Significance
Ketoreductases are the most commonly used enzymes in industrial pharmaceutical synthesis. We investigated the nature of enantioselectivity in closely related mutant ketoreductases that reduce almost-symmetrical 3-oxacyclopentanone and 3-thiacyclopentanone, which are difficult to reduce enantioselectively by other means. We present the efficiencies of select variants and their crystallographic structures. Our experimental and theoretical studies reveal how mutations modulate the stereoselectivity of the reduction. Molecular dynamics simulations of the Michaelis–Menten and transition state-bound complexes were used to rationalize the observed stereochemical outcomes. We discovered that the closed conformation of the flexible substrate binding loop is likely the catalytically active one imparting the stereochemical preferences. Our molecular dynamics approach reveals how each enzyme stabilizes the diastereomeric transition structures by altering the active site size.
Keywords: directed evolution, crystallographic structures, molecular dynamics, theozyme, enantioselectivity
Abstract
Mutants of Lactobacillus kefir short-chain alcohol dehydrogenase, used here as ketoreductases (KREDs), enantioselectively reduce the pharmaceutically relevant substrates 3-thiacyclopentanone and 3-oxacyclopentanone. These substrates differ by only the heteroatom (S or O) in the ring, but the KRED mutants reduce them with different enantioselectivities. Kinetic studies show that these enzymes are more efficient with 3-thiacyclopentanone than with 3-oxacyclopentanone. X-ray crystal structures of apo- and NADP+-bound selected mutants show that the substrate-binding loop conformational preferences are modified by these mutations. Quantum mechanical calculations and molecular dynamics (MD) simulations are used to investigate the mechanism of reduction by the enzyme. We have developed an MD-based method for studying the diastereomeric transition state complexes and rationalize different enantiomeric ratios. This method, which probes the stability of the catalytic arrangement within the theozyme, shows a correlation between the relative fractions of catalytically competent poses for the enantiomeric reductions and the experimental enantiomeric ratio. Some mutations, such as A94F and Y190F, induce conformational changes in the active site that enlarge the small binding pocket, facilitating accommodation of the larger S atom in this region and enhancing S-selectivity with 3-thiacyclopentanone. In contrast, in the E145S mutant and the final variant evolved for large-scale production of the intermediate for the antibiotic sulopenem, R-selectivity is promoted by shrinking the small binding pocket, thereby destabilizing the pro-S orientation.
Biocatalysis is a common method of stereoselective ketone reduction (1). This approach often replaces multistep syntheses and uses renewable, biodegradable, and nontoxic reagents and mild conditions (2). Ketoreductases (KREDs), the most commonly used enzymes in industrial pharmaceutical synthesis (3), reduce a wide range of ketones to alcohols with high chemoselectivity and stereoselectivity. These enzymes have been engineered to synthesize alcohols as intermediates for the production of atorvastatin (Lipitor), montelukast (Singulair), and atazanavir (Reyetaz) (4).
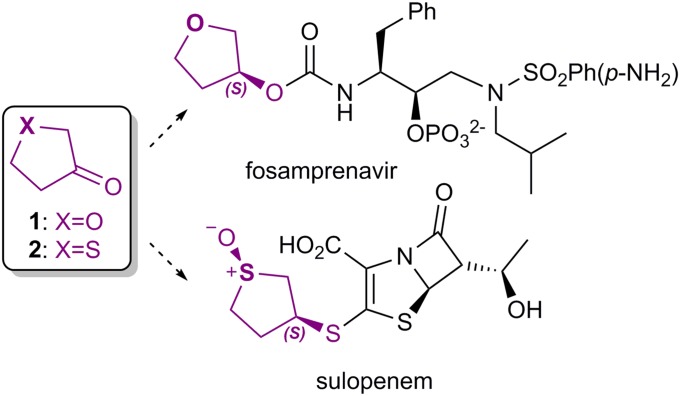
Small and almost symmetrical ketones, such as prochiral cyclopentanones, are attractive substrates that are difficult to reduce asymmetrically by chemical methods (5, 6). In particular, the enantiopure chiral alcohols derived from 3-oxacyclopentanone (1) and 3-thiacyclopentanone (2) are used in the synthesis of the pharmaceutical agents fosamprenavir and sulopenem, respectively (Fig. 1). Through a directed evolution (DE) program, Codexis, Inc. engineered a KRED obtained from Lactobacillus kefir for the reduction of 3-thiacyclopentanone (2) for the large-scale production of the antibiotic sulopenem. L. kefir KRED (WT) belongs to the short-chain dehydrogenase/reductase (SDR) family (7, 8). Via DE, a variant containing 10 mutations (so-called Sph) was obtained and used for large-scale synthesis of (R)-3-thiacyclopentanol.
Fig. 1.
Prochiral cyclopentanones 1 and 2 and their use in the synthesis of two pharmaceuticals.
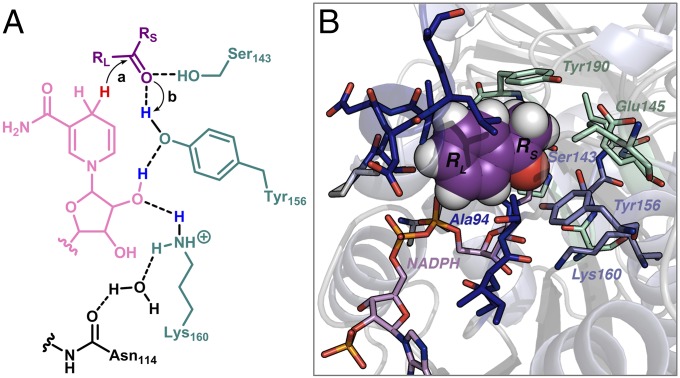
Sph and related KREDs use the NADPH cofactor as hydride reductant. In the industrial process, NADPH is regenerated by oxidation of glucose to glucono-1,5-lactone with glucose dehydrogenase. The proposed mechanism for carbonyl reduction and subsequent protonation of the intermediate (9) is shown in Fig. 2A. In L. kefir numbering, Ser143 and Tyr156 stabilize the alkoxide formed on hydride transfer from NADPH (10, 11). The alcohol is then liberated through a proton relay involving Tyr156, the cofactors’ ribofunarose, Lys160, a backbone carbonyl, and water. When the substrate’s substituents differ in size, the smaller (RS) and larger (RL) substituents bind to the small and large binding pockets, respectively (Fig. 2B) (12). It has been proposed that Tyr190 prevents binding of the large group in the small binding pocket (13).
Fig. 2.
(A) Reduction (a) and alkoxide protonation (b) steps in KREDs. The NADPH cofactor (in pink) is abbreviated, the substrate is in purple, and the catalytic residues are in teal. The reactive hydride and the hydrogens proposed to be involved in the proton relay are highlighted in red and blue, respectively. (B) X-ray structure of holo L. kefir WT with acetophenone (in purple) docked in. The cofactor (in pink) and residues in the active site are shown as sticks. The catalytic triad (Ser143-Tyr156-Lys160) is in blue, and the residues along the small and large binding pockets are shown in green and dark blue, respectively.
These enzymes are tetramers in their crystallographic and catalytically active forms, although they are purified as monomers (SI Appendix, Fig. S1). They feature a flexible substrate-binding loop spanning roughly residues 190–210, the most variable portion of SDRs, which has been implicated as crucial in determining the stereoselectivity of KREDs (14). In its closed conformation, this motif flanks one side of the active site and closes around the bound cofactor and substrate (11).
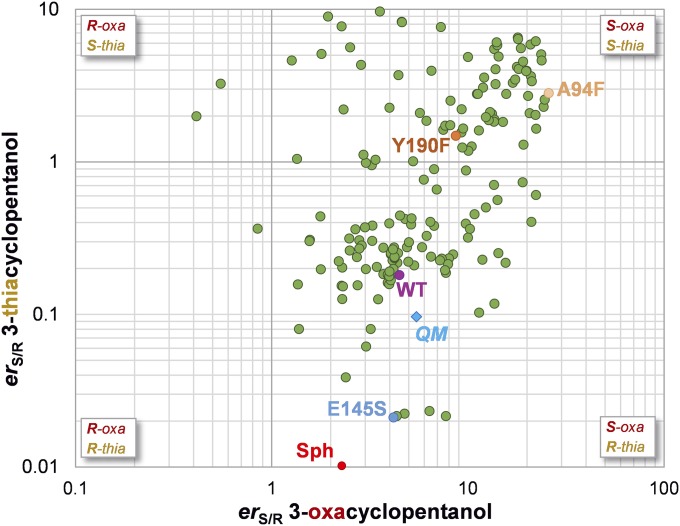
A plot of the enantioselectivities for the reduction of 1 and 2 given by 175 evolved KRED variants developed by Codexis is shown in Fig. 3. Despite the broad distribution of S/R enantiomeric ratios (erS/R) observed, some of these enzymes are highly enantioselective and are able to distinguish between a sulfur or oxygen atom and a methylene group, both located β to the reacting carbonyl group. Because the WT enzyme was evolved especially for 3-thiacylopentanone, the variants are not equally selective for both substrates. Nearly all of the mutants, including the WT, give (S)-3-oxacyclopentanol from 1. This enantioselectivity agrees with a qualitative model by which the 3-oxa and 4-methylene groups bind to the small and large binding pockets, respectively; however, the enantioselectivity in the reduction of 2 ranges from erR/S ∼50 to erS/R ∼10. These data indicate that in such small substrates, the typical distinction between “large” and “small” substituents is far from obvious.
Fig. 3.
Logarithmic graph of the experimental erS/R obtained by KRED-evolved variants for 3-oxacyclopentanol (horizontal axis) and 3-thiacyclopentanol (vertical axis). The values measured for selected variants (WT, A94F, Y190F, E145S, and Sph) and calculated for simplified QM models are highlighted.
The slight structural differences between ketones 1 and 2 were analyzed in terms of computed molar volumes and electrostatic potentials (SI Appendix, Figs. S2 and S3). O is slightly smaller but considerably more polar than a CH2 group. S is closer in size and polarity (slightly larger and more polar) to a CH2. Remarkably, the enzymes produced during DE—even single or double mutants—do recognize these very subtle differences, promoting different levels of enantioselectivity. To understand the origins of this exquisite stereocontrol achieved by evolved KREDs in the reduction of these small, almost symmetrical substrates, we performed crystallographic, activity, and computational studies of the WT, selected single mutants showing the largest differences in enantioselectivity, and the most evolved variant, Sph.
Results and Discussion
Enzymatic Activity and Theozyme Calculations.
The catalytic efficiencies (kcat/KM), enantiomeric ratios (erS/R), and associated differences in the activation barriers at 25 °C (ΔΔG‡S/R), assuming a rapid equilibration between pro-R and pro-S Michaelis–Menten complexes (i.e., Curtin–Hammett conditions), are summarized in Table 1 and in SI Appendix, Table S1. As these data show, the WT is the most efficient of these enzymes, given that the evolution program aimed to enhance enantioselectivity (>99% enantiomeric excess) in this already active enzyme. Moreover, because of the evolutionary pressure exerted toward 3-thiacyclopentanone, all of the variants are systematically better catalysts for reduction of 2 than of 1; the two more active and R-selective variants for 2 (E145S single mutant and Sph) are poorly S-selective and ∼200 times less active for 1. The decreased rate of reduction of 2 with A94F and Y190F, which favor formation of the S-enantiomer (er >1 and negative ΔΔG‡S/R), shows a significantly reduced rate of formation of the normally preferred R-enantiomer by these point mutations.
Table 1.
erS/R, ΔΔG‡S-R, and kcat/KM values determined experimentally for 3-oxacyclopentanol and 3-thiacyclopentanol obtained by KRED variants from ketones 1 and 2, respectively
| Variant | 3-oxacyclopentanol (from 1) | 3-thiacyclopentanol (from 2) | ||||
| erS/R | ΔΔG‡S-R, kcal mol−1 | kcat/KM, mM-1 min−1 | erS/R | ΔΔG‡S-R, kcal mol−1 | kcat/KM, mM-1 min−1 | |
| WT | 4.2 | −0.85 | 0.025 | 0.2 | 0.98 | 1.460 |
| A94F | 27.5 | −1.95 | 0.005 | 2.6 | −0.55 | 0.016 |
| Y190F | 9.3 | −1.31 | ND | 1.6 | −0.26 | ND |
| E145S | 4.2 | −0.84 | 0.003 | 0.02 | 2.34 | 0.772 |
| Sph | 1.3*,† | −0.14 | 0.004 | 0.004* | 3.33 | 0.703 |
ND, not determined.
Measured under the experimental conditions described in SI Appendix, Fig. S33.
Obtained for a related variant lacking the P194N mutation.
We assume that these differences in stereoselectivity emerge mainly from the chemical step (related to kcat), which was the main focus of our study, although contributions from other steps in the catalytic cycle (i.e., substrate binding, related to KM) cannot be ruled out.
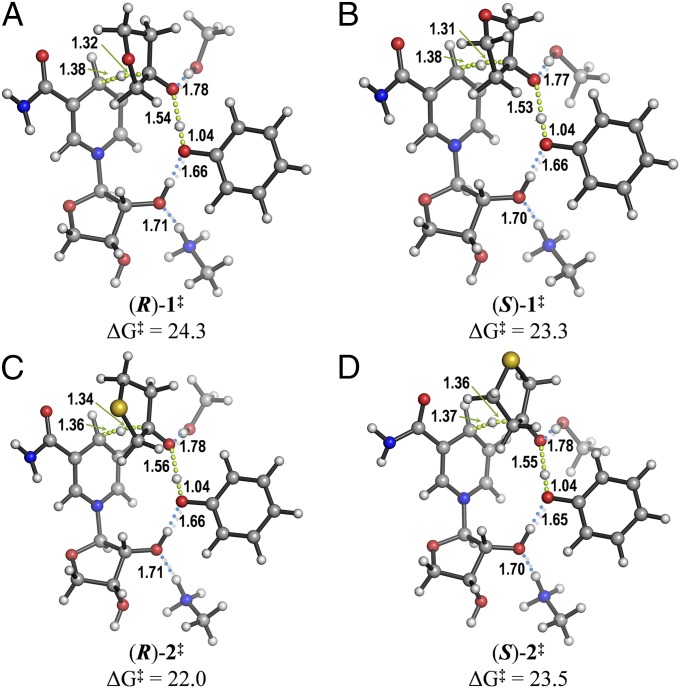
Enzymatic reduction was modeled quantum mechanically (Materials and Methods), using an abbreviated quantum mechanical (QM) model of the active site. The NADPH cofactor was truncated to a nicotinamide-ribofuranose complex, and the side chains of the catalytic Ser143, Tyr156, and Lys160 were abbreviated to methanol, phenol, and methylammonium, respectively. The calculated transition states (TSs) for the reduction of ketones 1 and 2 are shown in Fig. 4. As shown by their activation barriers, 3-thiacyclopentanone 2 is reduced more easily than 1 by ∼2 kcal mol−1, consistent with the higher efficiencies measured for 2 with all enzymes. This higher reactivity of 2 would not be expected if only inductive effects—which are greater for 3-oxacyclopentanone 1—were important. Conversely, a distortion/interaction analysis (15) showed that 3-thiacyclopentanone is easier to distort into the TS geometry, presumably owing to the longer C–S bonds (1.85 Å, compared with 1.43 Å in C–O) and consequent greater flexibility of the five-membered ring in 2 (SI Appendix, Table S4).
Fig. 4.
Transition structures leading to (R)-3-oxacyclopentanol (A), (S)-3-oxacyclopentanol (B), (R)-3-thiacyclopentanol (C), and (S)-3-thiacyclopentanol (D), calculated with CPCM(H2O)/M062X/6–311+G(d,p)//TPSS/6–31G(d). Activation free energies (ΔG‡) are given in kcal mol−1; distances, in Å.
In this highly simplified model, the calculated stereoselectivity is induced by the restricted positions of the cofactor and catalytic groups. Nevertheless, remarkably good agreement with the enantioselectivities measured for the WT with 1 (S-selective; erS/R = 5.4) and 2 (R-selective; erS/R = 0.1), was obtained. These calculations reveal the intrinsic preferences for formation of the (R)- or (S)-cyclopentanols for each starting ketone when all of the catalytic groups are arranged optimally. In fact, the calculated positions of the cofactor and catalytic side chains overlay almost perfectly with those in the X-ray structure of the WT KRED in the holo state (i.e., NADP+-bound; SI Appendix, Fig. S4), demonstrating the high preorganization of the active site achieved by natural evolution to stabilize the TS.
Analysis of X-Ray Structures.
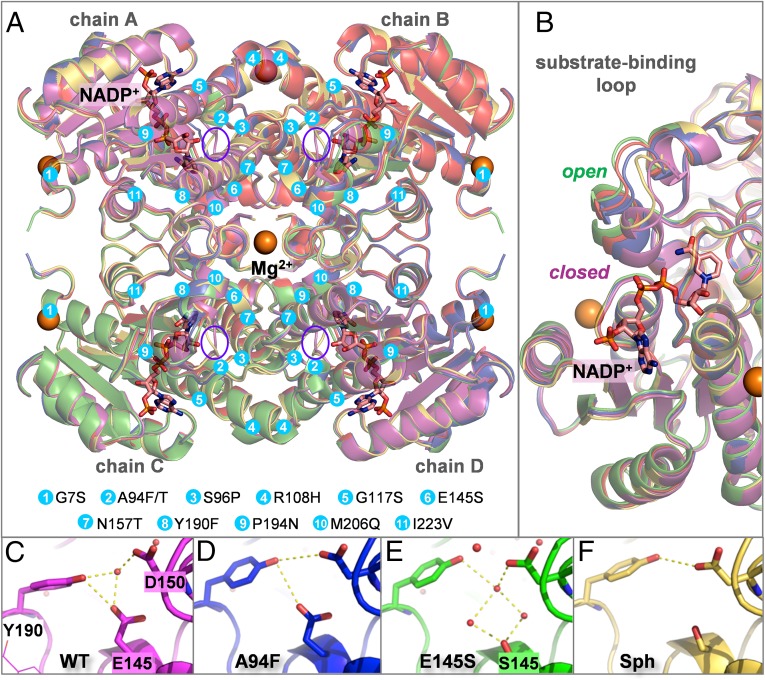
Five crystal structures of L. kefir KRED variants were obtained, four of them in the apo state (WT, E145S and A94F single mutants, and the final variant Sph), and the WT in the holo state, all crystallized as tetramers (Fig. 5A). Sph has 10 mutations, including two in the small binding pocket (E145S and N157T), two in the large binding pocket (A94T and S96P), two in the substrate-binding loop (P194N and M206Q), two in α-helix E10 (R108H and G117S), one in α-helix G (I223V), and one more in a loop (G7S) (SI Appendix, Table S7). The side chain conformations and hydrogen bonds established by these residues are obviously different in the Sph and WT crystal structures, but their backbone structures are very similar, with the exception of the P194N mutation, which results in a less-ordered helix turn in the substrate-binding loop in Sph. Further structural details are provided in SI Appendix, Table S5 and Figs. S5–S12.
Fig. 5.
(A) Overlay of the crystal structures obtained for apo WT (red), NADP+-bound WT (magenta), A94F (blue), E145S (green), and Sph (yellow). The cofactor is shown as pink sticks; the Mg2+ ions, as orange spheres. Mutations introduced in the studied variants are shown as cyan circles; the active sites, as purple ellipses. (B) The substrate/cofactor binding region showing the open and closed conformations of the loop. (C–F) The Tyr190-Glu/SerS145-Asp150 triad in different variants, showing the water and/or hydrogen bond network.
Of note, the structures of the holo WT and apo final mutant Sph are nearly identical (rmsd over all Cα, 0.35 Å; a per-residue comparison is shown in SI Appendix, Fig. S5) in both the protein backbone and the active site, a situation that we have described previously for another evolved enzyme also bearing a Ser-Tyr-Lys catalytic triad (16). It is frequently observed that in enzymes with such highly preorganized active sites based on complex hydrogen bond networks, DE does not perturb the optimized catalytic arrangement, at least in the solid state, and that the reasons for enhanced catalysis must be sought outside the active site.
Indeed, the most noticeable differences among these crystal structures lie in the conformation of the substrate-binding loop: either cofactor binding or DE mutations favor the closed conformation, whereas in the rest of the structures, the loop is more open (Fig. 5B). In the structure of the A94F single mutant, the active site is less solvent-exposed, and Phe94 makes a van der Waals contact with Leu195 on the substrate-binding loop; this promotes a slightly more closed conformation of the latter. This is significant, because this mutation alone decreases the reactivity and reverses the enantioselectivity in the reduction of 3-thiacyclopentanone with respect to the WT. Taken together, these observations suggest that not only the structure, but also the conformational dynamics, of the substrate-binding loop may be related to the observed reactivity and stereoselectivity of the reduction (see the discussion on molecular dynamics below).
Another minor difference observed in these structures is the different hydrogen bond and crystallographic water patterns exhibited by different variants for the noncatalytic Tyr190-Glu145-Asp150 triad. As shown in Fig. 5 C–F, the bridging water molecule trapped within this motif in the WT (Fig. 5C), is displaced by the hydrophobic A94F mutation present in both the single mutant (Fig. 5D) and the final Sph (Fig. 5F). In contrast, the E145S mutation (Fig. 5E) mimics the WT, in that two additional water molecules are cocrystallized to maintain a similar distribution of oxygen atoms.
Molecular Dynamics Simulations of Enzyme-Substrate Complexes in Water.
Given the lack of conclusive structural information on the effects of DE mutations on the variable catalytic profiles exhibited by the KRED variants, we analyzed the same proteins through molecular dynamics (MD) simulations in explicit water starting from the available crystal structures (Materials and Methods). These simulations predict the shape of the active site, including the conformational states of the enzyme.
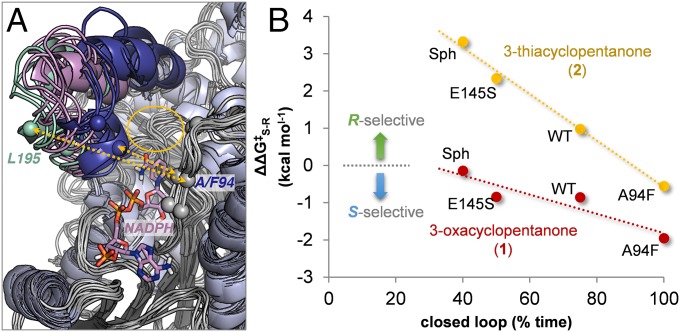
We first ran a series of unrestrained 100-ns trajectories of the Mg2+-bound tetramers with the cofactor (NADPH) and substrates (1 or 2) docked in the active site. Whereas the cofactor stays bound to the active site through multiple polar interactions, the very small substrates are usually unable to maintain a productive orientation in the active site, and in many cases the substrates diffuse to the solvent after a short simulation time (∼25 ns). As expected, the substrates are less mobile when the binding loop, which is the most dynamic region of the protein, is in a closed conformation. The closed-to-open transitions usually occur within ∼20 ns. When the loop is closed, residues Leu199 and Leu195 in this region establish van der Waals contacts with Ile93, Ala/Phe94, and Ser96 in the binding pocket, with Cα–Cα distances of ∼7–10 Å depending on the mutant; in the open conformation, these distances fluctuate within 12–18 Å. (SI Appendix, Figs. S15–S18 show open/closed population distributions.) Thus, the conformational states of this loop were monitored and classified according to the distance between the Cα of Leu195 and that of Ala94 (Fig. 6A). The results of per-residue fluctuation analysis are shown in SI Appendix, Figs. S13 and S14.
Fig. 6.
(A) Overlay of 10 snapshots derived from MD simulations of WT KRED in complex with 2. The substrate-binding loop is highlighted for the open (green), intermediate (purple), and closed (blue) conformations. One representative binding pose of the cofactor is shown as sticks, and the active site is shown as a yellow ellipse. The Cα of residues 93–96, 195, and 199 are shown as spheres, and the distances analyzed to define the conformation of the loop are shown as yellow arrows. (B) Experimental relative activation barriers leading to the S and R alcohols (ΔΔG‡S-R) from cyclopentanones 1 and 2, versus the accumulated population of the substrate-binding loop in the closed conformation (Cα–Cα distance between L195 and A/F94 <10 Å) derived from multiple 100-ns MD trajectories of different KRED variants in complex with substrates 1 and 2 in different orientations.
We found that the fraction of the simulation time during which the loop is closed for each mutant correlates well with the experimental relative activation barriers for the pro-R and pro-S ketone reduction pathways (ΔΔG‡S-R), especially for 3-thiacyclopentanone 2, for which the enzymes were evolved (Fig. 6B). These results confirm the key role of the binding loop structure and conformational dynamics in the activity and stereochemical outcome of the reduction reactions (14). This was also demonstrated by the crystal structures described above. Nevertheless, this seemingly very good correlation should not be overinterpreted, given the difficulty of clearly distinguishing between open and closed conformations in some very flexible variants, such as Sph (SI Appendix, Fig. S18).
The specific mechanism through which the closed loop induces asymmetry is not apparent. The closed conformation of the loop certainly increases the substrate-binding affinity and creates a more packed, reactive, and stereoselective environment in the active site; however, if the open conformation cannot be achieved, then certain substrates might be locked into nonproductive orientations, or catalyst turnover may be decreased by slow substrate binding and/or product release. This might be the case for the A94F single mutant, for which the reduction of 2 is much slower and with opposite stereoselectivity (S-selective) compared with WT (R-selective). With this variant, the substrate-binding loop always remains in the closed conformation (Fig. 7), and the pro-R reaction pathway is significantly slowed. In contrast, this blocking of the pro-R channel has an opposite effect on substrate 1, for which the WT is already S-selective, further enhancing S-selectivity.
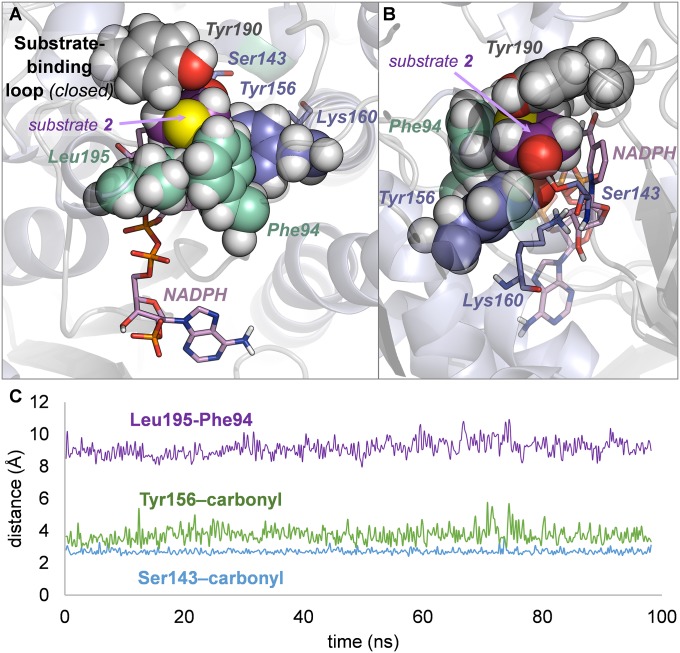
Fig. 7.
(A and B) An unproductive binding pose of 3-thiacyclopentanone 2 in A94F single mutant in two different orientations showing the hydrophobic cage surrounding the substrate (A) and the perpendicular orientation of 2 with respect to NADPH despite the conserved catalytic contacts (B). (C) Traces of selected interactions monitored along the MD trajectory: Cα–Cα distance between Leu195 and Phe94 (loop conformation, in purple) and O–O distances between the ketone carbonyl and the catalytic Ser143 (in blue) and Tyr156 (in green).
Careful inspection of the MD trajectory of the A94F single mutant in complex with NADPH and 3-thiacyclopentanone 2 corroborated this hypothesis. Throughout the entire simulation time, the substrate is sequestered in a nonproductive orientation imposed by four densely packed hydrophobic residues (Leu195 and Phe94, both involved in the loop closing, along with Tyr190 and catalytic Tyr156) (Fig. 7 A and B). This confined environment orients the cyclopentanone ring perpendicular to the cofactor, precluding the hydride transfer even though catalytic Ser143 and Lys160 are permanently hydrogen-bonded to the substrate carbonyl and NADPH ribose, respectively; the other catalytic residue, Tyr156, stays close to the substrate carbonyl but cannot hydrogen-bond to it (Fig. 7C).
On the other hand, the highly R-selective and more reactive E145S and Sph variants sample open-loop conformations to a greater degree than the WT (Fig. 6B). Although the crystal structure of Sph shows a more closed conformation of the loop, the MD simulations indicate that this region is quite flexible in solution, and that the distinction between open and closed conformations is not so apparent, likely providing a scaffold with balanced R-stereoselectivity and catalyst turnover.
Subsequent to our MD simulations of the Michaelis–Menten complexes, and to assess the ability of the active site to stabilize the pro-R and pro-S TSs, we developed a new MD-based methodology using the QM geometries of the theozymes.
Restricted MD Simulations with the Theozyme in Water.
Following Pauling’s postulate, it is now generally accepted that enzymes are complementary in structure to the TS of the reaction that they catalyze (17). In accordance with this idea, we used MD simulations to provide a detailed description of the active site near the reduction TSs for selected DE variants (16, 18). The activated complexes were built into the protein structure from the optimal QM geometries and atomic partial charges of the diastereomeric theozymes described above for substrates 1 and 2 (Fig. 4), following a similar protocol as described in the literature (19). The distances of the breaking CNADPH–H and forming Csubstrate–H bonds and the relative orientation of cofactor and substrates (pro-S or pro-R) were fixed by defining special TS parameters, and the systems were simulated for 160 ns in explicit water (Materials and Methods and SI Appendix, Materials and Methods).
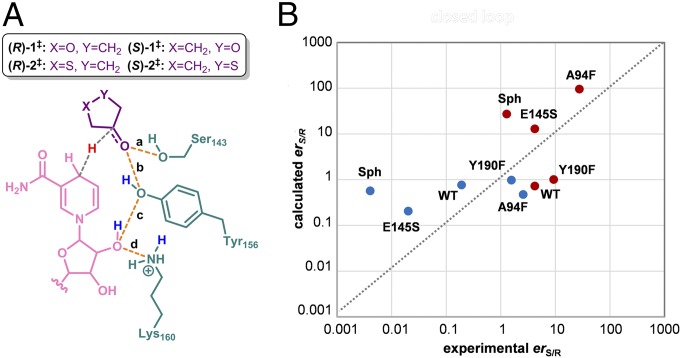
For each of the four diastereomeric TS complexes—(R)-1‡-, (S)-1‡, (R)-2‡, and (S)-2‡—the ability of each of the enzymes to maintain productive catalytic contacts with the substrate and cofactor along the simulations was analyzed by monitoring the four distances shown in Fig. 8A. These involve the interaction between hydrogen bond donors Ser143/Tyr156 and the developing alkoxide in the substrate (distances a and b), and between Tyr156/Lys160 and the cofactor ribofuranose (distances b and c) (SI Appendix, Table S8 presents average values). For each simulation step, the geometry was considered catalytically competent—i.e., to adequately lower the free energy of activation so that reaction occurs—only when all of the selected distances were simultaneously lower than an arbitrary value similar to that calculated for the theozymes (3.4 Å) (SI Appendix, Figs. S20 and S21 provide further details on the influence of this threshold value). This is an indirect way of estimating the number of reactive events occurring in a certain period, i.e., the rate constant for a particular reaction pathway. In these circumstances, and assuming rapidly equilibrating diastereoisomeric Michaelis–Menten complexes (i.e., Curtin–Hammett conditions), the relative populations of the reactive poses derived from the restrained pro-S and pro-R trajectories (calc. erS/R) can be compared with the experimental enantiomer ratios measured for each prochiral ketone (Table 1 and SI Appendix, Table S1).
Fig. 8.
(A) Catalytic distances (a–d) monitored in the MD simulations with the theozymes. (B) Logarithmic graph of the experimental vs. computed erS/R derived from the population of the catalytically competent poses in the MD trajectories [calc. erS/R = %(S)/%(R)]. The values for 3-oxacyclopentanone and 3-thiacyclopentanone are shown in red and blue, respectively.
Because a certain population of the closed conformation of the mobile substrate-binding loop is required for enantioselective catalysis, only those snapshots presenting the loop in a closed conformation (i.e., <10 Å distance between L195 and A/F94 Cα) were used to derive the calculated erS/R values. As shown in Fig. 8B, these estimates correlate roughly with the experimental observations; in nearly all cases, the favored enantiomer is predicted. In agreement with the crystallographic and MD results reported above, the erS/R values calculated when the loop is open show a poorer correlation with the experimental values (SI Appendix, Fig. S22). We examined how individual mutations alter the energetics of the diastereomeric active sites and modulate the enantioselectivity of the enzyme.
In the WT enzyme (Fig. 9 A and B), the small binding pocket is sterically hindered by the phenol group of Tyr190, disfavoring the orientation of the S atom (larger than CH2) in this region of the active site. In addition, the same phenol ring engages in a CH/π interaction with the β-methylene group (Fig. 9B). As a result, (R)-2‡ is slightly favored (calc. erS/R = 0.8). On the other hand, the smaller O atom fits better in the small pocket, increasing the contribution of the pro-S pathway for 3-oxacyclopentanone (calc. erS/R∼1) (Fig. 9A). Of note, the primary catalytic contacts—namely, the hydrogen bonds between the incipient alkoxide (COsubstrate) and Ser143/Tyr156—are perfectly maintained throughout the entire trajectories for the four systems, a finding frequently observed in other mutants as well. The subtle differences between the pro-R and pro-S pathways in favor of the latter arise from the contacts between the ribose hydroxyl (OHsugar) and Tyr156/Lys160. An unexpected and very persistent interaction between COsubstrate and OHsugar was also observed (SI Appendix, Figs. S23 and S28).
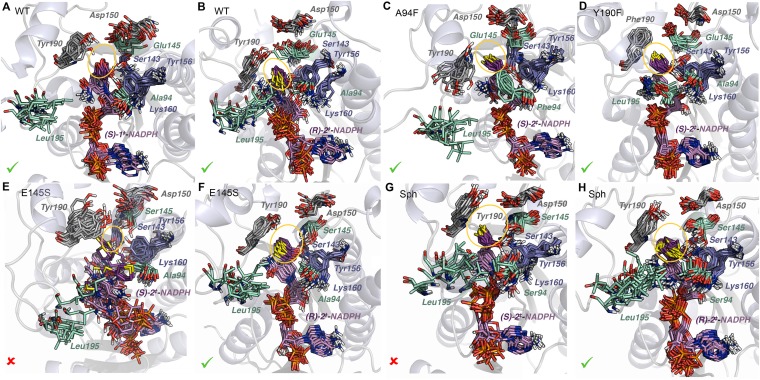
Fig. 9.
Overlays of 12 snapshots obtained along 160-ns MD simulations with TS complexes of selected KRED variants. (A) WT and (S)-1‡. (B) WT and (R)-2‡. (C) A94F and (S)-2‡. (D) Y190F and (S)-2‡. (E) E145S and (S)-2‡. (F) E145S and (R)-2‡. (G) Sph and (S)-2‡. (H) Sph and (R)-2‡. The cofactor and substrate are shown in pink and purple, respectively. The catalytic residues Ser143, Tyr156, and Lys160 are in blue; Tyr/Phe190 and Asp150 are in gray; and Leu195, Ala/Phe/Ser94, and Glu/Ser145 are in light green. The approximate extension of the active site is shown as a yellow ellipse. The stability of these TS complexes according to the preservation of the catalytic contacts throughout the MD trajectories is indicated as favored (✓) or disfavored (X).
In the A94F mutant (Fig. 9C), forcing the 3-thiacyclopentanone ring to adopt a reactive pro-S orientation in the presence of the bulky Phe94 in the large pocket induces a conformational change in Tyr190, which no longer binds to E145 and occasionally binds to the nicotinamide moiety. In this way, the formerly small binding pocket is significantly expanded to accommodate the larger S atom, and the primary catalytic contacts (COsubstrate with Ser143/Tyr156) are maintained very close to the optimal values, increasing S-enantioselectivity (SI Appendix, Figs. S24 and S29). The same effect is produced by the Y190F mutation, in which deletion of the OHTyr190 group disrupts its hydrogen bond network with Glu145, Tyr249, and Asp150 and opens up the small pocket to accommodate the endocyclic S and thus promote S-selectivity (Fig. 9D and SI Appendix, Figs. S26 and S31).
The hydrophobic cage formed by Y190, Tyr156, and especially Phe94 (Figs. 7 and 9C) interacts strongly with the 3-oxacyclopentanone ring through CH/π contacts in the pro-R TS complex. Consequently, the catalytic Ser143 is displaced from its optimal position and replaced by the OHsugar group, revealing the inability of this enzyme to stabilize this particular TS. This observation agrees with the fivefold decrease in activity of this mutant with respect to the WT and the enhancement of S-selectivity. In fact, the pro-S TS complex (S)-1‡ is able to accommodate the smaller endocyclic O atom in the encumbered small pocket without producing any conformational change in Tyr190, preserving all of the catalytic interactions during the simulation. In addition, an occasional hydrogen bond between the endocyclic O and OHTyr190, is observed. These geometric features are shown in SI Appendix, Figs. S24 and S29.
The E145S single mutation (Fig. 9 E and F) modifies the complex hydrogen bond network at the active site, disrupting the water-mediated (Fig. 5E) or direct interaction between Tyr190 and Glu145 and producing a new Tyr190–Asp150 interaction. This new contact shrinks the small binding pocket and precludes direct interaction of the OHTyr190 group with the endocyclic O of 3-oxacyclopentanone in the pro-S orientation (SI Appendix, Fig. S30). This reactive pose is also very much disfavored with 3-thiacyclopentanone, in which the larger S atom clashes with the aromatic sidechain of Tyr190 (Fig. 9E). The poorer binding properties of this variant are reflected by the departure of the TS complex from the active site observed in (S)-2‡ after ∼40 ns, which is the cause of the exceedingly high calculated R-enantioselectivity (SI Appendix, Figs. S25 and S30). In contrast, both substrates fit better in the pro-R orientation (Fig. 9F), in which the primary catalytic contacts stabilizing the incipient alkoxide are very well maintained, in agreement with the increased selectivity observed experimentally for 2. Again, the major source of the increased stereoselectivity in this mutant—which retains one-half the activity of the WT—seems to be the destabilization of one stereoselective pathway (pro-S in this case), with the other maintained or slightly stabilized.
The final Sph mutant (Fig. 9 G and H) is the most R-selective variant for 2 (experimental erS/R = 0.004), but our MD protocol largely underestimates this selectivity (calc. erS/R = 0.6). On the other hand, this variant is also the less S-selective one for 1 (experimental erS/R = 1.3), but in this case our method overestimates the selectivity (calc. erS/R = 27). These results show the limitations of the current protocol for highly evolved mutants, for which additional factors related to protein stability and catalyst turnover may play important roles in enantioselective catalysis.
Similar geometric features to those observed in the E145S mutant were observed in these systems, namely the formation of a persistent Tyr190–Asp150 interaction, although the contraction of the small pocket is less apparent. In fact, in the four analyzed systems, the cyclopentanone moieties of the TS complexes seem to fit in the active site without significant steric hindrance. Enhanced CH/π interactions between the β-CH2 group of the substrate and the phenyl ring of Tyr190 in (R)-2‡ (Fig. 9H) may be one source of the increased R-enantioselectivity with 3-thiacyclopentanone, given that the catalytic contacts are very well maintained in both approximations (SI Appendix, Figs. S27 and S32).
Conclusion
Enzyme design and engineering for enantioselective synthesis pose significant challenges for chemical biology that are receiving increasing attention (20). The highly enantioselective enzymes obtained through DE for the reduction of 3-oxacyclopentanone and 3-thiacyclopentanone are less active than the WT, however. Our computational studies of the origins of stereoselectivity in these evolved ketoreductases have revealed that the induction of asymmetry can be achieved by blocking certain reaction pathways rather than promoting others, yielding slower but more selective processes. Through crystallographic and MD simulations of selected variants, we have found that certain mutations can regulate the conformation and flexibility of the substrate-binding loop that can be transmitted to the active site with various stereochemical consequences depending on the substrate used in the reduction reaction.
Certain point mutations in the active site, such as A94F and E145S, can invert the enantioselectivity for 3-thiacyclopentanone reduction by further closing or opening of the substrate-binding loop and reconfiguring the space in the binding pockets. A94F generates a hydrophobic cage that traps both the reactant and TS in unproductive binding modes or enlarges the small binding pocket, whereas E145S shrinks the small binding pocket and amplifies the difference in size between an S atom and a CH2 group, making it more discriminating. These results have major implications for the computational design of new biocatalysts and demonstrate the importance of including appropriate protein conformational sampling in parallel with primary sequence design.
Materials and Methods
Generation and Screening of the Mutant Library.
Experiments were run following the Codexis standard KRED panel screening protocol (SI Appendix, Materials and Methods). A total of 175 KRED variants, including L. kefir KRED, with 1–16 mutations from L. kefir WT KRED, were mixed with each substrate, 1 and 2, in 96-well plates. Each well contained one substrate, cell lysate, which included one KRED variant, NADP+ sodium salt, and triethanolamine in isopropanol. Reactions were allowed to proceed at room temperature for 24 h. Gas chromatography was used to determine the conversion and the er of the resulting alcohol.
Crystallization Experiments.
Crystals of the WT KRED and variants were grown using a hanging-drop vapor diffusion method. Crystals of the WT KRED, both apo- and NADP+-bound, were grown at room temperature. Crystals of A94F and E145S were grown at 4 °C. Buffer compositions, concentrations, and other details are provided in SI Appendix, Materials and Methods.
Data Collection and Structure Determination.
X-ray diffraction data were collected at the Advanced Photon Source (Argonne National Laboratory), Beamline NECAT 24ID-C, using a DECTRIS PILATUS-6M pixel detector. Data reduction and scaling were performed using XDS for all crystals. Diffraction to 2.2-Å resolution was observed for the WT KRED, diffraction to 1.6-Å resolution was observed for A94F, and diffraction to 1.7-Å resolution was observed for E145S. X-ray diffraction data for the NADP+-bound WT KRED crystal were collected at the University of California Los Angeles, using a Rigaku FRE+ rotating anode and a Rigaku HTC image plate detector. Diffraction to 2.1-Å resolution was observed. All of the crystal structures were determined by the molecular replacement method using the PHASER program and using the KRED from L. brevis (Protein Data Bank ID code 1NXQ) as the search model. Specifics of the crystal preparation, data collection, and refinement are reported in SI Appendix, Materials and Methods.
Kinetics Experiments.
To measure the efficiency (kcat/KM), the concentration of enzyme was fixed at 0.05 mM, and the concentration was varied from 10 to 160 mM for 1 and from 0.2 to 1 mM for 2, because catalysis of its reduction is less efficient. DMSO was added to a final concentration of 2% (vol/vol) to facilitate solubilization of the substrates. The reactions took place in 150 mM phosphate buffer (pH 7.0) at 25 °C. The final reaction solution was 7% (vol/vol) glycerol with 2 mM MgSO4 and 1 mM NADPH. The rate of NADPH depletion was measured using a Tecan Infinite M200 Pro plate reader by recording the absorbance of the reaction solutions at 340 nm relative to an NADPH standard.
QM Calculations.
All calculations were performed with Gaussian 09 (21). The geometries were optimized with the TPSS functional (22) and 6–31G(d) basis set with the conductor-like polarizable continuum model (CPCM) (23) for water. Single-point energy calculations were performed on these geometries using the M06-2X method (24) and 6–311+G(d,p) basis set, and with CPCM in water. Further details of the methodology are provided in SI Appendix, Materials and Methods.
MD Simulations.
Crystallographic structures of the tetrameric proteins served as starting geometries. Parameters for the cofactor, substrates, and TS complexes were generated with the antechamber module of Amber 12 (25) using the general Amber force field (GAFF), and atomic partial charges were derived from QM electrostatic potentials. Topology and initial coordinates are available in Amber format in Datasets S1–S8. Each protein complex was immersed in a truncated octahedral box with a 10-Å buffer of TIP3P (26) water molecules, and was neutralized by adding explicit counterions (Na+Cl−). All subsequent simulations were performed using the Stony Brook modification of the Amber 99 force field (27). Equilibration and production procedures are described in SI Appendix, Materials and Methods.
Supplementary Material
Acknowledgments
We thank M. Collazo for help with protein crystallization and M. Sawaya for helpful discussions. We also thank M. Capel, K. Rajashankar, N. Sukumar, F. Murphy, and I. Kourinov of Northeastern Collaborative Access Team Beamline ID-24 at the Advanced Photon Source of Argonne National Laboratory, supported by National Institutes of Health Grants P41 RR015301 and P41 GM103403. Use of the Advanced Photon Source is supported by the US Department of Energy under Contract DE-AC02-06CH11357. This work was performed using the Extreme Science and Engineering Discovery Environment, which is supported by the National Science Foundation (Grant OCI-1053575) and the University of California, Los Angeles (UCLA) Institute for Digital Research and Education’s Hoffman2 computer cluster. The UCLA X-Ray Core Facility is supported in part by US Department of Energy Grant DE-FC03-02ER63421. This research was supported in part by the National Institutes of Health, National Institute of General Medical Sciences (Grants GM036700, GM097200, and GM075962). S.O. is supported by the Ministerio de Economía y Competitividad (Juan de la Cierva Postdoctoral Grant JCI-2012-14438) and the European Commission Career Integration Grants program (Grant PCIG14-GA-2013-630978). C.M.B. was supported by NIH, NIGMS Training Grant T32 GM067555-11.
Footnotes
The authors declare no conflict of interest.
Data deposition: The coordinates of the X-ray structures of variants apo WT, holo WT, apo E145S, and apo A94F have been deposited in the Protein Data Bank, www.rcsb.org/pdb/home/home.do (PDB ID codes 4RF4, 4RF2, 4RF5, and 4RF3, respectively).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1507910112/-/DCSupplemental.
References
- 1.Moore JC, Pollard DJ, Kosjek B, Devine PN. Advances in the enzymatic reduction of ketones. Acc Chem Res. 2007;40(12):1412–1419. doi: 10.1021/ar700167a. [DOI] [PubMed] [Google Scholar]
- 2.Strohmeier GA, Pichler H, May O, Gruber-Khadjawi M. Application of designed enzymes in organic synthesis. Chem Rev. 2011;111(7):4141–4164. doi: 10.1021/cr100386u. [DOI] [PubMed] [Google Scholar]
- 3.Huisman GW, Liang J, Krebber A. Practical chiral alcohol manufacture using ketoreductases. Curr Opin Chem Biol. 2010;14(2):122–129. doi: 10.1016/j.cbpa.2009.12.003. [DOI] [PubMed] [Google Scholar]
- 4.Bornscheuer UT, et al. Engineering the third wave of biocatalysis. Nature. 2012;485(7397):185–194. doi: 10.1038/nature11117. [DOI] [PubMed] [Google Scholar]
- 5.Quallich GJ, Woodall TM. Enantioselective oxazaborolidine reduction of ketones containing heteroatoms. Tetrahedron Lett. 1993;34(5):785–788. [Google Scholar]
- 6.Zhang X, et al. Asymmetric hydrogenation of cycloalkanones catalyzed by BINAP-iridium(I)-aminophosphine systems. J Am Chem Soc. 1993;115(8):3318–3319. [Google Scholar]
- 7.Oppermann U, et al. Short-chain dehydrogenases/reductases (SDR): The 2002 update. Chem Biol Interact. 2003;143-144(0):247–253. doi: 10.1016/s0009-2797(02)00164-3. [DOI] [PubMed] [Google Scholar]
- 8.Weckbecker A, Hummel W. Cloning, expression, and characterization of an (R)-specific alcohol dehydrogenase from Lactobacillus kefir. Biocatalysis Biotransform. 2006;24(5):380–389. [Google Scholar]
- 9.Filling C, et al. Critical residues for structure and catalysis in short-chain dehydrogenases/reductases. J Biol Chem. 2002;277(28):25677–25684. doi: 10.1074/jbc.M202160200. [DOI] [PubMed] [Google Scholar]
- 10.Blankenfeldt W, et al. Variation on a theme of SDR. dTDP-6-deoxy-L- lyxo-4-hexulose reductase (RmlD) shows a new Mg2+-dependent dimerization mode. Structure. 2002;10(6):773–786. doi: 10.1016/s0969-2126(02)00770-0. [DOI] [PubMed] [Google Scholar]
- 11.Thoden JB, Wohlers TM, Fridovich-Keil JL, Holden HM. Crystallographic evidence for Tyr 157 functioning as the active site base in human UDP-galactose 4-epimerase. Biochemistry. 2000;39(19):5691–5701. doi: 10.1021/bi000215l. [DOI] [PubMed] [Google Scholar]
- 12.Schlieben NH, et al. Atomic resolution structures of R-specific alcohol dehydrogenase from Lactobacillus brevis provide the structural bases of its substrate and cosubstrate specificity. J Mol Biol. 2005;349(4):801–813. doi: 10.1016/j.jmb.2005.04.029. [DOI] [PubMed] [Google Scholar]
- 13.Niefind K, Müller J, Riebel B, Hummel W, Schomburg D. The crystal structure of R-specific alcohol dehydrogenase from Lactobacillus brevis suggests the structural basis of its metal dependency. J Mol Biol. 2003;327(2):317–328. doi: 10.1016/s0022-2836(03)00081-0. [DOI] [PubMed] [Google Scholar]
- 14.Benach J, Atrian S, Gonzàlez-Duarte R, Ladenstein R. The catalytic reaction and inhibition mechanism of Drosophila alcohol dehydrogenase: Observation of an enzyme-bound NAD-ketone adduct at 1.4-Å resolution by X-ray crystallography. J Mol Biol. 1999;289(2):335–355. doi: 10.1006/jmbi.1999.2765. [DOI] [PubMed] [Google Scholar]
- 15.Medina JM, Mackey JL, Garg NK, Houk KN. The role of aryne distortions, steric effects, and charges in regioselectivities of aryne reactions. J Am Chem Soc. 2014;136(44):15798–15805. doi: 10.1021/ja5099935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jiménez-Osés G, et al. The role of distant mutations and allosteric regulation on LovD active site dynamics. Nat Chem Biol. 2014;10(6):431–436. doi: 10.1038/nchembio.1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pauling L. Nature of forces between large molecules of biological interest. Nature. 1948;161(4097):707–709. doi: 10.1038/161707a0. [DOI] [PubMed] [Google Scholar]
- 18.Smith AJT, et al. Structural reorganization and preorganization in enzyme active sites: Comparisons of experimental and theoretically ideal active site geometries in the multistep serine esterase reaction cycle. J Am Chem Soc. 2008;130(46):15361–15373. doi: 10.1021/ja803213p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang W, et al. Free-energy perturbation simulation on transition states and redesign of butyrylcholinesterase. Biophys J. 2009;96(5):1931–1938. doi: 10.1016/j.bpj.2008.11.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wijma HJ, et al. Enantioselective enzymes by computational design and in silico screening. Angew Chem Int Ed Engl. 2015;54(12):3726–3730. doi: 10.1002/anie.201411415. [DOI] [PubMed] [Google Scholar]
- 21.Frisch MJ, et al. 2009. Gaussian 09, revision D.01 (Gaussian, Inc., Wallingford, CT)
- 22.Tao J, Perdew JP, Staroverov VN, Scuseria GE. Climbing the density functional ladder: Nonempirical meta-generalized gradient approximation designed for molecules and solids. Phys Rev Lett. 2003;91(14):146401. doi: 10.1103/PhysRevLett.91.146401. [DOI] [PubMed] [Google Scholar]
- 23.Cossi M, Rega N, Scalmani G, Barone V. Energies, structures, and electronic properties of molecules in solution with the C-PCM solvation model. J Comput Chem. 2003;24(6):669–681. doi: 10.1002/jcc.10189. [DOI] [PubMed] [Google Scholar]
- 24.Zhao Y, Truhlar D. The M06 suite of density functionals for main group thermochemistry, thermochemical kinetics, noncovalent interactions, excited states, and transition elements: Two new functionals and systematic testing of four M06-class functionals and 12 other functionals. Theor Chem Acc. 2008;120(1-3):215–241. [Google Scholar]
- 25.Case DA, et al. 2012. Amber 12 (University of California, San Francisco)
- 26.Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML. Comparison of simple potential functions for simulating liquid water. J Chem Phys. 1983;79:926–935. [Google Scholar]
- 27.Wang J, Cieplak P, Kollman PA. How well does a restrained electrostatic potential (RESP) model perform in calculating conformational energies of organic and biological molecules? J Comput Chem. 2000;21(12):1049–1074. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.