Significance
An important question in evolution is: how does population structure affect the outcome of the evolutionary process? The theory of evolution in structured population has provided an impressive range of results, but an understanding of the computational complexity of even simple questions was missing. We prove that some fundamental problems in ecology and evolution can be precisely characterized by well-established computational complexity classes. This implies that the problems cannot be answered by simple equations. For example, there cannot be simple formulas for the fixation probability of a mutant given frequency-dependent selection in a structured population. We also show that, for example, calculating the molecular clock of neutral evolution in structured populations admit efficient algorithmic solutions.
Keywords: evolutionary games, fixation probability, complexity classes
Abstract
There are deep, yet largely unexplored, connections between computer science and biology. Both disciplines examine how information proliferates in time and space. Central results in computer science describe the complexity of algorithms that solve certain classes of problems. An algorithm is deemed efficient if it can solve a problem in polynomial time, which means the running time of the algorithm is a polynomial function of the length of the input. There are classes of harder problems for which the fastest possible algorithm requires exponential time. Another criterion is the space requirement of the algorithm. There is a crucial distinction between algorithms that can find a solution, verify a solution, or list several distinct solutions in given time and space. The complexity hierarchy that is generated in this way is the foundation of theoretical computer science. Precise complexity results can be notoriously difficult. The famous question whether polynomial time equals nondeterministic polynomial time (i.e., P = NP) is one of the hardest open problems in computer science and all of mathematics. Here, we consider simple processes of ecological and evolutionary spatial dynamics. The basic question is: What is the probability that a new invader (or a new mutant) will take over a resident population? We derive precise complexity results for a variety of scenarios. We therefore show that some fundamental questions in this area cannot be answered by simple equations (assuming that P is not equal to NP).
Evolution occurs in populations of reproducing individuals. Mutation generates distinct types. Selection favors some types over others. The mathematical formalism of evolution describes how populations change in their genetic (or phenotypic) composition over time. Deterministic models of evolution are based on differential equations. They assume infinitely large population size and ignore demographic and other stochasticity. The more precise descriptions of evolutionary dynamics, however, use stochastic processes, which take into account the intrinsic randomness of when and where individuals reproduce and how many of their offspring survive. They also describe populations of finite size.
A well-known stochastic process of evolution was formulated by Moran in 1958 (1). In any one-time step, a random individual is chosen proportional to fitness for reproduction and a random individual is chosen for death. The offspring of the first individual is added to the population. The total population size remains constant and is given by N. The original process was formulated for constant fitness, which means the fitness value of individuals does not depend on the relative abundance of various types in the population; it is a fixed number. The crucial question is: What is the probability that a newly introduced mutant will generate a lineage that takes over the entire population? This quantity is called the fixation probability. For the original Moran process, there is a simple formula. If the resident has fitness 1 and the mutant has fitness r, then the fixation probability of the mutant is given by .
The Moran process assumes that the biological population is well mixed. The offspring of any one individual can replace any other individual. If there is a spatial or social population structure, then such is not the case. The question arises as to which population structures affect the outcome of evolution, for example, by modifying the fixation probability. The classic studies of Maruyama (2) and Slatkin (3) showed that symmetrical population structures, such as regular lattices, do not change the fixation probability compared with the well-mixed population. The effect of population structure on evolution is also at the heart of the famous Wright–Fisher debate (4–6).
In 2005, evolutionary graph theory was introduced as a tool to study how generalized population structures affect evolutionary outcome (7), and it has been studied in many other works (8–12). The individuals occupy the vertices of the graph. The links (edges) determine who interacts with whom for receiving payoff and for reproduction. There can be a single graph for game dynamical interaction and evolutionary replacement, or the interaction and replacement graphs can be distinct (13). Often, the graph is held constant during evolutionary updating, but it is also possible to study dynamically changing graphs (14–22). The original Moran process is recovered as a special case given by the complete graph. It turns out that all isothermal graphs, where all vertices have the same rate of updating (the same “temperature”), have the same fixation probability as the well-mixed population (7). All symmetrical population structures lead to isothermal graphs, but the converse is not true.
Many evolutionary contests, however, are not fought out with constant fitness. Instead, the fitness of individual types depends on the frequency (i.e., relative abundance) of types in the population. A well-known approach to frequency-dependent selection is evolutionary game theory (23–27). Here, the fitnesses of individuals are linear functions of the frequencies. The coefficients of the linear function are the elements of the payoff matrix. Again, constant selection is a special case of frequency-dependent selection; for constant selection, all entries in a row of the payoff matrix are the same and this property holds for all rows. Evolutionary game theory has traditionally been investigated with deterministic equations describing infinitely large populations (23–26) and, more recently, has moved to finite population size and stochastic processes (27–38).
Evolutionary games have a long history of being studied on spatial lattices (28, 39–42) and, more recently, on graphs (7, 27, 43, 44). The crucial quantity that needs to be calculated to evaluate natural selection is the fixation probability, ρ, of a newly introduced mutant that arises at a random position on the graph. If , where N is the population size, then natural selection favors the fixation of the new mutant, because a neutral mutant would have a fixation probability of . In a contest between two strategies, another question would be if the fixation probability of the first strategy exceeds the fixation probability of the second strategy. If such is the case, then the first strategy would be more abundant in mutation-selection equilibrium for low mutation rate. The crucial problem is of the following form: Given a graph and a payoff matrix, what are the fixation probabilities of an individual of a new type arising in a population of the other type?
Spatial structure plays an important role in many cases. For example, spatial structure can affect the rate of neutral evolution (45), and there are results that describe which spatial structures do or do not affect the outcome of constant selection (46–48). Some population structures can be amplifiers or suppressors of constant selection (7, 49, 50), meaning that they modify the intensity of selective differences. Finally, for evolutionary games, spatial structure can favor evolution of cooperation (28, 51).
The study of spatial dynamics also has a long tradition in ecology (52–56). Here, the typical setting is that different species compete for ecological niches. Many evolutionary models are formally equivalent to ecological ones, especially if we consider only selection and not mutation. Then we can interpret the different types of evolutionary games as different species. Again, a crucial question is: What is the probability that a newly introduced species can get established or take over an ecological niche?
This paper is structured as follows. First, we give an intuitive account of the foundation of theoretical computer science. We describe classes of problems that can be solved by algorithms in certain time and space constraints. Subsequently, we present two simple problems of evolutionary dynamics in spatial settings. The first problem is motivated by a very simple ecological dynamic; the second problem is the general setting of evolutionary games on graphs. In both cases, the basic question is to calculate the takeover probability (or fixation probability) of a new type. That is, we introduce a new type in a random position in the population, and we ask what is the complexity of an algorithm that can characterize the probability that the new type takes over the population (becomes fixed). Unexpectedly, we are able to prove exact complexity results (Table 1).
Table 1.
Complexity results for various models and computational questions
| Model | Qualitative | Quantitative |
| Ecological scenario | NP-complete | P-complete |
| Linear fitness | PSPACE-complete | PSPACE-complete |
| Exponential fitness | P | PSPACE-complete |
The class PTIME (denoted as P) consists of problems whose solutions can be computed by an algorithm that uses polynomial time. Formally, an algorithm uses polynomial time if the running time of the algorithm grows as a polynomial function of the size of the input. In computer science, P represents the class of problems that can be solved efficiently.
The class nondeterministic polynomial time (denoted as NP) consists of problems for which solutions exist that are of polynomial length, and given a candidate for a solution of polynomial length, whether the candidate is indeed a solution can be checked in polynomial time. Therefore, an NP algorithm can verify a solution in polynomial time.
To proceed further, we need the notion of “reduction” between classes of problems. A reduction, from a given problem to a problem , is a translation such that a solution for can provide a solution for . More precisely, if there is a polynomial-time reduction from to , then a polynomial-time algorithm for implies a polynomial-time algorithm for .
A given problem is NP-hard if for every problem in NP, there is a polynomial reduction to the given problem. A problem is NP-complete if it is both NP-hard and there is an NP algorithm for the problem.
For example, consider a Boolean formula over variables, and the question of whether there exists an assignment to the variables such that the formula is true. A polynomial candidate solution is an assignment of truth values to variables, and given a candidate assignment, the formula can be evaluated in polynomial time. This question is the famous satisfiability (SAT) problem in computer science. The SAT problem is NP-complete.
The class P is contained in NP, and a major long-standing open question in computer science is whether P = NP. A polynomial-time algorithm for an NP-complete (or an NP-hard) problem would imply that P = NP, resolving the long-standing open problem.
The class sharp ( P) intuitively corresponds to counting the number of solutions. A problem is in P if it counts the number of distinct solutions such that (i) every possible candidate for a solution is of polynomial length, and (ii) given a candidate for a solution, it can be checked in polynomial time whether the candidate is a solution. For example, given a Boolean formula, the problem of whether there are at least k distinct satisfying assignments to the formula is a P-problem. A given problem is P-hard, if for every P-problem, there is a polynomial-time reduction to the given problem. A P-complete problem is a problem that is both P-hard and for which there is a P-solution. For example, counting the number of solutions in SAT is P-complete.
The class NP is contained in P because, given the enumeration of solutions for P, it is easy to check if there exists at least one solution. Intuitively, an NP problem asks whether there is at least one solution, whereas P is the counting version that asks if there are least k distinct solutions (and the special case of gives NP). Again, a major open question is whether NP = P. Note that a polynomial-time algorithm for a P-complete problem would be an even bigger result, because it would imply both P = NP and P = P.
The class PSPACE consists of problems that can be solved with polynomial space. Note that a polynomial space algorithm can reuse space and can, in general, require exponential time. Every P problem can be solved in PSPACE by simply enumerating each candidate for a solution and checking if it is a solution. Because we can reuse space to enumerate the candidates for solutions, the enumeration can be achieved in polynomial space. Moreover, every polynomial-time algorithm uses, at most, polynomial space. Hence, it follows that P is contained in PSPACE. The notion of PSPACE-hardness and PSPACE-completeness is similar to the notion of NP-hardness and NP-completeness, but with respect to the problems in PSPACE. Again, a long-standing open question in computer science is whether P = PSPACE, and a polynomial-time algorithm for a PSPACE-complete (or PSPACE-hard) problem would imply P = NP = P = PSPACE.
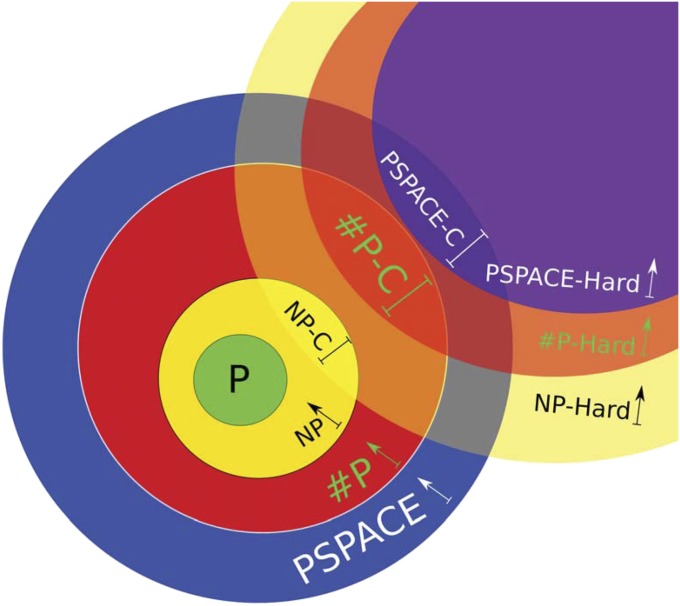
We have mentioned that the major questions about the equality of the complexity classes are open problems, but the widely believed conjecture is that P is strictly contained in NP, NP is strictly contained in P, and P is strictly contained in PSPACE. In other words, it is widely believed that NP-complete problems cannot be solved in polynomial time, P-complete problems are harder than NP-complete problems, and PSPACE-complete problems are harder than P-complete problems. A pictorial illustration of the complexity classes is shown in Fig. 1.
Fig. 1.
Pictorial illustration of the complexity classes P, NP, P, and PSPACE. The complexity class P is contained in NP, NP is contained in P, and P is contained in PSPACE. The widely believed conjecture is that these complexity classes are different. A problem is NP-hard if it is at least as hard as each problem in NP, and the case is similar for P-hardness and PSPACE-hardness. The intersection of NP and NP-hard gives the NP-complete problems, the intersection of P and P-hard gives the P-complete problems, and the intersection of PSPACE and PSPACE-hard gives the PSPACE-complete problems. A polynomial-time solution for an NP-hard or NP-complete problem would imply P = NP.
Results
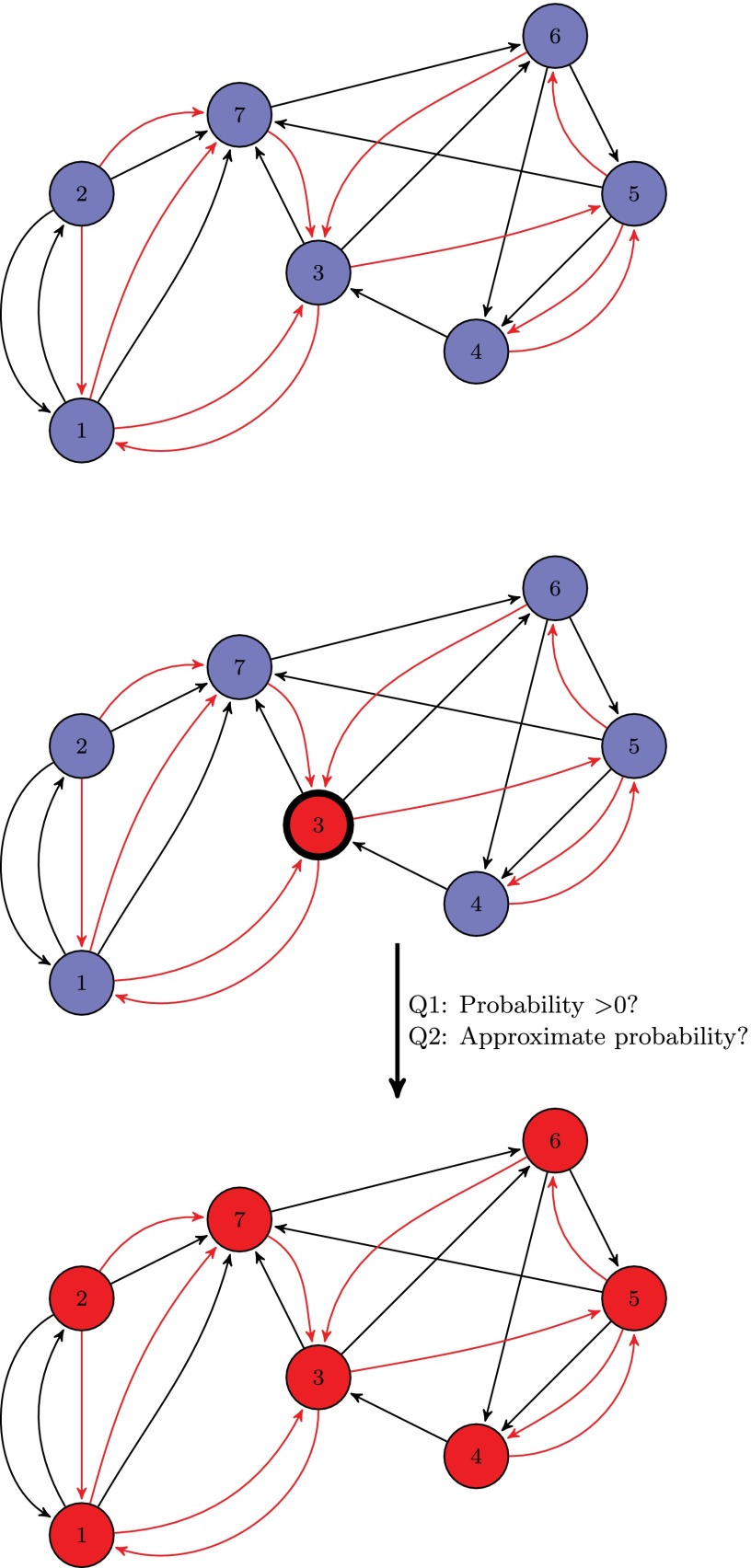
The first problem is motivated by ecological dynamics. There is an ecosystem occupied by resident species. The spatial structure of the ecosystem is given by a graph. An invading species is introduced (an illustration is provided in Fig. 2). We assume the invading species has a competitive advantage in the sense that once a position is occupied by the invading species, the resident cannot get it back. The invading species, however, has a density constraint: If the number of invaders around a focal invader is above a threshold, h, then the invader in the focal vertex cannot colonize another vertex.
Fig. 2.
Illustration of mutant introduction. The residents (type A) are colored blue, and the mutants (type B) are colored red. The black edges are the edges of the interaction graph and the red edges are the edges of the reproduction graph. The probability of introducing a mutant in a specific vertex is always 1 over the number of vertices. The computational questions of interest regarding the takeover probability are as follows: whether the probability is positive (qualitative question) and what is an approximation of the probability (quantitative question).
We are interested in the probability that the invader starting from a random initial position will take over the entire ecosystem (and therefore drive the resident to extinction). There are two types of questions. The “qualitative question” is whether the takeover probability is greater than 0. The “quantitative question” is concerned with computing the takeover probability subject to a small error. Fig. 2 gives a pictorial illustration. We prove the following results. The qualitative question is NP-complete (SI Appendix, Theorem 4). The quantitative question is P-complete (SI Appendix, Theorem 8).
The second problem is concerned with evolutionary games in structured populations. There are two types, A and B, whose reproductive rates depend on local interactions. We consider the setting of games on graphs. Each vertex is occupied by one individual, which is either A or B. Interactions occur pairwise with all neighbors. The payoff matrix is given by
| [1] |
The entries of the payoff matrix can be positive or negative (or 0). Each individual interacts with all of its neighbors on the graph to derive a payoff sum. The payoff sum is translated into reproductive success as follows. If the payoff sum is positive, then the fecundity equals the payoff sum. If the payoff sum is negative, then the fecundity is 0. We refer to this translation as linear fitness. In any one time step, a random individual is chosen for reproduction proportional to its fecundity. The offspring, which is of the same type as the parent, is placed into an adjacent position on the graph (illustrations are provided in Figs. 3 and 4).
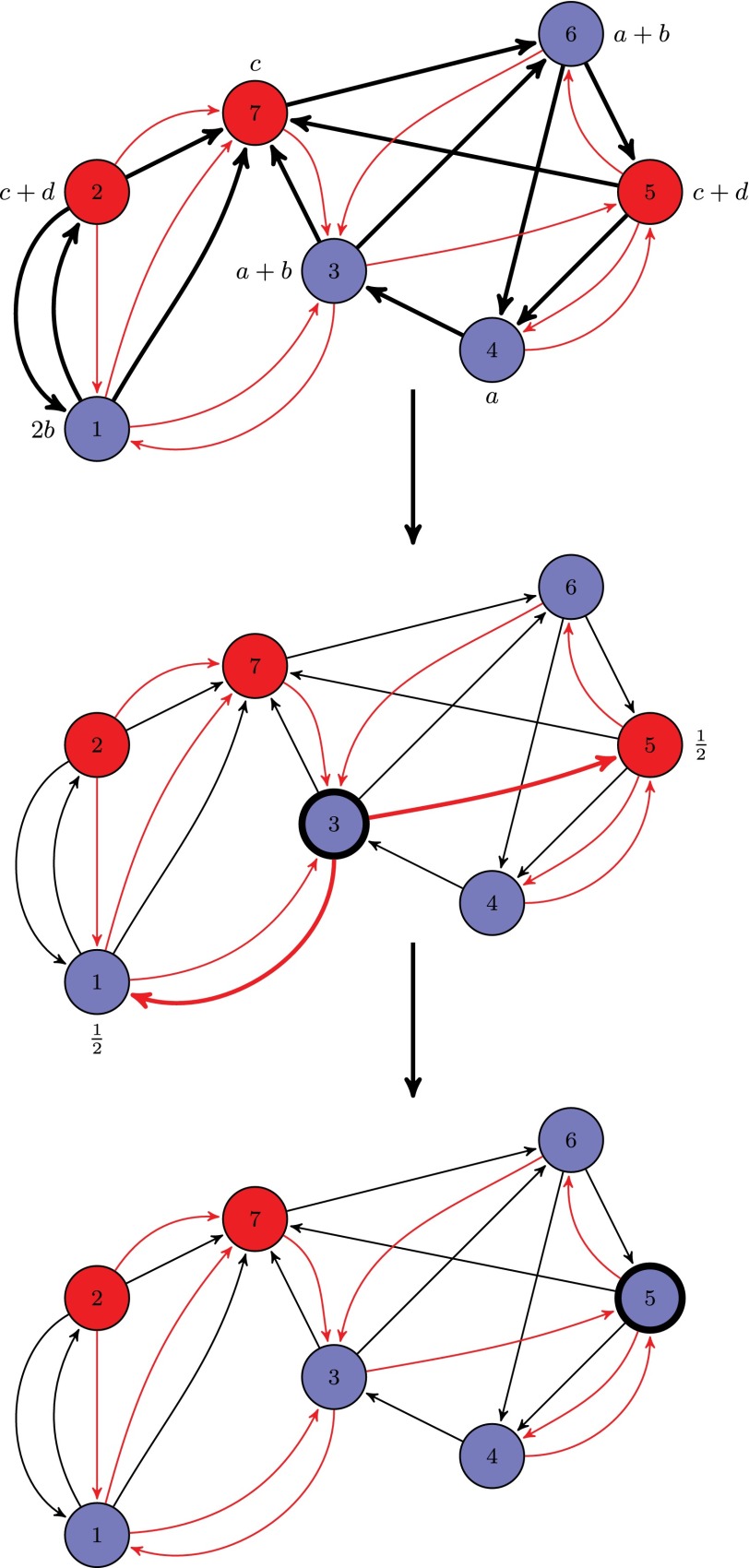
Fig. 3.
Illustration of reproduction with matrix . The residents (type A) are colored blue, and the mutants (type B) are colored red. The black edges are the edges of the interaction graph, and the red edges are the edges of the reproduction graph. In the first figure beside each vertex, the payoff of the vertex (which is the sum of the payoff of the interactions) is shown. Because the first figure shows the payoff computation, the interaction edges that are responsible for the payoff calculation are boldfaced. In the second figure, the vertex labeled 3 is selected for reproduction. The reproduction edges from vertex 3 are boldfaced, and each edge has the probability 1/2. Finally, the successor 5 is chosen for replacement (i.e., vertex 3 reproduces to vertex 5).
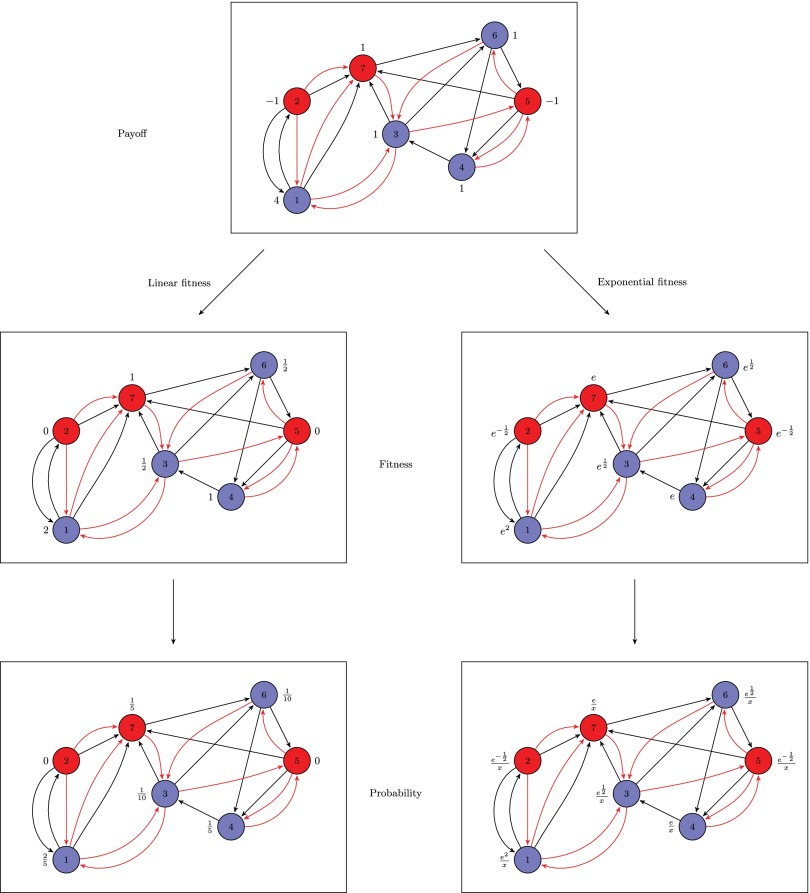
Fig. 4.
Illustration of different payoffs to fitness with . The residents (type A) are blue, and the mutants (type B) are red. The black edges are the edges of the interaction graph, and the red edges are the edges of the reproduction graph. In the figure of the first row, we show the payoff for every vertex. In the next row, we show the fitness, which is either a linear function of the payoff but at least 0 or an exponential function of the payoff. Finally, in the third row, with each vertex, we show the probability, which is the normalized fitness, that the vertex is selected for reproduction [in the last figure, the number x is the sum of the fitness (i.e., )].
We are interested in the probability that a single A individual starting in a random position on the graph generates a lineage that will take over the entire population; this probability is generally called fixation probability. As before, there are two types of questions. The qualitative question is whether the fixation probability is positive. The quantitative question is concerned with computing the fixation probability subject to a small error. We prove the following results. The qualitative question is NP-hard and in PSPACE. The quantitative question is P-hard and in PSPACE. The results follow from SI Appendix, Theorems 4, 8, and 15.
Note that the first problem can also be obtained as a special case of the second problem. In the payoff matrix (1), we can set, for example, , , . This “game” has the property that type B never reproduces and type A reproduces until half of its neighbors are also of type A. This parameter choice leads to the same qualitative behavior and the same complexity bounds as described in the first problem.
A generalization of games on graphs is the setting where the interaction graph and the replacement graph are distinct (13). Thus, each individual interacts with all of its neighbors on the interaction graph to receive payoff. Subsequently, an individual is chosen for reproduction proportional to its fecundity. The offspring is placed randomly among all neighbors of the focal individual on the replacement graph. In this case, both the qualitative and quantitative questions become PSPACE-complete (SI Appendix, Theorem 15).
We also consider a variation of the second problem. In particular, we change the mapping from payoff to fecundity. We now assume that fecundity is an exponential function of payoff, and refer to it as exponential fitness (an illustration is provided in Fig. 4). Therefore, the fecundity of an individual is always positive (even if its payoff sum is negative). In this setting, the qualitative question can be decided in polynomial time. The reason is that the fixation probability is positive if the graph is connected. Thus, to answer the qualitative question, the algorithm only needs to check whether the graph is connected; this problem is in P. However, the quantitative question has the same complexity as the previous problem (SI Appendix, Theorems 16 and 17).
A very special case of games on graphs is constant selection. Type A has constant fecundity a and type B has constant fecundity b independent of any interactions (i.e., fecundity is independent of the population structure). The qualitative question concerning the fixation probability of A is in P. The quantitative question is in PSPACE, but any nontrivial lower bound is an open question.
Finally, although we establish computational hardness for several problems, we also show that two classic problems can be solved in polynomial time (SI Appendix, section 7). First, we consider the molecular clock, which is the rate at which neutral mutations accumulate over time. The molecular clock is affected by population structure (13). We show that the molecular clock can be computed in polynomial time because the problem reduces to solving a set of linear equalities, which can be achieved in polynomial time using Gaussian elimination. Second, we consider evolutionary games in a well-mixed population structure, where the underlying structure is the complete graph (38). We show that the exact fixation probability can be computed in polynomial time. In this case, the problem can be reduced to computing absorption probabilities in Markov chains, where each state represents the number of mutants. Hence, the Markov chain is linear in the number of vertices of the graphs, and because absorption probabilities in Markov chains can be computed in polynomial time (by solving a set of linear equalities), we obtain the desired result.
Methods: Proof Ideas
We now present the key intuition and main ideas of our results. The most interesting and technically insightful results are the lower bounds (i.e., the hardness proofs), and we present the key ideas only for them.
NP-Hardness of the Qualitative Ecological Problem.
One of the most classic NP-complete problems is the 3SAT-problem, which is the SAT problem where every clause has exactly three literals (a literal is a Boolean variable x or a negation of a variable ). Given instances of the 3SAT problem, we construct instances of the ecological problem where we have a start vertex, where the mutant arises, followed by a sequence of vertices (i.e., each vertex can reproduce a mutant to the next), one for each clause. By means of our construction, a vertex in this sequence can reproduce, at most, three times, one of which must be the next vertex of the sequence, with the others corresponding to, at most, two literals of the clause (intuitively, these two literals represent the ones that are not set to true by a candidate-satisfying assignment). The last vertex of the sequence reproduces to a new sequence of vertices that corresponds to an assignment of truth values to the variables. Each vertex in this new sequence can reproduce twice, one to the next vertex of the sequence and other to a variable or its negation. The variables or the corresponding negation can then reproduce mutants to the corresponding literals of the clauses. After this sequence, all vertices that do not correspond to a literal in a clause become mutants. In essence, our construction ensures that if there is a satisfying assignment, then with positive probability, all vertices can become mutants, and, conversely, if there is no satisfying assignment, then the probability that all vertices become mutants is 0.
P-Hardness of the Quantitative Ecological Problem.
A P-complete problem is counting the number of perfect matchings in a bipartite graph (which also corresponds to computing the permanent of a Boolean matrix). A bipartite graph consists of two set of vertices, a set on the left side and a set on the right side, with edges from the left side to the right side. A perfect matching is a one-to-one mapping of each vertex of the left side to a vertex on the right side such that there is an edge between them. First, we argue that for the hardness proof, it suffices to consider bipartite graphs in which each vertex on the left side has an out-degree of for some integer k. A key idea in our construction is that in a full binary tree, if the root becomes a mutant and every vertex can reproduce exactly once, then the set of mutants will eventually consist of a path from the root to a leaf, chosen uniformly at random. Our construction is then as follows: We have a start vertex where the mutant arises, which reproduces to turn each of the vertices on the left side of the bipartite graph to mutants. Each of the vertices on the left side is the root of a full binary tree, where the leaves correspond to the right side of the bipartite graph. We show that the fixation process corresponds to a perfect matching (defined from the path in the full binary trees), and given an approximation of the fixation probability, the exact number of perfect matchings of the bipartite graph can be computed.
PSPACE-Hardness for the Game on Evolutionary Graph Problem.
Our PSPACE-hardness proof shows that the evolutionary process can solve the following concurrent-if problem, which we show is PSPACE-hard. The concurrent-if problem consists of a set of Boolean variables , with a given initial truth assignment to the variables, and a set of if-statements. Each if-statement is of the following form: If a conjunctive clause over the variables is true, then assign a truth value to a variable [e.g., if , then is assigned false]. The problem is to decide whether the first variable (which is the accepting variable) eventually becomes true. We show that each variable can be represented as four vertices, and each if-statement as a single vertex, in the evolutionary graph, and the evolutionary process can mimic the execution of the concurrent-if problem. Finally, if the accepting variable becomes true, then it corresponds to making a special vertex in the evolutionary graph as a mutant. There exists a part of the evolutionary graph that can only become mutants after the special vertex has become a mutant. Using this construction, we show that both the qualitative and quantitative problems are PSPACE-hard for evolutionary games on graphs.
Discussion
In summary, we have established computational complexity results for some fundamental problems of ecological and evolutionary dynamics in structured populations. Our main results are summarized in Table 1. We now discuss the significance of our findings.
Interdisciplinary Connection.
Although both computer science and biology examine the proliferation of information in time and space, the deep connection between them has been largely unexplored. Our work provides precise computational complexity results for several well-studied problems in biology and can be viewed as a step to establish a connection between the two disciplines.
Well-Studied Open Problem.
The problems we have considered are basic aspects of well-studied questions for ecological and evolutionary dynamics in structured populations (7, 28, 37, 42, 50, 51). Several reviews have been written on this topic (8, 11, 43, 57). We first discuss the significance of an algorithmic approach in evolutionary graph theory. An efficient algorithm, which considers all (even worst-case) graphs for evolutionary processes, is important for the following reasons.
First, it has been shown that some population structures (called amplifiers) can increase the effect of natural selection (7, 51), but amplifiers are rare and constructing them is difficult (7, 11, 50, 51, 58, 59). If there were an efficient algorithmic approach that worked for all graphs, then one could design candidates for amplifiers and efficiently check their fixation probabilities. Because there exists no algorithmic approach, research has to focus on special classes of graphs to identify simple formulas, such as calculating the fixation probabilities on star-like graphs (58).
Second, it is known that some population structures and evolutionary dynamics promote evolution of cooperation but that others do not (51). An important open problem is to characterize the set of graphs that promote cooperation. An efficient algorithmic approach would be useful to check candidate structures. Because no efficient algorithm exists, one has to study special cases, for example, by considering nearly regular graphs (51).
Thus, a general algorithmic approach is a very important problem for the well-studied question concerning the effect of population structures on evolutionary dynamics. An algorithmic approach has been studied for important special cases, such as for complete graphs (60), and NP-hardness was stated for the quantitative problem (7). The review by Shakarian et al. (57) identifies the complexity of computing fixation probabilities on evolutionary graphs as an important open question in the area. In that review, two open problems (2.1 and 2.2) are identified that ask for the complexity of computing the exact fixation probabilities for graphs and for games on graphs. Our results not only present answers to those crucial questions but also show that both the approximation problem and the qualitative question are computationally hard. The most interesting aspects of our results are the lower bounds, which show that there exists no efficient algorithm in most cases, under the widely believed conjecture that P is different from NP. A simple equation-based solution would give an efficient algorithm; thus, our result formally shows that for evaluating the fixation probability in spatial settings, there does not exist a simple equation-based solution in general. Our results are significant for the following reasons: (i) They establish the computational complexity for fundamental problems of ecological and evolutionary dynamics in structured populations (e.g., considered in 7, 8, 11, 28, 37, 42, 43, 50, 51, 57), and (ii) they significantly improve the complexity result of Lieberman et al. (7) and solve the computational complexity questions of the area as identified in the review by Shakarian et al. (57).
Methodological Insight.
Our proof ideas also reveal some important points. We show how evolutionary processes in structured populations can mimic aspects of computation. This insight could be useful for future research on understanding the computational complexities of other stochastic processes on population structures.
Supplementary Material
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1511366112/-/DCSupplemental.
References
- 1.Moran PAP. Random processes in genetics. Proc Camb Philos Soc. 1958;54(1):60–71. [Google Scholar]
- 2.Maruyama T. Effective number of alleles in a subdivided population. Theor Popul Biol. 1970;1(3):273–306. doi: 10.1016/0040-5809(70)90047-x. [DOI] [PubMed] [Google Scholar]
- 3.Slatkin M. Fixation probabilities and fixation times in a subdivided population. Evolution. 1981;35(3):477–488. doi: 10.1111/j.1558-5646.1981.tb04911.x. [DOI] [PubMed] [Google Scholar]
- 4.Wright S. Evolution in Mendelian populations. Genetics. 1931;16(2):97–159. doi: 10.1093/genetics/16.2.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wright S. Proceedings of the Sixth International Congress of Genetics. Ithaca; New York: 1932. The roles of mutation, inbreeding, crossbreeding and selection in evolution; pp. 356–366. [Google Scholar]
- 6.Fisher RA. The “Sewell Wright Effect”. Heredity (Edinb) 1950;4(1):117–119. doi: 10.1038/hdy.1950.8. [DOI] [PubMed] [Google Scholar]
- 7.Lieberman E, Hauert C, Nowak MA. Evolutionary dynamics on graphs. Nature. 2005;433(7023):312–316. doi: 10.1038/nature03204. [DOI] [PubMed] [Google Scholar]
- 8.Szabó G, Fáth G. Evolutionary games on graphs. Phys Rep. 2007;446(4-6):97–216. [Google Scholar]
- 9.Yang HX, Wu ZX, Du WB. Evolutionary games on scale-free networks with tunable degree distribution. Europhys Lett. 2012;99(1):10006. [Google Scholar]
- 10.Chen YT. Sharp benefit-to-cost rules for the evolution of cooperation on regular graphs. Ann Appl Probab. 2013;23(2):637–664. [Google Scholar]
- 11.Allen B, Nowak MA. Games on graphs. European Mathematical Society Surveys in Mathematical Sciences. 2014;1(1):113–151. [Google Scholar]
- 12.Débarre F, Hauert C, Doebeli M. Social evolution in structured populations. Nat Commun. 2014;5:3409. doi: 10.1038/ncomms4409. [DOI] [PubMed] [Google Scholar]
- 13.Ohtsuki H, Pacheco JM, Nowak MA. Evolutionary graph theory: Breaking the symmetry between interaction and replacement. J Theor Biol. 2007;246(4):681–694. doi: 10.1016/j.jtbi.2007.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Skyrms B, Pemantle R. A dynamic model of social network formation. Proc Natl Acad Sci USA. 2000;97(16):9340–9346. doi: 10.1073/pnas.97.16.9340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pacheco JM, Traulsen A, Nowak MA. Coevolution of strategy and structure in complex networks with dynamical linking. Phys Rev Lett. 2006;97(25):258103. doi: 10.1103/PhysRevLett.97.258103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fu F, Hauert C, Nowak MA, Wang L. Reputation-based partner choice promotes cooperation in social networks. Phys Rev E Stat Nonlin Soft Matter Phys. 2008;78(2 Pt 2):026117. doi: 10.1103/PhysRevE.78.026117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Antal T, Ohtsuki H, Wakeley J, Taylor PD, Nowak MA. Evolution of cooperation by phenotypic similarity. Proc Natl Acad Sci USA. 2009;106(21):8597–8600. doi: 10.1073/pnas.0902528106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tarnita CE, Antal T, Ohtsuki H, Nowak MA. Evolutionary dynamics in set structured populations. Proc Natl Acad Sci USA. 2009;106(21):8601–8604. doi: 10.1073/pnas.0903019106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Szolnoki A, Perc M. Resolving social dilemmas on evolving random networks. Europhys Lett. 2009;86(3):30007. [Google Scholar]
- 20.Cavaliere M, Sedwards S, Tarnita CE, Nowak MA, Csikász-Nagy A. Prosperity is associated with instability in dynamical networks. J Theor Biol. 2012;299(0):126–138. doi: 10.1016/j.jtbi.2011.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rand DG, Arbesman S, Christakis NA. Dynamic social networks promote cooperation in experiments with humans. Proc Natl Acad Sci USA. 2011;108(48):19193–19198. doi: 10.1073/pnas.1108243108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wu B, et al. Evolution of cooperation on stochastic dynamical networks. PLoS One. 2010;5(6):e11187. doi: 10.1371/journal.pone.0011187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Smith JM. Evolution and the Theory of Games. Cambridge Univ Press; Cambridge, UK: 1982. [Google Scholar]
- 24.Hofbauer J, Sigmund K, editors. The Theory of Evolution and Dynamical Systems: Mathematical Aspects of Selection. Cambridge Univ Press; Cambridge, UK: 1988. [Google Scholar]
- 25.Hofbauer J, Sigmund K. Evolutionary Games and Population Dynamics. Cambridge Univ Press; Cambridge, UK: 1998. [Google Scholar]
- 26.Cressman R. Evolutionary Dynamics and Extensive Form Games, Economic Learning and Social Evolution. MIT Press; Cambridge, MA: 2003. [Google Scholar]
- 27.Broom M, Rychtář J. 2013. Game-theoretical models in biology. Chapman & Hall/CRC Mathematical and Computational Biology (Chapman & Hall/CRC, Boca Raton, FL)
- 28.Nowak MA, May RM. Evolutionary games and spatial chaos. Nature. 1992;359(6398):826–829. [Google Scholar]
- 29.Ellison G. Learning, local interaction, and coordination. Econometrica. 1993;61(5):1047–1071. [Google Scholar]
- 30.Herz AV. Collective phenomena in spatially extended evolutionary games. J Theor Biol. 1994;169(1):65–87. doi: 10.1006/jtbi.1994.1130. [DOI] [PubMed] [Google Scholar]
- 31.Nakamaru M, Nogami H, Iwasa Y. Score-dependent fertility model for the evolution of cooperation in a lattice. J Theor Biol. 1998;194(1):101–124. doi: 10.1006/jtbi.1998.0750. [DOI] [PubMed] [Google Scholar]
- 32.Szabó G, Antal T, Szabó P, Droz M. Spatial evolutionary prisoner’s dilemma game with three strategies and external constraints. Phys Rev E Stat Phys Plasmas Fluids Relat Interdiscip Topics. 2000;62(1 Pt B):1095–1103. doi: 10.1103/physreve.62.1095. [DOI] [PubMed] [Google Scholar]
- 33.Kerr B, Riley MA, Feldman MW, Bohannan BJM. Local dispersal promotes biodiversity in a real-life game of rock-paper-scissors. Nature. 2002;418(6894):171–174. doi: 10.1038/nature00823. [DOI] [PubMed] [Google Scholar]
- 34.Helbing D, Yu W. Migration as a mechanism to promote cooperation. Adv Complex Syst. 2008;11(4):641–652. [Google Scholar]
- 35.Tarnita CE, Ohtsuki H, Antal T, Fu F, Nowak MA. Strategy selection in structured populations. J Theor Biol. 2009;259(3):570–581. doi: 10.1016/j.jtbi.2009.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Perc M, Szolnoki A. Coevolutionary games--a mini review. Biosystems. 2010;99(2):109–125. doi: 10.1016/j.biosystems.2009.10.003. [DOI] [PubMed] [Google Scholar]
- 37.van Veelen M, García J, Rand DG, Nowak MA. Direct reciprocity in structured populations. Proc Natl Acad Sci USA. 2012;109(25):9929–9934. doi: 10.1073/pnas.1206694109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nowak MA, Sasaki A, Taylor C, Fudenberg D. Emergence of cooperation and evolutionary stability in finite populations. Nature. 2004;428(6983):646–650. doi: 10.1038/nature02414. [DOI] [PubMed] [Google Scholar]
- 39.Killingback T, Doebeli M. Spatial evolutionary game theory: Hawks and doves revisited. Proc Biol Sci. 1996;263(1374):1135–1144. [Google Scholar]
- 40.Szabó G, Toke C. Evolutionary prisoner’s dilemma game on a square lattice. Phys Rev E Stat Phys Plasmas Fluids Relat Interdiscip Topics. 1998;58(1):69–73. doi: 10.1103/physreve.62.1095. [DOI] [PubMed] [Google Scholar]
- 41.Szabó G, Hauert C. Phase transitions and volunteering in spatial public goods games. Phys Rev Lett. 2002;89(11):118101. doi: 10.1103/PhysRevLett.89.118101. [DOI] [PubMed] [Google Scholar]
- 42.Hauert C, Doebeli M. Spatial structure often inhibits the evolution of cooperation in the snowdrift game. Nature. 2004;428(6983):643–646. doi: 10.1038/nature02360. [DOI] [PubMed] [Google Scholar]
- 43.Nowak MA, Tarnita CE, Antal T. Evolutionary dynamics in structured populations. Philos Trans R Soc Lond B Biol Sci. 2010;365(1537):19–30. doi: 10.1098/rstb.2009.0215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Allen B, Tarnita CE. Measures of success in a class of evolutionary models with fixed population size and structure. J Math Biol. 2012;68(1-2):109–143. doi: 10.1007/s00285-012-0622-x. [DOI] [PubMed] [Google Scholar]
- 45.Allen B, et al. The molecular clock of neutral evolution can be accelerated or slowed by asymmetric spatial structure. PLOS Comput Biol. 2015;11(2):e1004108. doi: 10.1371/journal.pcbi.1004108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Adlam B, Nowak MA. Universality of fixation probabilities in randomly structured populations. Sci Rep. 2014;4:6692. doi: 10.1038/srep06692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Maruyama T. A Markov process of gene frequency change in a geographically structured population. Genetics. 1974;76(2):367–377. doi: 10.1093/genetics/76.2.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Barton NH. The probability of fixation of a favoured allele in a subdivided population. Genet Res. 1993;62(2):149–157. [Google Scholar]
- 49.Nowak MA, Michor F, Iwasa Y. The linear process of somatic evolution. Proc Natl Acad Sci USA. 2003;100(25):14966–14969. doi: 10.1073/pnas.2535419100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nowak MA. Evolutionary Dynamics. Harvard Univ Press; Cambridge, MA: 2006. [Google Scholar]
- 51.Ohtsuki H, Hauert C, Lieberman E, Nowak MA. A simple rule for the evolution of cooperation on graphs and social networks. Nature. 2006;441(7092):502–505. doi: 10.1038/nature04605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Durrett R, Levin S. The importance of being discrete (and spatial) Theor Popul Biol. 1994;46(3):363–394. [Google Scholar]
- 53.Levin SA, Paine RT. Disturbance, patch formation, and community structure. Proc Natl Acad Sci USA. 1974;71(7):2744–2747. doi: 10.1073/pnas.71.7.2744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hassell MP, Comins HN, May RM. Species coexistence and self-organizing spatial dynamics. Nature. 1994;370(6487):290–292. [Google Scholar]
- 55.Tilman D, Kareiva PM. Spatial Ecology. The Role of Space in Population Dynamics and Interspecific Interactions. Princeton Univ Press; Princeton: 1997. [Google Scholar]
- 56.Dieckmann U, Law R, Metz JAJ, editors. The Geometry of Ecological Interactions. Cambridge Univ Press; Cambridge, UK: 2000. [Google Scholar]
- 57.Shakarian P, Roos P, Johnson A. A review of evolutionary graph theory with applications to game theory. Biosystems. 2012;107(2):66–80. doi: 10.1016/j.biosystems.2011.09.006. [DOI] [PubMed] [Google Scholar]
- 58.Adlam B, Chatterjee K, Nowak MA. 2015. Amplifiers of selection. Proc R Soc Lond A Math Phys Sci 471(2181):20150114.
- 59.Diaz J, et al. On the fixation probability of superstars. Proc R Soc Lond A Math Phys Sci. 2013;469(2156):20130193. [Google Scholar]
- 60.Diaz J, et al. Approximating fixation probabilities in the generalized Moran process. Algorithmica. 2014;69(1):78–91. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.