Significance
Inositol phosphate kinase 2 (Ipk2) is a conserved protein that initiates the production of inositol phosphate intracellular messengers that are critical for regulating a variety of cellular processes. Here we explore the developmental roles Ipk2 and its products in Drosophila. We report that Ipk2 kinase activity is required to develop adult body structures including eyes, legs, and wings, which are formed by tissue known as imaginal discs. Although Ipk2 mutant discs seem to pattern normally in embryogenesis, during larval development they fail to undergo normal expansion. We find defects in signaling pathways that control both cell death and proliferation. Our work demonstrates a specific role for Ipk2-produced intracellular messengers in regulating developmental pathways involved in tissue growth and stability.
Keywords: inositol phosphates, inositol phosphate multikinase, Ipk2, IPMK, Upd
Abstract
Inositol phosphate kinase 2 (Ipk2), also known as IP multikinase IPMK, is an evolutionarily conserved protein that initiates production of inositol phosphate intracellular messengers (IPs), which are critical for regulating nuclear and cytoplasmic processes. Here we report that Ipk2 kinase activity is required for the development of the adult fruit fly epidermis. Ipk2 mutants show impaired development of their imaginal discs, the primordial tissues that form the adult epidermis. Although disk tissue seems to specify normally during early embryogenesis, loss of Ipk2 activity results in increased apoptosis and impairment of proliferation during larval and pupal development. The proliferation defect is in part attributed to a reduction in JAK/STAT signaling, possibly by controlling production or secretion of the pathway’s activating ligand, Unpaired. Constitutive activation of the JAK/STAT pathway downstream of Unpaired partially rescues the disk growth defects in Ipk2 mutants. Thus, IP production is essential for proliferation of the imaginal discs, in part, by regulating JAK/STAT signaling. Our work demonstrates an essential role for Ipk2 in producing inositide messengers required for imaginal disk tissue maturation and subsequent formation of adult body structures and provides molecular insights to signaling pathways involved in tissue growth and stability during development.
At the confluence of numerous signaling pathways is phospholipase C (PLC)-mediated production of the second messenger inositol 1,4,5-trisphosphate (IP3), a key regulator of intracellular calcium release (1). IP3 also serves as an essential substrate for the synthesis of inositol phosphates (IPs) and pyrophosphates (PP-IPs) that are critical for eukaryotic cellular function in their own right (2–6). Phosphorylation of IP3 by conserved inositol phosphate kinases (IPKs) leads to the synthesis of complex pools of different IP species (6). Insights into the cellular functions of IPs have been gleaned by perturbing their synthesis through genetic manipulations of the IPKs. Consequently, defects in specific cellular processes were attributed to losses of particular IPs. This revealed that IPs are specific regulators of diverse cellular processes, such as transcription, chromatin remodeling, DNA repair, RNA editing, and RNA export (3–7).
An essential enzyme for the conversion of IP3 to the array of IP and PP-IP species found in cells is the evolutionarily conserved inositol phosphate kinase 2 (Ipk2, Fig. 1A), which is also known as IP multikinase (IPMK) (4). Ipk2 was first identified in yeast as a nuclear enriched protein whose activities toward the 6- and 3-positions on the inositol ring sequentially convert IP3 to IP4, and then IP5 (Fig. 1A) (8–10). Yeast Ipk2 mutants that fail to produce IP4 and IP5 exhibit defects in transcriptional responses and chromatin remodeling (3). Additional “multikinase” activities described for certain Ipk2 orthologs indicate an array of IP substrates, as well as the inositol lipid PIP2 (5, 11). Loss-of-function analysis of Ipk2 in a variety of species, cell types, and organisms demonstrates its requirement for proper metabolism and cellular functions (3, 12–14).
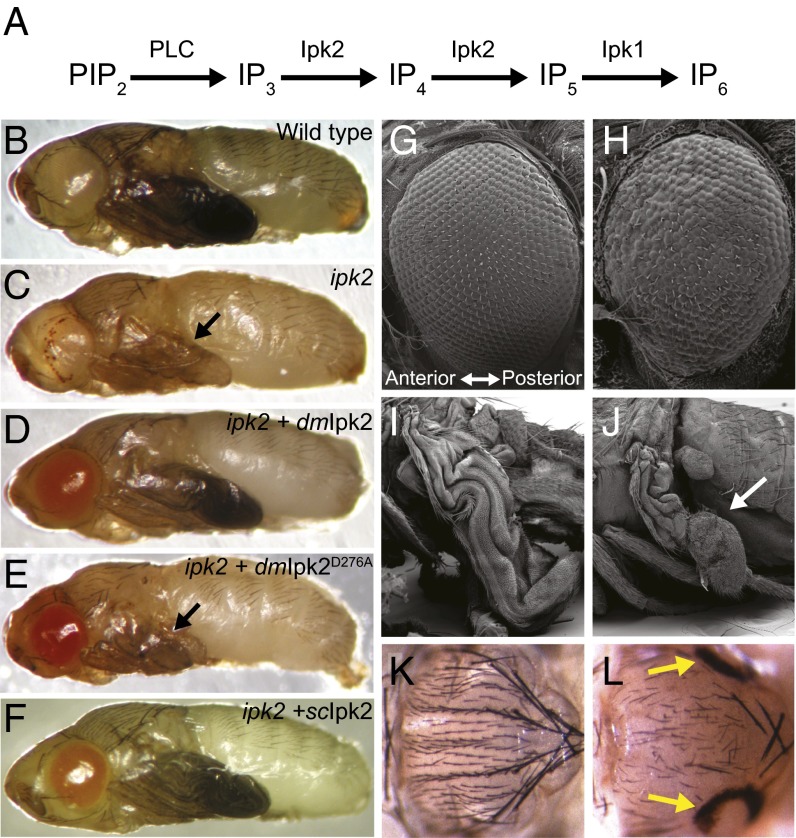
Fig. 1.
Ipk2 and its products are required for pupal development. (A) Synthesis pathway for producing the bulk of cellular IPs. PIP2 is hydrolyzed by PLC to produce soluble IP3. Ipk2 phosphorylates IP3 to IP5 that serves as a substrate for the production of IP6 by Ipk1. (B and C) Pupae were compared at the latest stage to which the ipk2 mutants could develop. Shown are (B) wild type and (C) ipk2. (D–F) ipk2 mutants with the following UAS transgenes expressed under control of Actin-GAL4: (D) UAS-dmIpk2, (E) UAS-dmipk2D276A, and (F) UAS-scIpk2. Black arrows point to defective wings. (G–J) Scanning electron microscope images of (G) wild-type and (H) ipk2 mutant eyes and (I) wild-type and (J) ipk2 mutant wings. The mutant wing is indicated with the white arrow. (K and L) Images of (K) wild-type (L) and ipk2 mutant notums. Degenerative holes are shown with yellow arrows.
As we determine how IPs are synthesized in cells and identify the processes that they regulate, it is also critical to understand how these messengers function in metazoans. The generation of mutant animals has revealed that IPKs are required for development and viability; however, the loss of a gene product alone, without complementation analysis, limits interpretation of the role of the inositide products in organismal physiology. To further our understanding of the roles of IP messengers, we report here our examination of the role of Ipk2 and its kinase activity in the development of a genetically tractable metazoan, Drosophila melanogaster. Building on previous studies establishing a molecular basis for IP metabolism in flies (15), we report that Ipk2-dependent production of IP messengers is critical for adult epidermal development, specifically through regulation of cellular proliferation and apoptosis. Our data link Ipk2 activity to the control of proliferation, in part through interactions with JAK/STAT signaling. This demonstrates a specific role for Ipk2-produced IPs in regulating developmental pathways involved in tissue growth and stability.
Results
IPs Are Required for Development and Viability.
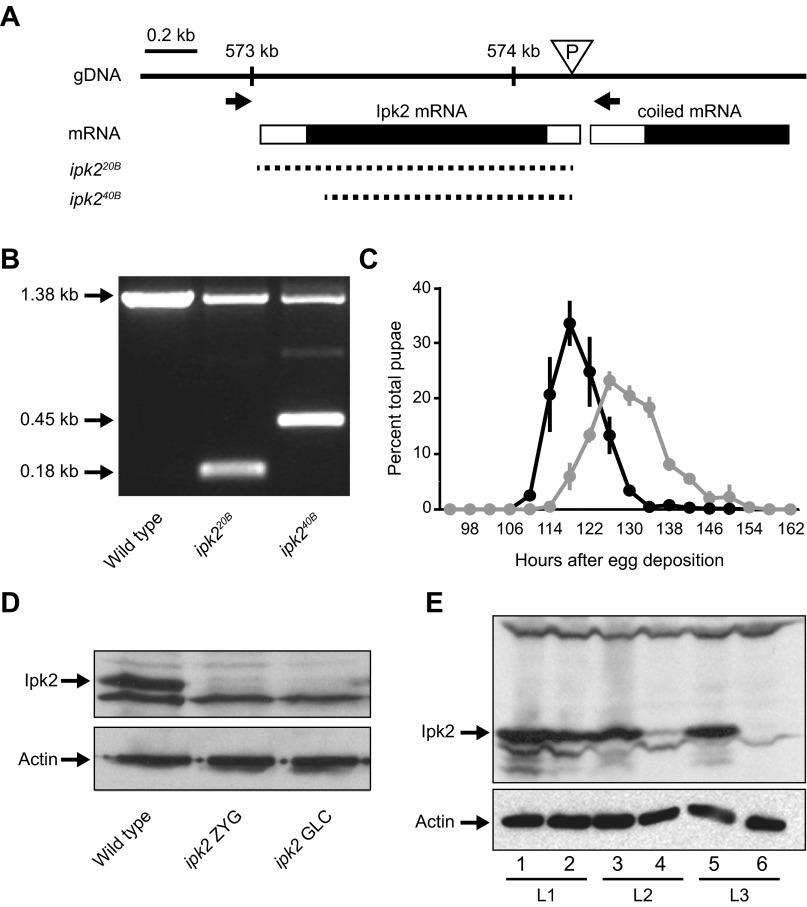
To identify roles for IPs in Drosophila, we generated Ipk2 deletion mutants. The ipk2G3545 line contains a P-element insertion within the 5′ untranslated region of the Ipk2 gene (Fig. S1A). Although these flies have near-complete losses of Ipk2 and its IP products, they are developmentally normal. We therefore generated Ipk2 null animals by mobilizing the ipk2G3545 P-element and obtained two independent imprecise excision alleles: ipk220B and ipk240B. The entire opening reading frame of Ipk2 is deleted in ipk220B, whereas ∼90% is excised in ipk240B (Fig. S1 A and B). Ipk2 protein was not detectible in homozygous ipk220B third-instar larvae, further confirming that the gene had been removed (Fig. S1D). Eighty-five percent of homozygous mutants for either ipk2 allele survived through the larval stages, but their pupation was delayed by about 12 h relative to wild type (Fig. S1C). Homozygotes and transheterozygotes (ipk220B/ipk240B) died during pupation, with no flies reaching eclosion. We focused on the ipk220B allele in subsequent experiments (hereafter referred as ipk2). This preliminary analysis of ipk2 mutants indicates that the gene is critical for progression through development, and for viability.
Fig. S1.
ipk2 excision mutants. (A) Schematic representation of the Ipk2 locus on the second chromosome (2L). The triangle represents the location of the ipk2G3545 P-element in the 5′ untranslated region that was used to generate the excision lines. Arrows show the locations of primers used to screen for unidirectional excisions. Dashed lines represent the missing genomic DNA in the excision or deficiency lines. ipk220B and ipk240B are the alleles generated in this study. (B) Single fly PCR of wild-type, ipk220B, and ipk240B heterozygous alleles using the primers indicated in A. The excision boundaries (shown in A) were determined based on sequence analysis of the PCR products. The 1.38-kb band indicates a wild-type ORF, whereas the 0.45- and 0.18-kb bands represent the shortened ORFs of the mutants. (C) Delayed pupation of the homozygous ipk220B mutant. The percent of total animals that pupated at each time point is plotted (mean ± SEM). (D) Western blot of ipk2 zygotic (ipk2 ZYG) and germ-line clone (ipk2 GLC) wandering third-instar larval mutants. (E) Perdurance of maternal Ipk2 protein during larval development in ipk2 zygotic mutants. Protein lysates from stage L1 (lanes 1 and 2), L2 (lanes 3 and 4), and L3 (lanes 5 and 6) of wild type (lanes 1, 3, and 5) and ipk2 (lanes 2, 4, and 6) were subject to SDS/PAGE and immunoblotted for Ipk2p (Upper). Compared with wild-type L1 larvae, L1 ipk2 null larvae have comparable levels of Ipk2 protein, whereas it drops to a lower level in L2 and is undetectable in L3.
Examination of the mutant pupae revealed defective eyes, wings, and notum (Fig. 1 B, C, and G–L). The “rough” eyes had missing or disordered omatidia and bristles that became progressively less defective from anterior to posterior (Fig. 1 G and H). ipk2 wings were small (Fig. 1 B, C, I, and J) and bristles on the notum were disordered and missing (Fig. 1 K and L). Fifty-six percent of ipk2 mutants formed symmetrical degenerative holes on their posterior notums (Fig. 1 K and L), with a similar percentage developing a single hole in the thoracic region of their ventral midline. These defects suggest an Ipk2 requirement in forming and/or maintaining external body structures.
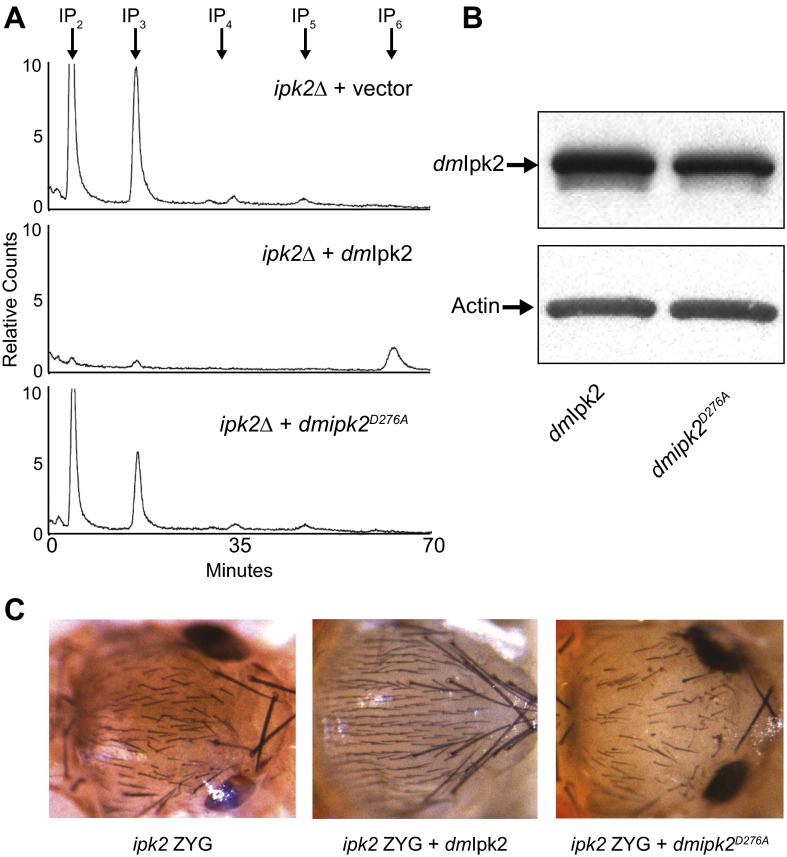
We tested the requirement of Ipk2 and its IP products for development and viability. The morphological phenotypes and lethality of the ipk2 mutants were fully rescued when an Ipk2 transgene (UAS-Ipk2) (15) was ubiquitously expressed in homozygous ipk2 mutants, demonstrating that the phenotypes were due to loss of the protein (Fig. 1D and Fig. S2). In contrast, a catalytically inactive point mutant (UAS-ipk2D276A) could not rescue any mutant phenotypes, indicating that kinase activity was critical for Ipk2 function (Fig. 1E and Fig. S2). Expression of catalytically conserved orthologs from Saccharomyces cerevisiae and Arabidopsis thaliana (scIpk2 and atIpk2, respectively) (10, 16) also rescued ipk2 lethality and developmental defects, despite their low sequence identities with Drosophila Ipk2 (22 and 31% identity, respectively; in Fig. 1F scIpk2 is shown). Thus, the kinase activity of Ipk2, and hence its IP products, are required for normal development and viability.
Fig. S2.
Characterization of kinase inactive Ipk2. (A) S. cerevisiae ipk2 null cells (ipk2Δ) were transformed with empty vector (Top), wild-type D. melanogaster Ipk2 (dmIpk2, Middle), or kinase-inactive Ipk2 (dmipk2D276A, Bottom). Cells were grown to late logarithmic phase in media containing 100 μCi/mL [3H]inositol. Soluble extracts were separated by Partisphere strong-anion exchange HPLC. (B) Western blot of ipk2 ZYG larvae expressing Drosophila Ipk2 or ipk2D276A transgenes under control of the actin promoter. (C) Notums of ipk2 ZYG mutants expressing the Drosophila Ipk2 transgenes described in B.
An Essential Role for Ipk2 in Body Morphogenesis.
Analysis of embryonic (17) and larval (Fig. S1E) gene expression revealed that maternally derived mRNA caused Ipk2 to persist into the second instar larval stages in ipk2 mutants. We hypothesized that this maternal contribution had partially rescued the ipk2 zygotic (ipk2 ZYG) mutants described above. Therefore, ipk2 germ-line clones (ipk2 GLCs) were generated using the flippase-dominant female sterile technique (18) to abolish the maternal mRNA and test the consequence of removing the gene product throughout development (Fig. S1D). Like the ZYG mutants, the ipk2 GLCs survived through larval development and arrested as pupae (Fig. 2 A–C). This suggested that larval tissues were largely unaffected in the GLC mutants, despite removal of maternal and zygotic Ipk2.
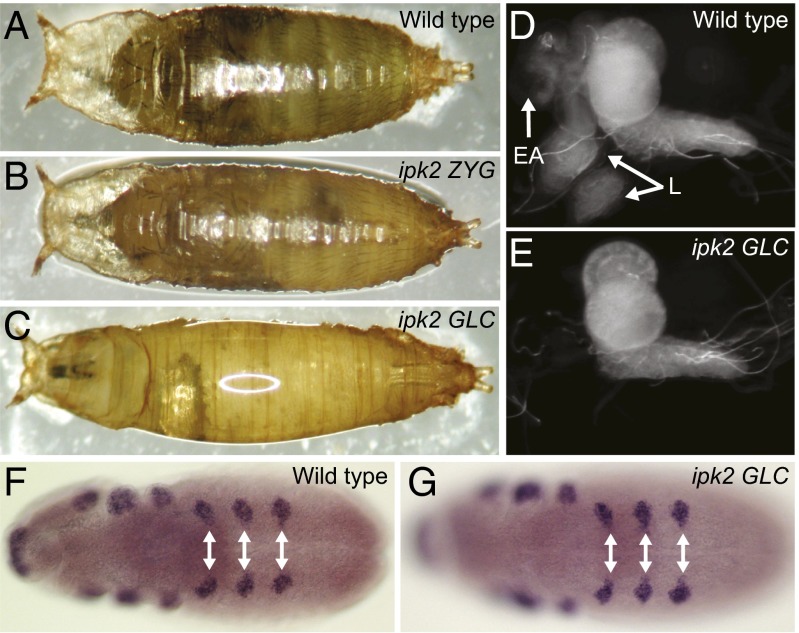
Fig. 2.
ipk2 germ-line clones fail to develop adult body structures. (A–C) Pupal images of (A) wild-type, (B) ipk2 ZYG, (C) and ipk2 germ-line GLC pupae. (D and E) Imaginal tissues are absent in the ipk2 GLC. Central nervous system and associated imaginal discs from (D) wild-type (E) and ipk2 GLC third-instar larvae. White arrows point to eye/antennal (EA) and leg discs (L). (F and G) Leg discs are specified normally in ipk2 GLCs. In situ hybridization using Distal-less probes on (F) wild-type and (G) ipk2 GLC germband-extended embryos. White arrows show leg disk primordial cells.
In contrast to ipk2 ZYG animals, the ipk2 GLCs failed to form an adult outer body (epidermis) during pupation, including the head, thorax, wings, abdomen, and legs (compare Fig. 2 B and C). ipk2 GLC larvae contained a central nervous system but lacked the larval precursor tissues that form the adult epidermis, the imaginal discs (Fig. 2 D and E). This revealed a critical role for Ipk2 in forming imaginal discs. One possible explanation for their absence in the ipk2 GLCs was their lack of specification as imaginal disk precursor cells in embryogenesis. To test this, we probed for the presence of Distal-less transcripts, which mark leg disk precursor cells (19). ipk2 GLCs and wild-type embryos had similar Distal-less expression patterns, indicating that the mutant embryos specified leg discs normally (Fig. 2 F and G). This indicates that the disk precursors formed in ipk2 GLCs and were either lost through cell death or failed to proliferate during early larval stages.
ipk2 Mutants Undergo Apoptosis.
We hypothesized that increased cell death could explain ipk2 defects, such as the “rough” eyes and small wings (Fig. 1 H and J), the degenerative holes on the thorax (Fig. 1L), and the lack of imaginal discs despite their specification in the GLCs (Fig. 2 C, E, and G). Therefore, we tested whether loss of Ipk2 leads to increased cell death. Indeed, ipk2 ZYG imaginal discs were prone to apoptosis as they showed increased DNA fragmentation (Fig. 3 A and B), a hallmark of programmed cell death (20). Increased cell death was found in all mutant imaginal disk tissues but was similar to wild type in other tissues such as salivary glands, gut, and fat bodies.
Fig. 3.
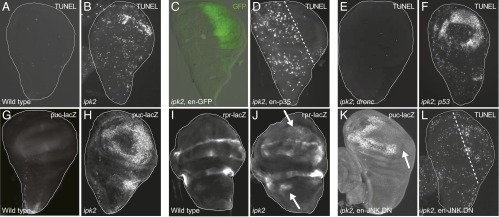
ipk2 imaginal discs undergo apoptosis. (A and B) TUNEL stain of wing imaginal discs from (A) wild-type and (B) ipk2 ZYG. (C and D) Suppression of apoptosis by expression of caspase inhibitor p35 in the posterior compartment of ipk2 ZYG wing discs. (C) Engrailed-GAL4 expression pattern (green) in the posterior compartment (ipk2, en-GAL4, UAS-GFP). (D) TUNEL stain of an ipk2 ZYG wing disk expressing p35 in the same cells (en-GAL4, UAS-p35). Area right of the dashed line expresses p35. (E) Suppression of apoptosis in ipk2, droncI29 double mutants (TUNEL stain). (F) No suppression of apoptosis with loss of p53. TUNEL stain of an ipk2 ZYG; p535A-1-4 double-mutant wing disk. (G and H) Expression of a Puckered reporter (puc-lacZ) in (G) wild-type or (H) ipk2 wing discs. Discs were stained using anti–β-galactosidase antibodies (G–K). (I and J) Expression of a Reaper reporter (rpr-lacZ) in (I) wild-type or (J) ipk2 wing discs. Arrows point to example areas of abnormal Reaper expression. (K) Suppression of puc-lacZ reporter expression in an ipk2 wing disk expressing JNK.DN in the posterior compartment. Arrow points to area of suppression. (L) No suppression of apoptosis in ipk2 wing discs by expressing dominant-negative JNK in the posterior compartment (ipk2, en-GAL4, UAS-JNK.DN). The area to the right of the dashed line would lack TUNEL staining had JNK.DN suppressed apoptosis.
We examined the requirement of the canonical apoptotic machinery in mediating cell death in ipk2 imaginal discs. Apoptosis requires the activation of caspases that cleave different substrates to induce an orderly dismantling of the cell (21). The baculovirus caspase inhibitor protein p35 is a specific genetically encoded tool capable of blocking apoptosis (22). Expression of p35 in ipk2 wing discs suppressed the abnormal cell death, indicating that it was caspase-mediated apoptosis (Fig. 3 C and D). Caspases are classified as either initiators or effectors, with initiators inducing apoptosis by cleaving and activating the effectors that bring about cell death (21). The major initiator caspase in Drosophila is Dronc, and therefore we examined the consequence of its removal in ipk2 mutants. ipk2 dronc double-null flies lacked increased cell death, indicating that the caspase is necessary for apoptosis in the ipk2 discs (Fig. 3E). These data are consistent with a role for IPs in regulating apoptosis via a caspase-dependent pathway.
We examined possible upstream signaling pathways that could initiate apoptosis in the ipk2 mutants. p53 initiates apoptosis during stress responses and development (23). Apoptosis was not suppressed in an ipk2 p53 double mutant (24), where the entire p53 gene was excised (Fig. 3F). Additionally, a transgene expressing a dominant-negative form of p53 (25) was unable to suppress apoptosis in the ipk2 mutant. Thus, p53 does not contribute to IP-mediated apoptosis.
We previously examined altered gene expression in ipk2 mutant imaginal discs and found that two transcripts linked to apoptosis, Puckered and Reaper, had increased expression relative to wild type, and could possibly be the cause of apoptosis. Puckered is a phosphatase whose expression increases to antagonize activated JNK signaling that can induce Reaper-mediated apoptosis (26). This prompted us to investigate the possibility that the JNK pathway induces apoptosis in ipk2 mutants. We generated ipk2 flies containing reporters of either Puckered or Reaper expression. ipk2 mutants showed increased expression of the Puckered reporter (Fig. 3 G and H), which indicated that JNK was activated (26). Additionally, Reaper expression was detected throughout the wing disk compared with its restricted pattern in a wild-type disk (Fig. 3 I and J). This suggested that IPs negatively regulate Reaper-induced apoptosis in imaginal discs through JNK signaling.
To further test whether JNK mediates apoptosis in the ipk2 mutant, we expressed a dominant negative form of JNK (JNK.DN) (27) in the posterior half of the ipk2 mutant wing disk. The JNK.DN transgene effectively blocked Puckered expression as judged by the loss of puc-lacZ staining (Fig. 3K), indicating that the increased JNK signaling in the ipk2 mutant discs was successfully suppressed. Although JNK.DN effectively suppressed JNK signaling as measured by decreased puc-LacZ expression, the transgene was unable to decrease the levels of apoptotic cells (Fig. 3L), suggesting that JNK is not required for initiation of apoptosis in ipk2 mutants. Therefore, apoptosis due to the loss of ipk2 occurs through a different pathway.
ipk2 Mutants Have Cellular Proliferation Defects.
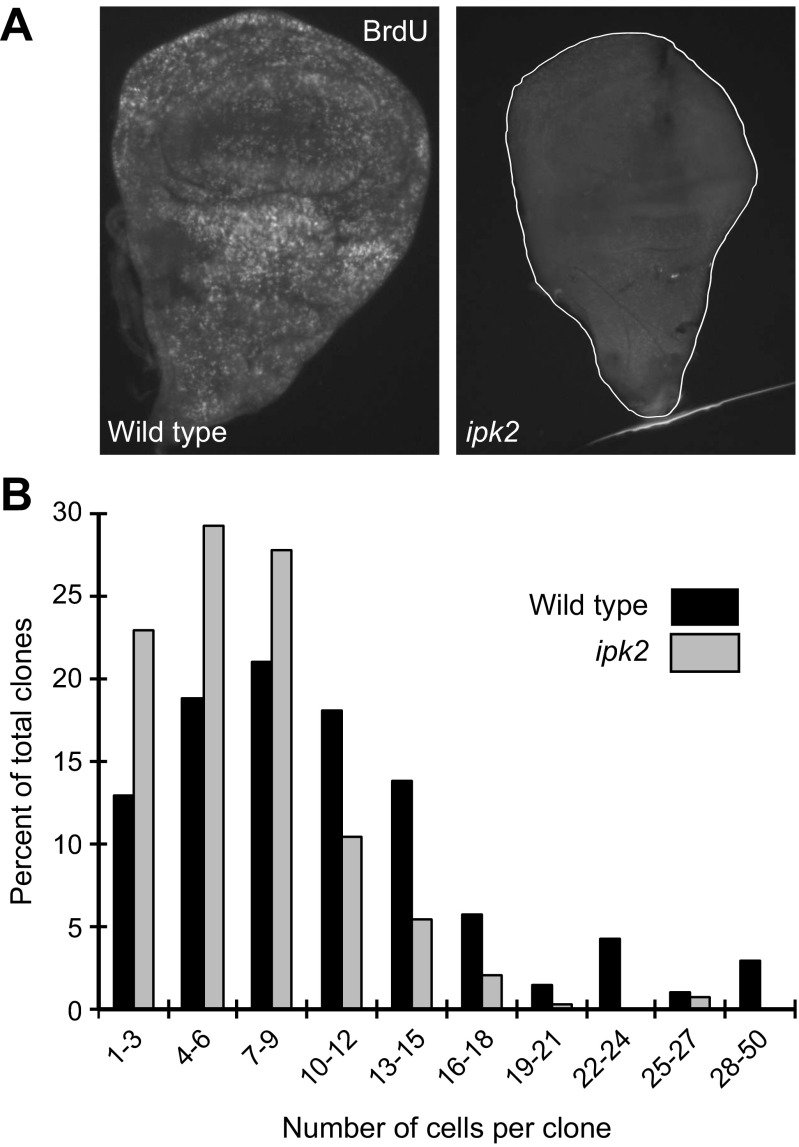
The finding that ipk2 GLC larvae lack imaginal discs may suggest that they fail to proliferate after being specified in embryogenesis. We examined whether ipk2 ZYG wing discs displayed cellular proliferation defects; 5′-bromo-2′-deoxyuridine (BrdU) incorporation analysis revealed that ipk2 ZYG discs from third-instar larvae had reduced DNA synthesis relative to wild type, suggesting that loss of Ipk2 leads to proliferation defects (Fig. S3A). We also used clonal analysis to compare the proliferation rates between wild-type and ipk2 wing discs (Fig. S3B). The median cell doubling time for wild-type and ipk2 mutants was 14.3 and 18 h, respectively. Relative to wild type, ipk2 clones contained fewer cells (on average six compared with nine cells per clone). About 30% of the wild-type clones consisted of greater than 13 cells, compared with less than 10% of ipk2 cells. These data demonstrate that IPs positively regulate cellular proliferation in imaginal discs.
Fig. S3.
Loss Ipk2 causes proliferation defects. (A) ipk2 ZYG mutant wing discs have defects in BrdU incorporation. Wing discs were incubated in the presence of BrdU for 30 min and then stained with anti-BrdU antibodies to measure the amount incorporated into cells. (B) Cell proliferation analysis in wing discs. GFP-positive clones were induced in wild-type and ipk2 ZYG larvae (SI Materials and Methods). The number of cells per clone as a percentage of the total number of clones analyzed for each genotype is plotted.
Ipk2 Mediates Proliferation Through JAK/STAT Signaling.
We next considered how IPs might mediate proliferation. Müller et al. (28) previously identified Ipk2 in an RNAi screen for regulators of JAK/STAT-mediated transcription. This signaling pathway consists of a secreted extracellular ligand Unpaired (Upd), its transmembrane receptor Domeless (Dome), a Janus tyrosine kinase Hopscotch (Hop), and the Stat92E transcription factor. In their assay, cultured cells were cotransfected with a luciferase transcriptional reporter containing Stat92E-binding sites and a plasmid that induces JAK/STAT signaling by constitutively expressing Upd. RNAi-mediated knockdown of Ipk2 in these cells caused reduced STAT reporter expression (28). This led us to examine the role of Ipk2 in JAK/STAT signaling, given that the pathway regulates cellular proliferation (29).
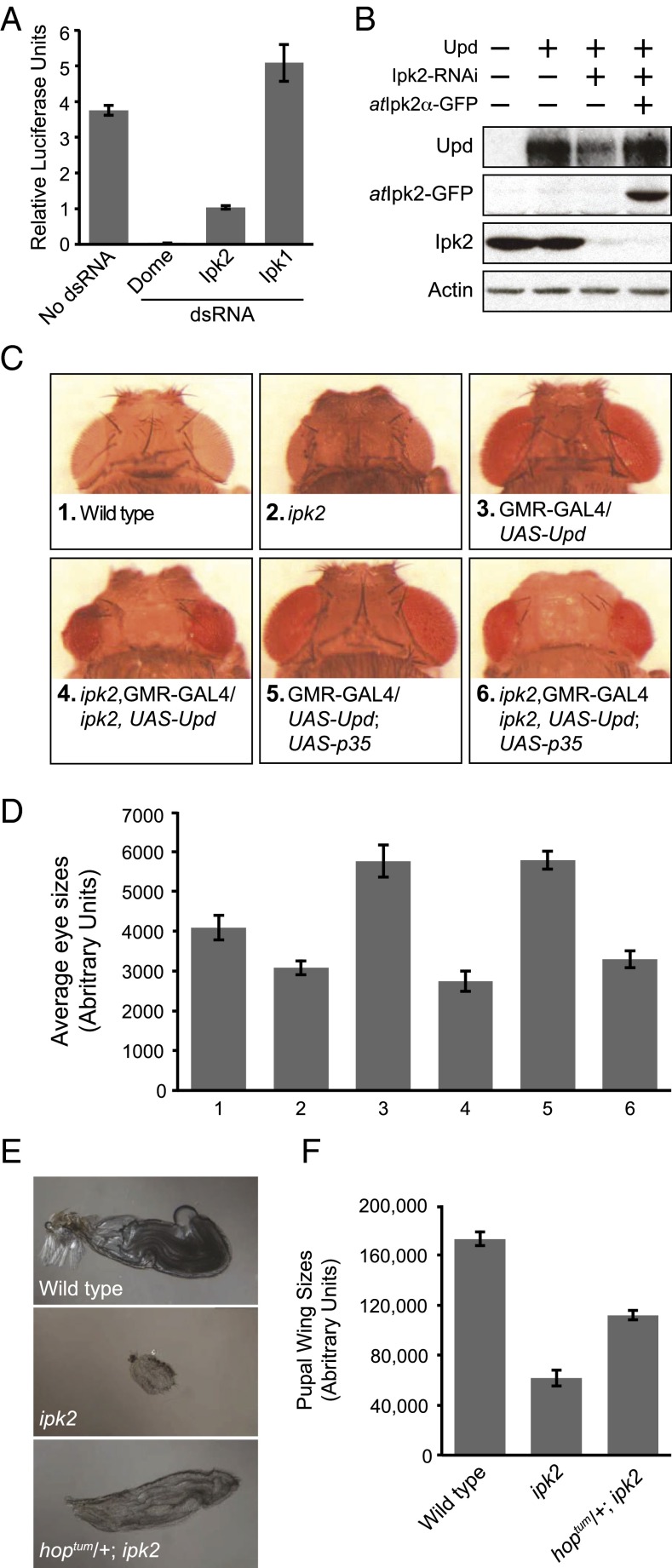
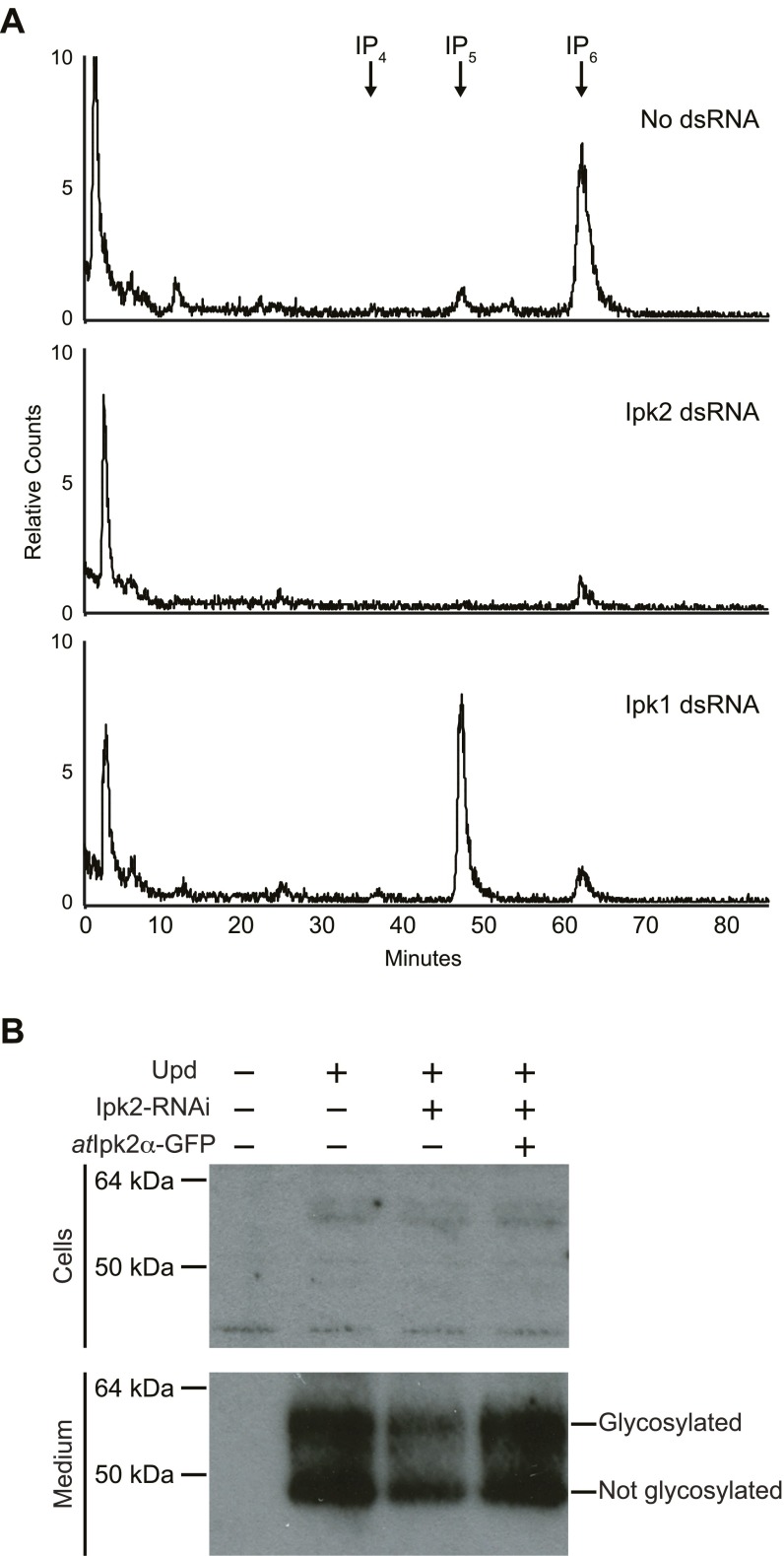
As previously reported, RNAi-mediated knockdown of Dome or Ipk2 in cell culture caused a loss of transcription from the STAT reporter (Fig. 4A). We addressed whether this defect was due to loss of Ipk2 products or downstream IPs. Ipk2 initiates the synthesis of the majority of IPs in flies by sequentially phosphorylating IP3 to IP4 and IP5, which then is phosphorylated by Ipk1 to produce IP6 (Fig. 1A). Consequently, RNAi knockdown of Ipk2 causes a reduction in cellular levels of IP6 (15) (Fig. S4A), which raises the possibility that it is the IP mediator of JAK/STAT signaling. However, RNAi knockdown of Ipk1 did not decrease transcription, but instead caused increased reporter activity (Fig. 4A). This increase might be explained by the observation that knockdown of Ipk1 causes a corresponding buildup of its IP5 substrate (15) (Fig. S4A), and therefore the increased IP5 levels might boost JAK/STAT-mediated transcription. Our data are most consistent with the conclusion that production of IP4 and/or IP5 is sufficient for JAK/STAT-mediated transcription, whereas IP6 seems dispensable. However, we cannot exclude that possibility that other potential downstream metabolites (e.g., PP-IP4) may also be involved as mediators of this transcription.
Fig. 4.
Interaction between Ipk2 and JAK/STAT. (A) Ipk2, but not Ipk1, is required for JAK/STAT-mediated transcription. Kc167 cells were treated with dsRNA targeted against Dome, Ipk2, and Ipk1 and assayed using a STAT transcriptional reporter. Relative luciferase units are plotted (mean ± SEM). (B) Ipk2 knockdown causes a reduction in secreted Upd. Cells were transfected with combinations of Upd plasmid, atIpk2-GFP plasmid, and treated with Ipk2 dsRNA, as indicated by the + and – signs. Protein extracts and conditioned medium were immunoblotted with antibodies against Upd, Ipk2, GFP, or actin. (C and D) ipk2 suppresses the eye overproliferation phenotype caused by ectopic expression of Upd. (C) Shown are representative images of heads from pharate adults of the genotypes indicated below the images. (D) Mean eye sizes of the genotypes indicated in C (mean ± SEM, n ≥ 6). (E and F) hopTum-l partially rescues the small wing phenotype of ipk2. (E) Representative images of pupal wings of the indicated genotypes. (F) Mean pupal wing sizes of these flies (mean ± SEM, n ≥ 40).
Fig. S4.
(A) Assessment IP levels in Kc167 cells that were treated with dsRNA for Ipk2 or Ipk1. Cells were labeled with [3H]inositol after being treated with dsRNA. Traces show no dsRNA treated control (Top), Ipk2 dsRNA (Middle), or Ipk1 dsRNA (Bottom). Cells were grown in media containing 50 μCi/mL [3H]inositol. Soluble extracts were separated by Partisphere strong-anion exchange HPLC. (B) Ipk2 knockdown causes a reduction in secreted Upd. Cells were transfected with combinations of Upd plasmid, atIpk2-GFP plasmid, and treated with Ipk2 dsRNA as indicated by the + and – signs. Cells were treated with heparin to release Upd from the extracellular matrix. Protein extracts from the treated cells and the conditioned medium were immunoblotted with antibodies against Upd. The top gel shows the cells and the bottom gel shows the conditioned medium. We were unable to detect appreciable amounts of Upd in the cell samples (Top), whereas the conditioned medium appeared to contain the bulk of the Upd (Bottom). The two bands of Upd correspond to the glycosylated and unglycosylated forms. The glycosylated form is shown in Fig. 4B.
Müller et al. (28) further demonstrated that the defect in STAT-mediated transcription caused by loss of Ipk2 could be suppressed by treating cells with Upd-conditioned culture medium. This suggests that Ipk2 functions upstream of the Upd receptor Dome, possibly by regulating the production or secretion of the Upd ligand. To investigate this further, we examined the levels of secreted Upd in cultured cells (30). Indeed, we found decreased secretion of Upd in cells treated with Ipk2 RNAi (Fig. 4B and Fig. S4B). This reduction could be rescued by cotransfection with atIpk2, indicating that the decreased Upd was due to the loss of Ipk2 activity (Fig. 4B and Fig. S4B). Thus, Ipk2 products positively regulate Upd production or secretion.
We next examined the role of Ipk2 in JAK/STAT-mediated proliferation. We used a sensitized transgenic assay to test whether loss of Ipk2 modulates JAK/STAT signaling in the eye (31). In this assay, Upd is ectopically expressed in developing eye discs under control of GMR-GAL4. This leads to hyperactivation of JAK/STAT-mediated proliferation, causing enlarged eyes in the pharate adults, compared with wild-type eyes (∼40% increase in eye surface area; Fig. 4 C and D, compare 3 and 1). Using eye size as a readout of JAK/STAT activation, we examined whether the Upd-mediated enlarged eyes could be suppressed in ipk2 mutants. We observed an average ∼50% decrease in eye size in the ipk2 mutants, compared with those expressing the Upd transgene in a wild-type background (Fig. 4 C and D, compare 3 and 4). This size decrease was unlikely due to the cell death that occurs in ipk2 mutants, because expression of p35 did not relieve the suppression (Fig. 4 C and D, compare 5 and 6). These results indicate that Ipk2 is required for efficient JAK/STAT-mediated proliferation.
We next considered whether JAK/STAT activation could suppress ipk2 developmental phenotypes. The loss of Ipk2 by RNAi causes a reduction in the levels of secreted Upd in cell culture (Fig. 4B) that can be bypassed by activating downstream components through addition of Upd-conditioned medium (28). Therefore, if the developmental defects in ipk2 mutants are due to a loss of JAK/STAT-mediated proliferation, then it may be possible to rescue these phenotypes by activating the JAK/STAT pathway downstream of Upd. To test this, we used a dominant gain-of-function mutant of the JAK kinase Hop (hopTum-l), which contains a mutation that causes hyperphosphorylation and hyperactivation of Stat92E (32, 33). hopTum-l partially rescued the small wing phenotype of the ipk2 mutant (Fig. 4 E and F), further suggesting that Ipk2 acts upstream of Hop. However, JAK/STAT activation could not completely rescue this wing phenotype, nor could it rescue ipk2 lethality. Taken together, these results demonstrate that Ipk2 products regulate JAK/STAT signaling to control proliferation in development, yet other processes are also likely involved.
Discussion
Inositol phosphate kinases are highly conserved, underscoring the importance of IP production for both single and multicelled eukaryotes. In particular, Ipk2 and its products serve an essential function for yeast adaptive responses and proper organismal development. Our study of fly Ipk2 provides molecular insights into how production of IP products contributes to developmental programs and cellular homeostasis. Our initial characterization of IP synthesis in Drosophila established that loss or gain of Ipk2 kinase activity had profound effects on cellular levels of IP4, IP5, and IP6 in flies (15). Here, we find that loss of these IP products through deletion of ipk2 results in inviability, and more specifically a “disc-less” phenotype, which causes major defects in the development and maintenance of adult epidermal tissues. Our data are most consistent with a role for proper spatiotemporal IP production during organismal development as a key regulator of proliferative and degenerative signaling pathways. The failure of imaginal disk primordial cells to proliferate and form an epidermis can be partially explained by defective JAK/STAT signaling. Although multiple different processes are likely affected by the loss of IPs to contribute to their essential functions, this work provides a foundation for determining the mechanisms of IP regulation in a metazoan.
Ipk2 Kinase Activity Plays a Key Role in Organismal Development.
An important goal of the complementation analysis and genetic studies reported here is to narrow down which of the “multikinase” activities of Ipk2, if any, play a role in development. Our data clearly demonstrate that kinase activity, and therefore the products of Ipk2, are required for imaginal disk development and regulation of signaling pathways. To distinguish which of the products of Ipk2 are responsible, for example water-soluble IP messengers versus lipophilic PIPs, we complemented mutant flies with Arabidopsis Ipk2 because it has been reported to only harbor water-soluble but not lipid kinase activity (11). From these analyses, we define a kinase-dependent role for water-soluble IP production, but not inositol lipids, in regulating Drosophila development. The ability of heterologous expression of either yeast or plant Ipk2 orthologs, despite their low sequence identity to Drosophila Ipk2, to completely rescue viability and imaginal disk development further supports our hypothesis that it is the conversion of IP3 to IP4 and IP5 that plays an important role in signaling. With respect to proliferation and regulation of JAK/STAT pathways, our data demonstrate that loss of IP6 production does not account for the phenotypes observed. Our work further pinpoints a role for Ipk2 products IP4 and IP5 as regulators of the JAK/STAT pathway; however, given that other inositide metabolites are produced downstream of Ipk2, such as PP-IPs, we leave open the possibility that such messengers may account for some of the phenotypes reported.
The production of IP4/IP5 in many cells requires the hydrolysis of PIP2 to produce IP3 and thus raises the question as to whether Ipk2-mediated development is dependent on phospholipase C activity. In budding yeast, it has been possible to delineate this through genetic and biochemical approaches because there is a single phospholipase C gene product Plc1. Drosophila possesses three different isoforms of PLC [NorpA, Plc21C, and Small wing (Sl)] (34), and although triple-deletion mutants have not been reported in the literature, individual PLC mutants show interesting phenotypes, including small wings and rough eyes (35), similar to the zygotic ipk2 mutant defects we report. Evidence in this study hints at a different mechanism but it is also quite likely that the different PLC orthologs can compensate for each other, confounding interpretation.
JAK/STAT-Mediated Signaling and Cell Proliferation.
Previous work implicated Ipk2 as necessary for JAK/STAT-mediated transcription in cultured cells (28), which prompted us to explore the relationship between this signaling pathway and IP production. We extended this finding by showing that JAK/STAT-mediated transcription requires the production of Ipk2 products, IP4 and IP5, whereas IP6 production by Ipk1 is dispensable. Further, IP regulation of JAK/STAT occurs through modulation of the production or secretion of the Upd ligand. This is in agreement with the finding that decreased JAK/STAT-mediated transcription caused by knockdown of Ipk2 can be relieved by addition of Upd-conditioned medium (28). An important next step will be to determine the mechanism by which IPs control the process that leads to Upd secretion.
A significant contribution of this work is the demonstration of a genetic interaction between Ipk2 and JAK/STAT in controlling cellular proliferation. ipk2 mutants suppress JAK/STAT-mediated proliferation during eye development, possibly because of decreased Upd secretion. Conversely, the partial rescue of the ipk2 mutant small-wing phenotype using hopTum-l indicates that up-regulation of the JAK/STAT pathway can partially overcome an ipk2 mutant phenotype. The gain-of-function hopTum-l mutant likely rescues ipk2 by bypassing the reduced levels of Upd. Thus, our results provide a partial explanation for why ipk2 mutants have disk proliferation defects.
The interaction between Ipk2 and JAK/STAT raises the question of whether IP production is always required for the signaling pathway. ipk2 phenotypes are reminiscent of the small or absent imaginal discs exhibited by some hop and stat92E mutants (36, 37). However, modulation of JAK/STAT activity by IPs may be restricted to the imaginal discs, because ipk2 mutants do not seem to have the embryonic segmentation or sex determination defects exhibited by JAK/STAT pathway mutants (38). Given the multitude of products generated by Ipk2 and the growing number of processes regulated by these products and downstream metabolites, such as inositol pyrophosphates, it is not surprising that ipk2 phenotypes cannot be fully rescued by up-regulation of the JAK/STAT pathway. Further genetic analyses in Drosophila of Ipk1, IP6K, and Vip1 mutants will greatly enhance our ability to further assign the roles of IP6 and PP-IPs, such as IP7 and IP8.
Tissue Degeneration and Apoptosis.
ipk2 mutants have tissue stability defects, which may be caused by increased apoptosis. For example, ipk2 ZYG eyes become increasingly disordered from posterior to anterior. This coincides with a gradual loss of maternally supplied Ipk2 to rescue the developing eye, because the anterior end of the eye differentiates later in development (39). Rough eye phenotypes have been previously linked to aberrant apoptosis (40). Additionally, holes form in the cuticle of ipk2 mutants, providing evidence of tissue degeneration. The apparent defects in tissue stability in ipk2 mutants led us to find that the imaginal discs undergo increased caspase-dependent apoptosis. Previous work in mammalian cells demonstrated that IP6 is protective from apoptosis (41), consistent with our results that loss of IP production leads to increased cell death.
Materials and Methods
Analysis of JAK/STAT-Mediated Signaling in Kc167 Cells.
Primers, ESTs, and protocols for generating dsRNA and performing RNAi in cells were the same as described previously (15, 28). For JAK/STAT reporter assays (28), Kc167 cells that were treated with dsRNA for 4 d were transfected with pAct-UpdGFP (Upd 1), 6 × 2x DrafLuc firefly luciferase reporter, and pAct-Renilla using Effectene (Qiagen) and treated with dsRNA for an additional 3 d. Assays were done using the Luciferase Assay System (Promega). Data are plotted (Fig. 4A) as relative luciferase units that were corrected for transfection efficiency by calculating the firefly versus renilla luciferase ratios.
For detection of Upd after Kc167 cells were treated with dsRNA for 4 d, the cells were transfected with Ubiquitin-GAL4, a combination of pUAST-Upd and pUAST-GFP-atIpk2α, as indicated in Fig. 4B, and pUAST (empty vector) to equalize the amount of total DNA input in each transfection. Cells were then treated with dsRNA for an additional 3 d. Twenty-four hours before harvesting, the cells were collected, washed, and plated with Sf-900 II Serum-Free Medium; 50 µg/mL heparin (Sigma H-9399) was added to the medium to release secreted Upd from the extracellular matrix (30). The conditioned medium for each sample was collected, centrifuged to remove cells, and concentrated to the same volume using a 10,000 molecular weight-cutoff concentrator (Sartorius). Cells were lysed in 1% Nonidet P-40, 150 mM NaCl, 10 mM Tris (pH 7.5) buffer, 1 mM PMSF, and complete protease inhibitor (Roche). Laemmli loading buffer (5×) was added to the cell lysates and concentrated conditioned medium and boiled for 5 min. Twenty micrograms of protein was loaded for the cell protein extracts, and 2 μL of the concentrated conditioned medium (0.8% total medium protein extracted) was loaded per lane onto a 10% (wt/vol) SDS/PAGE gel. Western blots were performed using a 1:5,000 dilution of rabbit anti-Upd from Doug Harrison, University of Kentucky, Lexington, KY.
Genetic Interaction Between Ipk2 and JAK/STAT.
Flies of the genotypes indicated in Fig. 4 C and E were raised on standard food at room temperature, under noncrowded conditions. Pharate adults were dissected from their pupal cases to expose the eyes (Fig. 4C). The pupae were placed on a glass slide in the same orientation. The wings were dissected and mounted on slides in 50% (vol/vol) glycerol. Heads and wings were imaged at 10× magnification. The eyes and wings in the images were segmented using manual tracing and show region statistics, and then region measurement tools were used to measure the areas (in arbitrary units) using Metamorph (Molecular Devices).
See SI Materials and Methods for additional materials and methods.
SI Materials and Methods
Fly Stocks.
Flies referred to as “wild-type” in this paper are w1118. FRT40A, FRT42 p535A-1-4, UAS-JNK.DN, and hopTum-l were obtained from the Bloomington Stock Center; Puc-lacZ was obtained from Rick Fehon, University of Chicago, Chicago, IL; rpr-11-lacZ and UAS-Puc was obtained from Donald McEwen, University of Texas Health Science Center, San Antonio, TX; UAS-Upd (Upd 1) was obtained from Martin Zeidler, University of Sheffield, Sheffield, UK; DroncI29 was obtained from Andreas Bergmann, University of Texas MD Anderson Cancer Center, Houston, TX; and rabbit anti-Upd was from Doug Harrison.
Generation of ipk2 Mutants.
The P-element of ipk2G3545 was mobilized to generate imprecise excisions by crossing the flies to a stable source of transposase (delta 2-3). About 100 white-eyed lines were screened by PCR for a unidirectional excision in the Ipk2 coding region. The primers used for the screen amplified from the 5′ UTR to immediately after the 3′ UTR (Fig. S1A) were as follows: forward, CCATC TCGCA CCTCT CATTC C and reverse, CCTTG AAATG ATTGC ATCTT GC. Excision lines that generated PCR products smaller than the full-length Ipk2 coding region were sequenced to determine the break points. Two independent lines were identified that contained deletions of the Ipk2 gene region (named ipk220B and ipk240B; Fig. S1 A and B). GLCs were generated using the dominant female sterile method (18). FRT sites were recombined onto the ipk220B and ipk240B mutant chromosomes. The strains were crossed to a heat shock-inducible FLP strain carrying a dominant female sterile (ovoD1) mutation and corresponding FRT sites. The resulting females were crossed with ipk2 mutant males carrying eGFP-marked balancer chromosome for selection of homozygous null flies. Double mutants were generated using standard fly genetics methods.
Measurement of Pupation Delay in ipk2 Mutants.
Eggs from wild-type (w1118) and ipk2/twi-GAL4, UAS-GFP were collected for 4 h at 25 °C. Newly hatched L1 larvae were transferred to vials in noncrowded conditions (30 larvae per vial) and aged at 25 °C. The numbers of newly formed pupae were recorded every 4 h from 92 to 164 h after egg deposition. ipk2 homozygous and heterozygous pupae were differentiated by the absence and presence of GFP signals, respectively. Data shown in Fig. S1C are the average percentages of pupae from three different experiments.
Generation of Kinase-Dead Ipk2 and Complementation Analysis.
Ipk2 is a member of a larger family of inositol phosphate kinases including IP3 3-kinases (IP3K) and IP6 kinases (IP6K) that share highly conserved catalytic motifs but possess different activities. Structural and biochemical studies on IP3K identified a critical asparagine residue that interacts with Mn2+ to coordinate ATP binding and, when mutated, completely ablated its kinase activity (42, 43). A D276A point mutation was made in the analogous site in Drosophila Ipk2 (ipk2D276A) using the QuikChange site-directed mutagenesis kit (Stratagene) and the following primers: 5′-GTCAA AATGA TCGCC TTTGC TCATG T-3′ and 5′-ACATG AGCAA AGGCG ATCAT TTTGA C-3′. We confirmed that the mutation had rendered the gene product catalytically inactive through its inability to complement IP production in yeast ipk2::G418 cells (Fig. S2A). Drosophila Ipk2 and ipk2D276A were cloned into pRS314-cup1-GFP-myc and transformed into S. cerevisiae ipk2::G418 cells (10). Expression was induced using 150 µM CuSO4 in CSM-tryptophan with 100 µCi/mL [3H]inositol. Soluble IPs were extracted and separated by Partisphere strong anion exchange HPLC as previously described (15).
pP[UAST]-GFP-dmIpk2, pP[UAST]-GFP-scIpk2, or pP[UAST]-GFP-ipk2D276A constructs and a P-element transposase plasmid were injected in w1118 embryos and germ-line transformants were selected with the w+ marker. Each insertion was then recombined onto the ipk220B mutant chromosome and crossed to ipk220B, Actin-GAL4/CyO recombinant flies and assessed for their abilities to rescue viability and developmental defects.
Scanning Electron Microscopy of Pupal Eyes and Wings.
w1118 and ipk220B zygotic pupae were dissected from their pupal cases and peripodial membranes, fixed in 4% (wt/vol) paraformaldehyde for 1 h, and dehydrated in a dilution series of ethanol (100% final). The samples were dried using a Pelco CPD2 critical point dryer and coated using an Anatech Hummer 6.2 sputter coater. Digital pictures were taken using a Philips XL 30 ESEM TMP equipped with AnalySiS image analysis and editing software (images shown in Fig. 1 G–J).
Developmental Western Blot for Ipk2.
Eggs from w1118 and ipk220B/twi-gal4, UAS-GFP were collected on grape juice plates for 4 h and aged at 25 °C on grape juice plates for L1 larvae and in food vials for L2 and L3 larvae. L1 and L2 were harvested 28 and 52 h after egg deposition, respectively, and wandering L3 larvae were picked after they left the food and were crawling along the wall of the vials. Larvae were squashed in lysis buffer [10 mM Tris (pH 7.5), 150 mM NaCl, and 1% Nonidet P-40] supplemented with complete protease inhibitor (Roche), 1 mM PMSF, 2 mM sodium orthovanadate, and 1 mM β-glycerolphosphate. Equal volumes of 2× Laemmli loading buffer [100 mM Tris⋅Cl (pH 6.8), 4% (wt/vol) SDS, 0.2% bromophenol blue, 20% (vol/vol) glycerol, and 5% (vol/vol) β−mercaptoethanol] were added to the lysates and then boiled for 5 min. The larval protein samples were loaded onto a 10% (wt/vol) SDS/PAGE gel. One hundred micrograms of protein was loaded per lane for detection of Ipk2 and 0.5 μg protein for detection of the actin loading control. Protein was then transferred to nitrocellulose membranes and subsequently immunoblotted with rabbit anti-Ipk2p and anti-actin antibodies (15).
In Situ Hybridization.
RNA probes were generated against Distal-less for in situ analysis of fly embryos, and hybridizations were performed as described in the Berkeley Drosophila Genome Project protocol (www.fruitfly.org/DGC/index.html). The following primers were used: Dll forward 5′-CGCAC TTTGT TCGTG TGCAG TCG, Dll reverse T7 5′-TTAAT ACGAC TCACT ATAGG GAGAC GTGCT GCTGC AGCTC CACG.
Staining of Wing Imaginal Discs.
Antibodies were anti–β-galactosidase (40a-1, 110; Developmental Studies Hybridoma Bank) and anti-BrdU antibodies (1:50; BD Biosciences). Late third-instar larval wing imaginal discs were used for immunofluorescence experiments. Discs were fixed in 4% (wt/vol) paraformaldehyde in PBS with 0.1% Triton X-100 (PBST) for 20 min. The discs were washed three times with PBST, blocked for 30 min with 5% (vol/vol) normal goat serum in PBST, and then incubated overnight with primary antibodies. The discs were next washed five times with PBST and incubated with FITC conjugated monoclonal secondary antibodies. After washing five times in PBST, the discs were mounted and visualized using a Nikon Eclipse TE-2000-E microscope.
For BrdU labeling experiments, wing and eye discs were dissected in PBS and incubated labeled with 0.1 mg/mL BrdU for 30 min. Then the discs were fixed in 4% (wt/vol) paraformaldehyde in PBS for 45 min and stained with anti-BrdU antibodies using staining methods described above.
TUNEL assay was performed on the discs using the ApopTag Red In Situ Apoptosis Detection Kit (Chemicon). Imaginal discs were dissected from L3 larvae, fixed in 4% (wt/vol) paraformaldehyde in PBS for 30 min at room temperature, washed three times with PBS, treated with 20 μg/mL proteinase K in PBS for 5 min, washed three times with PBS, and subsequently fixed again in 4% (wt/vol) paraformaldehyde in PBS for 20 min. The discs were then washed four times with PBS with 0.3% Triton X-100 and once with PBS with 0.1% Triton X-100, incubated in equilibration buffer for 1 h, and then in working strength TdT buffer at 37 °C for 13 h. The discs were washed with PBS with 0.1% Triton X-100 and stained with a rhodamine antibody for 45 min. After washing with PBS with 0.1% Triton X-100, the discs were mounted on slides with antifade and visualized using a Zeiss Axioscope equipped with Metamorph software.
Proliferation and Growth Rate Analysis.
Clones expressing GFP were induced using the flip-out technique that combines the FLP/FRT and UAS/GAL4 systems in wild-type (HS-FLP, [AyGal4]/[UAS-GFP]) and ipk2 null (HS-FLP, ipk220B, [AyGal4]/ipk220B[UAS-GFP]) (44). Eggs were collected for 4 h from crosses of wild-type (HS-FLP, [UAS-GFP] × w1118, [AyGal4]) or ipk2 (HS-FLP, ipk220B, [UAS-GFP] × w1118, ipk220B, [AyGal4]). Larvae were aged at 25 °C in food vials and heat-shocked for 30 min at 74 h after egg deposition. Wing discs were dissected from the GFP-positive larvae at 118 h and fixed in 4% (wt/vol) paraformaldehyde in PBS for 15 min. After washing in PBS, the discs were mounted on slides and the number of GFP-positive cells per clone were counted. Cell doubling time was calculated with the formula (log2/logn) hour, where n = median number of cells per clone and hour = time between heat shock and disk fixation. The experiment was performed twice.
Metabolic Labeling of Kc167 Cells.
Kc167 cell labeling experiments were done as described previously for S2 cells (15), with the following changes. Cells were treated with dsRNA for Ipk2 or Ipk1 for 4 d and then grown for one more day in media containing 50 μCi/mL [3H]inositol. Soluble extracts were separated by Partisphere strong-anion exchange HPLC.
Acknowledgments
We thank members of the J.D.Y. laboratory, especially Dr. Jessica Monserrate, for helpful discussions; Drs. Patrick Müller and Michael Boutros for sharing JAK/STAT pathway reagents; and Rick Fehon, Donald McEwen, Martin Zeidler, Andreas Bergmann, and Doug Harrison for fly stocks and reagents. This work is supported by funds from HHMI and NIH Grant HL-55672. J.D.Y. is an Alumni Investigator of HHMI.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1514684112/-/DCSupplemental.
References
- 1.Berridge MJ. Inositol trisphosphate and calcium signalling mechanisms. Biochim Biophys Acta. 2009;1793(6):933–940. doi: 10.1016/j.bbamcr.2008.10.005. [DOI] [PubMed] [Google Scholar]
- 2.Irvine RF, Schell MJ. Back in the water: The return of the inositol phosphates. Nat Rev Mol Cell Biol. 2001;2(5):327–338. doi: 10.1038/35073015. [DOI] [PubMed] [Google Scholar]
- 3.Seeds AM, York JD. Inositol polyphosphate kinases: Regulators of nuclear function. Biochem Soc Symp. 2007;74:183–197. doi: 10.1042/BSS0740183. [DOI] [PubMed] [Google Scholar]
- 4.Hatch AJ, York JD. SnapShot: Inositol phosphates. Cell. 2010;143(6):1030–1030.e1. doi: 10.1016/j.cell.2010.11.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Koldobskiy MA, Snyder SH. Inositol pyrophosphates in cell death and life. Cell Cycle. 2011;10(4):568–570. doi: 10.4161/cc.10.4.14771. [DOI] [PubMed] [Google Scholar]
- 6.Wilson MSC, Livermore TM, Saiardi A. Inositol pyrophosphates: Between signalling and metabolism. Biochem J. 2013;452(3):369–379. doi: 10.1042/BJ20130118. [DOI] [PubMed] [Google Scholar]
- 7.Lee J-Y, Kim Y-R, Park J, Kim S. Inositol polyphosphate multikinase signaling in the regulation of metabolism. Ann N Y Acad Sci. 2012;1271:68–74. doi: 10.1111/j.1749-6632.2012.06725.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.York JD, Odom AR, Murphy R, Ives EB, Wente SR. A phospholipase C-dependent inositol polyphosphate kinase pathway required for efficient messenger RNA export. Science. 1999;285(5424):96–100. doi: 10.1126/science.285.5424.96. [DOI] [PubMed] [Google Scholar]
- 9.Saiardi A, Caffrey JJ, Snyder SH, Shears SB. Inositol polyphosphate multikinase (ArgRIII) determines nuclear mRNA export in Saccharomyces cerevisiae. FEBS Lett. 2000;468(1):28–32. doi: 10.1016/s0014-5793(00)01194-7. [DOI] [PubMed] [Google Scholar]
- 10.Odom AR, Stahlberg A, Wente SR, York JD. A role for nuclear inositol 1,4,5-trisphosphate kinase in transcriptional control. Science. 2000;287(5460):2026–2029. doi: 10.1126/science.287.5460.2026. [DOI] [PubMed] [Google Scholar]
- 11.Maag D, et al. Inositol polyphosphate multikinase is a physiologic PI3-kinase that activates Akt/PKB. Proc Natl Acad Sci USA. 2011;108(4):1391–1396. doi: 10.1073/pnas.1017831108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim E, et al. Inositol polyphosphate multikinase is a coactivator for serum response factor-dependent induction of immediate early genes. Proc Natl Acad Sci USA. 2013;110(49):19938–19943. doi: 10.1073/pnas.1320171110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wickramasinghe VO, et al. Human inositol polyphosphate multikinase regulates transcript-selective nuclear mRNA export to preserve genome integrity. Mol Cell. 2013;51(6):737–750. doi: 10.1016/j.molcel.2013.08.031. [DOI] [PubMed] [Google Scholar]
- 14.Xu R, et al. Inositol polyphosphate multikinase is a transcriptional coactivator required for immediate early gene induction. Proc Natl Acad Sci USA. 2013;110(40):16181–16186. doi: 10.1073/pnas.1315551110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Seeds AM, Sandquist JC, Spana EP, York JD. A molecular basis for inositol polyphosphate synthesis in Drosophila melanogaster. J Biol Chem. 2004;279(45):47222–47232. doi: 10.1074/jbc.M408295200. [DOI] [PubMed] [Google Scholar]
- 16.Stevenson-Paulik J, Odom AR, York JD. Molecular and biochemical characterization of two plant inositol polyphosphate 6-/3-/5-kinases. J Biol Chem. 2002;277(45):42711–42718. doi: 10.1074/jbc.M209112200. [DOI] [PubMed] [Google Scholar]
- 17.Tomancak P, et al. Systematic determination of patterns of gene expression during Drosophila embryogenesis. Genome Biol. 2002;3(12) doi: 10.1186/gb-2002-3-12-research0088. research0088.1–88.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chou TB, Perrimon N. The autosomal FLP-DFS technique for generating germline mosaics in Drosophila melanogaster. Genetics. 1996;144(4):1673–1679. doi: 10.1093/genetics/144.4.1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cohen B, Simcox AA, Cohen SM. Allocation of the thoracic imaginal primordia in the Drosophila embryo. Development. 1993;117(2):597–608. doi: 10.1242/dev.117.2.597. [DOI] [PubMed] [Google Scholar]
- 20.Gavrieli Y, Sherman Y, Ben-Sasson SA. Identification of programmed cell death in situ via specific labeling of nuclear DNA fragmentation. J Cell Biol. 1992;119(3):493–501. doi: 10.1083/jcb.119.3.493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Parrish AB, Freel CD, Kornbluth S. Cellular mechanisms controlling caspase activation and function. Cold Spring Harb Perspect Biol. 2013;5(6):a008672. doi: 10.1101/cshperspect.a008672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hay BA, Wolff T, Rubin GM. Expression of baculovirus P35 prevents cell death in Drosophila. Development. 1994;120(8):2121–2129. doi: 10.1242/dev.120.8.2121. [DOI] [PubMed] [Google Scholar]
- 23.Mollereau B, Ma D. The p53 control of apoptosis and proliferation: Lessons from Drosophila. Apoptosis. 2014;19(10):1421–1429. doi: 10.1007/s10495-014-1035-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rong YS, et al. Targeted mutagenesis by homologous recombination in D. melanogaster. Genes Dev. 2002;16(12):1568–1581. doi: 10.1101/gad.986602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ollmann M, et al. Drosophila p53 is a structural and functional homolog of the tumor suppressor p53. Cell. 2000;101(1):91–101. doi: 10.1016/S0092-8674(00)80626-1. [DOI] [PubMed] [Google Scholar]
- 26.McEwen DG, Peifer M. Puckered, a Drosophila MAPK phosphatase, ensures cell viability by antagonizing JNK-induced apoptosis. Development. 2005;132(17):3935–3946. doi: 10.1242/dev.01949. [DOI] [PubMed] [Google Scholar]
- 27.Adachi-Yamada T, et al. p38 mitogen-activated protein kinase can be involved in transforming growth factor beta superfamily signal transduction in Drosophila wing morphogenesis. Mol Cell Biol. 1999;19(3):2322–2329. doi: 10.1128/mcb.19.3.2322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Müller P, Kuttenkeuler D, Gesellchen V, Zeidler MPM, Boutros M. Identification of JAK/STAT signalling components by genome-wide RNA interference. Nature. 2005;436(7052):871–875. doi: 10.1038/nature03869. [DOI] [PubMed] [Google Scholar]
- 29.Mukherjee T, Hombría JC-G, Zeidler MP. Opposing roles for Drosophila JAK/STAT signalling during cellular proliferation. Oncogene. 2005;24(15):2503–2511. doi: 10.1038/sj.onc.1208487. [DOI] [PubMed] [Google Scholar]
- 30.Harrison DA, McCoon PE, Binari R, Gilman M, Perrimon N. Drosophila unpaired encodes a secreted protein that activates the JAK signaling pathway. Genes Dev. 1998;12(20):3252–3263. doi: 10.1101/gad.12.20.3252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bach EA, Vincent S, Zeidler MP, Perrimon N. A sensitized genetic screen to identify novel regulators and components of the Drosophila janus kinase/signal transducer and activator of transcription pathway. Genetics. 2003;165(3):1149–1166. doi: 10.1093/genetics/165.3.1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Harrison DA, Binari R, Nahreini TS, Gilman M, Perrimon N. Activation of a Drosophila Janus kinase (JAK) causes hematopoietic neoplasia and developmental defects. EMBO J. 1995;14(12):2857–2865. doi: 10.1002/j.1460-2075.1995.tb07285.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Luo H, et al. Mutation in the Jak kinase JH2 domain hyperactivates Drosophila and mammalian Jak-Stat pathways. Mol Cell Biol. 1997;17(3):1562–1571. doi: 10.1128/mcb.17.3.1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Singer WD, Brown HA, Sternweis PC. Regulation of eukaryotic phosphatidylinositol-specific phospholipase C and phospholipase D. Annu Rev Biochem. 1997;66(1):475–509. doi: 10.1146/annurev.biochem.66.1.475. [DOI] [PubMed] [Google Scholar]
- 35.Murillo-Maldonado JM, Zeineddine FB, Stock R, Thackeray J, Riesgo-Escovar JR. Insulin receptor-mediated signaling via phospholipase C-γ regulates growth and differentiation in Drosophila. PLoS One. 2011;6(11):e28067. doi: 10.1371/journal.pone.0028067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Binari R, Perrimon N. Stripe-specific regulation of pair-rule genes by hopscotch, a putative Jak family tyrosine kinase in Drosophila. Genes Dev. 1994;8(3):300–312. doi: 10.1101/gad.8.3.300. [DOI] [PubMed] [Google Scholar]
- 37.Ekas LA, Baeg G-H, Flaherty MS, Ayala-Camargo A, Bach EA. JAK/STAT signaling promotes regional specification by negatively regulating wingless expression in Drosophila. Development. 2006;133(23):4721–4729. doi: 10.1242/dev.02675. [DOI] [PubMed] [Google Scholar]
- 38.Chen Q, Giedt M, Tang L, Harrison DA. Tools and methods for studying the Drosophila JAK/STAT pathway. Methods. 2014;68(1):160–172. doi: 10.1016/j.ymeth.2014.03.023. [DOI] [PubMed] [Google Scholar]
- 39.Baker NE. Cell proliferation, survival, and death in the Drosophila eye. Semin Cell Dev Biol. 2001;12(6):499–507. doi: 10.1006/scdb.2001.0274. [DOI] [PubMed] [Google Scholar]
- 40.Meier P, Silke J, Leevers SJ, Evan GI. The Drosophila caspase DRONC is regulated by DIAP1. EMBO J. 2000;19(4):598–611. doi: 10.1093/emboj/19.4.598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Verbsky J, Majerus PW. Increased levels of inositol hexakisphosphate (InsP6) protect HEK293 cells from tumor necrosis factor (alpha)- and Fas-induced apoptosis. J Biol Chem. 2005;280(32):29263–29268. doi: 10.1074/jbc.M503366200. [DOI] [PubMed] [Google Scholar]
- 42.González B, et al. Structure of a human inositol 1,4,5-trisphosphate 3-kinase: Substrate binding reveals why it is not a phosphoinositide 3-kinase. Mol Cell. 2004;15(5):689–701. doi: 10.1016/j.molcel.2004.08.004. [DOI] [PubMed] [Google Scholar]
- 43.Miller GJ, Hurley JH. Crystal structure of the catalytic core of inositol 1,4,5-trisphosphate 3-kinase. Mol Cell. 2004;15(5):703–711. doi: 10.1016/j.molcel.2004.08.005. [DOI] [PubMed] [Google Scholar]
- 44.Ito K, Awano W, Suzuki K, Hiromi Y, Yamamoto D. The Drosophila mushroom body is a quadruple structure of clonal units each of which contains a virtually identical set of neurones and glial cells. Development. 1997;124(4):761–771. doi: 10.1242/dev.124.4.761. [DOI] [PubMed] [Google Scholar]