Abstract
Candidate gene studies have reported CYP19A1 variants to be associated with endometrial cancer and with estradiol (E2) concentrations. We analyzed 2937 single nucleotide polymorphisms (SNPs) in 6608 endometrial cancer cases and 37 925 controls and report the first genome wide-significant association between endometrial cancer and a CYP19A1 SNP (rs727479 in intron 2, P=4.8×10−11). SNP rs727479 was also among those most strongly associated with circulating E2 concentrations in 2767 post-menopausal controls (P=7.4×10−8). The observed endometrial cancer odds ratio per rs727479 A-allele (1.15, CI=1.11–1.21) is compatible with that predicted by the observed effect on E2 concentrations (1.09, CI=1.03–1.21), consistent with the hypothesis that endometrial cancer risk is driven by E2. From 28 candidate-causal SNPs, 12 co-located with three putative gene-regulatory elements and their risk alleles associated with higher CYP19A1 expression in bioinformatical analyses. For both phenotypes, the associations with rs727479 were stronger among women with a higher BMI (Pinteraction=0.034 and 0.066 respectively), suggesting a biologically plausible gene-environment interaction.
Keywords: endometrial cancer, CYP19A1, estradiol
Introduction
Established risk factors for endometrial cancer include high BMI (IARC 2002), early menarche, late menopause, nulliparity, estrogen-only hormone replacement therapy (HRT) (Beral et al. 2005) and tamoxifen use (Cuzick et al. 2003), while cigarette smoking and the use of oral contraceptives or combined HRT (Beral et al. 2005) are associated with lower risks. It has been hypothesized that these factors alter endometrial cancer risk by increasing exposure to estrogens (Key & Pike 1988); indeed, higher concentrations of circulating estradiol (E2) in post-menopausal women have been associated with an increased risk of endometrial cancer (Zeleniuch-Jacquotte et al. 2001, Lukanova et al. 2004, Allen et al. 2008).
After the cessation of ovarian estrogen production at menopause, endogenous estrogens are primarily synthesized from testosterone (T) in adipose tissue via aromatase, encoded by CYP19A1. Candidate gene studies have found levels of E2 in pre-menopausal and post-menopausal women, and also in men, to be associated with genetic variants within or close to CYP19A1 (Dunning et al. 2004, Paynter et al. 2005, Haiman et al. 2007, Ahn et al. 2009, Eriksson et al. 2009, Kidokoro et al. 2009, Travis et al. 2009, Beckmann et al. 2011, Lundin et al. 2012, Prescott et al. 2012, Flote et al. 2014). Candidate studies have also identified associations between several different CYP19A1 variants and endometrial cancer (Paynter et al. 2005, Tao et al. 2007, Setiawan et al. 2009, Low et al. 2010), with some evidence of stronger associations in women with higher BMI (Setiawan et al. 2009).
None of the published studies have attempted a systematic assessment of all common CYP19A1 variants in order to determine i) which are most likely to be causal for endometrial cancer and/or E2 concentration, ii) whether multiple independent causal variants exist at this locus for either trait, and iii) whether the same variant or variants are responsible for both traits. The latter would help to address the question as to whether the reported association between E2 and endometrial cancer seen in epidemiological studies is causal or a consequence of confounding (Fig. 1). If the association is causal, then variants causally associated with E2 levels should also be associated with endometrial cancer, with a magnitude that can be predicted using a Mendelian randomization methodology (C-Reactive Protein Coronary Heart Disease Genetics Collaboration et al. 2011, Interleukin-6 Receptor Mendelian Randomisation Analysis Consortium 2012), a form of instrumental variable analysis in which the instrument is a genetic variant(s) known to be associated with the biomarker in a particular direction.
Figure 1.
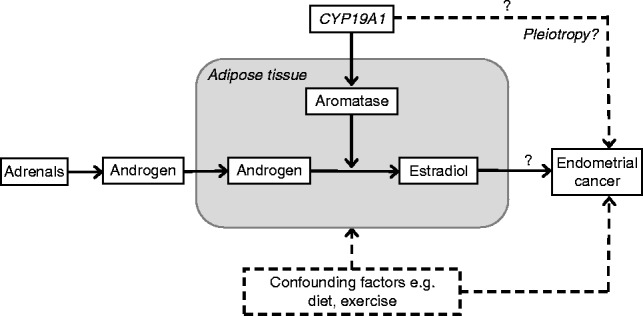
Factors potentially involved in the reported association between circulating post-menopausal E2 and endometrial cancer risk. Question marks highlight the issues to be addressed in this study.
To address the question of whether the same CYP19A1 variant(s) are associated with E2 levels and endometrial cancer with compatible effect sizes and directions, we used genotype information for 2937 single nucleotide polymorphisms (SNPs) across a 1.2 Mb region encompassing CYP19A1 in 6608 endometrial cancer cases and 37 925 controls of European ancestry, 1733 of whom (all controls) were post-menopausal and had measured E2 and T concentrations.
Materials and methods
Endometrial cancer case–control studies
The association between SNPs at the CYP19A1 locus and endometrial cancer was tested using data from four separate case-control studies:
The ANECS, SEARCH, and NSECG genomewide association studies
The results presented here are based on the ANECS, SEARCH, and NSECG genomewide association studies (GWAS) and country-matched datasets (McGregor et al. 1999, WTCCC 2007, Houlston et al. 2010, McEvoy et al. 2010, Painter et al. 2011, Spurdle et al. 2011), as shown in Supplementary Table 1A and B, see section on supplementary data given at the end of this article, and described in detail in (Painter et al. 2015). All cases and controls were of European ancestry.
The ECAC iCOGS study
The fourth dataset comprised 4402 women of European ancestry with a confirmed diagnosis of endometrial cancer (3535 with confirmed endometrioid histology), recruited via 11 separate studies in seven countries in the Endometrial Cancer Association Consortium (ECAC; Painter et al. 2015) and 28 758 healthy female controls from the same countries, all participating in the Breast Cancer Association Consortium (BCAC; Michailidou et al. 2013) or the Ovarian Cancer Association Consortium (OCAC; Pharoah et al. 2013), plus 282 Norwegian blood donor controls (see Supplementary Information, Supplementary Table 1A and B, see section on supplementary data given at the end of this article). Cases and controls were genotyped for 211 155 SNPs using a custom Illumina Infinium iSelect array (‘iCOGS’; Michailidou et al. 2013, Pharoah et al. 2013, Painter et al. 2015) designed by the Collaborative Oncological Gene-environment Study (‘COGS’). The iCOGS array includes 134 SNPs located within the ∼1.2 Mb region of chromosome 15 between 50 899 000–52 095 000 surrounding CYP19A1, 22 of which had been specifically selected for the study of post-menopausal E2 levels.
Post-genotyping quality control for all four studies was as described in (Spurdle et al. 2011) and (Painter et al. 2015). Individuals with <85% estimated European ancestry based on Identity-By-State (IBS) scores between study individuals and individuals in HapMap (http://hapmap.ncbi.nlm.nih.gov/) were excluded.
E2 datasets
The EPIC Norfolk study
Sex-hormone levels, including E2 and T concentrations, were measured on subsets of the ∼25 000 participants in the European Prospective Investigation of Cancer-Norfolk cohort study (see Day et al. (1999) for details). After recruitment, participants were invited to attend a first health check (HC1), at which a blood sample was taken. A second blood sample was taken from participants who attended the second health check (HC2), ∼3 years after the first. For each set of blood samples, a subset were randomly selected for hormone level measurement from among the women who were considered to be post-menopausal based on being >55 years, not having menstruated in the last year, and having not taken HRT for at least 3 months before sampling (Supplementary Table 1C, see section on supplementary data given at the end of this article). The plasma and serum samples collected from these women had been stored at −70 °C until analysis, and their whole blood samples had been stored at −30 °C before DNA extraction. 2368 of the women for whom hormone levels had been measured had also been genotyped using the iCOGS array as BCAC control subjects (and thus were also controls in our iCOGS endometrial cancer analysis). Of these, 1333 women had hormones measured from their HC1 blood sample and 1536 from their HC2 sample, of whom 501 had hormones measured from both samples. Where two measurements existed, we chose the measurement from HC2, when women would be further from the menopause. After excluding women within 2 years of the menopause at blood draw or with missing E2 levels or E2 values >300 pmol/l (i.e., outside the possible range for a post-menopausal woman) our analysis was based on 1500 genotyped women with HC2 E2 levels and 425 women with HC1 levels. Of the HC2 women, 1431 also had a valid T measurement (T was not measured as part of HC1).
Ethical approval was obtained from the Norwich Local Research Ethics Committee, LREC 98CN01. All study participants provided written informed consent.
The SIBS study
Participants in the Sisters in Breast Screening study (SIBS) were identified through the National Health Service breast screening program in the UK (Prescott et al. 2012). A subset of 905 SIBS women who were aged over 55 years at recruitment, 2 or more years since their last menstrual period, and not currently using HRT at the time of blood collection were selected for hormone measurement (Prescott et al. 2012). After excluding women with missing or extreme E2 levels (as above), 889 women were left, of whom 302 had been genotyped using the iCOGS array as BCAC control subjects (after quality control exclusions), and thus were also controls in our iCOGS endometrial cancer analysis (Supplementary Table 1C, see section on supplementary data given at the end of this article). All participants gave informed written consent. This study was approved by the Eastern Multicentre Research Ethics Committee (SIBS).
For the EPIC and SIBS studies, plasma E2 concentrations were measured at The Royal Marsden Hospital (London, UK), using an in-house RIA using a highly specific rabbit antiserum which had been raised against an E2-6-carboxymethyloxime- BSA conjugate and E2-6-carboxymethyloxime-[2−125I] iodohistamine (Dowsett et al. 1987). The detection limit was 3 pmol/l and values were replaced with this limit when they were reported as being undetectable. For the SIBS study, at a concentration of 25 pmol/l the within assay variation was 6.5% and the between assay variation was 16% (n=18). For the EPIC studies, at a concentration of 18 pmol/l the within assay and the between-batch coefficients of variation (CV) were 8.6 and 13% respectively.
Testosterone was measured in the EPIC and SIBS studies using a solid-phase RIA kit (Diagnostic Products, Gwynedd, UK), with within- and between-batch CV at a concentration of 3.1 nmol/l of 6.1 and 10% respectively and with a detection limit of 0.14 nmol/l.
Additional SIBS replication set
To increase statistical power we genotyped all of the 889 SIBS women with E2 measurements (described above) for rs727479 using a Custom Taqman Assay (Life Technologies, ThermoFisher Scientific, Waltham, MA, USA) according to manufacturer's instructions (details provided in Supplementary Table 5, see section on supplementary data given at the end of this article). After quality control exclusions described below, 813 women had measured E2 measurements and genotypes for rs727479, of whom 264 also had valid iCOGS genotyping i.e., 549 additional samples.
Regional imputation
We used IMPUTEv2 (Howie et al. 2009) to impute genotypes for SNPs in the 50 899 000–52 095 000 region of chromosome 15 in the 1000 Genomes dataset v3 (April 2012 release). We allowed the IMPUTE Software to select the most appropriate haplotypes from among the complete set of 1000 Genomes haplotypes (Howie et al. 2011). Imputation was conducted separately for the four datasets, and SNPs with imputation information score <0.7 and/or MAF <0.01 in any of the four studies were excluded.
Statistical analysis
The four imputed endometrial cancer datasets were analyzed separately using unconditional logistic regression with a per-allele (1 degree of freedom) model using SNPTEST v2 (Ferreira & Marchini 2011). For the iCOGS dataset, analyses were performed adjusting for country and for the first ten principal components, as in (Painter et al. 2015). The GWAS datasets were each analyzed as a single stratum, with adjustment for the first two (ANECS and NSECG) and three (SEARCH) principal components. Our ongoing genome-wide analyses have shown that the inclusion of these principal components is sufficient to control for population stratification (genomic control λ=1.002–1.038).
The endometrial cancer odds ratios (OR) for the four studies were combined using standard fixed-effects meta-analyses. The I2 statistic (Higgins & Thompson 2002) was used to estimate the proportion of the variance due to between-study heterogeneity. SNPs with significant between-study heterogeneity (P<0.05) were excluded. Analyses for all SNPs were repeated adjusting for the most significant SNP to assess whether multiple independent causal variants were present. A statistical significance cut-off of P≤10−4 was used for secondary and conditional analyses. To determine the most likely candidate causative SNPs, the log likelihoods of all tested SNPs were compared with that of the top SNP. SNPs with log-likelihood ratios of <1:100 of being the top SNP and which were correlated with the top SNP (linkage disequilibrium (LD) r2>0.2) were prioritized as potentially causal variants for follow-up in bioinformatic and functional analyses (Udler et al. 2010).
The analyses were repeated restricting the iCOGS and NSECG studies to those cases with endometrioid or non-endometrioid histology (the ANECS and SEARCH GWAS sample sets contained only endometrioid histology cases).
Associations between SNPs and E2 concentrations were tested using the natural logarithm transformed ratio of E2 to T concentrations, adjusting for laboratory batch, study (EPIC HC2 or SIBS), age and BMI at blood draw, prior HRT use (yes or no) and menopausal status (2–5 years since menopause or >5 years since menopause) using ProbABEL software (Aulchenko et al. 2010). Given the family-based design of the SIBS study, we used the matrix of kinship coefficients to adjust for the non-independence of relatives. This approach is also expected to avoid the effects of population stratification (Chen & Abecasis 2007).
For the analysis including the additional genotyping in the SIBS samples, the data were re-analyzed for all 2767 women, using the sandwich variance estimator to obtain standard errors robust to familial clustering (in the absence of a kinship coefficient matrix for the complete set).
The associations between the most significant SNP and the two phenotypes (endometrial cancer and E2 concentration) were repeated after stratifying the datasets according to quartiles of age diagnosis (cases) or interview (controls) or quartiles of BMI. These analyses were restricted to the iCOGS dataset (plus the SIBS replication set for the E2 analysis), as BMI was not available for all cases and controls in the GWAS sets. Since T concentration had not been measured in the EPIC HC1 women, the analyses stratified by BMI and age were based on log-transformed E2 concentrations uncorrected for T, in order to maximise the sample size and hence the statistical power. Quartiles were based on the variables' distributions in cases, to ensure roughly equivalent statistical power across the quartiles. The same categories were used for the E2 analysis to allow direct comparisons between the two phenotypes.
We used a Mendelian randomization style approach to compare the observed association of the top SNP with endometrial cancer with that predicted based on a SNP's effect on E2 levels. For this we re-estimated the effect of each effect allele on E2 (β) adjusting only for study and laboratory batch. Using a published estimate of the endometrial cancer OR associated with a doubling of post-menopausal E2 concentration (Lukanova et al. 2004), we multiplied the natural logarithm of this OR by the ratio (lnβ/ln2) to obtain a predicted endometrial cancer OR per effect allele. We then compared this predicted OR to that observed, to assess whether the observed association between the SNP and endometrial cancer is compatible with a causal association between higher post-menopausal E2 concentration and endometrial cancer. In the same way, we compared the predicted effect of the top SNP on breast cancer risk (based on a published estimate of the effect of doubling E2 concentration on breast cancer risk (Key et al. 2002)) with that observed in the iCOGS BCAC study of 45 290 breast cancer cases and 41 880 controls of European ancestry (Michailidou et al. 2013).
All statistical analyses used R software unless otherwise stated, and all statistical tests were two-sided. The association plot was produced using LocusZoom (Pruim et al. 2010).
Results
The CYP19A1 association with endometrial cancer is explained by a single signal
Genetic imputation of the ∼1.2 Mb region of chromosome 15 between 50 899 000–52 095 000 using the April 2012 release of the 1000 Genomes reference panel in four independent case–control sets yielded post-QC genotype information for 2937 SNPs in 6608 endometrial cancer cases and 37 925 controls (Supplementary Table 1A, see section on supplementary data given at the end of this article). Of these SNPs, 100 had been genotyped in the largest study (iCOGS), and 191, 201, and 187 in the three GWAS sets (SEARCH, ANECS, and NSECG GWASs respectively).
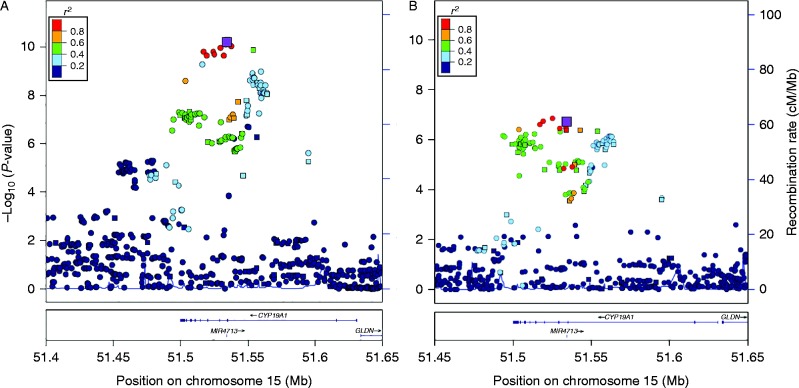
Combining results across the four studies, 171 SNPs had P<1×10−4, compared with an expected number of less than one under the null hypothesis (Supplementary Table 2, see section on supplementary data given at the end of this article). Fifty SNPs were significant at the conventional GWAS threshold of 5×10−8, of which rs727479 in intron 2 was the most significantly associated (OR per A allele=1.15, CI=1.11–1.21, P=4.81×10−11, Table 1, Fig. 2A). This SNP was directly genotyped in all four studies, and the strength of the association did not differ among studies (I2=0.0%, Phet=0.92). (Supplementary Table 2, see section on supplementary data given at the end of this article, Supplementary Fig. 1A). Conditioning on rs727479, no other SNPs reached P<10−4 (Supplementary Table 2).
Table 1.
SNP rs727479 A-allele associations with endometrial cancer and with circulating E2 levels
| Ncases/Nctrls | fcase/fctrl | OR (95% CI) | P value | Pinteraction | |
|---|---|---|---|---|---|
| Endometrial cancer | 6608/37 925 | 0.688/0.651 | 1.15 (1.11, 1.21) | 4.81×10−11 | |
| BMI Q1 (<24.3 kg/m2) | 715/5463 | 0.668/0.643 | 1.08 (0.95, 1.24) | 0.25 | |
| BMI Q2 (24.3–28.0 kg/m2) | 718/4372 | 0.677/0.648 | 1.12 (0.98, 1.29) | 0.10 | |
| BMI Q3 (28.0–33.2 kg/m2) | 702/2910 | 0.697/0.648 | 1.21 (1.04, 1.40) | 0.012 | |
| BMI Q4 (≥33.2 kg/m2) | 721/1254 | 0.709/0.651 | 1.25 (1.05, 1.49) | 0.012 | 0.047 |
| Age Q1 (<57.0 years) | 1005/14 106 | 0.679/0.653 | 1.10 (1.00, 1.22) | 0.060 | |
| Age Q2 (57.0–63.0years) | 1079/5309 | 0.679/0.652 | 1.10 (0.98, 1.23) | 0.10 | |
| Age Q3 (63.0–69.0 years) | 1145/4699 | 0.693/0.644 | 1.28 (1.15, 1.44) | 1.73×10−5 | |
| Age Q4 (≥69.0 years) | 1081/2948 | 0.701/0.651 | 1.24 (1.08, 1.42) | 0.0019 | 0.19 |
| Histology: | |||||
| Endometrioid | 5611/37 925 | 0.690/0.651 | 1.16 (1.11, 1.22) | 1.12×10−10 | |
| Non-endometrioid | 887/37 925 | 0.678/0.651 | 1.08 (0.98, 1.20) | 0.13 | 0.15 |
| Nsamples | f | β (se) | P value | ||
| E2 levels | 2767 | 0.656 | 0.096 (0.018) | 7.40×10−8 | |
| BMI Q1 (<24.3 kg/m2) | 868 | 0.656 | 0.052 (0.032) | 0.11 | |
| BMI Q2 (24.3–28.0 kg/m2) | 985 | 0.650 | 0.097 (0.031) | 0.0020 | |
| BMI Q3 (28.0–33.2 kg/m2) | 664 | 0.671 | 0.122 (0.036) | 8.45×10−4 | |
| BMI Q4 (≥33.2 kg/m2) | 250 | 0.642 | 0.096 (0.058) | 0.099 | 0.066 |
| Age Q1 (<57.0 years) | 287 | 0.656 | 0.095 (0.083) | 0.25 | |
| Age Q2 (57.0–63.0 years) | 802 | 0.661 | 0.104 (0.030) | 5.65×10−4 | |
| Age Q3 (63.0–69.0 years) | 789 | 0.653 | 0.080 (0.032) | 0.013 | |
| Age Q4 (≥69.0 years) | 889 | 0.654 | 0.105 (0.031) | 6.48×10−4 | 0.90 |
F, frequency of the rs727479 A allele; age is in years; Q1–Q4 are quartiles of the distribution of BMI or age in the endometrial cases. The endometrial cancer analysis by quartiles is adjusted for age. E2 concentrations are log transformed and adjusted for laboratory batch, study, age at blood draw, BMI, HRT use and menopausal status.
Figure 2.
Association of SNPs in the CYP19A1 region with (A) endometrial cancer and (B) E2:T, highlighting rs727479. Each point indicates the statistical significance of the association between a SNP and endometrial cancer (Fig. 2A) or between a SNP and the E2:T ratio (Fig. 2B). Squares denote SNPs directly genotyped by the iCOGS array; circles are SNPs for which genotypes were imputed. The larger purple square is rs727479, the SNP with the strongest evidence of association with endometrial cancer. Other colours show the strength of linkage disequilibrium between each SNP with rs7277479.
These results suggest that rs727479, or a SNP correlated with it, is causally related to disease. Based on a likelihood ratio threshold of 1:100 (Udler et al. 2010), 28 SNPs remain as possible causal variants (Supplementary Table 2, see section on supplementary data given at the end of this article); all are correlated with rs727479 at r2>0.2, and five were genotyped in iCOGS (rs7175531, rs727479, rs17601876, rs12050767, and rs749292).
Considering only the 5611 endometrioid–histology cases, 41 SNPs had P<5×10−8, of which rs727479 was the most significant (OR per A allele=1.16, CI=1.11–1.22, P=1.12×10−10, Table 1). The effect estimate was somewhat smaller in the analysis restricted to the 887 non-endometrioid cases (OR=1.08, CI=0.98–1.20, P=0.13), although the difference in allele frequencies between endometrioid and non-endometrioid cases was not significant (P=0.15, Table 1). No SNPs reached P<1×10−4 in the analysis restricted to the non-endometrioid histology cases (n=887) (Supplementary Table 2, see section on supplementary data given at the end of this article).
Stronger associations between rs727479 and endometrial cancer in women of older age and higher BMI
There was some suggestion of a stronger association between rs727479 and endometrial cancer among older women (OR=1.28 (1.15–1.44) P=1.7×10−5 and OR=1.24 (1.08–1.42) P=1.9×10−3 for the third and fourth quartiles of age respectively; Table 1), although the interaction between rs727479 and age was not significant (P=0.19).
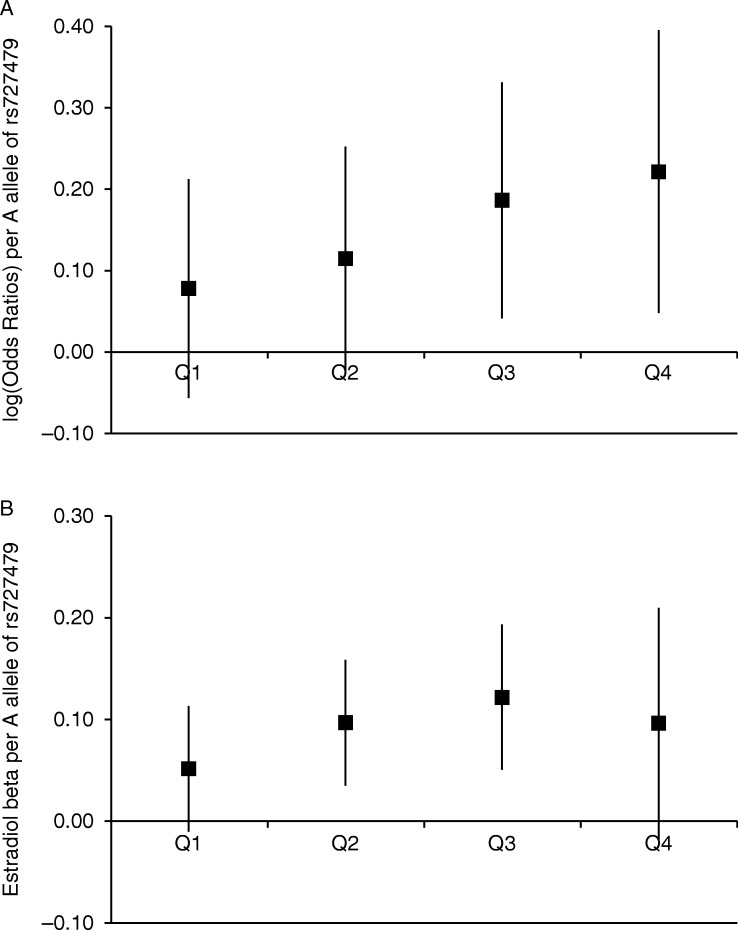
BMI was available for 2858 cases and 14 098 controls from the iCOGS studies. As expected, BMI was positively associated with endometrial cancer risk (OR=1.60 (1.54–1.66) per quartile, P<10−100). SNP rs727479 was not associated with BMI in previous analyses (P=0.94 in the >230 000 GIANT consortium participants (Locke et al. 2015))_ENREF_24 nor in the iCOGS controls (P=0.31). There was evidence of a stronger disease association for rs727479 among women with higher BMI (interaction P=0.034, which was slightly attenuated when adjusted for age, P=0.047), with the strongest association among women in the highest quartile (OR=1.25 (1.05–1.49), P=0.012, adjusting for age), (Table 1, Fig. 3A).
Figure 3.
Association of SNP rs727479 with (A) endometrial cancer and (B) E2 levels, by quartile of BMI distribution. In Fig. 3A the log(OR) of endometrial cancer associated with each A allele of SNP rs727479 is shown for each quartile of the BMI distribution, adjusting for age. There is a borderline significant interaction between genotype and BMI quartile (P=0.047). Figure 3B shows the regression coefficient (β) for the association between each A allele of rs727479 and log-transformed E2 levels (adjusted for laboratory batch, study, age at blood draw, BMI, HRT use and menopausal status), (Pinteraction=0.066). For both plots, the error bars are 95% CI, and the quartiles are based on the BMI distribution in endometrial cancer cases, to allow for comparability between plots, and to ensure sufficient cases in each quartile.
The set of correlated SNPs most significantly associated with endometrial cancer are all within the set of SNPs most significantly associated with the E2:T ratio
T is the substrate for aromatization to E2, and the ratio of E2 to T concentrations, in essence, corrects for the variation in T levels. This correction would be expected to lead to a more direct relationship with aromatase activity, hence we used the E2:T ratio as the hormonal phenotype in our initial fine-mapping of the CYP19A1 region. Circulating E2 and T concentrations were measured in 1733 healthy post-menopausal women from the EPIC Norfolk (N=1431) and SIBS (N=302) studies (Dunning et al. 2004, Prescott et al. 2012) who formed a subset of the controls in the iCOGS study. Imputation and post-imputation QC identical to that performed in the endometrial cancer analysis resulted in 1956 SNPs across the CYP19A1 region, of which 100 had been genotyped.
Adjusting for age, BMI, HRT use and menopausal status, 105 SNPs were associated with the E2:T ratio at P<1×10−4, including the lead endometrial cancer SNP rs727479 (P=2.06×10−7). Two imputed SNPs had very slightly smaller P values than rs727479 (rs12592697, P=1.46×10−7 and rs4775935, P=1.89×10−7), both of which were in near-complete LD with rs727479 (r2=0.99). Ninety four SNPs had odds of at least 1:100 compared with rs12592697 of being the causal E2:T SNP, and also have r2>0.2 with rs12592697 (Supplementary Table 2, see section on supplementary data given at the end of this article). Conditioning on rs12592697, no SNPs have P<1×10−4; hence there is no evidence of a second signal for E2:T in this region. The set of 95 SNPs contains all 28 non-excluded endometrial cancer candidate SNPs. Since rs727479 was the most significant of the genotyped SNPs and was statistically almost indistinguishable from the top two SNPs, rs727479 was used as the representative SNP for the set of 95 non-excluded SNPs. The rs727479 A allele was associated with higher E2 concentration (β=0.092, P=3.80×10−5) and, to a lesser extent, with lower T concentration (β=−0.045, P=0.057).
Including an additional 485 women from the EPIC-Norfolk cohort for whom E2 but not T concentrations had been measured (and therefore the E2:T ratio could not be computed), the association between rs727479 and E2 became stronger (β=0.094, P=3.1×10−6). To further increase the statistical power we genotyped rs727479 in the remaining 549 SIBS samples for whom E2 concentrations had been measured. In the full set of 2767 women, the association between E2 concentrations and rs727479 approached the genome-wide significance threshold (β=0.096, P=7.4×10−8) (Table 1).
There was no evidence of a difference in the association between rs727479 and E2 concentration with age (Pinteraction=0.90, Table 1). The rs727479-E2 association was the strongest among women with the highest BMIs, with borderline significant evidence of an interaction (Pinteraction=0.066, Table 1, Fig. 3B).
Evidence that higher E2 concentration is causal for endometrial cancer
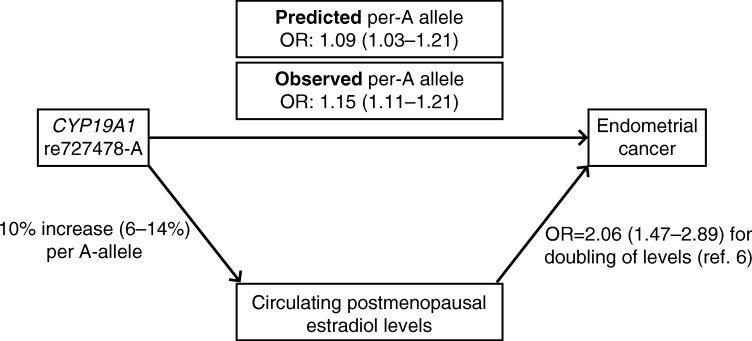
Following a Mendelian randomization argument, if elevated E2 concentration were causally associated with endometrial cancer (as opposed to an association produced by confounding), then we would expect any SNP which raises E2 to be proportionally associated with endometrial cancer. We observed an rs727429 per-A-allele increase in adjusted E2 concentration of 10% (95% CI=6–14%, from the regression coefficient in Table 1 for log-transformed levels). Lukanova et al. (2004) estimated that the odds ratio for endometrial cancer associated with a doubling of post-menopausal E2 concentration was 2.06 (CI=1.47–2.89; it was necessary to use an external estimate because hormone levels had only been measured in control subjects in our study). Based on this published estimate, the predicted per-allele OR for endometrial cancer would be 1.09 (CI=1.03–1.21), which is consistent with that observed in our study (OR=1.15, CI=1.11–1.21) (Fig. 4).
Figure 4.
The observed and predicted risks of endometrial cancer associated with each rs727479 A allele. The Observed per-A allele OR is that observed in this study of 6608 and 37 925 endometrial cancer cases and controls. The predicted per-A allele OR is estimated based on the observed association between rs727479 and E2 levels in 2767 healthy post-menopausal women, and on the endometrial cancer OR associated with a doubling of post-menopausal E2 levels reported by Lukanova et al. (2004).
Candidate causal variants may regulate CYP19 expression
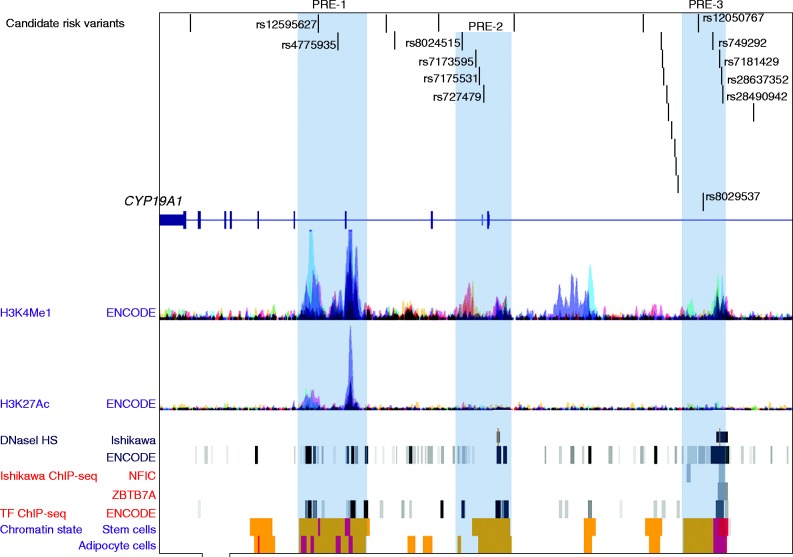
Bioinformatic analysis defined three putative regulatory elements (PREs) coincident with 12 of 28 candidate endometrial cancer causal variants prioritized by genetic analysis (Fig. 5). Altered binding of transcription factors was predicted for 10/12 candidates located within PREs, including top candidate rs727479 (Supplementary Table 3, see section on supplementary data given at the end of this article). For four of these (rs8024515, rs7181429, rs28637352, and rs28490942) there was experimental evidence for differential transcription factor (TF) binding in the cell types tested by ENCODE (Fig. 5) and SNPs rs7181429, rs28637352 overlap binding consensus sequences for NFIC and ZBTB7A in Ishikawa endometrial cancer cells (Fig. 5). Expression analysis identified nominal associations (P<0.05) between risk alleles for the 28 candidate causal variants and greater CYP19A1 expression in several tissues, with candidate SNP rs7181429's association with expression in blood passing a Bonferroni-corrected significance threshold (P=6.0×10−5; corrected P=1.62×10−4; Supplementary Table 4, see section on supplementary data given at the end of this article).
Figure 5.
Candidate endometrial risk variants coincide with three PREs. The 28 best candidate causal SNPs map towards the 3′ end of the CYP19A1 gene. The functional elements displayed were accessed through the UCSC Genome Browser and include: H3K4Me1 and H3K27Ac histone modifications measured by the Encyclopedia of DNA Elements (ENCODE) project in seven cell lines; open chromatin as delineated by DNaseI hypersensitivity sites (HS) in Ishikawa endometrial cancer cells (previously incorrectly named ECC-1)) and 125 other cell types; TF binding in Ishikawa cells and 91 cell lines within ENCODE: 21/28 candidates are predicted to overlap TF binding sites. Roadmap Epigenomics Project chromatin state segmentation of adipose-derived mesenchymal stem cells and adipocytes: orange bars represent enhancers and red bars represent regions flanking active transcription start sites. Twelve SNPs, marked by dbSNP rsIDs, are located in PREs: highlighted in blue. PREs were defined by the presence of histone modifications, DHS, TF binding and Roadmap enhancers.
Discussion
We conducted the largest comprehensive genetic study to date of SNPs across the CYP19A1 hormone metabolism gene and their associations with both endometrial cancer risk and circulating E2 concentration. Using genotype information on nearly 3000 SNPs we have, for the first time, identified GWAS-level significant associations between SNPs in this region and endometrial cancer. Our finding that rs727479 is the most significantly associated SNP in this region confirms the findings of a previous candidate-SNP study (Setiawan et al. 2009) and provides a list of 28 SNPs which cannot be excluded as causal on the basis of statistical analyses. We found no evidence for further causal variants outside of this set. For example, rs749292, previously reported as a possible second signal (Setiawan et al. 2009), was not significantly associated with risk in our analysis after conditioning on rs727479, given the number of SNPs included in the analysis (Pcond=0.017).
This is the first study to look at the CYP19A1 endometrial cancer association by histology. The most significant risk SNP, rs727479, appears to be more strongly associated with endometrioid histology endometrial tumors than with the rarer and poorer prognosis non-endometrioid cancers. However, the confidence intervals for the two ORs are not incompatible, and there was no significant difference in allele frequencies between the women with endometrioid and non-endometrioid tumors (P=0.15). Despite the common description of non-endometrioid tumours as ‘estrogen independent’, recent work has shown that the two subtypes largely share common risk factors, including those factors relating to endogenous or exogenous estrogen exposure (Setiawan et al. 2013), consistent with our findings.
We also confirmed at a borderline GWAS significance level (P=7.4×10−8) the association between rs727479 and E2 concentration previously reported in post-menopausal women (Haiman et al. 2007, Ahn et al. 2009, Beckmann et al. 2011, Prescott et al. 2012) and in males (Travis et al. 2009). It had been reported that rs749292 and rs727479 may act independently to alter levels (Haiman et al. 2007), but we found no association between rs749292 and E2 concentration after conditioning on rs727479 (P=0.20). Our sample set partially overlaps with that included in the GWAS of hormone levels reported by Prescott et al. (2012), (the additional 549 SIBS women genotyped here for rs727479 had all been included in the Prescott et al. GWAS), but a combination of nearly 2000 extra subjects, denser genotyping in and around the CYP19A1 gene and imputation to the 1000 Genomes reference panel allowed us to look in more detail at the region. Our results suggest the existence of a single causal variant in CYP19A1 underlying both E2 concentration and endometrial cancer, although we cannot exclude the possibility that there are instead multiple causal variants which are in sufficiently strong linkage disequilibrium that they are indistinguishable by epidemiological analysis.
We estimate that rs727479 accounts for 1.1% of the variance in post-menopausal E2 concentration (in contrast, BMI accounts for 16% of the variance). Given that the estimated heritability of post-menopausal E2 is around 40% (Varghese et al. 2012) it is clear that further genetic variants that affect E2 concentration remain to be found.
The predominant source of circulating estrogens in post-menopausal women is adrenal androgens (T), which are converted to estrogens in peripheral adipose tissues, with the final stage of this process requiring aromatase, the enzyme encoded by the CYP19A1 gene. Although E2 concentrations and endometrial cancer risks are both higher in women with larger BMI regardless of CYP19A1 genotype, there also appears to be a gene-environment interaction such that the associations of the rs727479 A allele with E2 concentration and also with endometrial cancer risk increase according to BMI, with BMI presumably serving as a proxy for the amount of adipose tissue (Fig. 1). Whole body aromatization is known to be directly associated with BMI and the aromatization rate per cell has been found to increase with increasing age (Cleland et al. 1985). Together these data suggest that the influence of the SNP may be more profound when the aromatization rate is already higher.
Twelve of the 28 candidate causal variants (including top candidate rs727429) lie in PREs. Further, the risk alleles of candidates located in PRE-3 associate with increased CYP19A1 expression, and ENCODE and other data (Eeckhoute et al. 2006, Lee & Maeda 2012) indicate that the NFIC and ZBTB7A TFs may affect PRE-3 repressor activity in endometrial cancer cells. Taken together these lines of evidence indicate that candidate causal variants within PRE3 should have high priority for follow-up studies to test their effects on CYP19A1 promoter activity through long-range chromatin interactions.
Using a Mendelian randomization argument with CYP19A1 genotype as the instrumental variable, we have shown that the endometrial cancer OR per A-allele of rs727479 predicted on the basis of the per-allele effect on E2 (1.09, CI=1.03–1.21) is in line with the directly observed effect of each A allele on endometrial cancer (OR=1.15, CI=1.11–1.21) (Fig. 4). Whereas previous epidemiological studies have observed a positive correlation between E2 concentration and risk, it has not been possible to distinguish between a causal relationship and one produced by confounding. By exploiting the random allocation of alleles to individuals at conception, Mendelian randomization mimics a randomized control trial, thus removing possible confounding. We have therefore found good evidence that higher post-menopausal E2 concentrations are indeed a causal risk factor for endometrial cancer, in line with other evidence such as the observed increase in risk associated with estrogen-only HRT but not with estrogen+progesterone HRT (Beral et al. 2005). Hence lowering E2 levels has the potential to be a useful strategy for reducing risk. Ideally we would like to be able to test this hypothesis in a prospective study, whereby E2 levels are measured at baseline for a cohort of healthy post-menopausal women who are then followed up for endometrial cancer incidence. However, such a study would need to be extremely large in order to accrue sufficient cancer cases within a reasonable time frame. It would also be interesting to repeat the study in a non-European setting in order to see whether the results are consistent across populations.
In Mendelian randomization, for a genetic variant to be a suitable instrument, in addition to being associated with the biomarker it must also be i) independent of the unobserved confounders of the biomarker-disease relationship and ii) associated with the disease only via the biomarker. Population stratification is the most obvious way in which condition i) can be violated. To guard against this we restricted our study to subjects of European ancestry and adjusted for principal components. Condition ii) can be broken by pleiotropy, or similarly if the variant is in LD with a separate disease-associated variant. It is impossible to be certain that this condition has been met, and our finding that the observed association between rs727479 and endometrial cancer is slightly stronger than that predicted according to rs727479's effect on E2 levels may in part be due to the SNP additionally acting on endometrial cancer risk via a pathway not involving circulating E2. A Mendelian randomization using an instrument consisting of multiple independent E2-associated SNPs, as and when they are reported (e.g., via larger GWAS) is one way to minimize the potential impact of pleiotropy on the results. We are confident that the results are not due to reverse causation, since E2 measurements were all carried out in women from the control arm of the endometrial cancer study.
Despite higher endogenous E2 concentration also being a known risk factor for breast cancer (Key et al. 2002), candidate SNP studies of CYP19A1 have not reported an association with breast cancer (Haiman et al. 2007). Based on the 45 290 European-ancestry breast cancer cases and 41 880 controls from the BCAC iCOGS study, none of the 171 CYP19A1 locus SNPs with P<10−4 for endometrial cancer were associated with breast cancer (Michailidou et al. 2013) (minimum P=0.0033, data not shown). This may in part be because E2 concentration is less strongly related to breast than to endometrial cancer; a doubling of E2 concentration has been reported to be associated with an OR of 1.29 (CI=1.15–1.44) for breast cancer (Key et al. 2002), from which we would predict a breast cancer OR of 1.03 (1.01–1.07) per A allele of rs727479. This predicted effect size is consistent with that observed for breast cancer in BCAC (OR=1.02 (1.00–1.04); P=0.10), but the effect size is too small to be confidently detected, even in a breast cancer study of this size.
In conclusion, we have confirmed at a genome-wide-level of significance the association between endometrial cancer and variants within the CYP19A1 gene, and shown that all of the reported associations can be explained by a single risk peak. We have also provided evidence that the same set of variants is associated with higher E2 concentration in post-menopausal women, supporting a causal role for E2 in endometrial cancer. For both traits, the SNP associations were stronger in women with a higher BMI, suggesting a biologically plausible gene-environment interaction.
Supplementary data
This is linked to the online version of the paper at http://dx.doi.org/10.1530/ERC-15-0386.
Author contribution statement
A M Dunning, D F Easton, K-T Khaw, G W Montgomery, P D P Pharoah, A B Spurdle, I Tomlinson, and P M Webb obtained funding for the study. D F Easton and A B Spurdle designed the study and A B Spurdle and D J Thompson drafted the manuscript. D J Thompson conducted all statistical analyses and genotype imputation. D M Glubb and T A O'Mara conducted bioinformatic analyses. T A O'Mara and J N Painter co-ordinated the endometrial cancer iCOGS genotyping, and associated data management. J Dennis, K Michailidou and J P Tyrer co-ordinated quality control and data cleaning for the iCOGS control datasets, and K Michailidou provided quality control for the SEARCH GWAS control set. M K Bolla, Q Wang, M Shah, and R Luben were responsible for data management. T A O'Mara and A B Spurdle co-ordinated the ANECS GWAS genotyping; A M Dunning co-ordinated the SEARCH GWAS genotyping; I Tomlinson co-ordinated the NSECG GWAS genotyping. E Folkerd, D Doody, and M Dürst carried out the hormone measurements. The remaining authors were involved in the co-ordination and/or extraction of phenotypic information for contributing studies. All authors provided critical review of the manuscript.
Acknowledgements
The authors thank many individuals who participated in this study and numerous institutions and their staff who supported recruitment, detailed in full in the Supplementary Text, which is linked to the online version of the paper at http://dx.doi.org/10.1530/ERC-15-0386.
Declaration of interest
The authors declare that there is no conflict of interest that could be perceived as prejudicing the impartiality of the research reported.
Funding
Fine-mapping analysis was supported by NHMRC project grant (ID#1031333) to A B Spurdle, D F Easton, and A M Dunning. A B Spurdle, P M Webb, G W Montgomery, and D R Nyholt are supported by the NHMRC Fellowship scheme. A M Dunning is supported by the Joseph Mitchell Trust. I Tomlinson is supported by Cancer Research UK and the Oxford Comprehensive Biomedical Research Centre. Funding for the iCOGS infrastructure came from: the European Community's Seventh Framework Programme under grant agreement no 223175 (HEALTH-F2-2009-223175) (COGS), Cancer Research UK (C1287/A10118, C1287/A 10710, C12292/A11174, C1281/A12014, C5047/A8384, C5047/A15007, C5047/A10692, C8197/A16565), the National Institutes of Health (CA128978), and Post-Cancer GWAS initiative (1U19 CA148537, 1U19 CA148065, and 1U19 CA148112 – the GAME-ON initiative), the Department of Defence (W81XWH-10-1-0341), the Canadian Institutes of Health Research (CIHR) for the CIHR Team in Familial Risks of Breast Cancer, Komen Foundation for the Cure, the Breast Cancer Research Foundation, and the Ovarian Cancer Research Fund. ANECS recruitment was supported by project grants from the NHMRC (ID#339435), The Cancer Council Queensland (ID#4196615) and Cancer Council Tasmania (ID#403031 and ID#457636). SEARCH recruitment was funded by a programme grant from Cancer Research UK (C490/A10124). Stage 1 2 case genotyping was supported by the NHMRC (ID#552402 and ID#1031333). This study makes use of data generated by the Wellcome Trust Case–Control Consortium (WTCCC). A full list of the investigators who contributed to the generation of the data is available from www.wtccc.org.uk. Funding for the project was provided by the Wellcome Trust under award 076113. We acknowledge use of DNA from the British 1958 Birth Cohort collection, funded by the Medical Research Council grant G0000934 and the Wellcome Trust grant 068545/Z/02 – funding for this project was provided by the Wellcome Trust under award 085475. NSECG was supported by the EU FP7 CHIBCHA grant and Wellcome Trust Centre for Human Genetics Grant 090532/Z/09Z, and CORGI by Cancer Research UK. Recruitment of the QIMR Berghofer controls was supported by the NHMRC. The University of Newcastle, the Gladys M Brawn Senior Research Fellowship scheme, The Vincent Fairfax Family Foundation, the Hunter Medical Research Institute, and the Hunter Area Pathology Service all contributed towards the costs of establishing the Hunter Community Study. The Bavarian Endometrial Cancer Study (BECS) was partly funded by the ELAN fund of the University of Erlangen. The Leuven Endometrium Study (LES) was supported by the Verelst Foundation for endometrial cancer. The Mayo Endometrial Cancer Study (MECS) and Mayo controls (MAY) were supported by grants from the National Cancer Institute of United States Public Health Service (R01 CA122443, P30 CA15083, P50 CA136393, and GAME-ON the NCI Cancer Post-GWAS Initiative U19 CA148112), the Fred C and Katherine B Andersen Foundation, the Mayo Foundation, and the Ovarian Cancer Research Fund with support of the Smith family, in memory of Kathryn Sladek Smith. MoMaTEC received financial support from a Helse Vest Grant, the University of Bergen, Melzer Foundation, The Norwegian Cancer Society (Harald Andersens legat), The Research Council of Norway and Haukeland University Hospital. The Newcastle Endometrial Cancer Study (NECS) acknowledges contributions from the University of Newcastle, The NBN Children's Cancer Research Group, Ms Jennie Thomas and the Hunter Medical Research Institute. RENDOCAS was supported through the regional agreement on medical training and clinical research (ALF) between Stockholm County Council and Karolinska Institutet (numbers: 20110222, 20110483, 20110141, and DF 07015), The Swedish Labor Market Insurance (number 100069) and The Swedish Cancer Society (number 11 0439). The Cancer Hormone Replacement Epidemiology in Sweden Study (CAHRES, formerly called The Singapore and Swedish Breast/Endometrial Cancer Study; SASBAC) was supported by funding from the Agency for Science, Technology and Research of Singapore (A*STAR), the US National Institutes of Health and the Susan G. Komen Breast Cancer Foundation. The Breast Cancer Association Consortium (BCAC) is funded by Cancer Research UK (C1287/A10118, C1287/A12014). The Ovarian Cancer Association Consortium (OCAC) is supported by a grant from the Ovarian Cancer Research Fund thanks to donations by the family and friends of Kathryn Sladek Smith (PPD/RPCI.07), and the UK National Institute for Health Research Biomedical Research Centres at the University of Cambridge. Additional funding for individual control groups is detailed in the Supplementary Information, see section on supplementary data given at the end of this article. EPIC-Norfolk was funded by research programme grant funding from Cancer Research UK and the Medical Research Council with additional support from the Stroke Association, British Heart Foundation, Department of Health, Research into Ageing and Academy of Medical Sciences. The SIBS study was supported by program grant C1287/A10118 and project grants from Cancer Research UK (grant numbers C1287/8459).
References
- Ahn J, Schumacher FR, Berndt SI, Pfeiffer R, Albanes D, Andriole GL, Ardanaz E, Boeing H, Bueno-de-Mesquita B, Chanock SJ. Quantitative trait loci predicting circulating sex steroid hormones in men from the NCI-Breast and Prostate Cancer Cohort Consortium (BPC3) Human Molecular Genetics. 2009;18:3749–3757. doi: 10.1093/hmg/ddp302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen NE, Key TJ, Dossus L, Rinaldi S, Cust A, Lukanova A, Peeters PH, Onland-Moret NC, Lahmann PH, Berrino F, et al. Endogenous sex hormones and endometrial cancer risk in women in the European Prospective Investigation into Cancer and Nutrition (EPIC) Endocrine-Related Cancer. 2008;15:485–497. doi: 10.1677/ERC-07-0064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aulchenko YS, Struchalin MV, van Duijn CM. ProbABEL package for genome-wide association analysis of imputed data. BMC Bioinformatics. 2010;11:134. doi: 10.1186/1471-2105-11-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckmann L, Husing A, Setiawan VW, Amiano P, Clavel-Chapelon F, Chanock SJ, Cox DG, Diver R, Dossus L, Feigelson HS, et al. Comprehensive analysis of hormone and genetic variation in 36 genes related to steroid hormone metabolism in pre- and postmenopausal women from the breast and prostate cancer cohort consortium (BPC3) Journal of Clinical Endocrinology and Metabolism. 2011;96:E360–E367. doi: 10.1210/jc.2010-0912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beral V, Bull D, Reeves G & Million Women Study Collaborators Endometrial cancer and hormone-replacement therapy in the Million Women Study. Lancet. 2005;365:1543–1551. doi: 10.1016/S0140-6736(05)66455-0. [DOI] [PubMed] [Google Scholar]
- Chen WM, Abecasis GR. Family-based association tests for genomewide association scans. American Journal of Human Genetics. 2007;81:913–926. doi: 10.1086/521580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleland WH, Mendelson CR, Simpson ER. Effects of aging and obesity on aromatase activity of human adipose cells. Journal of Clinical Endocrinology and Metabolism. 1985;60:174–177. doi: 10.1210/jcem-60-1-174. [DOI] [PubMed] [Google Scholar]
- C-Reactive Protein Coronary Heart Disease Genetics Collaboration , Wensley F, Gao P, Burgess S, Kaptoge S, Di Angelantonio E, Shah T, Engert JC, Clarke R, Davey-Smith G. Association between C reactive protein and coronary heart disease: Mendelian randomisation analysis based on individual participant data. British Medical Journal. 2011;342:d548. doi: 10.1136/bmj.d548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuzick J, Powles T, Veronesi U, Forbes J, Edwards R, Ashley S, Boyle P. Overview of the main outcomes in breast-cancer prevention trials. Lancet. 2003;361:296–300. doi: 10.1016/S0140-6736(03)12342-2. [DOI] [PubMed] [Google Scholar]
- Day N, Oakes S, Luben R, Khaw KT, Bingham S, Welch A, Wareham N. EPIC-Norfolk: study design and characteristics of the cohort. European Prospective Investigation of Cancer. British Journal of Cancer. 1999;80(Suppl 1):95–103. [PubMed] [Google Scholar]
- Dowsett M, Goss PE, Powles TJ, Hutchinson G, Brodie AM, Jeffcoate SL, Coombes RC. Use of the aromatase inhibitor 4-hydroxyandrostenedione in postmenopausal breast cancer: optimization of therapeutic dose and route. Cancer Research. 1987;47:1957–1961. [PubMed] [Google Scholar]
- Dunning AM, Dowsett M, Healey CS, Tee L, Luben RN, Folkerd E, Novik KL, Kelemen L, Ogata S, Pharoah PD, et al. Polymorphisms associated with circulating sex hormone levels in postmenopausal women. Journal of the National Cancer Institute. 2004;96:936–945. doi: 10.1093/jnci/djh167. [DOI] [PubMed] [Google Scholar]
- Eeckhoute J, Carroll JS, Geistlinger TR, Torres-Arzayus MI, Brown M. A cell-type-specific transcriptional network required for estrogen regulation of cyclin D1 and cell cycle progression in breast cancer. Genes and Development. 2006;20:2513–2526. doi: 10.1101/gad.1446006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eriksson AL, Lorentzon M, Vandenput L, Labrie F, Lindersson M, Syvanen AC, Orwoll ES, Cummings SR, Zmuda JM, Ljunggren O, et al. Genetic variations in sex steroid-related genes as predictors of serum estrogen levels in men. Journal of Clinical Endocrinology and Metabolism. 2009;94:1033–1041. doi: 10.1210/jc.2008-1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira T, Marchini J. Modeling interactions with known risk loci-a Bayesian model averaging approach. Annals of Human Genetics. 2011;75:1–9. doi: 10.1111/j.1469-1809.2010.00618.x. [DOI] [PubMed] [Google Scholar]
- Flote VG, Furberg A, McTiernan A, Frydenberg H, Ursin G, Iversen A, Lofteroed T, Ellison PT, Wist EA, Egeland T, et al. Gene variations in oestrogen pathways, CYP19A1, daily 17ss-E2 and mammographic density phenotypes in premenopausal women. Breast Cancer Research. 2014;16:499. doi: 10.1186/s13058-014-0499-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haiman CA, Dossus L, Setiawan VW, Stram DO, Dunning AM, Thomas G, Thun MJ, Albanes D, Altshuler D, Ardanaz E, et al. Genetic variation at the CYP19A1 locus predicts circulating estrogen levels but not breast cancer risk in postmenopausal women. Cancer Research. 2007;67:1893–1897. doi: 10.1158/0008-5472.CAN-06-4123. [DOI] [PubMed] [Google Scholar]
- Higgins JP, Thompson SG. Quantifying heterogeneity in a meta-analysis. Statistics in Medicine. 2002;21:1539–1558. doi: 10.1002/sim.1186. [DOI] [PubMed] [Google Scholar]
- Houlston RS, Cheadle J, Dobbins SE, Tenesa A, Jones AM, Howarth K, Spain SL, Broderick P, Domingo E, Farrington S, et al. Meta-analysis of three genome-wide association studies identifies susceptibility loci for colorectal cancer at 1q41, 3q26.2, 12q13.13 and 20q13.33. Nature Genetics. 2010;42:973–977. doi: 10.1038/ng.670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howie BN, Donnelly P, Marchini J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genetics. 2009;5:e1000529. doi: 10.1371/journal.pgen.1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howie B, Marchini J, Stephens M. Genotype imputation with thousands of genomes. G3. 2011;1:457–470. doi: 10.1534/g3.111.001198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Interleukin-6 Receptor Mendelian Randomisation Analysis (IL6R MR) Consortium. The interleukin-6 receptor as a target for prevention of coronary heart disease: a mendelian randomisation analysis. Lancet. 2012;379:1214–1224. doi: 10.1016/S0140-6736(12)60110-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Agency for Research on Cancer (IARC) Working Group on the Evaluation of Cancer-Preventive Strategies 2002 Weight control and physical activity. pp 1–315. Lyon, France: IARC Press.
- Key TJ, Pike MC. The dose-effect relationship between ‘unopposed’ oestrogens and endometrial mitotic rate: its central role in explaining and predicting endometrial cancer risk. British Journal of Cancer. 1988;57:205–212. doi: 10.1038/bjc.1988.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Key T, Appleby P, Barnes I, Reeves G, Endogenous Hormones and Breast Cancer Collaborative Group Endogenous sex hormones and breast cancer in postmenopausal women: reanalysis of nine prospective studies. Journal of the National Cancer Institute. 2002;94:606–616. doi: 10.1093/jnci/94.8.606. [DOI] [PubMed] [Google Scholar]
- Kidokoro K, Ino K, Hirose K, Kajiyama H, Hosono S, Suzuki T, Kawase T, Hiraki A, Hamajima N, Tanaka H, et al. Association between CYP19A1 polymorphisms and sex hormones in postmenopausal Japanese women. Journal of Human Genetics. 2009;54:78–85. doi: 10.1038/jhg.2008.11. [DOI] [PubMed] [Google Scholar]
- Lee SU, Maeda T. POK/ZBTB proteins: an emerging family of proteins that regulate lymphoid development and function. Immunological Reviews. 2012;247:107–119. doi: 10.1111/j.1600-065X.2012.01116.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locke AE, Kahali B, Berndt SI, Justice AE, Pers TH, Day FR, Powell C, Vedantam S, Buchkovich ML, Yang J, et al. Genetic studies of body mass index yield new insights for obesity biology. Nature. 2015;518:197–206. doi: 10.1038/nature14177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Low YL, Li Y, Humphreys K, Thalamuthu A, Li Y, Darabi H, Wedren S, Bonnard C, Czene K, Iles MM, et al. Multi-variant pathway association analysis reveals the importance of genetic determinants of estrogen metabolism in breast and endometrial cancer susceptibility. PLoS Genetics. 2010;6:e1001012. doi: 10.1371/journal.pgen.1001012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukanova A, Lundin E, Micheli A, Arslan A, Ferrari P, Rinaldi S, Krogh V, Lenner P, Shore RE, Biessy C, et al. Circulating levels of sex steroid hormones and risk of endometrial cancer in postmenopausal women. International Journal of Cancer. 2004;108:425–432. doi: 10.1002/ijc.11529. [DOI] [PubMed] [Google Scholar]
- Lundin E, Wirgin I, Lukanova A, Afanasyeva Y, Krogh V, Axelsson T, Hemminki K, Clendenen TV, Arslan AA, Ohlson N, et al. Selected polymorphisms in sex hormone-related genes, circulating sex hormones and risk of endometrial cancer. Cancer Epidemiology. 2012;36:445–452. doi: 10.1016/j.canep.2012.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McEvoy M, Smith W, D'Este C, Duke J, Peel R, Schofield P, Scott R, Byles J, Henry D, Ewald B, et al. Cohort profile: the hunter community study. International Journal of Epidemiology. 2010;39:1452–1463. doi: 10.1093/ije/dyp343. [DOI] [PubMed] [Google Scholar]
- McGregor B, Pfitzner J, Zhu G, Grace M, Eldridge A, Pearson J, Mayne C, Aitken JF, Green AC, Martin NG. Genetic and environmental contributions to size, color, shape, and other characteristics of melanocytic naevi in a sample of adolescent twins. Genetic Epidemiology. 1999;16:40–53. doi: 10.1002/(SICI)1098-2272(1999)16:1%3c40::AID-GEPI4%3e3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- Michailidou K, Hall P, Gonzalez-Neira A, Ghoussaini M, Dennis J, Milne RL, Schmidt MK, Chang-Claude J, Bojesen SE, Bolla MK. Large-scale genotyping identifies 41 new loci associated with breast cancer risk. Nature Genetics. 2013;45:353–361, 361e351–352. doi: 10.1038/ng.2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Painter JN, Anderson CA, Nyholt DR, Macgregor S, Lin J, Lee SH, Lambert A, Zhao ZZ, Roseman F, Guo Q, et al. Genome-wide association study identifies a locus at 7p15.2 associated with endometriosis. Nature Genetics. 2011;43:51–54. doi: 10.1038/ng.731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Painter JN, O'Mara TA, Batra J, Cheng T, Lose FA, Dennis J, Michailidou K, Tyrer JP, Ahmed S, Ferguson K, et al. Fine-mapping of the HNF1B multicancer locus identifies candidate variants that mediate endometrial cancer risk. Human Molecular Genetics. 2015;24:1478–1492. doi: 10.1093/hmg/ddu552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paynter RA, Hankinson SE, Colditz GA, Kraft P, Hunter DJ, De Vivo I. CYP19 (aromatase) haplotypes and endometrial cancer risk. International Journal of Cancer. 2005;116:267–274. doi: 10.1002/ijc.21041. [DOI] [PubMed] [Google Scholar]
- Pharoah PD, Tsai YY, Ramus SJ, Phelan CM, Goode EL, Lawrenson K, Buckley M, Fridley BL, Tyrer JP, Shen H. GWAS meta-analysis and replication identifies three new susceptibility loci for ovarian cancer. Nature Genetics. 2013;45:362–370, 370e361–362. doi: 10.1038/ng.2564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prescott J, Thompson DJ, Kraft P, Chanock SJ, Audley T, Brown J, Leyland J, Folkerd E, Doody D, Hankinson SE, et al. Genome-wide association study of circulating E2, T, and sex hormone-binding globulin in postmenopausal women. PLoS ONE. 2012;7:e37815. doi: 10.1371/journal.pone.0037815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruim RJ, Welch RP, Sanna S, Teslovich TM, Chines PS, Gliedt TP, Boehnke M, Abecasis GR, Willer CJ. LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics. 2010;26:2336–2337. doi: 10.1093/bioinformatics/btq419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Setiawan VW, Doherty JA, Shu XO, Akbari MR, Chen C, De Vivo I, Demichele A, Garcia-Closas M, Goodman MT, Haiman CA, et al. Two estrogen-related variants in CYP19A1 and endometrial cancer risk: a pooled analysis in the Epidemiology of Endometrial Cancer Consortium. Cancer Epidemiology, Biomarkers & Prevention. 2009;18:242–247. doi: 10.1158/1055-9965.EPI-08-0689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Setiawan VW, Yang HP, Pike MC, McCann SE, Yu H, Xiang YB, Wolk A, Wentzensen N, Weiss NS, Webb PM, et al. Type I and II endometrial cancers: have they different risk factors? Journal of Clinical Oncology. 2013;31:2607–2618. doi: 10.1200/JCO.2012.48.2596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spurdle AB, Thompson DJ, Ahmed S, Ferguson K, Healey CS, O'Mara T, Walker LC, Montgomery SB, Dermitzakis ET, Australian National Endometrial Cancer Study G , et al. Genome-wide association study identifies a common variant associated with risk of endometrial cancer. Nature Genetics. 2011;43:451–454. doi: 10.1038/ng.812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao MH, Cai Q, Zhang ZF, Xu WH, Kataoka N, Wen W, Xiang YB, Zheng W, Shu XO. Polymorphisms in the CYP19A1 (aromatase) gene and endometrial cancer risk in Chinese women. Cancer Epidemiology, Biomarkers & Prevention. 2007;16:943–949. doi: 10.1158/1055-9965.EPI-06-1012. [DOI] [PubMed] [Google Scholar]
- Travis RC, Schumacher F, Hirschhorn JN, Kraft P, Allen NE, Albanes D, Berglund G, Berndt SI, Boeing H, Bueno-de-Mesquita HB, et al. CYP19A1 genetic variation in relation to prostate cancer risk and circulating sex hormone concentrations in men from the Breast and Prostate Cancer Cohort Consortium. Cancer Epidemiology, Biomarkers & Prevention. 2009;18:2734–2744. doi: 10.1158/1055-9965.EPI-09-0496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udler MS, Tyrer J, Easton DF. Evaluating the power to discriminate between highly correlated SNPs in genetic association studies. Genetic Epidemiology. 2010;34:463–468. doi: 10.1002/gepi.20504. [DOI] [PubMed] [Google Scholar]
- Varghese JS, Smith PL, Folkerd E, Brown J, Leyland J, Audley T, Warren RM, Dowsett M, Easton DF, Thompson DJ. The heritability of mammographic breast density and circulating sex-hormone levels: two independent breast cancer risk factors. Cancer Epidemiology, Biomarkers & Prevention. 2012;21:2167–2175. doi: 10.1158/1055-9965.EPI-12-0789. [DOI] [PubMed] [Google Scholar]
- Wellcome Trust Case Control Consortium. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeleniuch-Jacquotte A, Akhmedkhanov A, Kato I, Koenig KL, Shore RE, Kim MY, Levitz M, Mittal KR, Raju U, Banerjee S, et al. Postmenopausal endogenous oestrogens and risk of endometrial cancer: results of a prospective study. British Journal of Cancer. 2001;84:975–981. doi: 10.1054/bjoc.2001.1704. [DOI] [PMC free article] [PubMed] [Google Scholar]



 This work is licensed under a
This work is licensed under a