Abstract
Light-driven ion-pumping rhodopsins are widely distributed in microorganisms and are now classified into the categories of outward H+ and Na+ pumps and an inward Cl− pump. These different types share a common protein architecture and utilize the photoisomerization of the same chromophore, retinal, to evoke photoreactions. Despite these similarities, successful pump-to-pump conversion had been confined to only the H+ pump bacteriorhodopsin, which was converted to a Cl− pump in 1995 by a single amino acid replacement. In this study we report the first success of the reverse conversion from a Cl− pump to a H+ pump. A novel microbial rhodopsin (MrHR) from the cyanobacterium Mastigocladopsis repens functions as a Cl− pump and belongs to a cluster that is far distant from the known Cl− pumps. With a single amino acid replacement, MrHR is converted to a H+ pump in which dissociable residues function almost completely in the H+ relay reactions. MrHR most likely evolved from a H+ pump, but it has not yet been highly optimized into a mature Cl− pump.
Keywords: bioenergetics, membrane protein, photobiology, photoreceptor, proton pump, microbial rhodopsin, retinal proteins
Introduction
Rhodopsins are ubiquitous membrane proteins that enable the cellular utilization of light as an information and energy source. According to their conserved residues, they are classified into two groups (1, 2). One is animal rhodopsin, represented by visual pigments in the eyes. The other is microbial rhodopsin, which shows divergent functions in unicellular microorganisms, such as light-driven ion pumps and light sensors for phototaxis and the regulation of gene expression. Different from animal rhodopsins, the microbial sensors represent a minority of the microbial rhodopsins. However, microbial sensors have individualities in their signal-transduction modes, which include interactions with other membrane proteins, interactions with cytoplasmic components, and light-gated ion channel activity.
Microbial rhodopsins were originally discovered in highly halophilic archaea in the early 1970s to 1980s (3). Since 1999 their homologues have begun to be identified in various microorganisms inhabiting a broad range of environments (4–6). It is now clear that in the microbial world, ion-pumping rhodopsins are abundant and widely distributed. In microorganisms, these ion pumps are probably the most convenient system for light energy utilization. The first and second ion pumps to be discovered were bacteriorhodopsin (BR)2 (7) and halorhodopsin (HR) (8) from halophilic archaea, which are an outward H+ pump and an inward Cl− pump, respectively. In 2000, an outward H+ pump named proteorhodopsin (PR) was discovered in a marine proteobacterium (9). Later, the PR homologues were identified in eubacteria living in the oceans worldwide (10). Recently, two groups of ion pumps, the outward Na+ pump and inward Cl− pump, were also discovered in eubacteria (11, 12). Their representatives are Krokinobacter eikastus rhodopsin 2 (KR2) (13) and Fulvimarina pelagi rhodopsin (FR) (14), respectively. The phylogenetic position of these ion pumps is shown in Fig. 1A. In contrast to their functional diversity, these ion pumps have essentially the same structural folds composed of seven helices and the chromophore retinal, which binds to a conserved lysine residue via a protonated Schiff base (PSB; deprotonated Schiff base is abbreviated as SB). In the dark, most ion-pumping rhodopsins dominantly contain retinal with the all-trans configuration, whereas some minorities such as BR and HR from archaeon Halobacterium salinarum (HsHR) can also accommodate 13-cis retinal (2). Regardless of this difference in the dark states, only the photoisomerization from all-trans to 13-cis can trigger the conformational changes for respective ion pumping functions. Thus, ion-pumping rhodopsins appear to share a common transport machinery where the transportable ions are probably determined by essential residues at appropriate positions. This scenario was partially demonstrated in 1995, when the H+ pump BR was converted to an inward Cl− pump by the single amino acid replacement of Asp-85 with Thr, the corresponding amino acid in HR (15). However, the success of pump-to-pump conversion was confined to this BR case. The reverse conversion, that is, from a Cl− pump to a H+ pump, has not been achieved (16–18) even after 10 mutations of HR (18). Here, we report a new class composed of an inward Cl− pump and its conversion to an outward H+ pump. Functional characterization was performed with a microbial rhodopsin from the cyanobacterium Mastigocladopsis repens, which was isolated from soil at Punta de la Mora, Tarragona in Spain. This microbial rhodopsin is designated as M. repens HR (MrHR) due to its similarities with HR. By a single amino acid replacement, MrHR begins to pump H+ outwardly. Thus, this is the first successful conversion from an inward Cl− pump to an outward H+ pump.
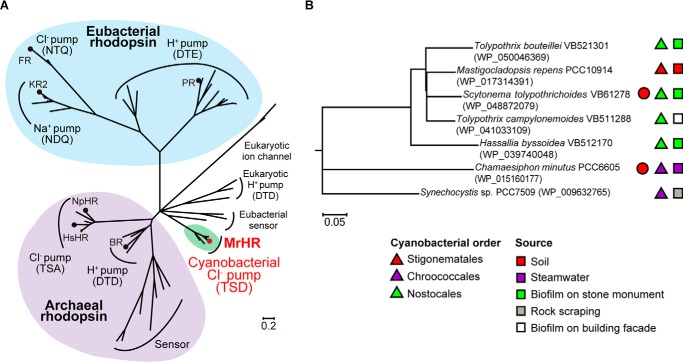
FIGURE 1.
Phylogenetic relationships of microbial rhodopsins. A, unrooted tree of microbial rhodopsins showing their functional diversity. The representative ion pumps are shown with the circles. Green is the clade consisting of the MrHR homologues. NpHR, HR from Natronomonas pharaonis. B, phylogenetic tree of the MrHR homologues. Their NCBI accession IDs are indicated in the parentheses. The strains labeled with closed circles also contain the Anabaena sensory rhodopsin gene. Other symbols indicate the cyanobacteria orders and the sources of the host strains. In both trees, nodes with <50% bootstrap percentages are collapsed.
Experimental Procedures
Phylogenetic Analysis
The 57-amino acid sequences were aligned using MUSCLE. All sequence data were obtained from the NCBI database. The phylogenetic tree was constructed by using the maximum likelihood method, with bootstrap percentages based on 1000 replications. Initial trees for the heuristic search were obtained by applying the neighbor-joining method (19). Evolutionary distances were calculated with the JTT matrix-based method (20). The branch lengths denote the number of amino acid substitutions. All analyses were performed using MEGA6.
Gene Preparation, Protein Expression, and Purification
Escherichia coli strain DH5α was used for DNA manipulation. An MrHR gene (NCBI accession ID: WP_017314391) with codons optimized for E. coli expression was chemically synthesized (Funakoshi, Japan) and inserted into the NdeI/XhoI site of the pET-21c(+) vector (Merck). This plasmid results in MrHR with additional amino acids in the C terminus (-LEHHHHHH). The T74D mutation was introduced using the QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, CA). The DNA sequences were confirmed by a standard method using an automated DNA sequencer (model 3100, Applied Biosystems, Foster City, CA). MrHR and the T74D mutant were expressed and purified from E. coli BL21(DE3) cells. The procedures were essentially the same as those previously described (21). The cells were grown at 37 °C in 2×YT medium supplemented with 50 μg/ml ampicillin. At the late exponential growth phase, the expression was induced by the addition of 1 mm isopropyl-β-d-thiogalactopyranoside in the presence of 10 μm all-trans retinal. After 3 h of induction, pink-colored cells were harvested by centrifugation (6,400 × g, 8 min at 4 °C) and washed once with buffer (50 mm Tris-HCl, pH 8.0) containing 5 mm MgCl2. Then the cells were broken with a French press (Ohtake, Tokyo, Japan) (100 megapascals × 4 times). After removing the undisrupted cells by centrifugation (5,600 × g, 10 min at 4 °C) the supernatant was ultracentrifuged (178,000 × g, 90 min at 4 °C). The collected cell membrane fraction was suspended with the same buffer containing 300 mm NaCl and 5 mm imidazole and was then solubilized with 1.5% n-dodecyl β-d-maltopyranoside (DDM) (Dojindo Lab, Kumamoto, Japan) at 4 °C overnight. After removal of the insoluble fraction by ultracentrifugation (178,000 × g, 60 min at 4 °C), the solubilized MrHR proteins were subjected to nickel-nitrilotriacetic acid-agarose (Qiagen, Hilden, Germany). The unbound proteins were removed by washing the column with 10 column volumes of wash buffer (50 mm sodium phosphate, pH 7.5) containing 400 mm NaCl, 50 mm imidazole, and 0.05% DDM. The bound protein was eluted with buffer (50 mm Tris-HCl, pH 7.0) containing 300 mm NaCl, 500 mm imidazole, and 0.1% DDM. The yield of MrHR was 15 mg from 1 liter of culture. The concentration was determined from the absorbance at 537 nm under an assumed extinction coefficient of 40,000 m−1cm−1. The purified samples were replaced with an appropriate buffer solution by passage over Sephadex G-25 in a PD-10 column (Amersham Bioscience).
Ion Pump Activity Measurements
The MrHR activity was measured in E. coli suspensions using a conventional pH electrode method (22) that detects the pH changes by the pump activity of H+ itself or passive H+ transfer in response to the membrane potential created by the pump activity for another ion. The E. coli suspensions were prepared as follows. The cells expressing MrHR were harvested at 3,600 × g for 5 min at 4 °C and washed twice with an unbuffered solution containing 200 mm salt (NaCl, choline chloride, NaBr, NaI, or NaNO3). They were resuspended in the same salt solution and gently shaken overnight at 4 °C in the presence of 10 μm carbonyl cyanide m-chlorophenylhydrazone (CCCP). Then the cells were washed twice with the same salt solution without CCCP and finally suspended at an A660 of 0.5. This cell density was ∼5% of the corresponding value for the original culture medium. As deduced from the purification yield, the original culture medium contained at least 15 μg/ml MrHR. Thus, the cell suspensions for the activity measurements contained at least 0.75 μg/ml MrHR. For the activation of MrHR, 530 ± 17.5 nm of green LED (LXHL-LM5C, Philips Lumileds Lighting Co., San Jose, CA) was used.
HPLC Analysis
The retinal configurations of MrHR were examined in both of the dark/light-adapted states. For dark adaptation, the MrHR samples were kept in the dark for 1 week in 10 mm MOPS, pH 6.5, containing 100 mm NaCl and 0.05% DDM. For light adaptation, the samples were irradiated for 2 min by green LED light as described above. MrHR underwent a prolonged photocycle as described below. To avoid the contamination of retinal in the photolysed state, the retinal oxime extraction was carried out after 1 min of incubation in the dark. The extraction and the following HPLC analysis was performed as previously described (22).
Absorption Spectra Measurements and Flash Photolysis
UV-visible spectra of MrHR samples were measured with a UV-1800 spectrometer (Shimadzu, Kyoto, Japan). Flash-induced absorbance changes were obtained in the 5-μs to 10-s time range on a single wavelength kinetic system. For the photoexcitation, the second harmonic (7 ns, 532 nm) of a Q-switched Nd:YAG laser (Surelite I-10, Continuum, Santa Clara, CA) was used. The details have been previously described (21). To improve the S/N ratio, 30 laser pulses were used at each measurement wavelength. All measurements were performed at 25 °C.
Results and Discussion
M. repens, isolated from the surface of soft powdery calcareous soil, is a blue-green alga belonging to the cyanobacterial order Stigonematales and was morphologically characterized according to the filaments it forms, which have many branches (23). In 2013 the existence of a gene encoding a microbial rhodopsin was revealed by whole genome sequencing of M. repens (24). We named this microbial rhodopsin “M. repens halorhodopsin (MrHR)” after its photochemical properties, described below. At present, six other homologues of MrHR are found in the NCBI database. Fig. 1A shows the phylogenetic tree of microbial rhodopsins, which revealed that MrHR homologues constitute a new clade that is distinct from the known microbial rhodopsins. The six homologues are also encoded in cyanobacteria and share 63–89% amino acid identities with MrHR. Meanwhile, the host strains belong to different cyanobacterial orders (Chroococcales and Nostocales) from M. repens, reflecting morphological differences. The phylogenetic relationships of MrHR homologues are shown in Fig. 1B together with the names, orders, and sources of their host strains. In addition to M. repens, all other genomes were sequenced (24–28). Cyanobacteria are a genetically diverse group and are widespread in fresh water, marine, terrestrial, and extreme environments such as hot springs and salt lakes. However, out of the seven strains of MrHR homologues, six were isolated from terrestrial environments such as soil and rock scrapings and biofilms on stone monuments and building facades (Fig. 1B) (23, 29, 30). Correspondingly, the desiccation resistance was experimentally confirmed for several strains (31). Two strains also contain the gene for Anabaena sensory rhodopsin, a putative sensor for chromatic adaptation that was originally discovered in the cyanobacterium Anabaena sp. PCC7120 (32). The other five strains do not contain other microbial rhodopsin genes.
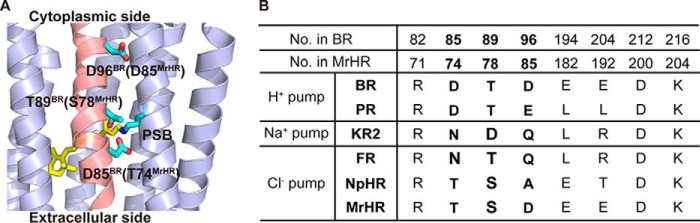
Microbial rhodopsins are largely categorized into ion pumps and sensors. All ion pumps are solely encoded in their respective operons in the genomes, whereas all sensors are encoded to be adjacent to the genes encoding the cognate transducers within the same operons. For MrHR homologues, their genes are solely encoded, implying that they function as ion pumps. Previous studies on ion pumps (12, 13) have indicated that three amino acids are responsible for determining the transportable ions (Fig. 2, A and B). For H+ pumps, a H+ from PSB is translocated during the photoreaction. This H+ is initially transferred to Asp-85 in BR (Asp-97 in PR), and SB subsequently accepts a H+ from Asp-96 in BR (Glu-108 in PR) (Fig. 2A). These residues are often referred to as the H+ acceptor and donor, respectively. For Cl− pumps, the acceptor is replaced with a Thr in HR and Asn in FR (Fig. 2B), which enables the binding of substrate Cl− as the counter ion of the PSB. Furthermore, for HR and KR2, another residue corresponding to Thr-89 in BR is known to play a crucial role (13, 21): these residues are Ser in HR and Asp in KR2, respectively (Fig. 2B). Thus, three residues (Asp-85, Thr-89, and Asp-96 in BR) are now designated as the “motif”: these are DTD and DTE for BR and PR, TSA for HR, and NTQ and NDQ for FR and KR2 (Fig. 2B). For MrHR, the motif is TSD (Fig. 2, A and B), which is close to TSA in HR. This implies that MrHR may pump Cl− inwardly despite a low amino acid identity of 22% for HR and 12% for FR. The identities for the H+ pumps are also low: 29% for BR and 17% for PR. However, the donor in the H+ pumps is conserved as Asp in MrHR, similar to BR(Asp) or PR(Glu). Furthermore, MrHR conserved Glu-194 and Glu-204 in BR, which consists of the H+ releasing complex to the extracellular medium (Fig. 2B). Thus, MrHR conserved the residues that are characteristic of both the Cl− pump (HR) and the H+ pump (BR and PR).
FIGURE 2.
A, positions of three amino acid residues consisting of the motif. X-ray crystal structure of BR (PDB code 1C3W) is shown from a view parallel to the membrane. The central part is magnified and the helix C is shown in pink. Three motif residues, Asp-85, Thr-89, and Asp-96, are shown in sticks together with the retinal bound to Lys-216 via PSB. The corresponding residues in MrHR, Thr-74, Ser-78, and Asp-85 are indicated in the parentheses. B, comparison of the important residues, especially the motifs among the ion pumps. NpHR, HR from Natronomonas pharaonis.
In this study we expressed MrHR in the cell membrane of E. coli and investigated the ion-pumping activity using the light-induced pH changes of the cell suspensions (Fig. 3). In a NaCl solution, light-induced alkalization was observed. This pH change was not abolished by the addition of the protonophore CCCP, which eliminates the electrochemical gradient of the proton (33). This means that the alkalization was not caused by the active proton transport but was caused by the passive proton influx in response to the interior negative membrane potential, which should be created by outward Na+ or inward Cl− translocation. In choline chloride, alkalization was also observed. Because choline is a large organic cation, microbial rhodopsin cannot transport it. This indicates that MrHR functions as a light-driven inward Cl− pump. The pump activity decreased in NaBr and disappeared in NaI and NaNO3. Thus, MrHR does not pump Na+ but does pump smaller anions. The transportable anions are severely restricted to only Cl− and Br−, different from HR and FR, which can even transport NO3− (14, 33).
FIGURE 3.
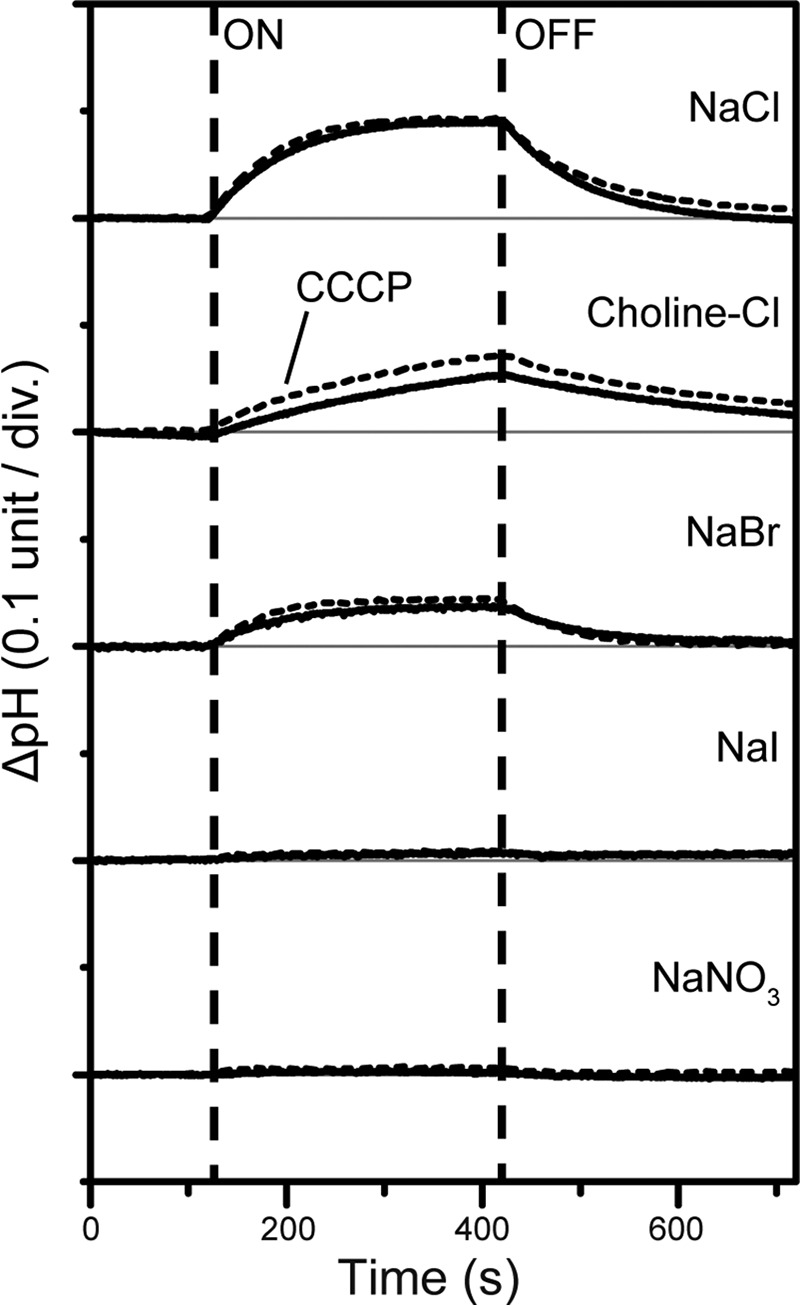
Light-induced pH changes by MrHR expressed in E. coli cells. The cells were suspended in a 200 mm salt solution without (solid lines) and with (broken lines) 10 μm CCCP. div., division.
The retinal isomer composition of MrHR was examined by HPLC analysis (Fig. 4). As described above, BR and HsHR in the dark states can accommodate both all-trans and 13-cis retinals and show so-called “light-dark adaptation” (2). In their light-adapted states after continuous illumination the unphotolysed states predominantly contain all-trans retinal. Upon dark adaptation, their 13-cis contents increased ∼50%. For MrHR (Fig. 4), the isomer composition did not depend upon the dark/light adaptation and was predominantly all-trans, similar to most ion-pumping rhodopsins. Thus, the all-trans retinal is responsible for the ion-pumping function of MrHR.
FIGURE 4.
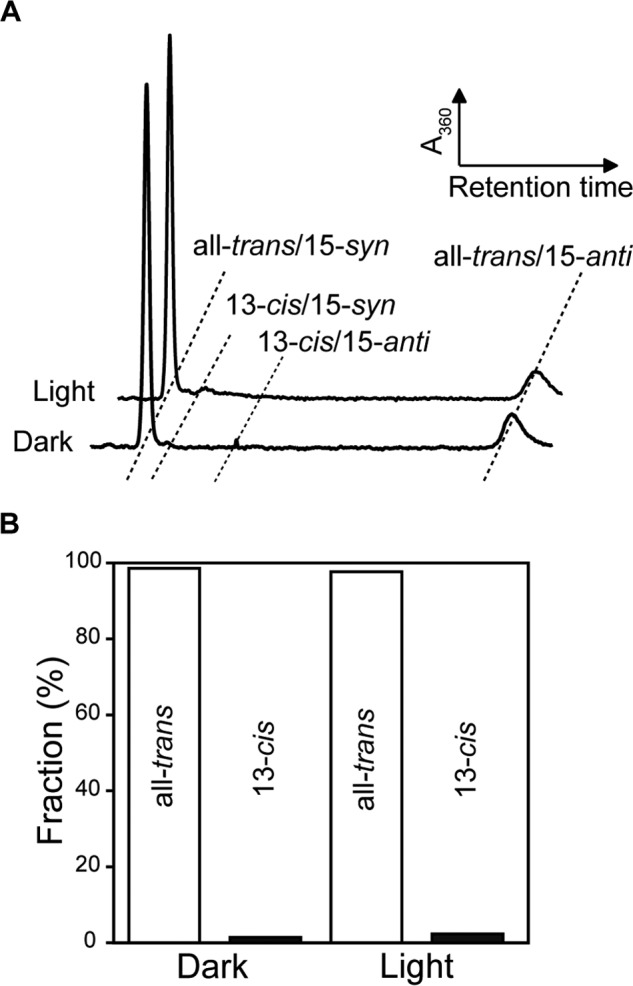
Retinal isomer compositions of MrHR in dark and light-adapted states. A, HPLC chromatographs of the retinal isomer extracted from MrHR. B, ratios of all-trans and 13-cis retinals determined from the peak areas. The all-trans configuration was dominant (> 98%), independent of the dark- and light-adapted states.
Anion binding near the PSB was monitored by the color change of retinal using purified MrHR (Fig. 5A). For other Cl− pumps, Cl− binding caused a blue shift (14, 34), except for HsHR (22). In contrast, a large red shift occurred for MrHR by its binding of Cl−, suggesting the differences in the Cl− binding site from other Cl− pumps. Similar red shifts were also observed for Br− and even for I− and NO3−. The dissociation constants (Kd) were determined from the absorbance changes at 550 nm (Fig. 5B), and the results are summarized in Table 1. The Kd values are close to those of HR, except for NO3− (171 mm); HR binds NO3− more strongly (Kd ∼ 16 mm) (35), whereas FR binds these ions more weakly (Kd = 40–130 mm) (14). These results indicate that MrHR can bind I− and NO3− but cannot transport them. These larger ions may be impossible to move toward a cytoplasmic half channel over the PSB region. Compared with HR and FR, the ion translocation pathway might be narrow and act as an ion-selective filter.
FIGURE 5.
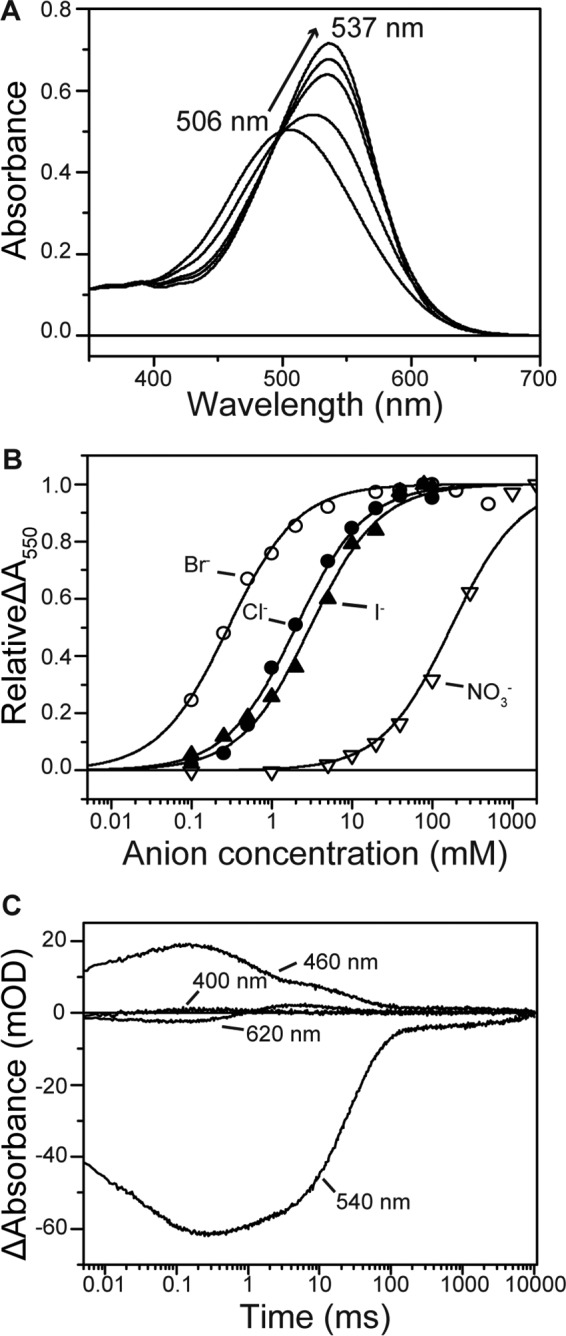
Spectroscopic characterization of MrHR. A, absorption spectral red shift by the Cl− binding to the unphotolysed state. The medium (pH 6.5) contained 33 mm Na2SO4, 0.05% DDM, and NaCl (0–100 mm). B, titration with various anions, which were added by sodium salts. The relative absorbance changes at 550 nm are plotted. C, flash-induced absorbance changes at selected wavelengths. The medium (pH 6.5) contained 0.05% DDM and 100 mm NaCl. mOD, milli-optical density.
TABLE 1.
Dissociation constants (Kd), absorption maxima (λmax), and their shifts (Δλmax) by the anion bindings
| Anions | Kda | λmaxb | Δλmaxc |
|---|---|---|---|
| mm | nm | nm | |
| Cl− | 1.99 ± 0.11 | 537 | 31 |
| Br− | 0.28 ± 0.08 | 538 | 32 |
| I− | 2.99 ± 0.21 | 535 | 29 |
| NO3− | 171 ± 20.6 | 532 | 26 |
a Kd values were determined by fitting analyses using Hill equation with n = 1. The best-fit curves are shown in Fig 5B.
b λmax values were the determined values at the following anion concentrations: Cl−, 0.5 m; Br−, 0.5 m; I−, 0.5 m; NO3−, 2 m. At these concentrations, the λmax shifts were almost saturated.
c Δλmax values denote the λmax shifts from the anion-free state (506 nm).
We also characterized the photocycle of MrHR by a flash photolysis method. Time-dependent absorbance changes at selected wavelengths are shown in Fig. 5C, which indicates that the photocycle of MrHR is very slow due to the prolonged decay of the last intermediate (540 nm). Ion-pumping rhodopsins transport one ion during a single turnover of photocycle and generally undergo fast photocycles (<100 ms), which enable the generation of a large transmembrane electrochemical potential under constant illumination. On the other hand, similar to HR and FR, no absorbance change at 400 nm was observed throughout the photocycle (14, 34, 36), indicating that there was no formation of an M intermediate, which is expected in H+ pumps and reflects the deprotonation of PSB. The observed intermediates and the order of their appearance resembles those of HR. These intermediates are (the estimated λmax are indicated in the parentheses): K (540 nm) → L (460 nm) → N (460 nm) + O (620 nm) → MrHR′ (540 nm). “N (460 nm) + O (620 nm)” means that the quasi-equilibrium between N and O appeared in the time period between 3 and 50 ms. A similar equilibrium was also observed for HR (34, 37). The last intermediate MrHR′ corresponded to HR′ in HR but had a substantially longer lifetime (∼6 s).
The slow photocycle of MrHR might reflect some differences in the physiological role from other Cl−-pumping rhodopsins. Although the details are not fully resolved, the Cl− pumps are believed to play roles in light-driven ATP production and/or maintaining the cellular osmotic balance during the volume increase of the growing cell (38–41). All host strains of MrHR homologues contain a photosynthetic apparatus. Thus, the slow photocycle of MrHR seems to contribute little to cellular ATP production under illumination. Instead, MrHR might relate to a regulation of osmotic pressure. Unlike aquatic cells harboring other Cl− pumps, most host strains of MrHR homologues inhabit terrestrial and non-aquatic environments, where the cells are occasionally exposed to drought stress. Thus, the cells might utilize MrHR homologues to survive in the non-aquatic habitats. Under desiccated conditions, the internal salt concentration should be increased to preserve the intracellular water content. Due to the interior negative membrane potential, cations could be passively transported inside. However, anions need an active transport system, such as MrHR. Thus, a simple scenario might be that MrHR elevates the intracellular osmotic pressure, which in turn preserves the intracellular water content under drought conditions. For further discussion, we must await future investigations.
MrHR conserves the H+ donor residue in the H+ pump, which is an essential residue in the H+ pump. Thus, we attempted the conversion of the MrHR to a H+ pump; a mutant T74D was made in which the Asp residue was introduced as a possible proton acceptor. The λmax of T74D is located at 522 nm (Fig. 6A) and did not change upon the addition of Cl−. This indicates that there is no chloride binding near the PSB. The HPLC analyses showed that the retinal isomer composition is predominantly all-trans (>95%) in both dark and light-adapted states, similar to wild-type MrHR. Next, we examined the ion-pumping activity using the pH change of the E. coli suspension. The results are shown in Fig. 6B, which indicates the opposite pH change to the wild-type MrHR. Even in the NaCl solution, light-induced acidification was observed showing proton extrusion. This pH change was decreased by the addition of CCCP, indicating that T74D actively pumps protons outward. Thus, a successful conversion from a Cl− pump to a H+ pump was effected by the single amino acid replacement. Corresponding to this conversion, T74D underwent a totally different photocycle from that of wild-type MrHR (Fig. 6C). Specifically, the M-formation (400 nm) was observed, indicating that the introduced Asp residue acts as a H+ acceptor from PSB. For HR, a Thr-to-Asp mutation was never observed to induce M-formation (16–18). For natural H+ pumps in the dark, SB is protonated, whereas the acceptor is deprotonated because the SB has a larger pKa than the acceptor. M-formation requires the inversion of the pKa values: The pKa of the acceptor must become larger than that of SB. T74D accomplishes these pKa changes. After M decay, two intermediates appear sequentially corresponding to N (470 nm) and O (590 nm) in BR. For H+ pumps, the donor facilitates both M-N and N-O transitions because the M-N transition reflects proton movement from the donor to SB, and the subsequent N-O transition reflects proton uptake from the cytoplasmic medium by the donor. Thus, the donor-lacking mutants of H+ pumps undergo substantially slower transitions after M formation (42, 43). Interestingly, both transitions in T74D are fast compared with natural H+ pumps, indicating that the donor functions very well. Moreover, M, N, and O appear sequentially without quasi-equilibrium. This reflects the strict “accessibility switch” of the donor, which communicates with SB during the M-N transition; then, the accessibility switches to the cytoplasmic medium to facilitate the N-O transition. In a H+ pump, this switch contributes to the one-way (irreversible) H+ transport (42, 43). These facts indicate that T74D contains the structural factors as well as the essential residues for an effective H+ pump.
FIGURE 6.
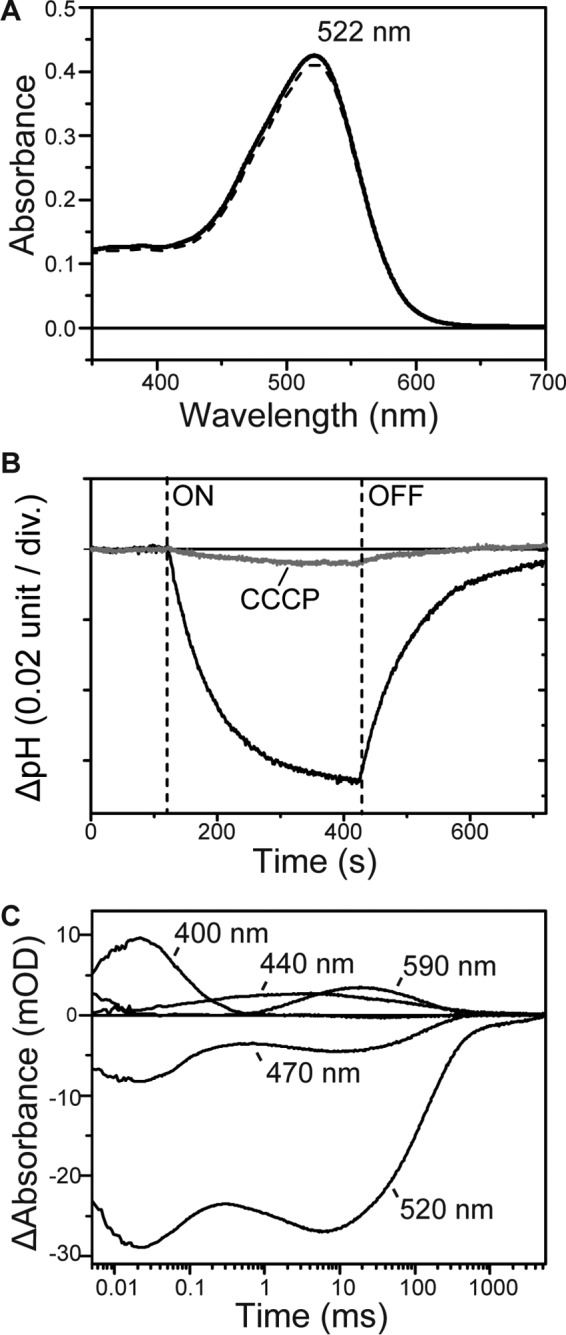
Characterizations of T74D MrHR. A, absorption spectra in the unphotolysed state. The medium (pH 6.5) contained 0.05% DDM and 33 mm Na2SO4 (solid line) or 500 mm NaCl (broken line). B, light-induced pH changes by T74D expressed in E. coli cells in 200 mm NaCl without and with 10 μm CCCP. div., division. C, flash-induced absorbance changes at selected wavelengths. The medium (pH 6.0) contained 0.05% DDM and 100 mm NaCl. mOD, milli-optical density.
The overall photocycle of T74D resembles that of natural H+ pumps but is prolonged due to the last intermediate (520 nm), which resembles MrHR′ in the wild-type photocycle. A similar intermediate was also observed in PR (44), but MrHR′ in T74D had a substantially long lifetime (∼3 s). Another difference from a natural H+ pump is the formation of an unknown intermediate (hereafter called X) at ∼440 nm. X is formed almost simultaneously with the M-decay, but it decays independently of N and O. Thus, X probably forms from M by a branching process. SB reprotonates during the M-decay. For the M-N transition, the donor provides this proton. For the M-X transition, the proton is probably provided through another pathway, implying no contribution of X to the H+ pumping activity. In natural H+ pumps, the antecedent deprotonation of SB triggers the accessibility switching of SB from the acceptor to donor, which enables the exclusive H+ transfer from the donor to SB (42, 43). In T74D, this switching might not be stringently controlled.
MrHR functions as an inward Cl− pump but undergoes a prolonged photocycle. The donor residue is not essential for Cl− pumping activity, which was confirmed by the alanine replacement mutant (data not shown). In contrast, in T74D, both the introduced acceptor and the conserved donor function are comparable with those in natural H+ pumps, which suggests that MrHR evolved from a H+ pump, but the residues and the structure have not been optimized into a mature Cl− pump. The mutant T74D has reduced H+ pumping activity as compared with natural H+ pumps because of the slow decay of the last intermediate and the formation of the X intermediate. These weaknesses are probably related to the lost mechanisms, which are important for a H+ pump but not for a Cl− pump. Thus, comparable studies of MrHR, Cl− pumps, and H+ pumps might provide insight into how sophisticated Cl− pumps evolved from H+ pumps.
Author Contributions
T. H. performed all experiments and wrote the paper. T. K. designed the study and wrote most of the paper. N. K. and M. D. provided technical assistance and contributed to the preparation of the manuscript. All authors reviewed the results and approved the final version of the manuscript.
This work was supported by Japan Society for the Promotion of Science KAKENHI (26440042; to T. K.). The authors declare that they have no conflicts of interest with the contents of this article.
- BR
- bacteriorhodopsin
- HR
- halorhodopsin
- PR
- proteorhodopsin
- KR2
- K. eikastus rhodopsin 2
- FR
- F. pelagi rhodopsin
- HsHR
- HR from H. salinarum
- PSB
- protonated Schiff base
- SB
- deprotonated Schiff base
- MrHR
- M. repens HR
- DDM
- n-dodecyl β-d-maltopyranoside
- CCCP
- carbonyl cyanide m-chlorophenylhydrazone.
References
- 1.Heberle J., Deupi X., and Schertler G. (2014) Retinal proteins: you can teach an old dog new tricks. Biochim. Biophys. Acta 1837, 531–532 [DOI] [PubMed] [Google Scholar]
- 2.Ernst O. P., Lodowski D. T., Elstner M., Hegemann P., Brown L. S., and Kandori H. (2014) Microbial and animal rhodopsins: structures, functions, and molecular mechanisms. Chem. Rev. 114, 126–163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Grote M., and O'Malley M. A. (2011) Enlightening the life sciences: the history of halobacterial and microbial rhodopsin research. FEMS Microbiol. Rev. 35, 1082–1099 [DOI] [PubMed] [Google Scholar]
- 4.Bieszke J. A., Braun E. L., Bean L. E., Kang S., Natvig D. O., and Borkovich K. A. (1999) The nop-1 gene of Neurospora crassa encodes a seven transmembrane helix retinal-binding protein homologous to archaeal rhodopsins. Proc. Natl. Acad. Sci. U.S.A. 96, 8034–8039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Spudich J. L., and Jung K.-H. (2005) Microbial rhodopsin: Phylogenetic and functional diversity. In Handbook of photosensory receptors (Briggs W. R., and Spudich J. L., eds) pp. 1–23, Wiley-VCH Verlag, Weinheim, Germany [Google Scholar]
- 6.Inoue K., Kato Y., and Kandori H. (2015) Light-driven ion-translocating rhodopsins in marine bacteria. Trends. Microbiol. 23, 91–98 [DOI] [PubMed] [Google Scholar]
- 7.Oesterhelt D., and Stoeckenius W. (1971) Rhodopsin-like protein from the purple membrane of Halobacterium halobium. Nat. New Biol. 233, 149–152 [DOI] [PubMed] [Google Scholar]
- 8.Matsuno-Yagi A., and Mukohata Y. (1977) Two possible roles of bacteriorhodopsin; a comparative study of strains of Halobacterium halobium differing in pigmentation. Biochem. Biophys. Res. Commun. 78, 237–243 [DOI] [PubMed] [Google Scholar]
- 9.Béjà O., Aravind L., Koonin E. V., Suzuki M. T., Hadd A., Nguyen L. P., Jovanovich S. B., Gates C. M., Feldman R. A., Spudich J. L., Spudich E. N., and DeLong E. F. (2000) Bacterial rhodopsin: evidence for a new type of phototrophy in the sea. Science 289, 1902–1906 [DOI] [PubMed] [Google Scholar]
- 10.Béjà O., Pinhassi J., and Spudich J. L. (2013) Proteorhodopsins: widespread microbial light-driven proton pumps. In Encyclopedia of Biodiversity (Levin S. A., ed.), 2nd Ed., pp. 280–285, Elsevier, New York [Google Scholar]
- 11.Jung K. H. (2012) 244th American Chemical Society National Meeting, August 22, 2012, Abstract 271, Philadelphia, PA [Google Scholar]
- 12.Yoshizawa S., Kumagai Y., Kim H., Ogura Y., Hayashi T., Iwasaki W., DeLong E. F., and Kogure K. (2014) Functional characterization of flavobacteria rhodopsins reveals a unique class of light-driven chloride pump in bacteria. Proc. Natl. Acad. Sci. U.S.A. 111, 6732–6737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Inoue K., Ono H., Abe-Yoshizumi R., Yoshizawa S., Ito H., Kogure K., and Kandori H. (2013) A light-driven sodium ion pump in marine bacteria. Nat. Commun. 4, 1678. [DOI] [PubMed] [Google Scholar]
- 14.Inoue K., Koua F. H., Kato Y., Abe-Yoshizumi R., and Kandori H. (2014) Spectroscopic study of a light-driven chloride ion pump from marine bacteria. J. Phys. Chem. B 118, 11190–11199 [DOI] [PubMed] [Google Scholar]
- 15.Sasaki J., Brown L. S., Chon Y. S., Kandori H., Maeda A., Needleman R., and Lanyi J. K. (1995) Conversion of bacteriorhodopsin into a chloride ion pump. Science 269, 73–75 [DOI] [PubMed] [Google Scholar]
- 16.Havelka W. A., Henderson R., and Oesterhelt D. (1995) Three-dimensional structure of halorhodopsin at 7 A resolution. J. Mol. Biol. 247, 726–738 [DOI] [PubMed] [Google Scholar]
- 17.Váró G., Brown L. S., Needleman R., and Lanyi J. K. (1996) Proton transport by halorhodopsin. Biochemistry 35, 6604–6611 [DOI] [PubMed] [Google Scholar]
- 18.Muroda K., Nakashima K., Shibata M., Demura M., and Kandori H. (2012) Protein-bound water as the determinant of asymmetric functional conversion between light-driven proton and chloride pumps. Biochemistry 51, 4677–4684 [DOI] [PubMed] [Google Scholar]
- 19.Saitou N., and Nei M. (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4, 406–425 [DOI] [PubMed] [Google Scholar]
- 20.Jones D. T., Taylor W. R., and Thornton J. M. (1992) The rapid generation of mutation data matrices from protein sequences. Comput. Appl. Biosci. 8, 275–282 [DOI] [PubMed] [Google Scholar]
- 21.Sato M., Kikukawa T., Araiso T., Okita H., Shimono K., Kamo N., Demura M., and Nitta K. (2003) Roles of Ser130 and Thr126 in chloride binding and photocycle of pharaonis halorhodopsin. J. Biochem. 134, 151–158 [DOI] [PubMed] [Google Scholar]
- 22.Yamashita Y., Kikukawa T., Tsukamoto T., Kamiya M., Aizawa T., Kawano K., Miyauchi S., Kamo N., and Demura M. (2011) Expression of salinarum halorhodopsin in Escherichia coli cells: solubilization in the presence of retinal yields the natural state. Biochim. Biophys. Acta 1808, 2905–2912 [DOI] [PubMed] [Google Scholar]
- 23.Hernández-Mariné M., Fernández M., and Merino V. (1992) Mastigocladopsis repens (Nostochopsaceae), a new cyanophyte from Spanish soils. Cryptogamie Algol. 13, 113–120 [Google Scholar]
- 24.Shih P. M., Wu D., Latifi A., Axen S. D., Fewer D. P., Talla E., Calteau A., Cai F., Tandeau de Marsac N., Rippka R., Herdman M., Sivonen K., Coursin T., Laurent T., Goodwin L., Nolan M., Davenport K. W., Han C. S., Rubin E. M., Eisen J. A., Woyke T., Gugger M., and Kerfeld C. A. (2013) Improving the coverage of the cyanobacterial phylum using diversity-driven genome sequencing. Proc. Natl. Acad. Sci. U.S.A. 110, 1053–1058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chandrababunaidu M. M., Singh D., Sen D., Bhan S., Das S., Gupta A., Adhikary S. P., and Tripathy S. (2015) Draft genome sequence of Tolypothrix boutellei strain VB521301. Genome Announc. 3, e00001–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Singh D., Chandrababunaidu M. M., Panda A., Sen D., Bhattacharyya S., Adhikary S. P., and Tripathy S. (2015) Draft genome sequence of cyanobacterium Hassallia byssoidea strain VB512170, isolated from Monuments in India. Genome Announc. 3, e00064–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Das A., Panda A., Singh D., Chandrababunaidu M. M., Mishra G. P., Bhan S., Adhikary S. P., and Tripathy S. (2015) Deciphering the genome sequences of the hydrophobic cyanobacterium Scytonema tolypothrichoides VB-61278. Genome Announc. 3, e00228–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Das S., Singh D., Madduluri M., Chandrababunaidu M. M., Gupta A., Adhikary S. P., and Tripathy S. (2015) Draft genome sequence of bioactive compound-producing cyanobacterium Tolypothrix campylonemoides Strain VB511288. Genome Announc. 3, e00226–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stanier R. Y., Kunisawa R., Mandel M., and Cohen-Bazire G. (1971) Purification and properties of unicellular blue-green algae (order Chroococcales). Bacteriol. Rev. 35, 171–205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rippka R., Deruelles J., Waterbury J. B., Herdman M., and Stanier R. Y. (1979) Generic assignments, strain histories and properties of pure cultures of cyanobacteria. J. Gen. Microbiol. 111, 1–61 [Google Scholar]
- 31.Keshari N., and Adhikary S. P. (2013) Characterization of cyanobacteria isolated from biofilms on stone monuments at Santiniketan, India. Biofouling 29, 525–536 [DOI] [PubMed] [Google Scholar]
- 32.Jung K.-H., Trivedi V. D., and Spudich J. L. (2003) Demonstration of a sensory rhodopsin in eubacteria. Mol. Microbiol. 47, 1513–1522 [DOI] [PubMed] [Google Scholar]
- 33.Schobert B., and Lanyi J. K. (1982) Halorhodopsin is a light-driven chloride pump. J. Biol. Chem. 257, 10306–10313 [PubMed] [Google Scholar]
- 34.Váró G., Brown L. S., Sasaki J., Kandori H., Maeda A., Needleman R., and Lanyi J. K. (1995) Light-driven chloride ion transport by halorhodopsin from Natronobacterium pharaonis. I. The photochemical cycle. Biochemistry 34, 14490–14499 [DOI] [PubMed] [Google Scholar]
- 35.Scharf B., and Engelhard M. (1994) Blue halorhodopsin from Natronobacterium pharaonis: wavelength regulation by anions. Biochemistry 33, 6387–6393 [DOI] [PubMed] [Google Scholar]
- 36.Váró G., Zimányi L., Fan X., Sun L., Needleman R., and Lanyi J. K. (1995) Photocycle of halorhodopsin from Halobacterium salinarium. Biophys. J. 68, 2062–2072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hasegawa C., Kikukawa T., Miyauchi S., Seki A., Sudo Y., Kubo M., Demura M., and Kamo N. (2007) Interaction of the halobacterial transducer to a halorhodopsin mutant engineered so as to bind the transducer: Cl− circulation within the extracellular channel. Photochem. Photobiol. 83, 293–302 [DOI] [PubMed] [Google Scholar]
- 38.Oesterhelt D. (1995) Structure and function of halorhodopsin. Isr. J. Chem. 35, 475–494 [Google Scholar]
- 39.Avetisyan A. V., Kaulen A. D., Skulachev V. P., and Feniouk B. A. (1998) Photophosphorylation in alkalophilic halobacterial cells containing halorhodopsin: chloride-ion cycle? Biochemistry 63, 625–628 [PubMed] [Google Scholar]
- 40.Ihara K., Narusawa A., Maruyama K., Takeguchi M., and Kouyama T. (2008) A halorhodopsin-overproducing mutant isolated from an extremely haloalkaliphilic archaeon Natronomonas pharaonis. FEBS Lett. 582, 2931–2936 [DOI] [PubMed] [Google Scholar]
- 41.Falb M., Pfeiffer F., Palm P., Rodewald K., Hickmann V., Tittor J., and Oesterhelt D. (2005) Living with two extremes: conclusions from the genome sequence of Natronomonas pharaonis. Genome Res. 15, 1336–1343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Balashov S. P. (2000) Protonation reactions and their coupling in bacteriorhodopsin. Biochim. Biophys. Acta 1460, 75–94 [DOI] [PubMed] [Google Scholar]
- 43.Lanyi J. K. (2004) Bacteriorhodopsin. Annu. Rev. Physiol. 66, 665–688 [DOI] [PubMed] [Google Scholar]
- 44.Váró G., Brown L. S., Lakatos M., and Lanyi J. K. (2003) Characterization of the photochemical reaction cycle of proteorhodopsin. Biophys. J. 84, 1202–1207 [DOI] [PMC free article] [PubMed] [Google Scholar]