Abstract
MLN4924 is an experimental cancer drug known as inhibitor of NEDD8-activating enzyme (NAE). This anti-tumor candidate is a selective small-molecule inhibitor of NAE which is conjugated to cullin protein on Cullin-RING ligases (CRLs). This covalent modification actives cullin complex to recruit an ubiquitin-charged E2 and leads to downstream target protein polyubiquitination and proteasomal degradation. MLN4924, which can form a covalent adduct with NEDD8, and block NAE at the first step in this pathway, has shown anti-tumor activity in many kinds of cancer cell lines and also xenograft models, including lung cancer, colon cancer, melanoma and lymphoma. The anti-tumor activity of MLN4924 results from inactivation of CLRs, which causes DNA re-replication and inhibition of nuclear factor (NF)-κB signaling, thus leading to cancer cell death. A mutation can reduce the enzyme’s sensitivity to MLN4924. Verma et al. in 2013 studied on molecular dynamics simulation of a mutant A171T and consequently found out that this mutation reduce MLN4924 interaction with DNA Binding site of enzyme as a result of reduction of enzyme affinity to ATP. One year later, in 2014, Wei Xu et al. carried out a research on inhibitor resistant cell lines and revealed that a couple of mutations so called Y352H and I310N leads to enzyme resistance to MLN4924 inhibitor, interestingly, the cause reported was the increase of enzyme affinity to ATP. As in Wei Xu et al. experiment the molecular dynamics simulation was not considered, present study is conducted to identify enzyme mutation mechanism by molecular dynamics approach using advantages of Gromacs software version 4.5.6.
Keywords: MLN4924, NEDD8-activating enzyme (NAE), Y352H, I310N, mutation, drug discovery
Introduction
Cancer cells are adopting essentially cell signaling not only for progressing of cell cycle but also for preventing programmed cell death, so-called apoptosis. This signaling requires specific regulatory proteins such as ubiquitin-proteasome system (UPS) that the check points on this process, include a family of E3 ubiquitin ligases called Cullin-RING Ligases (CRLs) [1,2]. The ligase activity of these complexes requires modification of the cullin protein, a family of hydrophobic proteins providing a scaffold for ubiquitin ligases (E3), situated at their core with a ubiquitin-like protein (ULP) called NEDD8 [1,2].
NEDD8 is a protein encoded in humans by the NEDD8 gene and in Saccharomyces cerevisiae this protein is known as Rub1 [3]. This ULP, which becomes covalently conjugated to a limited number of cellular proteins in a manner analogous to ubiquitination, an enzymatic post-translational modification (PTM) process in which a ubiquitin protein is attached to a substrate protein. Human NEDD8 shares 60% amino acid sequence identity to ubiquitin. The only known substrates of NEDD8 modification are the cullin subunits of SCF ubiquitin E3 ligases. The NEDDylation of cullins is critical for the recruitment of E2 to the ligase complex, thus facilitating ubiquitin conjugation. NEDD8 modification has therefore been implicated in cell cycle progression and cytoskeletal regulation [3].
The Ubiquitin-Proteasome System (UPS) is responsible for the bulk of protein turnover in eukaryotic cells and plays a major role in maintaining cellular protein homeostasis. Among UPS substrates are proteins involved in cell cycle regulation, cellular growth and proliferation, Intracellular signaling, DNA repair, membrane receptor regulation, and proapoptotic and antiapoptotic signaling [4-8].
The initial step required for attachment of NEDD8 to a cullin is catalyzed by the E1, NEDD8-activating enzyme (NAE). The NAE interaction with other proteins is illustrated in Figure 1. The inhibitor of NAE; MLN4924 has been revealed to block the activity of NAE and prevent the subsequent neddylation of cullins. Preclinical studies have demonstrated antitumor activity in various solid tumors and hematological malignancies, and preliminary clinical data have shown the anticipated pharmacodynamic effects in humans. MLN4924, a small molecule with Molecular Weight of 443.5193 g/mol (Table 1 and Figure 2). Inhibiting NEDD8-activating enzyme (NAE), is thought to be a potential anticancer drug [9] and previously in clinical trials has shown clinical activity in some solid tumor and hematologic malignancies [10] and recently in complications with Lymphoma or Multiple Myeloma [11].
Figure 1.
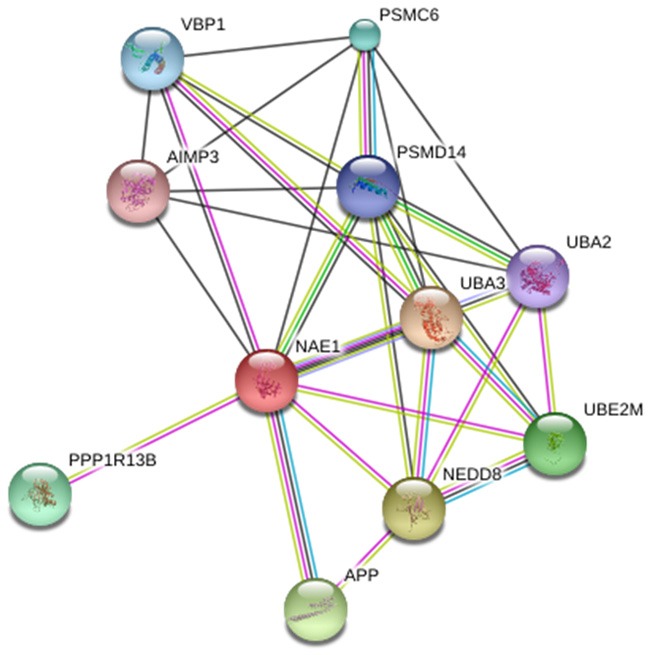
Interaction partners of NAE predicted by STRING Database (www.string-db.org/).
Table 1.
Computed Properties of MLN4924, Pfam database
| Molecular Weight | 443.5193 g/mol |
| Molecular Formula | C21H25N5O4S |
| XLogP3 | 1.7 |
| Hydrogen Bond Donor Count | 3 |
| Hydrogen Bond Acceptor Count | 8 |
| Rotatable Bond Count | 6 |
| Exact Mass | 443.162725 g/mol |
| Monoisotopic Mass | 443.162725 g/mol |
| Topological Polar Surface Area | 141 A^2 |
| Heavy Atom Count | 31 |
| Formal Charge | 0 |
| Complexity | 734 |
| Isotope Atom Count | 0 |
| Defined Atom Stereocenter Count | 4 |
| Undefined Atom Stereocenter Count | 0 |
| Defined Bond Stereocenter Count | 0 |
| Undefined Bond Stereocenter Count | 0 |
Figure 2.

Chemical structure of MLN4924 visualized by Pfam database (www.http://pfam.xfam.org/).
Furthermore, it exhibits potent in vitro cytotoxic activity against a variety of human tumor derived cell lines [12]. Treatment of tumor cells with MLN4924 increases the abundance of known CRL substrates. In the human cell lines studied, the mechanism of cell death seems to be a consequence of uncontrolled DNA synthesis in the S-phase of the cell cycle leading to DNA damage and induction of apoptosis. While still in the early stages of clinical development, the encouraging preclinical and clinical activity of MLN4924 supports investigation into the mechanisms of sensitivity and resistance to this drug [12]. It was shown that mutations in the nucleotide binding pocket and NEDD8 binding cleft of NAEb affect MLN4924 adduct formation and dissociation from NAEb. The NAEb mutations that have been detected in MLN4924-resistant cells derived in tissue culture or in vivo occur in two areas of the gene, the nucleotide binding pocket at Alanine 171 and at various residues that are within or close to the NEDD8 binding cleft [13]. The inhibition of NAE by MLN4924 occurs through the formation of a NEDD8-MLN4924 covalent adduct resembling NEDD8-AMP, a process that requires NAE catalytic activity [14]. Notably, the NEDD8-MLN4924 adduct is a tight binding inhibitor of NAE, and it has been proposed that the tight binding nature of the inhibitor-protein adduct is crucial for MLN4924 potency [15]. Inhibition of NAE has emerged as a highly promising approach to treat cancer through the adenosine sulfamate analog MLN4924. A mutation can reduce the enzyme’s sensitivity for MLN4924. Verma et al. in 2013 studied on molecular dynamics simulation of a mutant A171T and consequently found out that this mutation reduce MLN4924 interaction with DNA Binding site of enzyme as a result of reduction of enzyme affinity to ATP [16]. A year later, in 2014, Wei Xu et al. carried out a research on inhibitor resistant cell lines and revealed that a couple of mutations so called Y352H and I310N leads to enzyme resistance to MLN4924 inhibitor, interestingly, the reported cause was the increase of enzyme affinity to ATP. As in Wei Xu et al. experiment the molecular dynamics (MD) simulation was not considered, this study is conducted to identify mutation mechanism in enzyme using Gromacs software version 4.5.6.
Methodology
Prediction possible interaction NAE partners
Initially, in present study possible interaction partners of NAE partners predicted by taking advantages of STRING database (www.http://string-db.org/). This interactions include direct (physical) and indirect (functional) associations.
Computation properties and Chemical structure of MLN4924
Also, by Pfam database (www.http://pfam.xfam.org/), we evaluate Molecular Weight, Molecular Formula, XLogP3, Hydrogen Bond Donor Count, Hydrogen Bond Acceptor Count, Rotatable Bond Count and etc. beside Chemical structure of MLN4924.
Molecular dynamics simulation and molecular mechanics minimization
Molecular Dynamics (MD) simulation and molecular mechanics (MM) minimization were performed using Gromacs 4.5.6 package under anamber.ff99SB. The GAFF topologies for MLN4924 inhibitor were generated using the Antechamber software using partial charges calculated with the restrained electrostatic potential method. The GAFF topologies were converted into GROMACS format using the acpype tool. MD simulations were carried out with periodic boundary conditions. Van der Waals forces were treated with a cut-off of 10 Å. The Particle-Mesh Ewald method was used with a 10 Å cut-off. Also, crystallographic structure of NEDD8-activating enzyme (PDB code: 3GZN) was used for theoretical studies. Modeller program 9.13 was used to construct mutant Y352H and I310N.
The frequency to update the neighbor list was 5. The protonation state of Gromacs package was used to calculate the total charge of NEDD8-activating enzyme and mutants. MD simulation was accomplished in four steps. In the first step, the entire system was minimized using the steepest descent followed by conjugate gradients algorithms. In the second step or equilibration step, heavy atoms were restrained using a force constant of 1000 kJ/mol nm and the solvent and ions were allowed to evolve. This was done through minimization and molecular dynamics in the NVT ensemble for 100 ps and in the NPT ensemble for 100 ps. Then in order to obtain equilibrium geometry at 298 K and 1 atom, the temperature of the system was increased and the velocities at each step were reassigned according to the Maxwell-Boltzmann distribution at that temperature and equilibrated for 100 ps. Temperature coupling was set to 0.1 ps and pressure coupling to 2 ps. The Berendsen algorithm was used for thermostat and barostat during the equilibration step. All bonds were constrained via the LINCS algorithm. In the final step or production phase, a 20 ns MD simulation was performed under an NPT ensemble. In order to retain temperature and pressure stable in production step, Nosé-Hoover thermostat and Parrinello-Rahman barostat were used with removing position restraints. The temperature was at 298 K with a time step of 2 fs. In addition, the LINCS algorithm was used to constrain the lengths of hydrogen-containing bonds in this step. Na+ and Cl- ions were used to neutralize the system charge.
Result and discussion
Results of predicting interaction partners
Below figure represent how NAE may interact with other proteins, predicted by String database. In detail, NEDD8 activating enzyme E1 subunit 1; Regulatory subunit of the dimeric UBA3-NAE1 E1 enzyme. E1 activates NEDD8 by first adenylating its C-terminal glycine residue with ATP, thereafter linking this residue to the side chain of the catalytic cysteine, yielding a NEDD8-UBA3 thioester and free AMP. E1 finally transfers NEDD8 to the catalytic cysteine of UBE2M. Necessary for cell cycle progression through the S-M checkpoint. Overexpression of NAE1 causes apoptosis through deregulation of NEDD8 conjugation (534 aa).
Results of computation properties and Chemical structure of MLN4924
In next step, we computed properties and Chemical structure of MLN4924 and results are arranged in Table 1 and Figure 2. These data are useful to deep understanding physic-chemical properties of compound which will be used in MD.
Molecular dynamic simulations
After basic analysis, we precede molecular dynamic simulation. Figure 3 shows Nedd8-activating Enzyme in Complex with Nedd8 and Mln4924.
Figure 3.

3D structure of Nedd8-activating Enzyme in complex with Nedd8 and MLN4924 (http://www.ncbi.nlm.nih.gov/Structure/MMDB/mmdb.shtml).
In MD analysis, Root Mean Square Deviation (RSMD) of simulated structure upon time is an appropriated and common criterion to verify Molecular dynamics stimulation stability. Therefore, RSMD changes of main enzyme chain atoms for both natural and 2 mutants of that upon simulation period-which lasted 10 Nanoseconds-are calculated in comparison with experimental structure and the results are shown as following chart (Figure 4).
Figure 4.
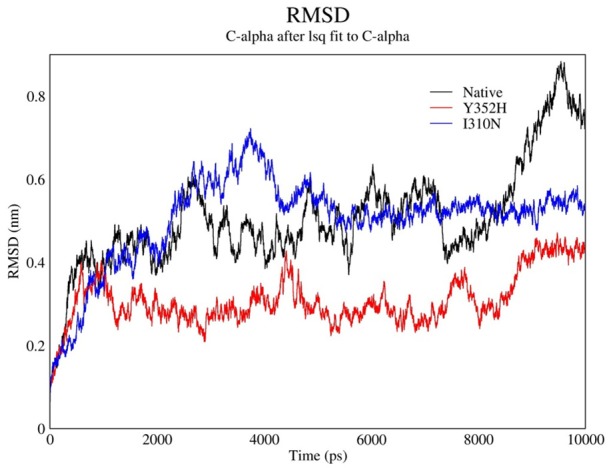
RMSD chart shows changes during time.
As it can be seen, RSMD after initiatory minimizing energy during first 2 nS has increased and it is the indicator of that the molecule based on surface has to be transferred to the places with lower energy level according to balance distance. After 2 nS this trend went downward whereas the fluctuation slightly decreased so that, they performed simulations are on acceptable conditions due to their stability and there is not any evidence of significant deviation of produced structure during simulation period from previous structures. Thus, to a large extent it can be claimed that the different regions of structures interact with each other and every structures met all the criteria to achieve ultimate conformation in simulation.
Furthermore, any modification in protein motif and shape can be evaluated by Gyration Radius (Figure 5). This parameter provides information about total protein volume distribution in spherical state. In other words, Gyration Radius demonstrates tensed molecular shape and as it is obviously indicated on following chart, this parameter is approximately constant on final section.
Figure 5.
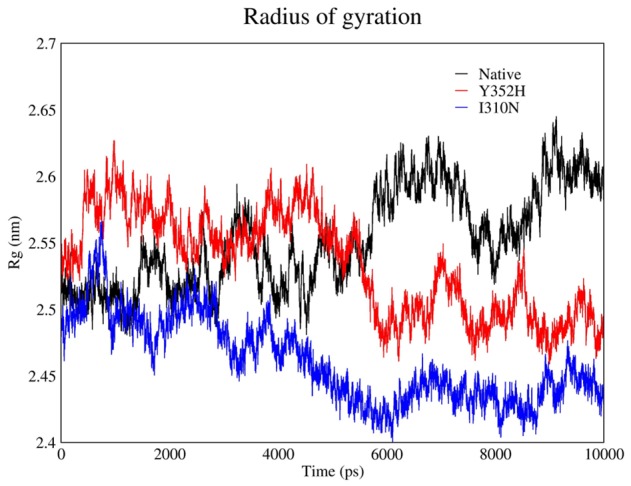
Radius of gyration obtained in MD analysis.
The results suggest that the Gyration Radius of mutants were downwards that is clue of constructed secondary structures while the natural Gyration radius was increased. Moreover, because of conformation changes to create more tensed structure compare to primary structure; the radius constantly showed fluctuation in simulation upon time. The evaluation of similarities between three structures revealed that Gyration Radius of natural sample was larger than the mutants and it seems that this is a result of limitation of main chain movement due to mutation. Besides, flexibility of interacting molecules specially those chains which are involved, have priority in importance of interaction process and eventually affect protein output efficiency.
In some region of these structures the flexibility change and the change pattern for those of natural make difference to those of mutants that can be considered on the comparing below chart (Figure 6).
Figure 6.
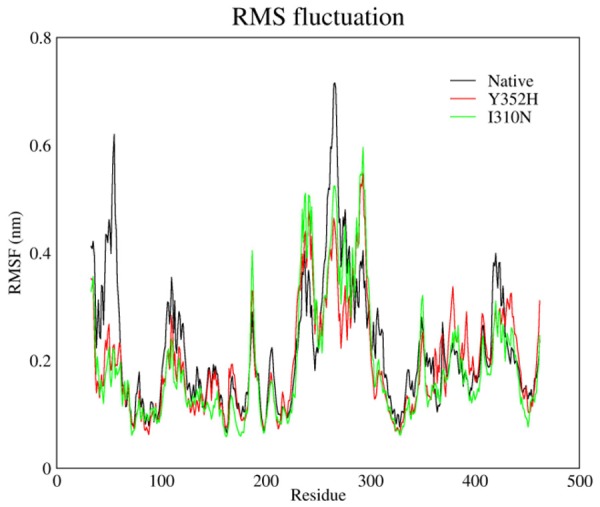
RMS fluctuation plot of Native, Y352H and I310N.
As it can be taken into account from above chart, both mutations in natural protein led to dramatic decline of flexibility on N-terminal end of this protein whereas, mutant’s flexibility slightly increased on two regions nearby mutation region at 230-240 and on other regions the flexibility decreased sharply.
The results suggested that this mutation that occurred in natural protein led to decreasing of structural flexibility as an effect of entire structure and eventually the mutant structure became tenser. The connectors’ energy for these states (natural and mutants) is summarized in Tables 2, 3 and 4.
Table 2.
Summary of natural protein and its connector energies
| Energy type | Calculated energy |
|---|---|
| Van der Waal energy | -207.177 ± 18.524 kJ/mol |
| Electrostattic energy | -129.819 ± 29.047 kJ/mol |
| Polar solvation energy | 282.588 ± 37.288 kJ/mol |
| SASA energy | -19.621 ± 1.176 kJ/mol |
| SAV energy | 0.000 ± 0.000 kJ/mol |
| WCA energy | 0.000 ± 0.000 kJ/mol |
| Binding energy | -74.030 ± 17.434 kJ/mol |
Table 3.
Summary of I310N Mutant and its connector energies
| Energy type | Calculated energy |
|---|---|
| Van der Waal energy | 67.683 ± 12.619 kJ/mol |
| Electrostattic energy | -1522.361 ± 62.344 kJ/mol |
| Polar solvation energy | 260.484 ± 40.990 kJ/mol |
| SASA energy | -1.675 ± 0.066 kJ/mol |
| SAV energy | 0.000 ± 0.000 kJ/mol |
| WCA energy | 0.000 ± 0.000 kJ/mol |
| Binding energy | -1195.869 ± 83.891 kJ/mol |
Table 4.
Summary of Y352H Mutantand its connector energies
| Energy type | Calculated energy |
|---|---|
| Van der Waal energy | -220.654 ± 18.622 kJ/mol |
| Electrostattic energy | -141.197 ± 28.372 kJ/mol |
| Polar solvation energy | 312.992 ± 33.450 kJ/mol |
| SASA energy | -19.995 ± 0.898 kJ/mol |
| SAV energy | 0.000 ± 0.000 kJ/mol |
| WCA energy | 0.000 ± 0.000 kJ/mol |
| Binding energy | -68.854 ± 42.452 kJ/mol |
Conclusions
The potential cancer drug, MLN4924 that inhibits NAE is affected by mutation and this can leads to reduce the enzyme’s sensitivity to MLN4924. Molecular dynamics simulation of a mutant A171T indicated that interaction with DNA Binding site of enzyme because of reduction of enzyme affinity to ATP is reduced. Y352H and I310N mutants leads to enzyme resistance to MLN4924 inhibitor and molecular dynamics simulation on this experiment identified enzyme mutation mechanism and related changes in natural protein which flexibility is reduced and the mutant became denser in its structure.
Acknowledgements
We would like to express our gratitude to Dr. Majid Taghdir, the assistant professor of Tarbiat Modares University, for sharing his pearls of wisdom with us during the course of this research. We are also immensely grateful to Mr. Ammar Mohseni for his meaningful contribution for conducting this research.
Disclosure of conflict of interest
None.
References
- 1.Soucy TA, Dick LR, Smith PG, Milhollen MA, Brownell JE. The NEDD8 Conjugation Pathway and Its Relevance in Cancer Biology and Therapy. Genes Cancer. 2010;1:708–716. doi: 10.1177/1947601910382898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gong L, Yeh ET. Identification of the activating and Conjugating enzymes of the NEDD8 conjugation pathway. J Biol Chem. 1999;274:12036–42. doi: 10.1074/jbc.274.17.12036. [DOI] [PubMed] [Google Scholar]
- 3.Kamitani T, Kito K, Nguyen HP, Yeh ET. Characterization of NEDD8, a developmentally downregulated ubiquitin-like protein. J Biol Chem. 1997;272:28557–62. doi: 10.1074/jbc.272.45.28557. [DOI] [PubMed] [Google Scholar]
- 4.Reinstein E, Ciechanover A. Narrative review. Protein degradation and human diseases: the ubiquitin connection. Ann Intern Med. 2006;145:676–84. doi: 10.7326/0003-4819-145-9-200611070-00010. [DOI] [PubMed] [Google Scholar]
- 5.Ciechanover A. The ubiquitin-proteasome pathway: on protein death and cell life. EMBO J. 1998;17:7151–60. doi: 10.1093/emboj/17.24.7151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ciechanover A, Schwartz AL. The ubiquitin-proteasome pathway: the complexity and myriad functions of proteins death. Proc Natl Acad Sci U S A. 1998;95:2727–30. doi: 10.1073/pnas.95.6.2727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ciechanover A. Intracellular protein degradation: from a vague idea thru the lysosome and the ubiquitin-proteasome system and onto human diseases and drug targeting. Cell Death Differ. 2005;12:1178–90. doi: 10.1038/sj.cdd.4401692. [DOI] [PubMed] [Google Scholar]
- 8.Hershko A. The ubiquitin system for protein degradation and some of its roles in the control of the cell division cycle. Cell Death Differ. 2005;12:1191–7. doi: 10.1038/sj.cdd.4401702. [DOI] [PubMed] [Google Scholar]
- 9.Yan ZH, Burkhardt A, Loke HK, Chen J, Xu Q, Brauer P, Ma J, Lin Y, Garcia K, Dick LR, Bembenek ME. Quantifiable analysis of cellular pathway inhibition of a Nedd8-activating enzyme inhibitor, MLN4924, using AlphaScreen. Anal Biochem. 2013;439:109–15. doi: 10.1016/j.ab.2013.04.016. [DOI] [PubMed] [Google Scholar]
- 10.Wang M, Medeiros BC, Erba HP, DeAngelo DJ, Giles FJ, Swords RT. Targeting protein neddylation: a novel therapeutic strategy for the treatment of cancer. Expert Opin Ther Targets. 2011;15:253–264. doi: 10.1517/14728222.2011.550877. [DOI] [PubMed] [Google Scholar]
- 11.Millennium Pharmaceuticals, Inc., Study of MLN4924, a Novel Inhibitor of Nedd8 Activating Enzyme, in Adult Patients With Lymphoma or Multiple Myeloma, ClinicalTrials.gov Identifier: NCT00722488. United States: Food and Drug Administration; 2013. [Google Scholar]
- 12.Xu GW, Toth JI, da Silva SR, Paiva SL, Lukkarila JL, Hurren R, Maclean N, Sukhai MA, Bhattacharjee RN, Goard CA, Medeiros B, Gunning PT, Dhe-Paganon S, Petroski MD, Schimmer AD. Mutations in UBA3 confer resistance to the NEDD8-activating enzyme inhibitor mln4924 in human leukemic cells. PLoS One. 2014;9:e93530. doi: 10.1371/journal.pone.0093530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Milhollen MA, Thomas MP, Narayanan U, Traore T, Riceberg J, Amidon BS, Bence NF, Bolen JB, Brownell J, Dick LR, Loke HK, McDonald AA, Ma J, Manfredi MG, Sells TB, Sintchak MD, Yang X, Xu Q, Koenig EM, Gavin JM, Smith PG. Treatment-Emergent Mutations in NAEb Confer Resistance to the NEDD8-Activating Enzyme Inhibitor MLN4924. Cancer Cell. 2012;21:388–401. doi: 10.1016/j.ccr.2012.02.009. [DOI] [PubMed] [Google Scholar]
- 14.Brownell JE, Sintchak MD, Gavin JM, Liao H, Bruzzese FJ, Bump NJ, Soucy TA, Milhollen MA, Yang X, Burkhardt AL, Ma J, Loke HK, Lingaraj T, Wu D, Hamman KB, Spelman JJ, Cullis CA, Langston SP, Vyskocil S, Sells TB, Mallender WD, Visiers I, Li P, Claiborne CF, Rolfe M, Bolen JB, Dick LR. Substrate-assisted inhibition of ubiquitin-like protein-activating enzymes: the NEDD8 E1 inhibitor MLN4924 forms a NEDD8-AMP mimetic in situ. Mol Cell. 2010;37:102–111. doi: 10.1016/j.molcel.2009.12.024. [DOI] [PubMed] [Google Scholar]
- 15.Brownell JE, Sintchak MD, Gavin JM, Liao H, Bruzzese FJ, Bump NJ, Soucy TA, Milhollen MA, Yang X, Burkhardt AL, Ma J, Loke HK, Lingaraj T, Wu D, Hamman KB, Spelman JJ, Cullis CA, Langston SP, Vyskocil S, Sells TB, Mallender WD, Visiers I, Li P, Claiborne CF, Rolfe M, Bolen JB, Dick LR. Substrate-assisted inhibition of ubiquitin-like protein-activating enzymes: the NEDD8 E1 inhibitor MLN4924 forms a NEDD8-AMP mimetic in situ. Mol Cell. 2010;37:102–11. doi: 10.1016/j.molcel.2009.12.024. [DOI] [PubMed] [Google Scholar]
- 16.Verma S, Singh A, Mishra A. Molecular dynamics investigation on the poor sensitivity of A171T mutant NEDD8-activating enzyme (NAE) for MLN4924. J Biomol Struct Dyn. 2014;32:1064–1073. doi: 10.1080/07391102.2013.804436. [DOI] [PubMed] [Google Scholar]


