Significance
We have used single-cell RNA sequencing to compare human cerebral organoids and fetal neocortex. We find that, with relatively few exceptions, cells in organoid cortex-like regions use genetic programs very similar to fetal tissue to generate a structured cerebral cortex. Our study is of interest, as it shows which genetic features underlying human cortical development can be accurately studied in organoid culture systems. This is important because although cerebral organoids have great promise for modeling human neurodevelopment, the extent to which organoids recapitulate neural progenitor proliferation and differentiation networks in vivo remained unclear.
Keywords: cerebral organoid, neocortex, corticogenesis, single-cell RNA-seq, stem cells
Abstract
Cerebral organoids—3D cultures of human cerebral tissue derived from pluripotent stem cells—have emerged as models of human cortical development. However, the extent to which in vitro organoid systems recapitulate neural progenitor cell proliferation and neuronal differentiation programs observed in vivo remains unclear. Here we use single-cell RNA sequencing (scRNA-seq) to dissect and compare cell composition and progenitor-to-neuron lineage relationships in human cerebral organoids and fetal neocortex. Covariation network analysis using the fetal neocortex data reveals known and previously unidentified interactions among genes central to neural progenitor proliferation and neuronal differentiation. In the organoid, we detect diverse progenitors and differentiated cell types of neuronal and mesenchymal lineages and identify cells that derived from regions resembling the fetal neocortex. We find that these organoid cortical cells use gene expression programs remarkably similar to those of the fetal tissue to organize into cerebral cortex-like regions. Our comparison of in vivo and in vitro cortical single-cell transcriptomes illuminates the genetic features underlying human cortical development that can be studied in organoid cultures.
Elucidating the cellular and molecular basis of human neocortex development and evolution has profound importance for understanding our species-specific cognitive abilities as well as our susceptibility to neurodevelopmental diseases. Neurons of the human neocortex originate during embryogenesis from cell divisions of a variety of neural progenitor cells (NPCs) located in compartmentalized germinal zones. NPC types differ in cell morphology, cell polarity, capacity to self-renew, lineage relationships, and location of mitosis (1). Apical progenitors (APs), including apical (or ventricular) radial glia (aRG), divide at the apical surface of the ventricular zone (VZ), whereas their derivative basal progenitors (BPs), including basal (or outer) radial glia (bRG) and basal intermediate progenitors (bIPs), lack apical contact and divide in the inner and outer subventricular zone (iSVZ and oSVZ) (2). In humans, both aRG and bRG are able to self-amplify by symmetric proliferative divisions. They also share the capacity to divide asymmetrically to self-renew while producing neurons either directly or via bIPs (3-6). In humans, bIPs further amplify the neuronal output of aRG and bRG by undergoing additional rounds of symmetric division before self-consuming into pairs of neurons (1, 7). Newborn neurons migrate radially from these germinal zones to finally establish in the cortical plate (CP) (8). The evolutionary expansion of the human neocortex has been linked to an increase in the proliferative potential of particular pools of NPCs, notably BPs, resulting in greater numbers of neocortical neurons (1, 2, 5, 9). Because of the challenges associated with primate experimentation, the mouse has been widely used as a model system for understanding human cortical neurogenesis. However, due to its evolutionary distance and divergent physiology, it is debatable how relevant the rodent genomic and developmental background is. Therefore, systems effectively recapitulating human cortical development are required.
Recently, self-organizing structures reminiscent of the developing human brain have been generated from pluripotent stem cells [embryonic stem cells (ESCs) or induced pluripotent stem cells (iPSCs)], presenting a unique opportunity to model human cerebral development in vitro (10). So-called cerebral organoids are generally heterogenous and allow the formation of a variety of brain-like regions including the cerebral cortex, ventral forebrain, midbrain–hindbrain boundary, and hippocampus (10). The protocol to grow cerebral organoids was designed to mimic early stages of forebrain development and then rely on the intrinsic self-organizational capacity of the cells to pattern, specify, and generate structured cerebral tissue (11). This protocol establishes cortical-like tissue with compartmentalized germinal zones including a VZ, where aRG-like cells line a lumen, express RG marker genes, undergo interkinetic nuclear migration, and divide at the apical surface, similar to their in vivo counterparts. In addition, time-lapse microscopy and immunostainings for bIP markers [e.g., TBR2; also known as (aka) EOMES] and neurons (TUJ1, aka TUBB3) revealed patterns of direct and indirect neurogenesis in an abventricular location, reminiscent of the SVZ. bRG-like cells, albeit more rarely than in the in vivo neocortex, were also observed. The observations of a cortical architecture and cellular behavior based largely on marker gene expression are generally consistent with in vivo patterns of human fetal cortical development. Bulk transcriptome analysis has provided a genome-wide perspective of the gene expression landscape in whole organoids (12). However, due to heterogeneity within cerebral organoids, bulk methods obscure cell-type diversity and provide coarse insight into organoid neurogenic programs. Moreover, the extent to which organoid cell composition and lineage progression are equivalent to fetal cortical tissue remains unresolved.
Single-cell RNA-seq (scRNA-seq) is a powerful method to unbiasedly identify molecularly distinct cell types in heterogeneous tissues (13–16). ScRNA-seq can also identify cells as intermediate between types, and cells can then be computationally aligned to delineate differentiation paths (15, 17). We sought to use single-cell transcriptomes to analyze the cellular composition of human cerebral organoids and fetal human cortical tissue to reconstruct the differentiation paths and gene networks underlying human corticogenesis. We find that, with some exceptions, the same genes used to build cortical tissue in vivo characterize corticogenesis in vitro. Our data thus show that genetic features underlying human cortical development can be accurately studied in organoid culture systems.
Results
Reconstructing Lineage Relationships in the Human Fetal Cerebral Cortex.
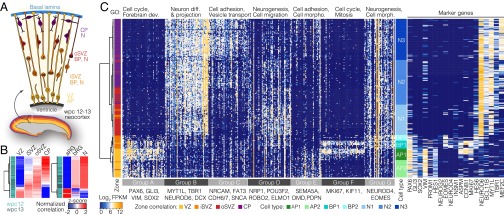
To compare corticogenesis in organoids with primary fetal tissue, we first dissected the cellular composition and lineage relationships in human fetal neocortex. We analyzed 226 single-cell transcriptomes from 12- and 13-wk postconception (wpc) human neocortex specimens (Fig. 1A) and performed principal components analysis (PCA) on all variable genes (variance > 0.5) expressed in more than two cells to survey cell diversity in this dataset (Fig. S1A). The genes correlating and anticorrelating with the first principal component from the PCA distinguished NPCs and neurons, respectively, and highlighted intermediates between the two (Fig. S1B). We identified a single cell expressing PECAM1 and many other endothelial markers and five interneurons derived from the ventral telencephalon (marked by GAD1, DLX1/2/5/6, and ERBB4; Fig. S1C), and we excluded these cells from further analyses. We next used all expressed genes to characterize cells based on their maximum correlation with bulk RNA-seq data from laser-dissected germinal zones, namely VZ (containing a majority of aRG), iSVZ and oSVZ (containing a majority of bIPs, bRG neurons, and migrating neurons), and CP (containing mature neurons) (18), or FACS-purified aRG, bRG, and neurons (19) (Fig. 1B). The transcriptome of each single fetal neocortical cell correlated maximally with one of these four different zones (CP, 100 cells; ISVZ, 24 cells; OSVZ, 62 cells; VZ, 39 cells) and with one of the purified cell populations representing diverse RG subtypes and neurons at different stages of maturation. Therefore, our scRNA-seq data likely include APs, BPs, and neurons, representing the range of neural cell types present in the human neocortex at this time point.
Fig. 1.
Deconstructing cell composition in the fetal human neocortex. (A) scRNA-seq was performed on cells from two human neocortex specimens at 12–13 wpc. Schematic shows NPC types (APs, BPs) and neurons (N) enriched in zones within the human neocortex at midneurogenesis. AP, apical progenitor; BP, basal progenitor; CP, cortical plate; iSVZ, inner subventricular zone; oSVZ, outer subventricular zone; VZ, ventricular zone. (B) Heat maps show normalized correlation (Z-score) of single-cell transciptomes from human wpc 12 (light green) and wpc 13 (dark green) cerebral cortex with bulk RNA-seq data from laser-microdissected zones (left, 18) or FACS-purified cell types (right, 19) from the human neocortex at the same developmental stage. (C) Hierarchical clustering of scRNA-seq data reveals cell types in the human fetal cortex. Each row represents a single cell and each column a gene. Genes were discovered using PCA (SI Methods). The maximum correlation to bulk RNA-seq data from germinal zones is shown in the left sidebar. Cell-type assignment is shown on the right sidebar. Expression of genes used to classify APs, BPs, newborn neurons (N1), and maturing neurons (N2 and N3) are show to the right of the cell-type assignment bar. Top GO enrichments are shown above the heat map, with representative genes listed below.
Fig. S1.
PCA of fetal cerebral cortex. (A) PCA on all variable genes and all 226 cells was used to identify genes describing cortical cell populations. Each dot represents a cell that is color-coded in shades of green representing different experiments. (B) Hierarchical clustering and heat map visualization showing the expression of genes that have highest correlation and anticorrleation with PC1 and highest correlation with PC2. Note that the genes correlating with PC2 identified a single endothelial cell. Cells are shown in rows and genes in columns. (C) Hierarchical clustering and heat map visualization showing the discovery of five interneurons based on marker gene expression.
We performed PCA again on all fetal transcriptomes (excluding the endothelial cell and interneurons), as well as on NPCs and neurons separately, to identify genes that are most informative for defining cell subpopulations. Using these genes (Dataset S1), we hierarchically clustered cells and identified seven major cell clusters that we classified as different populations of APs, BPs, and neurons based on the expression of known cell-type marker genes and correlations with bulk data (Fig. 1C). Two clusters (AP1 and AP2) had a high correlation with bulk VZ, expressed genes involved in forebrain development (group A) and cell adhesion (group E), and were marked by coexpression of RG markers PAX6, GLI3, SOX2, HES1, and VIM and the AP marker PROM1. AP1 cells (16 cells) expressed cell-cycle genes (group F) enriched during S/G2/M phase progression (e.g., MKI67), whereas AP2 cells (24 cells) did not. This suggests that AP1 and AP2 may contain APs in S/G2/M and G1, respectively. We identified a cell cluster (BP1, 11 cells) that had maximum correlations with bulk iSVZ and expressed RG markers PAX6, GLI3, and ASPM as well as early BP markers INSM1, EOMES, and HES6 but lacked expression of VIM and SOX2. In addition, these BP1 cells expressed a core of proneural genes, including NEUROD4, as well as many of the same S/G2/M cell-cycle regulators expressed in AP1 cells in the VZ (groups A, F, and G). This gene expression profile is compatible with AP-to-BP transition or an early BP. Furthermore, we identified a cluster (BP2, 10 cells) containing cells with highest correlation with bulk iSVZ and oSVZ and that was defined by coexpression of EOMES and early neuronal genes including NEUROD6 (group B) but did not express ASPM and other S/G2/M cell-cycle regulators. This cluster is likely to contain more mature BPs that are committed to the neurogenic fate and are in the G1 phase.
In addition to NPCs, we found three clusters (neuron clusters 1–3, N1–3) distinguished by the lack of an NPC signature. Each of these clusters expressed a large group of neuronal genes (group B, including MYT1L, BCL11B, and TBR1) enriched for GO (gene ontology) terms involved in neuronal differentiation. One neuron cluster (N1, 39 cells) correlated with iSVZ and oSVZ bulk data and expressed genes involved in various aspects of neurogenesis, including neuronal migration (group D). This cluster likely contains newborn neurons en route to the CP. A second neuron cluster (N3, 67 cells) correlated with CP bulk data and expressed genes involved in cell adhesion and vesicle transport (group C), indicative of a more mature stage of neuronal differentiation. The third neuron cluster (N2, 53 cells) appears to contain cells that are transitioning between these N1 and N3 neurons. The gene expression profiles of these clusters are therefore compatible with cortical neurons at different stages of maturation.
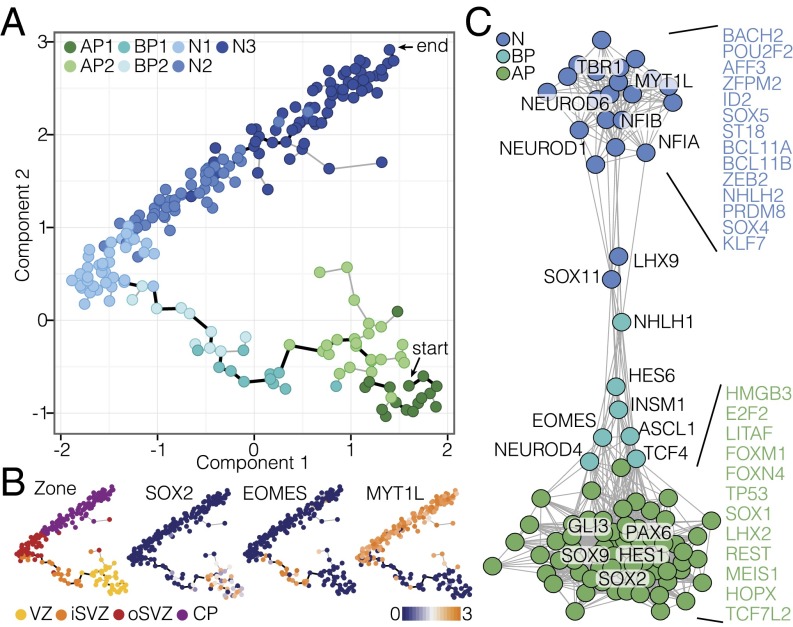
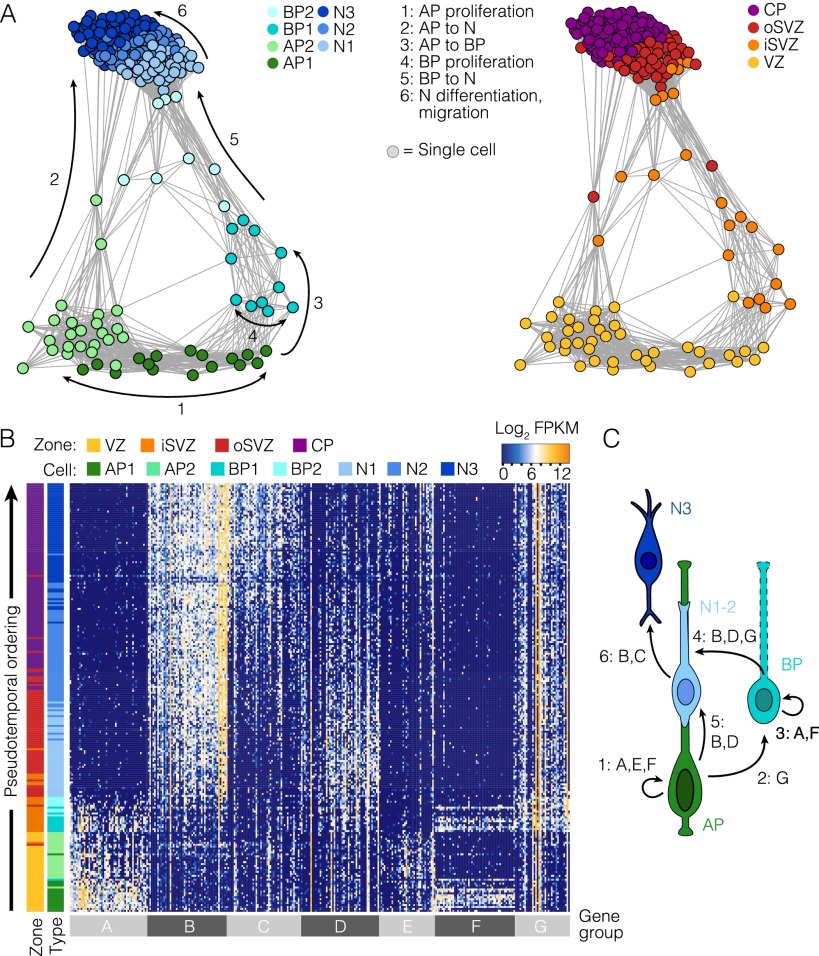
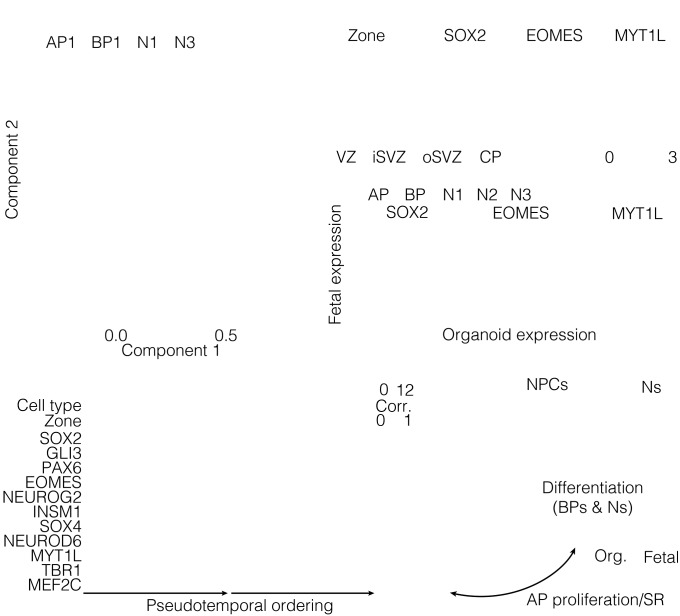
We next inferred lineage relationships among the cells using Monocle, an algorithm that combines differential gene expression, dimension reduction, and minimal spanning tree construction to link cells along a pseudotemporally ordered path (17) (Fig. 2A). This revealed a cortical lineage that linked APs in the VZ, through BPs and newborn neurons in the iSVZ and oSVZ, to CP neurons. This organization was corroborated using an adjacency network based on pairwise correlations between cells (Fig. S2A). Notably, expression of genes known to be enriched in APs (e.g., SOX2), BPs (e.g., EOMES), and neurons (e.g., MYT1L) exhibited restricted expression along the lineage path (Fig. 2B). Heat map visualization of gene expression with cells ordered according to their pseudotemporal position along the lineage revealed a temporal sequence of gene regulatory events during differentiation and many cells at intermediate stages (Fig. S2B).
Fig. 2.
Reconstructing lineage relationships in the fetal neocortex. (A) Monocle reveals an AP–BP–neuron lineage that correlates with the zones of the developing neocortex. Cells (circles, colored based on cell type) are arranged in the 2D independent component space based on genes used to classify cells in Fig. 1C. The minimal spanning tree (gray lines) connects cells, with the black line indicating the longest path. (B) Monocle plots with single cells are colored based on the maximum correlation with bulk RNA-seq data from cortical zones (Far Left) or gene expression that distinguishes the lineage transitions (Middle Left, Middle Right, and Far Right). (C) Transcription factor (TF) correlation network during lineage progression. Shown are nodes (TFs) with more than three edges, with each edge reflecting a high correlation (>0.3) between connected TFs.
Fig. S2.
Gene expression signatures controlling fetal corticogenesis. (A) Cell lineage network based on pairwise correlations between cells and using the same gene set used in the Monocle analysis (Fig. 2 A and B). Cells are color-coded based on cell-type classification (Left) or maximum correlation to cortical zones (Right). Arrows and numbers mark topological features of the network that reflect cell proliferation and differentiation events inferred from gene expression signatures. (B) Ordering of scRNA-seq expression data according to the pseudotemporal position along the lineage revealed a continuum of gene expression changes from NPCs to neurons. Genes from the heat map depicted in Fig. 1C are shown. Each row represents a single cell and each column a gene. The cell-type assignment and maximum correlation to cortical zone are shown as sidebars. (C) Schematic illustrating lineage decisions in the fetal cortex, labeled with groups of genes characterizing lineage progression. For example, APs are defined by the expression of group A genes and self-renew through the expression of cell-cycle regulators shown in gene groups E and F.
We constructed a transcription factor (TF) correlation network to understand the gene expression changes occurring during transitions through the AP–BP–neuron lineage (Fig. 2C). The network revealed two densely connected subnetworks regulating NPC proliferation and neuronal differentiation, respectively, linked by a series of TFs regulating the transition between the two major cell states. This unbiased approach highlighted known master regulators as central to each subnetwork. For example, HES1, SOX9, PAX6, and SOX2 are all tightly connected and highly expressed in most APs of the VZ and are down-regulated shortly after neuronal lineage commitment occurring in the SVZ. ASCL1, EOMES, NEUROD4, HES6, and INSM1 are all expressed concomitant with the AP-to-BP transition, consistent with their role in delamination and early neuronal specification. Another tightly connected subnetwork corresponds to genes such as TBR1, MYTL1, BCL11A/B, and NEUROD6, whose expressions are up-regulated concomitant with neuron differentiation and maintained throughout later stages of neuronal maturation. These data largely confirm the current knowledge of transcriptional regulation during cortical neurogenesis.
In conclusion, the analysis of single-cell transcriptomes in the developing human neocortex can faithfully reconstruct genetic and cellular networks involved in germinal zone organization, NPC proliferation, and NPC-to-neuron differentiation (Fig. S2C).
Dissecting Cell Composition in Human Cerebral Organoids.
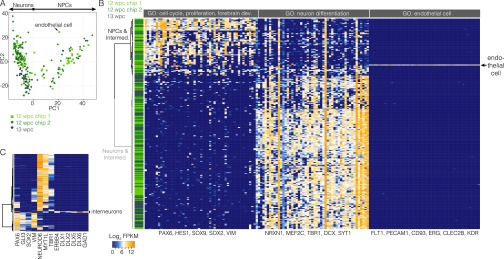
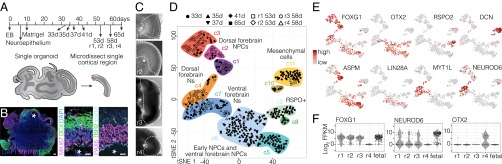
To survey the cellular composition of human cerebral organoids at different stages of development, we analyzed 333 single-cell transcriptomes from five whole organoids (days 33, 35, 37, 41, and 65, iPSC-derived) and 175 single-cell transcriptomes from four microdissected cortical regions (53 d, r1, r2, ESC-derived; 58 d, r3, r4, iPSC-derived) (Fig. 3 A–C and Fig. S3). We combined all single-cell transcriptomes and performed PCA to identify genes most informative for defining cell populations. Using these genes (Dataset S2), we used t-distributed Stochastic Neighbor Embedding (t-SNE) to reduce the complexity of the data and visualize cell relationships in a 2D space (Fig. 3D). This approach organized the cells into 11 transcriptionally distinct clusters, and we performed differential expression analysis to identify genes marking each cluster (Fig. 3 D and E, Figs. S3 and S4, and Dataset S2). In this way, we identified multiple types of progenitors, neurons, and mesenchymal cells from different regions of the cerebral organoid.
Fig. 3.
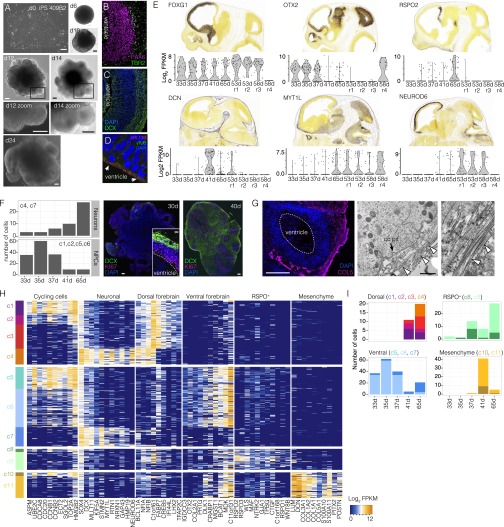
Dissecting cerebral organoid cell composition using scRNA-seq. (A) scRNA-seq was performed on whole organoids dissociated at 33, 35, 37, 41, and 65 d after EB culture and four microdissected regions surrounding single ventricles from two organoids (day 53, r1, r2, ESC-derived; day 58, r3, r4, iPSC-derived). (B) The 30 d iPSC-derived organoid immunostained with proliferation marker MKI67 (magenta), neuronal marker DCX (doublecortin) (green), and DAPI (blue). (Scale bar, 100 μm.) Zoom to ventricle (asterisk) shows MKI67, DCX, and DAPI (Left), and NPC marker PAX6 (magenta), BP marker TBR2/EOMES (green), and mitosis marker phospo-vimentin (pVim; cyan) (Right). (Scale bar, 20 μm.) (C) Images of microdissected cortical regions (r1 and r2 from 53 d ESC-derived organoid; r3 and r4 from 58 d iPSC-derived organoid). Dotted lines show microdissection boundaries; asterisks mark ventricles. (D) PCA and unbiased clustering using t-SNE reveals cell populations within organoids. Shapes indicate experiments, and colors represent significant clusters. See Dissecting Cell Composition in Human Cerebral Organoids and SI Results for cluster descriptions. (E) Marker genes for each cluster. Cells are colored based on expression level. Cerebral cortex cells (c1, c2, c3, and c4) have high expression of FOXG1 and low expression of OTX2. (F) Violin plot shows FOXG1, NEUROD6, or OTX2 expression from each microdissected region compared with fetal cortex.
Fig. S3.
Supporting data for dissecting cerebral organoid cell composition using scRNA-seq. (A) Images from various stages of cerebral organoid development. From left to right, iPSC colonies (d0), EB (d6), neural induction (d10), 2 d after matrigel embedding (d12), 4 d after matrigel embedding (d14), and early organoids after 10 d in cerebral organoid media containing vitamin A (d24). For d12 and d14, Insets show stratified epithelium surrounding a ventricle. Boxes indicate areas shown at greater magnification, as indicated. (Scale bar, 100 μm.) (B and C) Cortical region from 35 d organoid immunostained for PAX6 and TBR2 (B) and DCX and DAPI (C). (D) AP dividing perpendicular to the apical (ventricular surface) immunostained for the ciliary marker ARL13B (magenta) and the centrosomal marker γ-Tubulin (TUBBG; green); nuclei were counterstained with DAPI (blue). (E) Violin plots show distribution of the transcript levels of the representative marker genes across all experiments. In situ hybridizations in E11.5 (Foxg1, Otx2, and Rspo2) and E13.5 (Dcn, Myt1l, and Neurod6) mouse brains show the patterns of gene expression along the rostro–caudal or apical–basal axes. Data are from the Allen Brain Institute Gene Expression Atlas (courtesy of the Developing Mouse Brain, Allen Brain Atlas). (F) More neurons were detected per organoid over the time course relative to progenitors. Neuron and progenitor count represents clusters 4 and 7, and clusters 1, 2, and 5 from B, respectively. Representative image of a 30- and 40-d iPSC-derived organoid showing the relative increase in DCX+ neurons during this time window. (Scale bar, 100 μm.) Inset shows progenitor and neuron stratification within an individual neurogenic zone. (Scale bar, 20 μm.) (G) Immunohistochemistry and transmission electron microscopy validation that mesenchymal cells secrete collagen and form a fibrous ECM surrounding the periphery of individual cortical regions. Black arrow points to a clathrin-coated pit, and the white triangles point to collagen fibrils. (EM image scale, 1 μm.) (H) Heat map showing expression of genes that distinguish cell type and regional identity of the different t-SNE clusters. The cell type or region that is marked by a given gene set is shown above the heat map. Cells are in rows and genes in columns (Dataset S2 and SI Methods). (I) Bar plots show the number of cells in each cluster over time for all whole organoid experiments. The color- code of each bar corresponds to the cluster number.
Fig. S4.
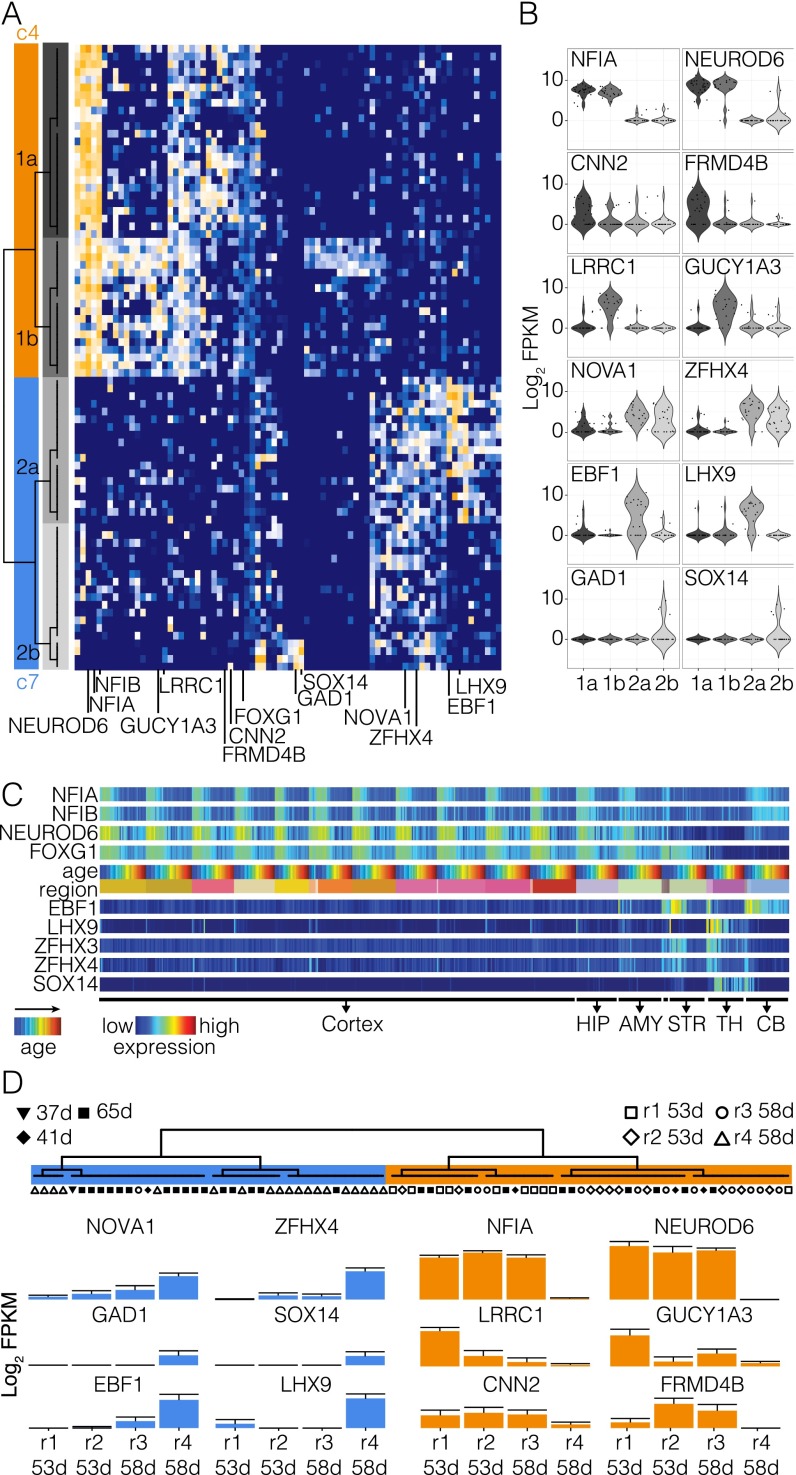
Characterizing organoid neuron heterogeneity. (A) Heat map visualization of variable genes describing neuronal heterogeneity in cerebral organoids. PCA on neuron clusters 4 (orange) and 7 (blue) from Fig. 3D was used to discover genes describing neuronal variation, and BackSPIN was used to cluster genes and cells to define different neuronal cell types. There were four major clusters of neurons (1a, 1b, 2a, and 2b), which are subpopulations of cluster 4 (1a and 1b) and 7 (2a and 2b), respectively. Note that four cells in cluster 2a coexpress GAD1, GAD2, SOX14, and ERBB4 and are likely interneurons derived from ventral telencephalon-like regions of the organoid. (B) Violin plots showing the distribution of representative marker gene expression for each neuron cluster. (C) Expression of representative markers from bulk RNAseq data along a time course in multiple regions of the developing human brain (sourced from the Allen Brain Atlas). (D) Gene expression averaged across all neuronal cells from each microdissected region shows that r1, r2, and r3 all express NFIA, NEUROD6, and the other cluster 4 markers, whereas r4 does not. These data support that r1, r2, and r3 contain neurons from organoid dorsal cortex-like regions.
We found that clusters c1, c2, and c3 are NPCs, and cluster c4 is neurons from organoid dorsal forebrain-like regions (cerebral cortex) based on enriched expression of genes expressed in nearly all fetal cerebral cortex NPCs or neurons (i.e., NPCs and Ns, FOXG1, NFIA, and NFIB; Ns, NEUROD6). Clusters c5 and c6 are NPCs, and c7 is neurons that lack expression of these genes that were expressed in the fetal cortex. Instead, many NPCs in c5 and c6 express OTX2 and are likely a mixture of ventral telencephalic or hippocampal NPCs and NPCs from immature dorsal telencephalic regions. c7 is composed of neurons from ventral forebrain-like structures and includes interneuron-like cells (Fig. S4). c8 and c9 contain cycling and noncycling cells that express R-spondin genes and WNT2B and are likely from the hem signaling center in the dorsal/ventral boundary region. c10 and c11 contain cycling and noncycling mesenchymal cells that express extracellular matrix (ECM) genes and surround the periphery of cortical regions (Fig. 3E and Fig. S3 G and H). Please see SI Results for a detailed analysis and discussion of organoid cell-type composition.
We observed that each microdissected cortical-like region contained NPCs and neurons (Fig. 3D). However, cells from three of the four regions were FOXG1-positive, were OTX2-negative, expressed cerebral cortex markers, and were contained within the dorsal cortex clusters in the t-SNE analysis (Fig. 3 D–F and Fig. S3). In contrast, cells in the fourth cortical region did not express FOXG1 or other fetal cortex markers (i.e., NFIA, NFIB, NEUROD6) but instead expressed OTX2 and were contained within ventral forebrain clusters (c5, c6, and c7). Thus, individual cerebral organoids contain cortical regions with different forebrain identities, which we could discriminate due to distinct signatures of NPC and neuron populations.
Reconstructing Lineages in the Organoid Cerebral Cortex.
We characterized organoid cortex-like cells (clusters 1, 2, 3, and 4; 157 cells in total) based on their maximum correlation with bulk RNA-seq data from laser-dissected germinal zones (18) or FACS-purified NPC subpopulations (Fig. S5A) (19). We found correlation patterns similar to the fetal tissue, suggesting that the organoid cells represent a similar range of cell types as in the fetal cortex data (CP, 27 cells; ISVZ, 3 cells; OSVZ, 19 cells; VZ, 110 cells). We next classified the organoid cells by determining the fetal cell type with which they correlate most strongly, resulting in 57 AP1, 57 AP2, 1 BP1, 6 BP2, 4 N1, 16 N2, and 16 N3 cells. Notably, we observed proportionally less BP cells (6% of NPCs) in the organoid than in the fetal tissue (34%) (Fig. S5).
Fig. S5.
Extended data showing similar gene expression profiles characterize lineage progression in organoid and fetal cerebral cortex. (A) Organoid cerebral cortex-like cells (c1, c2, c3, and c4 from Fig. 3) have differential correlation with bulk RNA-seq data from different laser-microdissected zones or FACS-purified cell types from fetal cerebral cortex (Fig. 1B). (B) Organoid cell lineage network based on pairwise correlations between cells. Cells are colored based on maximum correlation to cortical zones (Left) or cell type (Right). (C) Pseudotemporal cell ordering along the organoid lineage reveals gene expression changes from NPC to neuron. Genes with highest correlation and anticorrelation to PC1–3 are shown. Rows represent cells and columns genes. Maximum correlation to cortical zone and cell type is shown in the left sidebars. Top GO enrichments are shown above the heat map, with representative genes listed below. (D) Correlation network using the same TFs as in the fetal TF network reveals two highly connected subnetworks controlling AP proliferation/self-renewal and neuron differentiation. Shown are nodes (TFs) with more than two edges, with each edge reflecting a correlation between connected TFs that is greater than 0.3. (E) Bar plot shows the fraction of fetal and organoid cells that are assigned as AP, BP, or neuron. (F) The maximum and minimum correlation of each organoid cerebral cortex-like cell with any cell from the fetal neocortex plotted along the organoid pseudotime. The correlation is relatively even throughout the lineage. (G) Heat map visualization of fetal expression of the same genes discovered by PCA on organoid cells shown in C. (H) Heat map visualization of organoid expression of the same genes discovered by PCA on fetal cells, which was used in fetal cortex lineage analysis. For comparison, see Fig. 1C and Fig. S2B.
As with the fetal tissue, we next performed PCA on organoid cerebral cortex-like cells and used the genes with the highest PC1–3 loadings (Dataset S3) to infer lineage relationships using Monocle as well as construct an intercellular correlation network (Fig. 4 A and B and Fig. S5B). Similar to the fetal tissue, the minimal spanning tree revealed a lineage path from AP through BP to the neuron, with PAX6, EOMES, and MYT1L exhibiting restricted expression along the lineage. Also, a side branch from the main lineage suggested rare alternative paths to neuronal fate. The adjacency network graph revealed multiple connections from AP and BP to the neuron and also highlighted AP self-renewal and proliferation in cells correlating with VZ bulk data (Fig. S5B). In addition, BP cells correlating with iSVZ and oSVZ were organized as intermediates between AP cells in the VZ and CP neurons. In general, this analysis confirmed that cells in the organoid cortex are organized in a zonal and cellular hierarchy consistent with what we observed in the fetal tissue.
Fig. 4.
Similar gene expression profiles characterize lineage progression in organoid and fetal cerebral cortex. (A) Organoid AP–BP–neuron lineage. PCA on organoid dorsal cortex cells identified genes used for Monocle. The minimal spanning tree (gray lines) connects cells (circles, colored by cell type). Black line indicates the longest path (Dataset S3 and Fig. S5). (B) Monocle plot with cells colored by maximum correlation with bulk RNA-seq data from germinal zones (Far Left) or marker gene expression (Middle Left, Middle Right, and Far Right). (C) Scatter plot shows correlation (Pearson) between fetal and organoid average expression per cell type for marker TFs. (D) Heat map shows TF expression in organoid and fetal cells ordered by pseudotime. Top bars show cell type and maximum correlation with germinal zones. Each TF’s expression was averaged across cells of a given type (AP, BP, N1, N2, and N3), and the Pearson correlation between fetal and organoid cell types is shown to the right of the heat map. (E) Cell lineage network and dendrogram (Top Right) based on pairwise correlations between fetal (orange) and organoid (red) cells show that NPCs and neurons intermix.
Heat map visualization of cells ordered according to their pseudotemporal position along the lineage revealed a temporal sequence of gene regulatory events during differentiation, with many cells at intermediate stages (Fig. S5C). GO enrichments similar to the fetal tissue (cell cycle, mitosis, neuron projection and differentiation, forebrain development, synapse formation, and migration) were observed for gene groups that change expression along the lineage (Dataset S3). TFs involved in fetal neurogenesis showed a similar expression pattern and network topology in the organoid (Fig. 4 B–D and Fig. S5D). PCA and hierarchical clustering of organoid and fetal cells combined showed that organoid and fetal cells were distributed together within the two main subclusters representing NPCs and neurons (Fig. 4E). In addition, fetal and organoid cells intermix in a cell lineage network based on genes describing AP proliferation and AP–BP–neuron differentiation (Fig. 4E), with intercellular correlations being relatively constant along the lineage (Fig. S5). Thus, the major proportion of the variation in these data is not between in vitro and in vivo tissues but among cell states during neurogenesis.
Similarities in Neurogenic Programs Between Organoid and Fetal Cortex.
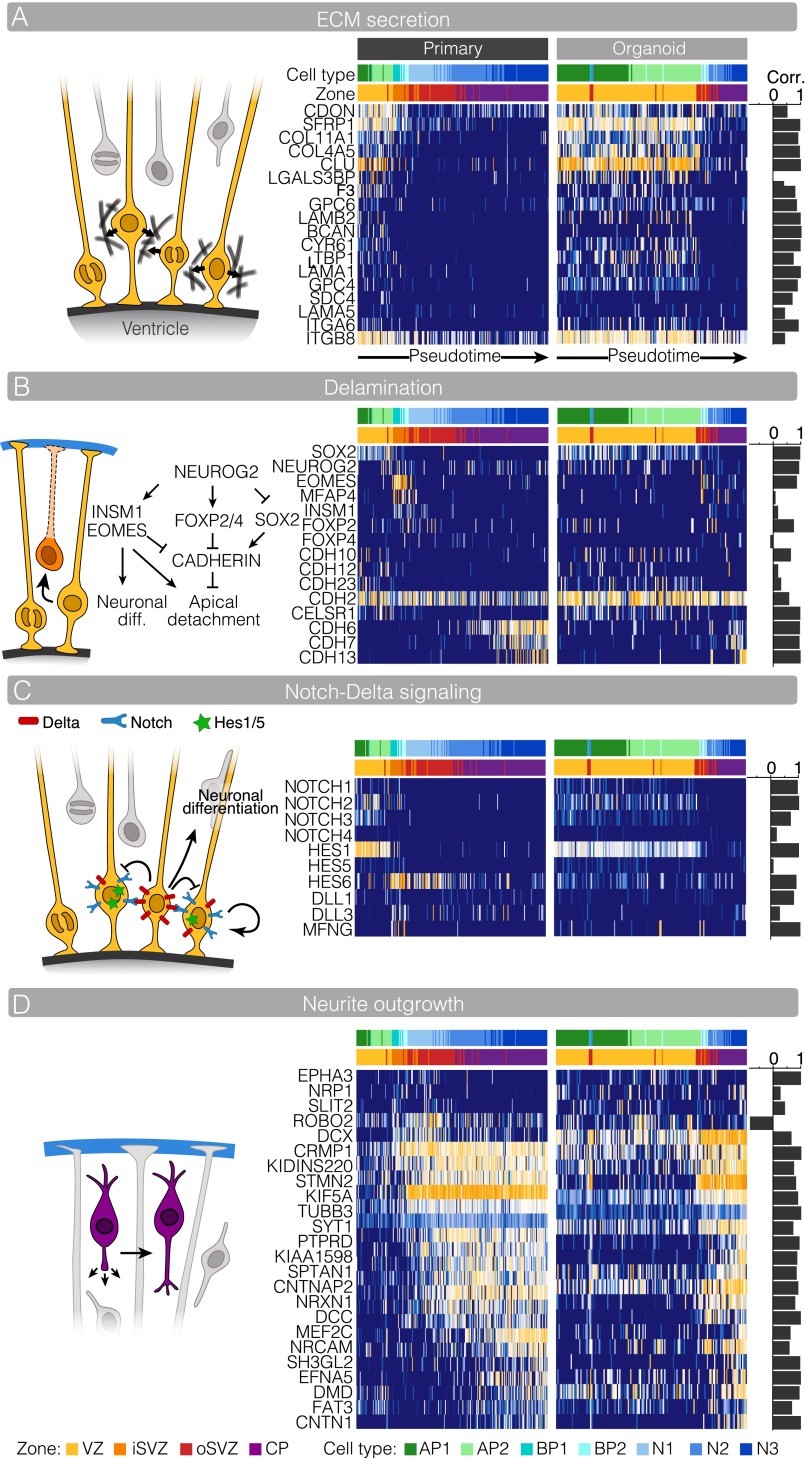
We compared fetal and organoid expression of genes involved in several important cell biological processes during cortex development. Extracelluar matrix (ECM) proteins are important for NPC self-renewal (18). We analyzed the expression of genes involved in ECM production and sensing in cells from the fetal tissue and organoids (Fig. S6A). We find that fetal APs express collagens (COL11A1, COL4A5), numerous glycoproteins implicated in ECM signaling (SDC4, LAMA1/5, BCAN), integrin receptors (ITGA6, ITGB8), and glypican coreceptors (GPC4/6). Most of these genes (16/18, 89%) have similar AP-specific expression patterns in organoid cells. Similarly, we found strongly positive correlations between fetal and organoid cell types for genes involved in transcription regulation (10/11; 90%), RG delamination (12/15, 80%), Notch/Delta signaling (7/10, 70%), and neurite outgrowth (24/25; 96%) (Fig. 4 D and E and Fig. S6 B–D).
Fig. S6.
Cerebral organoids recapitulate gene expression programs underlying cerebral cortex cell biology. (A) APs express genes involved in ECM production and sensing. The heat map shows expression for each ECM gene for organoid and fetal tissue cells in the order of the monocle lineage. For each gene, the correlation between organoid and fetal tissue lineages is plotted to the right of the organoid heat map. Note that the organoid dataset has proportionally more progenitors than the fetal tissue dataset, whereas the fetal tissue has more neurons. (B–D) A similar comparison as in A is shown for genes involved in aRG delamination (B), Delta/Notch signaling (C), and neurite outgrowth (D).
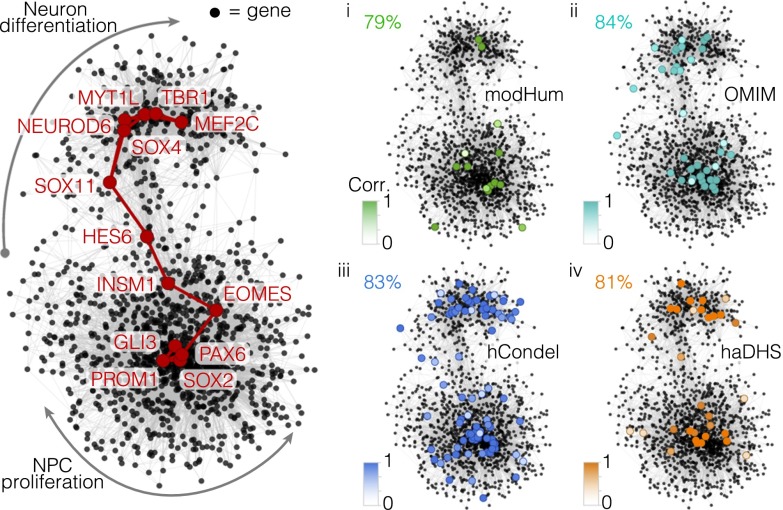
We next collected genes that (i) encode proteins with fixed amino acid changes in modern humans since divergence with Neanderthals (modHuman) (20), (ii) are mutated in human genetic disorders affecting neurogenesis (OMIM; omim.org/), (iii) are located nearby evolutionarily conserved sequences that have been specifically lost in the human lineage (hCondel) (21), and (iv) are nearby human-accelerated regions overlapping brain-accessible chromatin (haDHS) (22) (Dataset S4). We projected these changes onto a gene regulatory network inferred from correlations with the TFs that control the AP–BP–neuron lineages in the fetal cortex (Fig. 5). The expression of the majority of these genes (average, 82.5%) that may carry changes relevant to human cerebral cortex development and evolution was positively correlated between fetal and organoid cell types, suggesting these changes may be faithfully modeled in cerebral organoids.
Fig. 5.
Genomic scans of disease, evolutionary, and chromatin signatures highlight genetic aspects of human corticogenesis that can be modeled in vitro. Shown is the covariation network using genes that have high correlation (>0.3) with TFs controlling the AP–BP–neuron lineage from Fig. 2C. Select TF nodes are highlighted to delineate the path. (i–iv) Panels show genes that have amino acid changes that are modHum (i, green), OMIM (ii, turquoise), hCondel (iii, blue), or haDHS (iv, orange). The percentage of cells that have a positive correlation (>0.4) between fetal and organoid cells is shown, with nodes colored based on the correlation coefficient (Dataset S4).
Differences Between Organoid and Fetal Cerebral Cortex.
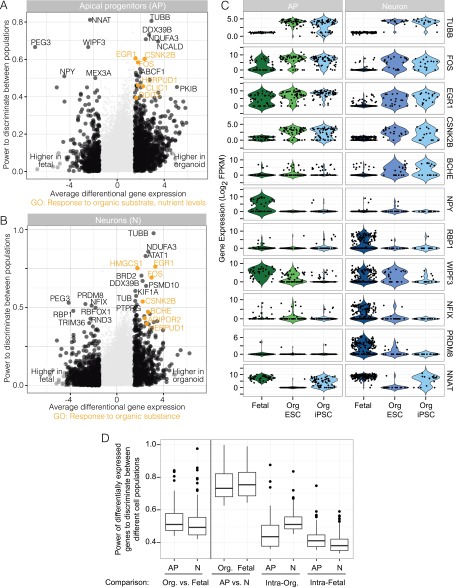
Finally, we searched for differences in gene expression between fetal and organoid APs (AP1 and AP2 combined) and neurons (N1, N2, and N3 combined). We excluded BPs from this analysis because of the small number of BPs in the organoid dataset. We combined fold difference and the median receiver operating characteristic (ROC) test (23) to identify genes that had a high average difference (>threefold) and power to discriminate (97th percentile) between fetal and organoid cells (Dataset S4 and Fig. S7). Among genes up-regulated in the organoid, the top GO category enriched was “response to organic substance,” which contained genes FOS, EGR1, CSNK2B, HMGCS1, BCHE, HERPUD1, CLIC1, and ADIPOR2. The immediate early genes FOS and EGR1 are Notch signaling targets recently reported to be expressed in human but not mouse radial glia (24). Among the genes down-regulated in organoid neurons was a transporter for vitamin A (RBP1), which might reflect a response to the inclusion of vitamin A in the culture medium. TUBB (tubulin, beta class I), a structural component of microtubules, was the most differentially expressed gene with higher expression in both progenitors and neurons in organoids. Other potentially relevant differences include PRDM8, which has a role in assembling neuronal circuits in upper layer cortical neurons (25), and NFIX, which is involved in progenitor cell differentiation (26), both of which have higher expression in fetal neuronal cells than in organoid cells. Most of these genes differed in both ESC- and iPSC-derived organoids relative to the fetal tissue. Differences between fetal and organoid cells are significant compared with differences between randomly selected fetal APs (or neurons), however <5% of differentially expressed genes between fetal and organoid cells reach the average classification powers seen between APs and neurons. We conclude that the major gene expression differences between organoid and fetal tissues seem to be a response to tissue culture environment and may not reflect fundamental differences in differentiation programs.
Fig. S7.
Gene expression differences between fetal and organoid cell types. (A and B) Volcano plots visualizing differential gene expression analysis between organoid and fetal APs (A) and organoid and fetal neurons (Ns; B). For each gene, the average difference between cells from each comparison is plotted against the power to discriminate between groups. Top-scoring genes for both metrics are labeled and genes in the top GO enrichment category “Response to organic substance” are highlighted in orange (Dataset S4). (C) Violin plots for some of the most differentially expressed genes show the distribution of gene expression across APs and neurons from fetal, ESC-derived organoid, and iPSC-derived organoid dorsal cortex. (D) Box plots show the distributions of the power of differentially expressed genes to discriminate between different cell populations. Shown in the two left-most box plots are comparisons between APs in organoid versus fetal tissue and neurons in organoid versus fetal tissue. As a reference, box plots are shown for comparisons between iPSC-derived and ESC-derived organoid APs and neurons, respectively. In addition, comparisons are shown for two randomly selected subgroups within fetal APs or fetal neurons, respectively.
SI Results
Gene abbreviations used in the manuscript are as follows: ASPM, abnormal spindle-like microcephaly-associated protein; BCL11B, B-cell CLL/lymphoma 11B; DLX, distal-less homeobox 6; EOMES, Eomesodermin; ERBB4, Erb-b2 receptor tyrosine kinase 4; FOXG1, forkhead box G1; GAD1, glutamate decarboxylase 1; GLI3, GLI family zink finger 3; HES1, hairy and enhancer of split-1; INSM1, insulinoma-associated 1; MKI67, marker of proliferation Ki-67; MYT1L, myelin transcription factor 1-like; NEUROD4, neuronal differentiation 4; NFIA, nuclear factor I/A; NFIB, nuclear factor I/B ; OTX2, orthodenticle homeobox 2; PAX6, paired box 6; PECAM1, platelet endothelial cell adhesion molecule 1; PROM1, prominin 1; SOX2, sex-determining region Y-box 2; TBR1, T-box brain; TUBB, tubulin beta class I; TUBB3, tubulin beta-3 chain; TUJ1, neuron-specific class III beta-tubulin; VIM, vimentin; WNT2B, wingless-type MMTV integration site family member 2B.
Cell Lineage Network Based on Pairwise Correlations Between Fetal Neocortical Cells.
Although Monocle uncovered a path linking APs to neurons through EOMES-expressing BPs, we expect that the true relationship between NPCs and neurons in the fetal cortex cannot be fully explained by a single path (3, 4). Indeed, a side branch from the minimal spanning tree indicated a link between APs in the VZ and cells expressing neuronal markers such as MYT1L yet lacking EOMES expression, suggesting the presence of rare alternate paths to a neuronal fate, such as direct neurogenesis. Also, the lineage revealed by Monocle does not account for proliferative or self-renewing cell divisions within the progenitor pools. Therefore, we applied an adjacency network based on pairwise correlations between cells to the same gene set used in Monocle. This graph’s topology revealed intercellular connections that reflect known modes of neurogenesis in the cerebral cortex (Fig. S2A). Specifically, most VZ APs directly connect with EOMES-expressing BPs, reflecting indirect neurogenesis via BPs. However, it also revealed a minority of AP-to-neuron connections that did not appear to transition through EOMES-positive BPs, potentially reflecting rare instances of direct neurogenesis. In addition, this network graph revealed bidirectional relationships between the APs in the VZ and BPs in the SVZ, consistent with proliferative and self-renewing modes of cell division of radial glial types. Finally, the network showed that the differentiation pathways of AP- and BP-derived newborn neurons coalesce to relatively homogeneous CP neurons.
Deconstructing Cell Composition in Cerebral Organoids.
We identified several NPC clusters (c1, c2, c5, and c6) that share high expression of genes involved in cell-cycle regulation but express different combinations of cell-type or regional marker genes. NPC clusters c5 and c6 are composed of cells from the earliest time points (33–37 d) and express NPC markers such as VIM, PAX6, and NES. In addition, these clusters show heterogeneous expression of genes involved in regional patterning of the developing telencephalon such as FOXG1 (enriched in mouse dorsal and ventral telencephalon and previously used to identify organoid forebrain regions) and OTX2 (enriched in ventral telencephalon and hippocampus and previously used to identify organoid forebrain/midbrain boundaries). These clusters lack expression of genes that we observed to be expressed in nearly all cells of the fetal cortex, such as NFIA and NFIB. In addition, some of these early NPCs also express the pluripotency marker LIN28A. Therefore, cells in these clusters are likely a mixture of ventral telencephalic or hippocampal NPCs and NPCs from immature dorsal telencephalic regions. In contrast, some NPCs from the later time points (41–65 d, cluster c1 and c2) highly express NPC markers PAX6, VIM, and NES, as well as genes expressed in nearly all fetal cortex cells such as FOXG1, NFIA, and NFIB, but do not express OTX2, suggesting these NPCs derive from cortical regions resembling the fetal cerebral cortex.
We identified two clusters of cells (c4 and c7) that shared high expression of certain neuronal markers such as MYT1L, DCX, NRXN1, TBR1, and MAP2 and were anticorrelated with genes highly expressed in NPCs (Fig. 3 and Fig. S4). Notably, the proportion of neuronal cells relative to NPCs increased over time in the organoids, an observation we validated by immunofluorescence (Fig. S3). Cluster c4 cells express FOXG1, NFIA, NFIB, and NEUROD6, suggesting that they are neurons in organoid regions resembling the fetal cortex (Fig. 3 and Fig. S4). Cells in cluster c7 instead express LHX9, EBF1, ZFHX3, ZFHX4, and/or SOX14, which are highly expressed in other fetal forebrain structures such as the striatum, hippocampus, and thalamus (Fig. S4). Notably, we found four cells from cluster c7 that coexpress GAD1, GAD2, SOX14, and ERBB4, suggesting these are interneuron-like cells derived from the ventral forebrain-like regions in the organoid. These data suggest that neurons within the organoid may be derived from germinal zones with identities reminiscent of different forebrain regions. To address this, we analyzed cellular relationships in each of the microdissected cortical regions. Although each region contained NPCs and neurons, cells from three of the four regions were FOXG1-positive, were OTX2-negative, and expressed cerebral cortex markers including NFIA, NFIB, PAX6, and NEUROD6 (Fig. 3F and Fig. S4D). In contrast, the fourth cortical region did not express FOXG1 or other cerebral cortex markers but instead expressed OTX2. Cells of the first three regions are contained within the dorsal cortex clusters from the t-SNE analysis (c1, c2, c3, and c4; Fig. 3), whereas cells from the fourth region are within ventral forebrain clusters (c5, c6, and c7). Thus, a single cerebral organoid contains cortical regions with dorsal as well as ventral forebrain identities, which we could discriminate due to distinct NPC and neuron populations.
We also find clusters with cycling (c8) and noncycling cells (c9) that express RSPO1 ,2, and 3, which may demarcate the boundary between dorsal and ventral telencephalon (Fig. 3 and Fig. S3). Further, we identified cycling and noncycling mesenchymal cells (c10 and c11) that express DCN, LUM, COL1A2, COL5A1, and many components of the ECM, suggesting these cells form connective tissue within the organoid (Fig. 3 and Fig. S3). When we stained cerebral organoids with an antibody against COL5A1, we observed expression surrounding the periphery of cortical-like regions (Fig. S3G). By electron microscopy, we observed collagen fibrils amid a thick ECM surrounding the cortex periphery. This suggests that the mesenchymal cells may form meninges-like connective tissue.
Similarities in Neurogenic Lineages Between Organoids and Fetal Tissue.
We analyzed a genetic pathway that controls RG delamination (30) (Fig. S6). Specifically, NEUROG2 up-regulation inhibits SOX2 and induces expression of INSM1 and EOMES, leading to neuronal differentiation. In parallel, NEUROG2 induces expression of FOXP2/4, which inhibits N-cadherin (CDH2) expression, leading to apical process detachment. Our fetal cortex data recapitulate a negative correlation between SOX2 and NEUROG2 expression and subsequent up-regulation of EOMES, INSM1, FOXP2, and FOXP4 transiently in cells directly adjacent to APs on the lineage path. However, we found that CDH2 is relatively highly expressed along the entire lineage, with only a slight decrease in expression during the transition from AP to neuron in the fetal cells. In contrast, cadherins CDH10, CDH12, CDH23, and CELSR1 are expressed in APs but more strongly decrease upon the transition to EOMES-expressing BPs. We also noted that a distinct set of cadherins (CDH6, CDH7, and CDH13) were up-regulated in CP neurons, confirming a dynamic pattern of calcium-dependent adherens junction formation during neurogenesis. The expression of most of these genes (12/15 genes, 80%) are positively correlated between corresponding fetal and organoid cell types. In addition, we found a strongly positive correlation between fetal and organoid cell types for genes involved in Notch/Delta signaling (7/10, 70%) and neurite outgrowth (24/25; 96%) (Fig. S6 C and D).
Discussion
Single-cell gene expression analysis is a powerful technique to deconstruct tissue heterogeneity and has recently been used to characterize NPC and neurons in the fetal brain (24, 27–29). Here we applied scRNA-seq to compare cell composition and lineage relationships in fetal and organoid cerebral cortex. We find that over 80% of genes implicated in neocortex disease or evolution and are differentially expressed along the fetal cortex lineage have similar expression profiles in organoid and fetal cerebral cortex. Organoid cells thus use similar sets of genes as their fetal counterparts to perform cortical processes such as NPC proliferation and self-renewal, production of ECM, migration, adherence, delamination, and differentiation that result in structured cerebral tissue. This has important implications for using the organoid system to model developmentally and evolutionarily relevant neocortical cell biology.
Our sampling of cells over development was relatively modest, and future higher throughput studies over an expanded time course are required. A study published while our manuscript was in review used scRNA-seq to survey the human cerebral cortex at gestational week 16–18 and reported the identification of a bRG (aka oRG) gene expression signature (29). Most of these bRG marker genes are expressed at 12–13 wpc in cells that we define as APs that highly correlate with VZ and purified aRG bulk RNA-seq data. This is consistent with the published results that the bRG signature emerges from the VZ after gestational week 13.5.
Many of the genes that differed in expression between fetal and organoid cells had relatively low expression in fetal tissue, suggesting that the differential expression observed may be due to noise in single-cell transcriptomics, whereas others seem to reflect responses to factors in the cerebral organoid culture media. One notable difference between fetal and organoid tissue was that the organoids had fewer BPs relative to APs. This might be explained by developmental time point or an underdeveloped SVZ intrinsic to the culture system. Future work will be required to understand if these differences have any bearing on corticogenesis and if culture conditions can be modified to further improve how organoid cultures reflect in vivo cerebral cortex development.
Methods
Research involving human tissue and human embryonic stem cells was approved by the Institutional Review Board of the Max Planck Institute of Molecular Cell Biology and Genetics, composed of the following people: Jussi Helppi (Chair, Head of the Animal Facility), Barbara Langen (designated veterinarian and animal welfare officer), Ronald Naumann (Head of the Transgenic Core Facility), Anke Münch-Wuttke (Animal Care Expert), and Ivan Baines (Chief Operating Officer, Max Planck Institute of Molecular Cell Biology and Genetics). In addition, the Universitätsklinikum Carl Gustav Carus of the Technische Universität Dresden Ethical Review Committee approved the research with human fetal tissue (see SI Methods, Dissociating fetal tissue).
Human fetal brain tissue (12–13 wpc) was obtained with ethical approval following elective pregnancy termination and informed written maternal consents. ESC and iPSC lines were grown in mTESR1 (Stem Cell Technologies) using standard protocols. Cerebral organoids were generated as described (10, 11), with the exception that mTESR1 was used during embryoid body (EB) formation. For immunohistochemistry and electron microscopy of organoids, see SI Methods. Fetal cortices were processed as described in ref. 19. Whole cerebral organoids and microdisseccted regions were dissociated in 2 mL of Accutase (StemPro) containing 0.2 U/µL Dnase I (Roche) for 45 min. Dissociated fetal and organoid cells were filtered through 40-, 30-, and 20-μm-diameter strainers to create a single-cell suspension. Cell viability (90–95%) was assessed using Trypan blue staining. Single-cell capture, lysis, and cDNA synthesis were performed with the Fluidigm C1 system using the SMARTer Ultra Low RNA Kit for Illumina (Clontech). cDNA size distribution was assessed by high-throughput capillary gelelectrophoresis (Advanced Analytical). Sequencing libraries were constructed in 96-well plates using the Illumina Nextera XT DNA Sample Preparation Kit (15). Up to 96 single-cell libraries were pooled, and each cell was sequenced 100 bp paired-end on Illumina HiSEq. 2500 to a depth of 2–5 million reads. A detailed description of methods and scRNAseq analyses is provided in SI Methods.
SI Methods
Cerebral Organoid Culture, Electron Microscopy, and Immunohistochemistry.
Organoid culture.
For the majority of the data, we used an iPSC line (PS409b2) (31), which we found to be competent to generate cerebral organoids with characteristic cortical zones (Fig. 3 and Fig. S3). In addition, two of the microdissected cortical regions (53 d, r1,and r2) were from an organoid derived from human embryonic stem cells (32) (Fig. 3). ESC and iPSC lines were passaged and maintained using standard protocols and grown in mTESR1 (Stem Cell Technologies). Cerebral organoids were generated using the same protocol as described (10, 11), with the exception that mTESR1 was used during EB formation.
Electron microscopy.
For conventional transmission electron microscopy, organoids were transferred into 2% (vol/vol) PFA and 1% (vol/vol) glutaraldehyde in 0.1 M Pipes, pH 7.2, containing 0.1 mM CaCl2 and fixed at least overnight. Aldehyde-fixed organoids were postfixed in 1% osmium tetroxide containing 1.5% potassium ferrocyanide for 1 h at room temperature (RT), stained with 0.4% tannic acid for 5 min, and subsequently contrasted with 0.5% uranyl acetate, dehydrated, and flat-embedded in Epon replacement (C.Roth). Semithin (1 µm) plastic sections through the embedded organoids were cut on a microtome (UCT6, Leica) and collected on glass slides, to be stained with toluidine blue O (0.5% in 1% sodium tetraborate solution) for judging when a ventricle has been reached. Ultrathin (70 nm) sections were cut through the ventricular level and poststained with uranyl acetate and lead citrate according to standard protocols. Images were taken on a Morgagni EM at 80 kV (FEI) with a Morada camera (Olympus) and ITEM software (Olympus).
Immunohistochemistry.
Organoids were fixed on ice for 20 min using 4% (vol/vol) PFA, equilibrated in 30% (vol/vol) sucrose at 4 °C overnight, mounted in optimal cutting temperature compound, and cryosectioned (10 μm thickness). The cryosections were rehydrated with 1× PBS, permeabilized with 0.3% TritonX 100 for 15 min, quenched with 0.1 M Glycine pH 7.4 for 15 min at RT, and washed with blocking solution containing 0.3% Triton X-100 and 0.2% gelatin in a 3:7 mixture of 1 M NaCl and PBS. The primary antibodies were incubated in the blocking solution overnight at 4 °C, washed with the same blocking solution, incubated with the secondary antibody in the blocking solution for 1 h at RT, washed with 1× PBS, and mounted using ProLong Gold. The following primary antibodies were used: rabbit anti-PAX6 (PRB-278P; Covance), chicken anti-TBR2 (AB15894; Millipore), rabbit anti-KI67 (ab15580; Abcam), goat anti-DCX (sc-8066; Santa Cruz), and rabbit anti- COLV (ab7046; Abcam). The secondary antibodies used were all donkey-derived Alexa antibodies from Life Technologies (488, 555, and 647).
ScRNA-Seq of Human Cortical Tissue and Cerebral Organoids.
Dissociating organoids.
Whole cerebral organoids were washed three times in 1× PBS and dissociated in 2 mL of Accutase (StemPro) containing 0.2 U/µL Dnase I (Roche) for ∼45 min, followed by serial filtering through 40-, 30-, and 20-μm-diameter strainers to create a single-cell suspension. Cells were collected by centrifugation at 300 × g for 5 min and resuspended in 500 μL of organoid culture media (11). Cell viability was assessed by Trypan blue staining (typically 85–95% viable) and counted using an automatic cell counter (Countess, ThermoFisher). For the single cortical region experiments, organoids were embedded in 4% low-melting-point agarose (Sigma) dissolved in Dulbecco’s PBS (Life Technologies) and sectioned at 4 °C to 150 μm thickness using a vibratome (Camden Instruments). Individual regions surrounding ventricles were then microdissected under a stereomicroscope and dissociated into single-cell suspension as described above using 200 μL of Accutase including DNase I (0.2 U/µL). Cells were collected at 300 × g for 5 min, resuspended in 15 μL of organoid culture media, and viable cells were counted as described above.
Dissociating fetal tissue.
Human fetal brain tissue was obtained from the Klinik und Poliklinik für Frauenheilkunde und Geburtshilfe, Universitätsklinikum Carl Gustav Carus of the Technische Universität Dresden, following elective pregnancy termination and informed written maternal consents and with approval of the local University Hospital Ethical Review Committees. Fetuses were 12–13 wpc as assessed by ultrasound measurements of crown-rump length and other standard criteria of developmental stage determination. Human fetuses were placed on ice immediately after abortion, and neocortices were dissected in ice-cold Tyrode’s Solution. Neocortices were microdissected under a stereomicroscope, and single-cell suspensions were produced using the MACS Neural Tissue Dissociation Kit according to the manufacturer’s instructions and as described (19). Briefly, cortices were digested with papain (Miltenyi Biotec, Enzyme Mix 1, 1 mL final volume) for 15 min at 37 °C on a rotating wheel, followed by addition of a papain inhibitor (Enzyme Mix 2, 15 μL final volume). Samples were then dissociated by ∼10 gentle triturations through a 1-mL pipette tip to obtain a single-cell suspension. Cells were collected by centrifugation at 300 × g for 3 min and resuspended in 600 μL of Tyrode’s Solution. Cell viability was assessed using Trypan blue staining and typically was 90–95%.
Capturing of single cells and preparation of cDNA.
Single cells were captured on a medium (10–17 µm cell diameter) or small (5–10 µm cell diameter) microfluidic chip for mRNA-seq (Fluidigm) using the Fluidigm C1 system. Cells were loaded onto the chip at a concentration of 400–500 cells/µL and imaged by phase contrast to assess the number of cells per capture site. cDNAs were prepared on chip using the SMARTer Ultra Low RNA Kit for Illumina (Clontech). External RNA Controls Consortium RNA Spike-In Mix (Ambion, Life Technologies) was added to the lysis reaction and processed in parallel to cellular mRNA.
RNA-seq library construction and cDNA sequencing.
Size distribution and concentration of single-cell cDNA was assessed by high-throughput capillary gelelectrophoresis (Fragment Analyzer, Advanced Analytical). Sequencing libraries were constructed in 96-well plates using the Illumina Nextera XT DNA Sample Preparation Kit according to the protocol supplied by Fluidigm and as described previously (15). Libraries were quantified by Agilent Bioanalyzer using High Sensitivity DNA Analysis Kit as well as fluorometrically using Qubit dsDNA HS Assay Kits and a Qubit 2.0 Fluorometer (Invitrogen, Life Technologies). Up to 96 single-cell libraries were pooled, and each cell was sequenced 100 bp paired-end on Illumina HiSEq. 2500 to a depth of 2–5 million reads. Base-calling, adaptor trimming, and de-multiplexing were performed as described (33, 34).
Processing, analysis, and graphic display of ScRNA-seq Data.
Raw reads were processed using a custom script and aligned to a Bowtie2 (35) indexed human genome (grch38 sourced from ENSEMBL) using TopHat (36) with default settings. Transcript levels were quantified as fragments per kilobase of mapped reads (FPKM) generated by Cufflinks (37) using gencode protein coding genes (grch38 v22 Havana). We excluded cells that did not express either of the two housekeeping genes ACTB and GAPDH. Transcript levels were converted to the log-space by taking the log2 (FPKM). R studio (https://www.rstudio.com/) was used to run custom R scripts to perform PCA (FactoMineR package) and hierarchical clustering (stats package) and to construct heat maps, correlation plots, scatter plots, violin plots, dendrograms, bar graphs, and histograms. Generally, ggplot2 and gplots packages were used to generate data graphs. The Seurat package (23) implemented in R was used to identify organoid cell clusters, and the BackSPIN algorithm was used to analyze organoid neuronal heterogeneity (16). The Monocle package (17) was used to analyze cell lineage relationships. Covariance network analysis and visualizations were done using igraph implemented in R (igraph.sf.net). Gene ontology enrichment analyses were performed using DAVID Informatics Resources 6.7 (38). A list of TFs was obtained from the online animal TF database, www.bioguo.org/AnimalTFDB/ (39).
Methods for Fig. 1: Deconstructing Cell-Type Heterogeneity in the Fetal Human Neocortex.
The endothelial cell (identified by PCA; Fig. S1) and the five interneurons (identified using a set of marker genes; Fig. S1) from the fetal cortex dataset were removed for the analyses presented in this figure.
For Fig. 1B, we calculated for each fetal cortex cell the Spearman correlation of its transcriptome (all genes) with bulk transcriptome data from each of four microdissected cortical zones (VZ, iSVZ, oSVZ, and CP; mean expression value of each gene across four replicates) (18) or three purified cell types (aRG, bRG, and neuron) (19). Both bulk RNAseq datasets were acquired from fetal tissue at 13 wpc. We hierarchically clustered (Pearson’s correlation distance metric) cells based on their correlation coefficient with germinal zones or cell types and visualized the clustering in a heat map showing correlation coefficients scaled across zones or cell types (mean-centering and dividing by SD). The scaling enables a better comparison between cells, as the maximum and minimum correlations for each cell are color-coded in the same way after scaling. We used this analysis to identify the zone or purified cell type with which each individual cell had a maximum correlation.
For Fig. 1C, we performed PCA on variable genes (variance > 0.5) expressed (>1 FPKM) in more than two cells. We extracted the genes correlating and anticorrelating with PC1–3, using an absolute PC loading threshold >0.2 with a maximum of 50 genes per PC to avoid individual PCs swamping the analysis, resulting in 265 genes. In addition, we performed PCA on NPCs (identified in Fig. S1) separately, and this identified a new set of genes (PC1–2 correlating and PC2 anticorrelating) describing a subpopulation of NPCs that did not become apparent when performing PCA on all cells. We hierarchically clustered cells based on the expression of these genes (316 genes in total) using Pearson correlation and plotted the genes in the order of correlation with each PC. We analyzed a set of known marker genes for RGs, IPs, and neurons in the same cell order as revealed by clustering of cells based on all PCA genes and used the marker genes to determine the positions in the tree to cut to define and name each cluster. This analysis resulted in the seven clusters reported in Fig. 1 (AP1, AP2, BP1, BP2, N1, N2, and N3). The specificity of the grouping was assessed using permutation analysis. The sum of squares within groups (SSW) was calculated for our chosen cell grouping as well as for 1,000 random permutations thereof, keeping the size of cell groups and the total number of groups constant. The SSWs for all 1,000 permutations were normally distributed, and the SSW for our chosen clustering was significantly lower than all other permutations (P value < 1 × 10–120).
Methods for Fig. 2: Reconstructing Lineage Relationships in the Fetal Cerebral Cortex.
The endothelial cell and the five interneurons from the fetal cortex dataset were removed from the analyses presented in this figure.
For Fig. 2 A and B, we used Monocle (17) (default settings) to perform dimensionality reduction by means of independent component analysis and to draw a minimal spanning tree connecting the 220 fetal cortical cells into a lineage. As input into Monocle, we used the expression levels of the PCA-derived genes used in Fig. 1C. Individual cells were color-coded based on the unbiased cell-type assignment (Fig. 1C), the zone assignment based on maximum correlation with bulk RNAseq data of microdissected cortical zones (Fig. 1B), or the expression level of representative marker genes.
For Fig. 2C, for the TF network analysis, we computed a pairwise correlation matrix for TFs annotated as such in the “Animal Transcription Factor Database” (www.bioguo.org/AnimalTFDB/) (39) and identified those TFs with a correlation of greater than 0.3 with at least three other TFs (99 TFs). We used a permutation approach to determine the probability of finding TFs meeting this threshold by chance. We randomly shuffled the columns (TFs) of each row (cells) 500 times and calculated the pairwise correlation matrix for each permutation of the input data frame. After each permutation, we counted the number of TFs meeting our threshold. The majority of randomized data frames (96%) resulted in 0 TFs that met our threshold. The maximum number of TFs that met our threshold was 2, which occurred in only 0.2% of the permutations. In contrast, our data resulted in 99 TFs that met this threshold, which suggests that our threshold was strict, but all nodes and connections that we present in the TF network are highly unlikely to be by chance. We used the pairwise correlation matrix for the selected TFs as input into the function graph.adjacency() of igraph implemented in R (igraph.sf.net) to generate a weighted network graph, in which the selected TFs are presented as vertices and all pairwise correlations >0.2 are presented as edges linking the respective vertices. The network graph was visualized using the fruchterman reingold layout. TF vertices were manually color-coded based on the expression pattern along the monocle lineage. Green, teal, and blue represent highest average expression in APs, BPs, and neurons, respectively.
Methods for Fig. 3: Dissecting Cerebral Organoid Composition Using ScRNA-Seq.
For Fig. 3D, the algorithm used to cluster organoid cells was implemented and performed using Seurat. We performed PCA to find genes that best described the heterogeneity in the data. To identify significant PCs, we performed 200 random samplings, each time randomly permuting 1% of the genes. This returned a P value for each gene in each PC based on how likely the gene/PC score would have been observed by chance. Based on this analysis, we identified highly significant PCs (P value < 1e-20) and selected genes from each PC with a P value cutoff < 1e-3 with a maximum of 50 genes per PC. We performed t-SNE to cluster the cells based on each gene-associated PC score using the perplexity parameter G = 5. We constructed a phylogenetic tree to identify relationships between the clusters and collapsed adjacent branches where we could not identify robust differences in gene expression. We used Seurat’s implementation of the ROC test (23) to perform differential gene expression analysis between the adjacent clusters that returns classification powers and fold-change differences. This analysis resulted in the 11 clusters reported in Fig. 3.
Methods for Fig. 4: Similar Genetic Programs Characterize Lineage Progression in the Organoid and Fetal Cerebral Cortex.
In this figure, the 157 organoid cells were included that were found in Fig. 3 to originate from dorsal cortex regions of the organoid (clusters c1, c2, c3, and c4).
For Fig. 4A, lineage analysis on the 157 organoid cerebral cortex cells was performed using the Monocle package, similar to the lineage analysis on fetal cells in Fig. 2A. To determine the gene set used as input into Monocle, we first performed PCA on variable genes (variance > 0.5) expressed (>1 FPKM) in at least two cells. We extracted the genes correlating and anticorrelating with PC1–3, using an absolute PC loading threshold >0.2 with a maximum of 50 genes per PC to avoid individual PCs swamping the analysis. A total of 266 genes resulted from this analysis and were used as input into Monocle to perform independent component analysis and to draw a minimal spanning tree connecting the 157 organoid cerebral cortex cells into a lineage. Next, we calculated the pairwise correlation between organoid and fetal cerebral cortex cells and assigned to each organoid cell the type of the fetal cell with which it most strongly correlated. We used this cell-type information to color-code the single cells.
For Fig. 4B, the same minimal spanning tree graph is shown as in Fig. 4A. Cells are color-coded based on the maximum correlation with bulk RNAseq data of microdissected cortical zones or the expression level of representative marker genes.
For Fig. 4 C and D, to compare the expression profiles of the selected TFs in fetal and organoid cells, we calculated the mean expression of each TF across the cells belonging to the same cell type [AP (AP1 + AP2), BP (BP1 + BP2), N1, N2, and N3] for fetal and organoid data and visualized the correlation between mean expression levels in fetal and organoid cell types using biplots (Fig. 4C) and heat map (Fig. 4D).
For Fig. 4E, for the dendrogram, we combined fetal and organoid cerebral cortex cells and performed PCA. We then used the top 50 genes correlating and anticorrelating with PC1 to draw a dendrogram of all cells with Spearman correlation as the distance metric. PC1-correlating genes were found to define NPCs, whereas PC1-anticorrelating genes defined neurons. Organoid and fetal single neurons as well as NPCs intermixed well in this dendrogram. To construct a combined fetal and organoid cellular network, we computed a pairwise correlation matrix for all organoid and fetal cerebral cortex cells and using genes discovered in the PCA of fetal tissue. We then generated a weighted adjacency network graph using the graph.adjacency() command and visualized cells as vertices connected to other cells via edges if the pairwise correlation between two cells was higher than 0.45. The fruchterman reingold layout was used to plot the network graph.
Methods for Fig. 5: Genomic Scans of Disease, Evolutionary, and Chromatin Signatures Highlight Genetic Aspects of Human Corticogenesis That Can Be Modeled in Vitro.
We expanded the TF network to non-TF genes by identifying genes that correlate with at least one TF shown in Fig. 2C with a Pearson correlation coefficient greater than 0.3. The network was drawn using igraph using the fruchterman reingold layout, and edges were shown that linked two or fewer vertices. We assessed the likelihood of obtaining a correlation coefficient of >0.3 between a TF and another gene by chance by calculating the correlation between the 99 selected TFs and all other variable genes for our data as well as randomly permuted data. We then calculated the number of genes that had a correlation coefficient of >0.3 with at least one TF. This analysis revealed that on average about 1% of the identified genes could have met the correlation threshold by chance.
For Fig. 5 i–iv, we collected genes that (i) encode proteins with fixed amino acid changes in modHum (20), (ii) are OMIM (omim.org/), (iii) are hCondel (21), or (iv) are haDHS (22) (Fig. 5 and Dataset S4). Genomic Regions Enrichment of Annotations Tool (GREAT) regulatory domains were used to link regulatory DNA to nearby genes as described (21). We calculated the correlation for each gene’s average expression pattern in organoid and fetal APs, N1s, N2s, and N3s similar to what is described in Fig. 4. The majority of these genes (average, 81.2%) had a correlation greater than 0.5 between organoid and fetal cells. We projected the changes onto the regulatory network described in Fig. 4A and color-coded each vertex using the correlation between fetal and organoid cells.
Methods for Fig. S2: Gene Expression Signatures Controlling Fetal Corticogenesis.
For Fig. S2A, to construct a cellular network, we computed a pairwise correlation matrix for all 220 fetal cortical cells using all PCA genes shown in Fig. 1C. We then generated a weighted adjacency network using the graph.adjacency() command and visualized cells as vertices connected to other cells via edges if the pairwise correlation between the two cells was higher than 0.45. The fruchterman reingold layout was used to plot the network graph. Vertices (cells) were color-coded based on the unbiased cell-type assignment (Fig. 1C) or the zone assignment based on maximum correlation with bulk RNAseq data of microdissected cortical zones (Fig. 1B).
Methods for Fig. S3: Characterizing Cerebral Organoid Cell Composition.
For Fig. S3H, for each of the 11 clusters from the t-SNE analysis presented in Fig. 3D, we searched for marker genes using Seurat’s implementation of the ROC test (23) by comparing the cells of a given cluster with all other cells. We used the10 genes with the highest classification power (Dataset S2) as marker genes for each cluster and plotted their expression profile across all organoid cells in the heat map presented in this panel.
Methods for Fig. S4: Characterizing Organoid Neuron Heterogeneity.
We performed PCA on variable genes (variance > 1) expressed (>1 FPKM) in more than three cells. We extracted the genes correlating and anticorrelating with PC1–2, using an absolute PC loading threshold >0.2 with a maximum of 25 genes per PC. We used the backSPIN algorithm to define cell groups by simultaneous cluster genes and cells using default settings. We then visualized the cell relationships by clustering cells using Euclidean distance on the first four levels (level 0, level 1, level 2, and level 3) identified by backSPIN, which resulted in the seven cell groups shown in Fig. S4A. Based on OTX2, GAD1, GAD2, ERBB4, and SOX14 expression, we concluded that the subgroup in cluster 2b is likely interneuron-like cells born in a ventral forebrain region.
Methods for Fig. S5: Extended Data Showing Similar Gene Expression Profiles Characterize Lineage Progression in Organoid and Fetal Cerebral Cortex.
For Fig. S5A, we used the same approach as applied to fetal cerebral cortex cells in Fig. 1B to determine the maximum correlation of each single organoid cerebral cortex cell transcriptome with the bulk transcriptome of microdissected fetal cortical zones or of FACS-purified cell types. Briefly, we calculated for each organoid cerebral cortex cell the Spearman correlation of its transcriptome (all genes) with bulk transcriptome data from each of four microdissected cortical zones (VZ, iSVZ, oSVZ, and CP; mean expression value of each gene across four replicates) (18) or three purified cell types (aRG, bRG, and neuron) (19). Both bulk RNAseq datasets were acquired from fetal tissue at 13 wpc. We hierarchically clustered (Pearson’s correlation distance metric) cells based on their correlation coefficient with germinal zones or cell types and visualized the clustering in a heat map showing correlation coefficients scaled across zones or cell types (mean-centering and dividing by SD). The scaling enables a better comparison between cells, as the maximum and minimum correlation for each cell is color-coded in the same way after scaling. We used this analysis to identify the zone or purified cell type with which each individual cell had a maximum correlation.
For Fig. S5B, to construct a cellular network, we computed a pairwise correlation matrix for all 157 organoid cerebral cortex cells using all PCA genes. We then generated a weighted adjacency network graph using the graph.adjacency() command and visualized cells as vertices connected to other cells via edges if the pairwise correlation between two cells was higher than 0.45. The fruchterman reingold layout was used to plot the network graph. Vertices (cells) were color-coded based on the zone assignment based on maximum correlation with bulk RNAseq data of microdissected cortical zones (Fig. S5A) or based on the cell-type assignment.
For Fig. S5C, we ordered the cells based on their pseudotime along the Monocle lineage and plotted the transcript levels of the PCA-derived genes in all cells in a heat map. We performed GO enrichment analysis for each of the six gene groups (Fig. S5 A–F, 50 genes per group) using DAVID.
For Fig. S5D, we used the list of TFs describing the fetal neocortex TF covariance network (Fig. 2C) to construct a TF network for the organoid cerebral cortex cells. We therefore used the pairwise correlation matrix for the selected TFs as input into the function graph.adjacency() of igraph implemented in R (igraph.sf.net) and generated a weighted network graph, in which the selected TFs are presented as vertices and all pairwise correlations >0.2 are presented as edges linking the respective vertices. The network graph was visualized using the fruchterman reingold layout. TF vertices were manually color-coded based on the expression pattern along the monocle lineage. Green, teal, and blue represent highest average expression in APs, BPs, and neurons.
For Fig. S5E, we calculated the fraction of all fetal neocortex cells or the fraction of all organoid cerebral cortex cells that were assigned AP, BP, or N and plotted the fractions in a bar graph to compare the relative abundance of each of these cell types in the fetal and organoid single-cell data.
For Fig. S5F, to assess how the organoid cerebral cortical lineage resembles the fetal lineage, we computed a pairwise correlation matrix between organoid and fetal cortical cells and determined for each organoid cell (corresponding to a pseudotimepoint) the maximum and minimum correlation with any of the fetal cells. We then plotted the maximum and minimum correlation of organoid cortical cells with fetal cortical cells as a function of the pseudotime. This shows that the correlation of gene expression in fetal and organoid cells along the inferred lineage is relatively constant.
Methods for Fig. S6: Cerebral Organoids Recapitulate Gene Expression Programs Underlying Cerebral Cortex Cell Biology.
Similar to the TF analysis in Fig. 4, to compare the expression profiles of the selected genes in fetal and organoid cells, we calculated the mean expression of each gene across the cells belonging to the same cell type [AP (AP1 + AP2), BP (BP1 + BP2), N1, N2, and N3) for fetal and organoid data. We then calculated the correlation between fetal and organoid for each gene based on the average expression in each cell type.
Methods for Fig. S7: Gene Expression Differences Between Fetal and Organoid Cell Types.
For Fig. S7 A and B, we searched for differences in gene expression between fetal and organoid tissues by calculating the difference in average expression for each gene in fetal and organoid APs (AP1 and AP2 combined) and in fetal and organoid neurons (N1, N2, and N3 combined). We excluded BPs from this analysis because of the small number of BPs in the organoid dataset. Seurat’s implementation of the median ROC test (23) was used to identify genes that had a high average difference (>threefold) and power to discriminate (97th percentile) between fetal and organoid cells (Dataset S4).
Supplementary Material
Acknowledgments
We thank E. Taverna for comments on the manuscript, M. Renner for cotutoring organoid culture, Y. J. Chang and M. T. Garcia for help with imunohistochemistry, and B. E. Daniel for discussions. We thank K. Köhler and A. Weigert for help with iPSC culture techniques and B. Höber, A. Weihmann, J. Kelso, G. Renaud, and M. Dannemann of Max Planck Institute for Evolutionary Anthropology for DNA sequencing and bioinformatics support. J.A.K. is funded by the Austrian Academy of Sciences, Austrian Science Fund Grants I_552-B19 and Z_153_B09, and an advanced grant of the European Research Council (ERC). S.P. was supported by the Allen Family Foundation. W.B.H. was supported by Deutsche Forschungsgemeinschaft (DFG) Grant Sonderforschungsbereich 655 A2, ERC Grant 250197, the DFG-funded Center for Regenerative Therapies Dresden, and the Fonds der Chemischen Industrie.
Footnotes
The authors declare no conflict of interest.
Data deposition: The sequences reported in this paper have been deposited in the NCBI Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE75140).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1520760112/-/DCSupplemental.
References
- 1.Florio M, Huttner WB. Neural progenitors, neurogenesis and the evolution of the neocortex. Development. 2014;141(11):2182–2194. doi: 10.1242/dev.090571. [DOI] [PubMed] [Google Scholar]
- 2.Dehay C, Kennedy H, Kosik KS. The outer subventricular zone and primate-specific cortical complexification. Neuron. 2015;85(4):683–694. doi: 10.1016/j.neuron.2014.12.060. [DOI] [PubMed] [Google Scholar]
- 3.Kriegstein A, Alvarez-Buylla A. The glial nature of embryonic and adult neural stem cells. Annu Rev Neurosci. 2009;32:149–184. doi: 10.1146/annurev.neuro.051508.135600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Taverna E, Götz M, Huttner WB. The cell biology of neurogenesis: Toward an understanding of the development and evolution of the neocortex. Annu Rev Cell Dev Biol. 2014;30:465–502. doi: 10.1146/annurev-cellbio-101011-155801. [DOI] [PubMed] [Google Scholar]
- 5.Lui JH, Hansen DV, Kriegstein AR. Development and evolution of the human neocortex. Cell. 2011;146(1):18–36. doi: 10.1016/j.cell.2011.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Betizeau M, et al. Precursor diversity and complexity of lineage relationships in the outer subventricular zone of the primate. Neuron. 2013;80(2):442–457. doi: 10.1016/j.neuron.2013.09.032. [DOI] [PubMed] [Google Scholar]
- 7.Kriegstein A, Noctor S, Martínez-Cerdeño V. Patterns of neural stem and progenitor cell division may underlie evolutionary cortical expansion. Nat Rev Neurosci. 2006;7(11):883–890. doi: 10.1038/nrn2008. [DOI] [PubMed] [Google Scholar]
- 8.Rakic P. Evolution of the neocortex: A perspective from developmental biology. Nat Rev Neurosci. 2009;10(10):724–735. doi: 10.1038/nrn2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stahl R, et al. Trnp1 regulates expansion and folding of the mammalian cerebral cortex by control of radial glial fate. Cell. 2013;153(3):535–549. doi: 10.1016/j.cell.2013.03.027. [DOI] [PubMed] [Google Scholar]
- 10.Lancaster MA, et al. Cerebral organoids model human brain development and microcephaly. Nature. 2013;501(7467):373–379. doi: 10.1038/nature12517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lancaster MA, Knoblich JA. Generation of cerebral organoids from human pluripotent stem cells. Nat Protoc. 2014;9(10):2329–2340. doi: 10.1038/nprot.2014.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mariani J, et al. FOXG1-dependent dysregulation of GABA/glutamate neuron differentiation in autism spectrum disorders. Cell. 2015;162(2):375–390. doi: 10.1016/j.cell.2015.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grün D, et al. Single-cell messenger RNA sequencing reveals rare intestinal cell types. Nature. 2015;525(7568):251–255. doi: 10.1038/nature14966. [DOI] [PubMed] [Google Scholar]
- 14.Patel AP, et al. Single-cell RNA-seq highlights intratumoral heterogeneity in primary glioblastoma. Science. 2014;344(6190):1396–1401. doi: 10.1126/science.1254257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Treutlein B, et al. Reconstructing lineage hierarchies of the distal lung epithelium using single-cell RNA-seq. Nature. 2014;509(7500):371–375. doi: 10.1038/nature13173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zeisel A, et al. Brain structure. Cell types in the mouse cortex and hippocampus revealed by single-cell RNA-seq. Science. 2015;347(6226):1138–1142. doi: 10.1126/science.aaa1934. [DOI] [PubMed] [Google Scholar]
- 17.Trapnell C, et al. The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat Biotechnol. 2014;32(4):381–386. doi: 10.1038/nbt.2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fietz SA, et al. Transcriptomes of germinal zones of human and mouse fetal neocortex suggest a role of extracellular matrix in progenitor self-renewal. Proc Natl Acad Sci USA. 2012;109(29):11836–11841. doi: 10.1073/pnas.1209647109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Florio M, et al. Human-specific gene ARHGAP11B promotes basal progenitor amplification and neocortex expansion. Science. 2015;347(6229):1465–1470. doi: 10.1126/science.aaa1975. [DOI] [PubMed] [Google Scholar]
- 20.Prüfer K, et al. The complete genome sequence of a Neanderthal from the Altai Mountains. Nature. 2014;505(7481):43–49. doi: 10.1038/nature12886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McLean CY, et al. Human-specific loss of regulatory DNA and the evolution of human-specific traits. Nature. 2011;471(7337):216–219. doi: 10.1038/nature09774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gittelman RM, et al. Comprehensive identification and analysis of human accelerated regulatory DNA. Genome Res. 2015;25(9):1245–1255. doi: 10.1101/gr.192591.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Macosko EZ, et al. Highly parallel genome-wide expression profiling of individual cells using nanoliter droplets. Cell. 2015;161(5):1202–1214. doi: 10.1016/j.cell.2015.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pollen AA, et al. Low-coverage single-cell mRNA sequencing reveals cellular heterogeneity and activated signaling pathways in developing cerebral cortex. Nat Biotechnol. 2014;32(10):1053–1058. doi: 10.1038/nbt.2967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ross SE, et al. Bhlhb5 and Prdm8 form a repressor complex involved in neuronal circuit assembly. Neuron. 2012;73(2):292–303. doi: 10.1016/j.neuron.2011.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhou B, et al. Loss of NFIX transcription factor biases postnatal neural stem/progenitor cells toward oligodendrogenesis. Stem Cells Dev. 2015;24(18):2114–2126. doi: 10.1089/scd.2015.0136. [DOI] [PubMed] [Google Scholar]
- 27.Darmanis S, et al. A survey of human brain transcriptome diversity at the single cell level. Proc Natl Acad Sci USA. 2015;112(23):7285–7290. doi: 10.1073/pnas.1507125112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Johnson MB, et al. Single-cell analysis reveals transcriptional heterogeneity of neural progenitors in human cortex. Nat Neurosci. 2015;18(5):637–646. doi: 10.1038/nn.3980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pollen AA, et al. Molecular identity of human outer radial glia during cortical development. Cell. 2015;163(1):55–67. doi: 10.1016/j.cell.2015.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rousso DL, et al. Foxp-mediated suppression of N-cadherin regulates neuroepithelial character and progenitor maintenance in the CNS. Neuron. 2012;74(2):314–330. doi: 10.1016/j.neuron.2012.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Koyanagi-Aoi M, et al. Differentiation-defective phenotypes revealed by large-scale analyses of human pluripotent stem cells. Proc Natl Acad Sci USA. 2013;110(51):20569–20574. doi: 10.1073/pnas.1319061110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thomson JA, et al. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282(5391):1145–1147. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- 33.Renaud G, Kircher M, Stenzel U, Kelso J. freeIbis: An efficient basecaller with calibrated quality scores for Illumina sequencers. Bioinformatics. 2013;29(9):1208–1209. doi: 10.1093/bioinformatics/btt117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Renaud G, Stenzel U, Maricic T, Wiebe V, Kelso J. deML: Robust demultiplexing of Illumina sequences using a likelihood-based approach. Bioinformatics. 2015;31(5):770–772. doi: 10.1093/bioinformatics/btu719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9(4):357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Trapnell C, Pachter L, Salzberg SL. TopHat: Discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25(9):1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Trapnell C, et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28(5):511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Huang W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4(1):44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 39.Zhang HM, et al. AnimalTFDB: A comprehensive animal transcription factor database. Nucleic Acids Res. 2012;40(Database issue):D144–D149. doi: 10.1093/nar/gkr965. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.