Significance
This work reveals the deeply conserved gene repertoire of animal stem cells, from sponges to mammals. This repertoire mostly contains ancient (premetazoan) genes and few novel (metazoan-specific) genes, but the latter point to the importance of genome protection in the origin of stem cells. Additionally, regulators of DNA transcription are only marginally represented among conserved stem-cell genes, whereas genes directly acting upon RNA predominate, including orthologues of RNA-binding proteins that control mammalian embryonic stem cells. Thus posttranscriptional regulation of gene expression has been crucial in animal stem-cell biology across hundreds of millions of years of animal evolution.
Keywords: Porifera, stem cells, evolution, uPriSCs, RNA binding
Abstract
Stem cells are pivotal for development and tissue homeostasis of multicellular animals, and the quest for a gene toolkit associated with the emergence of stem cells in a common ancestor of all metazoans remains a major challenge for evolutionary biology. We reconstructed the conserved gene repertoire of animal stem cells by transcriptomic profiling of totipotent archeocytes in the demosponge Ephydatia fluviatilis and by tracing shared molecular signatures with flatworm and Hydra stem cells. Phylostratigraphy analyses indicated that most of these stem-cell genes predate animal origin, with only few metazoan innovations, notably including several partners of the Piwi machinery known to promote genome stability. The ancestral stem-cell transcriptome is strikingly poor in transcription factors. Instead, it is rich in RNA regulatory actors, including components of the “germ-line multipotency program” and many RNA-binding proteins known as critical regulators of mammalian embryonic stem cells.
Stem cells are defined by their capacity to give rise to daughter cells that remain in the stem-cell state (self-renewal) as well as to progenitor(s) that can undergo cellular differentiation. In some animal lineages, there are adult stem cells that retain the potential to produce both the germ-line and at least several somatic cell types, and for that reason, they have been called “unlimited primordial stem cells” (uPriSCs) (1). uPriSCs have been documented in animal groups as diverse as colonial ascidians (2), acoels (3), flatworms (4, 5), cnidarians (6, 7), and sponges (8), leading to the suggestion that uPriSCs represent the ancestral type of animal stem cell (1).
To date, comprehensive transcriptomes of uPriSCs have been determined only for the hydrozoan cnidarian Hydra (7) and for several flatworm species (4, 9–12). The transcriptome of flatworm neoblasts is significantly enriched in genes encoding RNA-binding proteins (RBPs) whose bilaterian orthologs act in the maintenance of stem-cell identity (4, 9, 10), suggesting that posttranscriptional regulation might have been of prime importance for stem-cell regulation in the last common ancestor of Bilateria (“higher” animals). Transcriptome analysis of the uPriSCs of the nonbilaterian Hydra vulgaris (interstitial stem cells) revealed the existence of a set of transcription factors (TFs) with stem-cell functions conserved up to vertebrates (7). These data unveiled some conserved features of stem cells at the phylogenetic level of eumetazoans (the clade comprising cnidarians and bilaterians), but animal stem cells originated earlier, in a common ancestor of all extant Metazoa, and a comprehensive view of their ancestral gene toolkit is still lacking.
Sponges are widely thought to be the sister-group to all other metazoans (13). Although this view has recently been challenged (ctenophores rather than sponges being the basal-most animal phylum according to refs. 14 and 15; but see refs. 13 and 16), sponges uncontroversially represent the earliest diverging animal lineage in which uPriSCs have been characterized. As such, they are pivotal to unraveling the molecular mechanisms that underpinned the acquisition of stem cells in the last common metazoan ancestor. The stem-cell system of demosponges comprises totipotent archeocytes together with a specialized cell type, the choanocyte, the latter having the potency to produce other choanocytes and male gametes in demosponges (8) but also to change to archeocytes under certain conditions (17, 18). Ephydatia fluviatilis is a freshwater demosponge that reproduces both sexually and asexually. In asexual reproduction, the adult sponge produces small particles (gemmules) housing thousands of binucleated stem cells (thesocytes, a resting form of archeocytes). Under certain environmental conditions, gemmule hatching starts by the division of thesocytes into mononucleated totipotent archeocytes, which then migrate out from the gemmule, proliferate, self-renew, and give rise to all differentiated cell types of a functional juvenile sponge (8, 17, 19) (Fig. 1A).
Fig. 1.
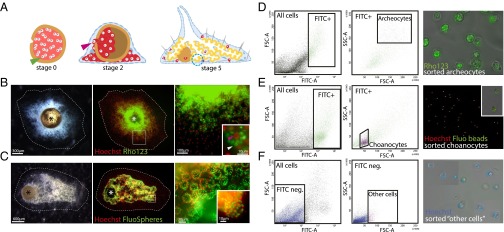
Staining of cells from juvenile Ephydatia and FACS sorting of archeocytes, choanocytes, and other cells. (A) Gemmule hatching process. Blue dotted circle, choanocyte chamber; green arrowhead, thesocyte; pink arrowhead, archeocyte. (B) Stage 2 juvenile E. fluviatilis (Left), Hoechst 33342 and Rho123 counterstaining (Middle), and magnified archeocytes (Right) containing Rhodamine-stained vitelline platelets (Inset, white arrowhead) and a large nucleolated nucleus (Inset, red arrowhead). (C) Stage 5 juvenile E. fluviatilis (Left), Hoechst 33342 and FluoSphere counterstaining (Middle), and magnified stained choanocyte chambers (Right and Inset) containing fluorescent beads (green). Asterisks in B and C indicate the gemmule coat. (D) FACS gate for archeocytes (Left panels) and photograph showing purified cells (Right panel) (large size and strong green fluorescence). (E) FACS gate for choanocytes (Left panels) and photograph showing purified cells (Right) (small size and strong green fluorescence). Inset shows two choanocytes with several ingested fluorescent beads. (F) FACS gate for other cells (Left panels) and purified cells (Right) (heterogeneous size and weak or no green fluorescence). Note that several fields have been combined to show the heterogeneity of cell sizes in that fraction. FITC-A, green fluorescence; FSC-A, forward scatter; SSC-A, side scatter.
In this study, we devised an original protocol to purify archeocytes and characterize their transcriptomic profile. We assembled a de novo transcriptome of juvenile Ephydatia fluviatilis and quantified the expression levels of transcripts in archeocytes versus choanocytes and “other cells” fractions. We identified the orthologs of archeocyte-specific genes in the uPriSC transcriptomes of Hydra and the flatworm Schmidtea (7, 10). From these comparisons we were able to construct a gene set representing the best inferred ancestral molecular repertoire to date of metazoan uPriSCs. The evolutionary and biological significance of this ancestral repertoire was evaluated by analyzing its gene content in terms of functional categories and of characterized stem-cell functions in animal models [notably, mammalian embryonic stem cells (ESCs)] and by mapping the origin of these genes on the tree of life.
Results
Transcriptomic Profile of Totipotent Archeocytes.
We first built a reference dataset of 17,419 assembled de novo E. fluviatilis transcripts obtained by cDNA sequencing of a mixture of juvenile sponges at different gemmule hatching stages (Methods; transcripts provided in Dataset S1). Next, archeocytes from stage 2 juvenile sponges were labeled with Rhodamine 123 (Rho123), a green fluorescent dye that we found to be incorporated into the abundant vitelline platelets known to be present in the archeocyte cytoplasm during the early stages of gemmule hatching (19) (Fig. 1B). At this stage we know that the vast majority of the sponge body is made of actively proliferating archeocytes. The food-entrapping choanocytes of stage 5 juveniles were labeled by feeding with green fluorescent beads as described previously (20) (Fig. 1C). Although it is known that several cell types have phagocytic activity in demosponges (e.g., pros-endopinacocytes), visual inspection of the sponges fed with fluorescent beads clearly shows that choanocytes are by far the most abundant and the most intensively stained cells. Then we sorted archeocytes and choanocytes by FACS based on their level of green fluorescence and their light scattering parameters (20) (Methods and Fig. 1 D and E). We also labeled stage 2 sponges with both Rho123 and fluorescent beads and collected a fraction, called other cells, lacking green fluorescence and possessing light scattering parameters outside the range of those of archeocytes or choanocytes (Fig. 1F). At this stage, we assessed cell fraction purity by controlling the sorted cells under light microscope. All observed cells stained by Rho123 had typical features of archeocytes: large in size, large nucleus with a single nucleolus, and many vitelline platelets (stained by Rho123) (Fig. 2D). Cells labeled by fluorescent beads had features of choanocytes: small in size, cytoplasm with ingested fluorescent beads (Fig. 2E), and in some cases, recognizable beating flagella. Unstained cells had diverse sizes and shapes, as expected for a mixture of multiple differentiated cell types (Fig. 2F). We cannot exclude an undetectable level of contamination, but we can confidently consider that we obtained two cell fractions respectively highly enriched in archeocytes and in choanocytes and a third fraction almost totally devoid of archeocytes and choanocytes.
Fig. 2.
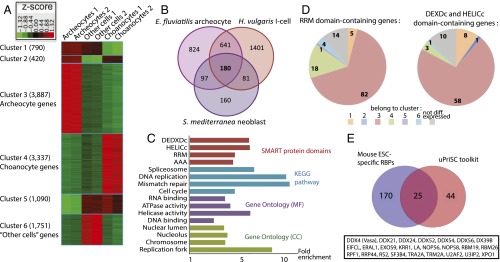
Transcriptomic signature of archeocyte and uPriSC genes. (A) Heat map of the 11,275 genes differentially expressed between archeocytes, choanocytes, and other cells. (B) Venn diagram showing the 180 OrthoMCL groups overexpressed in the interstitial stem cells of H. vulgaris, the neoblasts of S. mediterranea, and the archeocytes of E. fluviatilis. (C) Enriched functional categories in the uPriSC repertoire (FDR < 5e-2). In each of the four annotation sources depicted, terms are ordered by FDR value (with the lower values on top). The full enrichment study data are provided in Dataset S2, Table S5. (D) Number of putative RBPs belonging to each expression cluster. DEAD-box helicases are on the right and RRM-containing genes on the left. (E) Venn diagram showing the 25 OrthoMCL groups corresponding to RBPs from mouse ESCs that belong to the uPriSC repertoire.
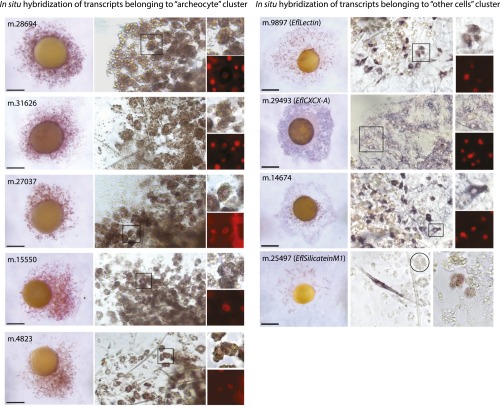
Around 25 million Illumina 100-bp single-end reads per biological duplicate were obtained and mapped onto the reference transcripts, leading to identification of 11,275 genes differentially expressed among the three cell fractions [Fisher’s exact test false discovery rate (FDR) < 0.05 and reads per kilobase per million mapped reads (RPKM) ≥ 1 in at least one fraction; see Methods] (Fig. 2A, Fig. S1, and Dataset S2, Tables S1–S3). Importantly, the following facts also support the purity of our cell fractions. All cell type-specific marker genes identified in previous studies consistently fell into the expected cluster (refs. 20–25; Dataset S2, Table S4 and Fig. S2), and as an additional control, in situ hybridization in juvenile sponges for genes belonging to the “archeocytes” and other cells clusters yielded the expected expression patterns (Fig. S2).
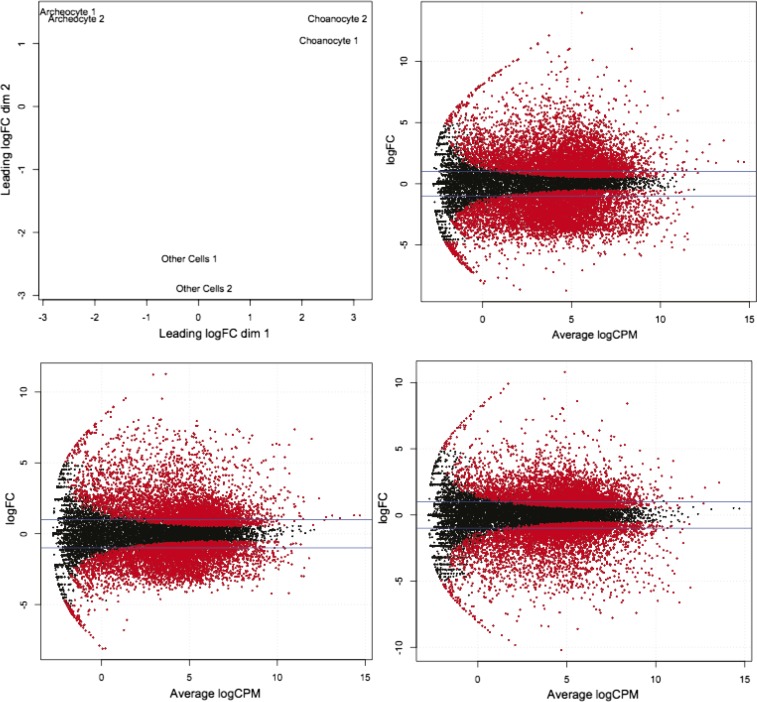
Fig. S1.
RNA-seq data analysis with EdgeR. (Top Left) Multidimensional scaling (MDS) plot from EdgeR analysis showing reproducibility between biological duplicates. FC, fold change; other49 and other44, cho46 and cho48, and arch43 and arch47 designate the biological duplicates for the samples of other cells, choanocytes, and archeocytes, respectively. Smear plots from EdgeR showing, in red, transcripts differentially expressed (FDR < 0.05) between, respectively, archeocytes and choanocytes (Top Right), archeocytes and other cells (Bottom Left), and choanocytes and other cells (Bottom Right). cpm, counts per million.
Fig. S2.
Controls of the specificity of the differential transcriptomic datasets with respect to corresponding cell types. Left panel, in situ hybridization for five genes belonging to the cluster archeocytes showing expression in cells with typical archeocyte morphology (nucleus with prominent nucleolus and many vitelline platelets). Images in the right column show enlarged views of stained archeocytes in the boxed areas in the middle column. (Scale bars, 300 µm.). Right panel, in situ hybridization for four genes belonging to the cluster other cells showing expression in diverse cell types (mesohyl cells for m.9897 and m.14674; basopinacocytes for m.29493; sclerocytes and archeocytes committed to sclerocyte fate for m.25497). (Scale bars, 400 µm.) Circle in m.25497 shows an unstained archeocyte.
A total of 3,350 transcripts were expressed at least twofold higher (“overexpressed” genes) in archeocytes than in choanocytes and other cells, including transcripts of the previously identified archeocyte marker EflMusashiA (21). Among these, 1,151 were expressed at least fivefold higher (“specific” genes) in archeocytes than in choanocytes and other cells, including the stem-cell markers EflPiwiA and EflPiwiB (22) (Dataset S2, Tables S2 and S3). The other cells cluster contained known markers for diverse differentiated cell types (e.g., EflSilicateinM1 and EflLectin, expressed in sclerocytes and cells involved in innate immunity, respectively) (23) and was enriched in developmental genes (cell communication, adhesion, and differentiation factors), probably reflecting the molecular programs used to construct the juvenile sponge body (Dataset S2, Table S5). The “choanocytes” cluster was significantly enriched in lysosome genes, as predicted from the role of these cells in intracellular food digestion, and also showed overexpression of the previously identified choanocyte marker EflAnnexin (20) (Dataset S2, Tables S4 and S5). The choanocyte cluster did not show enrichment in mitotic genes and showed low expression of EflPiwiA (transcripts of this gene were previously detected in choanocytes by in situ hybridization) (22), suggesting that the actively feeding choanocytes in stage 5 juvenile sponges we purified were in a nonproliferative state. The archeocyte cluster, in contrast, was enriched in DNA repair, DNA replication, and cell-cycle genes (Dataset S2, Table S5), as expected for highly proliferative cells. Thus, among the three cell fractions we compared here, only archeocytes showed stem-cell properties.
The Ancestral Stem-Cell Gene Repertoire Contains 180 Genes.
We reconstructed the genetic repertoire of ancestral metazoan uPRiSCs—that is, genes overexpressed in uPRiSCs across metazoan lineages—by compiling a list of orthology groups with transcripts overexpressed in the archeocytes of E. fluviatilis (present study), the interstitial stem cells of the cnidarian H. vulgaris (using data from ref. 7), and the neoblasts of the flatworm Schmidtea mediterranea (data from ref. 10) (Fig. 2B, Dataset S2, Table S6, and Methods). These species, the only ones for which RNA-sequencing (RNA-seq) data of uPRiSCs are available, represent three major early-diverging animal lineages: sponges, cnidarians, and bilaterians [ideally, we would have liked to compile comparable lists for ctenophores and placozoans, but ctenophores are thought to lack uPRiSCs (26) and placozoan stem cells have not yet been characterized]. The three compared stem-cell systems each have derived characteristics, but at the same time, they are also expected to share deep homologies at the molecular level, inherited from a genetic program that regulated uPRiSCs in the metazoan ancestor. The orthology groups were determined by matching transcripts of these three species against the OrthoMCL database (27).
The resulting combination is made up of 180 orthology groups (Fig. 2B and Dataset S2, Table S6) that can be taken as a proxy of the core transcriptome of the ancestral animal stem cell. As expected, this repertoire is strongly enriched in cell cycle, DNA replication, and DNA repair genes (e.g., pcna, G2/mitotic-specific cyclin B1) (Fig. 2C and Dataset S2, Table S6). Some genes overexpressed in archeocytes and with orthologs having known stem-cell functions in other animals do not appear among the 180 genes, due, for instance, to secondary gene loss in H. vulgaris or S. mediterranea (e.g., Myc is missing in flatworm) (28) or because they are also highly expressed in the unipotent epithelial stem cells of H. vulgaris (e.g., Piwi) or differentiated cells of S. mediterranea (e.g., GATA1–4). Interestingly, although the fold-change thresholds initially used to select stem-cell genes in the three species were permissive (Methods), the resulting 180 genes rank among the most strongly overexpressed in stem cells (in Ephydatia the median ratio of expression in archeocytes for these 180 genes is 4.3-fold over other cells and 9.5-fold over choanocytes).
TFs Are Underrepresented Among the Ancestral Stem-Cell Genes.
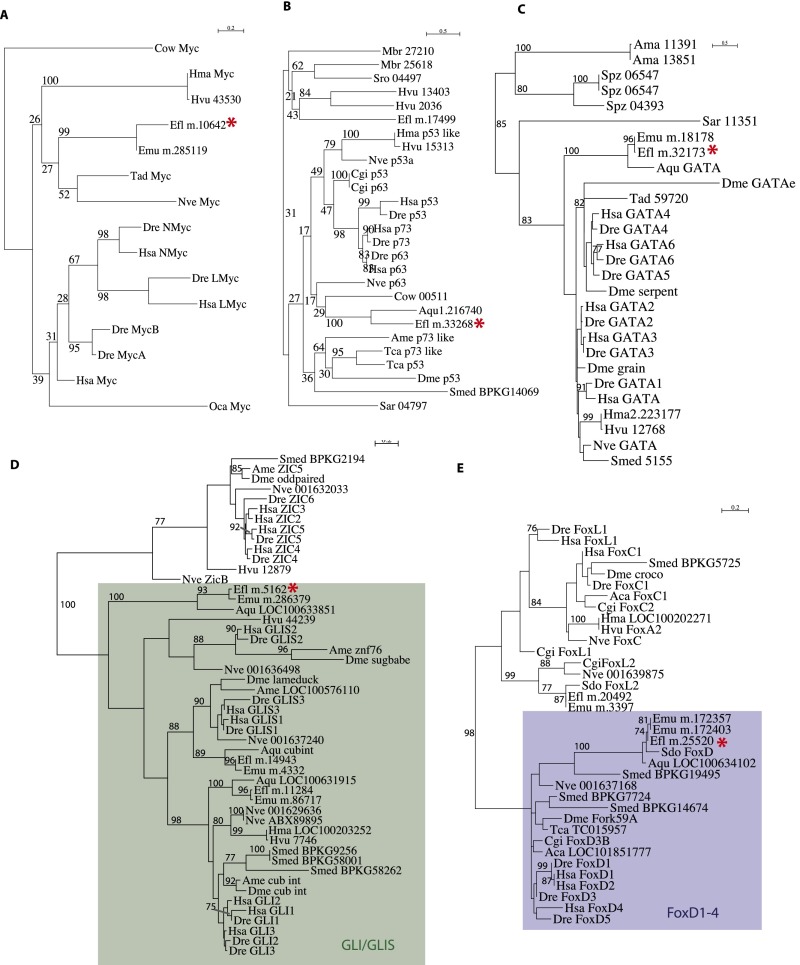
The first striking finding from examination of our data is that TFs are very poorly represented in both archeocytes and the ancestral uPriSC repertoire. Archeocytes overexpress only 67 of the 241 E. fluviatilis genes with “sequence-specific DNA binding TF activity” (Gene Ontology GO_0003700) (Dataset S2, Table S7), and thus, TFs are statistically underrepresented in archeocytes (χ2 P value = 9e-3). Nonetheless, among TFs overexpressed in archeocytes, a few notable genes have known functions in bilaterian stem cells and a very high expression level in archeocytes relative to the two other cell fractions. These include two members of the P53/63/73 family (transcripts m.33268 and m.17499 in Dataset S2, Table S1), one member of the Gata1–6 family (m.32173), a Gli/Glis family gene (m.5162), a FoxD1–4 subfamily gene (m.25520), and the Myc gene (m.10642) (see Fig. S3 for phylogenetic analyses). In contrast, both Ephydatia Sox1/2/3 paralogues [m.16180 and m.3390; EflSoxB1 specifically expressed in spicule-transporting cells (29)] and FoxO ortholog (m.32212) are expressed at a lower level in archeocytes than in the other two cell fractions (Dataset S2, Table S1). Underrepresentation of TFs appears even more striking among genes of the inferred metazoan uPriSC repertoire. In this list, only three genes encode TFs: a member of the high mobility group B family of cancer-associated genes (HMGB), a Sin3 family gene (tumor suppressors in mammals), and orthologs of multifunctional lysine (K)-specific demethylase 1A (KDM1A) (Dataset S2, Table S6).
Fig. S3.
ML phylogenetic analyses of TFs overexpressed in archeocytes. (A) Myc. (B) p53. (C) GATA. (D) Gli/Glis. (E) FoxD1–4. Red asterisks indicate Ephydatia transcripts overexpressed in archeocytes. The following is a list of abbreviations used for species names: Aae, Aedes aegypti; Aca, Aplysia californica; Ama, A. macrogynus; Ame, Apis mellifera; Aqu, Amphimedon queenslandica; Ath, Arabidopsis thaliana; Bde, Batrachochytrium dendrobatidis; Bmo, Bombyx mori; Cel, Caenorhabditis elegans; Cgi, Crassostrea gigas; Che, Clytia hemisphaerica; Cow, C. owczarzaki; Ddi, Dictyostelium discoideum; Dfa, Dictyostelium fasciculatum; Dja, Dugesia japonica; Dme, Drosophila melanogaster; Dre, Danio rerio; Efl, E. fluviatilis (from this study); Emu, E. muelleri; Fal, F. alba; Hma, Hydra magnipapillata; Hsa, Homo sapiens; Hvu HAEP, H. vulgaris (from ref. 8); Mbr, M. brevicollis; Mmu, Mus musculus; Mve, M. verticillata; Nve, Nematostella vectensis; Oca, Oscarella carmela; Pca, Podocoryne carnea; Pcr, Pseudourostyla cristata; Pte, Paramecium tetraurelia; Sar, S. arctica; Sme, S. mediterranea (from ref. 10); Sme BPKG, S. mediterranea (from ref. 12); Spu, Strongylocentrotus purpuratus; Spz, S. punctatus; Sro, S. rosetta; Tad, Trichoplax adherens; Tca, Tribolium castaneum; Tth, Tetrahymena sp.
Prominence of RBPs and Ancient Association of the “Germ-Line Multipotency Program” with Stem Cells.
In sharp contrast to TFs, RBPs are considerably enriched in Ephydatia archeocytes as well as in the reconstructed ancestral uPriSC repertoire. The archeocyte cluster is strongly enriched in RNA helicases and pre-mRNA splicing proteins (Dataset S2, Table S5). Domain annotation revealed that archeocytes overexpress 58 of the 81 DEXDc and HELICc domain-containing genes (which have a protein architecture typical of RNA helicases) present in the E. fluviatilis transcriptome (χ2 P value = 1e-13), with 25 of them expressed at least fivefold higher in archeocytes. Similarly, 82 of the 124 RNA recognition motif (RRM) domain-containing genes (χ2 P value = 1e-18) are overexpressed in archeocytes, including the archeocyte marker EflMsiA, with 25 of them expressed at least fivefold higher in archeocytes (Fig. 2D and Dataset S2, Table S8). In the inferred ancestral uPriSC repertoire, these protein domains (DEXDc, HELICc, and RRM) and functional categories related to posttranscriptional regulation are also strongly enriched (Fig. 2C and Dataset S2, Table S5). All DDX and DHX RNA helicases are overexpressed at high levels in Ephydatia, Hydra, and Schmidtea stem cells (Dataset S2, Table S6).
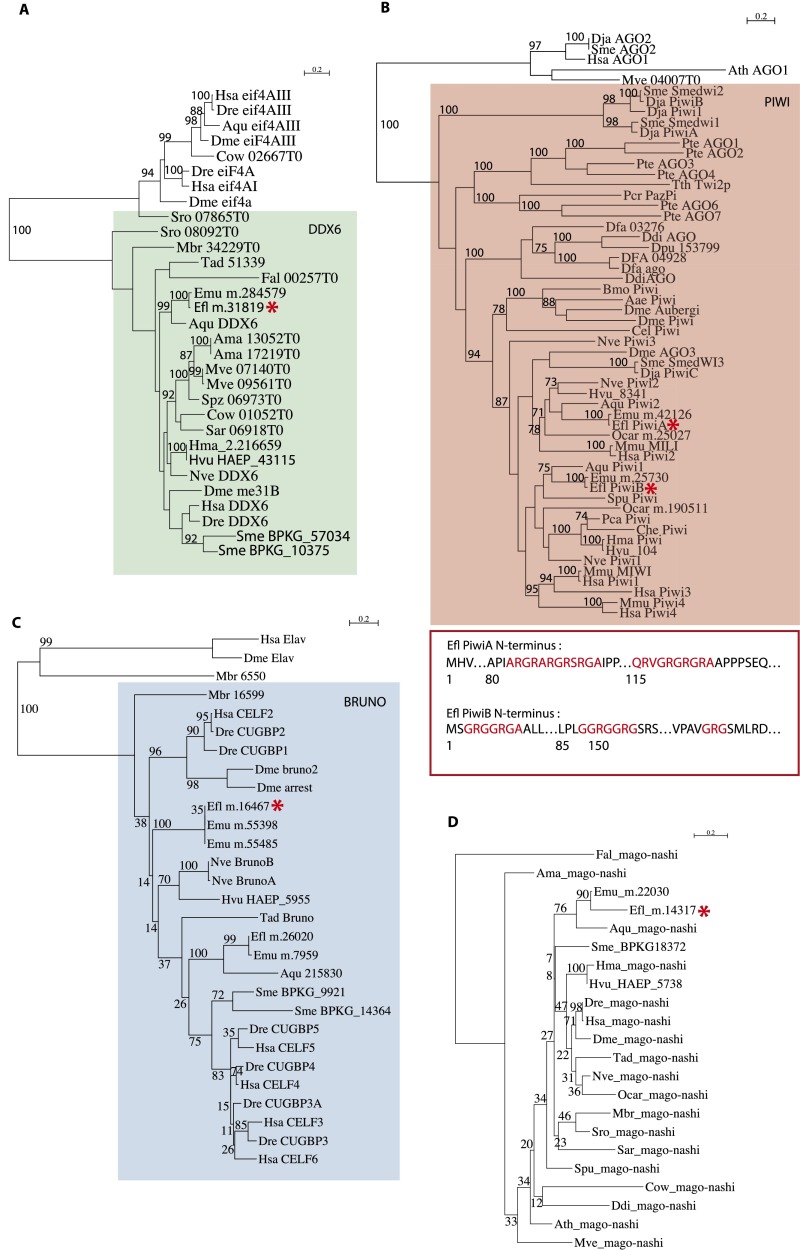
Prominent among RNA regulators specifically expressed in archeocytes is a particular suite of genes that make up the so-called germ-line multipotency program (GMP) (30). In addition to the two Piwi paralogues (EflPiwiA, m.15370; EflPiwiB, m.11401) already known to be expressed in archeocytes (21, 22), archeocytes strongly overexpress the RNA helicase Vasa (m.25201), the RRM domain-containing gene Bruno (m.16467), the RNA-binding genes Ddx6 (m.31819) and Mago-nashi (m.14317), the RNA helicase Pl-10 (m.25178), and all of the genes encoding Tudor domain-containing proteins (see Dataset S2, Table S1 and Fig. S4 for phylogenetic analyses).
Fig. S4.
ML phylogenetic analyses of GMP components (DDX6, Piwi, Bruno, and Mago-nashi) overexpressed in archeocytes. (A) DDX6. (B) Piwi. (C) Bruno. (D) Mago-Nashi. For Piwi, the diagram shows putative symmetrical dimethylation of arginine motifs (in red) in E. fluviatilis Piwi sequences. Red asterisks indicate Ephydatia transcripts overexpressed in archeocytes. For a list of abbreviations used for species names, see the Fig. S3 legend.
Similarities Between Archeocytes and Mammalian ESCs.
Archeocytes overexpress orthologs of several mammalian RBPs known to control the balance between ESC self-renewal and differentiation and/or to be specifically expressed in mouse ESCs. Using as a starting point the list of genes reviewed in Ye and Blelloch (2014) (31), we found 13 RBPs whose expression level in archeocytes suggests a possible conserved role in stem-cell regulation (Table 1). For instance, the alternative splicer RBFOX2, strongly expressed in mouse ESCs, is a master regulator for the production of ESC-specific isoforms. The Ephydatia RBFOX gene is expressed threefold higher in archeocytes than in the two other cell fractions. Conversely, in mouse, the alternative splicers MBNL1 and 2 are negative regulators of ESC renewal, and their expression is lower in ESCs than in differentiating cells. The Ephydatia MBNL ortholog is expressed approximately fivefold lower in archeocytes than in the two other cell fractions. Finally, among the 44 RBPs encoded in the ancestral uPriSC transcriptome, we found that 25 were previously identified as specifically expressed in mouse ES cells (32) (Fig. 2E and Dataset S2, Table S9).
Table 1.
Conserved expression features between sponge and mammalian RBPs
| Ephydatia | ||||||||
| Mammals | Expression value | |||||||
| Gene symbol | Positive/negative regulator of ESC renewal | mESC-specific in ref. 32 | OrthoMCL group | Transcript(s) in the same OrthoMCL group | Archeo | Other | Choano | Differential expression cluster (Fig. 2) |
| MBNL1 | Negative* | No | OG5_132352 | m.6274 | 29.835 | 164.2 | 139.68 | 5 |
| Pum1 | Negative† | Yes | OG5_126869 | m.145795 | 2.27 | 0.575 | 0.275 | 3 |
| ESRP1 | Negative‡ | Yes | OG5_130871 | m.33711 | 54 | 9.835 | 4.305 | 3 |
| Nat1 | Negative | Yes | OG5_132868 | m.25377 | 66.6 | 34.685 | 12.84 | 3 |
| RBFOX2 | Positive§ | Yes | OG5_135623 | m.28075 | 178.615 | 60.585 | 55.48 | 3 |
| ADAR | Positive¶ | No | OG5_138421 | m.34972 | 3.36 | 2.2 | 2.17 | No diff. exp. |
| SON | Positive# | No | OG5_133903 | m.27671 | 19.735 | 5.665 | 4.065 | 3 |
| Zcchc11 | Positive | No | OG5_128508 | m.33121 | 17.135 | 4.05 | 2.49 | 3 |
| Zcchc6 | Positive | No | OG5_128508 | m.33713 | 14.455 | 1.58 | 0.555 | 3 |
| Mettl3 | Positive | No | OG5_129618 | m.23877 | 26.545 | 12.54 | 13.2 | 3 |
| Mettl4 | Positive | No | OG5_132677 | m.28272 | 8.62 | 3.65 | 2.945 | 3 |
| Thoc2 | Positive|| | No | OG5_129528 | m.14726 | 116.27 | 13.425 | 7.13 | 3 |
| Thoc5 | Positive** | No | OG5_130349 | m.16805 | 97.675 | 17.915 | 11.99 | 3 |
| Ptbp1 | Positive | Yes | OG5_127719 | m.11907 | 132.735 | 54.79 | 16.635 | 3 |
No diff. exp., not differentially expressed.
Lower in mESC than in several differentiated tissues (61).
Increases during early differentiation of mESC and then comes back to the initial level (62).
Highly enriched in mESC (63).
Expressed in Oct4+, Nanog+, and Sox2+ mESCs, not in Oct4-negative fibroblasts (64).
No significant changes during hESC differentiation (65).
Increases during hESC reprogramming and decreases during ESC differentiation (66).
Decreases during mESC differentiation (67).
Decreases during mESC differentiation (67).
Stem Cells and Components of the piRNA Machinery Were Acquired Concomitantly.
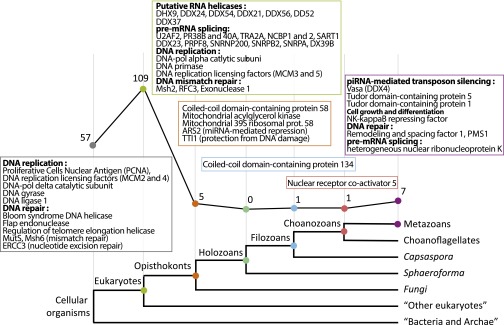
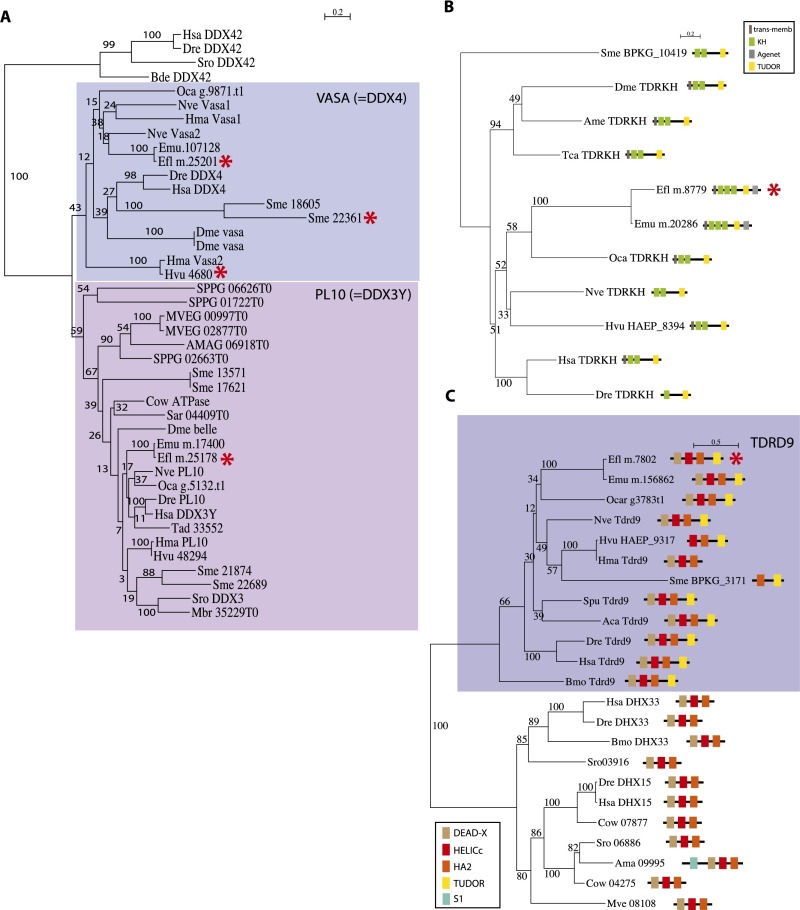
We mapped the acquisition of the 180 uPriSC genes on the tree of life by using the phyletic pattern (i.e., the presence or absence of orthologous genes across the tree of life) implemented in OrthoMCL, refined by additional reciprocal BLAST searches and additional phylogenetic analyses. The 180 genes were assigned to their phylostratum of acquisition (based on tree topology from ref. 33; Methods) (Fig. 3 and Dataset S2, Tables S6 and S10). Fifty-seven orthology groups are shared by eukaryotes and bacteria and/or archaea, comprising mainly DNA replication and DNA repair factors. The vast majority (109) of the orthology groups originated in the last common eukaryotic ancestor and include most of the RNA helicases and pre-mRNA splicing genes. Only five orthology groups originated in opisthokonts, one in filozoans, and one in choanozoans (Nuclear receptor coactivator 5, an important pluripotency factor in planarians) (34). Seven of the orthology groups are genetic innovations of a metazoan ancestor. Among the latter are Vasa [as previously shown by Kerner et al., 2011 (35)], Tdrd5, and Tdrd1, all known partners of the Piwi machinery. Moreover, two additional Tudor domain-containing proteins that belong to the Piwi machinery, Tdrd9 and TdrKH (36, 37), are metazoan innovations (see Fig. S5 for phylogenetic analyses). Both are specifically expressed in archeocytes and overexpressed in hydrozoan i-cells (but not specifically) (7), and Tdrd9 is specific to planarian neoblasts, whereas TdrKH is not (10).
Fig. 3.
The phylogenetic history of the uPriSC repertoire. Most of the 180 metazoan uPriSC repertoire genes are of ancient origin. The Bottom panel shows the phylostrata to which orthoMCL groups were assigned (adapted from ref. 33). The x axis of the graph corresponds to the phylostrata (branches of the tree). Numbers plotted along the y axis (and indicated next to dots of the graph) indicate how many orthology groups belong to each phylostratum. For phylostrata “cellular organisms” and “eukaryotes,” selected examples of acquired genes are indicated in boxes, whereas for the other phylostrata, all corresponding genes are given.
Fig. S5.
ML phylogenetic analyses of GMP components (Vasa, Tdrd9, and Tdrkh) overexpressed in archeocytes. (A) Vasa. (B) Tdrkh. (C) Tdrd9. For Trdrkh and Tdrd9, the domain composition is displayed and Ephydatia transcripts overexpressed in archeocytes are indicated by a red asterisk. In the Vasa tree, asterisks indicate genes from Ephydatia, Hydra, and Schmidtea that belong to the uPriSC gene repertoire. List of abbreviations used for species names is given in the Fig. S3 legend.
Piwi itself is a metazoan-specific gene. As previously reported in ref. 35 and confirmed here by our phylogenetic analysis using the most recently published Opisthokonta genomes, putative Piwi proteins of doubtful homology with typical metazoan Piwi are detected only in two relatively distant eukaryote lineages, amoebozoans and ciliates (Fig. S4). Thus, the most parsimonious scenario implies that Piwi is a metazoan innovation and that other eukaryote sequences of the Argonaute subfamily group artificially with animal Piwi genes. Interestingly, we found that Ephydatia Piwi proteins contain symmetrical dimethylation of arginine motifs, which are known to be recognized and bound by Tudor proteins and essential for Piwi localization and transposon repression (38) (Fig. S4).
Discussion
A necessary proviso of the origin of multicellularity in the last common animal ancestor was the acquisition of stem cells, as they are indispensable for body plan construction and for ensuring tissue homeostasis. In this study, we determined the molecular signature of uPriSCs in a sponge. Given the phylogenetic position of sponges and that uPriSCs likely represent the ancestral type of animal stem cells, the transcriptome of sponge archeocytes has the potential to inform us about the ancestral stem-cell gene toolkit of metazoans. However, this cannot be inferred from data on sponges only but rather requires comparison between sponges and distant eumetazoans. Therefore, we identified orthologous genes with conserved expression among uPriSCs of the sponge E. fluviatilis, the cnidarian H. vulgaris, and the flatworm S. mediterranea. This led to the identification of 180 genes (orthology groups) with conserved expression in animal stem cells across hundreds of millions of years of evolution.
TFs have strongly attracted the attention of biologists in the search for a stem-cell signature, at the level of eumetazoans (7, 39), exemplified for instance by the conserved role of FoxO in stem-cell maintenance between cnidarians and mammals (40, 41). Previous studies have also shown that the association of a given TF with stem cells in mammals is not always conserved on a wide evolutionary scale, as illustrated by lineage-specific recruitment for the mammalian pluripotency factors Nanog and Sox2 (7, 42, 43). We found very few TFs in the conserved repertoire of animal stem cells, but this does not mean that TFs were unimportant for the regulation of stem cells in the metazoan ancestor. Similarly in Ephydatia, even if the number of conserved TFs overexpressed in archeocytes is low, they are likely to play crucial roles in archeocyte regulation, together with lineage-restricted TFs (Ephydatia orphan genes), as in other metazoans.
Our comparative transcriptomic approach suggests that posttranscriptional regulators played a decisive role in the emergence of stem cells. The expression profile of most of the Ephydatia RBPs suggests that archeocytes intensively regulate gene expression through posttranscriptional mechanisms, as previously observed for flatworm neoblasts (9, 11). For instance, archeocytes overexpress all of the RBPs of the GMP (30) (except the Nanos ortholog, which showed similar expression levels in the three cell fractions). These GMP genes, initially characterized for their role in germ-line determination and maintenance in ecdysozoans and vertebrates (reviewed in ref. 30), are also expressed in somatic stem cells across multiple eumetazoan lineages (3, 6, 26, 37, 44). High expression of these genes in archeocytes demonstrates that deployment of the GMP in multipotent stem cells is an ancestral feature of animals and that the restriction of this program to the germ line in some bilaterians is a derived situation (26).
Whereas stem-cell acquisition in the metazoan ancestor was an evolutionary event of outstanding biological importance, our phylostratigraphy analysis indicates that the vast majority of ancestral uPriSC genes (including most RBPs) are of more ancient origin and have been later recruited in stem cells. It seems, however, that the finishing touch to the origination of the stem-cell gene repertoire consisted in the acquisition of a special genome protection machinery, the Piwi-interacting RNA (piRNA) pathway. Indeed, several metazoan-specific RBPs with conserved expression in animal stem cells are components of the Piwi machinery in bilaterian models, including Piwi itself, Vasa, Tdrd9, TdrKH, Tdrd1, and Tdrd5 (36–38, 45, 46). Recent studies have emphasized the highly conserved role of the piRNA pathway in retrotransposon silencing from Hydra to fly and vertebrates (47, 48). Amphimedon piRNAs possess the signature of the ping-pong cycle used to silence transposons (49), suggesting that genome protection by Piwi is active in sponges as well. Further experiments will be required to test whether transposon expression in Ephydatia stem cells is indeed regulated by the Piwi machinery.
The most unexpected of our findings is that there are conserved expression features between sponge orthologs of mammalian RBPs known to control the balance between renewal and differentiation of ESCs. In addition to the genes listed in Table 1, another example is Musashi-1, which is reported to segregate the Lin-28 protein into ESC nuclei and to silence the miRNA miR98, thereby controlling differentiation from stem cells (50). We previously showed that one of the Musashi paralogues (EflMsiA) is specifically expressed in archeocytes, with the protein localizing to the archeocyte nucleus (21). Our finding of the overrepresentation of many RBPs in the archeocyte transcriptome and in the ancestral uPriSC repertoire strongly supports the idea that posttranscriptional regulation could have been the dominant mode of gene expression control used in animal stem cells since their time of origin in the last common animal ancestor. We have to continue the ongoing efforts to establish gene manipulation in sponge laboratory models for a better understanding of the ancestral roles of these RBPs through future functional studies.
How can we explain that so many RBPs have conserved their stem-cell specificity over hundreds of millions of years of animal evolution? Studies on mammals have demonstrated that RBPs are embedded in so-called “RNA regulons,” whereby they coordinate splicing, export, decay, and translation of functionally related mRNAs. For instance, during the course of the cell cycle, RNA regulons are used to modulate nuclear export and/or translation of factors controlling cell proliferation and cell-cycle regulation (51). RNA regulons also orchestrate a timely balance between the proinflammatory response and tissue homeostasis via a mechanism involving cytoplasmic stress granules whose RBP composition alters the decay or translation of innate cell mRNAs (52). The ability of cells to rapidly change their protein repertoire might be of particular relevance in the context of Ephydatia gemmule hatching and for the extensive capacities of whole-body regeneration in planarians and hydrozoans. These processes rely on the stem cell entering the cell cycle, actively proliferating, and switching from self-renewal to differentiation to provide final cell types in the proper location at the proper time. We propose that the acquisition of stem cells in the last common metazoan ancestor was essentially supported by the use of ancient eukaryotic RBPs as an efficient tool to control the balance between self-renewal and differentiation and to orchestrate finely timed gene regulation within coexisting lineages of multiple cell types.
Methods
Specimen Collection and Cultivation.
Gemmule-bearing E. fluviatilis sponges were collected from the Ikusaka River in Okayama Prefecture, Japan. Gemmules attached to parental sponge tissue were stored in distilled water in the dark at 4 °C. Gemmules were isolated, and juvenile sponges were cultivated from them in M-medium (1 mM CaCl2·H2O, 0.5 mM MgSO4·7H2O, 0.5 mM NaHCO3, 0.05 mM KCl, 0.25 mM Na2SiO3·9H2O) as described previously (20).
RNA Isolation, Sequencing, and Assembly to Construct Reference Transcripts.
We extracted 450 μg of total RNA from sponges at different stages using an IsogenLS kit (Nippon Gene Co., Ltd.). Then cDNA was prepared for Illumina HiSEq. 2000 sequencing using TruSeq RNA-seq kits, and 300 million paired-end reads of 100 bp were obtained. Draft assembly using Oases software with default parameters produced 133,045 transcripts grouped into 39,484 loci with an average length of 1,216 nucleotides. Visual inspection of this assembly revealed that many predicted isoforms were in fact transcripts differing by only a few substitutions or gaps. Therefore, we decided to select a single transcript for each locus based on criteria of size and fold-coverage, using the publicly available Oases-to-csv python script (https://code.google.com/p/oases-to-csv/downloads/list). Then we kept only the ORFs over 270 nucleotides long using Transdecoder (transdecoder.sourceforge.net/) and ended up with 17,419 transcripts with an average length of 1,152 nt (provided in Dataset S1). This is of the same order of magnitude as the transcriptome of the close species Ephydatia muelleri, recently released on the Compagen platform (www.compagen.org/), containing 21,804 transcripts over 270 nt long with an average length of 1,242 nt. Raw sequencing data and assembled transcripts can be downloaded from the NCBI platform (BioProject PRJNA244851).
Transcript Annotation and Functional Enrichment Analysis.
A primary annotation (Dataset S1, Table S1) resulted from BLASTX against the human Uniprot database with 1e-5 as the cutoff e-value. Then we studied functional enrichment (KEGG, SMART domains, and Gene Ontology considered something as enriched if the modified Fisher FDR < 5e-2) through the DAVID online service (53). Conserved domains were identified with the hmmscan program from the HMMER 3.0 package against the SMART database (version of August 2012).
Sample Preparation and Cell Sorting (FACS).
Choanocytes were labeled following a protocol slightly modified from ref. 20: Stage 5 sponges (7 d old) were incubated in 2 μg⋅mL–1 Hoechst 33342 (in M-medium) for 30 min. During the last 8 min of this incubation, sponges were fed FluoSpheres [carboxylate-modified microspheres, 0.2 μm in size, yellow–green fluorescent (505/515 nm) (Molecular Probes)] at a concentration of 1 μL⋅mL–1 in M-medium. Then the sponges were washed three times for 5 min each with M-medium. Archeocytes were labeled as follows: Sponges were cultivated in M-medium with 2 μL⋅mL–1 Rho123 in the dark until they reached stage 1 or 2. Rho123 is a fluorescent dye that accumulates in the mitochondria due to their transmembrane potential (54), and we found that it was also incorporated into the abundant vitelline platelets present in the archeocyte cytoplasm (probably because the vitelline platelets contain mitochondria) (55). Then sponges were washed three times for 1 h each in normal M-medium and incubated in 2 μg⋅mL–1 Hoechst 33342 (in M-medium) for 30 min and washed three times for 5 min each in M-medium. Cells belonging to the other cells fraction were collected from stage 1–2 sponges labeled with Rho123 and Hoechst 33342 and fed FluoSpheres as described above. Gemmules used in these experiments were isolated from the same mother sponge. Before FACS sorting, sponges were rinsed with Ca2+, Mg2+-free medium (Na2SiO3, 22.48 g⋅L–1; NaHCO3, 13.34 g⋅L–1; KCl, 0.37 g⋅L–1) twice and scraped and pipetted to dissociate cells in 2 mL of Ca2+, Mg2+-free medium. Dissociated cells were filtered using a 35-μm nylon mesh (cell-strainer cap tube, Beckton-Dickinson Falcon) to remove spicules and then maintained on ice. Dissociated cells were stained with 2 μg⋅mL–1 propidium iodide (PI; Dojindo) on ice for 5 min and immediately subjected to cell sorting using a BD FACS Aria 2 (BD Bioscience). We empirically determined thresholds to collect Hoechst-positive, PI-negative viable cells. Archeocytes were isolated as large cells displaying Rho123 fluorescence (40,000 cells collected per biological replicate), choanocytes as small cells displaying FluoSphere fluorescence (100,000 cells collected per biological replicate), and other cells as cells having no fluorescence and side-scattered light (SSC)/forward-scattered light (FSC) values not overlapping with those of choanocytes or archeocytes (100,000 cells collected per biological replicate). The cell collection device was maintained at 4 °C throughout the procedure. The collected samples were centrifuged at 15,300 × g, 4 °C, 10 min, and the supernatant was removed. Samples were frozen at –80 °C. We tried to minimize the duration of each procedure, to minimize RNA degradation or expression changes. No more than 30 min elapsed between cell dissociation and sample freezing. For each cell type, the whole procedure was repeated twice to obtain biological duplicates of the experiment.
RNA Purification and Sequencing from Isolated Cells.
Total RNA from purified cells was extracted using an RNeasy Micro kit (Qiagen) following the instructions of the manufacturer. DNase digestion was performed. For each replicate, 10 ng of total RNA were used as the starting material to construct a cDNA library using a SMARTer Ultra Low RNA Kit for Illumina Sequencing (Clontech Takara Bio Inc.). RNA-seq was performed on an Illumina HiSeq2000 with single-end 100-bp reads. In total, 150 million reads were obtained—that is, 25 million per replicate. Raw sequencing data and expression analysis results for each sample can be downloaded from the NCBI-GEO platform (BioProject PRJNA244851). In addition, to facilitate access and analysis of the transcriptomic data presented in this study, all transcript sequences and annotation and expression values are accessible at the Compagen platform (www.compagen.org).
Differential Gene Expression Analysis.
Gene expression levels were calculated using RSEM-1.2.4 (56). Reads from each duplicate were mapped on the 17,419 reference transcripts using default Bowtie parameters (parameter bowtie-m 200, seed length of 25 nt). Resulting expected counts were analyzed using EdgeR 3.0.0 (57) (which assumes that read counts have a negative binomial distribution). Duplicate reproducibility was first confirmed by principal component analysis (Fig. S1), reads counts were TMM (trimmed mean of M-values) normalized, the common dispersion was calculated for each condition and gave similar values (between 0.01 and 0.08), tagwise dispersion was calculated, and pairwise exact tests were conducted. An FDR value of 5e10-2 and an RPKM ≥ 1 in at least one sample were chosen as thresholds to consider a transcript as differentially expressed. This led to 11,275 transcripts differentially expressed across the three cell fractions (i.e., statistically significantly differentially expressed between one cell fraction and each of the two other cell fractions) (Dataset S2, Table S1). A heat map of the differentially expressed transcripts was constructed with Java TreeView-1.1.6 (58).
Identification of Ancestral uPriSC Genes.
We identified a set of orthologous genes overexpressed in uPRiSCs across metazoans by comparing archeocyte-specific genes with the combination of genes overexpressed in neoblasts of the flatworm S. mediterranea (10) and in interstitial stem cells of the cnidarian H. vulgaris (7). Following the original authors’ criteria, interstitial stem cell-specific transcripts are overexpressed more than 2.5-fold higher in i-cells than in ectodermal and endodermal cells and are covered by at least ten 454 reads; neoblast-specific transcripts are those given in supplemental file 13 of Solana et al., 2012 (10). In our data, archeocyte genes were significantly overexpressed in archeocytes (Fisher exact test FDR < 0.05) at least twofold higher than in choanocytes and other cells, and their rpkm was ≥1 in archeocytes (Dataset S2, Table S2). These datasets of Ephydatia, Schmidtea, and Hydra transcripts were translated (best ORFs obtained using Transdecoder), matched to the OrthoMCL database (27), and then compared with each other to reconstruct the inferred uPriSC repertoire. This procedure yielded 2,303, 518, and 1742 orthology groups that contained one or more transcript(s) overexpressed in (respectively) archeocytes, neoblasts, and interstitial stem cells, with 180 orthology groups shared by the three species (Fig. 2B and Dataset S2, Table S10). This OrthoMCL annotation was also applied to identify conserved RBPs between human (from ref. 32) and our data.
In Situ Hydridization.
To prepare antisense cRNA probes, clones were cut with EcoRI, and digoxigenin-labeled probes were prepared according to the manufacturer’s instructions (Roche Diagnostics). Ephydatia sponges were grown on cover glasses, then fixed with 4% (wt/vol) paraformaldehyde in 1/4 Holtfreter’s solution (HS) overnight at 4 °C, washed with 1/4 HS, and then dehydrated with 50% methanol (MetOH) in 1/4 HS and 100% MetOH. The specimens were stored in 100% MetOH at −30 °C. Whole-mount in situ hybridization using a single probe for each sample with alkaline phosphatase staining was performed as described previously (23).
Phylogenetic Analyses.
For all analyses, we retrieved sequences by BLAST searches on published transcripts for the species H. vulgaris (sequences from ref. 7), S. mediterranea (sequences from ref. 10), and E. muelleri (sequences from Compagen platform at www.compagen.org/). Additionally, for each analysis, we enriched the sampling with diverse predicted protein sequences from other organisms. All abbreviated species names are given in the legend of Fig. S3. For TdrdKH, Tdrd9, and Vasa, the absence of these genes outside metazoans was further controlled by TBLASTN searches on the genomic sequences of multiple nonmetazoan species (fungi nucleotide collection on the NCBI database and all genomes are available on the Broad Institute’s “Origin of Multicellularity Project” at www.broadinstitute.org). Sequences were aligned using MAFFT v7.023b and were visually corrected. Ambiguous regions were identified by visual inspection and removed manually, and positions containing mostly missing data were deleted. Maximum likelihood (ML) analyses were performed using the PhyML program (59), with the LG (Le and Gascuel) model of amino acid substitution and a BioNJ tree as the input tree. A gamma distribution with four discrete categories was used in these ML analyses. The gamma shape parameter and the proportion of invariant sites were optimized during the searches. The statistical significance of the nodes was assessed by bootstrapping (100 replicates).
Phylostratigraphic Analysis of the uPriSC Repertoire.
Following the phylostratigraphy approach invented by Domazet-Loso et al. (60), we retrieved the phyletic pattern—that is, the presence or absence of orthologous genes across the tree of life—of each member of the 180 OrthoMCL group and first classified them as follows: shared by all cellular organisms if they are present in at least five bacteria or five archaea, originated in a eukaryote ancestor if they are absent from the latter and are present in at least five nonopisthokont eukaryotes, and originated in an opisthokont ancestor if they are absent from bacteria, archaea, and nonopithokont eukaryotes and are present in at least five fungi. From that point we performed reciprocal BLAST (e-value cutoff 1e-5) of Ephydatia members of the uPriSC repertoire against predicted protein sequences from the Broad Institute Multicellularity Project platform to identify orthologous genes in the fungi Spizellomyces punctatus, Allomyces macrogynus, and Mortierella verticillata; in the nonfungi opisthokont Fonticula alba; and the unicellular species Sphaeroforma arctica, Capsaspora owczarzaki, Salpingoeca rosetta, and Monosiga brevicollis (Dataset S2, Table S10). We subsequently assigned the corresponding OrthoMCL groups to their phylostrata.
Supplementary Material
Acknowledgments
We thank Dr. Kazuko Okamoto for discussions and for collecting sponges in the field. cDNA library construction was facilitated by Kouji Matsuzaki’s support. Access to the FACS machine was allowed by the kindness of Dr. Yoshinori Fujiyoshi and Dr. Yukihiro Tanimura. We are grateful to Dr. Elizabeth Nakajima for reading and improvement of the manuscript. We thank Dr. Norito Shibata, Dr. Takuya Imamura, Mr. Osamu Nishimura, Dr. Marcin Adamski, Dr. Maja Adamska, Dr. Hélène Hinaux, and Dr. Sylvie Rétaux for discussion. This research was supported by the Precursory Research for Embryonic Science and Technology (PREST) program of the Japan Science and Technology Agency (N.F.); the Japan Society for Promotion of Science Long-Term Postdoctoral Fellowship Program 2010-2012 (to A.A. and N.F.); and Grants-in-Aid for Scientific Research and a Grant-in-Aid for Scientific Research on Innovative Areas from the Ministry of Education, Culture, Sports, Science and Technology of Japan (to N.F.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequence reported in this paper has been deposited in the NCBI BioProject database (accession no. PRJNA244851).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1514789112/-/DCSupplemental.
References
- 1.Solana J. Closing the circle of germline and stem cells: The primordial stem cell hypothesis. Evodevo. 2013;4(1):2–16. doi: 10.1186/2041-9139-4-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tiozzo S, Brown FD, De Tomaso AW. Regeneration and stem cells in ascidians. In: Bosch TCG, editor. Stem Cells from Hydra to Man. Springer; Berlin: 2008. pp. 95–112. [Google Scholar]
- 3.De Mulder K, et al. Characterization of the stem cell system of the acoel Isodiametra pulchra. BMC Dev Biol. 2009;9:69. doi: 10.1186/1471-213X-9-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shibata N, et al. Comprehensive gene expression analyses in pluripotent stem cells of a planarian, Dugesia japonica. Int J Dev Biol. 2012;56(1-3):93–102. doi: 10.1387/ijdb.113434ns. [DOI] [PubMed] [Google Scholar]
- 5.Agata K, et al. Two different evolutionary origins of stem cell systems and their molecular basis. Semin Cell Dev Biol. 2006;17(4):503–509. doi: 10.1016/j.semcdb.2006.05.004. [DOI] [PubMed] [Google Scholar]
- 6.Leclère L, et al. Maternally localized germ plasm mRNAs and germ cell/stem cell formation in the cnidarian Clytia. Dev Biol. 2012;364(2):236–248. doi: 10.1016/j.ydbio.2012.01.018. [DOI] [PubMed] [Google Scholar]
- 7.Hemmrich G, et al. Molecular signatures of the three stem cell lineages in hydra and the emergence of stem cell function at the base of multicellularity. Mol Biol Evol. 2012;29(11):3267–3280. doi: 10.1093/molbev/mss134. [DOI] [PubMed] [Google Scholar]
- 8.Funayama N. The stem cell system in demosponges: Suggested involvement of two types of cells: archeocytes (active stem cells) and choanocytes (food-entrapping flagellated cells) Dev Genes Evol. 2013;223(1-2):23–38. doi: 10.1007/s00427-012-0417-5. [DOI] [PubMed] [Google Scholar]
- 9.Rouhana L, Shibata N, Nishimura O, Agata K. Different requirements for conserved post-transcriptional regulators in planarian regeneration and stem cell maintenance. Dev Biol. 2010;341(2):429–443. doi: 10.1016/j.ydbio.2010.02.037. [DOI] [PubMed] [Google Scholar]
- 10.Solana J, et al. Defining the molecular profile of planarian pluripotent stem cells using a combinatorial RNAseq, RNA interference and irradiation approach. Genome Biol. 2012;13(3):R19. doi: 10.1186/gb-2012-13-3-r19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Önal P, et al. Gene expression of pluripotency determinants is conserved between mammalian and planarian stem cells. EMBO J. 2012;31(12):2755–2769. doi: 10.1038/emboj.2012.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Labbé RM, et al. A comparative transcriptomic analysis reveals conserved features of stem cell pluripotency in planarians and mammals. Stem Cells. 2012;30(8):1734–1745. doi: 10.1002/stem.1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Philippe H, et al. Phylogenomics revives traditional views on deep animal relationships. Curr Biol. 2009;19(8):706–712. doi: 10.1016/j.cub.2009.02.052. [DOI] [PubMed] [Google Scholar]
- 14.Ryan JF, et al. The genome of the ctenophore Mnemiopsis leidyi and its implications for cell type evolution. Science. 2013;342(6164):1242592. doi: 10.1126/science.1242592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moroz LL, et al. The ctenophore genome and the evolutionary origins of neural systems. Nature. 2014;510(7503):109–114. doi: 10.1038/nature13400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Marlow H, Arendt D. Evolution: Ctenophore genomes and the origin of neurons. Curr Biol. 2014;24(16):R757–R761. doi: 10.1016/j.cub.2014.06.057. [DOI] [PubMed] [Google Scholar]
- 17.Connes R, Paris J, Artiges J. L’origine des cellules blastogénétiques chez Suberites domuncula Nardo. L’équilibre choanocyte-archeocytes chez les spongiaires. Ann Sci Natur Zool Paris. 1974;16:111–118. [Google Scholar]
- 18.Connes R. Contribution à l’étude de la gemmulogénèse chez la demosponge marine Suberites domuncula (Olivi) Nardo. Arch Zool Exp Gen. 1977;121:213–225. [Google Scholar]
- 19.Simpson TL. The Cell Biology of Sponges. Springer; New York: 1984. [Google Scholar]
- 20.Funayama N, Nakatsukasa M, Hayashi T, Agata K. Isolation of the choanocyte in the fresh water sponge, Ephydatia fluviatilis and its lineage marker, Ef annexin. Dev Growth Differ. 2005;47(4):243–253. doi: 10.1111/j.1440-169X.2005.00800.x. [DOI] [PubMed] [Google Scholar]
- 21.Okamoto K, et al. The active stem cell specific expression of sponge Musashi homolog EflMsiA suggests its involvement in maintaining the stem cell state. Mech Dev. 2012;129(1-4):24–37. doi: 10.1016/j.mod.2012.03.001. [DOI] [PubMed] [Google Scholar]
- 22.Funayama N, Nakatsukasa M, Mohri K, Masuda Y, Agata K. Piwi expression in archeocytes and choanocytes in demosponges: Insights into the stem cell system in demosponges. Evol Dev. 2010;12(3):275–287. doi: 10.1111/j.1525-142X.2010.00413.x. [DOI] [PubMed] [Google Scholar]
- 23.Funayama N, et al. Isolation of Ef silicatein and Ef lectin as molecular markers for sclerocytes and cells involved in innate immunity in the freshwater sponge Ephydatia fluviatilis. Zoolog Sci. 2005;22(10):1113–1122. doi: 10.2108/zsj.22.1113. [DOI] [PubMed] [Google Scholar]
- 24.Mohri K, Nakatsukasa M, Masuda Y, Agata K, Funayama N. Toward understanding the morphogenesis of siliceous spicules in freshwater sponge: Differential mRNA expression of spicule-type-specific silicatein genes in Ephydatia fluviatilis. Dev Dyn. 2008;237(10):3024–3039. doi: 10.1002/dvdy.21708. [DOI] [PubMed] [Google Scholar]
- 25.Rivera A, et al. The evolution and function of the Pax/Six regulatory network in sponges. Evol Dev. 2013;15(3):186–196. doi: 10.1111/ede.12032. [DOI] [PubMed] [Google Scholar]
- 26.Alié A, et al. Somatic stem cells express Piwi and Vasa genes in an adult ctenophore: Ancient association of “germline genes” with stemness. Dev Biol. 2011;350(1):183–197. doi: 10.1016/j.ydbio.2010.10.019. [DOI] [PubMed] [Google Scholar]
- 27.Fischer S, et al. Using OrthoMCL to assign proteins to OrthoMCL-DB groups or to cluster proteomes into new ortholog groups. Curr Protoc Bioinformatics. 2011;Chapter 6:1–19. doi: 10.1002/0471250953.bi0612s35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Resch AM, Palakodeti D, Lu YC, Horowitz M, Graveley BR. Transcriptome analysis reveals strain-specific and conserved stemness genes in Schmidtea mediterranea. PLoS One. 2012;7(4):e34447. doi: 10.1371/journal.pone.0034447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nakayama S, et al. Dynamic transport and cementation of skeletal elements build up the pole-and-beam structured skeleton of sponges. Curr Biol. 2015;25(19):2549–2554. doi: 10.1016/j.cub.2015.08.023. [DOI] [PubMed] [Google Scholar]
- 30.Juliano CE, Swartz SZ, Wessel GM. A conserved germline multipotency program. Development. 2010;137(24):4113–4126. doi: 10.1242/dev.047969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ye J, Blelloch R. Regulation of pluripotency by RNA binding proteins. Cell Stem Cell. 2014;15(3):271–280. doi: 10.1016/j.stem.2014.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kwon SC, et al. The RNA-binding protein repertoire of embryonic stem cells. Nat Struct Mol Biol. 2013;20(9):1122–1130. doi: 10.1038/nsmb.2638. [DOI] [PubMed] [Google Scholar]
- 33.Torruella G, et al. Phylogenetic relationships within the Opisthokonta based on phylogenomic analyses of conserved single-copy protein domains. Mol Biol Evol. 2012;29(2):531–544. doi: 10.1093/molbev/msr185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Böser A, et al. SILAC proteomics of planarians identifies Ncoa5 as a conserved component of pluripotent stem cells. Cell Reports. 2013;5(4):1142–1155. doi: 10.1016/j.celrep.2013.10.035. [DOI] [PubMed] [Google Scholar]
- 35.Kerner P, Degnan SM, Marchand L, Degnan BM, Vervoort M. Evolution of RNA-binding proteins in animals: Insights from genome-wide analysis in the sponge Amphimedon queenslandica. Mol Biol Evol. 2011;28(8):2289–2303. doi: 10.1093/molbev/msr046. [DOI] [PubMed] [Google Scholar]
- 36.Saxe JP, Chen M, Zhao H, Lin H. Tdrkh is essential for spermatogenesis and participates in primary piRNA biogenesis in the germline. EMBO J. 2013;32(13):1869–1885. doi: 10.1038/emboj.2013.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shoji M, et al. The TDRD9-MIWI2 complex is essential for piRNA-mediated retrotransposon silencing in the mouse male germline. Dev Cell. 2009;17(6):775–787. doi: 10.1016/j.devcel.2009.10.012. [DOI] [PubMed] [Google Scholar]
- 38.Siomi MC, Mannen T, Siomi H. How does the royal family of Tudor rule the PIWI-interacting RNA pathway? Genes Dev. 2010;24(7):636–646. doi: 10.1101/gad.1899210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Young RA. Control of the embryonic stem cell state. Cell. 2011;144(6):940–954. doi: 10.1016/j.cell.2011.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Boehm AM, et al. FoxO is a critical regulator of stem cell maintenance and immortality in Hydra. Proc Natl Acad Sci USA. 2012;109(48):9697–9702. doi: 10.1073/pnas.1209714109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Salih DA, Brunet A. FoxO transcription factors in the maintenance of cellular homeostasis during aging. Curr Opin Cell Biol. 2008;20(2):126–136. doi: 10.1016/j.ceb.2008.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jager M, Quéinnec E, Le Guyader H, Manuel M. Multiple Sox genes are expressed in stem cells or in differentiating neuro-sensory cells in the hydrozoan Clytia hemisphaerica. Evodevo. 2011;2:12. doi: 10.1186/2041-9139-2-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fortunato S, et al. Genome-wide analysis of the sox family in the calcareous sponge Sycon ciliatum: Multiple genes with unique expression patterns. Evodevo. 2012;3(1):14. doi: 10.1186/2041-9139-3-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gazave E, et al. Posterior elongation in the annelid Platynereis dumerilii involves stem cells molecularly related to primordial germ cells. Dev Biol. 2013;382(1):246–267. doi: 10.1016/j.ydbio.2013.07.013. [DOI] [PubMed] [Google Scholar]
- 45.Dufourt J, et al. Spatio-temporal requirements for transposable element piRNA-mediated silencing during Drosophila oogenesis. Nucleic Acids Res. 2014;42(4):2512–2524. doi: 10.1093/nar/gkt1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nishida KM, et al. Functional involvement of Tudor and dPRMT5 in the piRNA processing pathway in Drosophila germlines. EMBO J. 2009;28(24):3820–3831. doi: 10.1038/emboj.2009.365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lim RS, Anand A, Nishimiya-Fujisawa C, Kobayashi S, Kai T. Analysis of Hydra PIWI proteins and piRNAs uncover early evolutionary origins of the piRNA pathway. Dev Biol. 2014;386(1):237–251. doi: 10.1016/j.ydbio.2013.12.007. [DOI] [PubMed] [Google Scholar]
- 48.Juliano CE, et al. PIWI proteins and PIWI-interacting RNAs function in Hydra somatic stem cells. Proc Natl Acad Sci USA. 2014;111(1):337–342. doi: 10.1073/pnas.1320965111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Grimson A, et al. Early origins and evolution of microRNAs and Piwi-interacting RNAs in animals. Nature. 2008;455(7217):1193–1197. doi: 10.1038/nature07415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kawahara H, et al. Musashi1 cooperates in abnormal cell lineage protein 28 (Lin28)-mediated let-7 family microRNA biogenesis in early neural differentiation. J Biol Chem. 2011;286(18):16121–16130. doi: 10.1074/jbc.M110.199166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Blackinton JG, Keene JD. Post-transcriptional RNA regulons affecting cell cycle and proliferation. Semin Cell Dev Biol. 2014;34:44–54. doi: 10.1016/j.semcdb.2014.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kafasla P, Skliris A, Kontoyiannis DL. Post-transcriptional coordination of immunological responses by RNA-binding proteins. Nat Immunol. 2014;15(6):492–502. doi: 10.1038/ni.2884. [DOI] [PubMed] [Google Scholar]
- 53.Huang W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4(1):44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 54.Johnson LV, Walsh ML, Bockus BJ, Chen LB. Monitoring of relative mitochondrial membrane potential in living cells by fluorescence microscopy. J Cell Biol. 1981;88(3):526–535. doi: 10.1083/jcb.88.3.526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Simpson TL, Fell PE. Dormancy among the Porifera: Gemmule formation and germination in fresh-water and marine sponges. Trans Am Microsc Soc. 1974;94(4):544–577. [Google Scholar]
- 56.Li B, Dewey CN. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Robinson MD, McCarthy DJ, Smyth GK. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26(1):139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Saldanha AJ. Java Treeview--Extensible visualization of microarray data. Bioinformatics. 2004;20(17):3246–3248. doi: 10.1093/bioinformatics/bth349. [DOI] [PubMed] [Google Scholar]
- 59.Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 2003;52(5):696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- 60.Domazet-Loso T, Brajković J, Tautz D. A phylostratigraphy approach to uncover the genomic history of major adaptations in metazoan lineages. Trends Genet. 2007;23(11):533–539. doi: 10.1016/j.tig.2007.08.014. [DOI] [PubMed] [Google Scholar]
- 61.Han H, et al. MBNL proteins repress ES-cell-specific alternative splicing and reprogramming. Nature. 2013;498(7453):241–245. doi: 10.1038/nature12270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Leeb M, Dietmann S, Paramor M, Niwa H, Smith A. Genetic exploration of the exit from self-renewal using haploid embryonic stem cells. Cell Stem Cell. 2014;14(3):385–393. doi: 10.1016/j.stem.2013.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Fagoonee S, et al. The RNA binding protein ESRP1 fine-tunes the expression of pluripotency-related factors in mouse embryonic stem cells. PLoS One. 2013;8(8):e72300. doi: 10.1371/journal.pone.0072300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yeo GW, et al. An RNA code for the FOX2 splicing regulator revealed by mapping RNA-protein interactions in stem cells. Nat Struct Mol Biol. 2009;16(2):130–137. doi: 10.1038/nsmb.1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Osenberg S, et al. Alu sequences in undifferentiated human embryonic stem cells display high levels of A-to-I RNA editing. PLoS One. 2010;5(6):e11173. doi: 10.1371/journal.pone.0011173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lu X, et al. SON connects the splicing-regulatory network with pluripotency in human embryonic stem cells. Nat Cell Biol. 2013;15(10):1141–1152. doi: 10.1038/ncb2839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wang L, et al. The THO complex regulates pluripotency gene mRNA export and controls embryonic stem cell self-renewal and somatic cell reprogramming. Cell Stem Cell. 2013;13(6):676–690. doi: 10.1016/j.stem.2013.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.