Significance
Pasteur’s germ theory of disease initially seemed to have resolved the long-standing antagonism between the proponents of intrinsic and extrinsic disease mechanisms. However, by the turn of the 20th century, it had become clear that each microbe killed only a small minority of infected individuals. Infectious diseases killed half of all children before the age of 15 y, but this enormous burden was caused by the dazzling diversity of pathogens rather than by the potency of individual pathogens. The key problem concerning pediatric infectious diseases thus was identified: their pathogenesis. A human genetic theory of infectious diseases has emerged gradually from clinical and epidemiological studies, building on many elegant studies in plants and animals.
Keywords: human genetics, immunology, infectious diseases, pediatrics, primary immunodeficiency
Abstract
The key problem in human infectious diseases was posed at the turn of the 20th century: their pathogenesis. For almost any given virus, bacterium, fungus, or parasite, life-threatening clinical disease develops in only a small minority of infected individuals. Solving this infection enigma is important clinically, for diagnosis, prognosis, prevention, and treatment. Some microbes will inevitably remain refractory to, or escape vaccination, or chemotherapy, or both. The solution also is important biologically, because the emergence and evolution of eukaryotes alongside more rapidly evolving prokaryotes, archaea, and viruses posed immunological challenges of an ecological and evolutionary nature. We need to study these challenges in natural, as opposed to experimental, conditions, and also at the molecular and cellular levels. According to the human genetic theory of infectious diseases, inborn variants underlie life-threatening infectious diseases. Here I review the history of the field of human genetics of infectious diseases from the turn of the 19th century to the second half of the 20th century. This paper thus sets the scene, providing the background information required to understand and appreciate the more recently described monogenic forms of resistance or predisposition to specific infections discussed in a second paper in this issue.
For this Inaugural Article, I decided to deviate from the standard approach of providing an extensive review of the work carried out in my laboratory over the last 20 y or in the field of human genetics of infectious diseases more generally since the start of the 20th century. Instead, I thought it would be interesting to focus on the associated ideas, some responsible for shaping our work and others shaped by it. Indeed, the establishment of my laboratory was based on an idea that arose when we went back over previous work. My colleagues and I revisited a problem first posed at the turn of the 20th century, when Charles Nicolle discovered unapparent infections (1, 2). This discovery identified the key problem in the field of infectious diseases; paradoxically, the problem concerned their pathogenesis. A century later, although we are able to prevent many human infectious diseases through hygiene and vaccination and to cure others by drug treatment or surgery, we still do not fully understand the root causes of most infectious diseases and their clinical variability. Indeed, paradoxically, our ability to prevent and cure disease may have hindered our understanding. Microbes are the only living organisms that pose almost as much of a threat to mankind as mankind itself. In addition, eukaryotes were born into a world already populated by bacteria, archaea, and viruses, and small eukaryotes themselves rapidly acquired the ability to infect larger organisms. Therefore it is essential to consider the evolution of life from this immunological perspective. Predation, in its conventional sense, has constituted a less urgent and broad challenge to eukaryotes than infection, which triggered the development in eukaryotes first of cell-intrinsic immunity (cell-autonomous mechanisms), then of cell-extrinsic innate immunity (phagocytosis of pathogens by professional cells), and, finally, of cell-extrinsic adaptive immunity (somatic diversification of antigen-specific cells). Understanding the pathogenesis of infectious diseases, particularly those affecting humans, therefore is important from both clinical and biological standpoints.
Intrinsic and Extrinsic Theories of Disease
We should begin by considering the long period preceding the germ theory of disease. Throughout the history of medicine, there had been conflict between two opposing theories, one which saw disease as caused by extrinsic factors and the other that saw disease as an intrinsic disorder. With hindsight, it is easy to see that the microbial and genetic theories of disease underlie these two opposing views. These theories remained purely speculative for a very long period, serving only to fuel the controversy. In the absence of the solid scientific frameworks provided by microbiology and genetics, it is hardly surprising that the discussions were passionate and unfruitful. This topic has been excellently reviewed elsewhere (3, 4). It is sufficient here to highlight a point of historical importance that is relevant to this article. By painstakingly defeating vitalism, both the clinicopathological method that proved so useful in the construction of medicine and the experimental method that established the physicochemical basis of physiology and pathology—the two major medical breakthroughs of the late 18th and early 19th centuries (5, 6)—also delayed the emergence of the germ theory. Indeed, germs were reminiscent of “miasmas,” and the “milieu intérieur” was thought to isolate organisms from the environment. Organisms thus were seen as self-sustaining machines in which pathological conditions reflected the inner derailment of physiology (3, 6–10). In this context, the compelling proof of contagion provided by at least two superb investigators, Ignaz Semmelweiss in Vienna in the 1840s (11) and John Snow in London in the 1850s (12), was disregarded. The careers of these scientists lay in tatters, partly because their work incriminated humans, including rulers (in the case of Snow) and physicians (Semmelweiss), in the transmission of disease (11, 12). The proponents of the intrinsic theory of disease were in the ascendancy, but not for long. The establishment of the germ theory of diseases, between 1865 and 1882, marked a turning point, with the pendulum beginning to swing in the opposite direction.
The Germ Theory of Disease and Its Momentum
The legendary work that Pasteur began on silk worms in 1865 led to the germ theory of diseases and the identification of many pathogens of animals and humans (13, 14). This revolution culminated in Koch’s discovery of the agent of tuberculosis in 1882 (15). This advance was one of the most striking victories of the germ theory, because it dealt with the most feared condition of the time. It also was supported by Koch’s stringent postulates, a set of strict criteria that Koch thought were required to attribute the responsibility for disease to a microbe (15). These postulates include the notions that healthy individuals should not harbor the pathogen and that the pathogen should be found in all patients. At the time, these rigorous criteria were considered necessary to prove that microbes caused disease. However, even the most valid discoveries, or at least certain aspects of them, including some of the principles inferred from or supporting them, have a short life span. There is no such thing as an absolute truth outside the realm of mathematics; there are only causal relationships in given experimental or natural conditions that are subject to continual change, and this variance applies even more strongly to living organisms than to inanimate matter. Fast-forwarding to 1907–1911, it is easy to understand that the time for considering interindividual variability in the course of infection had not yet come. This niggling problem actually went against the emerging, prevailing current. It had taken nearly 20 y of painstaking effort for European microbiologists to convince the world, and the skeptical medical community in particular, that most diseases were infectious. Veterinarians were more open to the evidence supporting the germ theory (16). No matter how painful, the discoveries of Charles Nicolle rendered a partial revision of Koch’s postulates necessary for subsequent studies of the pathogenesis of infectious diseases (1, 2).
Latent and Unapparent Infections
Indeed, about 30 y after Koch formulated his criteria, it became apparent from several lines of observation, including Nicolle’s discovery of unapparent infections in 1911 (1, 17), that only a minority of infected individuals developed clinical disease, and an even smaller minority actually died from the disease. Charles Nicolle regarded his discovery of unapparent infections as his most important contribution to biomedical research (www.nobelprize.org). An unapparent infection is defined as the presence of replicating microbes in the tissues or bloodstream of an individual who has no symptoms but who nevertheless may be contagious. This differs from the silent incubation and convalescence periods seen in symptomatic individuals. These findings unequivocally showed that microbes are necessary, but not sufficient, for the development of infectious diseases. A number of previous observations, such as interindividual variability during epidemics or in endemic areas and variable clinical outcome in symptomatic individuals, already seemed to point in the same direction. Clemens von Pirquet, whose 1907 discovery of allergy to mycobacterial tuberculin can be seen as preliminary evidence of latent infections, also played an important role (18, 19). He and Charles Richet had independently discovered allergic and anaphylactic reactions. In the wake of the development, by Max von Gruber and Fernand Widal, of serological methods for diagnosing infectious diseases in the late 19th century, clinical serology was employed mostly for diagnostic purposes rather than for screening healthy individuals (20, 21). Von Pirquet’s tuberculin skin test not only confirmed the existence of survivors of an infectious disease that had killed other individuals but also led to the characterization of latent infections (18, 20–22). Indeed, in 1927 it led to the first proof of latency, with the growth of tubercle bacilli from the lungs of patients who had died of other causes (22), and to the subsequent demonstration, in 1954, that treating latent tuberculosis with antibiotics could prevent the development of disease by reactivation (23). A latent infection is defined as the presence of dormant, nonreplicating microbes in the tissues of an organism with no symptoms, without contagion, regardless of the previous occurrence of symptoms. The discoveries of unapparent and latent infections independently demonstrated that factors other than infection are required for disease development. They also made it clear that disease severity of any given infection varies greatly among patients.
Lethal Disease Is a Common Outcome of Infections Considered Collectively
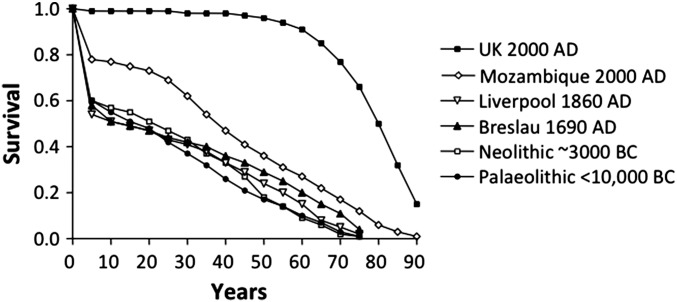
Interindividual variability undoubtedly has always been an element of infectious disease in humans, but it has largely been ignored for much of the history of medicine because it goes against germ theory and is counterintuitive from a historical standpoint. Indeed, the burden of infectious disease was colossal until the end of the 19th century, with half of all children dying of fever before the age of 15 y and with the mean life expectancy at birth being only about 20–25 y (Fig. 1) (24). This situation had prevailed worldwide since the dawn of mankind. Determinations of the ages at death of skeletons found in prehistoric burial sites and analyses of birth and death data recorded over the course of history have shown that human mortality curves did not begin to improve until the middle of the 19th century (24, 25). Infection was by far the greatest killer, with many more victims than war, famine, or predation. The black plague epidemic in the Middle Ages, which led to the loss of a third of the European population, followed an intriguing pattern of emergence followed by disappearance. This pattern is a key point, as we will see when we return to consider the evolutionary implications of the genetic theory of infectious diseases. The global infectious burden was caused mostly by primary infections, striking children and young adults. Clinical disease from reactivation or recurrent infection in the elderly is a problem that was almost unknown to mankind three or four generations ago. The burden of infection was gradually controlled by the successive development of an array of approaches, including hygiene, vaccines, aseptic surgery, and antibiotics, all of which were derived from the germ theory of disease. The approaches with the greatest epidemiological impact, causing rapid shifts in human mortality curves, were those such as hygiene and aseptic surgery that could be applied to many pathogens.
Fig. 1.
Human mortality curves. Mortality curves at various periods of human history, from the Paleolithic period (<10,000 BC) to modern times (2000 AD). Contemporary data for the United Kingdom and Mozambique are available from the WHO site (www.who.int/topics/global_burden_of_disease). Older data were obtained from ref. 24. Life tables for the Paleolithic and Neolithic periods are based on examinations of skeletons, assuming that 60% of newborn infants survived to the age of 5 y, because few very young skeletons were found in the burial grounds. For most of human prehistory and history worldwide (i.e., until the end of the 19th century), as many as half of all children died before the age of 15 y, and life expectancy at birth averaged only 20–25 y. Fever was by far the greatest killer. The gradual adjustment of the immune system by natural selection did not increase life expectancy because of the coevolution of microorganisms and the emergence of new infectious threats. Thus, the increase in life expectancy in the 20th century does not reflect the sudden and global natural selection of high-quality immune genes. It reflects the conquests that accompanied the germ theory of disease: hygiene, aseptic surgery, vaccines, and the development of drugs to treat infection. The area between the four ancient curves and the curve for the United Kingdom in 2000 corresponds to ∼65% of the individuals currently alive. Most of these individuals have retained immunodeficiencies against one or more infectious agents that are masked by medical progress. Reproduced from ref. 25.
Lethal Disease Is a Rare Outcome of Individual Infections
At the dawn of the 20th century, the idea that most of the individuals infected with a given microbe might remain healthy thus seemed to go against both the germ theory of disease and the infectious history of mankind. However, the historical burden of infection actually principally reflected the extraordinarily large number of infectious agents diversifying during the course of evolution. The burden of infection was huge, not because all microbes were pathogenic to a large proportion of individuals, but because there were countless types of microbes, most of which killed only a small proportion of people. We now know that, for the vast majority of microbes, including some of those responsible for the most dreadful scourges of mankind, such as Plasmodium, Mycobacterium, and influenza virus, most infected people remain well or develop self-healing disease (26). Plasmodium causes life-threatening malaria in only about one in 1,000 infected children. Less than 10% of individuals infected with Mycobacterium tuberculosis go on to develop tuberculosis, even counting benign forms. Even during the terrible 1918 pandemic, which killed more people than World War I, only about 1–10% of the individuals infected with the influenza virus died from the disease. A much smaller proportion of infected individuals go on to develop severe illness following infection with classic pathogens as diverse as the herpes simplex and varicella zoster viruses, the Streptococcus and Staphylococcus bacteria, Candida and Cryptococcus fungi, and Toxoplasma and Schistosoma parasites, to name a few examples. The same pattern held true for pathogens now controlled by vaccination, such as poliovirus, measles virus, Bordetella pertussis, and Corynebacterium diphtheriae. The Ebola and human immunodeficiency viruses, and a few others, appear to be exceptions, killing most of the individuals they infect, but at different rates. However, this situation may be temporary, and these diseases eventually may come to follow the typical pattern as the pandemics unfold and the human and viral populations evolve together.
The Birth of Immunology
Why does the clinical course of infection differ among individuals? In 1910 an explanation was already at hand, the immunological theory. Known as the “serological theory” during the “classic” period of immunology governed by immunochemistry (i.e., until the 1960s), but now more widely referred to as a “somatic cell theory” (during the “modern” period of immunology governed by immunobiology), its cornerstone is antigenic specificity. In jawed vertebrates, antigen-specificity is based on T and B lymphocytes, corresponding to the two arms of adaptive immunity, a phenomenon of such biological importance that it has evolved twice in vertebrates by convergent evolution, as beautifully shown by Max Cooper (27–30). Jawless vertebrates rely on a completely different mechanism to generate an equivalent level of clonally distributed somatic diversity. T cells themselves seem to have evolved similarly into two distinctive lineages, with αβ and γδ T cells (and their equivalents likewise are present in jawless fish). This adaptive immunity is perhaps the most extraordinary example of convergent evolution in extant and extinct organisms and arguably is the most thoroughly documented example at the molecular and cellular levels, with an entire physiology built in two different ways (31). The best illustration of the potential contribution of the immunological theory to interindividual variability in the course of infection, and, indeed, the first evidence in its favor, was Louis Pasteur’s demonstration of vaccination by attenuated microbes in 1880–1882, the birth of immunology (32).
An Immunological Theory of Infectious Diseases
By 1910, it was thus possible to surmise that interindividual clinical variability might result from selective protection conferred by a previous primary infection in some individuals. Individuals who had survived infections by a naturally occurring, less virulent related microbe or by a small inoculum of the same microbe would have developed acquired, specific immunity. The antigenic overlap between closely, or even remotely, related microbes (by cross-reaction) can be sufficient to influence the clinical response. Some infants also may benefit passively from maternal antibodies. However, this powerful concept is most applicable to adults; it is particularly applicable to the elderly, whose somatic immunity has had decades to diverge in response to different environmental or commensal microbial challenges and opportunities to decline differently with the effects of aging. Different outcomes following an outbreak in a population of individuals who have had 80 or 90 y to diverge somatically, both genetically and epigenetically, are understandable. It seems only logical that a greater diversity of vaccination and infection history is associated with greater clinical heterogeneity in the course of any current infection. Interindividual variability in the course of secondary infection at the population level is best explained by the immunological, somatic, adaptive theory: immunity acquired through a memory of primary infections. How, however, can we explain clinical heterogeneity in the course of primary infection with a completely new pathogen, unrelated to previous experiences, and typically affecting young children? It is difficult to accept the idea that the somatic imprint of past infections and immunizations is sufficiently idiosyncratic and powerful to account for matters of life and death relating to primary infection in childhood.
Acquired Immunodeficiency
Similarly, an acquired impairment of host defense, whether transient or permanent, might be expected to account for severe infections (33, 34). Some infections that themselves are benign can weaken the host for a few days. For example, deaths from pneumococcal disease often occur in the wake of influenza (35). Virus-induced immunosuppression may be long-lasting and underlie a broader susceptibility, as exemplified by measles (36). Other viruses, such as the HIV, weaken immunity in a more sustained and profound manner (37). Any chronic disease weakens the immunity of the body. A child with organ failure, whether of the kidney, lung, intestine, pancreas, liver, brain, or heart, is at risk for life-threatening infections through overt, subtle, or elusive mechanisms. Breaches of the skin or mucous membranes also pose an obvious threat of microbial infection. Needless to say, immunosuppressant drugs may underlie severe and even opportunistic infections. Iatrogenic infections result from a combination of these factors. Malnutrition in some ways mimics intestinal failure and has been shown to account for severe infections (38). The various types of organ failure associated with malnutrition may contribute to immunodeficiency (39). It also has been suggested that specific vitamin deficiencies favor infectious diseases. Few data are available, but these mechanisms are plausible, and medical common sense would suggest that the various forms of malnutrition are chronic diseases. As such, like other chronic conditions, they may precipitate the development of severe infectious diseases, even if some of their features are specific. Supplementation with micronutrients and vitamins does improve child health in developing countries (40). Nevertheless, in any group of malnourished or vitamin-deficient children, only a small proportion of those infected with a particular microbe develop disease. Admittedly, this proportion is larger than that in a comparable group of well-fed children. However, this hypothesis cannot account for the deaths from fever of Darwin’s son and two daughters and Pasteur’s three daughters, all of whom lived in upper-class clean environments with adequate nutrition and a good education (41). Understanding the deaths of these children and others living in similar conditions, which probably occured during a primary infection, can be seen as a holy grail in the field of infectious diseases.
A Radical Version of the Germ Theory
Are there alternative explanations for interindividual variability in the course of primary infection? A radical microbiological theory suggests that microbial variation, whether qualitative or quantitative, can account for the variability of manifestations. Qualitative differences resulting from microbial evolution have been shown to account for interpopulation differences. The acquisition or loss of a virulence genotype or phenotype may initiate or terminate an outbreak (42). However, it is more difficult to see how such qualitative variation could account for interindividual variability at a given time in a household, school, or village, because such variation in virulence is more likely to operate over time. Evidence for the importance of quantitative variation is provided by experimental infections in animal models (43–45). Infections of a given animal with one, 1,000, or one million viruses or bacteria certainly have very different impacts in vivo. How can this finding be translated to the conditions of natural infections? Has anyone ever measured the variability of infectious challenges? Are there huge differences among people infected with the influenza virus, pneumococcus, Cryptococcus, or Plasmodium? The microbial inoculum is comparable to the sting of a bee or a wasp: One can hurt, whereas 100 can kill. Unfortunately, it has not been possible to test this plausible theory, and we also know that one sting can kill in rare circumstances defined by Richet and von Pirquet as anaphylaxis or allergic. Interestingly, the tragic Lübeck accident in 1930 showed that inoculation with a given dose of a given virulent microbe (Mycobacterium tuberculosis, the agent of tuberculosis, instead of the bacillus Calmette–Guérin vaccine) did not erase interindividual variability, although questions remain concerning the homogeneity of the inoculum (46). Overall, although not yet documented scientifically in humans, the microbiological and immunological theories probably account for certain aspects of interindividual vulnerability in the course of primary infection.
The Microbiologists Had More Pressing Goals
These notions were actually seen more as implicit assumptions than as explicit theories, because neither microbiologists nor immunologists were particularly interested in the question of interindividual variability in the course of primary infection. Before discussing other theories, including the human genetic theory in particular, we should consider the reluctance of microbiologists and immunologists to tackle this problem, which I like to refer to as the “infection enigma,” immediately after World War I. There are various reasons of both historical and epistemological interest for this reticence. The microbiologists were busy discovering new pathogens and then new commensals and saprophytes and studying their taxonomy, metabolism, and genomes in rapid succession (47). Virology quickly emerged as a new discipline in the 1920s, also attracting considerable talent and resources. The study of bacteria and phages played a crucial role in the development of molecular biology. Microbiologists developed the fields of microbial pathogenesis and immunity to infection in animal models (mostly in vivo, with the development of inbred mice and other animals, from the 1930s onward) and later, building on the work of virologists, the field of cellular microbiology (mostly in vitro, at the crossroads of microbiology and cellular biology). They worked relentlessly to develop new vaccines and serotherapies. However, work on serotherapies gradually lost momentum as microbiologists managed to kill microbes directly with chemical compounds. Only recently has interest in this field increased again, with the possibility of developing mAbs against specific microbes. Following on from the purification of the antimalarial drug quinine by Joseph Pelletier and Joseph Caventou in the 1820s and Ehrlich’s synthesis of the antisyphilitic agent salvarsan in 1910, Domagk’s landmark discovery of sulfamides in 1931 (48) was followed rapidly by the discovery of various antibiotics by René Dubos, Alexander Fleming, Ernst Chain, and Howard Florey (49). These discoveries, in turn, opened substantial new avenues of research into antiviral, antifungal, and antiparasitic drugs and microbial resistance to them. From the 1980s onward, microbiologists increasingly turned their attention to immunological questions relating to infection, after decades in which immunologists had focused much of their work on noninfectious antigens. Microbiologists have clearly been extraordinarily busy and successful in this respect.
A Movement of Immunologists Away from Studies of Infection
Likewise, the immunologists were too busy to tackle the infection enigma. Their attention was understandably drawn to another, equally fascinating and challenging problem: the “antibody enigma” (50). How could humans and animals make specific antibodies not only against so many microbes but also against all sorts of noninfectious natural antigens and even against structures that did not exist naturally and were chemically designed? Paul Ehrlich had shown in 1897 that mammals could make antibodies against plant toxins, and in 1898 Jules Bordet demonstrated that antibodies could be mounted against xenogenic erythrocytes (reviewed in ref. 51). Then, from 1917 onward, Karl Landsteiner showed that animals could make antibodies against almost any type of newly synthesized chemical compound, provided it was bound to a carrier as a hapten (52). These observations were both fascinating and challenging. Given the well-known and discrete nature of these chemical antigens, these observations greatly facilitated the study of antigen-specific responses. Since the early days of immunology, based on immunochemistry focusing on antibodies and complement, and the emergence of immunology as an immunobiology, with the discovery of the T- and B-cell dichotomy by Jacques Miller and Max Cooper in the 1960s (53–58), antigen-specific responses and the mechanisms underlying them have been the main focus of immunologists. Granulocytes, and even macrophages, were left to hematologists. Macrophages were accepted by the immunological world only after they were shown to present antigens to T cells. Even well into the 1970s, at the Basel Institute of Immunology, Niels Jerne reportedly would fine anyone mentioning the name of a cell that was not a lymphocyte (59). Complement gained prestige in the early 1900s because of its alliance with antibodies. It was not recognized as a fundamental arm of innate immunity until immunologists rediscovered the importance of innate immunity and attempted to reappropriate it after the discovery of the role of Toll and the Toll-like receptors in host defense outside mainstream immunology (60–66). Immunology was born of vaccination and initially aimed to improve our understanding of vaccination, but the task of vaccinating people (and understanding how vaccination worked) was left to microbiologists. The greatest therapeutic achievement of immunologists has been the development of immunosuppressive medication, culminating in the clinical use of mAbs. The early and sustained focus on adaptive immunity to chemical antigens diverted the attention of immunologists from infectious agents, and it was not until the 1980s that a few immunologists, greatly outnumbered by microbiologists, realized that the study of immunity against chicken ovalbumin or hen egg lysozyme in inbred mice might not be entirely representative of immunity to the infectious threats confronting humans’ natural defenses.
Unconscious Trends?
Behind these rational and understandable motives, other deep trends may have been at work, perhaps unconsciously in most scientists. Here, I take the risk of offering a personal interpretation of the last 100 y of research in the fields of microbiology and immunology. Microbiologists worldwide, not only those from the French- and German-speaking communities, are the inheritors of the greatest medical theory ever, the germ theory of disease. This theory alone, with its direct implications, has saved billions of human lives and led to a swift increase in life expectancy from 20 to 80 y. It also has been enormously useful in agriculture and in veterinary medicine. Microbiologists are the guardians of the most epic and legendary conquest of mankind, a doctrine that has transformed the world. They were and have remained the “microbe hunters” (67). No other scientific community can claim to have saved larger numbers of people, not even immunologists, whose in-depth studies have not yet fundamentally changed the basic principles of vaccination and whose mAbs are not yet widely used to treat patients. Therefore, understandably, the vision of microbiologists has remained focused on microbes and, more specifically, on pathogens. They often discuss “host–pathogen” interactions, but, in reality, there is no such thing as a host—that term is a mere abstraction. There are only cells, tissues, organs, organisms, and populations. Likewise, there are no pathogens (34, 68). There are only viruses, bacteria, fungi, or parasites, which are tremendously diverse, each being pathogenic in some individuals and not in others. The convenience of referring to a “host” and a “pathogen” is reminiscent of typology and essentialism (69, 70), even though infinite diversity is an intrinsic part of the nature of micro- and macroorganisms. To some extent, these inner forces have diverted microbiologists from the key issue that any given microbe is a threat to only a small minority of humans.
Immunology vs. Immunity
The behavior of immunologists is a bit more difficult to decipher. My personal interpretation is that immunologists love the immune system, just as neurologists love the brain and cardiologists love the heart. In particular, since the founding of the field with the discovery of vaccination, they have been under the spell of adaptive immunity, a term coined much later [to my knowledge, by Robert Good (71) in 1963]. Most did not truly agree with Elie Metchnikoff and sided with Paul Ehrlich because the macrophage did not fit into the emerging paradigm of antigen-specific responses (72, 73). Phagocytes could not yet contribute to the understanding of acquired, adaptive, specific immunity. Immunologists’ notion of antigen specificity was so qualitative, so radical, that most, paradoxically, did not even appreciate Karl Landsteiner’s quantitative approach to the antibody enigma (72). Overall, immunologists have put much more effort into developing immunology than into studying immunity in general. Immunologists have trouble coping with the idea that the immune system as a whole and its hematopoietic component in particular is the weakest of all physiological systems at the individual level. In 2015, many publications in immunology still begin with a proud declaration that the “host immune system controls microorganisms.” Articles with self-contradictory titles, such as “Lethal infection in an immunocompetent individual” are still being published. Would anyone really consider death from respiratory failure to have occurred in a patient with a normal oxygen pressure in the blood? Death from infection is, by definition, death caused by immunodeficiency. However, immunologists still persist in designating immunodeficiencies not on the basis of the phenotype of the organism (e.g., death from infection) but purely on the basis of an identifiable biological phenotype (e.g., lack of antibodies). The immune system in its entirety, including both its hematopoietic and other components, actually is effective only at the population level, ensuring that enough individuals live long enough to reproduce. In natural conditions, it is not efficient at the individual level. This firm conclusion can be drawn from human mortality curves (Fig. 1) and does not require identification of the underlying immunological abnormalities. Even a short life expectancy is already a remarkable achievement, given the enormous difficulty of the task—defending the body against trillions of evolving microbes. It is clearly more challenging to deal with an infinite variety of evolving microbes than with atmospheric pressure or the concentration of oxygen in the air, which vary little and certainly not at such a rapid rate. Overall, although most of us are immunodeficient with respect to at least one microbe, this immunological “tour de force” is good enough for the species as a whole.
An Ecological Theory
The need to search for an explanation for interindividual variability in the course of primary infection outside the classical realms of microbiology and immunology was perceived by some scientists from these fields. In his famous 1955 article, “Second Thoughts on the Germ Theory” (74), René Dubos eloquently posed the problem of interindividual variability. He acknowledged the existence of well-known conditions, such as diabetes, that impair host defense. Between the lines, he also alluded to a fourth, more original theory, which I refer to here as the “ecological theory of infectious diseases.” René Dubos, a profound thinker and author of the most insightful intellectual biography of Louis Pasteur (75, 76), was an eminent microbiologist who purposefully discovered the first antibiotics. He also was one of the founders of the scientific, as opposed to obscurantist, branch of ecology. Although an experimentalist, he can be seen as one of the pioneers in the ecology of infectious diseases in wildlife (77) and the study of the microbiota of the gastrointestinal tract (78, 79). In his essay he suggested that the determinism of infection could be influenced by environmental variation other than that of the causal microbe, including variations in other microbes or their actions with a strong but transient impact on the infectious process. For example, it seems reasonable to assume that particular changes in diet or lifestyle (e.g., professional exposure, sexual behavior) would influence the infectious process. As discussed above, it now has been shown that some infections can impair host immunity and thus manifest as another infection. However, such variation would be predicted to be uniform in any given community; therefore it could hardly account for the death from pneumococcal disease, measles, or diphtheria, of a single child in a household, class, or school. The ecological theory could account plausibly for sudden or gradual changes in the course of an epidemic or a pandemic but not for the observation that only a small minority of children at a given location develop life-threatening disease. In his essay, Dubos implicated the vulnerability of a weak or weakened organism in the development of infectious disease, hence his “second thoughts,” but he did not use the term “immunodeficiency,” nor did he address, even remotely, the notion that an immunodeficiency could be inherited. He favored the notion that the immune response of certain individuals is weaker at certain times than at others because of well-delineated illnesses or more subtle environmental changes affecting host ecology. Surprisingly, his essay did not incorporate the findings of 50 y of fruitful research into the classical genetics of plants, animals, and humans, nor did it anticipate the outcome of the emerging molecular genetic revolution.
Plant Geneticists
It is perhaps unsurprising that the start of the 20th century saw investigators from a third group, geneticists, take up the gauntlet and decide to tackle the infection enigma. Geneticists may be seen as the heirs to the “intrinsic” theory of diseases, although their background is different. They also are inspired by the classical physiologists, who established the importance of the “milieu intérieur,” defeated the vitalists by showing that physiology obeys the laws of chemistry and physics, and showed that disease is an abnormal physiology. The early physiologists were reluctant to contemplate the possibility that microscopic organisms from the environment, reminiscent of miasmas, could kill. A century later, human, animal, and plant geneticists tackled the infection enigma as a problem of inborn resistance or susceptibility to infection. As early as 1905 very elegant studies in plants first established the importance of genetic make-up in the determinism of infection by demonstrating the Mendelian resistance or susceptibility of wheat to yellow rust (Table 1) (80). A series of superb studies then went on to prove, beyond any reasonable doubt, that infectious diseases in plants could also be genetic traits, often displaying Mendelian inheritance (81, 82). These data even supported Harold Flor’s 1942 gene-for-gene model, based on his study of rust in flax, in which a “resistance gene” in a plant counterbalanced a specific “effector gene” in a microbe (81, 83, 84). From 1993 onward, resistance genes against various specific pathogens were cloned in various species, and this model was adopted as a universal paradigm in plants (82, 85–87). The extraordinary achievements of plant genetics from Gregor Mendel onward are indeed admirable. Regrettably, these papers had much less influence in veterinary and human medicine than they deserved, providing an example of the lack of synergy and cross-fertilization (so to speak) between different branches of modern science.
Table 1.
Mendelian genetic determinism of infectious diseases in plants, mice, and humans
| Species | Mendelian infections | Gene identification | ||
| Broad | Specific | Expression | Linkage | |
| Plants (tomato) | – | 1905 (wheat) | – | 1993 |
| Mouse (Bcg/Lsh/Ity) | 1959 (Dh) | 1964 (Mx) | 1986 (Mx) | 1993 |
| Human (MSMD) | 1952 (XLA) | 1946 (EV) | 1993 (complement) | 1996 |
Mendelian infections (left two columns) can be broad (multiple infections) or specific (a single type of infection). Gene identification (right two columns) only corresponds to Mendelian infections to a specific infection. For mice and humans, a broad range of infections was associated with distinctive immunological phenotypes (e.g., asplenia for Dh and agammaglobulinemia for X-linked agammaglobulinemia (XLA), whereas no immunological phenotype was initially associated with myxovirus (Mx) and epidermodysplasia verruciformis (EV). In plants, the inheritance of susceptibility/resistance to infection appears to be often specific for a particular pathogen. The resistance genes typically have been mapped for plants, and the plant species, rather than the resistance loci or pathogens, are indicated in the table. MSMD, Mendelian susceptibility to mycobacterial disease. References are provided in the text.
Animal Geneticists
From the 1920s onward, the large-scale development of inbred mice led to a generalization of the observation that certain strains of rats, rabbits, or guinea pigs were more vulnerable than others to specific infectious challenges. The work of Max Lurie and Leslie Webster (43–45) was particularly important in this regard. This approach culminated, nearly 50 y later, in molecular studies of infections in inbred mice based on forward genetics (44, 88–91). The first mouse immunodeficiency, associated with broad susceptibility to infection, was attributed in 1959 to the Dh locus and syndromic asplenia (Table 1) (92). In 1964, Jean Lindenmann (93) attributed predisposition to severe influenza in the absence of detectable immunological abnormalities to the myxovirus resistance (Mx) locus, which was identified by expression cloning in 1986 (94). The Bcg, Lsh, and Ity loci, controlling predisposition to infections caused by Mycobacterium, Leishmania, and Salmonella, respectively, were implicated in these diseases in the 1970s and shown to be allelic in 1982 (95). The causal gene Nramp1, encoding natural resistance-associated macrophage protein 1, was identified by genome-wide linkage in 1993 (96). It is historically interesting that this heroic genetic achievement, which provided proof of principle for positional cloning in mice, concerned an infectious phenotype. It soon was followed by a series of spectacular discoveries in various fields of mouse genetics, including infectious diseases. The groundbreaking identifications of Tlr4 as the Lps gene (97) and Ly49h as the Cmv1 gene (98) arose from this approach. These and other studies provided a molecular basis for the Mendelian inheritance of resistance or susceptibility to a specific infectious agent.
Human Geneticists
Both clinical geneticists, including Archibald Garrod (99), and population geneticists, including Karl Pearson (100), working at the highest levels in human genetics, proposed a germline genetic theory of infectious diseases. Garrod described the first known Mendelian inborn error of mankind, alkaptonuria, in 1902 and was an archetypal Mendelist (101, 102). Pearson was the heir to the work of Francis Galton, with whom he founded the journal Biometrika in 1901, and as such was an emblematic biometrician (103). As often, the two perpetually competing factions of geneticists, the biometricians and Mendelists, were in agreement on the idea, although not of course on the specific genetic architecture. The amassed body of clinical and epidemiological evidence had become compelling by the 1940s. These studies ranged from the simple clinical description of multiplex pedigrees evocative of Mendelian transmission to more sophisticated genetic epidemiological studies. For example, appendicitis was shown to segregate as an apparently autosomal dominant trait in several multiplex kindreds (104). This disease, which is caused by commensal organisms of the gastrointestinal tract, is infectious but not contagious. Its familial segregation therefore was interpreted as reflecting host inheritance. Some beautiful twin studies on tuberculosis were carried out in Germany in the 1930s and in America in the 1940s, revealing concordance rates of up to 80% for monozygotic twins but only 20% for dizygotic twins (105, 106). These lines of observational investigation were pursued and culminated in the 1980s in an impressive Scandinavian epidemiological adoptee study, which showed that early death from infection was highly correlated with the early death from infection of the biological, but not the adoptive parents (107). Of all types of human disease tested, infection, paradoxically, appeared to be the most genetically determined. From this perspective, how can we account for the deaths from infection of the children of Pasteur and Darwin? Had Darwin been aware of the work of Mendel, he might have suspected, by 1865, that the childhood deaths from fever in his own family could be autosomal recessive, because he had married his first cousin, Emma, and lost 3 of his 10 children to fever (41). Admittedly, he also might have been side-tracked by the observation that one of the three children that died had Down syndrome, which is associated with an immunodeficiency.
The Birth of Primary Immunodeficiencies
The genetic theory of infectious diseases was not formulated until 1905 for plants, the early 1910s for humans, and the early 1920s for animals (108). By the 1940s this theory was supported in humans by clinical and epidemiological evidence. However, it did not really take off until the late 1940s and early 1950s, when cellular and molecular evidence was provided by the advent of tools for testing immunological hypotheses, such as electrophoresis for the detection of serum gamma globulins (109). The first Mendelian inborn errors of immunity, the first “primary immunodeficiencies,” were described in the late 1940s and early 1950s (Table 1) (110–113). Pediatricians focused their attention on children with multiple, recurrent infections, and they found that some of these children displayed immunological abnormalities, such as agammaglobulinemia. They noted that these rare clinical and immunological phenotypes cosegregated as X-linked or autosomal recessive traits in multiplex families. A key point to bear in mind here is that these clinical phenotypes were revealed thanks to the recent development of antibiotics. Ogden Bruton’s first patient with X-linked recessive agammaglobulinemia had suffered from 19 episodes of pneumococcal meningitis, all cured by antibiotics (110). Before the advent of antibiotics, he would have died during the first episode, as did nearly all children with agammaglobulinemia. From the 1950s the field was structured, imprinted with the notion that otherwise healthy children, including sporadic and familial cases, with a single severe infection, even if life-threatening or lethal, were not immunodeficient if the infection was isolated and did not recur. This trend was accentuated a few years later, with the development of the misleading notion of “opportunistic” infections—infections exclusively striking patients with detectable immunological abnormalities. This term was coined in the early 1960s when unusual fungal infections were diagnosed in patients receiving chemotherapy (114). The terms “immunodeficiency” and “immunocompromised host” were also defined in the 1960s as referring to patients with detectable immunological deficits, whether inherited or acquired (114). The self-contradictory idea of a lethal infection in an immunocompetent individual was reinforced during this period. This notion was, of course, an illusion, if not a delusion, but collective beliefs are difficult to break down. What was perfectly clear to plant biologists in 1905 remained totally unclear to human biologists 50 y later. Primary immunodeficiencies then were thought to be restricted to rare Mendelian disorders, fully penetrant, with an early onset and fatal outcome, underlying both detectable immunological abnormalities and multiple, recurrent, opportunistic infections (Table 2).
Table 2.
The evolution of the concept of monogenic inborn error of immunity to infection
| Infections | |
| 1952 | 1996 |
| Multiple | Single |
| Recurrent | Single (acute or chronic) |
| Early childhood | At any age |
| Opportunistic | Not necessarily |
| Rare | Rare or common |
| Familial | Sporadic |
The field of primary immunodeficiency was born with the description of patients with severe infectious diseases that met most of these six criteria, reflecting the advent of antibiotics and the Mendelian cosegregation of infectious and immunological phenotypes (109). The field evolved in numerous directions, with the description of a variety of noninfectious phenotypes caused by inborn errors of immunity. As far as infectious phenotypes are concerned, the paradigm shifted from 1996 onward, with increasing recognition that severe infectious diseases that did not fulfill these six criteria could be caused by novel types of primary immunodeficiencies. Mendelian infections (table 5 in ref. 161) are at variance from the initial paradigm for the first three criteria, and non-Mendelian monogenic infections are also at a variance for the remaining three criteria (table 6 in ref. 161).
Primary Immunodeficiencies: A Success Story
The field of primary immunodeficiencies grew more rapidly between 1960 and 1990. Genetic milestones included the discoveries of an autosomal dominant immunodeficiency in 1963 (115), spontaneous cure by somatic reversion in 1996 (116), and a gain-of-function mutation in 2001 (117). Other spectacular achievements have been made in the domain of therapy, including the first successful cases of hematopoietic stem cell transplantation in 1968 (118) and gene therapy in 2000 (119). Increasingly diverse clinical phenotypes, extending beyond infection and including various autoinflammatory, allergic, and autoimmune phenotypes, have been attributed to primary immunodeficiencies (120–124). Autoimmune phenotypes include systemic and organ-specific conditions such as the intriguingly related and occasionally allelic systemic lupus erythematosus and neurological Aicardi–Goutières syndrome, two type I interferonopathies (125). Autoinflammatory phenotypes include a tremendous variety of inflammatory conditions in which there is no evidence for the presence of autoreactive T and B cells (126). Allergic phenotypes have been attributed more recently to certain primary immunodeficiencies (127). The genetic causes of these conditions were discovered by candidate gene approaches from 1985 onward (adenosine deaminase deficiency) (128), by positional cloning from 1986 onward (chronic granulomatous disease) (129), and by next-generation sequencing from 2010 onward (fas-associated via death domain deficiency) (130). Genetic causes have been discovered for nearly 300 single-gene inborn errors of immunity. These discoveries clearly confirm that human monogenic lesions can account for rare phenotypes of multiple, recurrent, opportunistic infections, with or without noninfectious manifestations. There are hundreds of classical primary immunodeficiencies consistent with the definition used in the 1950s. Although these phenotypes are individually rare, they collectively provide proof of principle that life-threatening infection can have a genetic cause. Before discussing how the study of primary immunodeficiencies led to the discovery of monogenic lesions in otherwise healthy children with severe infections, we must turn our attention to two related questions to place these discoveries in a broader context.
Other Inborn Errors Underlying Death from Infection
Interestingly, primary immunodeficiencies were neither the only nor the first proof that lethal infections could have a genetic cause. Outside the realm of immunology, conditions such as sickle cell disease in hematology and cystic fibrosis in pulmonary medicine highlight the same phenomenon, although they were not interpreted as such. By 1949, even before the first clinical description of the primary immunodeficiencies, sickle cell disease was known to be an autosomal recessive disorder involving hemoglobin (Hb), i.e., a molecular disease (131). It is no coincidence that immunologists, hematologists, and pulmonologists still refuse to see sickle cell disease and cystic fibrosis as primary immunodeficiencies, even though most patients with either condition die from infection (132, 133). This refusal again reflects the evolution of this field since the 1950s. It also reflects the lack of interest in natural immunity displayed by immunologists from Landsteiner onward. Immunologists rarely considered immunity in the physiological and pathological context of a whole organism, in which each cell is endowed with some capacity to fend off invading microbes. This attitude is paradoxical, because circulating and resident hematopoietic cells populate all tissues, and all the nonhematopoietic cells of the organism work together to fend off infection. Cell-intrinsic immunity probably emerged along with the first cells, whereas cell-extrinsic innate immunity appeared with macrophages in multicellular organisms, and adaptive immunity developed much later, in vertebrates, with the somatic diversification of antigen-specific receptors. However, because the immunological and even general mechanisms underlying death from infection in patients with sickle-cell disease or cystic fibrosis have remained largely elusive, and the known cellular phenotypes of these conditions affect cells other than leukocytes, they generally are not seen as immunodeficiencies. By preventing fertile interactions between disciplines, this view may have hindered the elucidation of the mechanisms underlying infection in patients with these diseases.
The Sickle-Cell Trait and Protection from Malaria
We also need to place these findings in the context of the studies on the human genetics of infectious diseases subsequently carried out by population and epidemiological geneticists. Since the early 1990s, our own contribution to the field, and to the infection enigma, has been to take the plant, veterinary, and human Mendelian genetic studies from the start of the last century seriously and literally, leaving aside the immunodeficiency vs. immunocompetence paradigm defined in the 1950s and 1960s. We also have set aside the ideas of biometricians, by proposing a monogenic architecture for the genetic determinism of primary infections (134). This hypothesis was simple and testable but still is considered unlikely by many statistical geneticists, who tend to study infections from a polygenic angle. Their working model stems, in part, from the elegant discovery of a 10-fold increase in resistance to Plasmodium falciparum conferred by the sickle cell trait, published by Anthony Allison in 1954 (135, 136). Homozygotes for the sickle cell trait often die from infection, whereas heterozygotes do not have sickle cell disease and are protected against severe forms of malaria. This was a landmark discovery in evolutionary biology in two ways (137): It provided evidence that infections could shape the human genome by natural selection and showed that balancing selection and heterosis could account for the spread of sickle cell disease. However, it was never claimed that this observation could account for the pathogenesis of severe malaria, which strikes 1 in 1,000 or 1 in 10,000 individuals, depending on their HbB genotype. The genetic determinism of malaria remains unclear, regardless of HbB genotype. The landmark paper from 1954 remains the best paper in the field of the complex genetics of infectious diseases, highlighting the difficulties encountered with this population-based approach (138). Indeed, the chief failings of this approach have been the implicit assumption of phenotypic and genotypic homogeneity and the explicit use of a polygenic model of inheritance based on common alleles.
Common Variants and Infectious Diseases
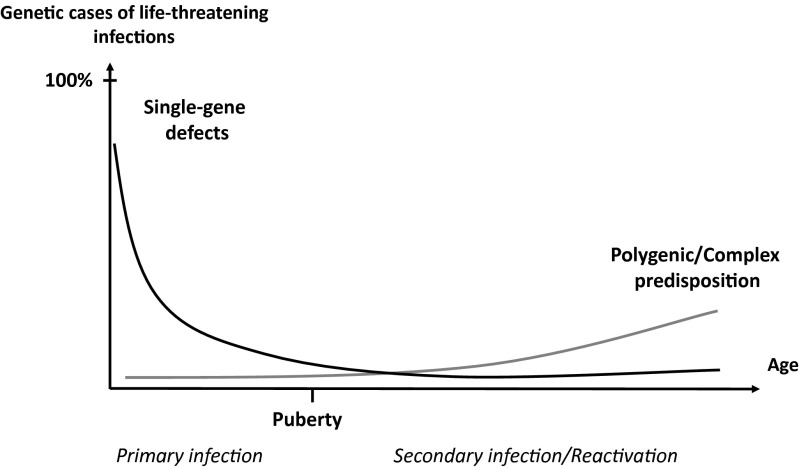
Experimental evidence for polygenic inheritance (at the individual level) garnered from inbred plants and animals drove the application of the polygenic theory to human conditions, including infectious diseases. However, the authors of these studies neglected the fundamental Darwinian notion that each living organism is unique, a phenomenon described by Archibald Garrod as “chemical individuality” and by Ernst Mayr as “population thinking” (69, 139). This Darwinian premise should have prevented the translation of polygenic inheritance from an inbred species, in which the multiple genes governing the phenotype are analyzed in numerous, albeit clonal, organisms and their crosses, to outbred populations, in which, by definition, the phenotype of each individual is unique and controlled by a unique genotype (regardless of the number of loci involved). Even the rarest Mendelian disorders, such as Fanconi anemia, display very high levels of genetic and phenotypic heterogeneity (140). How could this heterogeneity not hold true for infectious diseases as well? Interestingly, however, the polygenic theory provided evidence for natural selection driven by infectious agents in humans (141). It taught us more about the impact of infections on the genome of human populations than about the genetic determinism of infectious diseases. In this regard, the work of my colleague Laurent Abel and his collaborators, first detecting major genes by segregation studies (142–146) and then mapping these genes for conditions such as schistosomiasis, leishmaniasis, leprosy, and tuberculosis, has bridged the gap between population-based and patient-based studies of infectious diseases (147–154). Interestingly, the effect of most major genes decreases with age. Another remarkable discovery pertains to the role of genetic variants of type III interferons in the spontaneous and treatment-induced clearance of hepatitis C virus (155, 156). Likewise, heterozygous variants of apolipoprotein L-I (APOL1) increase resistance to trypanosomes in vitro. The selection of these APOL1 resistance alleles by trypanosomiasis may account for the higher rate of kidney sclerosis in African homozygotes, an evolutionary pattern that is reminiscent of sickle-cell disease being favored by malaria (157, 158). Finally, some polymorphisms have been shown to be associated with the rate of progression toward clinical disease in HIV-infected patients (159, 160). Globally however, the purely polygenic theory, inspired by the “common disease, common variant” model created some confusion between the impact of infection on the human genome and the genetic determinism of infectious disease. Initially with candidate gene association studies and then with genome-wide association studies, this approach nurtured the faulty idea that very modest relative risks, rarely exceeding two, were biologically and medically relevant. Both missing heritability and missing intelligibility characterize this approach. At odds with this particular polygenic model, we have suggested that life-threatening infections in the course of primary infection may result from single-gene inborn errors of immunity, displaying low, intermediate, or high but rarely complete penetrance (hence being polygenic in another way) (108, 134) (Fig. 2). In this model, severe infections can gradually shape the human genome by natural selection and the spread of common variants conferring relative resistance, but only rare or private variants actually cause the genetic forms of life-threatening disease. These ideas and data are discussed in the companion paper (161).
Fig. 2.
The genetic component of human infectious diseases, according to age. Schematic representation of a hypothetical, age-dependent, human genetic architecture of infectious diseases. We suggest that single-gene human variants make an important contribution to the determinism of life-threatening infectious diseases in the course of primary infection, which occurs most often in childhood. In this model, single-gene inborn errors of immunity are more often monogenic than Mendelian, with incomplete penetrance and variable expressivity accounting for infectious diseases being sporadic more often than familial. By contrast, predisposition to severe infectious diseases in the course of microbial reactivation from latency or secondary infection, typically in adults, is less influenced by germline human genetic variations, resulting in a more complex and less monogenic component. Somatic and epigenetic processes are likely to play a greater role in older individuals. Reproduced from ref. 134.
Acknowledgments
I am indebted to Gérard Orth, whose review of previous versions of this manuscript has been invaluable. I thank Laurent Abel, who leads the computational branch of The Laboratory of Human Genetics of Infectious Diseases; the researchers in the laboratory, including, in particular, Alexandre Alcais, Emmanuelle Jouanguy, Anne Puel, Capucine Picard, Jacinta Bustamante, Stéphanie Boisson-Dupuis, Bertrand Boisson, Guillaume Vogt, Michael Ciancanelli, Shen-Ying Zhang, and Aurélie Cobat; Yelena Nemirovskaya, without whom the laboratory in New York would never have taken off and grown; the other past and present members of the laboratory at the Necker Hospital for Sick Children and The Rockefeller University; the clinicians and scientists from around the world with whom we have been privileged to have formed links. Finally, I am humbled to thank the sick children and their parents, whose trust we did our best to deserve yet whose hope we too rarely satisfied. They are all too aware that pediatrics is mankind's greatest endeavor.
Footnotes
The author declares no conflict of interest.
See Profile on page 15533.
References
- 1.Nicolle C. Les infections inapparentes. Scientia. 1933:181–271. [Google Scholar]
- 2.Nicolle C. Destin des Maladies Infectieuses. 3rd Ed Alcan; Paris: 1937. [Google Scholar]
- 3.Lichtenthaeler C. 1987. Geschichte der Medizin: Die Reihenfolge ihrer Epochen-Bilder und die treibenden Kräfte ihrer Entwicklung: ein Lehrbuch für Studenten, Ärzte, Historiker, und geschichtlich Interessierte (Deutscher Ärzte-Verlag, Cologne, Germany) 4th Ed.
- 4.Gaudillière JP, Löwy I. Heredity and Infection. Routledge; London: 2001. [Google Scholar]
- 5.Ackerknecht EH. Medicine at the Paris Hospital, 1794-1848. Johns Hopkins Univ Press; Baltimore: 1967. [Google Scholar]
- 6.Olmsted JMD, Olmsted EH. Claude Bernard and the Experimental Method in Medicine. Abelard Schuman; New York: 1952. [Google Scholar]
- 7.Bernard C. An Introduction to the Study of Experimental Medicine. Dover Publications; New York: 1865. trans Green HC. [Google Scholar]
- 8.Olmsted JMD. 1st Ed Schuman's; New York: 1944. François Magendie. Pioneer in Experimental Physiology and Scientific Medicine in 19th Century France. [Google Scholar]
- 9.Lesh JE. 1st Ed Harvard Univ Press; Cambridge, MA: 1984. Science and Medicine in France. The Emergence of Experimental Physiology, 1790-1855. [Google Scholar]
- 10.Coleman W, Holmes FL. The Investigative Enterprise. Experimental Physiology in Nineteenth-Century Medicine. Univ of California Press; Berkeley, CA: 1988. [Google Scholar]
- 11.Céline L-F. 1924. The Life and Work of Semmelweis (Little, Brown, and Company, Boston); trans Parker RA (1937)
- 12.Hempel S. John Snow. Lancet. 2013;381(9874):1269–1270. doi: 10.1016/s0140-6736(13)60830-2. [DOI] [PubMed] [Google Scholar]
- 13.Pasteur L. Etudes sur la Maladie des Vers à Soie. La Pébrine et la Flacherie. 1870. (Gauthiers-Villars, Paris), 1st Ed.
- 14.Pasteur L. Masson et Cie; Paris: 1922–1939. Oeuvres Complètes de Louis Pasteur, Réunies par Pasteur Vallery-Radot. [Google Scholar]
- 15.Koch R. Die Aetiologie der Tuberkulose. Berl Klin Wochenschr. 1882;19:221–230. [Google Scholar]
- 16.Nocard E, Leclainche E. Les Maladies Microbiennes des Animaux. Masson; Paris: 1896. [Google Scholar]
- 17.Garnham P. Charles Nicolle and inapparent infections. Am J Trop Med Hyg. 1977;26(5):1101–1104. [Google Scholar]
- 18.von Pirquet C. Die Allergieprobe zur Diagnose der Tuberkulose im Kindersalter. Wien Med Wochenschr. 1907;57:1369. [Google Scholar]
- 19.Wagner R. Clemens von Irquet: His Life and Work. Univ Press, Baltimore: Johns Hopkins; 1968. [Google Scholar]
- 20.Widal GFI, Sicard A. Recherches de la réaction agglutinante dans le sang et le sérum dessechés des typhiques et dans la sérosité des vésicatoires. Bull Mém Soc Méd Hôp Paris series. 1896;3(13):681–682. [Google Scholar]
- 21.von Gruber M, Durham HE. Eine neue Methode zur raschen Erkennung des Choleravibrio und des Typhusbacillus. Munch Med Wochenschr. 1896;43:285–286. [Google Scholar]
- 22.Opie EL, Aronson JD. Tubercle bacilli in latent tuberculous lesions and in lung tissue without tuberculous lesions. Arch Pathol. 1927;4:1–21. [Google Scholar]
- 23.Lincoln EM. The effect of antimicrobial therapy on the prognosis of primary tuberculosis in children. Am Rev Tuberc. 1954;69(5):682–689. doi: 10.1164/art.1954.69.5.682. [DOI] [PubMed] [Google Scholar]
- 24.Cairns J. Matters of Life and Death. Princeton Univ Press; Princeton, NJ: 1997. [Google Scholar]
- 25.Casanova JL, Abel L. Inborn errors of immunity to infection: The rule rather than the exception. J Exp Med. 2005;202(2):197–201. doi: 10.1084/jem.20050854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Burnet M, White DO. Natural History of Infectious Diseases. 4th Ed Cambridge Univ Press; Cambridge, UK: 1972. [Google Scholar]
- 27.Cooper MD. A life of adventure in immunobiology. Annu Rev Immunol. 2010;28:1–19. doi: 10.1146/annurev-immunol-030409-101248. [DOI] [PubMed] [Google Scholar]
- 28.Guo P, et al. Dual nature of the adaptive immune system in lampreys. Nature. 2009;459(7248):796–801. doi: 10.1038/nature08068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hirano M, et al. Evolutionary implications of a third lymphocyte lineage in lampreys. Nature. 2013;501(7467):435–438. doi: 10.1038/nature12467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pancer Z, et al. Somatic diversification of variable lymphocyte receptors in the agnathan sea lamprey. Nature. 2004;430(6996):174–180. doi: 10.1038/nature02740. [DOI] [PubMed] [Google Scholar]
- 31.McGhee G. Convergent Evolution. Limited Forms Most Beautiful. MIT Press; Cambridge, MA: 2011. [Google Scholar]
- 32.Pasteur L. Maladies Virulentes, Virus-Vaccins, Prophylaxie de la Rage. Masson et Cie; Paris: 1922-1939. [Google Scholar]
- 33.Pirofski LA, Casadevall A. The damage-response framework of microbial pathogenesis and infectious diseases. Adv Exp Med Biol. 2008;635:135–146. doi: 10.1007/978-0-387-09550-9_11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nathan C. From transient infection to chronic disease. Science. 2015;350(6257):161. doi: 10.1126/science.aad4141. [DOI] [PubMed] [Google Scholar]
- 35.Rynda-Apple A, Robinson KM, Alcorn JF. Influenza and bacterial superinfection: Illuminating the immunologic mechanisms of disease. Infect Immun. 2015;83(10):3764–3770. doi: 10.1128/IAI.00298-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Avota E, Gassert E, Schneider-Schaulies S. Measles virus-induced immunosuppression: From effectors to mechanisms. Med Microbiol Immunol (Berl) 2010;199(3):227–237. doi: 10.1007/s00430-010-0152-3. [DOI] [PubMed] [Google Scholar]
- 37.Moir S, Chun TW, Fauci AS. Pathogenic mechanisms of HIV disease. Annu Rev Pathol. 2011;6:223–248. doi: 10.1146/annurev-pathol-011110-130254. [DOI] [PubMed] [Google Scholar]
- 38.Jones KD, Berkley JA. Severe acute malnutrition and infection. Paediatr Int Child Health. 2014;34(Suppl 1):S1–S29. doi: 10.1179/2046904714Z.000000000218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rytter MJ, Kolte L, Briend A, Friis H, Christensen VB. The immune system in children with malnutrition--a systematic review. PLoS One. 2014;9(8):e105017. doi: 10.1371/journal.pone.0105017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lassi ZS, et al. Essential interventions for child health. Reprod Health. 2014;11(Suppl 1):S4. doi: 10.1186/1742-4755-11-S1-S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Alcaïs A, Abel L, Casanova JL. Human genetics of infectious diseases: Between proof of principle and paradigm. J Clin Invest. 2009;119(9):2506–2514. doi: 10.1172/JCI38111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Falkow S. Molecular Koch’s postulates applied to microbial pathogenicity. Rev Infect Dis. 1988;10(Suppl 2):S274–S276. doi: 10.1093/cid/10.supplement_2.s274. [DOI] [PubMed] [Google Scholar]
- 43.Webster LT. Heredity in infectious disease. J Hered. 1939:365–370. [Google Scholar]
- 44.Vidal SM, Malo D, Marquis JF, Gros P. Forward genetic dissection of immunity to infection in the mouse. Annu Rev Immunol. 2008;26:81–132. doi: 10.1146/annurev.immunol.26.021607.090304. [DOI] [PubMed] [Google Scholar]
- 45.Lurie M. Heredity, constitution and tuberculosis: An experimental study. Am Rev Tuberc. 1941;44(suppl):1–125. [Google Scholar]
- 46.Fox G, Orlova M, Schurr E. Tuberculosis in newborns: The lessons of the “Lübeck disaster” (1929-1933) PLoS Pathog. 2015 doi: 10.1371/journal.ppat.1005271. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Foster WD. A History of Medical Bacteriology and Immunology. Heinemann; London: 1970. [Google Scholar]
- 48.Bovet D. Une Chimie Qui Guérit. Histoire de la Découverte des Sulfamides. Payot; Paris: 1988. [Google Scholar]
- 49.Moberg CL, Cohn Z. Launching the Antibiotic Era. The Rockefeller Univ Press; New York: 1990. [Google Scholar]
- 50.Kindt TJ, Capra JD. The Antibody Enigma. Plenum Publishing Company Limited; New York: 1984. [Google Scholar]
- 51.Silverstein AM. A History of Immunology. 2nd Ed Academic; London: 2009. [Google Scholar]
- 52.Landsteiner K. The Specificity of Serological Reactions. Dover Publications, Inc.; New York: 1936. [Google Scholar]
- 53.Cooper MD, Peterson RD, Good RA. Delineation of the thymic and bursal lymphoid systems in the chicken. Nature. 1965;205:143–146. doi: 10.1038/205143a0. [DOI] [PubMed] [Google Scholar]
- 54.Cooper MD, Raymond DA, Peterson RD, South MA, Good RA. The functions of the thymus system and the bursa system in the chicken. J Exp Med. 1966;123(1):75–102. doi: 10.1084/jem.123.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cooper MD. The early history of B cells. Nat Rev Immunol. 2015;15(3):191–197. doi: 10.1038/nri3801. [DOI] [PubMed] [Google Scholar]
- 56.Miller JF. Immunological function of the thymus. Lancet. 1961;2(7205):748–749. doi: 10.1016/s0140-6736(61)90693-6. [DOI] [PubMed] [Google Scholar]
- 57.Miller JF. Analysis of the thymus influence in leukaemogenesis. Nature. 1961;191:248–249. doi: 10.1038/191248a0. [DOI] [PubMed] [Google Scholar]
- 58.Miller JF. The golden anniversary of the thymus. Nat Rev Immunol. 2011;11(7):489–495. doi: 10.1038/nri2993. [DOI] [PubMed] [Google Scholar]
- 59.Soderqvist T. Science as Autobiography: The Troubled Life of Niels Jerne. Yale Univ Press; New Haven, CT: 2003. [Google Scholar]
- 60.Beutler B. Not “molecular patterns” but molecules. Immunity. 2003;19(2):155–156. doi: 10.1016/s1074-7613(03)00212-7. [DOI] [PubMed] [Google Scholar]
- 61.Beutler B. Inferences, questions and possibilities in Toll-like receptor signalling. Nature. 2004;430(6996):257–263. doi: 10.1038/nature02761. [DOI] [PubMed] [Google Scholar]
- 62.Lemaitre B. The road to Toll. Nat Rev Immunol. 2004;4(7):521–527. doi: 10.1038/nri1390. [DOI] [PubMed] [Google Scholar]
- 63.Lemaitre B, Hoffmann J. The host defense of Drosophila melanogaster. Annu Rev Immunol. 2007;25:697–743. doi: 10.1146/annurev.immunol.25.022106.141615. [DOI] [PubMed] [Google Scholar]
- 64.Janeway CA, Jr, Medzhitov R. Innate immune recognition. Annu Rev Immunol. 2002;20:197–216. doi: 10.1146/annurev.immunol.20.083001.084359. [DOI] [PubMed] [Google Scholar]
- 65.Takeda K, Kaisho T, Akira S. Toll-like receptors. Annu Rev Immunol. 2003;21:335–376. doi: 10.1146/annurev.immunol.21.120601.141126. [DOI] [PubMed] [Google Scholar]
- 66.Fearon DT, Locksley RM. The instructive role of innate immunity in the acquired immune response. Science. 1996;272(5258):50–53. doi: 10.1126/science.272.5258.50. [DOI] [PubMed] [Google Scholar]
- 67.de Kruif P. Microbe Hunters. 1 Ed Blue Ribbons Books; New York: 1926. [Google Scholar]
- 68.Casadevall A, Pirofski LA. Microbiology: Ditch the term pathogen. Nature. 2014;516(7530):165–166. doi: 10.1038/516165a. [DOI] [PubMed] [Google Scholar]
- 69.Mayr E. Toward a New Philosophy of Biology. Harvard Univ Press; Cambridge, MA: 1988. [Google Scholar]
- 70.Ayala FJ, Dobzhansky T. Studies in the Philosophy of Biology. Univ of California Press; Berkeley, CA: 1974. [Google Scholar]
- 71.Papermaster BW, Condie RM, Finstad JK, Good RA, Gabrielsen AE. Phylogenetic development of adaptive immunity. Fed Proc. 1963;22:1152–1155. [PubMed] [Google Scholar]
- 72.Mazumdar PMH. Species and Specificity. An Interpretation of the History of Immunology. Cambridge Univ Press; Cambridge, UK: 1995. [Google Scholar]
- 73.Besredka A. Histoire d’une Idée. L’oeuvre de Metchnikoff. Masson & Cie; Paris: 1921. [Google Scholar]
- 74.Dubos RJ. Second thoughts on the germ theory. Sci Am. 1955;192:31–35. [Google Scholar]
- 75.Dubos RJ. Louis Pasteur, Free Lance of Science. 1st Ed Little Brown; Boston: 1950. [Google Scholar]
- 76.Moberg CL. René Dubos. Friend of the Good Earth. Microbiologist, Medical Scientist, Environmentalist. ASM; Washington, D.C.: 2005. [Google Scholar]
- 77.Tompkins DM, Dunn AM, Smith MJ, Telfer S. Wildlife diseases: From individuals to ecosystems. J Anim Ecol. 2011;80(1):19–38. doi: 10.1111/j.1365-2656.2010.01742.x. [DOI] [PubMed] [Google Scholar]
- 78.Dubos R. The microbiota of the gastrointestinal tract. Gastroenterology. 1966;51(5):868–874. [PubMed] [Google Scholar]
- 79.Lee A, Gordon J, Lee CJ, Dubos R. The mouse intestinal microflora with emphasis on the strict anaerobes. J Exp Med. 1971;133(2):339–352. doi: 10.1084/jem.133.2.339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Biffen RH. Mendel’s laws of inheritance and wheat breeding. J Agric Sci. 1905;1(1):4–48. [Google Scholar]
- 81.Flor HH. Current status of the gene-for-gene concept. Annu Rev Phytopathol. 1971;9:275–296. [Google Scholar]
- 82.Dangl JL, Horvath DM, Staskawicz BJ. Pivoting the plant immune system from dissection to deployment. Science. 2013;341(6147):746–751. doi: 10.1126/science.1236011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Flor HH. Inheritance of pathogenicity in a cross between physiologic races 22 and 24 of Melampsora lini. Phytopathology. 1942;32:5. [Google Scholar]
- 84.Flor HH. Host-parasite interaction in flax rust - its genetics and other implications. Phytopathology. 1955;45:680–685. [Google Scholar]
- 85.Martin GB, et al. Map-based cloning of a protein kinase gene conferring disease resistance in tomato. Science. 1993;262(5138):1432–1436. doi: 10.1126/science.7902614. [DOI] [PubMed] [Google Scholar]
- 86.Grant MR, et al. Structure of the Arabidopsis RPM1 gene enabling dual specificity disease resistance. Science. 1995;269(5225):843–846. doi: 10.1126/science.7638602. [DOI] [PubMed] [Google Scholar]
- 87.Staskawicz BJ, Ausubel FM, Baker BJ, Ellis JG, Jones JD. Molecular genetics of plant disease resistance. Science. 1995;268(5211):661–667. doi: 10.1126/science.7732374. [DOI] [PubMed] [Google Scholar]
- 88.Casanova JL, Schurr E, Abel L, Skamene E. Forward genetics of infectious diseases: Immunological impact. Trends Immunol. 2002;23(10):469–472. doi: 10.1016/s1471-4906(02)02289-5. [DOI] [PubMed] [Google Scholar]
- 89.Shultz LD, Sidman CL. Genetically determined murine models of immunodeficiency. Annu Rev Immunol. 1987;5:367–403. doi: 10.1146/annurev.iy.05.040187.002055. [DOI] [PubMed] [Google Scholar]
- 90.Skamene E, Kongshawn PAL, Landy M. Genetic Control of Natural Resistance to Infection and Malignancy. Academic; New York: 1980. [Google Scholar]
- 91.Rosenstreich DL, Weinblatt AC, O’Brien AD. Genetic control of resistance to infection in mice. Crit Rev Immunol. 1982;3(4):263–330. [PubMed] [Google Scholar]
- 92.Searle AG. Hereditary absence of spleen in the mouse. Nature. 1959;184(Suppl 18):1419–1420. doi: 10.1038/1841419b0. [DOI] [PubMed] [Google Scholar]
- 93.Lindenmann J. Inheritance of resistance to influenza virus in mice. Proc Soc Exp Biol Med. 1964;116:506–509. doi: 10.3181/00379727-116-29292. [DOI] [PubMed] [Google Scholar]
- 94.Staeheli P, Haller O, Boll W, Lindenmann J, Weissmann C. Mx protein: Constitutive expression in 3T3 cells transformed with cloned Mx cDNA confers selective resistance to influenza virus. Cell. 1986;44(1):147–158. doi: 10.1016/0092-8674(86)90493-9. [DOI] [PubMed] [Google Scholar]
- 95.Skamene E, et al. Genetic regulation of resistance to intracellular pathogens. Nature. 1982;297(5866):506–509. doi: 10.1038/297506a0. [DOI] [PubMed] [Google Scholar]
- 96.Vidal SM, Malo D, Vogan K, Skamene E, Gros P. Natural resistance to infection with intracellular parasites: Isolation of a candidate for Bcg. Cell. 1993;73(3):469–485. doi: 10.1016/0092-8674(93)90135-d. [DOI] [PubMed] [Google Scholar]
- 97.Poltorak A, et al. Defective LPS signaling in C3H/HeJ and C57BL/10ScCr mice: Mutations in Tlr4 gene. Science. 1998;282(5396):2085–2088. doi: 10.1126/science.282.5396.2085. [DOI] [PubMed] [Google Scholar]
- 98.Brown MG, et al. Vital involvement of a natural killer cell activation receptor in resistance to viral infection. Science. 2001;292(5518):934–937. doi: 10.1126/science.1060042. [DOI] [PubMed] [Google Scholar]
- 99.Garrod AE. The Inborn Factors in Disease. Clarendon; Oxford: 1931. [Google Scholar]
- 100.Pearson K. Tuberculosis, Heredity and Environment. Dulau and Co., Ltd.; London: 1912. [Google Scholar]
- 101.Fernández-Cañón JM, et al. The molecular basis of alkaptonuria. Nat Genet. 1996;14(1):19–24. doi: 10.1038/ng0996-19. [DOI] [PubMed] [Google Scholar]
- 102.Garrod AE. The incidence of alkaptonuria: A study in clinical individuality. Lancet. 1902;2:1616–1620. [Google Scholar]
- 103.Pearson ES. Karl Peason, an Appreciation of Some Aspects of His Life and Work. Cambridge Univ Press; Cambridge, UK: 1938. [Google Scholar]
- 104.Flavia SM. A family pedigree of appendicitis. J Hered. 1940:113–115. [Google Scholar]
- 105.Puffer R. Familial Susceptibility to Tuberculosis; Its Importance as a Public Health Problem. Harvard Univ Press; Cambridge, MA: 1944. [Google Scholar]
- 106.Kallmann F, Reisner D. Twin studies on the significance of genetic factors in tuberculosis. Am Rev Tuberc. 1943;47:549–574. [Google Scholar]
- 107.Sørensen TI, Nielsen GG, Andersen PK, Teasdale TW. Genetic and environmental influences on premature death in adult adoptees. N Engl J Med. 1988;318(12):727–732. doi: 10.1056/NEJM198803243181202. [DOI] [PubMed] [Google Scholar]
- 108.Casanova JL, Abel L. The genetic theory of infectious diseases: A brief history and selected illustrations. Annu Rev Genomics Hum Genet. 2013;14:215–243. doi: 10.1146/annurev-genom-091212-153448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Etzioni A, Ochs H. Primary Immunodeficiency Disorders. A Historic and Scientific Perspective. Academic, Oxford; 2014. [Google Scholar]
- 110.Bruton OC. Agammaglobulinemia. Pediatrics. 1952;9(6):722–728. [PubMed] [Google Scholar]
- 111.Bruton OC. A decade with agammaglobulinemia. J Pediatr. 1962;60:672–676. doi: 10.1016/s0022-3476(62)80092-4. [DOI] [PubMed] [Google Scholar]
- 112.Cooper MD, et al. Classification of primary immunodeficiencies. N Engl J Med. 1973;288(18):966–967. doi: 10.1056/NEJM197305032881814. [DOI] [PubMed] [Google Scholar]
- 113.Fudenberg HH, et al. Classification of the primary immune deficiencies: WHO recommendation. N Engl J Med. 1970;283(12):656–657. doi: 10.1056/NEJM197009172831211. [DOI] [PubMed] [Google Scholar]
- 114.Armstrong D. History of opportunistic infection in the immunocompromised host. Clin Infect Dis. 1993;17(Suppl 2):S318–S321. doi: 10.1093/clinids/17.supplement_2.s318. [DOI] [PubMed] [Google Scholar]
- 115.Donaldson VH, Evans RR. A biochemical abnormality in herediatry angioneurotic edema: Absence of serum inhibitor of C′ 1-esterase. Am J Med. 1963;35:37–44. doi: 10.1016/0002-9343(63)90162-1. [DOI] [PubMed] [Google Scholar]
- 116.Hirschhorn R, et al. Spontaneous in vivo reversion to normal of an inherited mutation in a patient with adenosine deaminase deficiency. Nat Genet. 1996;13(3):290–295. doi: 10.1038/ng0796-290. [DOI] [PubMed] [Google Scholar]
- 117.Devriendt K, et al. Constitutively activating mutation in WASP causes X-linked severe congenital neutropenia. Nat Genet. 2001;27(3):313–317. doi: 10.1038/85886. [DOI] [PubMed] [Google Scholar]
- 118.Gatti RA, Meuwissen HJ, Allen HD, Hong R, Good RA. Immunological reconstitution of sex-linked lymphopenic immunological deficiency. Lancet. 1968;2(7583):1366–1369. doi: 10.1016/s0140-6736(68)92673-1. [DOI] [PubMed] [Google Scholar]
- 119.Cavazzana-Calvo M, et al. Gene therapy of human severe combined immunodeficiency (SCID)-X1 disease. Science. 2000;288(5466):669–672. doi: 10.1126/science.288.5466.669. [DOI] [PubMed] [Google Scholar]
- 120.Al-Herz W, et al. Primary immunodeficiency diseases: An update on the classification from the international union of immunological societies expert committee for primary immunodeficiency. Front Immunol. 2014;5:162. doi: 10.3389/fimmu.2014.00162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Bousfiha AA, et al. A phenotypic approach for IUIS PID classification and diagnosis: Guidelines for clinicians at the bedside. J Clin Immunol. 2013;33(6):1078–1087. doi: 10.1007/s10875-013-9901-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Zhang Y, Su HC, Lenardo MJ. Genomics is rapidly advancing precision medicine for immunological disorders. Nat Immunol. 2015;16(10):1001–1004. doi: 10.1038/ni.3275. [DOI] [PubMed] [Google Scholar]
- 123.Milner JD, Holland SM. The cup runneth over: Lessons from the ever-expanding pool of primary immunodeficiency diseases. Nat Rev Immunol. 2013;13(9):635–648. doi: 10.1038/nri3493. [DOI] [PubMed] [Google Scholar]
- 124.Pessach I, Walter J, Notarangelo LD. Recent advances in primary immunodeficiencies: Identification of novel genetic defects and unanticipated phenotypes. Pediatr Res. 2009;65(5 Pt 2):3R–12R. doi: 10.1203/PDR.0b013e31819dbe1e. [DOI] [PubMed] [Google Scholar]
- 125.Crow YJ. Type I interferonopathies: Mendelian type I interferon up-regulation. Curr Opin Immunol. 2015;32:7–12. doi: 10.1016/j.coi.2014.10.005. [DOI] [PubMed] [Google Scholar]
- 126.Masters SL, Simon A, Aksentijevich I, Kastner DL. Horror autoinflammaticus: The molecular pathophysiology of autoinflammatory disease. Annu Rev Immunol. 2009;27:621–668. doi: 10.1146/annurev.immunol.25.022106.141627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Williams KW, Milner JD, Freeman AF. Eosinophilia Associated with Disorders of Immune Deficiency or Immune Dysregulation. Immunol Allergy Clin North Am. 2015;35(3):523–544. doi: 10.1016/j.iac.2015.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Bonthron DT, Markham AF, Ginsburg D, Orkin SH. Identification of a point mutation in the adenosine deaminase gene responsible for immunodeficiency. J Clin Invest. 1985;76(2):894–897. doi: 10.1172/JCI112050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Royer-Pokora B, et al. Cloning the gene for an inherited human disorder--chronic granulomatous disease--on the basis of its chromosomal location. Nature. 1986;322(6074):32–38. doi: 10.1038/322032a0. [DOI] [PubMed] [Google Scholar]
- 130.Bolze A, et al. Whole-exome-sequencing-based discovery of human FADD deficiency. Am J Hum Genet. 2010;87(6):873–881. doi: 10.1016/j.ajhg.2010.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Pauling L, et al. Sickle cell anemia a molecular disease. Science. 1949;110(2865):543–548. doi: 10.1126/science.110.2865.543. [DOI] [PubMed] [Google Scholar]
- 132.Gilligan PH. Microbiology of airway disease in patients with cystic fibrosis. Clin Microbiol Rev. 1991;4(1):35–51. doi: 10.1128/cmr.4.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Booth C, Inusa B, Obaro SK. Infection in sickle cell disease: A review. Int J Infect Dis. 2010;14(1):e2–e12. doi: 10.1016/j.ijid.2009.03.010. [DOI] [PubMed] [Google Scholar]
- 134.Alcaïs A, et al. Life-threatening infectious diseases of childhood: Single-gene inborn errors of immunity? Ann N Y Acad Sci. 2010;1214(1):18–33. doi: 10.1111/j.1749-6632.2010.05834.x. [DOI] [PubMed] [Google Scholar]
- 135.Allison AC. Protection afforded by sickle-cell trait against subtertian malareal infection. BMJ. 1954;1(4857):290–294. doi: 10.1136/bmj.1.4857.290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Allison AC. Genetic control of innate immunity to malaria. Curr Opin Immunol. 2009;21(5):499–505. doi: 10.1016/j.coi.2009.04.001. [DOI] [PubMed] [Google Scholar]
- 137.Dobzhansky T. Mankind Evolving. Yale Univ Press; New Haven, CT: 1962. [Google Scholar]
- 138.Abel L, Alcaïs A, Schurr E. The dissection of complex susceptibility to infectious disease: Bacterial, viral and parasitic infections. Curr Opin Immunol. 2014;30:72–78. doi: 10.1016/j.coi.2014.07.002. [DOI] [PubMed] [Google Scholar]
- 139.Bearn AG. Archibald Garrod and the Individuality of Man. Clarendon Press; Oxford, UK: 1993. [Google Scholar]
- 140.Wang AT, Smogorzewska A. SnapShot: Fanconi anemia and associated proteins. Cell. 2015;160(1–2):354. doi: 10.1016/j.cell.2014.12.031. [DOI] [PubMed] [Google Scholar]
- 141.Kwiatkowski DP. How malaria has affected the human genome and what human genetics can teach us about malaria. Am J Hum Genet. 2005;77(2):171–192. doi: 10.1086/432519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Abel L, Demenais F. Detection of major genes for susceptibility to leprosy and its subtypes in a Caribbean island: Desirade Island. Am J Hum Genet. 1988;42(2):256–266. [PMC free article] [PubMed] [Google Scholar]
- 143.Abel L, Demenais F, Prata A, Souza AE, Dessein A. Evidence for the segregation of a major gene in human susceptibility/resistance to infection by Schistosoma mansoni. Am J Hum Genet. 1991;48(5):959–970. [PMC free article] [PubMed] [Google Scholar]
- 144.Abel L, Cot M, Mulder L, Carnevale P, Feingold J. Segregation analysis detects a major gene controlling blood infection levels in human malaria. Am J Hum Genet. 1992;50(6):1308–1317. [PMC free article] [PubMed] [Google Scholar]
- 145.Abel L, et al. Complex segregation analysis of leprosy in southern Vietnam. Genet Epidemiol. 1995;12(1):63–82. doi: 10.1002/gepi.1370120107. [DOI] [PubMed] [Google Scholar]
- 146.Alcaïs A, et al. Evidence for a major gene controlling susceptibility to tegumentary leishmaniasis in a recently exposed Bolivian population. Am J Hum Genet. 1997;61(4):968–979. doi: 10.1086/514882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Cobat A, et al. Two loci control tuberculin skin test reactivity in an area hyperendemic for tuberculosis. J Exp Med. 2009;206(12):2583–2591. doi: 10.1084/jem.20090892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Mira MT, et al. Susceptibility to leprosy is associated with PARK2 and PACRG. Nature. 2004;427(6975):636–640. doi: 10.1038/nature02326. [DOI] [PubMed] [Google Scholar]
- 149.Casanova JL, Abel L. Human genetics of infectious diseases: A unified theory. EMBO J. 2007;26(4):915–922. doi: 10.1038/sj.emboj.7601558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Baghdadi JE, et al. An autosomal dominant major gene confers predisposition to pulmonary tuberculosis in adults. J Exp Med. 2006;203(7):1679–1684. doi: 10.1084/jem.20060269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Alcaïs A, et al. Stepwise replication identifies a low-producing lymphotoxin-alpha allele as a major risk factor for early-onset leprosy. Nat Genet. 2007;39(4):517–522. doi: 10.1038/ng2000. [DOI] [PubMed] [Google Scholar]
- 152.Bucheton B, et al. A major susceptibility locus on chromosome 22q12 plays a critical role in the control of kala-azar. Am J Hum Genet. 2003;73(5):1052–1060. doi: 10.1086/379084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Dessein AJ, et al. Severe hepatic fibrosis in Schistosoma mansoni infection is controlled by a major locus that is closely linked to the interferon-gamma receptor gene. Am J Hum Genet. 1999;65(3):709–721. doi: 10.1086/302526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Marquet S, et al. Genetic localization of a locus controlling the intensity of infection by Schistosoma mansoni on chromosome 5q31-q33. Nat Genet. 1996;14(2):181–184. doi: 10.1038/ng1096-181. [DOI] [PubMed] [Google Scholar]
- 155.Ge D, et al. Genetic variation in IL28B predicts hepatitis C treatment-induced viral clearance. Nature. 2009;461(7262):399–401. doi: 10.1038/nature08309. [DOI] [PubMed] [Google Scholar]
- 156.Thomas DL, et al. Genetic variation in IL28B and spontaneous clearance of hepatitis C virus. Nature. 2009;461(7265):798–801. doi: 10.1038/nature08463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Genovese G, et al. Association of trypanolytic ApoL1 variants with kidney disease in African Americans. Science. 2010;329(5993):841–845. doi: 10.1126/science.1193032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Pays E, Vanhollebeke B, Uzureau P, Lecordier L, Pérez-Morga D. The molecular arms race between African trypanosomes and humans. Nat Rev Microbiol. 2014;12(8):575–584. doi: 10.1038/nrmicro3298. [DOI] [PubMed] [Google Scholar]
- 159.McLaren PJ, Carrington M. The impact of host genetic variation on infection with HIV-1. Nat Immunol. 2015;16(6):577–583. doi: 10.1038/ni.3147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Walker BD, Yu XG. Unravelling the mechanisms of durable control of HIV-1. Nat Rev Immunol. 2013;13(7):487–498. doi: 10.1038/nri3478. [DOI] [PubMed] [Google Scholar]
- 161.Casanova J-L. Severe infectious diseases of childhood as monogenic inborn errors of immunity. Proc Natl Acad Sci USA. 2015;112:E7128–E7137. doi: 10.1073/pnas.1521651112. [DOI] [PMC free article] [PubMed] [Google Scholar]