Significance
The key problem concerning pediatric infectious diseases, and more generally clinical diseases during primary infection, is their pathogenesis. A plausible and testable human genetic theory of primary infectious diseases has recently emerged, building on elegant studies in plants and animals. Three examples of monogenic resistance to common infections have been discovered. Moreover, a growing range of monogenic single-gene inborn errors of immunity, rarely Mendelian (with complete clinical penetrance) but more commonly non-Mendelian (with incomplete penetrance), have been found to underlie severe infectious diseases striking otherwise healthy children during primary infection. These findings provide a synthetic framework for inherited and infectious diseases and, more generally, for inborn and environmental conditions.
Keywords: human genetics, immunology, infectious diseases, pediatrics, primary immunodeficiency
Abstract
This paper reviews the developments that have occurred in the field of human genetics of infectious diseases from the second half of the 20th century onward. In particular, it stresses and explains the importance of the recently described monogenic inborn errors of immunity underlying resistance or susceptibility to specific infections. The monogenic component of the genetic theory provides a plausible explanation for the occurrence of severe infectious diseases during primary infection. Over the last 20 y, increasing numbers of life-threatening infectious diseases striking otherwise healthy children, adolescents, and even young adults have been attributed to single-gene inborn errors of immunity. These studies were inspired by seminal but neglected findings in plant and animal infections. Infectious diseases typically manifest as sporadic traits because human genotypes often display incomplete penetrance (most genetically predisposed individuals remain healthy) and variable expressivity (different infections can be allelic at the same locus). Infectious diseases of childhood, once thought to be archetypal environmental diseases, actually may be among the most genetically determined conditions of mankind. This nascent and testable notion has interesting medical and biological implications.
This paper reviews the monogenic inborn errors of immunity that have been shown to underlie resistance or vulnerability to human infectious diseases. These studies, first conducted in the 1970s, gained momentum from the mid-1990s onward. The analysis of Mendelian traits was followed by that of non-Mendelian monogenic traits, characterized by incomplete clinical penetrance and variable expressivity.
Mendelian Resistance to Malaria
An example of infection shaping the human genome was provided by the discovery of Mendelian resistance to Plasmodium vivax (1, 2). It is important to stress here that these studies concerned infection per se and not its manifestations (Table 1). As discussed in the companion paper (3), only a minority of infected individuals develop clinical disease, and an even smaller minority develop life-threatening disease in the course of primary infection. Moreover, not all exposed individuals become infected. In at least three cases, this phenomenon has been shown to be a Mendelian phenotype because of a well-defined monogenic genotype for a microbial receptor or coreceptor. Louis Miller carried out pioneering studies in this area. In 1976, he discovered that an autosomal recessive deficiency of the Duffy antigen and receptor for chemokines (DARC) in erythrocytes prevented the infection of these cells with Plasmodium vivax in vitro and the infection of individuals in vivo (1). An elegant set of follow-up studies demonstrated that this resistance could be attributed to a single nucleotide mutation in the DARC promoter, preventing the binding of the transcription factor GATA-binding protein 1 (GATA1), which is required for DARC expression in the erythrocyte lineage (4). Affected individuals express DARC normally in other cells. P. vivax infections are rarely lethal today, but the selective pressure exerted by this parasite may have favored the spread of this allele, which is more common in infested areas and is fixed in some African populations. These findings do not explain the pathogenesis of P. vivax malaria, but they do explain why some individuals are not infected despite repeated exposure to the parasite. They also show that a pathogen can favor the spread of a human allele conferring Mendelian protection against the pathogen concerned.
Table 1.
Mendelian resistance to human infection
| Infection | Gene | Inheritance | Year |
| P. vivax | DARC | Autosomal recessive | 1975 |
| HIV | CCR5 | Autosomal recessive | 1996 |
| Norovirus | FUT2 | Autosomal recessive | 2003 |
The three phenotypes are autosomal recessive. Biallelic mutations underlie complete deficiency of any of these three microbial receptors or coreceptors. These deficiencies can be common, especially in areas where the corresponding infection can select the resistance genotype. Clinical penetrance (resistance to infection per se) is complete or almost complete. The infections and resistance genes are described in the text, as are the references.
Other Forms of Mendelian Resistance to Infection
Another set of similar studies in 1996 explained why the HIV did not infect rare individuals in vivo, despite repeated exposure through sexual contact. The CD4+ T cells of these individuals were shown to be resistant to infection in vitro. An autosomal recessive chemokine (C-C motif) receptor 5 (CCR5) deficiency was rapidly identified in these subjects (5–7). The most common mutation underlying this phenotype probably originated in Scandinavia, and the evolutionary forces driving its spread toward Southern Europe have remained elusive. The third and last case we will consider here is autosomal recessive fucosyltransferase 2 (FUT2) deficiency, which was discovered in 2003. Individuals with this deficiency are naturally resistant to diarrhea caused by noroviruses (8, 9). This condition is typically benign in the developed world today but would have been potentially life-threatening in ancient times, as it still is in developing countries. With these three examples, we have true genetic determinism that is autosomal recessive with complete or almost complete penetrance. The monogenic lesions prevent infection through a causal mechanism (i.e., lack of cell infection by a specific microbe). These findings concern very large numbers of individuals and neatly illustrate the potential impact of Mendelian genetics at the population level. These findings have important biological implications, because these three sets of studies demonstrate the essential nature of a single specific gene for each infection. The clinical implications are equally important, because blocking these molecules in other individuals is a rational approach to prevention or therapy. It is surprising that this field has not blossomed. For almost all common microbes, seronegativity has been documented in at least a small proportion of the population, suggesting that there are naturally and perhaps genetically resistant individuals. The discovery of other forms of autosomal recessive resistance to infection by other microorganisms, especially common and virulent pathogens, would not be surprising.
Mendelian Susceptibility to Mycobacterial Diseases
We now can discuss the central theme of single-gene variants underlying heterogeneities in clinical outcome in infected individuals. Our work proceeded in two successive steps. We first focused on Mendelian infectious diseases, the mirror image of the aforementioned Mendelian forms of resistance. Indeed, from the 1940s onward, clinicians had reported rare infectious diseases displaying true Mendelian segregation (Tables 2 and 3). In the absence of detectable immunological abnormalities, these conditions were not considered to be primary immunodeficiencies (10, 11). Our attention was drawn immediately to the cases of “idiopathic” disease caused by live bacillus Calmette–Guérin vaccines, which probably were described first in 1951 (12–14). Patients selectively prone to clinical disease caused by environmental mycobacteria were reported later, when these mycobacterial species emerged as opportunistic agents in other settings (15). The patients are selectively susceptible to weakly virulent mycobacteria and, more rarely, to other intramacrophagic pathogens, including Salmonella in particular (16). The clinical association of disease caused by weakly virulent Mycobacterium and Salmonella is almost pathognomonic for this condition. Analysis of the frequently multiplex and/or consanguineous affected families suggested that these patients displayed Mendelian susceptibility to mycobacterial disease (MSMD) (15). MSMD comprises a group of truly Mendelian conditions—monogenic disorders displaying complete or at least very high levels of clinical penetrance. The proportion of familial, as opposed to sporadic, cases therefore is high, at least in large kinships and kindreds. These patients were long considered to suffer from idiopathic infections, rather than primary immunodeficiencies, despite Mendelian inheritance, because there was no immunological phenotype segregating with disease.
Table 2.
Mendelian genetic determinism of infectious diseases in plants, mice, and humans
| Species | Mendelian infections | Gene identification | ||
| Broad | Specific | Expression | Linkage | |
| Plants | – | 1905 (wheat) | – | 1993 (tomato) |
| Mouse | 1959 (Dh) | 1964 (Mx) | 1986 (Mx) | 1993 (Bcg/Lsh/Ity) |
| Human | 1952 (XLA) | 1946 (EV) | 1993 (complement) | 1996 (MSMD) |
Mendelian infections (left two columns) can be broad (multiple infections) or specific (a single type of infection). Gene identification (right two columns) only corresponds to Mendelian infections to a specific infection. For mice and humans, a broad range of infections were associated with distinctive immunological phenotypes (asplenia for Dh and agammaglobulinemia for XLA), but no immunological phenotype was initially associated with Mx and EV. In plants, the inheritance of susceptibility/resistance to infection appears to be often specific for a particular pathogen. The resistance genes typically have been mapped for plants, and the plant species, rather than the resistance loci or pathogens, are indicated in the table. References are provided in the text.
Table 3.
The evolution of the concept of monogenic inborn error of immunity to infection
| Infections | |
| 1952 | 1996 |
| Multiple | Single |
| Recurrent | Single (acute or chronic) |
| Early childhood | At any age |
| Opportunistic | Not necessarily |
| Rare | Rare or common |
| Familial | Sporadic |
The field of primary immunodeficiency was born with the description of patients with severe infectious diseases that met most of these six criteria, reflecting the advent of antibiotics and the Mendelian cosegregation of infectious and immunological phenotypes (144). The field evolved in numerous directions, with the description of a variety of noninfectious phenotypes caused by inborn errors of immunity. As far as infectious phenotypes are concerned, the paradigm shifted from 1996 onward, with increasing recognition that severe infectious diseases that did not fulfill these six criteria could be caused by novel types of primary immunodeficiencies. Mendelian infections (Table 5) are at variance from the initial paradigm for the first three criteria, whereas non-Mendelian monogenic infections are also at variance for the remaining three criteria (Table 6).
Inborn Errors of IFN-γ Immunity
We thus searched for the genetic basis of MSMD by a combination of positional cloning and candidate gene approaches (Tables 3 and 4). Michael Levin followed a similar approach in London. In 1996, we jointly reported the first genetic etiology of MSMD: autosomal recessive complete IFN-γ receptor 1 (IFN-γR1) deficiency (17, 18). Over the next 20 y, we and others pursued this forward genetic approach and discovered a group of 17 inborn errors of IFN-γ immunity involving nine genes (19–23). Some MSMD-causing genes control the production of IFN-γ; others govern its action. These nine MSMD-causing genes display high levels of allelic heterogeneity. However, these deficiencies display physiological homogeneity, because all defects impair IFN-γ immunity. Moreover, human IFN-γ immunity, as a quantitative trait, controls host defense against mycobacteria. The clinical features of MSMD depend strongly on the levels of IFN-γ immunity, being most severe in patients with complete IFN-γ deficiency (24). The clinical implications of this work are obvious. These studies identified the genetic defects underlying these Mendelian infections, providing molecular genetic proof that mycobacterial disease could be Mendelian. The patients are treated with IFN-γ and antibiotics or by hematopoietic stem cell transplantation when the receptor is completely nonfunctional. The immunological implications are not negligible: These studies confirmed Carl Nathan’s suggestion that IFN-γ is much more a macrophage-activating factor than an antiviral agent (25). They also showed that the Th1 arm of CD4+ T cells (IL-12 being the Th1-inducing signature cytokine and IFN-γ the Th1 effector signature cytokine), which in inbred mice had been reported to control experimental infections with intracellular microbes, had a narrower spectrum of action in outbred humans in conditions of natural infection (with environmental mycobacteria) or iatrogenic infection (with the bacillus Calmette–Guérin vaccine). If IL-12 and IFN-γ are essential for the control of such a small range of microbes, which genes are essential for the control of other microbes? The genetic dissection of MSMD was the first study of Mendelian predisposition to a narrow, specific group of microbes, and it remains the most comprehensive study of this type. This work is continuing, because genetic etiologies have been identified for only about half the known MSMD patients.
Table 4.
Mendelian infectious diseases
| Condition | Year | Genes | Year |
| EV | 1946 | TMC6, TMC8 | 2002 |
| MSMD | 1951 | IFNGR1, IFNGR2, and others | 1996 |
| Dermatophytic disease | 1957 | CARD9 | 2013 |
| CMC | 1968 | IL17F, IL17RA, and others | 2011 |
| XLP | 1974 | SAP | 1998 |
From the middle of the 20th century these infectious diseases were shown to segregate as Mendelian traits, with complete or nearly complete penetrance. They are listed in the order of their clinical description (left columns), which differs from the discovery of their molecular genetic basis (right columns). Resistance to the idea of Mendelian infections is best exemplified by EV, which was the first inborn error of immunity to infection to be described clinically but the last to be accepted as such by immunologists. It finally was accepted as an inborn error of immunity to infection in 2002, when its disease-causing genes were discovered. TMC6 and TMC8 are also known as EVER1 and EVER2.
A Neglected Connection: Neisseria and Complement
Before turning to other examples of Mendelian infections, I wish to discuss the case of complement, because it is interesting to try to understand why it did not play a more important role in the field (Table 5). Mutations of genes encoding the terminal components of complement (C5–C9) or its activating protein properdin (encoded by the X chromosome) were described between 1993 and 1998 (26–31). A mutation in a gene encoding another activating protein, factor D, was first reported in 2001 (32). Strikingly, these recessive defects underlie selective, late-onset, recurrent, and invasive disease caused by Neisseria (typically meningococcal meningitis). However, the discovery of these defects did not result from the identification of a familial predisposition to Neisseria in the absence of overt immunological abnormalities, as was the case for MSMD. Instead, impaired global complement activity in a sporadic case of Neisseria meningitis led to the discovery of defects in C6 and C8 activity in 1976 (33, 34) and, two decades later, to the identification of mutations of the C6, C8B, and C8A genes. Likewise, defective factor D activity was first reported in a sporadic case of meningitis in 1989 (35), and the first causal mutation for this condition was described a decade later. C5 and C9 defects were first identified in 1976 and 1981, in a lupus patient and a healthy individual, respectively (36, 37). Such defects then were detected in sporadic cases of Neisseria disease in 1979 and 1989 (38, 39), leading to the discovery of C5 and C9 mutations. Only C7, properdin, and factor D deficiencies were first discovered in multiplex kindreds with Neisseria disease, in 1978 and 1982 (40, 41). These studies, in turn, led to the study of other patients with Neisseria infections. In other words, kindreds with Mendelian infections caused by Neisseria had never been described in the medical literature as idiopathic or as having a Mendelian predisposition to Neisseria meningitis, probably because Neisseria was thought to be sufficiently pathogenic to cause disease by itself (despite the presence of this microbe in the nostrils of almost all children). These discoveries were not made through a genetic genome-wide approach or even through the testing of a genetic hypothesis by a candidate gene approach. Instead, they were based on the serendipitous observation of impaired total complement activity. Nevertheless, with hindsight, the defects of properdin, factor D, and the terminal components of complement are fascinating. These findings unambiguously demonstrate that mutations in any one of several genes controlling a common component of a major arm of immunity can manifest as invasive diseases caused by a unique type of common pathogen. Moreover, a significant proportion of children with invasive meningococcal disease have been found to display one or other of these complement defects (42). However, these findings did not have as great an impact as the study of MSMD on the definition of candidate genetic architectures for infectious diseases, possibly because of the way in which they were obtained.
Table 5.
Inborn errors of complement and Neisseria infections
| Gene | First patient (reference) | First Neisseria infection (reference) | First mutation (reference) |
| C5 | 1976, SLE, S (36) | 1979, S (38) | 1995 (26) |
| C6 | 1976, S (33) | 1976, S (33) | 1995 (27) |
| C7 | 1978, F (40) | 1978, F (40) | 1996 (28) |
| C8b | 1976, S (34) | 1976, S (34) | 1993 (29) |
| C8a | 1998 (30) | ||
| C9 | 1981, Healthy, S (37) | 1989, S (39) | 1997 (145) |
| Properdin | 1982, F (41) | 1982, F (41) | 1995 (31) |
| Factor D | 1989, S (35) | 1989 S (35) | 2001 (32) |
The importance of the chronology of the history of these inborn errors of complement is explained in the text. The salient point is that these inborn errors were not discovered by testing the hypothesis that invasive Neisseria infections could be inherited as Mendelian traits. As shown in this table, they were discovered serendipitously, even sometimes in individuals without Neisseria infection. F, familial; Healthy, healthy individual; S, sporadic; SLE, systemic lupus erythematosus.
Epidermodysplasia Verruciformis
Epidermodysplasia verruciformis (EV) is a unique example in human genetics, immunology, infectious diseases, and oncology (Table 4). EV is an exceedingly rare condition, as suggested by its having no common name, and it is undoubtedly unknown to most readers (43). It is, however, arguably the most interesting condition, at least from a historical perspective, reviewed here. Patients with EV develop flat warts and other skin lesions in the first decade of life. From the age of 20 onward, these lesions tend to degenerate into nonmelanoma skin cancers. They are caused by a specific group of human papillomaviruses (HPV), the EV-associated oncogenic β-HPV (EV-HPV), which are harmless commensals of the general population. The detection of HPV-5 in EV cancers provided the first evidence of the oncogenic role of a papillomavirus in a human cancer (44). The clinical manifestations of this disease were first described in 1922 (45), its genetic nature was described in 1933 (46), and its viral cause was described in 1946 (47). As such, EV should have been considered the first primary immunodeficiency. However, it was not, probably because EV patients displayed no detectable immunological phenotype. Not until 2004, 2 y after Gérard Orth and colleagues elucidated the first single-gene inborn errors underlying autosomal recessive EV—mutations of transmembrane channel-like 6 and 8 (TMC6 and TMC8) (48)—was this condition finally included, not without hesitation, in the international classification of primary immunodeficiencies (49, 50). Ironically, mild immunological abnormalities that probably were more a consequence than a cause of EV were subsequently discovered in these patients (51). The cellular and molecular mechanism of EV remains elusive, because the functions of TMC6 and TMC8 are largely unknown (52). One plausible hypothesis is that the TMC complex governs cell-intrinsic immunity to oncogenic EV-specific HPV in keratinocytes. The search for new genetic etiologies of EV in patients without TMC mutations should help resolve this important issue.
X-Linked Lymphoproliferative Disease
A clinical description of a large kindred with X-linked recessive lymphoproliferative disease (XLP) was reported in 1974–1977 (53–56), leading to the immediate recognition of this disease as a primary immunodeficiency (Table 4) (57). Three distinct phenotypes in response to EBV infection were identified in maternally related males, providing a clear example of complete penetrance with variable expressivity: hemophagocytosis, lymphoma, and hypogammaglobulinemia. The three phenotypes were encompassed by the domain of immunology. However, they were the consequences of the inborn error and did not even correspond to an intermediate phenotype in the absence of EBV infection. Nevertheless, in contrast to the lack of recognition of EV, this triad was, paradoxically, considered relevant enough for the inclusion of this condition in the 1978 international classification of primary immunodeficiencies (57). Three groups determined the first genetic basis of XLP in 1998 (58–60). The disease-causing gene encodes signaling lymphocytic activation molecule-associating protein (SAP), an intracellular molecule expressed in T and natural killer (NK) cells. The pathophysiology of XLP was elegantly clarified more than a decade later through somatic genetic studies of heterozygous female and revertant male subjects (61, 62). This somatic genetic approach to tackling the cellular basis of a germline disorder was very fruitful. SAP-expressing CD8+ T cells were shown to be absolutely required for the control of EBV-infected B cells. Their absence explains the occurrence of two of the three B-cell phenotypes: polyclonal B-cell proliferation (precipitating hemophagocytosis via T-cell activation) and B-cell lymphoma. The third phenotype, B-cell exhaustion (hypogammaglobulinemia), may be caused by impaired CD4+ T-cell function. The mechanisms governing the development of a particular exclusive phenotype in individual patients remain elusive (63) but may involve modifier genes or other factors. It is tempting to speculate that the viral inoculum plays a key role, with high viral load during primary infection leading to the most common phenotype (hemophagocytosis), intermediate inoculum levels being controlled at the cost of impaired maintenance of memory B cells (hypogammaglobulinemia), and lower levels of inoculum controlled with the elimination of a subset of EBV-infected B cells; surviving clones subsequently undergo transformation to become malignant (lymphoma). At any rate, XLP provides a fine example of a life-threatening infection or virus-induced cancer that also is strictly Mendelian. It paved the way for the discovery of other related recessive disorders, X-linked inhibitor of apoptosis (XIAP) deficiency and X-linked magnesium transporter 1 (MAGT1) deficiency, conditions that also often manifest clinically upon EBV infection (64–67).
Chronic Mucocutaneous Candidiasis
We also deciphered the genetic basis of another Mendelian “hole” in host defense, familial chronic mucocutaneous candidiasis (CMC), which was first described clinically in the late 1960s and early 1970s (Table 4) (68, 69). Unexpectedly, whereas investigations of MSMD led us to defects of Th1 cells, our studies of CMC led to the discovery of mutations in another arm of the effector CD4+ T-cell response, Th17 cells. Rare MSMD patients with IL-12 receptor β1 (IL-12Rβ1) deficiency who also displayed CMC bridged the two conditions (70). Patients with CMC display persistent or recurrent mucocutaneous infections with the commensal fungus Candida albicans. In multiplex families, the condition segregates as a recessive or dominant trait. Impaired IL-17A/F immunity in patients with autosomal dominant hyper-IgE syndrome or autosomal recessive autoimmune polyendocrinopathy syndrome 1, who display various phenotypes, accounts for CMC being common to them (71). Following on from this discovery, from 2011 onward (72–74), we identified loss-of-function mutations of the IL17F, IL-17 receptor A (IL17RA), IL17RC, and actin-related gene 1 (ACT1) genes in patients with isolated CMC. Interestingly, patients with mutations of IL17RA and ACT1, who do not respond to IL-17E, are also susceptible to staphylococcal infections. More surprisingly, in about half the patients we also found gain-of-function mutations of STAT1 impairing the development of IL-17A/F–expressing T cells (75). These patients also presented various other infectious and autoimmune features. Surprisingly, we identified biallelic mutations of RAR-related orphan receptor C (RORC) in patients with both CMC and mycobacterial disease (76). RORC encodes the isoforms and transcription factors ROR-γ and ROR-γT, which govern Th17 development in mice. Therefore the occurrence of CMC in these patients was predictable, and indeed they have no circulating IL-17A/F T cells. However, their most severe phenotype is mycobacterial disease. In-depth studies revealed that RORC also controlled the production of IFN-γ by γδ T cells and by a newly described subset of memory CD4+ T cells, Th1* cells, which are particularly abundant among Mycobacterium-reactive T cells (77). The genetic dissection of CMC led to the discovery that human IL-17A and F are essential for mucocutaneous immunity against C. albicans but otherwise are mostly redundant. These findings were at odds with the results obtained for the corresponding inbred mice, which were susceptible to a broad range of experimental infections. Clinically, studies of MSMD and CMC provided insight into the fundamental mechanism of mycobacteriosis and candidiasis in other settings, such as AIDS and immunosuppression: impaired IFN-γ and IL-17A/F immunity. These studies thus paved the way for immunotherapy with IFN-γ for patients with mycobacteriosis and with IL-17A/F or related cytokines for patients with candidiasis.
Invasive Dermatophytosis
Dermatophytosis is a banal superficial infection, commonly known as athlete’s foot. In very rare cases, the dermatophytes can penetrate the skin, invade the dermis and draining lymph nodes, and disseminate within the body, in a condition known as invasive (or deep) dermatophytosis or dermatophytic disease (Table 4). Patients with invasive dermatophytic disease can live for decades, well into their 70s, with no other notable infections. From 1957 onward, this condition was shown to segregate as an autosomal recessive trait, often in kindreds from North Africa (78, 79). We identified the genetic lesion underlying this striking example of Mendelian infection. All patients tested carried biallelic mutations of the gene encoding caspase recruitment domain family member 9 (CARD9) (80). Deficiency of this intracellular molecule had previously been identified in a large kindred with invasive and peripheral candidiasis (81). These studies led to the determination of the genetic basis of other invasive fungal infections. Thus it has become apparent that CARD9 deficiency manifests as invasive fungal disease in children, adolescents, and adults. Many different fungi can be involved, including dermatophytes, Candida, Phialophora, and Exophialia (82, 83). The central nervous system often is affected. However, each patient (and typically each kindred) suffers from a single type of invasive fungal disease. This condition is another example of complete penetrance with variable expressivity. The molecular and cellular mechanisms underlying the specificity of the phenotype in the patient are unknown. The broader question of the molecular and cellular basis of predisposition to invasive fungal disease in CARD9-deficient patients also remains unresolved. CARD9 may be a critical component acting downstream of the surface receptors for these fungi on myeloid cells. These studies showed that an isolated, invasive fungal disease in a previously healthy child, adolescent, or even an adult can have strict Mendelian determinism in humans.
Toward Non-Mendelian Monogenic Infections: Tuberculosis
We now turn our attention to the discovery of non-Mendelian monogenic infections (Table 6). Surprisingly, our studies of MSMD revealed that not all genetic etiologies were fully penetrant for the case-definition phenotype of MSMD. IL-12Rβ1 deficiency is a case in point (84, 85). Only about half the genetically affected siblings of MSMD index cases also had MSMD. In addition to incomplete penetrance, MSMD patients with IL-12Rβ1 deficiency had variable expressivity: They displayed bacillus Calmette–Guérin disease (in only a fraction of vaccinated patients) or environmental mycobacteriosis (typically in individuals not vaccinated with bacillus Calmette–Guérin), but not both, implying that live bacillus Calmette–Guérin vaccination protected against mycobacteriosis. These observations raise fundamental questions about the mechanisms underlying incomplete penetrance and variable expressivity that have yet to be answered. In this context, we serendipitously discovered the first cases (to our knowledge) of monogenic tuberculosis, in patients and even in families with IL-12Rβ1 deficiency manifesting solely as severe tuberculosis (86–91). These patients were normally resistant to live bacillus Calmette–Guérin vaccine and environmental mycobacteria. However, IL-12Rβ1 deficiency rendered them vulnerable to the more virulent Mycobacterium tuberculosis. Perhaps not coincidentally, these reports were the ones we had the most difficulty publishing: It turned out to be much more difficult to report genetic etiologies of tuberculosis than of MSMD. Perhaps the community was not ready to accept the idea that severe tuberculosis could be monogenic, or monogenic tuberculosis might have been seen purely as an exception of little interest. I still find our identification of truly monogenic cases of tuberculosis one of the most encouraging pieces of evidence we have obtained to date in support of a human genetic theory of primary infectious diseases. We hypothesize that severe forms of tuberculosis are governed by high levels of genetic heterogeneity that are likely to be deciphered only with specific study designs (15, 86, 92). The study of tuberculosis led us to consider in greater depth the possibility that severe primary infections may be determined by non-Mendelian monogenic inborn errors of immunity, single-gene lesions with incomplete penetrance. Indeed, the incomplete penetrance of single-gene lesions conferring susceptibility to infection would account for the great majority of infectious diseases not appearing to be Mendelian traits.
Table 6.
Monogenic infectious diseases
| Infection | Gene | Year |
| Tuberculosis | IL12RB1 | 2001 |
| Invasive pneumococcal disease | IRAK4, etc. | 2003 |
| Herpes simplex encephalitis | UNC93B1, etc. | 2006 |
| Trypanosomiasis (T. evansi) | APOL1 | 2006 |
| Invasive fungi (e.g., Candida) | CARD9 | 2009 |
| Kaposi sarcoma | OX40 | 2013 |
| Severe influenza | IRF7 | 2015 |
The typically sporadic nature of these infectious diseases prevented them from being clinically considered as Mendelian, as monogenic traits, or even as genetic disorders in the first place. However, they have been shown, in a growing number of patients, to be caused by monogenic lesions of various degrees of penetrance (from low to high penetrance) and expressivity (manifesting as one or another infectious disease). The relative risk conferred by these lesions is sufficiently high for them to be considered monogenic. For single-patient studies (APOL1, OX40, IRF7), the observed penetrance was complete, and the identification of new patients will determine whether these disorders are Mendelian. I do not list as causal genes those genes that first might be identified as defective for any of these infections but that sooner or later would be associated with other infections [e.g., STIM1 deficiency in Kaposi sarcoma (106)]. These monogenic infections typically strike otherwise healthy patients. They are listed according to the date of identification of the first gene mutations.
Invasive Pneumococcal Disease
We assessed the broader significance of these encouraging results by studying children with sporadic infections or with infections that had not been described as displaying Mendelian segregation or that had been reported to display such segregation only anecdotally (Table 6). These children therefore were not thought to carry single-gene inborn errors of immunity. We studied children with invasive pneumococcal disease (IPD), hoping to identify the equivalent of the complement disorders that selectively underlie invasive meningococcal disease. Pneumococcus is a well-known commensal of the nasopharynx, present in almost all children. It commonly causes otitis media and, more rarely, lobar pneumonia. It is also known to cause IPD in patients with various inherited or acquired immunodeficiencies, particularly those with impaired complement- and Ab-mediated opsonization and destruction by splenic macrophages (93). We began our search by focusing on a rare syndromic primary immunodeficiency known as anhidrotic ectodermal dysplasia with immunodeficiency (EDA-ID) (94). Patients with this condition have dysmorphic features and suffer from various infections, including IPD and, less commonly, mycobacterial disease. EDA-ID and congenital asplenia are the known primary immunodeficiencies conferring the highest risk of IPD, much greater, for example, than that conferred by agammaglobulinemia or complement defects. We and others discovered that boys with X-linked recessive EDA-ID carried hypomorphic mutations of NF-κB essential modulator (NEMO) that impaired NF-κB responses (95, 96). Heterozygous gain-of-function mutations of the gene encoding the inhibitor IκBα were soon discovered in patients with an autosomal dominant form of EDA-ID (97). By spotlighting NF-κB immunity, these studies paved the way for investigations of children with IPD but without EDA. We found mutations of the genes encoding IL-1R–associated kinase-4 (IRAK-4) and myeloid differentiation primary response gene 88 (MyD88), controlling the canonical Toll-like receptor (TLR) and IL-1R signaling pathways, in children with IPD displaying little clinical and biological inflammation (98, 99). These children are otherwise healthy, although they also are vulnerable to invasive staphylococcal disease (100–103). However, it remains unclear which TLR and IL-1R pathways are defective and specifically underlie IPD or staphylococcal disease. Another interesting aspect of these conditions is that they improve with age. The penetrance of IRAK4 and MyD88 deficiencies for IPD appears to be high by the age of 10 y (almost Mendelian), although some children suffer only from staphylococcal disease, but IPD recurrence subsequently decreases markedly from adolescence onward (typically non-Mendelian). As a result, the mortality of these two genetic disorders decreases with age, contrary to observations for almost all previously described inborn errors of immunity. It is plausible that acquired immunity to pneumococcus gradually compensates for the inborn defects. Studies of cohorts of children with IPD are required to identify new genetic etiologies and to estimate the proportion of children with monogenic inborn errors, which may be Mendelian in rare cases or, more commonly, non-Mendelian (104).
Miscellaneous Endeavors
Rare children with IPD are born without a spleen, a condition known as isolated congenital asplenia (here referred to simply as “asplenia”). The risk of IPD seems to be as high in children with asplenia as it is in children with EDA-ID (105). We found that asplenia was caused by haploinsufficiency at the RPSA locus, which encodes ribosomal protein SA (106). This finding was surprising, because mutations of genes encoding many other ribosomal proteins underlie Blackfan–Diamond anemia, a multiorgan developmental disorder that does not affect the spleen. These observations suggested a role for the translational regulation of gene expression by ribosomes. Inspired by the study of EV, we also studied childhood Kaposi sarcoma, which is driven by human herpes virus 8. This endothelial cell cancer is rare in children without HIV infection or immunosuppression. The classic form, in patients from the Mediterranean basin, is even rarer than the endemic form in sub-Saharan Africa. We discovered the first (to our knowledge) primary immunodeficiencies underlying Kaposi sarcoma in children with other infectious and tumoral phenotypes. We then determined the genetic basis of rare cases of isolated, classic KS in childhood: mutations of the stromal interaction molecule 1 (STIM1) and OX40 genes (107, 108). These studies, which we are currently pursuing, served as a platform for a much more difficult and innovative study, that of herpes simplex virus encephalitis (HSE). Indeed, the infections described above—tuberculosis, IPD, and Kaposi sarcoma—were all known to be potentially opportunistic, because they were seen more frequently in patients with known inherited or acquired immunodeficiency (e.g., AIDS). Etienne Pays and colleagues made a similar observation in their elegant studies of trypanosomiasis. They found that a patient suffered from clinical disease caused by poorly virulent Trypanosoma evansi because of autosomal recessive apolipoprotein L-I (APOL1) deficiency (109). APOL1 is also necessary, but not sufficient, for host defense against more virulent species of trypanosome (Trypanosoma brucei) (110). We therefore needed to study an infection not known to be Mendelian, or even genetic in general (i.e., purely sporadic and not familial), and not considered to be opportunistic.
HSE: A Model Disease
We selected childhood HSE for study because there was no evidence to suggest that it had a genetic basis, only an intuition, and—the strongest clue—the lack of an alternative plausible hypothesis (Table 6). HSE is the most common sporadic viral encephalitis in the Western world. This life-threatening disease strikes otherwise healthy children once (recurrence is rare) in the course of primary infection with the herpes simplex virus, which is ubiquitous and typically innocuous (111). The arguments against a genetic hypothesis and against a monogenic hypothesis in particular included the sporadic nature of the disease, with only four multiplex families having been reported since its first description in 1941 (112). Moreover, none of the known primary immunodeficiencies, including severe combined immunodeficiency, and none of the known acquired immunodeficiencies of childhood, including AIDS, had been identified as risk factors. Finally, this infection is strictly limited to the central nervous system, at odds with the disseminated infections commonly seen in the context of immunodeficiency. We felt that there could be no better infection on which to test our non-Mendelian monogenic model. One observation guided the entire project in the right direction: We found that 12% of French children with HSE were born to consanguineous parents (112). This finding was surprising, because all cases in this 20-y nationwide epidemiological study were considered to be sporadic. Indeed, there was not a single multiplex family. This simple clinical survey therefore suggested that HSE probably is caused by monogenic inborn errors, including autosomal recessive traits, albeit generally with incomplete clinical penetrance, resulting in a non-Mendelian pattern of inheritance.
HSE: A Genetic Disease
We thus searched for HSE-causing genes. Two children with a unique phenotype of HSE and mycobacterial disease were referred to us serendipitously. These two children remain the only children with this phenotype that I have come across. These children played a key role in subsequent studies, because their mycobacterial susceptibility phenotype enabled us to identify their genetic lesions, based on our previous studies of MSMD. They were found to have particular mutations of the genes encoding STAT1 (homozygous and loss of function) and NEMO (hypomorphic by reinitiation of translation) (113, 114) not previously seen in children who had MSMD but did not have HSE. STAT1 mutations underlying MSMD are heterozygous and affect IFN-γ but not IFN-α/β responses (23), whereas hemizygous NEMO mutations underlying MSMD selectively affect the production of IL-12 and IFN-γ (115). This finding led us to hypothesize that HSE in otherwise healthy children might be caused by inborn errors of antiviral IFN-α/β immunity. We analyzed genome-wide linkage data and both blood and fibroblastic responses to herpes simplex virus and various viral intermediates in children with isolated HSE from this IFN-α/β–based angle, and we discovered mutations affecting five genes governing the TLR3-mediated IFN-α/β pathway (116–121). Another group recently identified a mutation in a sixth gene (122). TLR3 can recognize viral dsRNA intermediates. With the aid of induced pluripotent stem cell (iPSC) technology, we and our colleagues Lorenz Studer and Luigi Notarangelo demonstrated that the mutations in these patients impaired intrinsic antiviral immunity in neurons and oligodendrocytes, accounting for the brain-tropic nature of HSE (123). The central nervous system-restricted nature of HSE is explained by the TLR3-independence of most other cell types, including leukocytes and keratinocytes, tested for responses to dsRNAs (117, 121). The lack of herpes labialis in HSE patients may reflect the presence of higher levels of herpes simplex virus-specific T cells around the trigeminal ganglia and in the blood, a hypothesis that we are currently testing. Of course, these findings have opened up further questions: What is the clinical penetrance of these genetic lesions? What governs incomplete penetrance? What proportion of cases is caused by variants of the TLR3–IFN–α/β pathway? Are there other types of HSE-causing genetic lesions? Overall, these studies provided proof of principle that an isolated, common, organ-specific, life-threatening infection of childhood can result from single-gene inborn errors of immunity not displaying full penetrance.
Influenza as Another Genetic Disease
Although HSE is not very rare, striking about 1 child in 10,000, it is largely unknown outside the patients’ families and the medical community. Could severe infections of childhood that are not necessarily more common but are better known to the lay public also be caused by single-gene inborn errors of immunity? Could other organ-specific infections be genetically determined? We decided to tackle some emblematic fulminant viral infections, such as viral hepatitis, viral myocarditis, and pulmonary influenza. Influenza provides a particularly interesting historical example (Table 6). Through 80 y of superb research on influenza viruses, we have learned much about the pathogenicity of these viruses in cells in vitro, in animal models in vivo, and in humans in natura, as well as the epidemiological course of seasonal and pandemic infections (124–126). However, we still know little about the determinism of life-threatening influenza, which occurs in only a minority of infected individuals in the course of epidemic or pandemic influenza (127). The best-known risk factors are preexisting pulmonary conditions. In this context, we recently discovered the first (to our knowledge) genetic etiology of acute respiratory distress syndrome in a previously healthy child infected for the first time with the influenza virus. This patient, one of only four children with life-threatening influenza during primary infection (i.e., without detectable serum antibodies against any influenza virus) enrolled in our whole-exome sequencing program, carried two loss-of-function mutations of the gene encoding IFN regulatory factor 7 (IRF7), a transcription factor required for the amplification of antiviral IFN-α/β (128). We showed that IRF7 was indeed the gene responsible for influenza in this child, because the patient’s leukocytes failed to produce IFNs other than IFN-β in response to influenza virus. The only cells that normally express IRF7 constitutively, plasmacytoid dendritic cells, displayed a complete functional deficiency of this protein. Moreover, the patient’s fibroblasts and iPSC-derived pulmonary epithelial cells also produced only small amounts of the various types of IFN, resulting in enhanced viral replication. This study raises multiple questions, including the proportion of genetic cases among children with severe influenza and the nature of the cells responsible for influenza in patients with IRF7 deficiency (126). We currently are testing the hypothesis that other children with severe influenza carry mutations impairing antiviral immunity. The identification of new mutations should help to address these questions.
Severe Infections as Inborn Errors: Clinical Implications
The discovery of single-gene inborn errors of immunity in children with tuberculosis, IPD, HSE, or pulmonary influenza is important from a clinical perspective. These findings provide proof of principle for a monogenic (but not Mendelian) theory of life-threatening primary infectious diseases, including infections not previously thought to be opportunistic (Fig. 1). These rare alleles act in a monogenic manner that is rarely Mendelian (i.e., monogenic with complete penetrance). Indeed, incomplete penetrance seems to be the general rule, with the exception of the Mendelian infections discussed above (Tables 2–5). If more, most, or all human infections were Mendelian, their genetic basis would have been suspected a century ago (phenotype) and discovered 20 y ago (genotype). Incomplete penetrance goes hand-in-hand with genetic heterogeneity, because it allows hypomorphs and hypermorphs of genes for which null alleles would be lethal even in the absence of infection to govern infectious diseases in at least some patients. There actually seems to be considerable genetic heterogeneity, which is not surprising because there are many cell layers between the invading microbe and a fatal outcome in the host, and, of course, there are many molecular pathways, each depending on many genes. The genetic dissection of infectious diseases therefore will progress slowly, almost patient by patient (129). However, there also seems to be some physiological homogeneity because, for each infection, the products of the disease-causing genes appear to be related. The implications, in terms of diagnosis, genetic counseling, and prognosis, merit more detailed consideration. These studies also explain the pathogenesis of infections in other settings (as discussed for IFN-γ and IL-17A/F) and pave the way for the prevention or treatment of acute infections with cytokines, such as IFN-α/β or IFN-γ, in addition to small anti-infectious compounds. Moreover, as we develop treatments for genetic diseases, they should also have an impact on infectious diseases that have a genetic basis, just as CRISPR can prevent viral infections in bacteria. That said, we currently have a genetic and immunological understanding of only a minute proportion of severe childhood infections in a minute proportion of sick children. Admittedly, the field of human genetics of infectious diseases is still in its infancy. Beyond pediatric infections, these findings also pose questions relevant to internal medicine, as we have seen for fungal infections. Primary infections can occur in adults, or their incubation period can be particularly long.
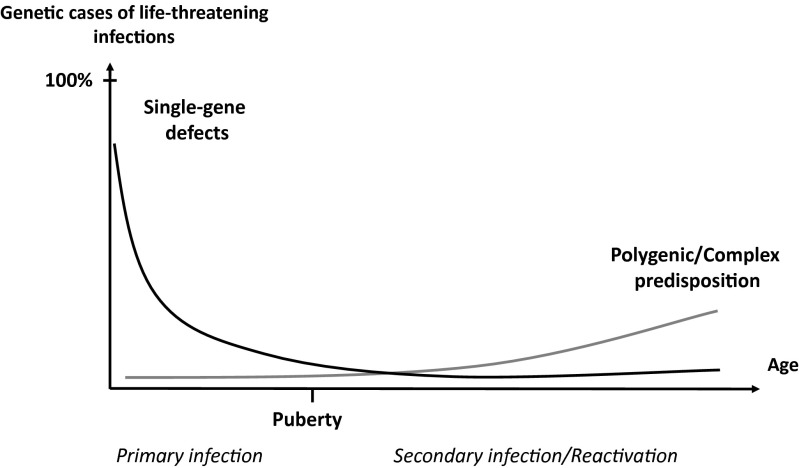
Fig. 1.
The genetic component of human infectious diseases, according to age. Schematic representation of a hypothetical, age-dependent, human genetic architecture of infectious diseases. We suggest that single-gene human variants make an important contribution to the determinism of life-threatening infectious diseases in the course of primary infection, which occurs most often in childhood. In this model, single-gene inborn errors of immunity are more often monogenic than Mendelian, with incomplete penetrance and variable expressivity accounting for infectious diseases being more often sporadic than familial. By contrast, predisposition to severe infectious diseases in the course of microbial reactivation from latency or secondary infection, typically in adults, is less influenced by germline human genetic variations, resulting in a more complex and less monogenic component. Somatic and epigenetic processes are likely to play a greater role in older individuals. Reproduced from ref. 143.
Immunological Implications
We also should consider the immunological implications of these genetic studies, which can be seen as a fortunate by-product. These studies have demonstrated that intrinsic immunity in nonhematopoietic cells can be life-saving, as exemplified for HSE and perhaps for pulmonary influenza or EV. Other studies, dealing with molecules as diverse as IFN-γ, IL-17, TLRs, and IL-1R, also have turned up a number of immunological surprises, in particular revealing a much greater degree of redundancy in outbred humans in natural conditions of infection than in inbred mice in experimental conditions of infection (130–132). The specificity of the infectious phenotypes seen with each inborn error actually results from its redundancy in protective immunity against other infectious agents. There are profound differences between immunity in natural conditions and immunity in experimental conditions, in terms of the pathogen, the route and dose of infection, the host, and the manifestations of infection (130–132). Influenza in a child living in a small French town (128) has little in common with the injection of influenza virus (which is not rodent-tropic) into an inbred B6 mouse lacking functional myxovirus resistance 1 (Mx1), the key gene product conferring resistance to orthomyxoviruses in rodents (126). The real surprise is the robustness of the infectious phenotypes in mice. The human phenotypes are mostly replicated in mice, but the converse is not true, because of the much greater redundancy of the mechanisms at work in natura. For this reason we have never claimed any general infectious specificity for a monogenic lesion or pathway. Specificity is unlikely to be absolute across all the individuals of a given species, in light of population thinking and chemical individuality. It is enough for specificity to be observed in a given patient, and perhaps then in another, and another. It can be predicted from genetic heterogeneity and incomplete penetrance that several infections may be specific in individual patients but allelic at the population level. This individual specificity is neatly illustrated by the impact of inherited IL-12Rβ1 and CARD9 deficiencies, which display variable expressivity, each patient typically suffering from only one type of mycobacterial or fungal disease.
A Genetic Theory of Infectious Diseases: Challenges Ahead
It remains perplexing that, in plants, the monogenic inheritance of specific infectious diseases was documented as early as 1905, proposed as a general model in 1942, and documented as a paradigm at the molecular level from 1993 onward, but this notion has failed to gain a firm foothold in human medicine more than a century later (Table 2). This lack of acceptance attests to the strength of the paradigm positing a fundamental dichotomy between disease transmission by heredity and by infection (133). Nevertheless, the human genetic theory of infectious diseases has moved forward recently with the discovery that a number of severe infections of childhood result from inborn errors of immunity. Only a tiny proportion of cases and infections are currently understood genetically. Gargantuan efforts, on a par with those used in microbiology in the late 19th century, are required to test this model rigorously and at a larger scale. In all probability, not all primary infectious diseases will be found to be genetic traits. A much smaller proportion of infectious diseases are likely to be found to be genetic in adults, particularly elderly adults, in the course of secondary infection or reactivation. Indeed, adaptive immunity blurs and buffers the impact of the germline, which controls both cell-intrinsic (autonomous) and cell-extrinsic (innate) immunity. This evolutionary raison d’être of adaptive immunity explains its dependence on so few genes, germline mutations of which underlie B- and/or T-cell deficiencies with a broad and lethal infectious phenotype. Consistently, a recent twin study showed that adaptive immunity parameters diverge much more rapidly with age than do other hematological parameters in identical twins (134, 135). The somatic (as opposed to germline) failure of adaptive responses, whether genetic or epigenetic, may account for a proportion of primary infections. Somatic variation may make an even greater contribution to the genetic architecture of infections during secondary infection or reactivation, particularly in the elderly population.
Concluding Remarks: A Testable Model
We can propose the following tentative and testable model for the human genetic architecture of infectious diseases in the course of primary infection (Fig. 1). Emerging or reemerging infectious agents that can kill a large proportion of infected individuals gradually select for human resistance alleles, which spread and become common alleles through natural selection. They can act in a monogenic or polygenic model, as illustrated by the spread of mutant resistance alleles in areas in which P. vivax and Plasmodium falciparum malaria, respectively, are endemic. When infections threaten a smaller proportion of infected individuals (e.g., HSE or invasive fungal disease), genetically determined death is more likely to be the result of rare mutant alleles that may be of variable expressivity, with a penetrance that is not necessarily complete. The nature of the rare genetic lesions may depend on the more common variants modifying the proportion of cases among infected individuals. This non-Mendelian monogenic determinism of primary infectious diseases, polygenic in its own way, is plausible and testable. The proportion of cases following this form of genetic determinism is likely to decrease with age, both in diseases in which genetics plays only a partial role and in diseases that are purely genetic. For secondary infections and infections in aging individuals, the proportion of genetic cases is likely to be smaller and the proportion of monogenic cases even smaller. This model also may turn out to apply to various noninfectious immunological phenotypes (129, 136–142). It is tempting to speculate that the death of children, adolescents, and young adults may be monogenic as a rule, or at least caused by a single, rare genetic event, even for cancer and infection, the standard examples of somatic and environmental diseases, respectively. This model of rare monogenic lesions underlying heritable phenotypes with incomplete penetrance or variable expressivity may be valid in diverse related areas, such as infection, allergy, autoimmunity, autoinflammation, and certain cancers.
Acknowledgments
I am indebted to Gérard Orth, whose review of previous versions of this manuscript has been invaluable. I thank Laurent Abel, who leads the computational branch of The Laboratory of Human Genetics of Infectious Diseases; the researchers in the laboratory, including, in particular, Alexandre Alcais, Emmanuelle Jouanguy, Anne Puel, Capucine Picard, Jacinta Bustamante, Stéphanie Boisson-Dupuis, Bertrand Boisson, Guillaume Vogt, Michael Ciancanelli, Shen-Ying Zhang, and Aurélie Cobat; Yelena Nemirovskaya, without whom the laboratory in New York would never have taken off and grown; the other past and present members of the laboratory at the Necker Hospital for Sick Children and The Rockefeller University; the clinicians and scientists around the world with whom we have been privileged to have formed links. Finally, I am humbled to thank the sick children and their parents, whose trust we did our best to deserve yet whose hope we too rarely satisfied. They are all too aware that pediatrics is mankind's greatest endeavor.
Footnotes
The authors declare no conflict of interest.
References
- 1.Miller LH, Mason SJ, Clyde DF, McGinniss MH. The resistance factor to Plasmodium vivax in blacks. The Duffy-blood-group genotype, FyFy. N Engl J Med. 1976;295(6):302–304. doi: 10.1056/NEJM197608052950602. [DOI] [PubMed] [Google Scholar]
- 2.Miller LH, Mason SJ, Dvorak JA, McGinniss MH, Rothman IK. Erythrocyte receptors for (Plasmodium knowlesi) malaria: Duffy blood group determinants. Science. 1975;189(4202):561–563. doi: 10.1126/science.1145213. [DOI] [PubMed] [Google Scholar]
- 3.Casanova J-L. Human genetic basis of interindividual variability in the course of infection. Proc Natl Acad Sci USA. 2015;112:E7118–E7127. doi: 10.1073/pnas.1521644112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tournamille C, Colin Y, Cartron JP, Le Van Kim C. Disruption of a GATA motif in the Duffy gene promoter abolishes erythroid gene expression in Duffy-negative individuals. Nat Genet. 1995;10(2):224–228. doi: 10.1038/ng0695-224. [DOI] [PubMed] [Google Scholar]
- 5.Samson M, et al. Resistance to HIV-1 infection in caucasian individuals bearing mutant alleles of the CCR-5 chemokine receptor gene. Nature. 1996;382(6593):722–725. doi: 10.1038/382722a0. [DOI] [PubMed] [Google Scholar]
- 6.Liu R, et al. Homozygous defect in HIV-1 coreceptor accounts for resistance of some multiply-exposed individuals to HIV-1 infection. Cell. 1996;86(3):367–377. doi: 10.1016/s0092-8674(00)80110-5. [DOI] [PubMed] [Google Scholar]
- 7.Dean M, et al. Genetic restriction of HIV-1 infection and progression to AIDS by a deletion allele of the CKR5 structural gene. Hemophilia Growth and Development Study, Multicenter AIDS Cohort Study, Multicenter Hemophilia Cohort Study, San Francisco City Cohort, ALIVE Study. Science. 1996;273(5283):1856–1862. doi: 10.1126/science.273.5283.1856. [DOI] [PubMed] [Google Scholar]
- 8.Marionneau S, et al. Norwalk virus binds to histo-blood group antigens present on gastroduodenal epithelial cells of secretor individuals. Gastroenterology. 2002;122(7):1967–1977. doi: 10.1053/gast.2002.33661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lindesmith L, et al. Human susceptibility and resistance to Norwalk virus infection. Nat Med. 2003;9(5):548–553. doi: 10.1038/nm860. [DOI] [PubMed] [Google Scholar]
- 10.Cooper MD, et al. Classification of primary immunodeficiencies. N Engl J Med. 1973;288(18):966–967. doi: 10.1056/NEJM197305032881814. [DOI] [PubMed] [Google Scholar]
- 11.Fudenberg HH, et al. Classification of the primary immune deficiencies: WHO recommendation. N Engl J Med. 1970;283(12):656–657. doi: 10.1056/NEJM197009172831211. [DOI] [PubMed] [Google Scholar]
- 12.Casanova JL, et al. Idiopathic disseminated bacillus Calmette-Guérin infection: A French national retrospective study. Pediatrics. 1996;98(4 Pt 1):774–778. [PubMed] [Google Scholar]
- 13.Casanova JL, Jouanguy E, Lamhamedi S, Blanche S, Fischer A. Immunological conditions of children with BCG disseminated infection. Lancet. 1995;346(8974):581. doi: 10.1016/s0140-6736(95)91421-8. [DOI] [PubMed] [Google Scholar]
- 14.Mimouni J. Notre expérience de trois années de vaccination à Constantine; étude de 25 cas de complications. Alger Med. 1951;55(8):1138–1147. [PubMed] [Google Scholar]
- 15.Casanova JL, Abel L. Genetic dissection of immunity to mycobacteria: The human model. Annu Rev Immunol. 2002;20:581–620. doi: 10.1146/annurev.immunol.20.081501.125851. [DOI] [PubMed] [Google Scholar]
- 16.Heyne K. Generalisatio BCG familiaris semibenigna, Osteomyelitis salmonellosa und Pseudotuberculosis intestinalis--Folgen eines familiären Makrophagendefektes. Eur J Pediatr. 1976;121(3):179–189. doi: 10.1007/BF00445481. [DOI] [PubMed] [Google Scholar]
- 17.Newport MJ, et al. A mutation in the interferon-gamma-receptor gene and susceptibility to mycobacterial infection. N Engl J Med. 1996;335(26):1941–1949. doi: 10.1056/NEJM199612263352602. [DOI] [PubMed] [Google Scholar]
- 18.Jouanguy E, et al. Interferon-gamma-receptor deficiency in an infant with fatal bacille Calmette-Guérin infection. N Engl J Med. 1996;335(26):1956–1961. doi: 10.1056/NEJM199612263352604. [DOI] [PubMed] [Google Scholar]
- 19.Bustamante J, Boisson-Dupuis S, Abel L, Casanova JL. Mendelian susceptibility to mycobacterial disease: Genetic, immunological, and clinical features of inborn errors of IFN-γ immunity. Semin Immunol. 2014;26(6):454–470. doi: 10.1016/j.smim.2014.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Altare F, et al. Impairment of mycobacterial immunity in human interleukin-12 receptor deficiency. Science. 1998;280(5368):1432–1435. doi: 10.1126/science.280.5368.1432. [DOI] [PubMed] [Google Scholar]
- 21.Bogunovic D, et al. Mycobacterial disease and impaired IFN-γ immunity in humans with inherited ISG15 deficiency. Science. 2012;337(6102):1684–1688. doi: 10.1126/science.1224026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.de Jong R, et al. Severe mycobacterial and Salmonella infections in interleukin-12 receptor-deficient patients. Science. 1998;280(5368):1435–1438. doi: 10.1126/science.280.5368.1435. [DOI] [PubMed] [Google Scholar]
- 23.Dupuis S, et al. Impairment of mycobacterial but not viral immunity by a germline human STAT1 mutation. Science. 2001;293(5528):300–303. doi: 10.1126/science.1061154. [DOI] [PubMed] [Google Scholar]
- 24.Dupuis S, et al. Human interferon-gamma-mediated immunity is a genetically controlled continuous trait that determines the outcome of mycobacterial invasion. Immunol Rev. 2000;178:129–137. doi: 10.1034/j.1600-065x.2000.17810.x. [DOI] [PubMed] [Google Scholar]
- 25.Nathan CF, Murray HW, Wiebe ME, Rubin BY. Identification of interferon-gamma as the lymphokine that activates human macrophage oxidative metabolism and antimicrobial activity. J Exp Med. 1983;158(3):670–689. doi: 10.1084/jem.158.3.670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang X, et al. Inherited human complement C5 deficiency. Nonsense mutations in exons 1 (Gln1 to Stop) and 36 (Arg1458 to Stop) and compound heterozygosity in three African-American families. J Immunol. 1995;154(10):5464–5471. [PubMed] [Google Scholar]
- 27.Würzner R, et al. Molecular basis of subtotal complement C6 deficiency. A carboxy-terminally truncated but functionally active C6. J Clin Invest. 1995;95(4):1877–1883. doi: 10.1172/JCI117868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nishizaka H, Horiuchi T, Zhu ZB, Fukumori Y, Volanakis JE. Genetic bases of human complement C7 deficiency. J Immunol. 1996;157(9):4239–4243. [PubMed] [Google Scholar]
- 29.Kaufmann T, et al. Genetic basis of human complement C8 beta deficiency. J Immunol. 1993;150(11):4943–4947. [PubMed] [Google Scholar]
- 30.Kojima T, et al. Genetic basis of human complement C8 alpha-gamma deficiency. J Immunol. 1998;161(7):3762–3766. [PubMed] [Google Scholar]
- 31.Westberg J, Fredrikson GN, Truedsson L, Sjöholm AG, Uhlén M. Sequence-based analysis of properdin deficiency: Identification of point mutations in two phenotypic forms of an X-linked immunodeficiency. Genomics. 1995;29(1):1–8. doi: 10.1006/geno.1995.1208. [DOI] [PubMed] [Google Scholar]
- 32.Biesma DH, et al. A family with complement factor D deficiency. J Clin Invest. 2001;108(2):233–240. doi: 10.1172/JCI12023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lim D, et al. Absence of the sixth component of complement in a patient with repeated episodes of meningococcal meningitis. J Pediatr. 1976;89(1):42–47. doi: 10.1016/s0022-3476(76)80924-9. [DOI] [PubMed] [Google Scholar]
- 34.Petersen BH, Graham JA, Brooks GF. Human deficiency of the eighth component of complement. The requirement of C8 for serum Neisseria gonorrhoeae bactericidal activity. J Clin Invest. 1976;57(2):283–290. doi: 10.1172/JCI108279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hiemstra PS, et al. Complete and partial deficiencies of complement factor D in a Dutch family. J Clin Invest. 1989;84(6):1957–1961. doi: 10.1172/JCI114384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rosenfeld SI, Kelly ME, Leddy JP. Hereditary deficiency of the fifth component of complement in man. I. Clinical, immunochemical, and family studies. J Clin Invest. 1976;57(6):1626–1634. doi: 10.1172/JCI108433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Harriman GR, et al. The role of C9 in complement-mediated killing of Neisseria. J Immunol. 1981;127(6):2386–2390. [PubMed] [Google Scholar]
- 38.Snyderman R, Durack DT, McCarty GA, Ward FE, Meadows L. Deficiency of the fifth component of complement in human subjects. Clinical, genetic and immunologic studies in a large kindred. Am J Med. 1979;67(4):638–645. doi: 10.1016/0002-9343(79)90247-x. [DOI] [PubMed] [Google Scholar]
- 39.Nagata M, et al. Inherited deficiency of ninth component of complement: An increased risk of meningococcal meningitis. J Pediatr. 1989;114(2):260–264. doi: 10.1016/s0022-3476(89)80793-0. [DOI] [PubMed] [Google Scholar]
- 40.Lee TJ, Utsinger PD, Snyderman R, Yount WJ, Sparling PF. Familial deficiency of the seventh component of complement associated with recurrent bacteremic infections due to Neisseria. J Infect Dis. 1978;138(3):359–368. doi: 10.1093/infdis/138.3.359. [DOI] [PubMed] [Google Scholar]
- 41.Sjöholm AG, Braconier JH, Söderström C. Properdin deficiency in a family with fulminant meningococcal infections. Clin Exp Immunol. 1982;50(2):291–297. [PMC free article] [PubMed] [Google Scholar]
- 42.Owen EP, et al. A complement C5 gene mutation, c.754G>A:p.A252T, is common in the Western Cape, South Africa and found to be homozygous in seven percent of Black African meningococcal disease cases. Mol Immunol. 2015;64(1):170–176. doi: 10.1016/j.molimm.2014.11.010. [DOI] [PubMed] [Google Scholar]
- 43.Orth G. Host defenses against human papillomaviruses: Lessons from epidermodysplasia verruciformis. Curr Top Microbiol Immunol. 2008;321:59–83. doi: 10.1007/978-3-540-75203-5_3. [DOI] [PubMed] [Google Scholar]
- 44.Orth G, et al. Epidermodysplasia verruciformis: A model for the role of papillomaviruses in human cancer. Cold Spring Harbor Conferences on Cell Proliferation. 1980;7:259–282. [Google Scholar]
- 45.Lewandowsky F, Lutz W. Ein Fall einer bisher nicht beschriebenen Hauterkrankung (Epidermodysplasia verruciformis) Arch Derm Syphilol. 1922;141:193–203. [Google Scholar]
- 46.Cockayne EA. Inherited Abnormalities of the Skin and Its Appendages. Oxford Univ Press; London: 1933. Epidermodysplasia verruciformis; p. 156. [Google Scholar]
- 47.Lutz W. A propos de l’epidermodysplasie verruciforme. Dermatologica. 1946;92(1):30–43. [PubMed] [Google Scholar]
- 48.Ramoz N, et al. Mutations in two adjacent novel genes are associated with epidermodysplasia verruciformis. Nat Genet. 2002;32(4):579–581. doi: 10.1038/ng1044. [DOI] [PubMed] [Google Scholar]
- 49.Chapel H, Geha R, Rosen F. IUIS PID (Primary Immunodeficiencies) Classification committee Primary immunodeficiency diseases: An update. Clin Exp Immunol. 2003;132(1):9–15. doi: 10.1046/j.1365-2249.2003.02110.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Notarangelo L, et al. International Union of Immunological Societies Primary Immunodeficiency diseases classification committee Primary immunodeficiency diseases: An update. J Allergy Clin Immunol. 2004;114(3):677–687. doi: 10.1016/j.jaci.2007.08.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Crequer A, et al. EVER2 deficiency is associated with mild T-cell abnormalities. J Clin Immunol. 2013;33(1):14–21. doi: 10.1007/s10875-012-9749-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lazarczyk M, et al. Regulation of cellular zinc balance as a potential mechanism of EVER-mediated protection against pathogenesis by cutaneous oncogenic human papillomaviruses. J Exp Med. 2008;205(1):35–42. doi: 10.1084/jem.20071311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Purtilo DT. Pathogenesis and phenotypes of an X-linked recessive lymphoproliferative syndrome. Lancet. 1976;2(7991):882–885. doi: 10.1016/s0140-6736(76)90542-0. [DOI] [PubMed] [Google Scholar]
- 54.Purtilo DT, Cassel C, Yang JP. Letter: Fatal infectious mononucleosis in familial lymphohistiocytosis. N Engl J Med. 1974;291(14):736. doi: 10.1056/nejm197410032911415. [DOI] [PubMed] [Google Scholar]
- 55.Purtilo DT, Cassel CK, Yang JP, Harper R. X-linked recessive progressive combined variable immunodeficiency (Duncan’s disease) Lancet. 1975;1(7913):935–940. doi: 10.1016/s0140-6736(75)92004-8. [DOI] [PubMed] [Google Scholar]
- 56.Purtilo DT, et al. Variable phenotypic expression of an X-linked recessive lymphoproliferative syndrome. N Engl J Med. 1977;297(20):1077–1080. doi: 10.1056/NEJM197711172972001. [DOI] [PubMed] [Google Scholar]
- 57.Anonymous Immunodeficiency: Report of a WHO scientific group. World Health Organ Tech Rep Ser. 1978;(630):3–80. [PubMed] [Google Scholar]
- 58.Coffey AJ, et al. Host response to EBV infection in X-linked lymphoproliferative disease results from mutations in an SH2-domain encoding gene. Nat Genet. 1998;20(2):129–135. doi: 10.1038/2424. [DOI] [PubMed] [Google Scholar]
- 59.Sayos J, et al. The X-linked lymphoproliferative-disease gene product SAP regulates signals induced through the co-receptor SLAM. Nature. 1998;395(6701):462–469. doi: 10.1038/26683. [DOI] [PubMed] [Google Scholar]
- 60.Nichols KE, et al. Inactivating mutations in an SH2 domain-encoding gene in X-linked lymphoproliferative syndrome. Proc Natl Acad Sci USA. 1998;95(23):13765–13770. doi: 10.1073/pnas.95.23.13765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Palendira U, et al. Expansion of somatically reverted memory CD8+ T cells in patients with X-linked lymphoproliferative disease caused by selective pressure from Epstein-Barr virus. J Exp Med. 2012;209(5):913–924. doi: 10.1084/jem.20112391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Palendira U, et al. Molecular pathogenesis of EBV susceptibility in XLP as revealed by analysis of female carriers with heterozygous expression of SAP. PLoS Biol. 2011;9(11):e1001187. doi: 10.1371/journal.pbio.1001187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tangye SG. XLP: Clinical features and molecular etiology due to mutations in SH2D1A encoding SAP. J Clin Immunol. 2014;34(7):772–779. doi: 10.1007/s10875-014-0083-7. [DOI] [PubMed] [Google Scholar]
- 64.Rigaud S, et al. XIAP deficiency in humans causes an X-linked lymphoproliferative syndrome. Nature. 2006;444(7115):110–114. doi: 10.1038/nature05257. [DOI] [PubMed] [Google Scholar]
- 65.Aguilar C, Latour S. X-linked inhibitor of apoptosis protein deficiency: More than an X-linked lymphoproliferative syndrome. J Clin Immunol. 2015;35(4):331–338. doi: 10.1007/s10875-015-0141-9. [DOI] [PubMed] [Google Scholar]
- 66.Li FY, et al. XMEN disease: A new primary immunodeficiency affecting Mg2+ regulation of immunity against Epstein-Barr virus. Blood. 2014;123(14):2148–2152. doi: 10.1182/blood-2013-11-538686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Li FY, et al. Second messenger role for Mg2+ revealed by human T-cell immunodeficiency. Nature. 2011;475(7357):471–476. doi: 10.1038/nature10246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chilgren RA, Quie PG, Meuwissen HJ, Hong R. Chronic mucocutaneous candidiasis, deficiency of delayed hypersensitivity, and selective local antibody defect. Lancet. 1967;2(7518):688–693. doi: 10.1016/s0140-6736(67)90974-9. [DOI] [PubMed] [Google Scholar]
- 69.Wells RS, Higgs JM, Macdonald A, Valdimarsson H, Holt PJ. Familial chronic muco-cutaneous candidiasis. J Med Genet. 1972;9(3):302–310. doi: 10.1136/jmg.9.3.302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.de Beaucoudrey L, et al. Mutations in STAT3 and IL12RB1 impair the development of human IL-17-producing T cells. J Exp Med. 2008;205(7):1543–1550. doi: 10.1084/jem.20080321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Puel A, et al. Inborn errors of mucocutaneous immunity to Candida albicans in humans: A role for IL-17 cytokines? Curr Opin Immunol. 2010;22(4):467–474. doi: 10.1016/j.coi.2010.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Puel A, et al. Chronic mucocutaneous candidiasis in humans with inborn errors of interleukin-17 immunity. Science. 2011;332(6025):65–68. doi: 10.1126/science.1200439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ling Y, et al. Inherited IL-17RC deficiency in patients with chronic mucocutaneous candidiasis. J Exp Med. 2015;212(5):619–631. doi: 10.1084/jem.20141065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Boisson B, et al. An ACT1 mutation selectively abolishes interleukin-17 responses in humans with chronic mucocutaneous candidiasis. Immunity. 2013;39(4):676–686. doi: 10.1016/j.immuni.2013.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Liu L, et al. Gain-of-function human STAT1 mutations impair IL-17 immunity and underlie chronic mucocutaneous candidiasis. J Exp Med. 2011;208(8):1635–1648. doi: 10.1084/jem.20110958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Okada S, et al. Impairment of immunity to Candida and Mycobacterium in humans with bi-allelic RORC mutations. Science. 2015;349(6248):606–613. doi: 10.1126/science.aaa4282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Becattini S, et al. T cell immunity. Functional heterogeneity of human memory CD4⁺ T cell clones primed by pathogens or vaccines. Science. 2015;347(6220):400–406. doi: 10.1126/science.1260668. [DOI] [PubMed] [Google Scholar]
- 78.Hadida E, Schousboe A. [Generalized trichophytosis with dermohypodermal & lymphnodal localizations due to Trichophyton of faviform culture (Trichophyton verrucosum)] Minerva Dermatol. 1959;34(3):225–231. [PubMed] [Google Scholar]
- 79.Blank F, Schopflocher P, Poirier P, Riopelle JL. Extensive Trichophyton infections of about fifty years’ duration in two sisters. Dermatologica. 1957;115(1):40–51. doi: 10.1159/000255985. [DOI] [PubMed] [Google Scholar]
- 80.Lanternier F, et al. Deep dermatophytosis and inherited CARD9 deficiency. N Engl J Med. 2013;369(18):1704–1714. doi: 10.1056/NEJMoa1208487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Glocker EO, et al. A homozygous CARD9 mutation in a family with susceptibility to fungal infections. N Engl J Med. 2009;361(18):1727–1735. doi: 10.1056/NEJMoa0810719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lanternier F, et al. Inherited CARD9 deficiency in 2 unrelated patients with invasive Exophiala infection. J Infect Dis. 2015;211(8):1241–1250. doi: 10.1093/infdis/jiu412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Lanternier F, et al. Inherited CARD9 deficiency in otherwise healthy children and adults with Candida species-induced meningoencephalitis, colitis, or both. J Allergy Clin Immunol. 2015;135(6):1558–1568. doi: 10.1016/j.jaci.2014.12.1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Fieschi C, et al. Low penetrance, broad resistance, and favorable outcome of interleukin 12 receptor beta1 deficiency: Medical and immunological implications. J Exp Med. 2003;197(4):527–535. doi: 10.1084/jem.20021769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.de Beaucoudrey L, et al. Revisiting human IL-12Rβ1 deficiency: A survey of 141 patients from 30 countries. Medicine (Baltimore) 2010;89(6):381–402. doi: 10.1097/MD.0b013e3181fdd832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Alcaïs A, Fieschi C, Abel L, Casanova JL. Tuberculosis in children and adults: Two distinct genetic diseases. J Exp Med. 2005;202(12):1617–1621. doi: 10.1084/jem.20052302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Altare F, et al. Interleukin-12 receptor beta1 deficiency in a patient with abdominal tuberculosis. J Infect Dis. 2001;184(2):231–236. doi: 10.1086/321999. [DOI] [PubMed] [Google Scholar]
- 88.Boisson-Dupuis S, et al. IL-12Rβ1 deficiency in two of fifty children with severe tuberculosis from Iran, Morocco, and Turkey. PLoS One. 2011;6(4):e18524. doi: 10.1371/journal.pone.0018524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Caragol I, et al. Clinical tuberculosis in 2 of 3 siblings with interleukin-12 receptor beta1 deficiency. Clin Infect Dis. 2003;37(2):302–306. doi: 10.1086/375587. [DOI] [PubMed] [Google Scholar]
- 90.Özbek N, et al. Interleukin-12 receptor beta 1 chain deficiency in a child with disseminated tuberculosis. Clin Infect Dis. 2005;40(6):e55–e58. doi: 10.1086/427879. [DOI] [PubMed] [Google Scholar]
- 91.Tabarsi P, et al. Lethal tuberculosis in a previously healthy adult with IL-12 receptor deficiency. J Clin Immunol. 2011;31(4):537–539. doi: 10.1007/s10875-011-9523-9. [DOI] [PubMed] [Google Scholar]
- 92.Abel L, El-Baghdadi J, Bousfiha AA, Casanova JL, Schurr E. Human genetics of tuberculosis: A long and winding road. Philos Trans R Soc Lond B Biol Sci. 2014;369(1645):20130428. doi: 10.1098/rstb.2013.0428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Picard C, Puel A, Bustamante J, Ku CL, Casanova JL. Primary immunodeficiencies associated with pneumococcal disease. Curr Opin Allergy Clin Immunol. 2003;3(6):451–459. doi: 10.1097/00130832-200312000-00006. [DOI] [PubMed] [Google Scholar]
- 94.Abinun M. Ectodermal dysplasia and immunodeficiency. Arch Dis Child. 1995;73(2):185. doi: 10.1136/adc.73.2.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Döffinger R, et al. X-linked anhidrotic ectodermal dysplasia with immunodeficiency is caused by impaired NF-kappaB signaling. Nat Genet. 2001;27(3):277–285. doi: 10.1038/85837. [DOI] [PubMed] [Google Scholar]
- 96.Jain A, et al. Specific missense mutations in NEMO result in hyper-IgM syndrome with hypohydrotic ectodermal dysplasia. Nat Immunol. 2001;2(3):223–228. doi: 10.1038/85277. [DOI] [PubMed] [Google Scholar]
- 97.Courtois G, et al. A hypermorphic IkappaBalpha mutation is associated with autosomal dominant anhidrotic ectodermal dysplasia and T cell immunodeficiency. J Clin Invest. 2003;112(7):1108–1115. doi: 10.1172/JCI18714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Picard C, et al. Pyogenic bacterial infections in humans with IRAK-4 deficiency. Science. 2003;299(5615):2076–2079. doi: 10.1126/science.1081902. [DOI] [PubMed] [Google Scholar]
- 99.von Bernuth H, et al. Pyogenic bacterial infections in humans with MyD88 deficiency. Science. 2008;321(5889):691–696. doi: 10.1126/science.1158298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Picard C, et al. Clinical features and outcome of patients with IRAK-4 and MyD88 deficiency. Medicine (Baltimore) 2010;89(6):403–425. doi: 10.1097/MD.0b013e3181fd8ec3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Casanova JL, Abel L, Quintana-Murci L. Human TLRs and IL-1Rs in host defense: Natural insights from evolutionary, epidemiological, and clinical genetics. Annu Rev Immunol. 2011;29:447–491. doi: 10.1146/annurev-immunol-030409-101335. [DOI] [PubMed] [Google Scholar]
- 102.Ku CL, et al. Selective predisposition to bacterial infections in IRAK-4-deficient children: IRAK-4-dependent TLRs are otherwise redundant in protective immunity. J Exp Med. 2007;204(10):2407–2422. doi: 10.1084/jem.20070628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Frans G, et al. PID in disguise: Molecular diagnosis of IRAK-4 deficiency in an adult previously misdiagnosed with autosomal dominant hyper IgE syndrome. J Clin Immunol. October 15, 2015 doi: 10.1007/s10875-015-0205-x. [DOI] [PubMed] [Google Scholar]
- 104.Gaschignard J, et al. Invasive pneumococcal disease in children can reveal a primary immunodeficiency. Clin Infect Dis. 2014;59(2):244–251. doi: 10.1093/cid/ciu274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Mahlaoui N, et al. Isolated congenital asplenia: A French nationwide retrospective survey of 20 cases. J Pediatr. 2011;158(1):142–148. doi: 10.1016/j.jpeds.2010.07.027. [DOI] [PubMed] [Google Scholar]
- 106.Bolze A, et al. Ribosomal protein SA haploinsufficiency in humans with isolated congenital asplenia. Science. 2013;340(6135):976–978. doi: 10.1126/science.1234864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Byun M, et al. Whole-exome sequencing-based discovery of STIM1 deficiency in a child with fatal classic Kaposi sarcoma. J Exp Med. 2010;207(11):2307–2312. doi: 10.1084/jem.20101597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Byun M, et al. Inherited human OX40 deficiency underlying classic Kaposi sarcoma of childhood. J Exp Med. 2013;210(9):1743–1759. doi: 10.1084/jem.20130592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Vanhollebeke B, et al. Human Trypanosoma evansi infection linked to a lack of apolipoprotein L-I. N Engl J Med. 2006;355(26):2752–2756. doi: 10.1056/NEJMoa063265. [DOI] [PubMed] [Google Scholar]
- 110.Pays E, Vanhollebeke B, Uzureau P, Lecordier L, Pérez-Morga D. The molecular arms race between African trypanosomes and humans. Nat Rev Microbiol. 2014;12(8):575–584. doi: 10.1038/nrmicro3298. [DOI] [PubMed] [Google Scholar]
- 111.Steiner I, Benninger F. Update on herpes virus infections of the nervous system. Curr Neurol Neurosci Rep. 2013;13(12):414. doi: 10.1007/s11910-013-0414-8. [DOI] [PubMed] [Google Scholar]
- 112.Abel L, et al. Age-dependent Mendelian predisposition to herpes simplex virus type 1 encephalitis in childhood. J Pediatr. 2010;157(4):623–629, 629.e1. doi: 10.1016/j.jpeds.2010.04.020. [DOI] [PubMed] [Google Scholar]
- 113.Dupuis S, et al. Impaired response to interferon-alpha/beta and lethal viral disease in human STAT1 deficiency. Nat Genet. 2003;33(3):388–391. doi: 10.1038/ng1097. [DOI] [PubMed] [Google Scholar]
- 114.Puel A, et al. The NEMO mutation creating the most-upstream premature stop codon is hypomorphic because of a reinitiation of translation. Am J Hum Genet. 2006;78(4):691–701. doi: 10.1086/501532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Filipe-Santos O, et al. X-linked susceptibility to mycobacteria is caused by mutations in NEMO impairing CD40-dependent IL-12 production. J Exp Med. 2006;203(7):1745–1759. doi: 10.1084/jem.20060085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Casrouge A, et al. Herpes simplex virus encephalitis in human UNC-93B deficiency. Science. 2006;314(5797):308–312. doi: 10.1126/science.1128346. [DOI] [PubMed] [Google Scholar]
- 117.Guo Y, et al. Herpes simplex virus encephalitis in a patient with complete TLR3 deficiency: TLR3 is otherwise redundant in protective immunity. J Exp Med. 2011;208(10):2083–2098. doi: 10.1084/jem.20101568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Herman M, et al. Heterozygous TBK1 mutations impair TLR3 immunity and underlie herpes simplex encephalitis of childhood. J Exp Med. 2012;209(9):1567–1582. doi: 10.1084/jem.20111316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Pérez de Diego R, et al. Human TRAF3 adaptor molecule deficiency leads to impaired Toll-like receptor 3 response and susceptibility to herpes simplex encephalitis. Immunity. 2010;33(3):400–411. doi: 10.1016/j.immuni.2010.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Sancho-Shimizu V, et al. Herpes simplex encephalitis in children with autosomal recessive and dominant TRIF deficiency. J Clin Invest. 2011;121(12):4889–4902. doi: 10.1172/JCI59259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Zhang SY, et al. TLR3 deficiency in patients with herpes simplex encephalitis. Science. 2007;317(5844):1522–1527. doi: 10.1126/science.1139522. [DOI] [PubMed] [Google Scholar]
- 122.Andersen LL, et al. Functional IRF3 deficiency in a patient with herpes simplex encephalitis. J Exp Med. 2015;212(9):1371–1379. doi: 10.1084/jem.20142274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Lafaille FG, et al. Impaired intrinsic immunity to HSV-1 in human iPSC-derived TLR3-deficient CNS cells. Nature. 2012;491(7426):769–773. doi: 10.1038/nature11583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.García-Sastre A, Palese P. Genetic manipulation of negative-strand RNA virus genomes. Annu Rev Microbiol. 1993;47:765–790. doi: 10.1146/annurev.mi.47.100193.004001. [DOI] [PubMed] [Google Scholar]
- 125.Medina RA, García-Sastre A. Influenza A viruses: New research developments. Nat Rev Microbiol. 2011;9(8):590–603. doi: 10.1038/nrmicro2613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Palese P. Influenza: Old and new threats. Nat Med. 2004;10(12) Suppl:S82–S87. doi: 10.1038/nm1141. [DOI] [PubMed] [Google Scholar]
- 127.Ciancanelli M, Abel L, Zhang SY, Casanova JL. Host genetics of influenza: From mouse Mx to human IRF7. Curr Opin Immunol. 2015 doi: 10.1016/j.coi.2015.12.002. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Ciancanelli MJ, et al. Life-threatening influenza and impaired interferon amplification in human IRF7 deficiency. Science. 2015;348(6233):448–453. doi: 10.1126/science.aaa1578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Casanova JL, Conley ME, Seligman SJ, Abel L, Notarangelo LD. Guidelines for genetic studies in single patients: Lessons from primary immunodeficiencies. J Exp Med. 2014;211(11):2137–2149. doi: 10.1084/jem.20140520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Casanova JL, Abel L, Quintana-Murci L. Immunology taught by human genetics. Cold Spring Harb Symp Quant Biol. 2013;78:157–172. doi: 10.1101/sqb.2013.78.019968. [DOI] [PubMed] [Google Scholar]
- 131.Casanova JL, Abel L. The human model: A genetic dissection of immunity to infection in natural conditions. Nat Rev Immunol. 2004;4(1):55–66. doi: 10.1038/nri1264. [DOI] [PubMed] [Google Scholar]
- 132.Quintana-Murci L, Alcaïs A, Abel L, Casanova JL. Immunology in natura: Clinical, epidemiological and evolutionary genetics of infectious diseases. Nat Immunol. 2007;8(11):1165–1171. doi: 10.1038/ni1535. [DOI] [PubMed] [Google Scholar]
- 133.Gaudillière JP, Löwy I. Heredity and Infection. Routledge; London, New York: 2001. p. 383. [Google Scholar]
- 134.Brodin P, et al. Variation in the human immune system is largely driven by non-heritable influences. Cell. 2015;160(1-2):37–47. doi: 10.1016/j.cell.2014.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Casanova JL, Abel L. Disentangling inborn and acquired immunity in human twins. Cell. 2015;160(1-2):13–15. doi: 10.1016/j.cell.2014.12.029. [DOI] [PubMed] [Google Scholar]
- 136.Anderson MS, Casanova JL. More than Meets the Eye: Monogenic Autoimmunity Strikes Again. Immunity. 2015;42(6):986–988. doi: 10.1016/j.immuni.2015.06.004. [DOI] [PubMed] [Google Scholar]
- 137.Rieux-Laucat F, Casanova JL. Immunology. Autoimmunity by haploinsufficiency. Science. 2014;345(6204):1560–1561. doi: 10.1126/science.1260791. [DOI] [PubMed] [Google Scholar]
- 138.Crow YJ, Casanova JL. STING-associated vasculopathy with onset in infancy--a new interferonopathy. N Engl J Med. 2014;371(6):568–571. doi: 10.1056/NEJMe1407246. [DOI] [PubMed] [Google Scholar]
- 139.Casanova JL, Abel L. Revisiting Crohn’s disease as a primary immunodeficiency of macrophages. J Exp Med. 2009;206(9):1839–1843. doi: 10.1084/jem.20091683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Pritchard JK. Are rare variants responsible for susceptibility to complex diseases? Am J Hum Genet. 2001;69(1):124–137. doi: 10.1086/321272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Pritchard JK, Cox NJ. The allelic architecture of human disease genes: Common disease-common variant...or not? Hum Mol Genet. 2002;11(20):2417–2423. doi: 10.1093/hmg/11.20.2417. [DOI] [PubMed] [Google Scholar]
- 142.McClellan J, King MC. Genetic heterogeneity in human disease. Cell. 2010;141(2):210–217. doi: 10.1016/j.cell.2010.03.032. [DOI] [PubMed] [Google Scholar]
- 143.Alcaïs A, et al. Life-threatening infectious diseases of childhood: Single-gene inborn errors of immunity? Ann N Y Acad Sci. 2010;1214(1):18–33. doi: 10.1111/j.1749-6632.2010.05834.x. [DOI] [PubMed] [Google Scholar]
- 144.Etzioni A, Ochs H. Primary Immunodeficiency Disorders. A Historic and Scientific Perspective. Academic; Oxford: 2014. pp. 376–389. [Google Scholar]
- 145.Witzel-Schlömp K, et al. The human complement C9 gene: Identification of two mutations causing deficiency and revision of the gene structure. J Immunol. 1997;158(10):5043–5049. [PubMed] [Google Scholar]