Dear Editor,
Stenotrophomonas maltophilia has received considerable attention owing to its high infection rate and resistance to multiple antibiotic classes [1].Trimethoprim/sulfamethoxazole (SXT) is the recommended first-line therapy for S. maltophilia infection; however, resistance ratesto SXT of S. maltophilia have increased worldwide; in Anhui province of China, the resistance rate has increased from 10% to 40% over the past five years [2]. Accordingly, it is important to study the mechanisms underlying SXT resistance and prevent the spread of resistance. We found that sul1 in combination with sul2 confers high-level resistance to SXT; sul1 was always found as part of the 3' end of the class 1 integron, in Anhui province [3]; however, no association between sul2 and integrons was found.
In the present report, 41 sul2-positive S. maltophilia samples were collected in one month (September) every year (from 2010 to 2012) from different patients across 31 hospitals in Anhui, China. All S. maltophilia isolates were identified using the MicroScan WalkAway 40 System (Dade Behring, Deerfield, IL, USA). Pseudomonas aeruginosa ATCC 27853, Escherichia coli ATCC 35218, and E. coli ATCC 25922 were used as quality control strains for antimicrobial susceptibility testing. Minimal inhibitory concentrations (MICs) of SXT were determined for each isolate by the agar dilution method according to the Clinical and Laboratory Standards Institute [4]. Presence of sul1, sul2, and dfrA in each strain was assessed by PCR as described previously [3].
Of the 41 sul2-positive isolates, 30 (73.2%) were resistant to SXT, 28 (68.3%) carried sul1, eight (19.5%) carried dfrA12, and 10 (24.4%) carried dfrA17 (Table 1). Twenty-eight of the 30 SXT-resistant and sul2-positive isolates contained sul1 and/or dfrA. Sul1 and dfrA, which lead to an increase in the MIC of SXT or SXT resistance in S. maltophilia, have been described previously [3].
Table 1. Distribution of the sul and dfrA genes in 41 sul2-positive Stenotrophomonas maltophilia isolates.
| Gene | N of isolates | MIC of SXT (µg/mL) | |||||||
|---|---|---|---|---|---|---|---|---|---|
| 0.25/4.95 | 1/19 | 2/38 | 4/76 | 8/152 | 16/304 | 32/608 | > 32/608 | ||
| sul2 | 13 | 1 | 6 | 4 | 2 | ||||
| sul2, sul1 | 10 | 2 | 1 | 7 | |||||
| sul2, sul1, dfrA12 | 8 | 3 | 1 | 4 | |||||
| sul2, sul1, dfrA17 | 10 | 1 | 1 | 3 | 5 | ||||
Abbreviations: MIC, minimal inhibitory concentration; SXT, trimethoprim/sulfamethoxazole.
Sul2 genes linked to insertion sequence common region (ISCR) elements were previously reported [5]. We investigated whether sul2 could be linked to other mobile elements in this region by thermal asymmetric interlaced PCR (TAIL-PCR), which was developed for amplification of unknown DNA fragments flanked by known sequences. In TAIL-PCR, three nested, sequence-specific primers are utilized in consecutive reactions together with arbitrary degenerate (AD) primers to enhance the amplification efficiency of specific products [6]. Three specific primers (sul2-SP1: AAAGAACGCCGCAATGTGATCC, sul2-SP2: GGATAGAACGCAGCGTCTGGAAAA, and sul2-SP3: CATCTGCCTTGAGCGCGTCC) and four AD primers (AD1, AD2, AD3, and AD4) from the Genome Walking Kit (TaKaRa Bio, Dalian, China) were used to amplify the 3' flanking sequences of sul2. Three specific primers (sul2-SP4: GTGGGAATGAGTTGGAAGAA, sul2-SP5: GTTGCCGTTGTGGGTGCTGT, and sul2-SP6: TGCGCTATCGTGGGGGAATG) and the four AD primers were used to amplify the 5' flanking sequences of sul2. The specific primers were designed to have a melting temperature (60 to 65℃) that was higher than that of the AD primers (45℃). The flanking sequences of sul2 were amplified by PCR according to the protocol of the Genome Walking Kit (TaKaRa Bio). PCR products were cloned into the pMD19-T vector and sequenced.
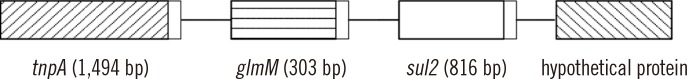
According to results of TAIL-PCR and sequence analysis of the 41 sul2-positive isolates, in 27 isolates resistant to SXT, a 2-kb nucleotide sequence encoding transposase (tnpA2) and phosphoglucosamine mutase (glmM) was obtained for the 5' end of sul2; in the same 27 isolates, a 3.2-kb nucleotide sequence encoding a conserved hypothetical protein was obtained for the 3' end of sul2. The genetic structure of the flanking nucleotides linked to sul2 is shown in Fig. 1 and has been deposited in GenBank.
Fig. 1. Schematic diagram of nucleotides flanking the sul2 gene.
Epidemiological genotyping of the 27 isolates was performed by randomly amplified polymorphic DNA PCR, as described previously [7]. Twenty-three isolateshad unrelated genotypes, and the other four isolateswere found to share the same genotype. After tracking clinical data, these four isolates were determined to be from the same department of a hospital, indicating that clonal spread was responsible for dissemination of sul2 among S. maltophilia isolates. Presence of sul2 has increased SXT resistance, when it is associated with transposons; sul2 could be further disseminated among bacteria through horizontal gene transfer in S. maltophilia isolates.
In summary, this is the first report showing the close association of the SXT resistance gene sul2 with transposase in S. maltophilia isolates. These results suggest that acquired resistance has an important role in the resistance of S. maltophilia to SXT, which may increase by means of mobile elements.
Nucleotide sequence accession numbers
Sequences of the tnpA2-glmM-sul2-hypothetical protein have been deposited in GenBank and were assigned the accession number JX869967.
Acknowledgments
This work was supported by a research grant from the National Natural Science Foundation of China (No. 81101313).
Footnotes
Authors' Disclosures of Potential Conflicts of Interest: No potential conflicts of interest relevant to this article were reported.
References
- 1.Brooke JS. Stenotrophomonas maltophilia: an emerging global opportunistic pathogen. Clin Microbiol Rev. 2012;25:2–41. doi: 10.1128/CMR.00019-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hu LF, Gao LP, Ye Y, Chen X, Zhou XT, Yang HF, et al. Susceptibility of Stenotrophomonas maltophilia clinical strains in China to antimicrobial combinations. J Chemother. 2014;26:282–286. doi: 10.1179/1973947814Y.0000000168. [DOI] [PubMed] [Google Scholar]
- 3.Hu LF, Chang X, Ye Y, Wang ZX, Shao YB, Shi W, et al. Stenotrophomonas maltophilia resistance to trimethoprim/sulfamethoxazole mediated by acquisition of sul and dfrA genes in a plasmid-mediated class 1 integron. Int J Antimicrob Agents. 2011;37:230–234. doi: 10.1016/j.ijantimicag.2010.10.025. [DOI] [PubMed] [Google Scholar]
- 4.Clinical and Laboratory Standards Institute. Performance standards for antimicrobial susceptibility testing. Twenty-third informational supplement, M100-S23. Wayne, PA: CLSI; 2013. [Google Scholar]
- 5.Toleman MA, Bennett PM, Bennett DM, Jones RN, Walsh TR. Global emergence of trimethoprim/sulfamethoxazole resistance in Stenotrophomonas maltophilia mediated by acquisition of sul genes. Emerg Infect Dis. 2007;13:559–565. doi: 10.3201/eid1304.061378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Trinh Q, Zhu P, Shi H, Xu W, Hao J, Luo Y, et al. A-T linker adapter polymerase chain reaction for determining flanking sequences by rescuing inverse PCR or thermal asymmetric interlaced PCR products. Anal Biochem. 2014;466:24–26. doi: 10.1016/j.ab.2014.07.022. [DOI] [PubMed] [Google Scholar]
- 7.Nazik H, Ongen B, Erturan Z, Salcioğlu M. Genotype and antibiotic susceptibility patterns of Pseudonas aeruginosa and Stenotrophomonas maltophilia isolated from cystic fibrosis patients. Jpn J Infect Dis. 2007;60:82–86. [PubMed] [Google Scholar]