E/L273S or K85E/L276D by quantifying the reversal potentials of mutant channel currents in a calcium-containing solution. Reversal potentials recorded from cells expressing V102A Orai1-SS channels with wild-type NBD/CBD or with a single mutation in either the NBD (K85E) or CBD (L273S or L276D) were of similar values of about 40–50 mV (Fig. 4, A and B). In contrast, even when directly linked to CAD, the double NBD/CBD mutations (K85/L273S and K85/L276D) displayed non-selective currents with a reversal potential of 19.5 ± 4 mV for K85E/L273S and 6.2 ± 1.2 mV for K85E/L276D. These findings indicate that, as shown above for gating, the regulation of ion selectivity of constitutively active Orai1 channels by STIM1 also involves non-linear interaction with the NBD and CBD of Orai1.
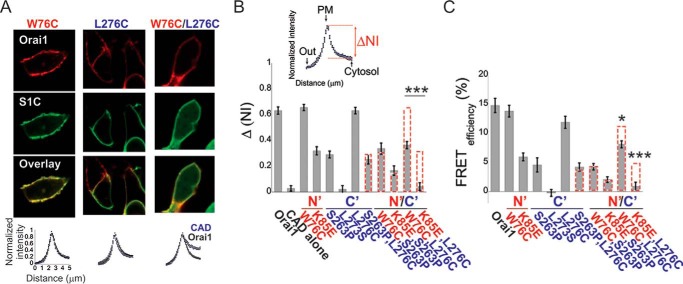
An Ensemble of Orai1 NBD and CBD Forms a Distinct Interaction Site for S1C
Having studied functional coupling between S1C and the Orai1 NBD and CBD, we turned to investigate their molecular association. We expressed WT Orai1 or Orai1 with mutations in either the NBD (W76C or K85E), TM4-CBD linker region (S263P), CBD (L276C), or with combination NBD/linker/CBD mutations (W76C/S263P, K85E/S263P, W76C/L276C, or K85E/L276C) and performed co-localization and FRET analyses. We found that two of the single mutants, W76C (NBD) and L276C (CBD), did not significantly change the interaction with S1C, whereas the K85E (NBD) mutation and S263P (TM4-CBD linker) moderately weakened this interaction (Fig. 5, A–C). Combining either NBD mutation W76C or K85E or CBD mutation L276C with the linker mutation S263P yielded an additive reduction in S1C-Orai1 association in each double mutant. In contrast, the double mutant Orai1 W76C/L276C, which combined an NBD mutant with a CBD mutant that individually had no significant effect, exhibited a substantially reduced S1C-Orai1 association (Fig. 5, A–C). Similarly, interaction was nullified in the Orai1 K85E/L276C double mutant (Fig. 5B-C). The results were also compared with the co-localization and FRET values predicted for the additive effects of these combined mutations (Fig. 5, B and C, dashed red columns). Therefore, just as we observed supralinear effects on gating and on regulation of ion selectivity of combining NBD and CBD mutations, as above, so too did combining NBD and CBD mutations have a supralinear effect on association with S1C.
FIGURE 5.
Cooperative binding of S1C to the N and C termini of Orai1. A and B, representative fluorescence images (top panel) and summary of normalized intensity values (bottom panel) across the plasma membrane and intracellular regions of cells expressing EGFP-S1C together with the indicated mCherry-Orai1 mutants (n = 12–22 cells). B, quantitative summary of EGFP-S1C fraction (ΔNI = NIPM − NICyto, see inset) that is localized to the plasma membrane (PM). C, summary of FRET efficiency values measured in cells expressing EGFP-S1C together with mCherry-Orai1 or with the indicated single or double mCherry-Orai1 mutants (n = 15–35 cells). Dashed red columns show the predicted additive effect for the double mutants. *, p < 0.05; ***, p < 0.001.
Discussion
Our finding that STIM1-Orai1 protein association, channel activation, as well as modulation of ion selectivity are governed by cooperative interactions between STIM1 and the Orai1 N- and C-terminal regions sheds new light on important aspects of CRAC channel gating and permeation mechanisms. First, it implies that cooperativity in Orai1 channel activation by STIM1 begins in the binding step and is imparted to the subsequent gating and permeation processes. Second, it provides new evidence to support directs roles for both the Orai1 N- and C-terminal domains in channel gating. Importantly, they also suggest that CRAC channel activation involves simultaneous binding of STIM1 to both the Orai1 N- and C-terminal domains. Third, although alternative scenarios can be considered, as detailed below, taken together with earlier reports (1, 13, 18, 19, 22), the findings from this work support the idea that the Orai1 NBD and CBD assemble to form a distinct binding site for STIM1 and that STIM1 binding to this site controls channel gating and modulation of ion selectivity.
A recent NMR structure of the STIM1 CC2 and the Orai1 CBD regions added structural insights on the interacting residues within the CAD-CBD complex (25). Notably, several of these residues have been shown to play important roles in channel activation. The exact regions in the Orai1 NBD and the corresponding residues in CAD that contribute to NBD-CAD interactions remain, however, poorly defined. Although our results suggest that CAD directly interacts with the Orai1 NBD through interactions with Trp-76 and Lys-85, we cannot rule out other interpretations. First, through allosteric coupling between the Orai1 NBD and CBD, a conformational change in the NBD, induced by the mutations employed in this work, may affect the affinity of the CBD for CAD, and second, the Orai1 NBD may indirectly contribute to communication with CAD through interactions with additional regulatory components, like calmodulin (31) or CRACR2A (26), that have been shown previously to involve the implicated NBD residues Trp-76 and Lys-85, respectively. Ultimately, further structural work is needed to shed light on and resolve this fundamental issue.
Author Contributions
R. P and E. Y. I. designed the research and wrote the paper, and R. P. performed the research and analyzed the data.
Acknowledgments
We thank Cherise Stanley and Holly Aaron (University of California Berkeley Molecular Imaging Center) for technical assistance.
This work was supported by American Heart Association Postdoctoral Fellowship 13POST14000008 (to R. P.) and National Institutes of Health Grant R01 NS35549 (to E. Y. I.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
- ER
- endoplasmic reticulum
- CRAC
- Ca2+ release-activated Ca2+
- CC
- coiled coil
- NBD
- N-terminal binding domain
- CBD
- C-terminal binding domain
- CAD
- CRAC activation region.
References
- 1.Park C. Y., Hoover P. J., Mullins F. M., Bachhawat P., Covington E. D., Raunser S., Walz T., Garcia K. C., Dolmetsch R. E., and Lewis R. S. (2009) STIM1 clusters and activates CRAC channels via direct binding of a cytosolic domain to Orai1. Cell 136, 876–890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yuan J. P., Zeng W., Dorwart M. R., Choi Y.-J., Worley P. F., and Muallem S. (2009) SOAR and the polybasic STIM1 domains gate and regulate Orai channels. Nat. Cell Biol. 11, 337–343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Luik R. M., Wang B., Prakriya M., Wu M. M., and Lewis R. S. (2008) Oligomerization of STIM1 couples ER calcium depletion to CRAC channel activation. Nature 454, 538–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhou Y., Srinivasan P., Razavi S., Seymour S., Meraner P., Gudlur A., Stathopulos P. B., Ikura M., Rao A., and Hogan P. G. (2013) Initial activation of STIM1, the regulator of store-operated calcium entry. Nat. Struct. Mol. Biol. 20, 973–981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Covington E. D., Wu M. M., and Lewis R. S. (2010) Essential role for the CRAC activation domain in store-dependent oligomerization of STIM1. Mol. Biol. Cell 21, 1897–1907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yu F., Sun L., Courjaret R., and Machaca K. (2011) Role of the STIM1 C-terminal domain in STIM1 clustering. J. Biol. Chem. 286, 8375–8384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yu F., Sun L., Hubrack S., Selvaraj S., and Machaca K. (2013) Intramolecular shielding maintains the ER Ca2+ sensor STIM1 in an inactive conformation. J. Cell Sci. 126, 2401–2410 [DOI] [PubMed] [Google Scholar]
- 8.Muik M., Fahrner M., Schindl R., Stathopulos P., Frischauf I., Derler I., Plenk P., Lackner B., Groschner K., Ikura M., and Romanin C. (2011) STIM1 couples to ORAI1 via an intramolecular transition into an extended conformation. EMBO J. 30, 1678–1689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhou Y., Meraner P., Kwon H. T., Machnes D., Oh-hora M., Zimmer J., Huang Y., Stura A., Rao A., and Hogan P. G. (2010) STIM1 gates the store-operated calcium channel ORAI1 in vitro. Nat. Struct. Mol. Biol. 17, 112–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liou J., Kim M. L., Heo W. D., Jones J. T., Myers J. W., Ferrell J. E. Jr., and Meyer T. (2005) STIM is a Ca2+ sensor essential for Ca2+-store-depletion-triggered Ca2+ influx. Curr. Biol. CB. 15, 1235–1241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wu M. M., Buchanan J., Luik R. M., and Lewis R. S. (2006) Ca2+ store depletion causes STIM1 to accumulate in ER regions closely associated with the plasma membrane. J. Cell Biol. 174, 803–813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Prakriya M., Feske S., Gwack Y., Srikanth S., Rao A., and Hogan P. G. (2006) Orai1 is an essential pore subunit of the CRAC channel. Nature 443, 230–233 [DOI] [PubMed] [Google Scholar]
- 13.McNally B. A., Somasundaram A., Yamashita M., and Prakriya M. (2012) Gated regulation of CRAC channel ion selectivity by STIM1. Nature 482, 241–245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Luik R. M., Wu M. M., Buchanan J., and Lewis R. S. (2006) The elementary unit of store-operated Ca2+ entry: local activation of CRAC channels by STIM1 at ER-plasma membrane junctions. J. Cell Biol. 174, 815–825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xu P., Lu J., Li Z., Yu X., Chen L., and Xu T. (2006) Aggregation of STIM1 underneath the plasma membrane induces clustering of Orai1. Biochem. Biophys. Res. Commun. 350, 969–976 [DOI] [PubMed] [Google Scholar]
- 16.Sharma S., Quintana A., Findlay G. M., Mettlen M., Baust B., Jain M., Nilsson R., Rao A., and Hogan P. G. (2013) An siRNA screen for NFAT activation identifies septins as coordinators of store-operated Ca2+ entry. Nature 499, 238–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lioudyno M. I., Kozak J. A., Penna A., Safrina O., Zhang S. L., Sen D., Roos J., Stauderman K. A., and Cahalan M. D. (2008) Orai1 and STIM1 move to the immunological synapse and are up-regulated during T cell activation. Proc. Natl. Acad. Sci. U.S.A. 105, 2011–2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Palty R., Stanley C., and Isacoff E. Y. (2015) Critical role for Orai1 C-terminal domain and TM4 in CRAC channel gating. Cell Res. 25, 963–980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McNally B. A., Somasundaram A., Jairaman A., Yamashita M., and Prakriya M. (2013) The C- and N-terminal STIM1 binding sites on Orai1 are required for both trapping and gating CRAC channels. J. Physiol. 591, 2833–2850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gudlur A., Quintana A., Zhou Y., Hirve N., Mahapatra S., and Hogan P. G. (2014) STIM1 triggers a gating rearrangement at the extracellular mouth of the ORAI1 channel. Nat. Commun. 10.1038/ncomms6164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Palty R., Raveh A., Kaminsky I., Meller R., and Reuveny E. (2012) SARAF inactivates the store operated calcium entry machinery to prevent excess calcium refilling. Cell 149, 425–438 [DOI] [PubMed] [Google Scholar]
- 22.Soboleski M. R., Oaks J., and Halford W. P. (2005) Green fluorescent protein is a quantitative reporter of gene expression in individual eukaryotic cells. FASEB J. 19, 440–442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Muik M., Frischauf I., Derler I., Fahrner M., Bergsmann J., Eder P., Schindl R., Hesch C., Polzinger B., Fritsch R., Kahr H., Madl J., Gruber H., Groschner K., and Romanin C. (2008) Dynamic coupling of the putative coiled-coil domain of ORAI1 with STIM1 mediates ORAI1 channel activation. J. Biol. Chem. 283, 8014–8022 [DOI] [PubMed] [Google Scholar]
- 24.Derler I., Plenk P., Fahrner M., Muik M., Jardin I., Schindl R., Gruber H. J., Groschner K., and Romanin C. (2013) The extended transmembrane Orai1 N-terminal (ETON) region combines binding interface and gate for Orai1 activation by STIM1. J. Biol. Chem. 288, 29025–29034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stathopulos P. B., Schindl R., Fahrner M., Zheng L., Gasmi-Seabrook G. M., Muik M., Romanin C., and Ikura M. (2013) STIM1/Orai1 coiled-coil interplay in the regulation of store-operated calcium entry. Nat. Commun. 10.1038/ncomms3963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Srikanth S., Jung H.-J., Kim K.-D., Souda P., Whitelegge J., and Gwack Y. (2010) A novel EF-hand protein, CRACR2A, is a cytosolic Ca2+ sensor that stabilizes CRAC channels in T cells. Nat. Cell Biol. 12, 436–446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li Z., Liu L., Deng Y., Ji W., Du W., Xu P., Chen L., and Xu T. (2011) Graded activation of CRAC channel by binding of different numbers of STIM1 to Orai1 subunits. Cell Res. 21, 305–315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zheng H., Zhou M.-H., Hu C., Kuo E., Peng X., Hu J., Kuo L., and Zhang S. L. (2013) Differential roles of the C and N termini of Orai1 protein in interacting with stromal interaction molecule 1 (STIM1) for Ca2+ release-activated Ca2+ (CRAC) channel activation. J. Biol. Chem. 288, 11263–11272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Navarro-Borelly L., Somasundaram A., Yamashita M., Ren D., Miller R. J., and Prakriya M. (2008) STIM1-Orai1 interactions and Orai1 conformational changes revealed by live-cell FRET microscopy. J. Physiol. 586, 5383–5401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hoover P. J., and Lewis R. S. (2011) Stoichiometric requirements for trapping and gating of Ca2+ release-activated Ca2+ (CRAC) channels by stromal interaction molecule 1 (STIM1). Proc. Natl. Acad. Sci. 108, 13299–13304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mullins F. M., Park C. Y., Dolmetsch R. E., and Lewis R. S. (2009) STIM1 and calmodulin interact with Orai1 to induce Ca2+-dependent inactivation of CRAC channels. Proc. Natl. Acad. Sci. U.S.A. 106, 15495–15500 [DOI] [PMC free article] [PubMed] [Google Scholar]