Abstract
Histone lysine methylation is important in early zebrafish development; however, the role of histone arginine methylation in this process remains unclear. H3R2me2a, generated by protein arginine methyltransferase 6 (Prmt6), is a repressive mark. To explore the role of Prmt6 and H3R2me2a during zebrafish embryogenesis, we identified the maternal characteristic of prmt6 and designed two prmt6-specific morpholino-oligos (MOs) to study its importance in early development, application of which led to early epiboly defects and significantly reduced the level of H3R2me2a marks. prmt6 mRNA could rescue the epiboly defects and the H3R2me2a reduction in the prmt6 morphants. Functionally, microarray data demonstrated that growth arrest and DNA damage-inducible, α, a (gadd45αa) was a significantly up-regulated gene in MO-treated embryos, the activity of which was linked to the activation of the p38/JNK pathway and apoptosis. Importantly, gadd45αa MO and p38/JNK inhibitors could partially rescue the defect of prmt6 morphants, the downstream targets of Prmt6, and the apoptosis ratios of the prmt6 morphants. Moreover, the results of ChIP quantitative real time PCR and luciferase reporter assay indicated that gadd45αa is a repressive target of Prmt6. Taken together, these results suggest that maternal Prmt6 is essential to early zebrafish development by directly repressing gadd45αa.
Keywords: apoptosis, c-Jun N-terminal kinase (JNK), histone methylation, p38, zebrafish, gadd45αa, prmt6
Introduction
During embryogenesis, the embryo undergoes dramatic developmental changes. After fertilization, the embryo is engaged in a rapid and synchronous series of cleavages, which are absent of gene transcription and instead regulated by maternal mRNA and proteins. At a certain critical period thereafter, the cell cycle becomes longer and asynchronous, zygotic genome activation starts, and maternal materials begin to degrade (1, 2). This process is known as the maternal-zygotic transition in all animals, paralleling the midblastula transition (MBT)2 of zebrafish. At the end of this transition, epiboly, which is the spreading and thinning of blastomere cells to enclose the yolk cell and which has been reported to be controlled by many maternal factors (3, 4), is initiated and plays an essential role in the progress of gastrulation. Therefore, maternal factors drive early embryogenesis, and a long standing issue in developmental biology is the identity of the maternal transcripts that are vital for early embryogenesis (5–7).
Early embryo development comes down to transcriptional activation and repression, which are precisely controlled by the state of the local chromatin (9). In this respect, histone methylation, including lysine and arginine methylation, might be an important post-translational modification involved in transcriptional regulation during early embryogenesis. In animal sperm, developmental genes are marked by histone lysine methylation, such as H3K4me2/3 and H3K27me3 in human and mouse spermatozoa (10) and H3K4me2/3, H3K27me3, H3K36me3, and H3K9me3 in zebrafish sperm (11). The modified chromatin in sperm could be transmitted to embryo through fertilization (8, 11, 12). Before the MBT, H3K4me3, H3K9me3, and H3K27me3 mark the zebrafish embryonic genome; they are transcriptionally silent and predict a propensity for transcriptional activation after the MBT (8). After the MBT, H3K4me3 levels increase and mark more than 80% of genes, including many inactive developmental regulatory genes that are also marked by H3K27me3, poising genes for their transcription (13).
Histone arginine methylations are generated by proteinarginine methyltransferases (Prmts) (14). It is known that H3R2me2a is methylated primarily by Prmt6, preventing the binding of the SET1 methyltransferase complex or WDR5 (15, 16). Therefore, H3R2me2a mutually antagonizes H3K4me3 (17) and is known as a transcription-repressive mark. Prmt6 has been reported to regulate numerous biological process, including transcription (18–21), DNA repair (22), DNA replication (23), and signal transduction (24). These functions agree with the nuclear localization of Prmt6 (25). To date, many target genes of Prmt6 have been reported, including HoxA2 (17), thrombospondin-1 (26), p21 (20), p27 (27), p53 (21), Oct4 and Nanog (28), CD41 (29), and IL-6 (24).
The literature discussed above highlights the importance of histone lysine methylation for embryonic development; however, the roles of histone arginine methylation in early embryogenesis remain unknown. The goal of this study was to explore the role of Prmt6 and H3R2me2a during zebrafish embryogenesis. Our results indicate that Prmt6 is essential for early zebrafish development and acts by repressing growth arrest and DNA damage-inducible, α, a (gadd45αa).
Experimental Procedures
Zebrafish Maintenance and Injection
Zebrafish (Danio rerio) of the AB strain were provided by the Zebrafish Core Facility at the Shanghai Institute of Biochemistry and Cell Biology, and all experimental protocols were approved by the Institutional Animal Care and Use Committee. The embryos were harvested from the breeding tanks using a sieve and used for microinjection at the one- to two-cell stage (3 nl/embryo).
Antibodies and Other Reagents
The following antibodies were used for Western blotting: anti-H3R2me2a (1:500; 04-808, Millipore), anti-H3K4me3 (1:2,000; ab8580, Abcam), anti-H3 (1:5,000; p30266, Abmart), anti-Tp53 Z-FISH® (1:500; AS-55925s, AnaSpec), anti-PhosphoPlus® c-Jun (Ser-73) (1:1000; Duet 8222, Cell Signaling Technology), anti-β-actin (1:10,000; KC-5A08, KangChen Bio-tech), and horseradish peroxidase-conjugated secondary antibody (1:4,000; goat anti-rabbit; sc2030, Santa Cruz Biotechnology). The following antibodies (2 μg of each) were used for the ChIP experiments: anti-Myc (M20002, Abmart), anti-H3 (p30266, Abmart), anti-H3R2me2a and anti-H3K4me3 from Dr. Degui Chen at our institution, and normal mouse IgG (sc-2025, Santa Cruz Biotechnology). The following kits and reagents were also used: EZ ChIP kit 22 assays (17-371RF, Millipore), p38/JNK inhibitors SB203580/SP600125 (Selleck Chemicals, Houston, TX), mMESSAGE mMACHINE® kit (AM1340, Ambion), ReverTra Ace qPCR-RT kit (FSQ-101, Toyobo), UltraSYBR Mixture (with ROX Reference Dye; CW0956, CWBIO); radioimmune precipitation assay lysis buffer (PL005, Sangon Biotech), Fast Mutagenesis System (FM111-01, TransGen Biotech), and Annexin V-FITC/PI Apoptosis Detection kit (CW2574, CWBIO).
Plasmid Construction
The coding sequences of zebrafish prmt6 and gadd45αa were subcloned into the EcoRI and XbaI sites in the pCS2+ vector. The catalytically inactive Prmt6 (VLD to KLA) was generated by the Fast Mutagenesis System using the prmt6 plasmids as reported previously (23). The primer sequences are listed in Table 1. All mRNAs were synthesized using the mMESSAGE mMACHINE kit and used to induce overexpression or co-injected with the prmt6 morpholino-oligos (MOs) for the rescue experiments.
TABLE 1.
The sequences of MOs and primers
F, forward; R, reverse.
| Name | Sequence (5′–3′) | Amplicon length | Analysis |
|---|---|---|---|
| bp | |||
| prmt6-MO1 | ggccatgcgtctttagtttttccac | Translation blocking | |
| prmt6-control MO1 | ggcgatccctctttactttttgcac | 5-bp mismatch | |
| prmt6-MO2 | caagtcgcttccgtttctgaaaggc | Translation blocking | |
| prmt6-control MO2 | caactcgattccctttctcaaagcc | 5-bp mismatch | |
| hsp70-MO | tagcgattccttttggagaagacat | Translation blocking | |
| p53-MO | gcgccattgctttgcaagaattg | Translation blocking | |
| gadd45αa-MO | gttcttcaaaagtcatgttgcatgt | Translation blocking | |
| tbpl2-MO | gtaagcatcaaactccagacctgct | Splice blocking | |
| Full prmt6 F | atggccaacttgtgtaaaatgtca | 1068 | RT-PCR |
| Full prmt6 R | ttatttaacttctaaatactggtca | RT-PCR | |
| Mutational prmt6 F | gaggggaaagtgaagctagctgtgggagccggtacc | 1068 | RT-PCR |
| Mutational prmt6 R | ggtaccggctcccacagctagcttcactttcccctc | RT-PCR | |
| Probe of prmt6 F | aggacagtacggcgtggaca | 752 | RT-PCR |
| Probe of prmt6 R | cccagacacttaacatacaactcca | RT-PCR | |
| prmt6-5′-UTR F | gcctttcagaaacggaagcgact | 256 | RT-PCR |
| prmt6-5′-UTR R | ccggctcccacatctagcacc | RT-PCR | |
| gadd45αa-5′-UTR F | agttattcaagtccgttctggat | 262 | RT-PCR |
| gadd45αa-5′-UTR R | agagtgaagtgaatctgaagc | RT-PCR | |
| Full gadd45αa F | atgacttttgaagaacttaacggag | 492 | RT-PCR |
| Full gadd45αa R | tcatcgctctggaaggttgataatg | RT-PCR | |
| gadd45αa promoter F | ttctagttggaagggattgc | 1053 | RT-PCR |
| gadd45αa promoter R | gactttgcgaggcgttacat | RT-PCR | |
| prmt6 F | ccgagctggacttgaacacg | 150 | RT-PCR |
| prmt6 R | agagcaccagggccttctcc | RT-PCR | |
| β-Actin F | cgagcaggagatgggaacc | 102 | RT-PCR |
| β-Actin R | caacggaaacgctcattgc | RT-PCR | |
| tbpl2-p1 | ctggtcctcactcatcagcag | RT-PCR | |
| tbpl2-p2 | tgataccggacgcagaat | RT-PCR | |
| tbpl2-p3 | agacacaaaaatcagcagcaca | RT-PCR | |
| c-jun F | ggagcgcaaaacttcactcg | 167 | qPCR |
| c-jun R | tcgcgtccctgttttactcc | qPCR | |
| gadd45αa F | gaatggaagtgagcacctgagt | 118 | qPCR |
| gadd45αa R | ggaagccccctttggaaaac | qPCR | |
| hsp70 F | cccaccattgaagaggtggat | 120 | qPCR |
| hsp70 R | gaaccaacacactgtgcaataaga | qPCR | |
| tbpl2 F | ggagccaagagtgaagagca | 127 | qPCR |
| tbpl2 R | agacgtcacaactcccaacc | qPCR | |
| rx3 F | ctgtgtgtcccccgtatcag | 82 | qPCR |
| rx3 R | gcgatgcttgtgtttcgtgg | qPCR | |
| gadd45αa (−107) F | caggcgcgaaaaccaatcaa | 91 | qPCR |
| gadd45αa (−107) R | tgactttgcgaggcgttaca | qPCR | |
| gadd45αa (−887) F | tgatgccatgtgttgatttgtcc | 92 | qPCR |
| gadd45αa (−887) R | cgcaatccgctcgtctttg | qPCR | |
| tbpl2 (−1107) F | ggcaacatatatttcggtctacaca | 161 | qPCR |
| tbpl2 (−1107) R | cagctgatccaaataatcgggc | qPCR | |
| tbpl2 (−172) F | cagcgaaaaaccaaacactgaac | 110 | qPCR |
| tbpl2 (−172) R | cgcatgagcggaaaacaactg | qPCR | |
| eef1a1l1 F | taccctcctcttggtcgctt | 108 | qPCR |
| eef1a1l1 R | ctttgtgaccttgccagcac | qPCR |
Morpholino Construction and Validation
MOs were obtained from Gene Tools and are listed in Table 1. To validate the efficacy of the prmt6 MOs, a fragment of the prmt6 mRNA (NM_001163988.2) from 40 to 295 (containing prmt6 MO target sequence) was inserted into the BamHI and EcoRI sites of pCS2+-GFP to construct the plasmid pCS2+-prmt6-5′-UTR-GFP. The plasmid pCS2+-prmt6-5′-UTR-GFP (100 pg) was injected alone or co-injected with the prmt6 MOs or the prmt6 control MOs (cMOs). GFP fluorescence was detected at the late gastrulation stage. In the same way, the efficacy of the gadd45αa MO was validated using the plasmid pCS2+-gadd45αa-5′-UTR-GFP, which was inserted into a fragment of the gadd45αa mRNA (NM_200576.2) from 74 to 335. To test the efficacy of the tbpl2 MO, three primers were designed in exon 5 (P1), intron 5 (P2), and exon 6 (P3) of tbpl2, respectively. The splice of intron 5 of tbpl2 was analyzed with β-actin as a loading control. The primers are listed in Table 1.
RNA Extraction, RT-PCR, and Quantitative Real Time PCR (qPCR)
Total RNA was isolated from embryos using TRIzol (15596-018, Invitrogen/Life Technologies), and cDNAs were prepared using a ReverTra Ace qPCR-RT kit. RT-PCRs were performed with 2× Taq PCR Master Mix (PT102-02, Shanghai Lifefeng Biotechnology) with the following conditions: 95 °C for 5 min followed by 26 cycles of 95 °C for 20 s, 58/62 °C (β-actin/prmt6) for 20 s, 72 °C for 30 s, and a final extension step of 72 °C for 5 min. β-Actin was used as a reference gene. The PCR products were visualized using a 2% agarose gel electrophoresis. For qPCR, an Applied Biosystems 7500 Fast Real-Time PCR System was used using UltraSYBR Mixture (with ROX) following the manufacturer's instructions. eef1a1l1 was used for gene expression normalization, and the primer sequences are shown in Table 1. All experimental results are representative of three replicates.
Whole Mount in Situ Hybridization
A fragment sequence targeting bases 656–1407 of prmt6 mRNA was subcloned with pGEM®-T Easy Vector Systems using the primers listed in Table 1. Digoxigenin-UTP-labeled antisense RNA probes were synthesized using the DIG RNA labeling kit (11 175 025 910, Roche Applied Science) after linearization with NotI, and whole mount in situ hybridization was performed as described (30).
Western Blotting Analysis
For Western blotting analysis, embryos collected at the indicated times were lysed in radioimmune precipitation assay lysis buffer (with 1% PMSF and protease inhibitor mixture) and sonicated 10 times using a Bioruptor UCD-200 (Diagenode; high, 30 s with a 30-s interval). The supernatants of the lysate were denatured by boiling for 10 min in 1% SDS after centrifugation (13,200 rpm, 4 °C, 10 min). The proteins were then resolved using 15% SDS-PAGE and transferred to a nitrocellulose membrane. The membranes were blocked with 5% nonfat milk or 1% BSA, according to the instructions for each antibody, for 1 h at room temperature and subsequently incubated with the corresponding antibodies overnight at 4 °C. The next day, the blots were washed three times with 1× TBS-Tween 20 (5 min/wash) and subsequently incubated with peroxidase-conjugated secondary antibodies for 1 h at room temperature. Antibody binding was then detected using enhanced chemiluminescence.
Apoptosis Assay
To assess apoptosis, embryos were dechorionated at 6 h postfertilization (hpf) and disaggregated into single cell suspensions in 500 μl of DMEM with 10% FCS. Next, the samples were washed and prepared for analysis of apoptosis using an Annexin V-FITC/PI Apoptosis Detection kit according to the manufacturer's instructions. Approximately 20 embryos were analyzed from each treatment group.
Microarray Analysis
RNAs were extracted by TRIzol, and to ensure the RNA integrity, the RNA integrity number was checked using an Agilent Bioanalyzer 2100 (Agilent Technologies, Santa Clara, CA). Qualified RNAs were then purified using a RNeasy Micro kit (74004, Qiagen) and an RNase-Free DNase Set (79254, Qiagen). Biotin-labeled cRNAs were obtained using a GeneChip 3′IVT Express kit (901229, Affymetrix, Santa Clara, CA). Array hybridization and washing were carried out using a GeneChip® Hybridization, Wash and Stain kit (900720, Affymetrix) in Hybridization Oven 645 (00-0331-220V, Affymetrix) and Fluidics Station 450 (00-0079, Affymetrix) following the manufacturer's instructions. Scanning of slides was performed with a GeneChip Scanner 3000 (00-00212, Affymetrix) and Command Console Software 3.1 (Affymetrix). The raw data were normalized using an MAS 5.0 algorithm with Gene Spring Software 11.0 (Agilent Technologies). A total of 15,502 probe sets were used for statistical analysis (p < 0.05, -fold change >2), and heat maps were generated using Cluster 3.0 (Michael Eisen). Gene ontology analysis was carried out using the Database for Annotation, Visualization and Integrated Discovery (DAVID) (31) v6.7.
ChIP-qPCR
A six-repeat myc epitope-coding sequence was tagged at the 5′-end of the prmt6 sequence. The myc-prmt6 mRNA was injected into one- to two-cell embryos (25 pg/embryo). At 5.3 hpf, 50 embryos were collected for the ChIP experiment as described previously (32). Cross-linking of DNA and proteins was conducted by addition of 16% formaldehyde to a final concentration of 1% for 8 min at 37 °C and then quenched by addition of 10× glycine for 5 min on ice. The samples were then sonicated using a Bioruptor UCD-200 (high, 25 times, 10 s each with 40-s interval) on ice. The ChIP procedure was performed using EZ ChIP kit 22 assays following the manufacturer's instructions. The enriched DNA samples were used for qPCR analysis using UltraSYBR Mixture (with ROX), and the primer sequences are listed in Table 1. There are only two regions on the promoter of gadd45αa that were suitable for qPCR primer design and analyzed by qPCR. The results are presented as percent input (for anti-Myc and IgG) and over anti-H3 (for anti-H3R2me2a and anti-H3K4me3). ChIP experiments were repeated three times, and the data are shown as average values ±S.D.
Luciferase Reporter Assay
The 1503-bp promoter of gadd45αa was obtained using the primers listed in Table 1 and inserted into the KpnI and HindIII sites of the pGL3-Basic vector (Promega). The pGL3-gadd45αa promoter vector (50 pg/embryo) and pRL-TK (Promega; as control reporter vector; 5 pg/embryo) were co-injected with the prmt6 MO1/cMO1 (0.3/0.3 mm) or with the pCS2+-prmt6 vector/pCS2+ vector (100/100 pg/embryo) at the one- to two-cell stage (3 nl/embryo). After 10 h, 40 embryos were collected and assayed using a Dual-Luciferase Reporter Assay System (Promega) in SynergyNEO (BioTek). Three biological repeats were performed, and the data are expressed as average values ±S.D.
Hydrogen Peroxide (H2O2) Treatment
H2O2 (A501976, Sangon Biotech) was diluted to 5 mm to produce an oxidative stress solution. The embryos were treated with 0 or 5 mm H2O2 for 2 h before sample collection. Three time points were used for analysis: 2.25, 4, and 6 hpf. ChIP experiments were repeated three times, and the data are shown as average values ±S.D.
Statistical Analysis
Statistical analyses were performed with a Student's two-tailed t test, and a p value <0.05 was considered to reflect a statistically significant difference. All data are presented as the mean ± S.D.
Results
Prmt6 Is a Maternal Transcript That Is Expressed throughout Early Zebrafish Development
The H3R2me2a level during early zebrafish development was examined by Western blotting. The H3R2me2a modification was detected between 1.25 and 6 hpf with an increasing level from 4 hpf (Fig. 1A). Although Prmt6 is responsible for the generation of the H3R2me2a mark, its expression at the protein level could not be assessed because of a lack of an effective antibody. Instead, we used RT-PCR to gauge the prmt6 expression at the mRNA level, detecting its expression in the unfertilized egg and between 2 and 10 hpf (Fig. 1B). Interestingly, the expression of prmt6 was highest before 6 hpf (Fig. 1B). To confirm this result, whole mount in situ hybridization was also performed at the indicated time points, revealing high embryonic prmt6 expression between 0.2 and 4.5 hpf (Fig. 1C). These data demonstrate that prmt6 is a maternally derived transcript, suggesting it has a role during early zebrafish development.
FIGURE 1.
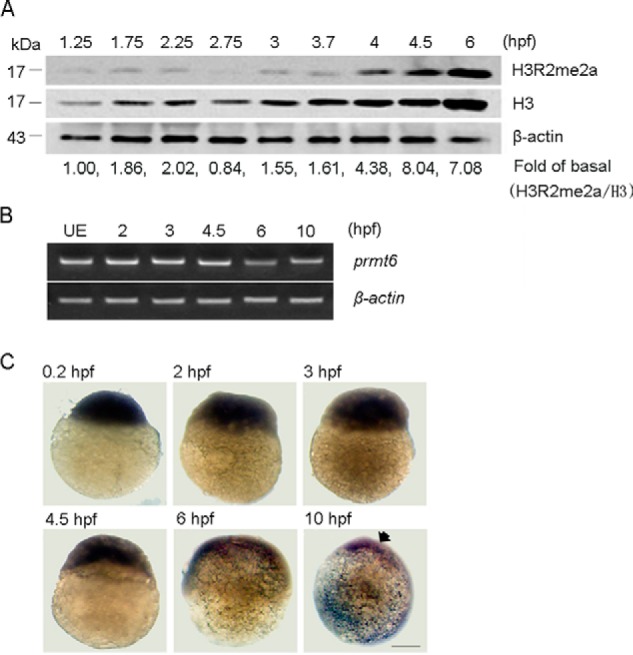
Prmt6 is expressed throughout early zebrafish development. A, a representative Western blot showing the level of H3R2me2a in normal embryos during early zebrafish development with H3 and β-actin as loading controls. -Fold of basal, the H3R2me2a ratios were determined by the densitometric value of each treatment relative to that of the control (1.25 hpf) after normalizing to the H3 densitometric values. Protein load per lane, 40 μg. B, a representative agarose gel showing the mRNA expression level of prmt6 in unfertilized eggs (UE) and during early zebrafish development with β-actin as a loading control. C, the expression pattern of prmt6 was also determined by whole mount in situ hybridization at 0.2 (lateral view), 2 (lateral view), 3 (lateral view), 4.5 (lateral view), 6 (lateral view), and 10 hpf (lateral view with dorsal toward right). Scale bar, 200 μm.
Prmt6 Is Essential for Early Zebrafish Development
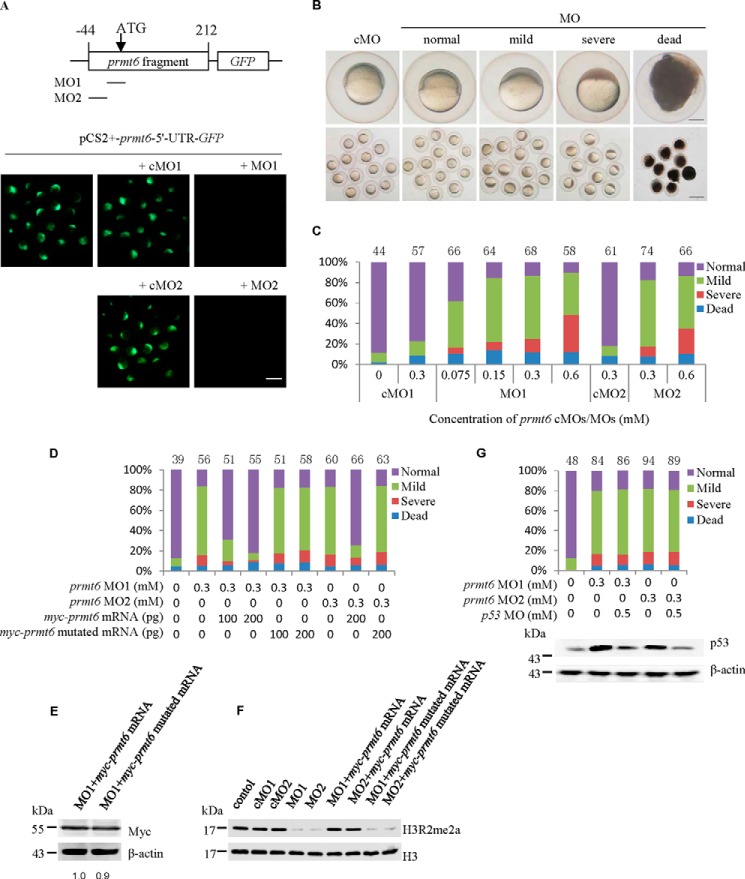
Having established that prmt6 is detectable in the developing zebrafish embryo, we decided to examine its physiological role using a morpholino-mediated gene knockdown strategy. Two pairs of prmt6-specific MOs were designed to block prmt6 translation as follows: one targeted the prmt6 ATG site (MO1) and its 5-base mismatch control (cMO1); the other targeted the prmt6 5′-UTR region (MO2) and its 5-base mismatch control (cMO2) (Fig. 2A). First, to test the effectiveness of the prmt6 MOs, the pCS2+-prmt6-5′-UTR-GFP was constructed and injected alone or co-injected with the prmt6 MOs/cMOs. Using this approach, we observed GFP fluorescence at the late gastrulation stage of embryos injected with the pCS2+-prmt6-5′-UTR-GFP that was unaffected by the prmt6 cMO1 and cMO2 but blocked by the prmt6 MO1 and MO2 (Fig. 2A). These data indicated that the prmt6 MOs could efficiently block the translation of prmt6.
FIGURE 2.
Prmt6 is essential for early zebrafish development. A, successful suppression of prmt6 expression was confirmed in embryos injected with the plasmid pCS2+-prmt6-5′-UTR-GFP and prmt6 MO1/MO2. B, knockdown of Pmrt6 expression using the prmt6 MOs resulted in a normal, mild, or severe phenotype characterized by defective epiboly as shown at 6 hpf. The single enlarged embryo is shown from lateral views with the animal pole on the top. C, a quantification of the relative distribution of the phenotype of prmt6 morphants at 6 hpf. D, a quantification of the relative distribution of the phenotype of prmt6 morphants rescued with myc-prmt6 mRNA (containing only the coding sequence) and mutated myc-prmt6 mRNA (containing only the coding sequence) that codes an inactive form of Prmt6 (VLD to KLA). E, a representative Western blot showing the protein expression level of the rescue constructs by Myc tag. -Fold of basal, the Myc ratios were determined by the densitometric value of each construct relative to that of the control (myc-prmt6 mRNA) after normalizing to the β-actin densitometric values. myc-prmt6 mRNA/myc-prmt6 mutated mRNA, 200/200 pg. F, a representative Western blot showing the effect of the prmt6 MOs on the H3R2me2a level. cMOs/MOs, 0.3/0.3 mm; myc-prmt6 mRNA/myc-prmt6 mutated mRNA, 200/200 pg. G, a quantification of the relative distribution of the phenotype of prmt6 morphants and a representative Western blot showing the p53 protein level rescued with p53 MO. Protein loads per lane for E, F, and G, 37.5 μg. In C, D, and G, the number of embryos in each group is indicated above the relevant column. Scale bar in A and B, 200 μm.
Next, the prmt6 MO1 was injected into one- to two-cell embryos at doses ranging between 0.075 and 0.6 mm. Compared with normal embryos, prmt6 MO1/cMO1-treated embryos had no obvious phenotype before and during the MBT (data not shown). However, after the MBT, the epiboly of embryos injected with prmt6 MO1 was severely compromised (Fig. 2B). We classified this effect into three subtypes (normal, mild, and severe) based on the degree of epiboly at 6 hpf. The amount and severity of the defective epiboly positively correlated with the administered dose of prmt6 MO1 (Fig. 2C). Embryos with severe epiboly arrest died between 8 and 10 hpf, whereas the survival percentage of the mildly affected embryos was 66.7, 41.5, 38.7, and 32.7% with injection of 0.075, 0.15, 0.3, and 0.6 mm prmt6 MO1, respectively. As predicted, prmt6 MO2 injection led to phenotypes similar to those of prmt6 MO1 (Fig. 2C). From these data, we determined that 0.3 mm prmt6 MOs was the optimal dose for generating an effective phenotype. The severe phenotype following MO treatment suggests that Prmt6 has an important role in early zebrafish development.
Co-injection of Prmt6 Messenger RNA Rescues the Prmt6 Morphants
To confirm that the effect of the prmt6 MOs was specific, embryos were co-injected with myc-prmt6 mRNA (containing only the coding sequence), which had only 6 bases matching those of prmt6 MO1 and was not targeted by prmt6 MO2. This experiment showed that prmt6 mRNA could rescue the prmt6 morphants and that the extent of the rescue effect was dose-dependent (Fig. 2D). Furthermore, to determine whether Prmt6 function was enzyme-dependent, a rescue experiment was performed using myc-prmt6 mRNA (containing only the coding sequence) mutated to a catalytically inactive form (VLD to KLA) by site-specific mutagenesis. This mutated mRNA was expressed at the same level as the WT myc-prmt6 mRNA as shown by Myc tag (Fig. 2E) but could not rescue the prmt6 morphants (Fig. 2D). To further study the effect of prmt6 MOs, the level of H3R2me2a marks was measured in the prmt6 morphants at 6 hpf, showing a reduction over 80% compared with that of cMO treatment that could be rescued by myc-prmt6 mRNA but not by the mutated mRNA (Fig. 2F). Besides, potential off-target effects of MO were of concern (33). To exclude this possibility, embryos were co-injected with p53 MO and prmt6 MO1/MO2. These embryos showed a decrease of p53 protein compared with the prmt6 morphants but displayed a phenotype similar to that of the prmt6 morphants (Fig. 2G). These results indicate that the phenotype of prmt6 morphants is specific and depends on the enzymatic activity of Prmt6.
Gadd45αa Is Up-regulated and Acts as a Mediator in the Prmt6 Morphants
Based on the remarkable decrease of H3R2me2a in the prmt6 morphants, we speculated that the genome of these embryos might be dramatically changed. To verify this hypothesis, a gene microarray was performed in control embryos and in the prmt6 morphants with normal, mild, and severe phenotypes. Compared with the cMO1-injected embryos, microarray analysis of the prmt6 morphant with severe phenotype identified 1,878 misregulated probe sets (824 up-regulated and 1,054 down-regulated, p < 0.05, -fold change >2), which accounted for 12% of the total 15,502 probe sets. These differential probe sets mapped to 724 up-regulated and 858 down-regulated genes (supplemental Table 1A). The differentially expressed genes were enriched for the following biological processes: pattern specification process, regionalization, embryonic morphogenesis, dorsal/ventral pattern formation, gastrulation, heart development, digestive tract morphogenesis, gut development, cell migration involved in gastrulation, sensory organ development, and some others (see supplemental Table 1B). This indicates that prmt6 MO1 altered global gene expression and development processes in the prmt6 severe morphant embryo.
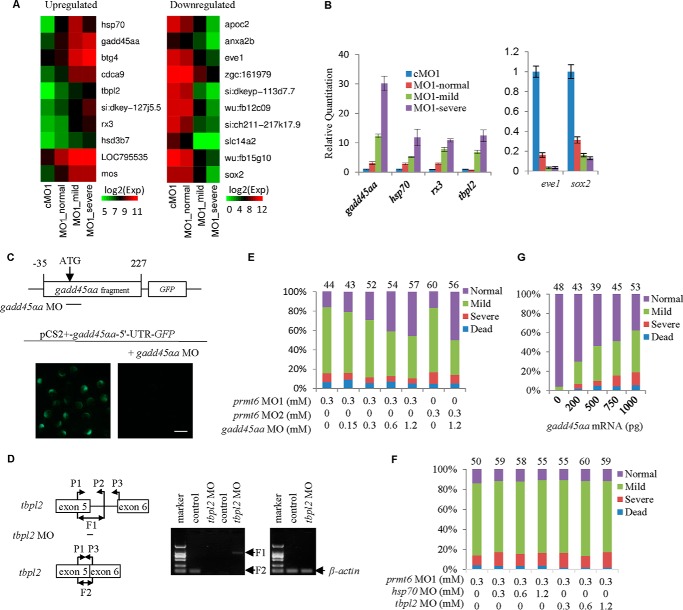
To explore the Prmt6 candidate targets, the top 10 up-regulated and down-regulated genes (see supplemental Table 1A) were studied using the three prmt6 subtypes (Fig. 3A). To verify the microarray data, four genes were selected from the top 10 up-regulated list, namely gadd45αa, hsp70, tbpl2, and rx3, and two genes were selected from the top 10 down-regulated list, namely eve1 and sox2. Our RT-qPCR analysis of these genes confirmed the microarray data with the exception of hsp70 in the severe phenotype group (Fig. 3B). These data suggest a relationship between the phenotype of the prmt6 morphants and the degree of dysregulation of the top Prmt6 targets.
FIGURE 3.
Inhibition of the up-regulation of gadd45αa partially rescues the prmt6 morphants. A, a microarray analysis of the prmt6 morphants at 6 hpf was conducted, and the top 10 up-regulated and down-regulated genes are shown. B, microarray data were validated by RT-qPCR, confirming that prmt6 MO1 induced the expression of gadd45αa, hsp70, tbpl2, and rx3 and decreased the expression of eve1 and sox2. C, a gadd45αa MO was designed and validated in embryos injected with the plasmid pCS2+-gadd45αa-5′-UTR-GFP and the gadd45αa MO. D, a splice-blocking tbpl2 MO was designed and validated according to the strategy described under “Experimental Procedures” with β-actin as a loading control. Co-injection of a gadd45αa MO partially rescues the prmt6 morphants in a dose-dependent manner (E), whereas co-injection of hsp70 MO or tbpl2 MO has no obvious effects on the prmt6 morphants (F). G, interestingly, a quantification of the relative distribution of embryonic phenotypes at 6 hpf showed that overexpression of gadd45αa phenocopies the prmt6 morphants. In E, F, and G, the number of embryos in each group is indicated above the relevant column. Scale bar in C, 200 μm. Error bars represent S.D.
Given the reported suppressive function of Prmt6 (19–21) and the decrease of repressive modifications in the prmt6 morphants (Fig. 2F), we hypothesized that the prmt6 morphants might be a result of up-regulation of Prmt6 target genes. Interestingly, gadd45αa, a member of the Gadd45 stress sensor genes involved in activation of the p38/JNK pathway and apoptosis via activation of MEKK4 (34–36), was the top up-regulated gene in the list (Fig. 3, A and B). Besides, some reports have demonstrated that Prmt6 is involved with proliferation and apoptosis (19–21). Based on this, we hypothesized that the up-regulation of gadd45αa might be responsible for the prmt6 morphants. To test this hypothesis, we designed a gadd45αa translation-blocking MO, which was validated using the plasmid pCS2+-gadd45αa-5′-UTR-GFP (Fig. 3C). It was used to rescue the defects of the prmt6 MOs, showing that the gadd45αa MO could partially rescue the prmt6 morphants in a dose-dependent manner (Fig. 3E). We further analyzed the roles of hsp70 and the maternal and zygotic tbpl2 in the top 10 up-regulated list using a designed and validated tbpl2 MO (Fig. 3D) and a reported hsp70 MO (37, 38). However, neither tbpl2 MO nor hsp70 MO could rescue the prmt6 morphants (Fig. 3F). We also found that one- to two-cell embryos injected with gadd45αa mRNA for a gain of function analysis displayed a phenotype similar to that of prmt6 MO-treated embryos in a dose-dependent manner (Fig. 3G). These data suggest that the up-regulation of gadd45αa is responsible for the prmt6 morphants.
The Up-regulation of Gadd45αa in the Prmt6 Morphants Is Associated with Activation of the p38/JNK Pathway and Apoptosis
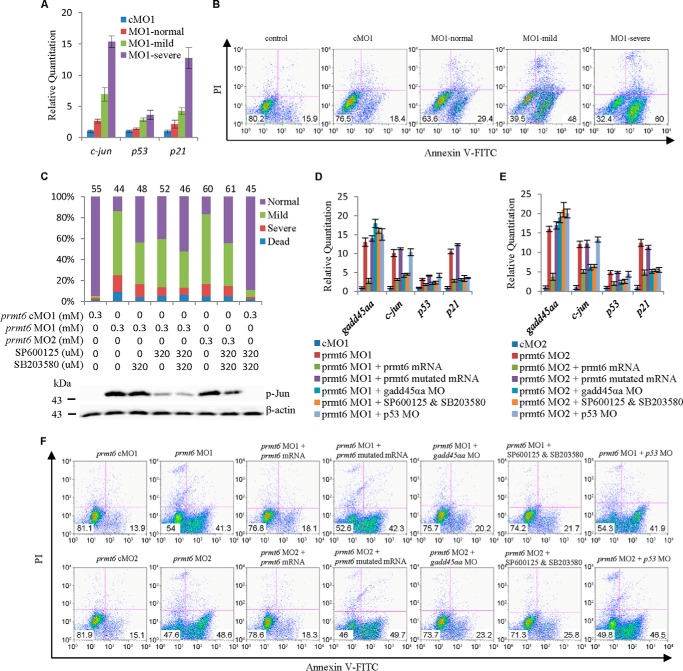
To test whether the up-regulation of stress-inducible gadd45αa in the prmt6 morphants could lead to the activation of the p38/JNK pathway, qPCR analysis of p38/JNK targets was carried out, showing that the transcription of c-jun, p53, and p21 were up-regulated in the prmt6 morphants (Fig. 4A). Furthermore, analysis of apoptosis showed that early apoptosis (Annexin V+/PI−) increased in the prmt6 morphants, particularly in the severe phenotype group, with an apoptosis ratio of 60 versus 18.6% in cMO1-injected embryos (Fig. 4B). Therefore, prmt6 MO1 might activate the p38/JNK pathway through up-regulation of gadd45αa, thereby causing apoptosis.
FIGURE 4.
The p38/JNK pathway is activated in the prmt6 morphants. A, downstream targets of the p38/JNK pathway (c-jun, p53, and p21) were up-regulated in the prmt6 morphants. B, the extent of apoptosis was measured in the prmt6 morphants by flow cytometry. C, co-injection with inhibitors of the p38/JNK pathway partially rescued the prmt6 morphants. The inhibition of the phosphorylation of c-Jun by SP600125 was shown by Western blotting. Representative quantitative analyses of targets of Prmt6 by RT-qPCR following rescue are shown (D and E). F, in agreement with the above data, the prmt6 mRNA, gadd45αa MO, and p38/JNK pathway inhibitors decreased the ratios of early apoptosis (Annexin V+/PI−) in the prmt6 morphant, whereas no obvious effects of the prmt6 mutated mRNA and p53 MO were observed. In C, the number of embryos in each group is indicated above the relevant column. Protein load per lane, 37.5 μg. Error bars represent S.D.
Based on the above data, we performed a series of experiments to determine the importance of the p38/JNK signaling pathway in mediating the effect of the prmt6 MOs. To confirm the importance of the p38/JNK pathway in this scenario, embryos were treated with the p38/JNK inhibitors SB203580/SP600125 (39, 40). The efficacy of SP600125 was validated by analyzing the phosphorylation level of c-Jun (Fig. 4C). However, the efficacy of SB203580 could not be tested because of a lack of an effective antibody. As expected, p38/JNK inhibitors partially rescued the prmt6 morphants with the two inhibitors having a mild additive effect (Fig. 4C).
To detect whether the Prmt6 downstream targets could be rescued, gadd45αa, c-jun, p53, and p21 were analyzed in the embryos of rescue experiments using qPCR. This showed that the prmt6 mRNA could rescue the expression level of gadd45αa, c-jun, p53, and p21 and that the gadd45αa MO and the p38/JNK inhibitors could rescue the expression level of c-jun, p53, and p21 (Fig. 4, D and E). Accordingly, analysis of apoptosis showed that early apoptosis increased in the prmt6 MO1 morphants with an apoptosis ratio of 41.3 versus 13.9% in cMO1-injected embryos. This could be rescued by the prmt6 mRNA, the gadd45αa MO, and the p38/JNK inhibitors to 18.1, 20.2, and 21.7%, respectively, but not by the prmt6 mutated mRNA or the p53 MO (Fig. 4F). The analysis of apoptosis in the rescue experiments showed a similar trend using prmt6 MO1 and MO2 (Fig. 4F). Taken together, these data show that activation of the p38/JNK pathway by up-regulation of gadd45αa is responsible for the phenotype of prmt6 morphants.
Gadd45αa Is a Repressive Target of Prmt6
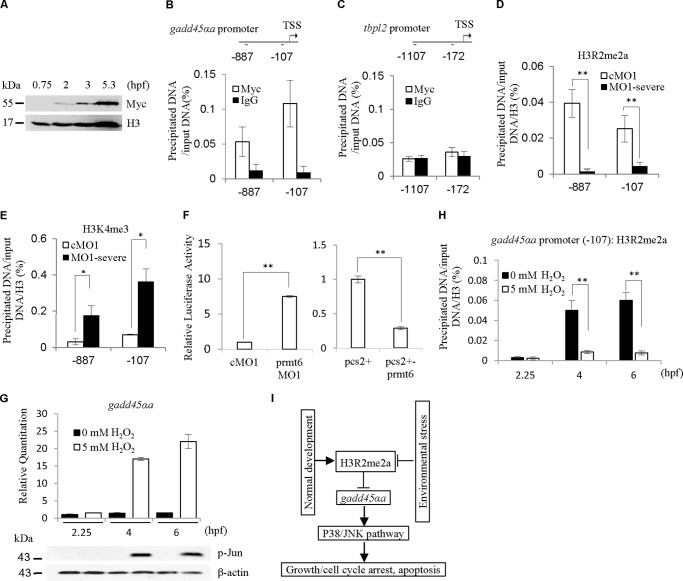
To check whether Prmt6 directly regulates gadd45αa expression, we performed a ChIP analysis with 5.3-hpf embryos. The myc-prmt6 mRNA was injected into one- to two-cell embryos (25 pg/embryo) without abnormal phenotypes, and the embryos were collected for the ChIP experiment. The myc-prmt6 translation could be detected by Western blotting from 2 to 5.3 hpf (Fig. 5A). The data show that Prmt6 binds to the gadd45αa promoter (Fig. 5B). As a control, the maternal and zygotic tbpl2 gene was also assayed but was found to display no significant enrichment at the tbpl2 promoter (Fig. 5C).
FIGURE 5.
Gadd45αa is a direct target of Prmt6. A, a representative Western blot showing the protein expression level of the myc-prmt6 mRNA using Myc tag. myc-prmt6 mRNA, 25 pg. Our ChIP data revealed that Prmt6 binds to the promoter of gadd45αa (B) but not tbpl2 (C). Furthermore, prmt6 MO1 decreases the modification of H3R2me2a on the promoter of gadd45αa (D) but increases the modification of H3K4me3 (E). F, in agreement with the above data, the luciferase activity assay demonstrated that prmt6 MO1 enhances the activity of the promoter of gadd45αa, whereas prmt6 mRNA represses the activity of the promoter of gadd45αa. Under H2O2 treatment, the transcription of gadd45αa was induced, the JNK pathway was activated (G), and the H3R2me2a level at the gadd45αa promoter (−107) was decreased (H) at 4 and 6 hpf. I, a diagram illustrating the requirement of H3R2me2a for early embryogenesis through preventing the activation of the p38/JNK pathway by directly suppressing gadd45αa. Protein load per lane for A and G, 37.5 μg. *, p < 0.05; **, p < 0.01. Error bars represent S.D. TSS, transcriptional start site.
H3R2me2a is generated by Prmt6 and antagonizes H3K4me3 (15, 16). We analyzed the changes of H3R2me2a and H3K4me3 in the gadd45αa promoter of the prmt6 severe morphant using antibodies for H3R2me2a, H3K4me3, and H3. The results showed that the prmt6 MO1 remarkably reduced H3R2me2a levels and significantly increased H3K4me3 levels on the promoter of gadd45αa (Fig. 5, D and E). To confirm the direct regulation of the promoter of gadd45αa by Prmt6, the pGL3-gadd45αa promoter vector was constructed for the luciferase reporter assay, which showed that prmt6 MO1 significantly enhanced the activity of the promoter of gadd45αa, whereas prmt6 mRNA markedly suppressed the activity of the promoter of gadd45αa (Fig. 5F).
Based on the above data, we tried to explore the effects on the functional transcription of gadd45αa of environmental stress caused by 5 mm H2O2 treatment. The data from three time points (2.25, 4, and 6 hpf) showed that the transcription level of gadd45αa during early zebrafish development is low (Fig. 5G). Under the H2O2 treatment, the transcription of gadd45αa was induced, and the JNK pathway was activated at 4 and 6 hpf (Fig. 5G). Furthermore, we also observed that the H3R2me2a level at gadd45αa promoter, which is higher at 4 and 6 hpf than at 2.25 hpf during normal development, was decreased at 4 and 6 hpf by the H2O2 treatment (Fig. 5H). These data suggest that the response of gadd45αa to stress might be through the modification of H3R2me2a levels at gadd45αa promoter. Taken together, and as summarized in Fig. 5I, these data indicate that gadd45αa is directly suppressed through the modification of H3R2me2a generated by Prmt6 to prevent apoptosis induced by the activation of the p38/JNK pathway during early zebrafish development.
Discussion
In this study, knockdown of Prmt6 using MO decreased the global level of H3R2me2a and led to early epiboly arrest, highlighting an important role for maternal Prmt6 in early zebrafish development. Our mechanistic studies revealed that the prmt6 MO directly up-regulated gadd45αa and subsequently activated p38/JNK pathway signaling and induced apoptosis, thereby explaining the phenotype of prmt6 morphants.
Previous studies have demonstrated that Prmt6 plays important roles in numerous biological process (19–21, 26, 29). However, the importance of the H3R2me2a and Prmt6 in embryogenesis remains unknown. In this study, we initially detected the existence of the H3R2me2a mark during early zebrafish development (Fig. 1A). This result is consistent with a previous report that four-cell-stage mouse embryos are characterized by high levels of H3R2me2a (41). In addition, we generated a zebrafish catalytically inactive Prmt6 form (VLD to KLA) by means of the reported human methylase-inactive PRMT6 mutant (23). This mutant could not increase the level of H3R2me2a in the prmt6 morphants as expected (Fig. 2F). This is evidence of the conservation of Prmt6 between zebrafish and human (14).
Many direct targets of Prmt6 have been reported (19–21, 26). In this study, we determined the misregulated genes in the prmt6 morphants through microarray analysis and proved that the up-regulation of gadd45αa (Fig. 3E), not the up-regulation of hsp70 and tpbl2 (Fig. 3F), is responsible for the prmt6 morphants. Although the Gadd45αa protein level could not be detected because of a lack of an effective antibody, the results from qPCR (Fig. 3B), quantitative ChIP (Fig. 5, B, D, and E), and the luciferase reporter assay (Fig. 5F) confirmed that gadd45αa is directly regulated by Prmt6. The up-regulation of hsp70 might be a response to the activation of the p38/JNK pathway induced by the up-regulation of gadd45αa, and the up-regulation of tpbl2 might be the result of the failure of degradation of maternal mRNA. This needs to be confirmed by further studies.
Of note, gadd45αa MO could only partially rescue the prmt6 morphants, suggesting that additional targets of Prmt6 exist. Interestingly, there were 858 down-regulated genes in the prmt6 severe morphant that included zygotic genes and demonstrated the failure of zygotic genome activation. Therefore, whether Prmt6 directly contributes to zygotic genome activation through generating active modifications, such as H4R3me2a and H3R42me2a (14, 18), deserves to be investigated. Additional ChIP-sequencing studies are required to determine whether the misregulated genes (see supplemental Table 1A) are directly regulated by Prmt6.
The timing and the mediators involved at the start of apoptosis in different animals have been identified (42–44). In zebrafish, the apoptotic response is activated during development at 8 hpf (45) as zBik, a key factor in regulating apoptosis, is not expressed until 8 hpf (46). In this study, apoptosis was observed at 6 hpf in the prmt6 morphants (Fig. 4, B and F) that was the result of the up-regulation of gadd45αa by knockdown of Prmt6. These results reveal that the high maternal level of Prmt6 might be one of the reasons for the absence of apoptosis in early zebrafish embryos.
Given the specificities of SP600125 (47), JNK-IN-8, another JNK inhibitor, was also used and found to partially rescue the prmt6 morphants with results similar to SP600125 (data not shown). A high mortality rate of embryos incubated in 0.8 μm SP600125 has been reported (48). However, this was not seen in our experiment (Fig. 4C), which suggests that the amount of SP600125 injected (320 μm; 3 nl/embryo) was low. These results indicated that the rescue by SP600125 is specific.
In conclusion, this study identified that maternal Prmt6 is required for early embryogenesis by preventing apoptosis through direct suppression of gadd45αa. Other reports demonstrate that PRMT6 is up-regulated in diverse cancer types, promoting growth and suppressing apoptosis, which is of potential significance for cancer therapeutics (19, 20, 26). These studies suggest that the balance of Prmt6 plays an important role in proliferation and apoptosis and that further study of this protein may prove of importance to developmental biology and anticancer research.
Author Contributions
X.-X. Z., P.-L. N., and Z.-L. W. conceived and performed the experiments. X.-X. Z., Y.-B. Z., and Y.-P. L. analyzed the data. X.-X. Z. and Y.-B. Z. prepared the figures. X.-X. Z., Y.-B. Z., Y.-C. Y., and Y.-P. L. wrote the manuscript. All authors reviewed the results and approved the final version of the manuscript.
Acknowledgments
We appreciate the members of Germ Cell and Embryo Development Research Group, Shanghai Institute of Biochemistry and Cell Biology (SIBCB) for the kind help during the progress of this work. We thank Prof. Degui Chen Lab (SIBCB) for the generous gift of antibodies, the data analysis, and the valuable suggestions for manuscript preparation.
This work was supported by Major State Basic Research Development Program of China (973 Program) Grants 2011CB966301 and 2011CB915502 and Science and Technology Commission of Shanghai Municipality Grant 11140900100. The authors declare that they have no conflict of interest.
The microarray data have been deposited in the NCBI Gene Expression Omnibus (GEO) with GEO Series Accession number GSE63735.

This article contains supplemental Table 1.
- MBT
- midblastula transition
- Prmt
- protein arginine methyltransferase
- gadd45αa
- growth arrest and DNA damage-inducible
- α
- a
- MO
- morpholino-oligo
- hpf
- hours postfertilization
- qPCR
- quantitative real time PCR
- cMO
- control MO
- PI
- propidium iodide.
References
- 1.Kane D. A., and Kimmel C. B. (1993) The zebrafish midblastula transition. Development 119, 447–456 [DOI] [PubMed] [Google Scholar]
- 2.Schier A. F. (2007) The maternal-zygotic transition: death and birth of RNAs. Science 316, 406–407 [DOI] [PubMed] [Google Scholar]
- 3.Zhang W. W., Zhang Y. B., Zhao X. X., Hua Y., Wu Z. L., Yan Y. C., and Li Y. P. (2015) Prmt7 regulates epiboly by facilitating 2-OST and modulating actin cytoskeleton. J. Mol. Cell Biol. 7, 489–491 [DOI] [PubMed] [Google Scholar]
- 4.Song S., Eckerle S., Onichtchouk D., Marrs J. A., Nitschke R., and Driever W. (2013) Pou5f1-dependent EGF expression controls E-cadherin endocytosis, cell adhesion, and zebrafish epiboly movements. Dev. Cell 24, 486–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Miyamoto K., Teperek M., Yusa K., Allen G. E., Bradshaw C. R., and Gurdon J. B. (2013) Nuclear Wave1 is required for reprogramming transcription in oocytes and for normal development. Science 341, 1002–1005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee M. T., Bonneau A. R., Takacs C. M., Bazzini A. A., DiVito K. R., Fleming E. S., and Giraldez A. J. (2013) Nanog, Pou5f1 and SoxB1 activate zygotic gene expression during the maternal-to-zygotic transition. Nature 503, 360–364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Leichsenring M., Maes J., Mössner R., Driever W., and Onichtchouk D. (2013) Pou5f1 transcription factor controls zygotic gene activation in vertebrates. Science 341, 1005–1009 [DOI] [PubMed] [Google Scholar]
- 8.Lindeman L. C., Andersen I. S., Reiner A. H., Li N., Aanes H., Østrup O., Winata C., Mathavan S., Müller F., Aleström P., and Collas P. (2011) Prepatterning of developmental gene expression by modified histones before zygotic genome activation. Dev. Cell 21, 993–1004 [DOI] [PubMed] [Google Scholar]
- 9.Kolasinska-Zwierz P., Down T., Latorre I., Liu T., Liu X. S., and Ahringer J. (2009) Differential chromatin marking of introns and expressed exons by H3K36me3. Nat. Genet. 41, 376–381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brykczynska U., Hisano M., Erkek S., Ramos L., Oakeley E. J., Roloff T. C., Beisel C., Schübeler D., Stadler M. B., and Peters A. H. (2010) Repressive and active histone methylation mark distinct promoters in human and mouse spermatozoa. Nat. Struct. Mol. Biol. 17, 679–687 [DOI] [PubMed] [Google Scholar]
- 11.Wu S. F., Zhang H., and Cairns B. R. (2011) Genes for embryo development are packaged in blocks of multivalent chromatin in zebrafish sperm. Genome Res. 21, 578–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Arico J. K., Katz D. J., van der Vlag J., and Kelly W. G. (2011) Epigenetic patterns maintained in early Caenorhabditis elegans embryos can be established by gene activity in the parental germ cells. PLoS Genet. 7, e1001391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vastenhouw N. L., Zhang Y., Woods I. G., Imam F., Regev A., Liu X. S., Rinn J., and Schier A. F. (2010) Chromatin signature of embryonic pluripotency is established during genome activation. Nature 464, 922–926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang Y. C., and Li C. (2012) Evolutionarily conserved protein arginine methyltransferases in non-mammalian animal systems. FEBS J. 279, 932–945 [DOI] [PubMed] [Google Scholar]
- 15.Guccione E., Bassi C., Casadio F., Martinato F., Cesaroni M., Schuchlautz H., Lüscher B., and Amati B. (2007) Methylation of histone H3R2 by PRMT6 and H3K4 by an MLL complex are mutually exclusive. Nature 449, 933–937 [DOI] [PubMed] [Google Scholar]
- 16.Kirmizis A., Santos-Rosa H., Penkett C. J., Singer M. A., Vermeulen M., Mann M., Bähler J., Green R. D., and Kouzarides T. (2007) Arginine methylation at histone H3R2 controls deposition of H3K4 trimethylation. Nature 449, 928–932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hyllus D., Stein C., Schnabel K., Schiltz E., Imhof A., Dou Y., Hsieh J., and Bauer U. M. (2007) PRMT6-mediated methylation of R2 in histone H3 antagonizes H3 K4 trimethylation. Genes Dev. 21, 3369–3380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Casadio F., Lu X., Pollock S. B., LeRoy G., Garcia B. A., Muir T. W., Roeder R. G., and Allis C. D. (2013) H3R42me2a is a histone modification with positive transcriptional effects. Proc. Natl. Acad. Sci. U.S.A. 110, 14894–14899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stein C., Riedl S., Rüthnick D., Nötzold R. R., and Bauer U. M. (2012) The arginine methyltransferase PRMT6 regulates cell proliferation and senescence through transcriptional repression of tumor suppressor genes. Nucleic Acids Res. 40, 9522–9533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Phalke S., Mzoughi S., Bezzi M., Jennifer N., Mok W. C., Low D. H., Thike A. A., Kuznetsov V. A., Tan P. H., Voorhoeve P. M., and Guccione E. (2012) p53-independent regulation of p21Waf1/Cip1 expression and senescence by PRMT6. Nucleic Acids Res. 40, 9534–9542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Neault M., Mallette F. A., Vogel G., Michaud-Levesque J., and Richard S. (2012) Ablation of PRMT6 reveals a role as a negative transcriptional regulator of the p53 tumor suppressor. Nucleic Acids Res. 40, 9513–9521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.El-Andaloussi N., Valovka T., Toueille M., Steinacher R., Focke F., Gehrig P., Covic M., Hassa P. O., Schär P., Hübscher U., and Hottiger M. O. (2006) Arginine methylation regulates DNA polymerase β. Mol. Cell 22, 51–62 [DOI] [PubMed] [Google Scholar]
- 23.Boulanger M. C., Liang C., Russell R. S., Lin R., Bedford M. T., Wainberg M. A., and Richard S. (2005) Methylation of Tat by PRMT6 regulates human immunodeficiency virus type 1 gene expression. J. Virol. 79, 124–131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Di Lorenzo A., Yang Y., Macaluso M., and Bedford M. T. (2014) A gain-of-function mouse model identifies PRMT6 as a NF-κB coactivator. Nucleic Acids Res. 42, 8297–8309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Frankel A., Yadav N., Lee J., Branscombe T. L., Clarke S., and Bedford M. T. (2002) The novel human protein arginine N-methyltransferase PRMT6 is a nuclear enzyme displaying unique substrate specificity. J. Biol. Chem. 277, 3537–3543 [DOI] [PubMed] [Google Scholar]
- 26.Michaud-Levesque J., and Richard S. (2009) Thrombospondin-1 is a transcriptional repression target of PRMT6. J. Biol. Chem. 284, 21338–21346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kleinschmidt M. A., de Graaf P., van Teeffelen H. A., and Timmers H. T. (2012) Cell cycle regulation by the PRMT6 arginine methyltransferase through repression of cyclin-dependent kinase inhibitors. PLoS One 7, e41446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee Y. H., Ma H., Tan T. Z., Ng S. S., Soong R., Mori S., Fu X. Y., Zernicka-Goetz M., and Wu Q. (2012) Protein arginine methyltransferase 6 regulates embryonic stem cell identity. Stem Cells Dev. 21, 2613–2622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Herglotz J., Kuvardina O. N., Kolodziej S., Kumar A., Hussong H., Grez M., and Lausen J. (2013) Histone arginine methylation keeps RUNX1 target genes in an intermediate state. Oncogene 32, 2565–2575 [DOI] [PubMed] [Google Scholar]
- 30.Thisse C., and Thisse B. (2008) High-resolution in situ hybridization to whole-mount zebrafish embryos. Nat. Protoc. 3, 59–69 [DOI] [PubMed] [Google Scholar]
- 31.Huang da W., Sherman B. T., and Lempicki R. A. (2009) Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44–57 [DOI] [PubMed] [Google Scholar]
- 32.Lindeman L. C., Vogt-Kielland L. T., Aleström P., and Collas P. (2009) Fish'n ChIPs: chromatin immunoprecipitation in the zebrafish embryo. Methods Mol. Biol. 567, 75–86 [DOI] [PubMed] [Google Scholar]
- 33.Robu M. E., Larson J. D., Nasevicius A., Beiraghi S., Brenner C., Farber S. A., and Ekker S. C. (2007) p53 activation by knockdown technologies. PLoS Genet. 3, e78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Miyake Z., Takekawa M., Ge Q., and Saito H. (2007) Activation of MTK1/MEKK4 by GADD45 through induced N-C dissociation and dimerization-mediated trans autophosphorylation of the MTK1 kinase domain. Mol. Cell. Biol. 27, 2765–2776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Takekawa M., and Saito H. (1998) A family of stress-inducible GADD45-like proteins mediate activation of the stress-responsive MTK1/MEKK4 MAPKKK. Cell 95, 521–530 [DOI] [PubMed] [Google Scholar]
- 36.Abell A. N., Granger D. A., and Johnson G. L. (2007) MEKK4 stimulation of p38 and JNK activity is negatively regulated by GSK3β. J. Biol. Chem. 282, 30476–30484 [DOI] [PubMed] [Google Scholar]
- 37.Evans T. G., Yamamoto Y., Jeffery W. R., and Krone P. H. (2005) Zebrafish Hsp70 is required for embryonic lens formation. Cell Stress Chaperones 10, 66–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bruns A. F., Yuldasheva N., Latham A. M., Bao L., Pellet-Many C., Frankel P., Stephen S. L., Howell G. J., Wheatcroft S. B., Kearney M. T., Zachary I. C., and Ponnambalam S. (2012) A heat-shock protein axis regulates VEGFR2 proteolysis, blood vessel development and repair. PLoS One 7, e48539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hsu R. J., Lin C. C., Su Y. F., and Tsai H. J. (2011) dickkopf-3-related gene regulates the expression of zebrafish myf5 gene through phosphorylated p38a-dependent Smad4 activity. J. Biol. Chem. 286, 6855–6864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yabu T., Shiba H., Shibasaki Y., Nakanishi T., Imamura S., Touhata K., and Yamashita M. (2015) Stress-induced ceramide generation and apoptosis via the phosphorylation and activation of nSMase1 by JNK signaling. Cell Death Differ. 22, 258–273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Torres-Padilla M. E., Parfitt D. E., Kouzarides T., and Zernicka-Goetz M. (2007) Histone arginine methylation regulates pluripotency in the early mouse embryo. Nature 445, 214–218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Coucouvanis E., and Martin G. R. (1995) Signals for death and survival: a two-step mechanism for cavitation in the vertebrate embryo. Cell 83, 279–287 [DOI] [PubMed] [Google Scholar]
- 43.Nakajima K., Takahashi A., and Yaoita Y. (2000) Structure, expression, and function of the Xenopus laevis caspase family. J. Biol. Chem. 275, 10484–10491 [DOI] [PubMed] [Google Scholar]
- 44.Yamashita M. (2003) Apoptosis in zebrafish development. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 136, 731–742 [DOI] [PubMed] [Google Scholar]
- 45.Ikegami R., Hunter P., and Yager T. D. (1999) Developmental activation of the capability to undergo checkpoint-induced apoptosis in the early zebrafish embryo. Dev. Biol. 209, 409–433 [DOI] [PubMed] [Google Scholar]
- 46.Zhong J. X., Zhou L., Li Z., Wang Y., and Gui J. F. (2014) Zebrafish Noxa promotes mitosis in early embryonic development and regulates apoptosis in subsequent embryogenesis. Cell Death Differ. 21, 1013–1024 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 47.Bain J., McLauchlan H., Elliott M., and Cohen P. (2003) The specificities of protein kinase inhibitors: an update. Biochem. J. 371, 199–204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xiao Y., Zhou Y., Xiong Z., Zou L., Jiang M., Luo Z., Wen S., Liu W., Liu S., and Li W. (2013) Involvement of JNK in the embryonic development and organogenesis in zebrafish. Mar. Biotechnol. 15, 716–725 [DOI] [PubMed] [Google Scholar]