Denisova Cave in the Altai region of Siberia had been known to Russian scientists for some 40 y, but the site only achieved wide attention in 2010 with the publication of a very distinctive mtDNA sequence (dubbed “Lineage X”) from a human finger bone fragment [Denisova 3 (1)]. The sequence suggested that “Denisovans” diverged from the lineages of modern humans and Neanderthals close to 1 Mya ago. Later in the same year, a draft whole genome was published from the finger bone, which, in contrast, suggested the Denisovans were quite closely related to Neanderthals, and also indicated the presence of portions of similar DNA in extant Melanesians, presumably from ancient introgression from a Denisovan-like population (2). The early work left many questions about the Denisovans unanswered; however, in PNAS, Sawyer et al. (3) present full mitochondrial and partial nuclear genomes for two other Denisovan individuals represented by adult molar teeth (Denisova 4 and 8). The new study indicates that Denisovans had lower diversity than extant Europeans, and suggests that they occupied the cave through a considerable period.
The description of the new Denisova 8 molar matches the molar of Denisova 4 in its very large size, and the absence of crown traits typical of either Neanderthal or modern human upper molars. Adding evidence of the large splayed roots preserved on Denisova 4, these teeth look very primitive for specimens dated within the past 150,000 y. Sandstone layer 11 in the main East gallery of Denisova Cave appears to be a palimpsest containing archaeological material attributable to modern humans, some of which has been directly dated to ∼40 ka or less; the Denisova 3 finger fragment (in layer 11.2, which is dated to at least 50 ka); a Neanderthal toe phalanx [identified from its genome (4)] in layer 11.4; and the Denisova 8 molar at the junction of layer 11.4 and the underlying layer 14. The Denisova 4 molar was found in the upper part of layer 11 (11.1) in the separate South gallery. Its stratigraphic position and associated radiocarbon dating suggest it is comparable in age to Denisova 3, and younger than Denisova 8. This antiquity is supported by molecular clock analyses, which provide estimates that Denisova 8 is older than Denisova 3 and 4 by at least 60,000 y. Additionally, Gibbons (5) reported on presentations at a London conference, where ancient DNA from a further deciduous molar (Denisova 2) was also discussed. This molar derives from the even deeper layer 22, dated by luminescence to ∼170 ka, and mtDNA branch shortening suggests it is of comparable age to Denisova 8. Thus, there may well have been alternations or lacunae in the Denisovan and Neanderthal presence in the cave over many millennia, followed by a modern human occupation.
Since the late 1980s, researchers have been recovering and characterizing DNA from archaeological specimens using the PCR assay. PCR has many useful attributes for this type of work, but it is difficult to scale up to high-throughput, genome-wide analyses. In addition, it is challenging to apply PCR to the very short DNA fragments that are characteristic of ancient samples, especially when PCR is preferentially prone to recover chemically unaltered, modern contaminant DNA from excavators and laboratory workers. In the past decade, the development of next-generation sequencing technologies has driven a renewed rush of interest into the application of ancient DNA, of which the Denisovan genome is one of the most notable stories. However, Denisovan samples are not easy to come by, and so the opportunity to obtain new data is especially important, even when samples are poorly preserved. Denisova 8 is reconstructed from four fragments, raising the possibility that modern, exogenous DNA may be a significant component of the overall sequence yield.
Of the big three problems of ancient DNA work—base modification, fragment size, and contamination, it is the latter that remains the most difficult to resolve. Here, Sawyer et al. (3) identify significant contamination in both of the specimens studied, using three different approaches. In the mitochondrial genome, the authors make use of the fact that they have the greatest depth of sequence coverage—on average, 72 unique reads for each base position for Denisova 4 and 119 for Denisova 8. They identify a small proportion of individual sequence reads (5% and 3%, respectively) that are in disagreement with the overall consensus, and therefore likely to be contaminants. In the nuclear genome, they find both a significant excess of X chromosome reads and that the two new samples are significantly closer to modern European genomes than would be expected from the existing high-quality Denisova 3 genome. These analyses suggest much greater contamination, with values for the autosomal genome as high as 66% for Denisova 4 and 15% for Denisova 8. The differences between the estimates given by these three methods may seem surprising but, as the authors note, are presumably due to different sources of exogenous DNA in the data. Until we have a better idea of where and how contamination enters the system, our expectations of its quantity and nature are unresolved.
Filtering by retention of only those DNA fragments that show a pattern characteristic of ancient DNA sequences (the replacement of Cyt bases by Thy bases at the ends of sequence strands) reduces the data by over 90% for each sample. This step is necessary, although extreme, as it will undoubtedly have resulted in the loss of many ancient, but undamaged, strands of DNA. Even so, the authors were left with 1 million bases (1 Mb) of sequence for Denisova 4 and 24.1 Mb for Denisova 8, numbers that far exceed the total lifetime sequencing ambitions of those 1980s researchers.
The paper by Sawyer et al. (3) bridges two fields, paleoanthropology and ancient DNA, that have been near-continuous sources of the extraordinary over the past few years. The Denisovan story provides one such example because, since 2010, paleoanthropologists have found themselves in an unexpected position: awareness of a recent hominin group for which the only significant source of information has been the genome sequence. In this context, the publication of two further, very partial Denisovan genomic datasets could therefore be seen as confirmatory, rather than revelatory. However, such an interpretation ignores the important contribution that these data make. Even the comparatively small amounts of genomic information given here place the Denisovans as a monophyletic group, with low autosomal diversity, more comparable to Neanderthals than to modern humans. They appear to have been present in the region over tens of millennia, although details of their history will require significantly greater sampling. As we approach a period where the genomes of the best-preserved samples have been thoroughly characterized, this paper provides us with some important new ideas on how to identify and deal with contamination in samples of poorer quality.
The detailed discussions of dental morphology and ancient DNA provided by Sawyer and colleagues (3) nevertheless still leave many questions about the Denisovans unanswered. For example, how can the contrasting signals of mtDNA and nuclear DNA regarding their relationship with
The paper by Sawyer et al. bridges two fields, paleoanthropology and ancient DNA, that have been near-continuous sources of the extraordinary over the past few years.
Neanderthals be reconciled? One possibility is that detected archaic introgression into the Denisova nuclear genome (4), additional to detected archaic introgression from Neanderthals, was accompanied by the introduction of more “ancient” mtDNA lineages. However, the similarity of the Denisova mtDNAs to the mtDNAs recovered from the ∼400-ka Sima de los Huesos fossils in Spain (6) suggests another possibility, which is that the Sima and Denisovan samples retain diversity in “ancestral” mtDNA lineages, which was lost in later Neanderthal and modern human evolution. Perhaps older mtDNA lineages were even replaced in modern humans and Neanderthals within the past 400 ka by new variants shared between them, consonant with their much younger mtDNA coalescence dates.
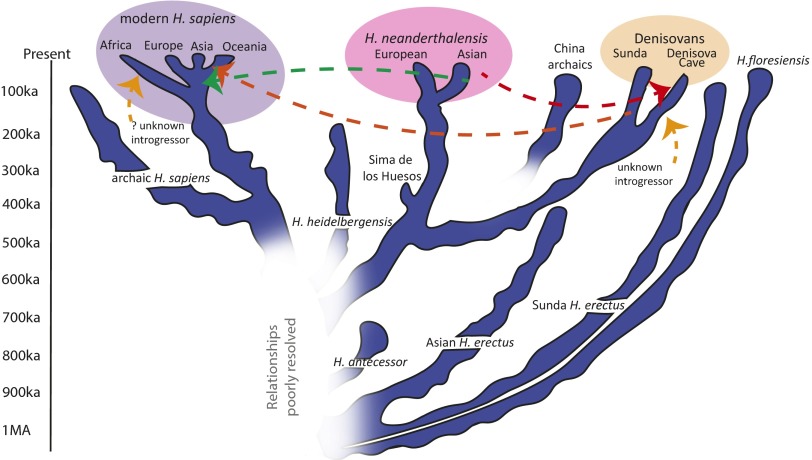
A second and even more fundamental question is how the Denisovans fit into the wider picture of human evolution during the past 1 million y (Fig. 1). As already mentioned, their nuclear DNA strongly suggests that they are part of a wider Neanderthal clade, but this assignment is contradicted by their divergent mtDNAs and their primitive dental morphology. It would be valuable in this regard to have larger fossil and DNA samples of the Denisovans, who are still only known from fragmentary fossils in a single cave site. However, the fact that Denisovan-like introgression in extant humans is concentrated in Oceania suggests that the introgressing population (“Sunda Denisovans” in Fig. 1) was located far south of the Altai, and perhaps even over the Wallace Line (7). Thus, Denisovan-like humans probably ranged over a region from which we already have a reasonable fossil record, so which of the known finds might actually represent Denisovans? So far, none of the later archaic Chinese fossils (China archaics in Fig. 1) or equivalent material from Sunda or Sahul has yielded publishable ancient DNA sequences to compare with those ancient DNA sequences from the Denisova Cave, and the samples show great morphological diversity. In China, fossils such as the Penghu mandible [dredged from the sea near Taiwan (8)] and the Xujiayao material (9) do contain comparably large molar teeth, but the associated mandibles suggest that these two specimens are unlikely to represent the same kind of human. In addition, as Sawyer et al. (3) note, there are similarly large molar teeth in the Romanian early modern Oase 2 fossil and in the archaic Obi-Rakhmat 1 specimen from Uzbekistan, which should remind us that the Denisovans or their past genetic influence may have extended westward as well. Until more complete Denisovan material is recovered from the cave itself, or further Denisovan-like ancient DNA is extracted from other fossils in Asia or Oceania, Lineage X seems as good a name as any for this enigmatic human group.
Fig. 1.
Representation of human evolution during the past 1 million y. Diagnosable units from morphology or DNA are shown, but some lineages (e.g., “archaic H. sapiens” and China archaics) are almost certainly amalgams of fossils with differing affinities. How many of the lineages deserve specific distinction is an open question, given levels of morphological variation and the growing evidence for interlineage gene flow (indicated by dashed arrows).
Acknowledgments
We thank Ali Freyne for her help in preparing the figure and Tom Booth for comments on the manuscript. The research of C.B.S. is supported by the Calleva Foundation and the Human Origins Research Fund of the Natural History Museum.
Footnotes
The authors declare no conflict of interest.
See companion article on page 15696.
References
- 1.Krause J, et al. The complete mitochondrial DNA genome of an unknown hominin from southern Siberia. Nature. 2010;464(7290):894–897. doi: 10.1038/nature08976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Reich D, et al. Genetic history of an archaic hominin group from Denisova Cave in Siberia. Nature. 2010;468(7327):1053–1060. doi: 10.1038/nature09710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sawyer S, et al. Nuclear and mitochondrial DNA sequences from two Denisovan individuals. Proc Natl Acad Sci USA. 2015;112:15696–15700. doi: 10.1073/pnas.1519905112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Prüfer K, et al. The complete genome sequence of a Neanderthal from the Altai Mountains. Nature. 2014;505(7481):43–49. doi: 10.1038/nature12886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gibbons A. HUMAN EVOLUTION. Cave was lasting home to Denisovans. Science. 2015;349(6254):1270–1271. doi: 10.1126/science.349.6254.1270-b. [DOI] [PubMed] [Google Scholar]
- 6.Meyer M, et al. A mitochondrial genome sequence of a hominin from Sima de los Huesos. Nature. 2014;505(7483):403–406. doi: 10.1038/nature12788. [DOI] [PubMed] [Google Scholar]
- 7.Cooper A, Stringer CB. Paleontology. Did the Denisovans cross Wallace’s Line? Science. 2013;342(6156):321–323. doi: 10.1126/science.1244869. [DOI] [PubMed] [Google Scholar]
- 8.Chang CH, et al. The first archaic Homo from Taiwan. Nat Commun. 2015;6:6037. doi: 10.1038/ncomms7037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xing S, Martinón-Torres M, Bermúdez de Castro JM, Wu X, Liu W. Hominin teeth from the early Late Pleistocene site of Xujiayao, Northern China. Am J Phys Anthropol. 2015;156(2):224–240. doi: 10.1002/ajpa.22641. [DOI] [PubMed] [Google Scholar]