Abstract
Genetic risk for breast cancer is conferred by a combination of multiple variants of small effect. To better understand how risk loci might combine, we examined whether risk-associated genes share regulatory mechanisms. We created a breast cancer gene regulatory network between transcription factors (TFs) and putative target genes (regulons) and asked whether specific regulons are enriched for genes associated with risk loci via eQTLs. We identified 36 overlapping regulons that were enriched and formed a distinct cluster within the network, suggesting shared biology. The risk-TFs driving these regulons are frequently mutated in cancer and lie in two opposing subgroups, which relate to ER+ luminal A/B and to ER− basal-like cancers and to different, luminal epithelial cell populations in the adult mammary gland. Our network approach provides a foundation to reveal the regulatory circuits governing breast cancer, to identify targets for intervention, and is transferable to other disease settings.
Introduction
Polygenic disease susceptibility results in a distribution of risk within the population. Given the large number of known risk loci there is a huge number of possible combinations of genotypes associated with high risk. Therefore, in parallel with the ongoing analysis of individual loci, a framework is needed to understand how multiple risk variants can combine at the cellular level, and indicate whether they work through many different mechanisms or – which would be more tractable for understanding and intervention – whether they converge on just a few. Germline variants will interact not only with each other, but with exposures and with acquired somatic events. Ideally, the framework should be able to capture these interactions.
Systems biology approaches may be able provide such a framework1. Protein-protein interaction networks have been derived in attempts to shed light on the pathways underlying risk2, but most of these networks remain sparse and have only yielded limited insight into cancer risk. Most germline risk variants are thought to affect gene expression. Therefore regulatory networks may be an appropriate starting point to understand the combinatorial effect of risk variants.
Here, we model breast cancer as such a gene regulatory network3 onto which the loci relating to risk can be mapped to identify key regulators4. We extend our previous analysis4 to map onto the network all genes that are associated with the known breast cancer GWAS loci5. We found that the transcription factors (TFs) regulating the genes linked to risk loci cluster within the network, suggesting potential commonality of mechanisms. We also show that the same TFs are frequently mutated in breast cancer. Our analysis provides insight into the gene regulatory circuits operating in breast cancer and has implications for treatment and for the identification of novel therapeutic targets. The approach can be applied in any other settings where data from GWAS, large-scale genotyping and gene expression are available.
Results
Mapping of breast cancer risk loci to regulatory networks
Briefly, our analysis builds a regulatory network and then asks for each regulon in the network whether the genes within it are linked to more risk loci than would be expected by chance. In a subsequent step we examine whether the risk regulons, and the TFs driving them, cluster in the overall network.
First we created a regulatory network for breast cancer using the ARACNe algorithm3,4 which defines regulons (possible target genes) for a set of curated TFs. Each TF-regulon is composed of all those genes whose gene expression data display significant mutual information with that of a given TF and are therefore likely to be regulated by that TF. We previously validated the functional significance of these regulons using ChIP-seq data and TF-knock-down studies4. Regulatory networks were inferred using separate analyses on gene expression data from the METABRIC cohort I (n=997) and II (n=995)6. Within each network regulons overlap because many genes are regulated by more than one TF. We confirmed that copy number variation does not significantly impact the network structure (Supplementary Note, Supplementary Fig. 1).
Secondly, we identified regulons enriched for genes associated with risk loci using EVSE (eQTL-conditioned variant set enrichment)4. GWAS identify risk loci, marked by tagging SNPs that may themselves not be causative. Therefore each tagging SNP was expanded into an associated variant set (AVS)7 that includes all SNPs in strong linkage disequilibrium (methods). We then used variation in gene expression to determine which risk loci can be assigned to a given regulon using eQTL4 (expression quantitative trait loci; SNPs where allelic differences determine expression of a target gene). We used a multivariate eQTL analysis to test the association between the genotypes of the SNPs in each AVS, and, for each regulon separately, the expression of all the genes that lay within a +/− 250kb window around the AVS. If such an association was found, the locus was counted towards a mapping tally of the number of GWAS loci associated with genes in the regulon. Finally the statistical significance of the mapping tally was assessed by permutation analysis (methods, Supplementary Fig. 2). We refer to TFs whose regulons were significantly enriched as “risk-TFs”.
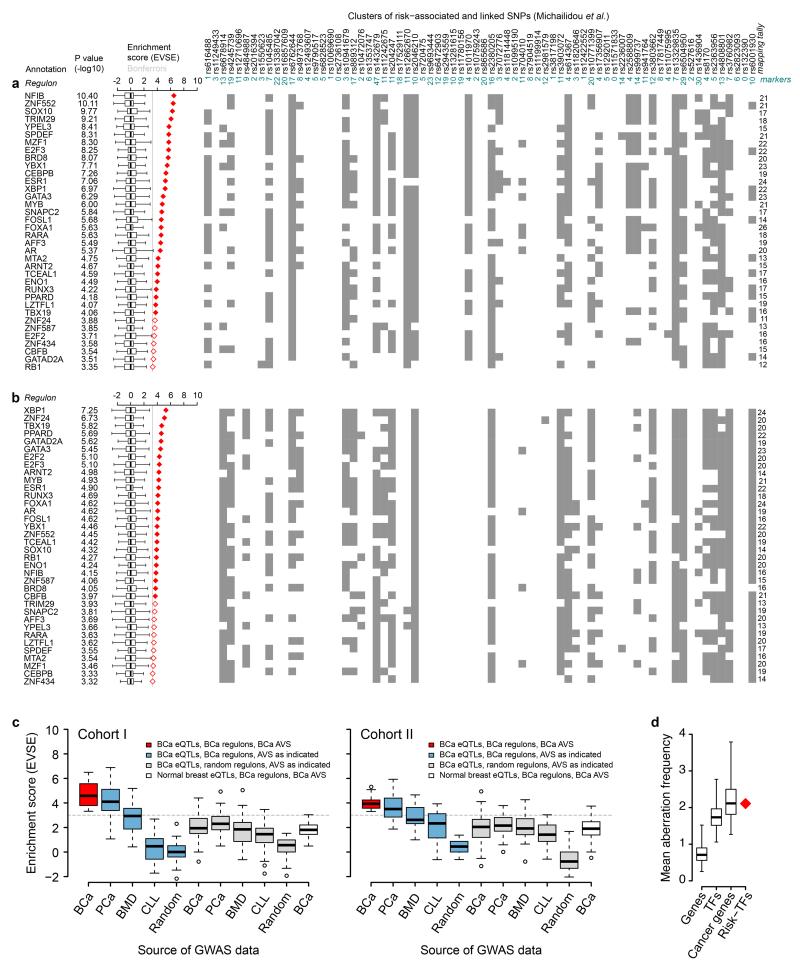
We carried out the EVSE analysis independently for cohort I and II of the METABRIC cancer data set and identified 63 and 61 TFs, respectively, with significant enrichment scores, but none using the much smaller data set from normal tissue (Supplementary Fig. 3). Frequently, a single risk locus contributes to the mapping tally of many regulons. This can be driven by a single gene that is part of many regulons or by multiple distinct genes encoded at that locus contributing to the association with different regulons (Supplementary Note, Supplementary Figure 4). The regulons for 36 TFs were significant in both cohorts (Fig. 1a,b).
Figure 1. EVSE-based identification of 36 risk-TFs.
Lists of 36 TF-regulons identified in the EVSE analysis, showing enrichment score and p-value based rank order of risk-TFs for METABRIC cohort I (a) and II (b). The tagging SNP for each breast cancer GWAS hit5 is listed above the panel, together with the number of markers (SNPs in the AVS for which genotypes were available in METABRIC) for each locus. The matrix shows each multivariate eQTL test with a significant result as a grey box. Mapping tallies are summed on the right of the matrix. Box plots show the normalized null distributions of the enrichment scores (box: 1st–3rd quartiles; bars: extremes). Solid and open red diamonds highlight enrichment scores that satisfy a Bonferroni-corrected threshold for significance of P<0.01 and P<0.05, respectively. P-values are based on null distributions from 1,000 random AVSs. (c) Computational validation of EVSE analysis for cohort I and II. Averages of enrichment scores obtained in the EVSE analysis using different GWAS data sets (breast cancer (BCa), prostate cancer (PCa), bone mineral density (BMD) or chronic lymphocytic leukaemia (CLL) or random SNPs) shown along the x-axis, using different regulons and eQTLs (origin indicated by colour). The grey dotted line highlights the Bonferroni-corrected threshold (P<0.05). (d) Mean aberration frequency of the 36 risk-TFs compared to sets of 36 random genes (empirical p<0.001, boxplot whiskers extend to the 99th percentile of the random distribution with 10,000 random sets). Aberration frequencies for sets of random TFs and cancer genes12 are also shown.
Validation of the risk-TFs
To gain confidence in the identification of the 36 risk-TFs, we tested the effects of changing the input GWAS data or regulons on the resultant enrichment score. The red box plots in Figure 1c show the average enrichment score for the 36 risk-TFs using eQTLs and regulons from METABRIC. When replacing the breast cancer GWAS data, we found that GWAS hits for bone mineral density (BMD), chronic lymphocytic leukaemia (CLL) or random SNPs did not give significant enrichment scores (Fig. 1c blue box plots). For prostate cancer GWAS loci the scores obtained were lower but still significant, probably reflecting similarities in these two hormone driven cancers8. When we replaced the regulons calculated from METABRIC with random regulons of similar size (Fig. 1c, grey box plots) none of the associations were significant. These results support the specificity and validity of the EVSE analysis. Our results were not confounded by population stratification (Supplementary Note, Supplementary Fig. 5). We did not find enrichment when using normal breast samples from METABRIC to calculate eQTLs (white box plot). This is possibly surprising, as one might expect inherited risk to be expressed in normal tissue. However, eQTL discovery is dependent on sample size9 and only 144 normal tissue samples were available in this data set.
Comparison of ARACNe/EVSE to other methods
We compared our analysis to alternative methods for the derivation of the network structure and expansion of tagging SNPs into AVSs and obtained very similar results (Supplementary Note, Supplementary Figures 6-8). We also compared our EVSE algorithm to analyses in which the multivariate eQTL step was replaced by a distance-based gene selection, or by using ‘pre-defined’ eQTLs10 from the same sample set (Supplementary Note, Supplementary Figures 9-12). EVSE identified more risk-TFs and showed better reproducibility than the other tested methods.
Risk-TFs are frequently mutated in breast cancers
To ask whether somatic and germline variation are associated with the same regions of the network, we examined the frequency of mutations and/or copy number changes affecting TF genes in data from the Cancer Genome Atlas (TCGA)11. Collectively our 36 risk-TFs have a significantly increased frequency of alterations compared to random genes (Fig. 1d; Supplementary Table 1) and are mutated at a similar frequency as annotated cancer genes for which mutations have been causally implicated in cancer12.
Confirmation of risk association using ChIP-seq data
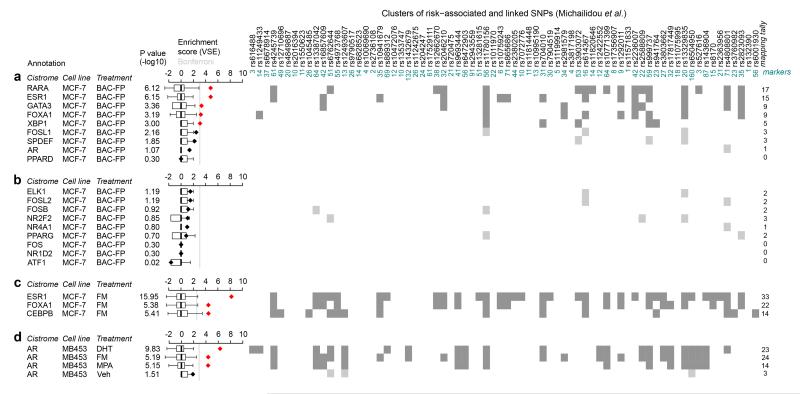
To validate that our risk-TFs are indeed associated with the regulation of GWAS loci we examined ChIP-seq data13 that was generated for TF-eGFP fusion proteins, driven from endogenous sequences in MCF-7 cells. We used these data in a variant set enrichment (VSE) analysis7 to test whether risk-TF binding sites are enriched at risk SNPs. Our analysis correlated the position of TF binding sites with risk AVSs. ChIP-seq data were available for 9 of our 36 risk-TFs and were compared to 9 low-risk TFs chosen from the EVSE analysis. 5 out of the 9 high-risk TFs, but none of the low-risk TFs (Fig. 2a,b), yielded a significant enrichment score. The signal in this analysis is likely to be relatively low since fusion proteins rather than the native TFs were assayed. When we used ChIP-seq data obtained with anti-FOXA1 and anti-ESR1 antibodies, much higher enrichment scores were obtained (Fig. 2c), corroborating previous results7. CEBPB binding was also enriched at breast cancer risk loci (Fig. 2c). Some of the TFs, such as AR and PPARD, are expressed at very low levels in MCF-7 cells. We therefore tested whether AR binding sites were significantly enriched for GWAS hits in the cell line MDAMB453, which belongs to the molecular apocrine subclass14 and expresses high levels of AR. Figure 2d shows that after AR activation, AR targets yield significant enrichment scores in this cell line. Collectively, the ChIP-seq experiments strongly support our conclusion that the risk-TFs play a role in regulating transcription at risk SNPs.
Figure 2. Enrichment of risk-TF binding sites at breast cancer GWAS loci.
VSE analysis7 of the cistrome of (a) 9 risk-TFs and (b) 9 non-risk TFs defined in the EVSE analysis for which ChIP-seq data was available13 in MCF-7 cells. Cells were transfected with BAC-fusion proteins (BAC-FP) of the relevant TF and eGFP and grown in full medium. Antibodies against eGFP were used in these ChIP experiments. VSE tallies that yielded a significant enrichment score are shown in dark grey, those that did not in light grey. (c) VSE analysis of ChIP-seq experiments using α-ESR1, α-FOXA1 and α-CEBPB antisera in MCF-7 cells. Cells were grown in full medium (FM). (d) VSE analysis of ChIP-seq data for AR using the molecular apocrine cell line MDAMB453 stimulated as indicated with DHT: 5αdihydro-testosterone; MPA: medroxyprogesterone acetate or vehicle treated or grown in full medium. Box plots show the normalized null distributions (box: 1st–3rd quartiles; bars: extremes). Diamonds show the corresponding VSE scores, either in black or in red for mapping tallies that satisfy a Bonferroni-corrected threshold for significance (P<0.01). P-values are based on null distributions from 1,000 random AVSs.
Confirmation of risk association by master regulator analysis
Estrogen and FGFR2 signalling pathways are known to be associated with breast cancer risk. We examined differential gene expression in response to estrogen and FGFR2 signalling in three ER+ breast cancer cell lines: MCF-7, T47D and ZR751. Using a master regulator analysis (MRA)15 (methods) we identified MRs consistently associated with these responses (Supplementary Note, Supplementary Table 2) and found a high prevalence of risk-TFs amongst the MRs, providing further support that our risk-TFs are indeed functionally related to breast cancer risk.
Clustering of risk-TFs and clues to function
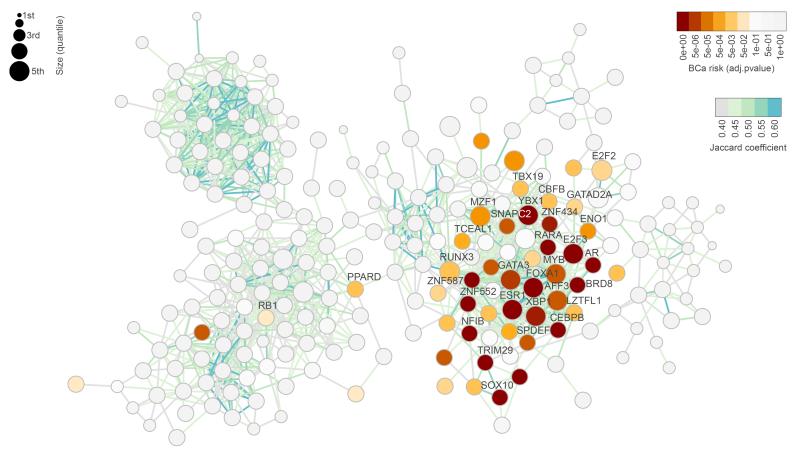
To examine whether the different risk-TFs converge on common mechanisms, we used ARACNe to calculate the breast cancer regulatory network and mapped onto this network the p-values for risk-association (shown in orange to red) using METABRIC cohort I. The network was visualised by the degree of overlap of regulons (Fig. 3, Supplementary Fig. 13). The enriched regulons mostly cluster together, suggesting that the risk-TFs share biological function.
Figure 3. Regulatory network for breast cancer, showing clustering of breast cancer risk.
The network is depicted on the basis of the overlap of regulons, with risk association shown in yellow to red (based on cohort I of METABRIC). The 36 consensus risk-TFs identified in both cohorts are labelled. The colouring of the edges (shown in light green to blue) indicates the overlap as measured by the Jaccard coefficient (JC) and the size of circles represents the size of each regulon. Only regulons with JC ≥ 0.4 are shown in the diagram. All regulons and a heatmap depicting the overlap of the regulons of risk-TFs is shown in Supplementary Figure 13. BCa risk: Bonferroni adjusted p-values obtained for each regulon when calculating the enrichment with breast cancer GWAS loci in the EVSE analysis, using cohort I for calculating the network and eQTLs. P-values are based on null distributions from 1,000 random AVSs.
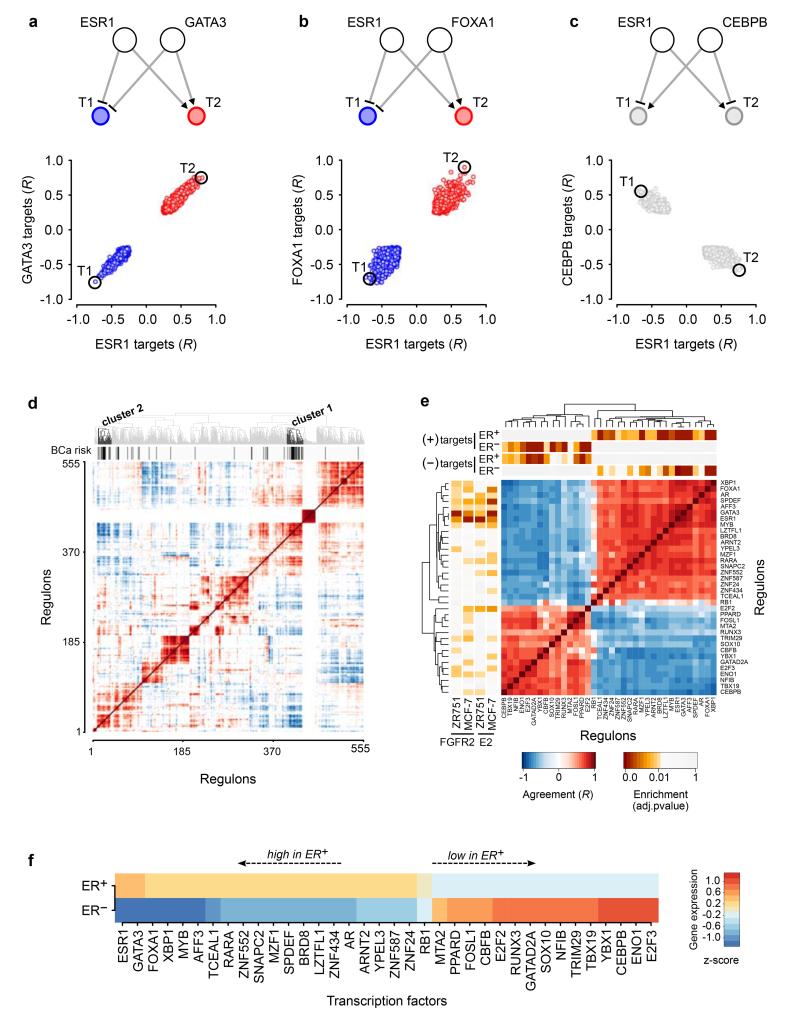
To refine the clustering analysis and look for clues to biological function, we extended the RTN16 package (methods) to include the direction of association between any TF-target gene pair using Pearson correlation. For all pairs of TFs with a target gene in common, the correlation values were used to assess whether the TFs regulated shared target gene in the same direction (up or down), or in different (opposite) directions (Fig. 4a-c). This analysis was carried out for all TFs in our regulatory network, and the correlation heat map was used in unsupervised clustering to generate the dendrogram depicted above the matrix (Fig. 4d). The position of the 36 risk-TFs is highlighted by the black bars below the dendrogram.
Figure 4. Correlation of expression of targets shared between TF pairs in breast tumours.
(a-c) Correlations of gene expression between a given TF and its targets were plotted for three different TF-TF pairs as indicated. Above each panel a cartoon depicts the observed interactions. Red circles indicate co-activation, blue circles co-repression. Targets are shown in grey if the two TFs have opposing effects on the target. (d) Heat map of the correlation of gene expression for targets shared by any pair of the 555 TFs (cohort I, METABRIC) whose regulons were of sufficient size to be analysed in the EVSE pipeline. Unsupervised clustering was applied to this correlation heat map resulting in the dendrogram shown at the top of the plot. The black bars depict the 36 risk-TFs, which fall into two distinct clusters. (e) Enlargement of the correlation heat map for the risk-TFs only. Above the matrix a bar with yellow to red colouring depicts the results (BH adjusted p-values) of a MRA analysis for the enrichment within each regulon of positive and negative targets that are upregulated in ER+ or ER− tumours, respectively, in cohort I of the METABRIC samples. The panel to the left of the matrix shows the master regulators identified for the FGFR2 and E2 responses. (f) Relative gene expression levels of the risk-TFs in ER+ or ER− tumours in cohort I of the METABRIC samples: expression levels were averaged in all ER+ and all ER− tumours and compared to expression levels averaged across all samples. TFs are shown ranked by differential gene expression between ER+ and ER− tumours.
Figure 4e shows an enlargement of the analysis for just the 36 risk-TFs. They fall into two distinct groups with high correlation within each group: Gene targets shared between two TFs in the same group are regulated in the same direction by both TFs, whereas gene targets shared between a TF in one group and a TF in the other, are regulated in opposite directions, suggesting the existence of two distinct regulatory groups of TFs able to oppose the effects of the other. The two groups of TFs are highly expressed in ER+ and ER− tumours respectively (Fig. 4f). Bootstrap analysis demonstrated that the split into two distinct groups is extremely stable (Supplementary Fig. 14). The behaviour of shared gene targets was mirrored in the correlation between the expression of the TFs themselves, but with much weaker signals (Supplementary Fig. 15a-c). This may reflect the difference between the regulatory activity of a TF, influenced by post-translational regulation and the presence of interacting factors, and the level of TF expression.
With respect to the intrinsic breast cancer subtypes, group 1 TFs are highly expressed in luminal A and B subclasses, while group 2 TFs are highly expressed in basal tumours. Her2 and normal-like tumours showed more heterogeneous gene expression patterns (Supplementary Fig. 16). Given this distribution, we tested the enrichment of each regulon for genes upregulated in ER+ or ER− tumours using MRA (methods). We split each regulon into activated and repressed targets and found that group 1 positive targets were enriched in the ER+ gene signature, whilst the negative targets were enriched in the ER− signature (Fig. 4e, bar above the matrix). Group 2 generated the opposite pattern, demonstrating that each group of TFs is associated with gene expression changes in both tumour subtypes, but with opposite effects.
Identification of clusters associated with known breast cancer subtypes
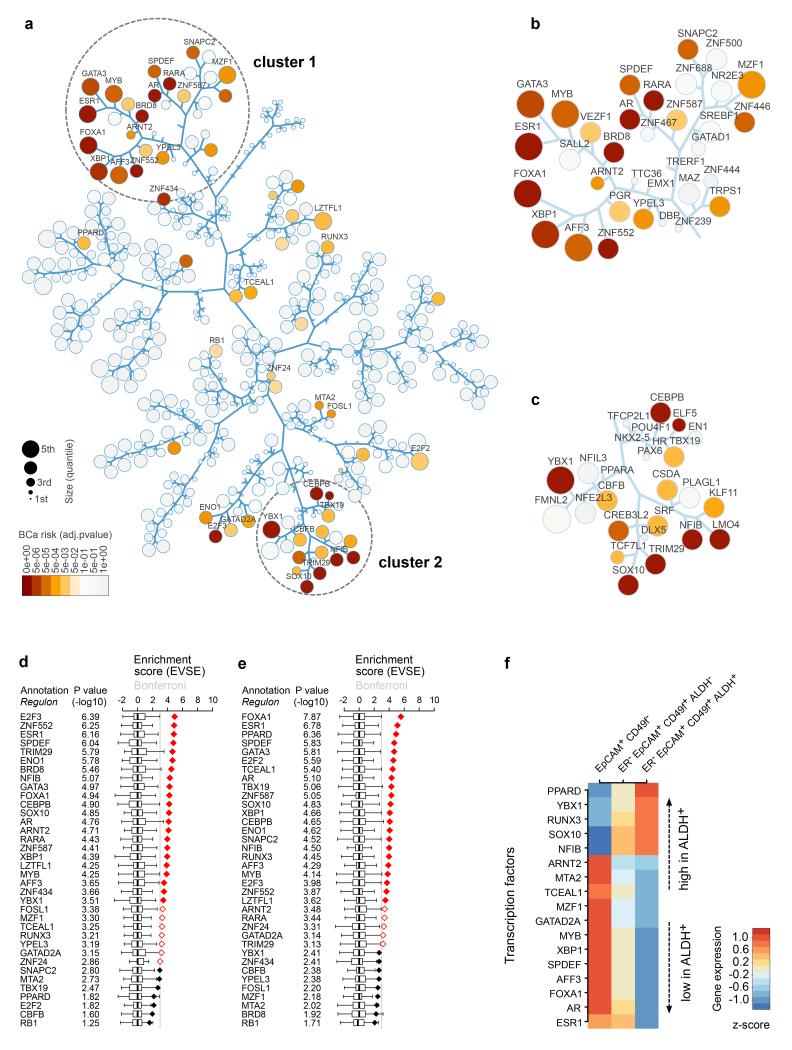
The dendrogram generated in Figure 4d was used to draw a tree and leaf diagram (Fig. 5a) representing the 555 TFs whose regulons in cohort I were of sufficient size to be analysed in the EVSE pipeline. Colouring of regulons indicates p-values for risk association. While some risk-TFs occur scattered throughout the diagram, two distinct clusters emerge: cluster 1 (enlarged in Fig. 5b) corresponds closely to group 1 in the previous analysis. These TFs include those important for the FGFR2 and estrogen response and also correlate with the TFs highly expressed in luminal A and B subtypes. Group 2 TFs are somewhat more dispersed throughout the tree, but there is clear clustering around the TFs YBX1, CBFB, NFIB, TRIM29 and SOX10, labelled as cluster 2 (Fig. 5c). Another branch in this node contains the risk-TFs CEBPB and TBX19.
Figure 5. Tree and leaf representation of correlation matrix reveals two clusters of risk-TFs.
(a) Tree and leaf representation of the dendrogram depicted in Figure 4d, where branches represent the arms in the dendrogram. The size of regulons is represented by circle size as indicated and Bonferroni adjusted p-values for EVSE enrichment of regulons for breast cancer GWAS loci in cohort I are shown in colour. Only consensus risk-TFs are labelled. (b) Enlargement of cluster 1 and (c) cluster 2 of the correlation heat map. All TFs present in these clusters are labelled, independent of risk association. (d-e) EVSE analysis showing enrichment score and p-value based rank order of the 36 risk-TFs for cohort I (d) and cohort II (e) using only ER+ tumours from the METABRIC dataset. The mapping tallies are shown in Supplementary Figure 17. (f) Relative gene expression levels of the risk-TFs that are differentially expressed in a comparison of three primary human luminal mammary cell populations18 as listed (P<0.05; BH adjusted from limma comparisons, see methods). Expression (Z-score) in each subpopulation is calculated relative to the average in the three populations analysed, and ranked by differential expression between the ALDH+ and ALDH− cell population.
A literature survey confirmed that TFs in cluster 2 are primarily associated with basal-like breast cancer. We therefore tested whether a gene signature for basal tumours17 was linked to cluster 2 using MRA. Of the six consensus MRs for basal-like cancers obtained from the METABRIC cohorts (Supplementary Table 3), the two most strongly associated TFs map to this cluster (SOX10, TRIM29). PLAGL1 also maps to cluster 2, but none of the basal-like cancer MRs fall within cluster 1.
Given the high differential expression of these clusters of TFs in ER+ and ER− tumours, we carried out the EVSE analysis separately in ER+ and ER− tumours. Risk-TFs for ER+ tumours map to both cluster 1 and 2 (Fig. 5 d-e, Supplementary Fig. 17), reinforcing our previous observation that both groups of risk-TFs can play a role in ER+ and ER− tumours, most likely with opposite effects. Both clusters were also marked by a VSE analysis using pre-defined eQTLs for ER+ tumours or using different network construction tools (Supplementary Fig. 18, Supplementary Table 4). An EVSE analysis with ER− tumours found very few, non-reproducible risk-TFs (not shown).
Activity of cluster 1 and 2 TFs in primary cells
Next we examined the expression patterns of our risk-TFs in primary cell populations isolated from the normal human mammary gland. Gene expression patterns for three luminal cell populations have previously been described18: an EpCAM+ CD49f− population highly enriched in ER+ cells that express high levels of luminal cell differentiation markers; ER− EpCAM+ CD49f+ ALDH+ cells that function as alveolar precursor cells; and ER− EpCAM+ CD49f+ ALDH− luminal cells that have a phenotype intermediate between the EpCAM+ CD49f− and the ER− EpCAM+ CD49f+ ALDH+ subpopulations. Figure 5f lists the risk-TFs that showed differential gene expression across these three populations (adj p-value <0.05). Eight cluster 1 TFs were overexpressed in the ER+-enriched population while several cluster 2 TFs (Fig. 5f, Supplementary Fig. 19a) were overexpressed in ER− EpCAM+ CD49f+ ALDH+ alveolar progenitors. The ALDH− population showed an intermediate pattern. Myoepithelial and stromal cells showed no clear expression pattern for the clusters (Supplementary Fig. 19b). The gene expression patterns seen in the ALDH+ versus the ER+-enriched primary cell population are reminiscent of that seen in basal-like versus luminal cancers.
Functional analysis of cluster 2 TFs
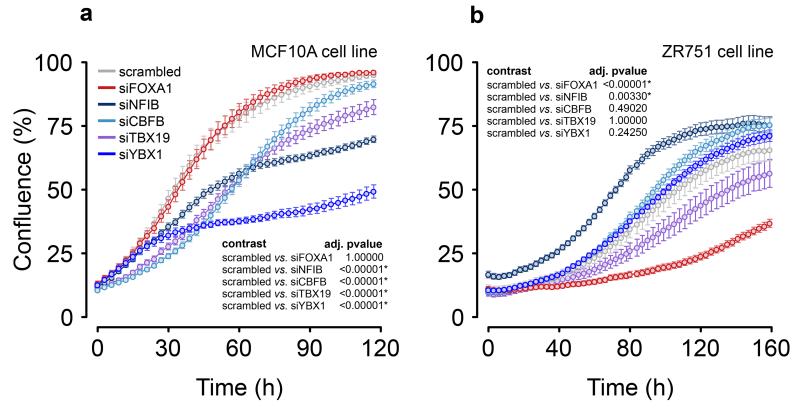
We examined the effect of siRNA knock-down of cluster 2 risk-TFs (NFIB, YBX1, CBFB and TBX19) in the ER− (MCF10A) and ER+ (ZR751) cell lines. In MCF10A cells, siRNA-targeting of YBX1 strongly reduced proliferation (Fig. 6a), and targeting CBFB, NFIB, TBX19 and LMO4 (Fig. 6a, Supplementary Fig. 20) all had a significant anti-proliferative effect. In contrast, repression of the cluster 2 TFs in ZR751 cells had either no or little effect on proliferation, whilst repression of FOXA1 strongly inhibited growth (Fig. 6b). Interestingly in ZR751 cells siNFIB led to a slight, but significant increase in proliferation, in keeping with the hypothesis that members of the two clusters have opposing effects.
Figure 6. Effects of risk-TF knock-down on cell proliferation.
Growth curves for (a) ER− cell line MCF10A and (b) the ER+ cell line ZR751 after transient transfection of the siRNAs as indicated. Cells transfected with a scrambled siRNA were included as a control. Error bars depict the standard error of the mean of 8 wells each in a minimum of two independent experiments (methods). The statistical analysis (insets) compares the growth curves using 100,000 simulations, with p-values adjusted by the BY correction method.
Whilst a group of ESR1-cooperating factors was already well defined, our analysis has extended the ESR1-cluster and revealed a group of TFs opposing ESR1 function, likely to be important in regulating basal-like cancers and their precursors.
Regulon activity as prognostic read-out
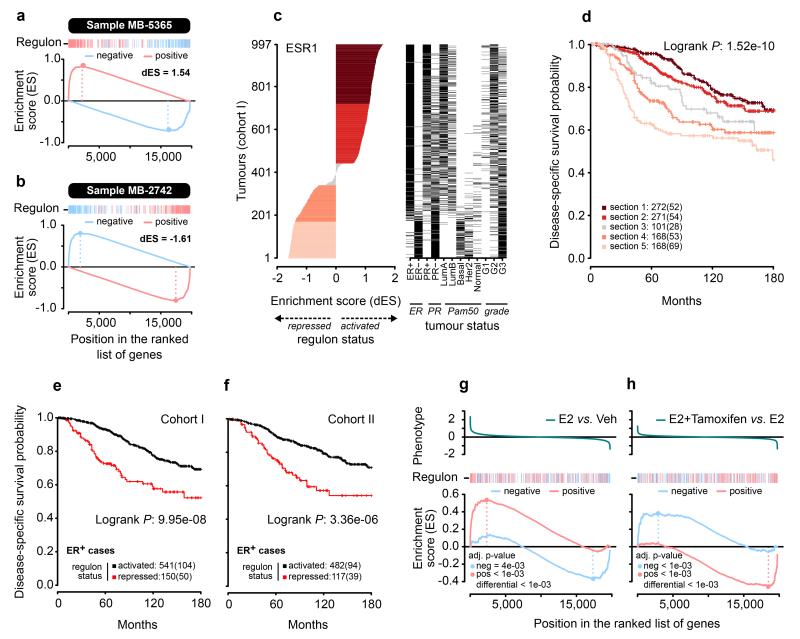
The ESR1 regulon consists of estrogen-induced and estrogen-repressed genes in approximately equal proportions.4 Our current analysis suggests that the relative activity of these two groups of genes may be important for determining the phenotype of the cell. We therefore devised a 2-tailed GSEA (Fig. 7a,b; methods) in which positive and negative targets of the ESR1 regulon are considered separately to generate a differential enrichment score (dES) (methods) representing the activity of the regulon. We used this in a stratified survival analysis in the METABRIC data (Fig. 7c,d). We found a continuous spectrum of dES across the tumors, except near the transition between the active and repressed state of the ESR1 regulon, which was characterized by an abrupt change. There was a strong trend for better survival with a high dES. Interestingly, we identified a set of patients with histochemically ER+ tumours that had a repressed ESR1 regulon and a significantly worse outcome than those with tumours with an active ESR1 regulon (Fig. 7e,f, Supplementary Fig. 21). This is not apparent when stratifying by ESR1 gene expression alone (Supplementary Fig. 22). We also tested the effect of tamoxifen treatment on the activity of the ESR1 regulon in MCF-7 cells using 2-tailed GSEA. As expected, we found that estrogen induction of steroid-starved MCF-7 cells led to a strong activation of the ESR1 regulon (Fig. 7g). However with estrogen plus tamoxifen treatment, the ESR1 regulon was shifted towards a more repressed state than with estrogen alone (Fig. 7h). This finding suggests that tamoxifen, while inhibiting proliferation, may also push luminal tumours to a more basal-like state19.
Figure 7. The ESR1 regulon as read-out of cell state.
(a and b) Examples of two tumours for which 2-tailed GSEA was carried out. The ESR1 regulon is split into targets activated by ESR1 (red bars) and targets repressed by ESR1 (blue bars). GSEA is carried out for each group. The running enrichment scores are shown. The differential enrichment scores (dES) are obtained by subtracting the maximal deviation from zero for the running enrichment score for repressed targets from that obtained for activated targets. (c) dES calculated for all tumours in METABRIC cohort I. Black bars indicate the ER status, PAM50 subclass and tumour grade for each of the tumours analysed. (d) Kaplan-Meier survival curve for disease-specific survival for each of the tumour subgroups highlighted in c. The number of patients in each section is listed, with the number of patients who died in brackets. (e and f) Kaplan-Meier survival curves for immunohistochemically ER+ tumours in cohort I and II, respectively, of the METABRIC patients comparing those in which the ESR1 regulon is in an activated state (dES>0 and ESA>0 and ESB<0) to those with repressed ESR1 regulons (dES<0 and ESA<0 and ESB>0). (g and h) 2-tailed GSEA in MCF-7 cells activated by estrogen (E2) or estrogen plus tamoxifen. Phenotypes were defined as differential gene expression between estrogen and vehicle treated cells (g) or between estrogen plus tamoxifen and estrogen (h) treatment.
Discussion
Our goal was to develop a network-based approach to understand how the effects of multiple GWAS loci combine to influence susceptibility. We derived a TF-centric regulatory network for breast cancer and asked by eQTL analysis which regulons were enriched for an association with confirmed breast cancer GWAS loci. We identified 36 regulons that were enriched in both of two separate analyses. The TFs controlling these regulons are frequently mutated in breast cancer, implying a convergence of germline and somatic events in the etiology of breast cancer. Many of the risk-TFs are master regulators of pathways associated with breast cancer risk, such as estrogen and FGFR2 signalling. Within the regulatory network, almost all of the risk-TFs clustered around a group of TFs already known to be central to breast cancer risk: ESR1, FOXA1, GATA3 and SPDEF4,7. This clustering supports the functional significance of the newly identified risk-TFs and suggests that risk-TFs share regulatory mechanisms.
The validity of the ARACNe/EVSE analysis was confirmed through extensive comparisons to other methods. The 36 identified risk-TFs were specific for hormone-driven cancer and could be validated experimentally. The EVSE analysis avoids the multiple testing problems of unrestrained eQTL calling and was therefore able to identify more risk-TFs than other methods. However, as in other analyses10, we identified eQTLs for only a minority of GWAS loci. Our method utilised gene expression data from breast tumours. Yet, our hypothesis is that inherited variation exerts its effects on normal tissue, and indeed on specific cell types within that tissue. To detect this, improved, context-specific methods for eQTL identification20,21 are required. The EVSE analysis we have developed can provide a general approach to interpret GWAS data in the context of regulatory networks.
Considering the direction (up or down) of the response of shared target genes revealed two distinct clusters of risk-TFs: those in cluster 1 whose positive targets were overexpressed in ER+ cancers, and those in cluster 2, whose positive targets were overexpressed in basal-like ER− cancers. However, the inverse also holds true: cluster 2 TFs repress genes associated with ER+ cancers, and cluster 1 TFs repress those associated with ER− cancers. Therefore both clusters of TFs are likely to be important for the establishment of ER+ and ER− tumours, albeit with opposing effects. This is supported by GWAS results, where the majority of loci confer risk for both ER+ and ER− disease22. Furthermore, our EVSE analysis using only ER+ tumours identified risk-TFs from both clusters.
Some cluster 1 TFs have previously been reported as critical for ER+ disease23-25 (ESR1, FOXA1, GATA3, SPDEF). We confirmed these and added more validated risk-TFs: XBP1, RARA and AR. XBP1 and ESR1 gene expression is highly correlated in laser microdissected breast tissue26 and RARA cooperates with ESR1 to drive estrogen-induced transcription27. Recent data suggest that in ER− apocrine tumours AR is able to replace the function of ESR1, leading to a luminal-like gene expression profile28. The identification of XBP1, RARA and AR as risk-TFs fits the overall framework that estrogen-driven gene expression is the predominant determinant of luminal breast cancer risk.
Cluster 2 comprises YBX1, CBFB, NFIB, TRIM29, SOX10, CEBPB and TBX19, all highly expressed in ER− tumours. Of these, our functional assays identified YBX1, NFIB and CBFB as important for proliferation in ER− cells in culture. Existing literature links individual TFs in cluster 2 to basal-like breast cancer29-32, which is associated with increased aggressiveness, metastasis and epithelial-to-mesenchymal transition (EMT). Here we suggest a network of cooperating TFs important in determining this cancer subtype. The link of cluster 2 TFs to basal-like breast cancer is further supported by increased binding at GWAS loci by CEBPB, a TF required for lobuloalveolar development33 whose loss is associated with EMT34.
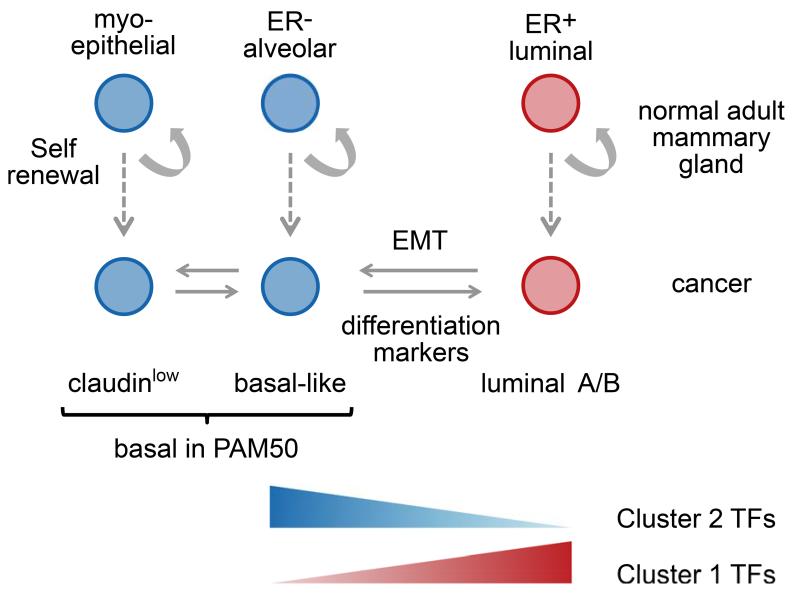
The most striking aspect of cluster 1 and 2 TFs is the opposing regulatory effect they exert on their target genes. We postulate that this mutually exclusive activity reflects the decision of a progenitor to commit to either an ER+ ductal or an ER− alveolar cell fate. In line with this hypothesis we find that in primary human mammary cell populations18, those representative of ER− alveolar progenitors show differential upregulation of cluster 2 TFs, whilst ER+ luminal cells display higher expression of cluster 1 TFs (Fig. 8). Recent genetic tracing experiments have shown that the ER+ ductal progenitors and ER− alveolar progenitors are self-renewing in the mouse mammary gland35,36,37. The differential expression of risk-TFs in these two self-renewing populations may suggest that these are the populations where risk genes are effective and cell transformation occurs. In line with this, transcriptional profiles of basal-like tumours most resemble that of ER− alveolar progenitors38,39, while luminal A and B tumours phenocopy ER+ ductal cells39-41,18. Furthermore, the ER− alveolar progenitor population is expanded in BRCA1 mutation carriers39, which are predisposed to develop ER− breast cancer.
Figure 8. Schematic model of mammary gland cell populations.
In this model we show the predominant expression of cluster 1 versus cluster 2 risk-TFs with respect to the cell populations found in the mammary gland and the cancer subtypes that arise from them. In the normal mammary gland all three populations have self-renewal capacity. Claudinlow tumours were originally classified as basal in the PAM signature, but are likely to represent a separate lineage arising from myoepithelial cells40. Basal-like cancer is thought to arise from alveolar progenitor cells, (The somewhat misleading term ‘basal-like’ reflects the fact these tumours not only express epithelial, but also mesenchymal cell surface markers that are also highly expressed in the myoepithelial lineage located near the basal membrane.) and luminal A/B cancer from ER+ precursors.
The opposing activity of two distinct networks of TFs has not previously been reported, but is consistent with studies carried out for individual TFs. For example, ELF5, an important inducer of alveolar differentiation42 can reduce estrogen sensitivity in ER+ cell lines43. FOXA1 in combination with GATA3 and ESR1 can specify an estrogen-responsive phenotype24, and, conversely, is able to repress the basal phenotype44. The concept of antagonism between TFs, led us to the 2-tailed GSEA analysis of the ESR1 regulon (Fig. 7). Of potential clinical relevance, the analysis identifies a subgroup of histochemically ER+ patients in whom the ESR1 regulon is functionally in a repressed state and in whom anti-estrogen treatment might not be effective. Our results also highlight the possibility that repression of cluster 1 TFs may lead to a shift in cell state towards more basal-like cancer, that is potentially associated with a more aggressive tumour phenotype and resistance to therapy. Better understanding of the interplay of the key regulators will be critical for optimal therapeutic strategies.
In summary, we have shown that EVSE analysis, together with gene regulatory networks, can identify key regulators that may influence disease risk. The analysis can be applied to any combination of GWAS loci for which eQTLs can be interrogated, not just those for which the causative SNPs and genes are already known. For breast cancer, the risk-enriched regulons include many driven by TFs already implicated in breast cancer, but many others that were not. The mutual antagonism of the two identified clusters of risk-TFs provides novel insights into their interactions, with potential clinical implications.
Online Methods
Computational Analysis
ARACNe/EVSE analysis
Regulons were calculated based on mutual information using the ARACNe algorithm3. Of the 809 TFs3 tested, we were able to assign regulons to 555 TFs in cohort I and 635 in cohort II of the METABRIC data set. The EVSE analysis has been described before4 and here we extended our previous computational pipeline (RTN16) to allow the testing of all regulons defined in the network. Supplementary Figure 2 illustrates the steps and data sets used in this analysis. In more detail, EVSE was carried out using the 72 breast cancer risk SNPs identified by Michailidou et al.5. For most of these GWAS loci neither the causative SNP nor the potential target genes are known. To deal with the former, the top hit at each locus (tagging SNP) was expanded into an AVS including all SNPs with a D′> 0.99 and a LOD>3.0 (Supplementary Fig. 2a), following the previously published VSE method7. This approach gave similar results to those obtained using r2 to expand the tagging SNP into the AVS (Supplementary Fig. 8). To identify potential target genes at each GWAS locus, we used gene expression and genotyping data in a multivariate eQTL analysis4. When considering multiple GWAS loci in a single analysis, the number of potential target genes may vary strongly for each GWAS locus to be analysed, making statistical comparisons between them difficult. For this reason we carried out a single multivariate eQTL analysis at each GWAS locus, asking whether there is an association of any of the SNPs in the AVS and the expression of any of the genes in a given regulon in a window of +/− 250kb around the AVS (Supplementary Fig 2b,c). (For each AVS only those SNPs for which genotyping data was available in METABRIC were considered in the analysis.) If a positive association was found, the locus was counted towards a mapping tally (Supplementary Fig. 2d) as described by Cowper Sal Iari et al7. In a subsequent step statistical significance was assessed (Supplementary Fig. 2e). To reduce the cost of the computational analysis when interrogating many regulons, we ran a low resolution analysis to remove obviously non-significant regulons (RTN package16). For all remaining regulons the EVSE analysis using breast cancer GWAS hits was tested against a null distribution based on random permutations of the AVS (that is, matched random variant sets). These distributions were normalised and centred around the null to obtain the enrichment score, which is the number of standard deviations that the observed mapping tally deviates from the null mapping mean. From these null distributions p-values were calculated. To gain confidence in our results we used cohort I and II of the METABRIC data set separately and only considered regulons that were significant in both cohorts. Where different GWAS results were tested (BMD, prostate cancer and CLL), each GWAS set was controlled with the appropriate number of random SNPs. As threshold for significance a Bonferroni correction was applied.
eQTL analysis
We performed a cis-eQTL analysis for cohort I and cohort II breast cancer samples generated by the METABRIC study6. The analysis largely followed that by Li et al10. We required probes to map to one of the RefSeq genes according to the annotation data obtained from the R package illuminaHumanv3.db45. Probes that map to genes in the highly polymorphic human leukocyte antigen region were excluded from the analysis. Genes with low expression levels (within 10% quantile of all expression values) were removed. Probes mapping to the same gene were treated independently in the eQTL analysis.
Copy number values for each gene of each sample were estimated from the segmented copy numbers by averaging the copy number of all segments that fall into the region of the gene while using the length of the copy number segments as weights. Gene expression levels were adjusted for copy number effects, using the equation
where, Ti is the measured gene expression, CNi is the copy number value, βi is the regression coefficient and εi is the residual gene expression level of gene i.
The eQTL analysis was performed using MatrixEQTL in R46 by correlating the genotypes of all remaining SNPs with the residual expression levels of proximal genes, i.e. genes within 1 Mb of the SNP. In the case that multiple probes map to a gene, all probes of that gene were tested separately. Finally, significant associations were selected based on a Benjamini-Hochberg False Discovery Rate (FDR) threshold of 0.1. Only SNPs with minor allele frequency (MAF) greater than 0.05 were tested. This is necessary because the effect of different genotypes on transcript levels cannot be evaluated if the genotypes at a given SNP locus are very homogeneous.
MRA analysis
The master regulator analysis uses a hypergeometric test to assess whether a gene list is enriched in a given regulon15. If significant the TF controlling the regulon is likely to be involved in the regulation of the gene list. Our experimental design compares resting with cycling cells and we therefore removed TFs that were also enriched with the Meta-PCNA signature47 (Supplementary Note).
VSE analysis
The variant set enrichment analysis was carried out as previously described7 using publically available data4,13,23 (GSE48930, GSE41995, E-MTAB-223, GSM1010889, E-MTAB-986). Briefly, VSE analysis tests enrichment of a chromosomal annotation, here TF-binding sites, at the AVSs. An overlap between a ChIP-seq peak and a SNP in the AVS is counted towards a mapping tally that is tested against random SNPs as in the EVSE.
Differential gene expression
Differential gene expression was assessed using limma48. Z-scores were obtained by comparing the gene expression values averaged across all cell populations in the analysis against averages of subgroups tested in each case. When determining significant differences in gene expression across primary cell populations, the following contrasts were examined: ALDH+ versus ALDH− cells, ALDH+ versus EpCAM+ CD49f− cells and ALDH− versus EpCAM+ CD49f− cells.
Two-tailed gene set enrichment analysis (GSEA)
GSEA49 assesses the skewed distribution of a selected gene set (S), here the ESR1 regulon, in a list of genes (L) ranked by a particular phenotype, in this case the differential gene expression observed when comparing a given tumour with the average expression for all METABRIC tumours. The enrichment score (ES) was calculated by walking down the list L, increasing by 1/|S| a running-sum statistic when encountering a gene in S and decreasing it by 1/(|L|-|S|) when encountering a gene not in S. The ES is the maximum deviation from zero. The two-tailed GSEA method is based on the Connectivity Map (CMAP) procedure50. The ESR1 regulon was derived by ARACNe from METABRIC cohort I and filtered using genefilter in Bioconductor51 to remove uninformative genes, about 15% of the regulon mostly of low variance. Feature selection is performed on cohort II and used to filter the regulon in cohort I and vice versa. The resultant regulon was split into two subgroups, positive targets (A) and negative targets (B) using Pearson’s correlation to assign directionality. The distribution of A and B was then tested by the GSEA statistics in the ranked phenotype, producing independent enrichment scores for each subgroup. An additional step calculated the differential enrichment (dES=ESA-ESB). The two-tailed GSEA was performed in R using the function tni.gsea2 in the RTN package4,16 with 1000 permutations.
Survival data6 were used to plot Kaplan-Meier curves and p-values were calculated using the log-rank statistics. On the basis of dES values the patients fell into three groups: those with an active ESR1 regulon (dES>0 and ESA>0 and ESB<0), those with a repressed ESR1 regulon (dES<0 and ESA<0 and ESB>0) and a small group in which the dES values were around zero (inconclusive, with ESA and ESB distributions skewed to the same side). The two large groups were further subdivided in half.
We tested the response of the ESR1 regulon to estrogen or estrogen plus tamoxifen treatment by applying the two-tailed GSEA to gene expression data from Hurtado et al.23 using the differential gene expression (estrogen versus vehicle and estrogen versus estrogen plus tamoxifen, GSE25316) as the phenotype to rank the gene list (L).
Cell culture
The human breast cancer cells MCF-7 and MDA-MB-453 (HTB 131; ATCC, USA) were cultured in DMEM (Invitrogen), ZR751 and T47D were cultured in RPMI (Invitrogen), all supplemented with 10% FBS and antibiotics, and MCF10A in DMEM, 5% horse serum, 5ug/mL insulin, 1 ug/mL hydrocortisone, 100 ng/mL cholera toxin, 20 ng/mL EGF and L-glutamine. Unless otherwise stated all cells were from the CRUK Cambridge Institute biorepository and maintained at 37°C, 5% CO2.
Chromatin immunoprecipitation (ChIP)
ChIP-seq experiments were carried out as previously described52. Cells were seeded at ~70% confluence into 15-cm tissue culture dishes (4 per treatment). Following overnight attachment, cells were starved using base media containing 5% steroid-stripped FBS. To ensure steroid depletion prior to treatment, media was changed every day for 3 days; then cells were treated for 4 hours with vehicle control (ethanol), 5α-dihydrotestosterone (DHT; 10nM), medroxyprogesterone acetate (MPA; 10nM). Cells were cross-linked and ChIP-seq performed using an AR antibody (N20; sc-816; Santa Cruz Biotech; 10 μg/IP) with subsequent data processing as previously described28. Two independent experiments were performed in each cell line and consensus AR chromatin binding events determined for each treatment condition.
Gene expression analysis after estrogen and FGF10 signalling
MCF-7 cells were plated at 5×105 cells/well in 6-well dishes and left in complete medium overnight. Cell synchronisation via estrogen-starvation was then carried out for three days in estrogen-free media (phenol red-free media supplemented with 5% charcoal dextran-treated FBS and 2 mM L-glutamine), with media changed every 24 hours. Estrogen-deprived cells were stimulated with 1 nM β-estradiol (E2; Sigma) or 100 ng/ml FGF10 (Invitrogen) in combination with 1 nM E2. 6 hours after cell treatment, total RNA was isolated from 3 biological replicates, quality controlled and used for cRNA amplification and labeling using the Illumina TotalPrep-96 kit (Ambion). cRNA was hybridised to HumanHT-12 v4 Expression BeadChips according to the manufacturer’s protocol (Illumina WGGX DirectHyb Assay Guide 11286331 RevA). Raw image files were processed and analysed using the beadarray package from Bioconductor.
Transient transfection of siRNA
Cell lines were transfected with ON-TARGETplus SMARTpool siRNA (Dharmacon) directed against risk-TFs NFIB (L-008456-00), YBX1 (L-010213-00), CBFB (L-011602-00), LMO4 (L-012124-00), ELF5 (L-011265-02), TBX19 (L-011910-00) and SOX10 (L-017192-00). CEBPB was not included in the analysis as multiple distinct isoforms with opposing function may be present in the cell. A custom siRNA was used against FOXA123. Knock-down of mRNA was confirmed for each cell line by RT-PCR of cDNA 48 hours after transfection (Supplementary Fig. 20c) using the primer pairs shown in Supplementary Table 5. 1 μg of total RNA was reverse transcribed using the High Capacity cDNA Reverse Transcription Kit (Applied Biosystems) and qRT-PCR performed using cDNA obtained from 10 ng of total RNA. qRT-PCR was performed using an ABI 9800HT Sequence Detection System (Applied Biosystems) with SDS software version 2.3. Amplification and detection were carried out in 384-well Optical Reaction Plates (Applied Biosystems) with Power SYBR Green Fast 2× qRT-PCR Mastermix (Applied Biosystems). All expression data were normalised to DGUOK expression. Primer-specificity was confirmed at the end of each qRT-PCR run through the generation of single peaks in melt-curve analysis. siRNA against SOX10 did not cause a reduction in mRNA levels and was not examined further. A control non-targeting pool of siRNAs (D-001810-01-05) was included in each experiment. Transfections were carried out using Lipofectamine RNAiMax Reagent (Invitrogen), according to manufacturer’s protocol. Growth was measured in 96-well plates using the IncuCyte (Essen BioScience) system every 3 hours. 8 wells were averaged for each experiment and at least two repeats were carried out for each cell line (MCF10A n=3, ZR751 n=2). The results of knock-downs for TFs in cluster 2 that are not consensus risk-TFs are shown in Supplementary Fig. 20). Statistical analysis was carried out using the compareGrowthCurves command in the statmod package53 in R, generating BY54 adjusted p-values.
Code availability
The source code developed in this study is publicly available from the Bioconductor55 in the R packages RTN4 and RedeR56 and the relevant URLs are listed in the appropriate section.
Supplementary Material
Acknowledgements
We thank Andrea Califano for helpful discussions and John Stingl for advice and critical reading of the manuscript. We are grateful to the genomics and bioinformatics cores at the CRUK Cambridge Institute for their support. This work was funded by Cancer Research UK and the Breast Cancer Research Foundation. MAAC is funded by the National Research Council (CNPq) of Brazil. TEH held a fellowship from the US DOD Breast Cancer Research Program (W81XWH-11-1-0592) and is currently supported by an RAH Career Development Fellowship (Australia). TEH and WDT are funded by the NHMRC of Australia (NHMRC) (ID: 1008349 WDT; 1084416 WDT, TEH) and Cancer Australia/National Breast Cancer Foundation (ID 627229; WDT, TEH). BAJP is a Gibb Fellow of Cancer Research UK. We would like to acknowledge the support of The University of Cambridge, Cancer Research UK and Hutchison Whampoa Limited.
Footnotes
Accession Codes
ChIP-seq data reported in this paper has been deposited at the Gene Expression Omnibus (GEO) under accession GSE74069; all microarray gene expression data under GSE70759.
URLs
http:/www.genome.gov/gwastudies
http://cancer.sanger.ac.uk/census
The authors have no competing financial interests to declare.
References
- 1.Califano A, Butte AJ, Friend S, Ideker T, Schadt E. Leveraging models of cell regulation and GWAS data in integrative network-based association studies. Nat Genet. 2012;44:841–7. doi: 10.1038/ng.2355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Leiserson MD, Eldridge JV, Ramachandran S, Raphael BJ. Network analysis of GWAS data. Curr Opin Genet Dev. 2013;23:602–10. doi: 10.1016/j.gde.2013.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Basso K, et al. Reverse engineering of regulatory networks in human B cells. Nat Genet. 2005;37:382–90. doi: 10.1038/ng1532. [DOI] [PubMed] [Google Scholar]
- 4.Fletcher MN, et al. Master regulators of FGFR2 signalling and breast cancer risk. Nat Commun. 2013;4:2464. doi: 10.1038/ncomms3464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Michailidou K, et al. Large-scale genotyping identifies 41 new loci associated with breast cancer risk. Nat Genet. 2013;45:353–61. 361e1–2. doi: 10.1038/ng.2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Curtis C, et al. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature. 2012;486:346–52. doi: 10.1038/nature10983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cowper-Sal lari R, et al. Breast cancer risk-associated SNPs modulate the affinity of chromatin for FOXA1 and alter gene expression. Nat Genet. 2012;44:1191–8. doi: 10.1038/ng.2416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Risbridger GP, Davis ID, Birrell SN, Tilley WD. Breast and prostate cancer: more similar than different. Nat Rev Cancer. 2010;10:205–12. doi: 10.1038/nrc2795. [DOI] [PubMed] [Google Scholar]
- 9.Schliekelman P. Statistical power of expression quantitative trait loci for mapping of complex trait loci in natural populations. Genetics. 2008;178:2201–16. doi: 10.1534/genetics.107.076687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li Q, et al. Integrative eQTL-based analyses reveal the biology of breast cancer risk loci. Cell. 2013;152:633–41. doi: 10.1016/j.cell.2012.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Forbes SA, et al. COSMIC: exploring the world’s knowledge of somatic mutations in human cancer. Nucleic Acids Res. 2015;43:D805–11. doi: 10.1093/nar/gku1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kittler R, et al. A comprehensive nuclear receptor network for breast cancer cells. Cell Rep. 2013;3:538–51. doi: 10.1016/j.celrep.2013.01.004. [DOI] [PubMed] [Google Scholar]
- 14.Hickey TE, Robinson JL, Carroll JS, Tilley WD. Minireview: The androgen receptor in breast tissues: growth inhibitor, tumor suppressor, oncogene? Mol Endocrinol. 2012;26:1252–67. doi: 10.1210/me.2012-1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Carro MS, et al. The transcriptional network for mesenchymal transformation of brain tumours. Nature. 2010;463:318–25. doi: 10.1038/nature08712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Castro M, Wang X, Fletcher M, Markowetz F, Meyer K. RTN: reconstruction of transcriptional networks and analysis of master regulators. R package. 2014 http://bioconductor.org/packages/release/bioc/html/RTN.html. [Google Scholar]
- 17.Bertucci F, et al. Gene expression profiling shows medullary breast cancer is a subgroup of basal breast cancers. Cancer Res. 2006;66:4636–44. doi: 10.1158/0008-5472.CAN-06-0031. [DOI] [PubMed] [Google Scholar]
- 18.Shehata M, et al. Phenotypic and functional characterisation of the luminal cell hierarchy of the mammary gland. Breast Cancer Res. 2012;14:R134. doi: 10.1186/bcr3334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Haughian JM, et al. Maintenance of hormone responsiveness in luminal breast cancers by suppression of Notch. Proc Natl Acad Sci U S A. 2012;109:2742–7. doi: 10.1073/pnas.1106509108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fu J, et al. Unraveling the regulatory mechanisms underlying tissue-dependent genetic variation of gene expression. PLoS Genet. 2012;8:e1002431. doi: 10.1371/journal.pgen.1002431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Montgomery SB, Dermitzakis ET. From expression QTLs to personalized transcriptomics. Nat Rev Genet. 2011;12:277–82. doi: 10.1038/nrg2969. [DOI] [PubMed] [Google Scholar]
- 22.Fachal L, Dunning AM. From candidate gene studies to GWAS and post-GWAS analyses in breast cancer. Curr Opin Genet Dev. 2015;30:32–41. doi: 10.1016/j.gde.2015.01.004. [DOI] [PubMed] [Google Scholar]
- 23.Hurtado A, Holmes KA, Ross-Innes CS, Schmidt D, Carroll JS. FOXA1 is a key determinant of estrogen receptor function and endocrine response. Nat Genet. 2011;43:27–33. doi: 10.1038/ng.730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kong SL, Li G, Loh SL, Sung WK, Liu ET. Cellular reprogramming by the conjoint action of ERalpha, FOXA1, and GATA3 to a ligand-inducible growth state. Mol Syst Biol. 2011;7:526. doi: 10.1038/msb.2011.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Marcotte R, et al. Essential gene profiles in breast, pancreatic, and ovarian cancer cells. Cancer Discov. 2012;2:172–89. doi: 10.1158/2159-8290.CD-11-0224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Andres SA, Wittliff JL. Relationships of ESR1 and XBP1 expression in human breast carcinoma and stromal cells isolated by laser capture microdissection compared to intact breast cancer tissue. Endocrine. 2011;40:212–21. doi: 10.1007/s12020-011-9522-x. [DOI] [PubMed] [Google Scholar]
- 27.Ross-Innes CS, et al. Cooperative interaction between retinoic acid receptor-alpha and estrogen receptor in breast cancer. Genes Dev. 2010;24:171–82. doi: 10.1101/gad.552910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Robinson JL, et al. Androgen receptor driven transcription in molecular apocrine breast cancer is mediated by FoxA1. EMBO J. 2011;30:3019–27. doi: 10.1038/emboj.2011.216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Davies AH, et al. YB-1 transforms human mammary epithelial cells through chromatin remodeling leading to the development of basal-like breast cancer. Stem Cells. 2014;32:1437–50. doi: 10.1002/stem.1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cimino-Mathews A, et al. Neural crest transcription factor Sox10 is preferentially expressed in triple-negative and metaplastic breast carcinomas. Hum Pathol. 2013;44:959–65. doi: 10.1016/j.humpath.2012.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moon HG, et al. NFIB is a potential target for estrogen receptor-negative breast cancers. Mol Oncol. 2011;5:538–44. doi: 10.1016/j.molonc.2011.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ai L, et al. TRIM29 suppresses TWIST1 and invasive breast cancer behavior. Cancer Res. 2014;74:4875–87. doi: 10.1158/0008-5472.CAN-13-3579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Seagroves TN, et al. C/EBPbeta, but not C/EBPalpha, is essential for ductal morphogenesis, lobuloalveolar proliferation, and functional differentiation in the mouse mammary gland. Genes Dev. 1998;12:1917–28. doi: 10.1101/gad.12.12.1917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Johansson J, et al. MiR-155-mediated loss of C/EBPbeta shifts the TGF-beta response from growth inhibition to epithelial-mesenchymal transition, invasion and metastasis in breast cancer. Oncogene. 2013;32:5614–24. doi: 10.1038/onc.2013.322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Van Keymeulen A, et al. Distinct stem cells contribute to mammary gland development and maintenance. Nature. 2011;479:189–93. doi: 10.1038/nature10573. [DOI] [PubMed] [Google Scholar]
- 36.Lafkas D, et al. Notch3 marks clonogenic mammary luminal progenitor cells in vivo. J Cell Biol. 2013;203:47–56. doi: 10.1083/jcb.201307046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rodilla V, et al. Luminal Progenitors Restrict Their Lineage Potential during Mammary Gland Development. PLoS Biol. 2015;13:e1002069. doi: 10.1371/journal.pbio.1002069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lim E, et al. Aberrant luminal progenitors as the candidate target population for basal tumor development in BRCA1 mutation carriers. Nat Med. 2009;15:907–13. doi: 10.1038/nm.2000. [DOI] [PubMed] [Google Scholar]
- 39.Molyneux G, et al. BRCA1 basal-like breast cancers originate from luminal epithelial progenitors and not from basal stem cells. Cell Stem Cell. 2010;7:403–17. doi: 10.1016/j.stem.2010.07.010. [DOI] [PubMed] [Google Scholar]
- 40.Perou CM, Borresen-Dale AL. Systems biology and genomics of breast cancer. Cold Spring Harb Perspect Biol. 2011;3 doi: 10.1101/cshperspect.a003293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lim E, et al. Transcriptome analyses of mouse and human mammary cell subpopulations reveal multiple conserved genes and pathways. Breast Cancer Res. 2010;12:R21. doi: 10.1186/bcr2560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Oakes SR, et al. The Ets transcription factor Elf5 specifies mammary alveolar cell fate. Genes Dev. 2008;22:581–6. doi: 10.1101/gad.1614608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kalyuga M, et al. ELF5 suppresses estrogen sensitivity and underpins the acquisition of antiestrogen resistance in luminal breast cancer. PLoS Biol. 2012;10:e1001461. doi: 10.1371/journal.pbio.1001461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bernardo GM, et al. FOXA1 represses the molecular phenotype of basal breast cancer cells. Oncogene. 2013;32:554–63. doi: 10.1038/onc.2012.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
Methods-only References
- 45.Dunning M, Lynch A, Eldridge M. illuminaHumanv3.db: Il lumina HumanHT12v3 annotation data. R package version 1.18.0. 11. [Google Scholar]
- 46.Shabalin AA. Matrix eQTL: ultra fast eQTL analysis via large matrix operations. Bioinformatics. 2012;28:1353–8. doi: 10.1093/bioinformatics/bts163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Venet D, Dumont JE, Detours V. Most random gene expression signatures are significantly associated with breast cancer outcome. PLoS Comput Biol. 2011;7:e1002240. doi: 10.1371/journal.pcbi.1002240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ritchie ME, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015 doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Subramanian A, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–50. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lamb J, et al. The Connectivity Map: using gene-expression signatures to connect small molecules, genes, and disease. Science. 2006;313:1929–35. doi: 10.1126/science.1132939. [DOI] [PubMed] [Google Scholar]
- 51.Gentleman R, Carey V, Huber W. genefilter: methods for filtering genes from high-throughput experiments. 2015. R package version 1.48.1. R package version 1.48.1. [Google Scholar]
- 52.Schmidt D, et al. ChIP-seq: using high-throughput sequencing to discover protein-DNA interactions. Methods. 2009;48:240–8. doi: 10.1016/j.ymeth.2009.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Smyth G, Hu Y, Dunn P, Phipson B, Chen Y. statmod: Statistical Modeling. 2014 R package version 1.4.20. http://CRAN.R-project.org/package=statmod.
- 54.Benjamini Y, Yekutieli D. The control of the false discovery rate in multiple testing under dependency. Ann. Satist. 2001;29:1165–1188. [Google Scholar]
- 55.Gentleman RC, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Castro MA, Wang X, Fletcher MN, Meyer KB, Markowetz F. RedeR: R/Bioconductor package for representing modular structures, nested networks and multiple levels of hierarchical associations. Genome Biol. 2012;13:R29. doi: 10.1186/gb-2012-13-4-r29. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.