Abstract
TGF-β is a pleiotropic cytokine that regulates a wide range of cellular actions and pathophysiological processes. TGF-β signaling is spatiotemporally fine-tuned. As a key negative regulator of TGF-β signaling, Smad7 exerts its inhibitory effects by blocking receptor activity, inducing receptor degradation or interfering with Smad-DNA binding. However, the functions and the molecular mechanisms underlying the actions of Smad7 in TGF-β signaling are still not fully understood. In this study we report a novel mechanism whereby Smad7 antagonizes TGF-β signaling at the Smad level. Smad7 oligomerized with R-Smad proteins upon TGF-β signaling and directly inhibited R-Smad activity, as assessed by Gal4-luciferase reporter assays. Mechanistically, Smad7 competes with Smad4 to associate with R-Smads and recruits the E3 ubiquitin ligase NEDD4L to activated R-Smads, leading to their polyubiquitination and proteasomal degradation. Similar to the R-Smad-Smad4 oligomerization, the interaction between R-Smads and Smad7 is mediated by their mad homology 2 (MH2) domains. A positive-charged basic region including the L3/β8 loop-strand module and adjacent amino acids in the MH2 domain of Smad7 is essential for the interaction. These results shed new light on the regulation of TGF-β signaling by Smad7.
Keywords: cell signaling, protein degradation, protein-protein interaction, SMAD transcription factor, transforming growth factor beta (TGF-B), ubiquitylation (ubiquitination), Smad7, signaling regulation
Introduction
Transforming growth factor-β (TGF-β) is a prototype of the secreted polypeptide cytokine superfamily that consists of 33 members in mammalians, including TGF-β, activins, nodal, bone morphogenetic proteins (BMPs)3 and others (1, 2). TGF-β family cytokines regulate a wide range of cellular actions, such as cell proliferation, differentiation, apoptosis, and movements in addition to extracellular matrix rearrangement, angiogenesis, etc. (3–6). Therefore, TGF-β plays a pivotal role in embryonic development and adult homeostasis maintenance. To achieve it, TGF-β signaling is finely controlled via multiple modes, and its deregulation has been associated with various human diseases like embryonic defects, cancer development, tissue fibrosis, autoimmune diseases, and skeletal disorders (1–3, 7, 8).
The TGF-β signaling pathway has been well documented (4, 9–12). TGF-β family cytokines initiate signal transduction by binding to the receptor complex, wherein the type II receptor TβRII activates the type I receptor TβRI via phosphorylation. Then TβRI in turn phosphorylates the C-terminal Ser-Xaa-Ser motif of receptor-regulated Smads (R-Smads, Smad2/3 for TGF-β, and Smad1/5/8 for BMPs), leading to their oligomerization with the common Smad (Co-Smad, Smad4). The oligomeric Smad complex is then accumulated in the nucleus and controls target gene expression, in collaboration with other transcription factors or co-factors. Structurally, both R-Smads and Co-Smad consist of a conserved MH1 domain that mediates DNA binding (except Smad2) and a MH2 domain that mediates Smad oligomerization, Smad-receptor interaction, etc. The two domains are bridged by a proline-rich linker region that is divergent in length and amino acid sequence (9, 11). Besides this canonical Smad pathway, other signaling molecules are reported to transduce signals from TGF-β and its receptors, such as mitogen-activated protein kinases (MAPKs, ERK, JNK, and p38), PI3K/Akt, RhoA GTPase, PAK2 and others in a context-dependent manner (13, 14).
Smad7 is a key negative regulator of TGF-β signaling (8, 15, 16). It was first discovered to inhibit TGF-β signaling by binding to the TGF-β type I receptor TβRI through its MH2 domain and blocking R-Smad activation (17, 18). Subsequent studies showed that Smad7 regulates the activity or stability of TβRI by recruiting protein phosphatase, E3 ubiquitin ligases, or deubiquitinating enzymes (1, 8, 15, 19, 20). In addition, Smad7 can also interfere with the R-Smad-Smad4-DNA complex formation by binding to DNA in the nucleus (21), and Yin Yang 1 (YY1) is able to synergize with Smad7 to impede TGF-β/Smad-driven transcription (22). Smad7 deficiency in mice leads to cardiac defects, renal dysfunction, immune-suppression, or growth retardation, accompanied with augmented TGF-β signaling as indicated by enhanced phopho-Smad2/3 levels (16, 23). Emerging evidence also indicates that Smad7 can regulate Wnt/β-catenin, NF-κB, interleukin-1/Toll-like receptor, EGF/MAPK signaling pathways (24–27). Consistent with its important function, altered expression of Smad7 has been associated with inflammatory bowel disease or tissue fibrosis, wherein low or high activity of TGF-β signaling is observed, respectively (28–30). Moreover, Smad7 is highly expressed in several cancers, such as colorectal, pancreatic, skin, breast, liver, and prostate cancers and either inhibits or promotes cancer development depending on cancer types and contexts (23–26, 29).
Although the function of Smad7 in TGF-β signaling has been extensively studied, the underlying molecular mechanisms by which Smad7 exerts its regulatory roles are not fully understood. In this study we uncover a novel mechanism whereby Smad7 directly inhibits R-Smad activity. Upon TGF-β treatment, Smad7 forms a heteromeric complex with R-Smads through the MH2 domain and hence interferes with R-Smad-Smad4 oligomerization in a competitive manner. In addition, Smad7 recruits the E3 ubiquitin ligase NEDD4L to the activated R-Smads, resulting in their polyubiquitination and degradation. Together, these results advance our understanding of the molecular functions of Smad7 in regulating TGF-β signaling.
Experimental Procedures
Plasmids and Reagents
Gal4-Smad2 was constructed based on the vector pcDNA3.1(+) by insertion of cDNAs encoding Gal4 DNA binding domain (DBD) and Smad2 in-frame. Constructs encoding Gal4-Smad3, Gal4-Smad1, Gal4-DBD, and the Gal4 reporters (Gal4-TK-luciferase and pFR-luciferase) were described previously (31). The EYFP (N)- and Venus (C)-expressing control constructs and those encoding the fusion proteins were generated based on pcDNA3.1(+) as described (32). Smad7 and NEDD4L shRNAs were based on pSUPER-puro, targeting GAGGCTGTGTTGCTGTGAA and GCTAGACTGTGGATTGAGT, respectively (33, 35). Mutations of Smad3 or Smad7 were accomplished by PCR-based strategy. Other plasmids encoding NEDD4L and wild-type and mutant Smads have been described before (21, 30, 33–35).
Recombinant human TGF-β1 and BMP2 proteins were purchased from R&D Systems Inc. Cycloheximide, MG132, and anti-FLAG antibody (M2) were from Sigma. Antibodies recognizing NEDD4L, phospho-Smad2, phospho-Smad3, Smad2/3, or TβRI were from Cell Signaling, and antibodies against Smad7 and Smad4 were generated by immunizing rabbits with proteins of Smad7 N terminus (1–259 amino acids) and Smad4-linker (144–316 amino acids), respectively. Other antibodies, including anti-Myc, anti-HA, anti-GAPDH, and anti-tubulin antibodies were all obtained from Santa Cruz Biotechnology.
Cell Culture and Transfection
Human embryonic kidney epithelial HEK293FT cells were maintained in Dulbecco's minimum essential medium (DMEM) (Corning) supplemented with 10% of fetal bovine serum (Gibco) at 37 °C in a humidified, 5% CO2 incubator. Human hepatocellular carcinoma Hep3B and TβRI-deficient mink lung epithelial R1B/L17 cells were maintained in minimum essential medium (Corning), and MDA-MB-231 cells were maintained in RPMI 1640 (Corning) by the addition of 2 mm l-glutamine. Cell transfection was conducted with VigoFect (Vigorous Biotechnology, Beijing) or Lipofectamine 2000 (Invitrogen).
Stable Cell Line Establishment
The construct encoding FLAG-tagged Smad7 under the control of CMV promoter and carrying the puromycin-resistant gene was transfected into MDA-MB-231 cells, paralleled by transfection of the control empty vector. After puromycin selection, the drug-resistant cells were pooled as stable cells.
Luciferase Reporter Assay, Total RNA Extraction, Reverse Transcription (RT), and Quantitative RT-PCR
Cells were plated in 24-well plates one night before transfection. Transfection was performed as described above, and empty vectors were used to equalize the total amounts of plasmids in each sample. Luciferase activity was measured at 40 h post-transfection by using the dual luciferase reporter assay system (Promega, Madison, WI) following the manufacturer's protocol. The experiments were repeated in triplicate, and the data are presented as the means ± S.D. after normalization to Renilla activity. Total cell RNA extraction, reverse transcription, and quantitative RT-PCR were described previously (30). The primers used were as follows: for human GAPDH (glyceraldehyde-3-phosphate dehydrogenase), 5′-ACCACAGTCCAT GCCATCAC-3′ and 5′-TCCACCACCCTGTTGCTGTA-3′; for human Smad7, 5′-CCAACTGCAGACTGTCCAGA-3′ and 5′-TTCTCCTCCCAGTATGCCAC-3′.
Immunoprecipitation and Immunoblotting
Cells for immunoprecipitation (IP) were lysed on ice with lysis solution (50 mm Tris-Cl (pH 7.5), 150 mm NaCl, 1 mm EDTA, 0.5% Nonidet P-40, 10 mm NaF, 10 mm sodium pyrophosphate, 1 mm sodium orthovanadate dodecahydrate, and protease inhibitors) and rotated for more than 10 min at 4 °C. After an aliquot was taken for protein expression analyses, the left cell lysates containing equivalent amounts of total proteins were precleared for 2 h with protein A-Sepharose (GE Healthcare) at 4 °C. Immunoprecipitation was carried out by the addition of appropriate antibodies and protein A-Sepharose followed by incubation at 4 °C overnight with gentle rotation. Then the immune complex was isolated by centrifugation and repeated washes with lysis buffer, analyzed by SDS-PAGE and immunoblotting, and detected with the enhanced chemiluminescent substrate (Pierce) according to the manufacturer's instructions.
Protein Turnover Analysis
HEK293FT cells were transfected with indicated plasmids in 12-well plates, and the amounts of plasmids in each sample were equalized by the addition of empty vectors. The cells were treated with 50 μg/ml cycloheximide for the indicated time periods before harvest. Then cells were lysed for protein level analyses by immunoblotting.
In Vivo Ubiquitination Assay
HEK293FT cells were transfected with His-Myc-ubiquitin plasmid and other constructs as indicated. At 40 h post-transfection, cells were treated with 10 μm MG132 for 4 h then collected and lysed in 1% SDS followed by 5 min of boiling. The lysates were immediately diluted 10-fold with lysis buffer (20 mm Tris-HCl (pH 7.4), 2 mm EDTA, 10 mm NaF, 10 mm sodium pyrophosphate, 1 mm sodium orthovanadate dodecahydrate, 1% Triton X-100) plus protease inhibitors and rotated for 30 min at 4 °C. After centrifugation at 13,000 rpm for 10 min, an aliquot of the supernatants was taken for protein expression analyses, and the remaining lysates were subjected to immunoprecipitation and immunoblotting detection.
Homology Modeling of Smad7 MH2 Domain Structure
The structural model of Smad7 MH2 domain was constructed based on the three-dimensional structure of C-terminally phosphorylated Smad3 MH2 domain (RCSB Protein Data Bank (PDB) code 1U7F) using SWISS-MODEL.
Statistic Analysis
All the experiments were repeated at least three times. The values were presented as the mean ± S.D., and the significance between the means was calculated using Student's t test. A p value <0.05 was considered as statistically significant.
Results
Smad7 Inhibits the Signaling Activity of R-Smads
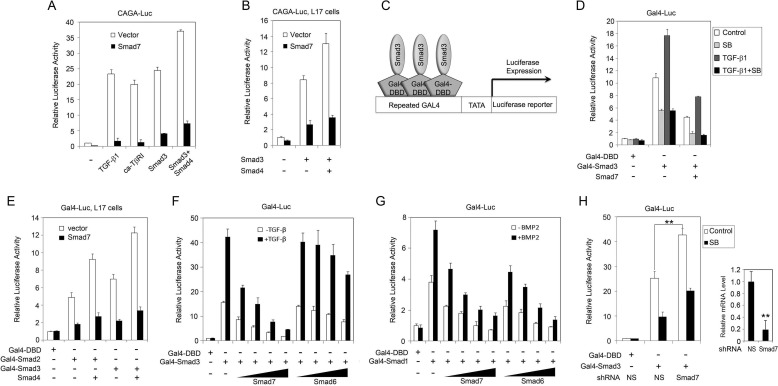
Although Smad7 has been established as a pivotal negative regulator of TGF-β signaling, the underlying molecular mechanisms still need a better understanding (8, 15). To clarify at what levels Smad7 acts in TGF-β signaling, we first carried out a reporter assay in HEK293FT cells using the Smad-responsive CAGA-luciferase reporter (36). As shown in Fig. 1A, treatment with TGF-β1, overexpression of the constitutively active (ca-) type I TGF-β receptor TβRI, Smad3 alone, or Smad3 together with Smad4 stimulated reporter expression. Co-expression of Smad7 blocked all of these effects, indicating that Smad7 can function at both the receptor level and the Smad level. In addition, Smad7 also inhibited the expression of CAGA-luciferase induced by ectopic expression of Smad3 with or without Smad4 in the TβRI-deficient mink lung epithelial R1B/L17 cells (Fig. 1B), further indicating that Smad7 is capable of inhibiting TGF-β signaling at the Smad level.
FIGURE 1.
Smad7 antagonizes R-Smad activity. A and B, HEK293FT cells (A) or R1B/L17 cells (B) were transfected with constructs encoding CAGA-luciferase reporter (200 ng) together with Renilla luciferase (20 ng), ca-TβRI (50 ng), Smad3 (50 ng), Smad4 (50 ng), and Smad7 (100 ng), or empty vector as indicated. At 20 h post-transfection, cells were treated with or without 100 pm TGF-β1 for another 20 h before harvested for luciferase activity measurement. C, schematic representation of a Gal4-luciferase reporter system. The reporter is driven by a promoter that contains repeated Gal4-binding elements. D and E, HEK293FT cells (D) or R1B/L17 cells (E) transfected with plasmids encoding Gal4-TK-Luc (200 ng) together with Renilla luciferase (20 ng), Gal4-DBD (50 ng), Gal4-Smad2 (50 ng), Gal4-Smad3 (50 ng), Smad4 (50 ng), and Smad7 (100 ng) were treated with 100 pm TGF-β1 or/and 10 μm SB431542 overnight as indicated and harvested for luciferase activity determination. F and G, Gal4-TK-luciferase reporter assays were done similarly as in D in HEK293FT cells, and different amounts of plasmids encoding Smad7 or Smad6 were used (10, 25, 50, and 100 ng). At 20 h post transfection, cells were treated with 100 pm TGF-β1 (F) or 25 ng/ml BMP2 (G) overnight. H, HEK293FT cells were transfected with plasmids encoding pFR-luciferase, nonspecific (NS) shRNA (100 ng), or Smad7-tergeting shRNA (100 ng) and then treated with or without 10 μm SB431542 (SB) overnight before harvested for luciferase assay (left). Smad7 knockdown efficiency was tested by transfection of NS and Smad7-specific shRNAs into HEK293FT cells followed by total mRNA extraction and RT-quantitative RT-PCR analysis (right). Smad7 mRNA expression level was normalized to that of GAPDH. **, p < 0.01. In all the reporter assays, empty vectors were used to equalize the total amounts of plasmids in each sample. Each experiment was performed in triplicate, and the data are presented as the mean ± S.D. after normalization to Renilla activity.
Smad7 has been demonstrated to inhibit Smad-driven transcription by competing with the R-Smad-Smad4 complex for DNA binding (21). To address whether Smad7 hampers the binding of R-Smad-Smad4 to DNA or directly inhibits R-Smad activity, we employed the Gal4-luciferase reporter in which luciferase expression is driven by a promoter that includes repeated Gal4 binding elements, and fusion proteins of Gal4 DBD with Smad3 or other transcription activators are able to induce the reporter expression (Fig. 1C) (31). The Gal4-luciferase reporter was activated by Gal4-Smad3 overexpression, and TGF-β1 further promoted the activation (Fig. 1D). However, Smad7 attenuated the Smad3-induced Gal4-luciferase expression even in the presence of the TβRI kinase inhibitor SB431542, strongly suggesting that the effect of Smad7 is independent of receptor activity. Notably, SB431542 attenuated the activity of Smad3, indicating that the stimulating effect of Smad3 is partially due to autocrine TGF-β activity. A similar Gal4-luciferase reporter assay in R1B/L17 cells showed that both the Gal4-Smad2 and Gal4-Smad3 fusion proteins activated the reporter expression, and the activations were reinforced by Smad4 but inhibited by Smad7 (Fig. 1E). Furthermore, Smad7 inhibited Smad3 activity in a dose-dependent manner (Fig. 1F). Moreover, Smad7 was able to inhibit Smad1 activity in a similar manner (Fig. 1G). Although Smad6 was less effective in attenuating Smad3 activity, it efficiently blocked Smad1 activity (Fig. 1, F and G). Finally, to confirm the physiological role of endogenous Smad7 in regulating Smad3 activity, Smad7 was knocked down with a specific shRNA (Fig. 1H, right) (33). Consistently, silencing of Smad7 expression enhanced the Gal4-Smad3-induced expression of the Gal4 reporter (Fig. 1H, left). Together, these results indicate that Smad7 can inhibit the signaling activity of R-Smads downstream of the receptors, in addition to interfering with the binding of R-Smad-Smad4 complex to DNA.
Smad7 Interacts with R-Smads in Response to TGF-β Signaling
To explore how Smad7 inhibits Smad2/3 activity, we first examined whether Smad7 could oligomerize with R-Smad proteins. Hep3B cells treated with or without TGF-β1 ligands were subjected to co-IP. Indeed, TGF-β induced the association of Smad2/3 not only with Smad4 but also with Smad7 (Fig. 2A). To further characterize their interactions, we expressed Smad7, R-Smad proteins (Smad2/3 and Smad1/8), Smad4, and the other inhibitory Smad Smad6 in HEK293FT cells and performed co-IP experiments. Ectopic expression of Smad7 interacted with R-Smads involved in both TGF-β and BMP pathways at the basal level, and these interactions were greatly reinforced by co-expression of ca-TβRI (Fig. 2B) or ca-ALK6/BMPRIB (Fig. 2C). Furthermore, we also noticed the existence of Smad7-Smad6 and Smad7-Smad4 associations (Fig. 2, B–C), which is consistent with a previous study reporting that Smad7 facilitates Smad4 degradation (37). To determine whether TGF-β-induced carboxyl phosphorylation of R-Smads is required for their interaction with Smad7, we compared the binding of Smad7 to wild-type (WT) Smad3, Smad3(2D) mutant in which the last two serine residues at the C-terminal tail are mutated to aspartic acid to mimic phosphorylation (38), or Smad3(3A) mutant that contains three serine-to-alanine mutations at the C-terminal tail and cannot be activated by TGF-β. As shown in Fig. 2D, Smad7 interacted strongly with ca-TβRI-phosphorylated wild-type Smad3 and 2D mutant but weakly with non-activated wild-type Smad3 or 3A mutant. Accordantly, wild-type Smad3 was able to induce the CAGA-luciferase reporter expression, and TGF-β1 enhanced the effect of wild-type Smad3, comparable with that of 2D mutant (Fig. 2E). Furthermore, Smad7 attenuated the expression of the reporter induced by Smad3 (Fig. 2E).
FIGURE 2.
Smad7 associates with R-Smads. A, Hep3B cells were treated with or without 100 pm TGF-β1 for 1 h and then lysed for IP with control rabbit IgG or anti-Smad2/3 antibody followed by anti-Smad7 or anti-Smad4 immunoblotting (IB). Total protein expression was examined by immunoblotting of whole cell lysates (WCL). B and C, HEK293FT cells transfected with indicated constructs were harvested at 40 h post-transfection for anti-FLAG immunoprecipitation and anti-Myc immunoblotting. D and E, co-IP experiment (D) was done in HEK293FT cells as in B. The CAGA-luciferase reporter assay (E) was done as in Fig. 1A. Plasmids encoding Smad3 (50 ng) and Smad7 (100 ng) were transfected to HEK293FT cells. After treatment with or without 100 pm TGF-β1 or 10 μm SB431542 for 20 h, the cells were subjected to luciferase activity measurement. F, schematic diagram of bimolecular fluorescence complementation and its application for detection of protein interaction. The N terminus of EYFP and C terminus of Venus are fused to protein A and B, respectively. The interaction of protein A and B in cells brings EYFP(N) and Venus(C) together and generates green fluorescence. G, Venus(C)-fused Smad4/7 and EYPF(N)-fused Smad3 were expressed in HEK293FT cells as indicated. At 40 h post-transfection, cells were treated with 100 pm TGF-β1 for 1 h before being fixed for fluorescence examination by an Olympus confocal microscope (FV10i-Oil) and analyzed using the OLYMPUS FLUOVIEW software. H, MDA-MB-231 cells stably expressing FLAG-Smad7 were treated with 100 pm TGF-β1, harvested at different time points, and subjected for co-IP examination.
Then we further confirmed the Smad7-Smad3 interaction using bimolecular fluorescence complementation approach. As demonstrated previously (32) and illustrated in Fig. 2F, if protein A and B could associate with one another in vivo, the protein A-fused N-terminal part of EYPF (EYFP(N)) and protein B-fused C-terminal part of Venus (Venus(C)) would be brought together and reconstitute functional fluorescent proteins, emitting green fluorescence. Indeed, co-expression of EYFP(N)-Smad3 fusion protein with Venus(C)-Smad4 gave rise to green fluorescence in the nucleus upon TGF-β treatment (Fig. 2G, upper panels), whereas co-expression of EYFP(N) with Venus(C)-Smad4 had no such effect (data not shown), indicative of in vivo specific interaction between Smad3 and Smad4. In contrast, TGF-β-induced complex of EYFP(N)-Smad3 and Venus(C)-Smad7 were mainly localized in the cytoplasm, although a weak signal could also be detected in the nucleus (Fig. 2G, lower panels).
As TGF-β and BMP induce both the R-Smad-Smad4 association that transduces signals and the R-Smad-Smad7 interaction that interferes with signal transduction, the two events seem to contradict each other. To elucidate the physiological consequence of these interactions, we examined the kinetics of the R-Smad-Smad7 interaction in MDA-MB-231 cells that stably express FLAG-Smad7. The cells were treated with TGF-β1, harvested at different time points, and subjected to anti-Smad2/3 immunoprecipitation and anti-Smad4 or anit-Smad7 immunoblotting. The Smad2/3-Smad4 association appeared quickly after ligand treatment, peaked at 1 h, and then decreased slowly (Fig. 2H). The kinetics was very similar to the one of phospho-Smad2. Similarly, Smad7 also associated with Smad2/3 at 0.5 h and lasted 1 h, then their associations quickly weakened. Based on these results, we reasoned that Smad4 and Smad7 might compete in interacting with the phosphorylated form of R-Smads, and the balance between Smad4-binding and Smad7-binding of R-Smads would determine the intensity or duration of TGF-β signaling.
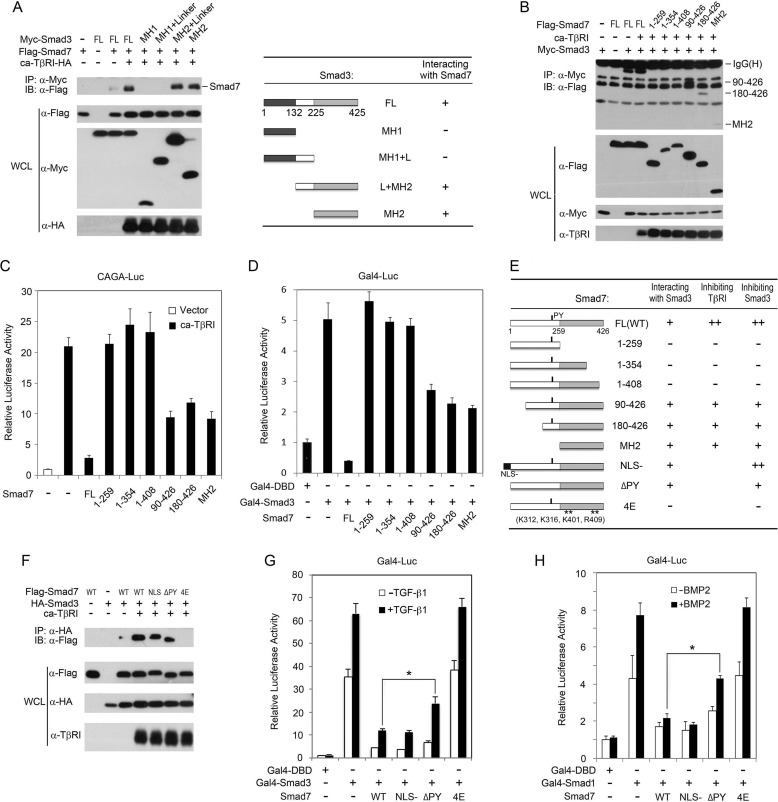
The MH2 Domains of Smad7 and Smad3 Mediate Their Interaction
To test our hypothesis that Smad7 competes with Smad4 in binding to activated R-Smads, we first characterized the interaction between Smad7 and Smad3 by mapping the interacting domains. As shown in Fig. 3A, both the full-length (FL) Smad3 and its truncations containing the MH2 domain were able to interact with Smad7, suggesting that the MH2 domain of Smad3 mediates its interaction with Smad7. Similarly, the MH2 domain of Smad7 was also involved in Smad3 interaction (Fig. 3, B and E). Functionally, the Smad7 truncations containing the MH2 domain (including 90–426 amino acids, 180–426 amino acids, and the MH2 domain (260–426 amino acids)) were capable of inhibiting the ca-TβRI-induced CAGA-luciferase reporter expression and the Gal4-Smad3-mediated Gal4-luciferase reporter activation (Fig. 3, C–E). These results together demonstrate that the MH2 domains of Smad3 and Smad7 not only physically but also functionally mediate their interaction.
FIGURE 3.
The MH2 domains mediate the Smad3-Smad7 interaction. A, HEK293FT cells transfected with constructs encoding Myc-tagged FL Smad3 or truncations, FLAG-Smad7, and ca-TβRI-HA were subjected to co-IP analyses (left). Smad3 truncations were shown diagrammatically (right). IB, immunoblot; WCL, whole cell lysate. B, plasmids were transfected as indicated into HEK293FT cells for co-IP analysis. C and D, CAGA-luciferase or Gal4-TK-luciferse reporter assays were carried out as in Fig. 1, A and B, respectively. E, diagrammatic representation of Smad7 mutants and summary of Smad7 domain-mapping results. F, HA-Smad3 and ca-TβRI plasmids were transfected into HEK293FT cells as indicated, whereas FLAG-Smad7 proteins (WT, NLS-, ΔPY, and 4E) and control empty vector were independently expressed. After cell lysis, the cell lysates were mixed together as indicated and subjected to co-IP experiments. G and H, pFR-luciferase reporter assays were performed in HEK293FT cells as described in Fig. 1D. Cells were treated with 100 pm TGF-β1 (G) or 25 ng/ml BMP2 (H) for 20 h before harvested. *, p < 0.05.
To consolidate the above conclusion, we then examined the interaction of Smad3 with various Smad7 mutants, including a nuclear localization signal (NLS)-tagged Smad7 that guides Smad7 to stay in the nucleus (21), a PY motif deletion mutant (ΔPY) that loses the ability to associate with WW-HECT type E3 ubiquitin ligases like NEDD4L, Smurf1/2, or WWP1 (39), and the 4E mutant that bears the mutations (K312E, K316E, K410E, and R409E) in the MH2 domain and affects the binding of Smad7 to TGF-β receptors (40). Smad7 could inhibit Smad3 activation by blocking receptor activity, thereby indirectly affecting Smad7-Smad3 interaction. To exclude this possibility, we co-expressed ca-TβRI and HA-Smad3 in HEK293FT cells, whereas various FLAG-Smad7 constructs were separately expressed in different set of cells. After lysing the cells the lysates were mixed as indicated and subjected to co-IP assay. Interestingly, only Smad7 4E mutant lost the ability to associate with Smad3, whereas the other two mutants still retained the Smad3 binding activity (Fig. 3, E and F). Consistently, the Gal4-luciferase reporter assays showed that the 4E mutant was unable to inhibit Smad3 or Smad1 activity, whereas the ΔPY mutant exhibited an attenuated inhibitory effect, in comparison to those of the wild-type or NLS-tagged Smad7 (Fig. 3, E, G, and H). These results indicate that the four amino acids Lys-312, Lys-316, Lys-410, and Arg-409 are important for Smad7 to interact with both TβRI and R-Smads. The data also suggest that the Smad7-Smad3 interaction could occur in the nucleus and that the binding of WW-HECT type E3 ubiquitin ligases could play a role in Smad7-mediated inhibition of R-Smad activity.
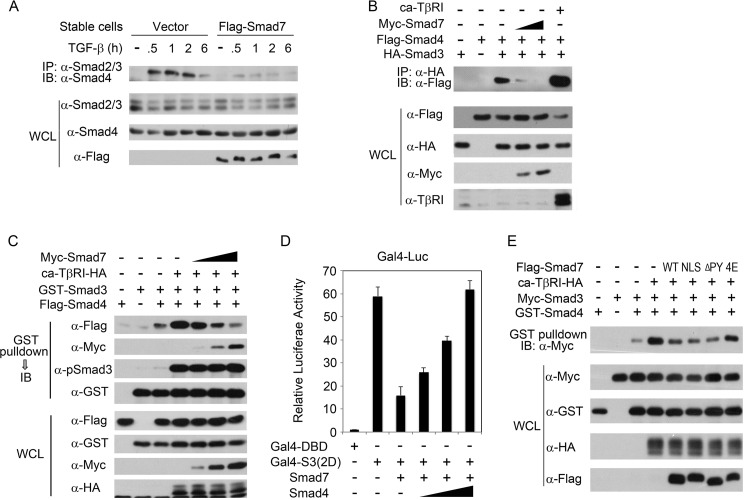
Smad7 Interferes with R-Smad-Smad4 Complex Formation
As homo-oligomerization of R-Smads or Smad4 and hetero-oligomerization of R-Smads with Smad4 or Smad7 are all mediated by their MH2 domains, we examined whether Smad7 could interfere with the complex formation between R-Smads and Smad4. As shown in Fig. 4A, TGF-β stimulated robust Smad2/3-Smad4 interaction in the control MDA-MB-231 cells, but the interaction was dramatically reduced in the FLAG-Smad7-expressing stable cells. To dissect whether Smad7 could interfere with the R-Smad-Smad4 interaction directly or indirectly due to the impaired receptor activity, co-IP experiment was carried out in TβRI-deficient R1B/L17 cells. Ectopic interaction between Smad3 and Smad4 was detected, and the interaction was attenuated by co-expression of Smad7 (Fig. 4B). To further confirm the direct interference of Smad7 on the Smad2/3-Smad4 complex formation, we performed a co-IP experiment similarly as done in Fig. 3F. Smad3 and Smad4 co-expressed with or without ca-TβRI in HEK293FT cells were mixed with separately expressed Myc-Smad7, and then protein interactions were examined by co-IP. Again, the Smad3-Smad4 interaction was attenuated by Smad7 in a dose-dependent manner (Fig. 4C). In accordance, Smad4 was able to overcome the inhibitory effect of Smad7 on Smad3-induced Gal4-luciferase reporter expression (Fig. 4D).
FIGURE 4.
Smad7 interferes with the R-Smad-Smad4 hetero-complex formation. A, MDA-MB-231 cells stably expressing FLAG-Smad7 or control empty vector were treated with 100 pm TGF-β1, harvested at the indicated time points, and subjected for co-IP analyses. IB, immunoblot. WCL, whole cell lysate, B, R1B/L17 cells were transfected with the indicated plasmids, and co-IP was carried out at 40 h post-transfection. C, GST-Smad3, FLAG-Smad4, and ca-TβRI-HA were expressed in HEK293FT cells, whereas Myc-Smad7 (0.5, 1, or 2 μg of DNA) were expressed independently. Upon cell lysis, the lysates were mixed together as indicated and subjected to co-IP experiments. D, plasmids encoding pFR-luciferase, Renilla, Gal4-DBD (50 ng), Gal4-Smad3 (2D) (50 ng), Smad7 (100 ng), and Smad4 (25, 50, 100 ng, respectively) were transfected into HEK293FT cells for luciferase assay. E, co-IP was similarly done as above. GST-Smad4, Myc-Smad3, and ca-TβRI-HA proteins were ectopically expressed in HEK293FT cells as indicated, whereas Smad7 proteins (WT, NLS, PY mutant, and 4E mutant) and control empty vector were separately expressed. Cell lysates were mixed as indicated before immunoprecipitation.
Finally, in line with the above observation that the 4E Smad7 mutant no longer interacts with Smad3, this mutant was unable to interfere with Smad3-Smad4 association in co-IP assay (Fig. 4E). However, the PY motif-deleted (ΔPY) and the NLS-tagged Smad7 that retained the ability to associate with Smad3 decreased the Smad3-Smad4 interaction (Fig. 4E). These data together strongly support the notion that Smad7 can directly interfere with R-Smad-Smad4 hetero-oligomerization.
Smad7 Promotes NEDD4L-induced Polyubiquitination and Degradation of Smad3
Several WW-HECT-type E3 ubiquitin ligases including NEDD4L (NEDD4–2), Smurf1, Smurf2, and WWP1 have been shown to associate with both Smad7 and R-Smad proteins via the PY motif in the linker regions (1, 15, 39, 41, 42). Among them, NEDD4L and Smurf1 have been demonstrated to bind TGF-β-activated Smad2/3 and BMPs-activated Smad1, respectively, and target them for degradation (43).
Although deletion of the PY motif (ΔPY) has little effect on the ability of Smad7 to associate with Smad3 (Fig. 3, E and F) or to interfere with the R-Smad-Smad4 complex formation (Fig. 4E), inhibition of the ΔPY mutant on Smad3 or Smad1 signaling activity was less effective than wild-type Smad7 (Figs. 3, E, G, and H, and 5, A and B). Together these results suggest that the recruitment of WW-HECT type E3 ubiquitin ligases may contribute to Smad7-mediated inhibition of R-Smad activity.
FIGURE 5.
Smad7 promotes NEDD4L-elicited polyubiquitination and degradation of Smad3. A and B, pFR-luciferase reporter assays were performed as in Fig. 1D. HEK293FT cells were transfected with plasmids encoding pFR-Luc (200 ng), Renilla (20 ng), Gla4-DBD (50 ng), Gal4-Smad1/3 (50 ng), wild-type Smad7 (25, 50, 100 ng), and the ΔPY mutant (25, 50, 100 ng) and treated with (A) or without (B) 100 pm TGF-β1 overnight before harvested for luciferase activity measurement. *, p < 0.05. C, HEK293FT cells transfected with constructs encoding FLAG-NEDD4L, Myc-Smad7, or empty vector were pretreated with or without 10 μm MG132 for 3 h and then stimulated with 100 pm TGF-β1 in the absence or presence of MG132 before harvested at the indicated time points. D and E, HEK293FT cells were transfected with the indicated constructs, treated with 50 μg/ml of cycloheximide (CHX) for the indicated time periods, and then harvested for immunoblotting (D). GAPDH was served as a leading control. The relative Smad3(2D) expression levels were quantified (E). F, pFR-Luc reporter assay was done in HEK293FT, which were transfected with plasmids encoding Gal4-DBD (50 ng), Gal4-Smad3 (2D) (50 ng), Smad7 (10 ng), wild-type (10, 25, 50, 100 ng), or the ligase-deficient C821A mutant (50, 100 ng) NEDD4L. G, pFR-luciferase reporter assay was similarly done as above. NEDD4L shRNA (N4L, 100 ng), Smad7 shRNA (S7, 100 ng) and a nonspecific (NS) control shRNA were transfected into HEK293T cells. Cells were treated with 100 pm TGF-β1 overnight before harvest. NEDD4L gene silence efficiency was detected by immunoblotting. *, p < 0.05; **, p < 0.01. H, HEK293FT cells transfected with FLAG-Smad7 (wild-type and ΔPY) or control vector were treated with 100 pm TGF-β1 for 1 h and subjected to co-IP. IB, immunoblot; WCL, whole cell lysate. I, HA-NEDD4L (CA), Myc-Smad2/3, and ca-TβRI were overexpressed in HEK293FT cells, and FLAG-Smad7 (1 μg, 2 μg) or the empty vector were expressed separately. Cell lysates were mixed as indicated, and protein interaction was analyzed by co-IP. J, HEK293FT cells expressing the indicated proteins were treated with 10 μm MG132 for 4 h before harvested for anti-Myc immunoprecipitation and anti-HA immunoblotting.
As Smad7 preferentially interacts with activated R-Smads, we tested whether Smad7 could modulate NEDD4L-elicited degradation of phospho-Smad3. As shown in Fig. 5C, NEDD4L was able to decrease TGF-β1-induced C-terminally phosphorylated Smad3 (p-Smad3) level, and this effect was augmented by co-expression of Smad7. Moreover, the p-Smad3 level was restored in the presence of the proteasome inhibitor MG132, suggesting the contribution of Smad7 to NEDD4L-induced proteasomal degradation of p-Smad3. Next, we examined whether Smad7 could promote NEDD4L-induced turnover of Smad3(2D), the active Smad3 mimic. Indeed, expression of NEDD4L alone accelerated the turnover of Smad3(2D), and Smad7 remarkably enhanced the effect (Fig. 5, D and E). Functionally, wild-type NEDD4L, but not its ligase-deficient C821A derivative (35), inhibited Smad3(2D) activity in a dose-reliant manner, and the inhibition was greatly facilitated by Smad7 (Fig. 5F). Accordantly, silencing of NEDD4L or Smad7 gene expression enhanced Smad3(2D) activity, and silencing of either of the two genes was capable of attenuating the inhibitory effect of the other (Fig. 5G), again demonstrating that Smad7 and NEDD4L act in concert to inhibit R-Smad activity.
Next we explored how Smad7 and NEDD4L cooperate with one another to regulate R-Smad stability. Co-IP experiments revealed that ectopic wild-type Smad7 inhibited endogenous Smad2/3-Smad4 interaction while enhancing the Smad2/3-NEDD4L binding, whereas deletion of the PY motif exhibited an attenuated effect (Fig. 5H). Furthermore, Smad7 also reinforced the ca-TβRI-induced interactions between Smad2/3 and NEDD4L when overexpressed (Fig. 5I), and wild-type Smad7, but not the ΔPY mutant, enhanced NEDD4L-elicited polyubiquitination of Smad3 (Fig. 5J). Taken together, these results demonstrated that Smad7 promotes polyubiquitination and degradation of active R-Smads through recruiting NEDD4L.
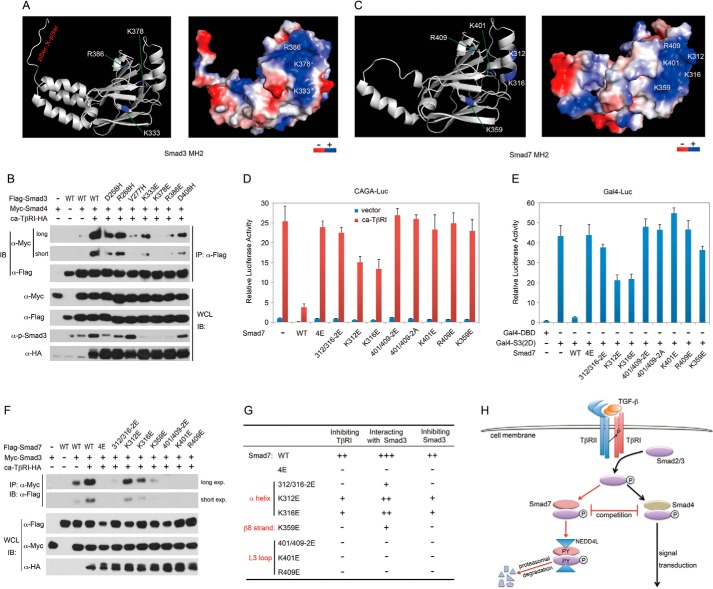
Identification of Amino Acids Mediating the Smad7-Smad3 Interaction
The MH2 domains mediate both Smad-Smad and the Smad-TβRI interactions (9, 11). In these complexes the positively charged basic region in MH2 domains interact directly with the C-terminal Ser(P)-Xaa-Ser(P) of adjacent R-Smad molecules or the Thr(P)-Ser(P) motif in the GS region of TβRI (9, 11, 44). As shown in the structure of C-terminally phosphorylated Smad3 MH2 domain (Fig. 6A) (9, 44), the basic region comprises the L3 loop (Lys-378 and Arg-386) and β8 strand (Lys-333). In addition, several other residues including Asp-258, Arg-268, Val-277, and Asp-408 on the other side of the basic region also play important roles through intermolecular and intramolecular interactions. Indeed, mutation of these residues impaired the Smad3-Smad4 interaction as examined by co-IP (Fig. 6B), and this is in accordance with previous structural studies (9, 11, 44). Similarly, the modeling structure of the Smad7 MH2 domain based on that of Smad3 also contains a basic region that includes the L3 loop (Lys-401, Arg-409), β8 strand (Lys-359, equivalent to Lys-333 in Smad3), and an adjacent α-helix that contains Lys-312 and Lys-316 (Fig. 6C). Among the residues, four (Lys-312, Lys-316, Lys-401, and Arg-409) have been reported to mediate binding of Smad7 to TβRI (40), and simultaneous mutation of the four residues to glutamic acids (4E mutant) disabled the Smad7-Smad3 association (Fig. 3, D–G). In fact, the double mutation of Lys-312/Lys-316–2E, Lys-401/Arg-409–2E, and Lys-401/Arg-409–2A or either of the K359E, K401E, and R409E single mutations was able to disable Smad7 in inhibiting the activity of TβRI or Smad3, whereas the K312E and K316E single mutation exhibited attenuated inhibitory effects (Fig. 6, D and E). In accordance with above data, the 4E, Lys-312/Lys-316–2E and Lys-401/Arg-409–2E as well as the K359E, K401E, R409E mutants were unable to associate with Smad3, and the binding ability of K312E and K316E single mutants were also partially impaired (Fig. 6, F and G). Together these results demonstrated that the basic region including the five key residues in the MH2 domain is essential for Smad7 to associate with Smad3 and inhibit its activity.
FIGURE 6.
Identification of amino acids mediating the Smad7-Smad3 interaction. A, a ribbon drawing (left) and surface electrostatic potential (right) of C-terminally phosphorylated Smad3 MH2 domain (PDB code 1U7F). The positively charged regions and amino acids are shown in blue, and negatively charged regions are in red. B, co-IP experiment was carried out in HEK293FT cells as in Fig. 2B. IB, immunoblot; WCL, whole cell lysate. C, Smad7 MH2 domain modeling based on that of Smad3. D and E, effects of wild-type and mutant Smad7 on the activity of ca-TβRI or Smad3. CAGA-luciferase (D) and pFR-luciferase (E) reporter assays were performed in HEK293FT cells. F, Myc-Smad3 and ca-TβRI-HA were expressed as shown in HEK293FT cells, whereas wild-type or mutant Smad7 were separately expressed. The lysates were mixed as indicated before co-IP analysis. G, summary of the functions of Smad7 mutants. H, a schematic diagram showing regulation of R-Smad activity and TGF-β/Smad signaling by Smad7.
Discussion
TGF-β signaling is finely controlled both in strength and in duration in normal contexts and is deregulated in a variety of human diseases (1, 3, 5, 7, 8, 13). Smad7, an inhibitory Smad, has been found to play a vital role in restricting TGF-β signaling (15, 24, 25, 29). It has been reported to exert inhibitory effects at the receptor level or the transcription level (1, 8, 15). Here we report a novel mechanism whereby Smad7 inhibits TGF-β/Smad signaling by forming hetero-complexes with and inhibiting the activity of R-Smads directly (Fig. 6H).
Smad7 is able to form a stable complex with the activated TGF-β receptors, thereby inhibiting their activity or protein stability (1, 8, 17, 18). In the nucleus, Smad7 binds to DNA directly via its MH2 domain and impedes bindings of the functional R-Smad-Smad4 complex to DNA (21). Here we found that Smad7 was capable of inhibiting the expression of Gal4-luciferase reporter induced by Gal4-Smad3 and Gal4-Smad2 independent of TGF-β receptor activity, as assessed by the addition of the TβRI inhibitor SB431542 or in TβRI-deficient R1B/L17 cells. Because R-Smads were brought to the Gal4 reporter by the Gal4 DNA binding domain and activated the reporter, our results demonstrate that Smad7 executes its inhibitory effect at another layer by directly inhibiting R-Smad activity.
The MH2 domains of Smad proteins mediate homo-oligomerization and hetero-oligomerization of R-Smads or Co-Smad and interaction of R-Smads or Smad7 to the receptors (9, 11). Similarly, Smad7 also associates with R-Smads through the MH2 domain in response to TGF-β or BMP signaling, and mutation of four basic amino acids (Lys-312, Lys-316, Lys-401, and Arg-409) in the Smad7 MH2 domain impairs the Smad3-Smad7 interaction. These four amino acids have been reported to mediate the binding of Smad7 to activated TβRI (40), suggesting that the R-Smad-Smad7 interaction shares a structural similarity as the TβRI-Smad interaction or the R-Smad-Smad4 interaction. Indeed, structure modeling of the Smad7 MH2 domain revealed the existence of a highly positively charged region including the L3 loop and the β8 strand, similar to that of the Smad3 MH2 (9, 11, 44). Intriguingly, different from those of R-Smads or Smad4, an α helix containing Lys-312 and Lys-316 also contributes to the basic region in Smad7 MH2.
In addition to interacting with R-Smads, Smad7 was also shown to associate with Smad4 upon TGF-β or BMP signaling. This is consistent with a previous study, which showed that the Smad4-Smad7 interaction depends on ligand-activated R-Smads (37). It is possible that Smad7 interacts with either Smad3 or Smad4 and thus prevents the R-Smad-Smad4 complex formation. However, we cannot rule out other possibilities, as Smad7 can form a homodimer (data not shown). Moreover, Smad6, the other inhibitory Smad that is more specifically involved in BMP pathways, was also able to oligomerize with Smad7 in the presence or absence of BMP signaling, with unknown biological significance. Together these results implicate that oligomerization could be an intrinsic and general nature of Smad family proteins.
In line with the importance of MH2 domains in mediating the interactions of both Smad7 and Smad4 with active R-Smads, Smad7 competed with Smad4 for R-Smad binding in a dose-dependent manner. Interestingly, as observed in the Smad7-expressing stable cells, TGF-β induced a rapid association of Smad2/3 with Smad4 or with Smad7, and then both interactions decreased gradually along with the decreasing phosphorylated R-Smad level. As Smad7 is a target gene of TGF-β/BMP signaling, it is possible that endogenous R-Smad-Smad7 interaction would increase along with signaling activation and duration to serve as a negative feedback loop. Taken into account that Smad6 also completes with Smad4 to interact with BMPs-activated Smad1 (45), it could be a general mechanism for the two inhibitory Smads to oligomerize with R-Smads, leading to inhibition of their signaling activity.
Although deletion of the PY motif in Smad7 had little effect on the interaction of Smad7 with Smad3, it alleviated Smad7-mediated inhibition of Smad3 activity substantially. The PY motif is important for recruitment of WW-HECT type E3 ubiquitin ligases such as Smurf1/2, NEDD4L (NEDD 4–2) and WWP1 (1, 8, 15, 42). NEDD4L has been shown to associate with and degrade both Smad2 and Smad3 upon TGF-β signaling (41, 43). In this study we found that Smad7 enhanced the interaction of Smad2/3 with NEDD4L and facilitated NEDD4L-induced polyubiquitination and proteasomal degradation of phospho-Smad3 and the turnover of Smad3(2D), a phosphorylation mimicking-mutant of Smad3. Therefore, Smad7 could act as an adaptor to recruit NEDD4L not only to the receptors but also to active R-Smads, and NEDD4L degrades R-Smads either directly or indirectly via Smad7. Moreover, Smurf2, which has been demonstrated to associate with Smad2/3 upon TGF-β signaling and target them for ubiquitination (46, 47), could also cooperate with Smad7 to inhibit Smad2/3 activity in Gal4-luciferase reporter assays (data not shown), suggesting that NEDD4L, Smurf2, and other potential WW-HECT type E3 ubiquitin ligases could be engaged in Smad7-mediated inhibition of R-Smad activity via a similar mechanism.
Author Contributions
Y.-G. C. and X. Y. designed the study and wrote the manuscript. X. Y. carried out most experiments and analyzed the data. H. L. modeled the structure of Smad7 MH2 domain. M. C. generated some of the Smad3 mutant constructs. X. S. prepared plasmids for the bimolecular fluorescence complementation experiment. X.-H. F. and X. L. prepared plasmids for the Gal4-luciferase reporter system, established the MDA-MB-231 stable cells, and revised the manuscript.
Acknowledgment
We are grateful to Dr. Wei Wu (Tsinghua University) for affording us with bimolecular fluorescence complementation-related constructs.
This work was supported by National Natural Science Foundation of China (NSFC) Grants 31401198 (to X. Y.) and 31330049 and the Ministry of Science and Technology-973 Program (2013CB933700; to Y.-G. C.). The authors declare that they have no conflicts of interest with the contents of this article.
- BMP
- bone morphogenetic protein
- ca-
- constitutively active
- Co-Smad
- common Smad
- IP
- immunoprecipitation
- MH
- mad homology
- R-Smad
- receptor-regulated Smad
- TβRI
- type I TGF-β receptor
- TβRII
- type II TGF-β receptor
- DBD
- DNA binding domain
- FL
- full-length
- NLS
- nuclear localization signal.
References
- 1.Xu P., Liu J., and Derynck R. (2012) Post-translational regulation of TGF-β receptor and Smad signaling. FEBS Lett. 586, 1871–1884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Moustakas A., and Heldin C. H. (2009) The regulation of TGFβ signal transduction. Development 136, 3699–3714 [DOI] [PubMed] [Google Scholar]
- 3.Massagué J. (2008) TGFβ in Cancer. Cell 134, 215–230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wakefield L. M., and Hill C. S. (2013) Beyond TGFβ: roles of other TGFβ superfamily members in cancer. Nat. Rev. Cancer 13, 328–341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ikushima H., and Miyazono K. (2010) TGFβ signalling: a complex web in cancer progression. Nat. Rev. Cancer 10, 415–424 [DOI] [PubMed] [Google Scholar]
- 6.Massagué J. (2012) TGFβ signalling in context. Nat. Rev. Mol. Cell Biol. 13, 616–630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pardali E., Goumans M. J., and ten Dijke P. (2010) Signaling by members of the TGF-β family in vascular morphogenesis and disease. Trends Cell Biol. 20, 556–567 [DOI] [PubMed] [Google Scholar]
- 8.Lönn P., Morén A., Raja E., Dahl M., and Moustakas A. (2009) Regulating the stability of TGFβ receptors and Smads. Cell Res. 19, 21–35 [DOI] [PubMed] [Google Scholar]
- 9.Macias M. J., Martin-Malpartida P., and Massagué J. (2015) Structural determinants of Smad function in TGF-β signaling. Trends Biochem. Sci. 40, 296–308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Feng X. H., and Derynck R. (2005) Specificity and versatility in TGF-β signaling through Smads. Annu. Rev. Cell Dev. Biol. 21, 659–693 [DOI] [PubMed] [Google Scholar]
- 11.Shi Y., and Massagué J. (2003) Mechanisms of TGF-β signaling from cell membrane to the nucleus. Cell 113, 685–700 [DOI] [PubMed] [Google Scholar]
- 12.Massagué J. (1998) TGF-β signal transduction. Annu. Rev. Biochem. 67, 753–791 [DOI] [PubMed] [Google Scholar]
- 13.Zhang L., Zhou F., and ten Dijke P. (2013) Signaling interplay between transforming growth factor-β receptor and PI3K/AKT pathways in cancer. Trends Biochem. Sci. 38, 612–620 [DOI] [PubMed] [Google Scholar]
- 14.Zhang Y. E. (2009) Non-Smad pathways in TGF-β signaling. Cell Res. 19, 128–139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yan X., and Chen Y. G. (2011) Smad7: not only a regulator, but also a cross-talk mediator of TGF-β signalling. Biochem. J. 434, 1–10 [DOI] [PubMed] [Google Scholar]
- 16.Beppu H. (2013) Smad7-modified alleles by various gene-targeting strategies. J. Biochem. 153, 399–401 [DOI] [PubMed] [Google Scholar]
- 17.Hayashi H., Abdollah S., Qiu Y., Cai J., Xu Y. Y., Grinnell B. W., Richardson M. A., Topper J. N., Gimbrone M. A. Jr., Wrana J. L., and Falb D. (1997) The MAD-related protein Smad7 associates with the TGFβ receptor and functions as an antagonist of TGFβ signaling. Cell 89, 1165–1173 [DOI] [PubMed] [Google Scholar]
- 18.Nakao A., Afrakhte M., Morén A., Nakayama T., Christian J. L., Heuchel R., Itoh S., Kawabata M., Heldin N. E., Heldin C. H., and ten Dijke P. (1997) Identification of Smad7, a TGFβ-inducible antagonist of TGF-β signalling. Nature 389, 631–635 [DOI] [PubMed] [Google Scholar]
- 19.Zhang J., Zhang X., Xie F., Zhang Z., van Dam H., Zhang L., and Zhou F. (2014) The regulation of TGF-β/SMAD signaling by protein deubiquitination. Protein Cell 5, 503–517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Herhaus L., and Sapkota G. P. (2014) The emerging roles of deubiquitylating enzymes (DUBs) in the TGFβ and BMP pathways. Cell. Signal. 26, 2186–2192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang S., Fei T., Zhang L., Zhang R., Chen F., Ning Y., Han Y., Feng X. H., Meng A., and Chen Y. G. (2007) Smad7 antagonizes transforming growth factor β signaling in the nucleus by interfering with functional Smad-DNA complex formation. Mol. Cell. Biol. 27, 4488–4499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yan X., Pan J., Xiong W., Cheng M., Sun Y., Zhang S., and Chen Y. (2014) Yin Yang 1 (YY1) synergizes with Smad7 to inhibit TGF-β signaling in the nucleus. Sci. China Life Sci. 57, 128–136 [DOI] [PubMed] [Google Scholar]
- 23.Zhu L., Chen S., and Chen Y. (2011) Unraveling the biological functions of Smad7 with mouse models. Cell Biosci. 1, 44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bian L., Han G., Zhao C. W., Garl P. J., and Wang X. J. (2015) The role of Smad7 in oral mucositis. Protein Cell 6, 160–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Luo L., Li N., Lv N., and Huang D. (2014) SMAD7: a timer of tumor progression targeting TGF-β signaling. Tumour Biol. 35, 8379–8385 [DOI] [PubMed] [Google Scholar]
- 26.Stolfi C., Marafini I., De Simone V., Pallone F., and Monteleone G. (2013) The dual role of Smad7 in the control of cancer growth and metastasis. Int. J. Mol. Sci. 14, 23774–23790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ha Thi H. T., Kim H. Y., Choi S. W., Kang J. M., Kim S. J., and Hong S. (2015) Smad7 modulates epidermal growth factor receptor turnover through sequestration of c-Cbl. Mol. Cell. Biol. 35, 2841–2850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Monteleone G., Pallone F., and MacDonald T. T. (2004) Smad7 in TGF-β-mediated negative regulation of gut inflammation. Trends Immunol. 25, 513–517 [DOI] [PubMed] [Google Scholar]
- 29.Briones-Orta M. A., Tecalco-Cruz A. C., Sosa-Garrocho M., Caligaris C., and Macías-Silva M. (2011) Inhibitory Smad7: emerging roles in health and disease. Curr. Mol. Pharmacol. 4, 141–153 [PubMed] [Google Scholar]
- 30.Yan X., Zhang J., Pan L., Wang P., Xue H., Zhang L., Gao X., Zhao X., Ning Y., and Chen Y. G. (2011) TSC-22 promotes transforming growth factor β-mediated cardiac myofibroblast differentiation by antagonizing Smad7 activity. Mol. Cell. Biol. 31, 3700–3709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Feng X. H., Liang Y. Y., Liang M., Zhai W., and Lin X. (2002) Direct interaction of c-Myc with Smad2 and Smad3 to inhibit TGF-β-mediated induction of the CDK inhibitor p15(Ink4B). Mol. Cell 9, 133–143 [DOI] [PubMed] [Google Scholar]
- 32.Ding Y., Su S., Tang W., Zhang X., Chen S., Zhu G., Liang J., Wei W., Guo Y., Liu L., Chen Y. G., and Wu W. (2014) Enrichment of the β-catenin-TCF complex at the S and G2 phases ensures cell survival and cell cycle progression. J. Cell Sci. 127, 4833–4845 [DOI] [PubMed] [Google Scholar]
- 33.Yan X., Lin Z., Chen F., Zhao X., Chen H., Ning Y., and Chen Y. G. (2009) Human BAMBI cooperates with Smad7 to inhibit transforming growth factor-β signaling. J. Biol. Chem. 284, 30097–30104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yan X., Zhang J., Sun Q., Tuazon P. T., Wu X., Traugh J. A., and Chen Y. G. (2012) p21-Activated kinase 2 (PAK2) inhibits TGF-β signaling in Madin-Darby canine kidney (MDCK) epithelial cells by interfering with the receptor-Smad interaction. J. Biol. Chem. 287, 13705–13712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ding Y., Zhang Y., Xu C., Tao Q. H., and Chen Y. G. (2013) HECT domain-containing E3 ubiquitin ligase NEDD4L negatively regulates Wnt signaling by targeting dishevelled for proteasomal degradation. J. Biol. Chem. 288, 8289–8298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dennler S., Itoh S., Vivien D., ten Dijke P., Huet S., and Gauthier J. M. (1998) Direct binding of Smad3 and Smad4 to critical TGF-β-inducible elements in the promoter of human plasminogen activator inhibitor-type 1 gene. EMBO J. 17, 3091–3100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Morén A., Imamura T., Miyazono K., Heldin C. H., and Moustakas A. (2005) Degradation of the tumor suppressor Smad4 by WW and HECT domain ubiquitin ligases. J. Biol. Chem. 280, 22115–22123 [DOI] [PubMed] [Google Scholar]
- 38.Zhao X., Nicholls J. M., and Chen Y. G. (2008) Severe acute respiratory syndrome-associated coronavirus nucleocapsid protein interacts with Smad3 and modulates transforming growth factor-β signaling. J. Biol. Chem. 283, 3272–3280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Aragón E., Goerner N., Xi Q., Gomes T., Gao S., Massagué J., and Macias M. J. (2012) Structural basis for the versatile interactions of Smad7 with regulator WW domains in TGF-β Pathways. Structure 20, 1726–1736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mochizuki T., Miyazaki H., Hara T., Furuya T., Imamura T., Watabe T., and Miyazono K. (2004) Roles for the MH2 domain of Smad7 in the specific inhibition of transforming growth factor-β superfamily signaling. J. Biol. Chem. 279, 31568–31574 [DOI] [PubMed] [Google Scholar]
- 41.Kuratomi G., Komuro A., Goto K., Shinozaki M., Miyazawa K., Miyazono K., and Imamura T. (2005) NEDD4–2 (neural precursor cell expressed, developmentally down-regulated 4–2) negatively regulates TGF-β (transforming growth factor-β) signalling by inducing ubiquitin-mediated degradation of Smad2 and TGF-β type I receptor. Biochem. J. 386, 461–470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Komuro A., Imamura T., Saitoh M., Yoshida Y., Yamori T., Miyazono K., and Miyazawa K. (2004) Negative regulation of transforming growth factor-β (TGF-β) signaling by WW domain-containing protein 1 (WWP1). Oncogene 23, 6914–6923 [DOI] [PubMed] [Google Scholar]
- 43.Gao S., Alarcón C., Sapkota G., Rahman S., Chen P. Y., Goerner N., Macias M. J., Erdjument-Bromage H., Tempst P., and Massagué J. (2009) Ubiquitin ligase Nedd4L targets activated Smad2/3 to limit TGF-β signaling. Mol. Cell 36, 457–468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chacko B. M., Qin B. Y., Tiwari A., Shi G., Lam S., Hayward L. J., De Caestecker M., and Lin K. (2004) Structural basis of heteromeric smad protein assembly in TGF-β signaling. Mol. Cell 15, 813–823 [DOI] [PubMed] [Google Scholar]
- 45.Hata A., Lagna G., Massagué J., and Hemmati-Brivanlou A. (1998) Smad6 inhibits BMP/Smad1 signaling by specifically competing with the Smad4 tumor suppressor. Genes Dev. 12, 186–197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tang L. Y., Yamashita M., Coussens N. P., Tang Y., Wang X., Li C., Deng C. X., Cheng S. Y., and Zhang Y. E. (2011) Ablation of Smurf2 reveals an inhibition in TGF-β signalling through multiple mono-ubiquitination of Smad3. EMBO J. 30, 4777–4789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wrighton K. H., and Feng X. H. (2008) To TGFβ or not to TGFβ: fine-tuning of Smad signaling via post-translational modifications. Cell. Signal. 20, 1579–1591 [DOI] [PMC free article] [PubMed] [Google Scholar]