Abstract
Oral squamous cell carcinoma (OSCC), which is malignant tumors in oral cavity, is the fourth most common male cancer in Taiwan. EZH2 plays a key role in transcriptional repression through chromatin remodeling and in cancer development. Although the EZH2 expression in OSCC is highly correlated with tumorigenesis, it has not been determined if specific EZH2 genetic variants are associated with OSCC risk. The aim of this study was to investigate the relationship between genetic polymorphisms of EZH2 and susceptibility to OSCC in Taiwan. Here, four SNPs of EZH2 (rs6950683, rs2302427, rs3757441, and rs41277434) were analyzed by a real-time PCR genotyping in 576 patients with oral cancer and 552 cancer-free controls. After adjusting for other co-variants, we found that carrying CC genotype at EZH2 rs6950683 and rs3757441 had a lower risk of developing OSCC than did wild-type carriers. The CCCA or CCTA haplotype among the four EZH2 sites was also associated with a reduced risk of OSCC. Furthermore, OSCC patients who carried CC genotype at EZH2 rs6950683 had a higher methylation than TC genotype. Our results suggest that the two SNPs of EZH2 (rs6950683 and rs3757441) might contribute to the prediction of OSCC susceptibility. Moreover, rs6950683 CC genotype exhibits hypermethylation in EZH2 promoter. This is the first study to provide insight into risk factors associated with EZH2 variants and epigenetic changes in carcinogenesis of OSCC in Taiwan.
Keywords: EZH2, oral squamous cell carcinoma, single-nucleotide polymorphism (SNP), bisulfite sequencing, methylation
Introduction
Oral squamous cell carcinoma (OSCC) represents the fourth common male cancer of cancer-related deaths per year in Taiwan [1]. The development of OSCC is a multistep process modulated by the accumulation of endogenous and environmental factors including alcohol drinking, tobacco smoking, betel nut chewing, and chronic inflammation [2-4]. The current treatment for OSCC is surgery combined with radiotherapy and chemotherapy [5]. However, the prognosis of OSCC is poor due to aggressive local metastasis and invasion. The percentage of OSCC recurrence rate was 32.7% and 5-year survival for patients was 54.5% [6].
Polycomb group (PcG) proteins are important epigenetic regulators of embryonic development, cell-fate determination, proliferation, stem cell pluripotency and self-renewal [7]. EZH2 is a subunit of the multi-enzyme complex polycomb repressive complex 2 and functions as a histone H3 Lys27 (H3K27) trimethyltransferase [8]. Recent findings implicate that EZH2 is overexpressed in a wide range of cancer types, including brain [9], breast [10], and liver cancer [11]. Overexpression of EZH2 has been correlated with advanced stages of human cancer progression and poor prognosis [12]. Moreover, EZH2 promotes epithelial-to-mesenchymal transition (EMT) and enhanced cell migration and invasion [13]. Although EZH2 contributes to the formation of many types of cancer, the association between EZH2 variants and OSCC risk and prognosis has been poorly investigated.
Recently, several studies suggested that genetic variation will affect cancer susceptibility and response to environmental carcinogens [14]. Among these genetic factors, SNPs are one of the most common types of genetic variation and have a significant impact on the diagnosis of the causes, treatment and prevention of human genetic diseases [15-17]. We therefore performed a case-control study of four SNPs of EZH2 to clarify the associations between these SNPs and OSCC susceptibility and clinicopathologic features.
Epigenetic regulation is an important genetic modification in silencing of tumor suppressor genes and activation of oncogenes [18]. It is commonly known that hypermethylation within the gene promoter regions cause inactivation of certain tumor-suppressor genes and numerous studies have demonstrated DNA methylation is associated with gene silencing in different cancer types [19]. In this study, we analyzed associations among SNPs of EZH2, epigenetic changes, and OSCC susceptibility. To our knowledge, this is the first study that has used a bisulfite DNA sequencing approach to examine methylation status with EZH2 gene SNPs in oral carcinogenesis.
Materials and methods
Ethics statement
Study protocols were approved by the institutional review boards of the Taichung Chung Shan Medical University Hospital. All methods were carried out in accordance with the approved guidelines. All subjects provided written informed consent to participate in the study.
Study subjects and specimen collection
This hospital-based case-control study recruited 576 OSCC patients between 2010 and 2014 from the Chung Shan Medical University Hospital in Taichung and the Changhua Christian Hospital and Show Chwan Memorial Hospital in Changhua, Taiwan, to serve as the case group. The diagnosis of OSCC was made according to the criteria specified in the national guidelines for OSCC. An additional 552 race- and ethnic group-matched non-cancer patients who entered the hospital for health check-ups were enrolled as the control group. OSCC patients were clinically staged at the time of diagnosis according to the tumor/node/metastasis staging system of the American Joint Committee on Cancer. The patients’ clinicopathological characteristics, including clinical staging, lymph node metastasis, and histopathologic grading levels, were verified by chart review. Whole-blood specimens collected from the controls and OSCC patients were placed in tubes containing EDTA, immediately centrifuged, and stored at -80°C.
Selection of EZH2 polymorphisms
A total of four SNPs in EZH2 (NM_004456) were selected from the International HapMap Project data for this study. We included the non-synonymous SNP rs2302427 (D185H in exon 6) in the coding sequence of the gene. The other SNPs (rs6950683, rs3757441, and rs41277434) were selected in this study because they have been found in cancer patients.
Genomic DNA extraction
Genomic DNA was extracted using QIAamp DNA blood mini kit reagents (Qiagen, Valencia, CA). DNA was dissolved in buffer containing 10 mM Tris (pH 7.8) and 1 mM EDTA and then quantified by measurement of the optical density at 260 nm. Final DNA preparations were stored at -20°C and used as templates for PCR.
Real-time PCR
Allelic discrimination of the EZH2 polymorphisms rs6950683, rs2302427, rs3757441, and rs41277434 was assessed using an ABI StepOneTM Real-Time PCR System (Applied Biosystems), SDS v3.0 software (Applied Biosystems), and the TaqMan assay. The final volume for each reaction was 5 μL, containing 2.5 μL TaqMan Genotyping Master Mix, 0.125 μL TaqMan probes mix, and 10 ng genomic DNA. The reaction conditions included an initial denaturation step at 95°C for 10 min followed by 40 cycles at 95°C for 15 sec and 60°C for 1 min.
Bisulfite modification
Genomic DNA (0.5 μg) was treated with sodium bisulfite via the EZ DNA Methylation-Gold Kit (Zymo Research, Irvine, CA). Bisulfite treatments changed unmethylated cytosines into uracils while leaving methylated cytosines unmodified. Bisulfite-treated genomic DNA was used for further analysis of methylation status of CpG sites via bisulfite DNA sequencing.
Bisulfite DNA sequencing
Bisulfite-modified DNA was amplified by PCR with primer designed to amplify a 412-bp-long promoter region of the EZH2 gene. Bisulfite primer sequences were: 5’-AGTTTTGAATTGGTTTAAATTTGGT-3’ (forward) and 5’-CCTCCAATCACAAAACCC-3’ (reverse). Purified PCR products were cloned into the pGEM vector (GeneMark, Taichung, Taiwan) by using the manufacturer’s standard protocol. Seven clones were submitted for DNA sequencing at the Genomics BioSci & Tech, Ltd. (New Taipei City, Taiwan).
Statistical analysis
Continuous variables were expressed as mean ± standard deviation (SD), and categorical variables were expressed as number (%). Hardy-Weinberg equilibrium was assessed using a χ2 goodness-of-fit test for biallelic markers. A Mann-Whitney U-test and a Fisher’s exact test were used to compare differences of age and demographic characteristics distributions between controls and OSCC patients. The odds ratios (ORs) with 95% confidence intervals (CIs) were estimated by logistic regression models. The adjusted odds ratios (AORs) with 95% CIs of the association between genotype frequencies and OSCC risk as well as clinical pathological characteristics were estimated by multiple logistic regression models after controlling for other covariates. The haplotype-based analysis was carried out using the Phase program. Linkage disequilibrium (LD) coefficients, D’=D/Dmax (or D/Dmin if the D’ value was negative), were assessed for pairs of alleles between the two sites of EZH2 polymorphisms. All P values < 0.05 were considered significant. The data were analyzed using SAS statistical software (Version 9.1, 2005; SAS Institute Inc., Cary, NC).
Results
Population characteristics
A total of 1128 subjects were enrolled in this case-control study, which containing 576 OSCC patients and 552 healthy controls (Table 1). The healthy controls were comparable with the OSCC patients in regard to significantly different distributions of age (P < 0.001), gender (P < 0.001), betel nut chewing (P < 0.001), alcohol consumption (P < 0.001) and tobacco consumption (P < 0.001) between healthy controls and OSCC patients. To reduce possible interference of confounding variables, AORs with 95% CIs were estimated by multiple logistic regression models after controlling for age, gender, betel nut chewing, tobacco and alcohol consumption in each comparison.
Table 1.
The distributions of demographical characteristics in 552 healthy controls and 576 patients with oral cancer
| Variable | Controls (N=552) | Patients (N=576) | p value |
|---|---|---|---|
| Age (yrs) | Mean ± S.D. | Mean ± S.D. | |
| 51.65 ± 14.62 | 54.37 ± 11.28 | P < 0.001* | |
| Gender | n (%) | n (%) | |
| Male | 449 (81.3%) | 554 (96.2%) | |
| Female | 103 (18.7%) | 22 (3.8%) | P < 0.001* |
| Betel nut chewing | |||
| No | 460 (83.3%) | 136 (23.6%) | |
| Yes | 92 (16.7%) | 440 (76.4%) | P < 0.001* |
| Alcohol consumption | |||
| No | 345 (62.5%) | 235 (40.8%) | |
| Yes | 207 (37.5%) | 341 (59.2%) | P < 0.001* |
| Tobacco consumption | |||
| No | 336 (60.9%) | 87 (15.1%) | |
| Yes | 216 (39.1%) | 489 (84.9%) | P < 0.001* |
Mann-Whitney U test or Fisher’s exact test was used between healthy controls and patients with oral cancer.
P value < 0.05 as statistically significant.
Association between EZH2 SNPs and OSCC risk
In our recruited control group, the frequencies of EZH2 genes were in Hardy-Weinberg equilibrium. The reconstructed linkage disequilibrium plot for the four SNPs was shown in Figure 1. We found that rs6950683 and rs3757441 show a high degree of D’ in our study. Table 2 shows the genotype distributions and the association between OSCC and EZH2 polymorphisms. The alleles with the highest distribution frequency at EZH2 rs6950683, rs2302427, rs3757441, and rs41277434 in both OSCC patients and controls were homozygous T/T, homozygous C/C, homozygous T/T, and homozygous A/A, respectively. After adjusting variables, people with rs6950683 CC showed a 0.492-fold (95% CI: 0.274-0.884), and rs3757441 CC showed a 0.524-fold (95% CI: 0.284-0.968) lower risk of OSCC, respectively, compared with individuals carrying the wild-type allele. Individuals with polymorphisms at rs2302427 and rs41277434 showed no reduction in OSCC risk compared with wild-type individuals.
Figure 1.
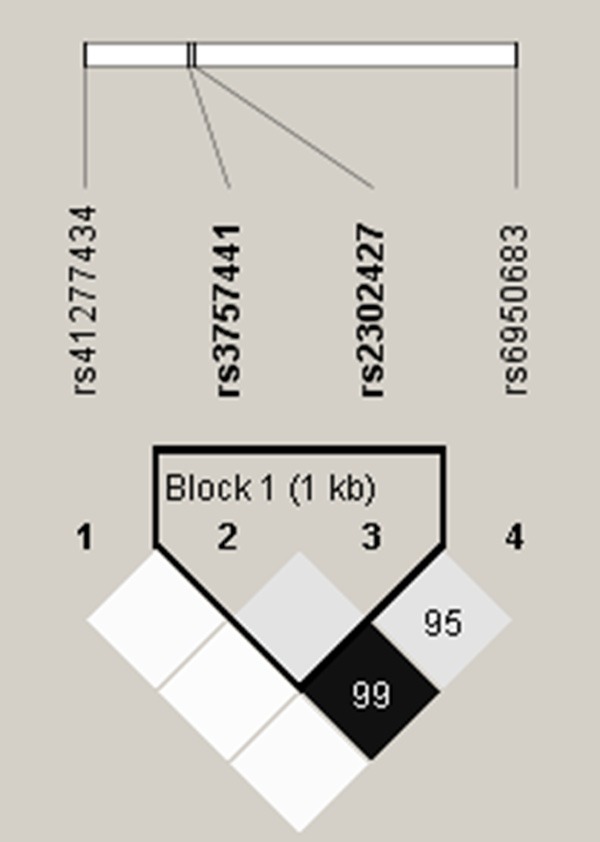
EZH2 pairwise linkage disequilibrium (LD) patterns. Schematic presentation of the EZH2, indicating the locations of the SNP polymorphism and the pairwise linkage disequilibrium measures D’. The measure of D’ of SNP is shown graphically according to a grey scale, where white represents low D’ and dark represents high D’.
Table 2.
Distribution frequency of EZH2 genotypes in 552 controls and 576 oral cancer patients
| Variable | Controls (N=552) n (%) | Patients (N=576) n (%) | OR (95% CI) | AOR (95% CI) |
|---|---|---|---|---|
| rs6950683 | ||||
| TT | 264 (47.8%) | 308 (53.5%) | 1.00 | 1.00 |
| TC | 220 (39.9%) | 221 (38.4%) | 0.861 (0.671-1.104) | 0.893 (0.622-1.280) |
| CC | 68 (12.3%) | 47 (8.2%) | 0.592 (0.395-0.890)* | 0.492 (0.274-0.884)* |
| TC+CC | 288 (52.2%) | 268 (46.5%) | 0.798 (0.631-1.008) | 0.790 (0.563-1.108) |
| rs2302427 | ||||
| CC | 346 (62.7%) | 356 (61.8%) | 1.00 | 1.00 |
| CG | 171 (31.0%) | 200 (34.7%) | 1.137 (0.883-1.463) | 0.930 (0.646-1.405) |
| GG | 35 (6.3%) | 20 (3.5%) | 0.555 (0.314-1.002) | 0.611 (0.265-1.405) |
| CG+GG | 206 (37.3%) | 220 (38.2%) | 1.038 (0.816-1.321) | 0.883 (0.623-1.250) |
| rs3757441 | ||||
| TT | 271 (49.1%) | 312 (54.2%) | 1.00 | 1.00 |
| TC | 223 (40.4%) | 221 (38.4%) | 0.861 (0.672-1.102) | 0.887 (0.620-1.269) |
| CC | 58 (10.5%) | 43 (7.5%) | 0.644 (0.420-0.987)* | 0.524 (0.284-0.968)* |
| TC+CC | 281 (50.9%) | 264 (45.8%) | 0.816 (0.646-1.031) | 0.804 (0.573-1.127) |
| rs41277434 | ||||
| AA | 517 (93.6%) | 540 (93.7%) | 1.00 | 1.00 |
| AC | 34 (6.2%) | 35 (6.1%) | 0.986 (0.606-1.604) | 1.546 (0.784-3.048) |
| CC | 1 (0.2%) | 1 (0.2%) | 0.957 (0.060-15.347) | 3.341 (0.157-70.898) |
| AC+CC | 35 (6.4%) | 36 (6.3%) | 0.985 (0.609-1.592) | 1.590 (0.814-3.104) |
The odds ratios (ORs) and with their 95% confidence intervals (CIs) were estimated by logistic regression models. The adjusted odds ratios (AORs) with their 95% confidence intervals (CIs) were estimated by multiple logistic regression models after controlling for age, gender, betel nut chewing, tobacco and alcohol consumption.
The distribution of clinical status/EZH2 genotypes in OSCC patients was estimated to clarify the role of EZH2 polymorphisms in the clinicopathologic state of OSCC patients. Clinical status assessments included TNM clinical stage, primary tumor size, lymph node involvement, distant metastasis. Compared with control subjects having the wild-type genotype, patients with at least one polymorphic C allele at EZH2 rs6950683 showed significant differences in EZH2 genotypic frequencies (Table 3), clinical stage and tumor size (Table 4); however, patients with at least one polymorphic C allele at EZH2 rs3757441 showed no significant differences in the clinicopathologic state.
Table 3.
Distribution of the allele frequencies of EZH2 genotypes in controls and oral cancer patients
| Variable | Controls (N=1104) n (%) | Patients (N=1152) n (%) | OR (95% CI) | p value |
|---|---|---|---|---|
| rs6950683 | ||||
| T | 748 (67.8%) | 837 (72.7%) | 1.00 | |
| C | 356 (32.2%) | 315 (27.3%) | 0.791 (0.660-0.948) | P=0.011* |
| rs2302427 | ||||
| C | 863 (78.2%) | 912 (79.2%) | 1.00 | |
| G | 241 (21.8%) | 240 (20.8%) | 0.942 (0.770-1.153) | P=0.564 |
| rs3757441 | ||||
| T | 765 (69.3%) | 845 (73.4%) | 1.00 | |
| C | 339 (30.7%) | 307 (26.6%) | 0.820 (0.683-0.984) | P=0.033* |
| rs41277434 | ||||
| A | 1068 (96.7%) | 1115 (96.8%) | 1.00 | |
| C | 36 (3.3%) | 37 (3.2%) | 0.984 (0.617-1.570) | P=0.948 |
The odds ratios (ORs) and with their 95% confidence intervals (CIs) were estimated by logistic regression models.
Statistically significant at P < 0.05.
Table 4.
Clinical status and EZH2 genotypic frequencies in 576 oral cancer patients
| Variable | rs6950683 | rs2302427 | rs3757441 | rs41277434 | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
| ||||||||||||
| TT (N=308) | TC + CC (N=268) | p value | CC (N=356) | CG + GG (N=220) | p value | TT (N=312) | TC + CC (N=264) | p value | AA (N=540) | AC + CC (N=36) | p value | |
| Clinical Stage | ||||||||||||
| Stage I/II | 122 (39.6%) | 133 (49.6%) | 0.016* | 159 (44.7%) | 96 (43.6%) | 0.810 | 127 (40.7%) | 128 (48.5%) | 0.061 | 241 (44.6%) | 14 (38.9%) | 0.502 |
| Stage III/IV | 186 (60.4%) | 135 (50.4%) | 197 (55.3%) | 124 (56.4%) | 185 (59.3%) | 136 (51.5%) | 299 (55.4%) | 22 (61.1%) | ||||
| Tumor size | ||||||||||||
| ≤T2 | 178 (57.8%) | 177 (66.0%) | 0.042* | 224 (62.9%) | 131 (59.5%) | 0.418 | 183 (58.7%) | 172 (65.2%) | 0.110 | 336 (62.2%) | 19 (52.8%) | 0.259 |
| >T2 | 130 (42.2%) | 91 (34.0%) | 132 (37.1%) | 89 (40.5%) | 129 (41.3%) | 92 (34.8%) | 204 (37.8%) | 17 (47.2%) | ||||
| Lymph node metastasis | ||||||||||||
| No | 191 (62.0%) | 174 (64.9%) | 0.469 | 228 (64.0%) | 137 (62.3%) | 0.668 | 192 (61.5%) | 173 (65.5%) | 0.322 | 344 (63.7%) | 21 (58.3%) | 0.517 |
| Yes | 117 (38.0%) | 94 (35.1%) | 128 (36.0 %) | 83 (37.7%) | 120 (38.5%) | 91 (34.5%) | 196 (36.3 %) | 15 (41.7%) | ||||
| Distant metastasis | ||||||||||||
| No | 304 (98.7%) | 264 (98.5%) | 0.843 | 353 (99.2%) | 215 (97.7%) | 0.154 | 308 (98.7%) | 260 (98.5%) | 0.812 | 533 (98.7%) | 35 (97.2%) | 0.462 |
| Yes | 4 (1.3%) | 4 (1.5%) | 3 (0.8 %) | 5 (2.3%) | 4 (1.3%) | 4 (1.5%) | 7 (1.3 %) | 1 (2.8%) | ||||
T2: tumor size > 2 cm in the greatest dimension.
P value < 0.05 as statistically significant.
Haplotypes analysis of EZH2 SNPs with OSCC risk
We further explored the haplotypes to evaluate the combined effect of the four polymorphisms on oral-cancer susceptibility. The haplotype distributions of EZH2 rs6950683, rs2302427, rs3757441, and rs41277434 were further evaluated, and seven haplotypes were derived from these four SNPs in our recruited individuals. The most common haplotype in the control group was TCTA (42.4%), and it was, therefore, chosen as the reference. Compared with this reference, two minor haplotypes, CCCA and CCTA, significantly reduced the risk of OSCC by 0.763-fold (95% CI: 0.626-0.930) and 0.250-fold (95% CI: 0.100-0.627), respectively (Table 5).
Table 5.
Distribution frequency of EZH2 haplotype in controls and oral cancer patients
| Variable | Controls (N=1104) n (%) | Patients (N=1152) n (%) | OR (95% CI) | p value | |||
|---|---|---|---|---|---|---|---|
|
| |||||||
| rs6950683 | rs2302427 | rs3757441 | rs41277434 | ||||
| T/C | C/G | T/C | A/C | ||||
| T | C | T | A | 468 (42.4%) | 562 (48.8%) | Reference | |
| C | C | C | A | 335 (30.3%) | 307 (26.6%) | 0.763 (0.626-0.930) | 0.007* |
| T | G | T | A | 240 (21.7%) | 238 (20.7%) | 0.826 (0.665-1.026) | 0.084 |
| T | C | T | C | 36 (3.3%) | 37 (3.2%) | 0.856 (0.532-1.376) | 0.520 |
| C | C | T | A | 20 (1.8%) | 6 (0.5%) | 0.250 (0.100-0.627) | 0.001* |
| T | C | C | A | 4 (0.4%) | 0 (0.0%) | ---- | |
| C | G | T | A | 1 (0.1%) | 2 (0.2%) | 1.665 (0.151-18.425) | 0.674 |
P value < 0.05 as statistically significant.
Methylation level of EZH2 rs6950683
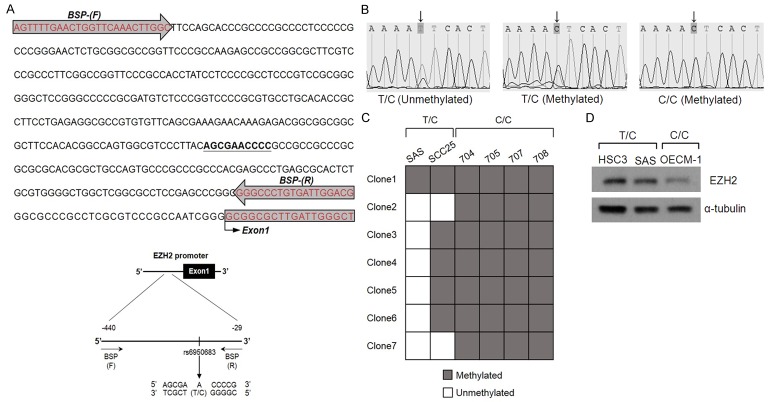
DNA methylation plays an important role in regulating gene expression. To explore the methylation status of rs6950683 on EZH2 promoter, we first examined the 2 OSCC cell lines (TC genotype) and 4 OSCC patient samples (CC genotype), followed by detailed investigation of the methylation status of rs6950683 via bisulfite sequencing analysis. Therefore, the EZH2 promoter further upstream spanning nucleotides -440 to -29 bp that include the rs6950683 was sequenced after bisulfite modification of genomic DNA from cell lines and patient samples. As shown in Figure 2, individual cell colonies derived from patients were fully methylated at rs6950683. In contrast, cell lines SAS and SCC25 were both methylated and unmethylated at rs6950683. Moreover, we also found that EZH2 expression from total cell lysates were much lower in C/C genotype than T/C genotype. These results suggested that the rs6950683 C/C genotype was more easily methylated than the T/C genotype and associated with low EZH2 expression in OSCC.
Figure 2.
Bisulfite sequencing of the rs6950683. A. Positions of bisulfite sequencing PCR (BSP) primers. BSP primers (-440 to -29) were designed to include the rs6950683. B. Bisulfite sequence analyses of the EZH2 promoter reveal TC (two signal) or CC genotype (one signal) at the rs6950683 location at which the arrow points. C. Bisulfite sequencing results from oral cancer patient samples (704, 705, 707 and 708) and oral cancer cell lines (SAS and SCC25). Open squares, unmethylated status; gray squares, methylated status. Each square represents an individual sequenced DNA strand. D. Western blot analysis of EZH2 in oral cancer cell lines. α-tubulin was used as a loading control.
Discussion
The present study focused on the association of genetic polymorphisms and haplotypes of the EZH2 gene with oral cancer risk. EZH2 is a member of the polycomb group of genes and is important in cell cycle regulation. EZH2 is also associated with cell proliferation, apoptosis, metastasis and invasion in OSCC development [20]. In previous studies, the results suggested that EZH2 acts as an oncogene and correlated with malignant potential and poor prognosis in OSCC [21].
Epidemiological studies have provided evidences that genetic variation of humans can affect how humans develop diseases [14]. Analyzing these DNA polymorphisms may help in identifying progression and susceptibility of malignancy. Therefore, EZH2 polymorphisms may be associated with OSCC development. However, the predictive value of EZH2 for susceptibility to OSCC has not previously been investigated. In this study, we provide novel information of SNPs of EZH2 on the oral cancer susceptibility and clinicopathologic statuses association.
In our hospital-based case-control study, 4 polymorphisms of EZH2 were examined in 552 healthy controls and 576 patients with oral cancer. Patients recruited among the oral cancer at our hospital showed a higher interference of environmental factors than did the control cases, such as alcohol drinking, tobacco smoking and betel-nut chewing (Table 1), which indicates environmental factors are highly associated to the increased risk of oral cancer. Exposure to environmental carcinogen might involve the process of tumorigenesis, but more evidence indicates that genetic variants might more helpful in predicting cancer risk [22,23]. Therefore, the association between the SNPs and the risk of oral cancer was analyzed by controlling for environmental influences.
Data in Table 2 show that individuals with the EZH2 polymorphisms rs6950683 CC and rs3757441 CC genotype have lower risks for OSCC compared to the TT genotype. Although the functional importance of these two SNPs has not been tested experimentally, clinical research from previous studies also shows that these two SNPs carrying CC genotype have a protective effect of risk than those carrying TT wild type in lung cancer and colorectal cancer [24,25]. We also observed in the EZH2 gene haplotypes “CCCA” and “CCTA” that were associated with a decreased the risk of developing OSCC. These results suggest that EZH2 gene polymorphisms rs6950683 and rs3757441 have a strong impact on oral cancer susceptibility.
Epigenetic regulations including DNA methylation and histone modification, which can lead to genomic instability, control gene expression and play important roles during cancer development [19,26]. DNA methylation typically occurs in the promoter CpG-rich regions of tumor suppressor genes, which mediates gene silencing. SNP rs6950683 is located in a tissue-specific CpG island within the EZH2 promoter region upstream of the EZH2 coding sequence, several studies have shown promoter might play an important role in regulating the process of transcription and protein expression. One of the intriguing findings of our study is that rs6950683 remains hypermethylation status when patient sample carrying CC genotype. This result raises the possibility that methylation of rs6950683 most likely impact EZH2 expression by affecting promoter function, and consequently reduce the risk of OSCC. Further functional studies are needed to confirm the specific mechanisms by epigenetic regulation of EZH2 polymorphisms influence the development of OSCC. However, the number of case subjects in this study was relatively small and only considers a Taiwanese population, which may limit the application of these findings. Therefore, additional studies with larger sample sizes are needed to validate the epigenetic regulation of EZH2 polymorphisms to OSCC. Further investigations should focus on epigenetic changes of EZH2 polymorphisms and their biological function in OSCC patients.
In conclusion, this is the first study to associate EZH2 polymorphisms and epigenetic change with risk of OSCC. Results showed genotypes of “CC” of rs6950683 and rs3757441 in the EZH2 gene that decreased OSCC susceptibility. Moreover, OSCC patients with the rs6950683 CC genotype have a hypermethylation status in EZH2 promoter. These results suggest that EZH2 variants might play important roles in the susceptibility to OSCC.
Acknowledgements
This study was supported by funding from Ministry of Science and Technology, Taiwan (MOST103 2320-B-039-052-MY3; MOST104-2321-B-039-005); Ministry of Health and Welfare (MOHW104-TDU-B-212-124-002) and National Health Research Institutes, Taiwan (NHRI-EX102-10245BI).
Disclosure of conflict of interest
None.
References
- 1.Chung TT, Pan MS, Kuo CL, Wong RH, Lin CW, Chen MK, Yang SF. Impact of RECK gene polymorphisms and environmental factors on oral cancer susceptibility and clinicopathologic characteristics in Taiwan. Carcinogenesis. 2011;32:1063–1068. doi: 10.1093/carcin/bgr083. [DOI] [PubMed] [Google Scholar]
- 2.Nagaraj NS, Beckers S, Mensah JK, Waigel S, Vigneswaran N, Zacharias W. Cigarette smoke condensate induces cytochromes P450 and aldo-keto reductases in oral cancer cells. Toxicol Lett. 2006;165:182–194. doi: 10.1016/j.toxlet.2006.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sharan RN, Mehrotra R, Choudhury Y, Asotra K. Association of betel nut with carcinogenesis: revisit with a clinical perspective. PLoS One. 2012;7:e42759. doi: 10.1371/journal.pone.0042759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Feller L, Altini M, Lemmer J. Inflammation in the context of oral cancer. Oral Oncol. 2013;49:887–892. doi: 10.1016/j.oraloncology.2013.07.003. [DOI] [PubMed] [Google Scholar]
- 5.Liao CT, Wang HM, Ng SH, Yen TC, Lee LY, Hsueh C, Wei FC, Chen IH, Kang CJ, Huang SF, Chang JT. Good tumor control and survivals of squamous cell carcinoma of buccal mucosa treated with radical surgery with or without neck dissection in Taiwan. Oral Oncol. 2006;42:800–809. doi: 10.1016/j.oraloncology.2005.11.020. [DOI] [PubMed] [Google Scholar]
- 6.Wang B, Zhang S, Yue K, Wang XD. The recurrence and survival of oral squamous cell carcinoma: a report of 275 cases. Chin J Cancer. 2013;32:614–618. doi: 10.5732/cjc.012.10219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zeng J, Kirk BD, Gou Y, Wang Q, Ma J. Genome-wide polycomb target gene prediction in Drosophila melanogaster. Nucleic Acids Res. 2012;40:5848–5863. doi: 10.1093/nar/gks209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schuettengruber B, Chourrout D, Vervoort M, Leblanc B, Cavalli G. Genome regulation by polycomb and trithorax proteins. Cell. 2007;128:735–745. doi: 10.1016/j.cell.2007.02.009. [DOI] [PubMed] [Google Scholar]
- 9.Kim E, Kim M, Woo DH, Shin Y, Shin J, Chang N, Oh YT, Kim H, Rheey J, Nakano I, Lee C, Joo KM, Rich JN, Nam DH, Lee J. Phosphorylation of EZH2 activates STAT3 signaling via STAT3 methylation and promotes tumorigenicity of glioblastoma stem-like cells. Cancer Cell. 2013;23:839–852. doi: 10.1016/j.ccr.2013.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chang CJ, Yang JY, Xia W, Chen CT, Xie X, Chao CH, Woodward WA, Hsu JM, Hortobagyi GN, Hung MC. EZH2 promotes expansion of breast tumor initiating cells through activation of RAF1-beta-catenin signaling. Cancer Cell. 2011;19:86–100. doi: 10.1016/j.ccr.2010.10.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sasaki M, Ikeda H, Itatsu K, Yamaguchi J, Sawada S, Minato H, Ohta T, Nakanuma Y. The overexpression of polycomb group proteins Bmi1 and EZH2 is associated with the progression and aggressive biological behavior of hepatocellular carcinoma. Lab Invest. 2008;88:873–882. doi: 10.1038/labinvest.2008.52. [DOI] [PubMed] [Google Scholar]
- 12.Sauvageau M, Sauvageau G. Polycomb group proteins: multi-faceted regulators of somatic stem cells and cancer. Cell Stem Cell. 2010;7:299–313. doi: 10.1016/j.stem.2010.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Blumenberg M, Gao S, Dickman K, Grollman AP, Bottinger EP, Zavadil J. Chromatin structure regulation in transforming growth factor-beta-directed epithelial-mesenchymal transition. Cells Tissues Organs. 2007;185:162–174. doi: 10.1159/000101317. [DOI] [PubMed] [Google Scholar]
- 14.Shastry BS. SNP alleles in human disease and evolution. J Hum Genet. 2002;47:561–566. doi: 10.1007/s100380200086. [DOI] [PubMed] [Google Scholar]
- 15.Kwok PY, Chen X. Detection of single nucleotide polymorphisms. Curr Issues Mol Biol. 2003;5:43–60. [PubMed] [Google Scholar]
- 16.Wang DG, Fan JB, Siao CJ, Berno A, Young P, Sapolsky R, Ghandour G, Perkins N, Winchester E, Spencer J, Kruglyak L, Stein L, Hsie L, Topaloglou T, Hubbell E, Robinson E, Mittmann M, Morris MS, Shen N, Kilburn D, Rioux J, Nusbaum C, Rozen S, Hudson TJ, Lipshutz R, Chee M, Lander ES. Large-scale identification, mapping, and genotyping of single-nucleotide polymorphisms in the human genome. Science. 1998;280:1077–1082. doi: 10.1126/science.280.5366.1077. [DOI] [PubMed] [Google Scholar]
- 17.Collins FS, Patrinos A, Jordan E, Chakravarti A, Gesteland R, Walters L. New goals for the U. S. Human Genome Project: 1998-2003. Science. 1998;282:682–689. doi: 10.1126/science.282.5389.682. [DOI] [PubMed] [Google Scholar]
- 18.Kulis M, Queiros AC, Beekman R, Martin-Subero JI. Intragenic DNA methylation in transcriptional regulation, normal differentiation and cancer. Biochim Biophys Acta. 2013;1829:1161–1174. doi: 10.1016/j.bbagrm.2013.08.001. [DOI] [PubMed] [Google Scholar]
- 19.Kulis M, Esteller M. DNA methylation and cancer. Adv Genet. 2010;70:27–56. doi: 10.1016/B978-0-12-380866-0.60002-2. [DOI] [PubMed] [Google Scholar]
- 20.Zhao L, Yu Y, Wu J, Bai J, Zhao Y, Li C, Sun W, Wang X. Role of EZH2 in oral squamous cell carcinoma carcinogenesis. Gene. 2014;537:197–202. doi: 10.1016/j.gene.2014.01.006. [DOI] [PubMed] [Google Scholar]
- 21.Kidani K, Osaki M, Tamura T, Yamaga K, Shomori K, Ryoke K, Ito H. High expression of EZH2 is associated with tumor proliferation and prognosis in human oral squamous cell carcinomas. Oral Oncol. 2009;45:39–46. doi: 10.1016/j.oraloncology.2008.03.016. [DOI] [PubMed] [Google Scholar]
- 22.Lindstrom S, Schumacher FR, Cox D, Travis RC, Albanes D, Allen NE, Andriole G, Berndt SI, Boeing H, Bueno-de-Mesquita HB, Crawford ED, Diver WR, Gaziano JM, Giles GG, Giovannucci E, Gonzalez CA, Henderson B, Hunter DJ, Johansson M, Kolonel LN, Ma J, Le Marchand L, Pala V, Stampfer M, Stram DO, Thun MJ, Tjonneland A, Trichopoulos D, Virtamo J, Weinstein SJ, Willett WC, Yeager M, Hayes RB, Severi G, Haiman CA, Chanock SJ, Kraft P. Common genetic variants in prostate cancer risk prediction-results from the NCI Breast and Prostate Cancer Cohort Consortium (BPC3) Cancer Epidemiol Biomarkers Prev. 2012;21:437–444. doi: 10.1158/1055-9965.EPI-11-1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wacholder S, Hartge P, Prentice R, Garcia-Closas M, Feigelson HS, Diver WR, Thun MJ, Cox DG, Hankinson SE, Kraft P, Rosner B, Berg CD, Brinton LA, Lissowska J, Sherman ME, Chlebowski R, Kooperberg C, Jackson RD, Buckman DW, Hui P, Pfeiffer R, Jacobs KB, Thomas GD, Hoover RN, Gail MH, Chanock SJ, Hunter DJ. Performance of common genetic variants in breast-cancer risk models. N Engl J Med. 2010;362:986–993. doi: 10.1056/NEJMoa0907727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Crea F, Fornaro L, Paolicchi E, Masi G, Frumento P, Loupakis F, Salvatore L, Cremolini C, Schirripa M, Graziano F, Ronzoni M, Ricci V, Farrar WL, Falcone A, Danesi R. An EZH2 polymorphism is associated with clinical outcome in metastatic colorectal cancer patients. Ann Oncol. 2012;23:1207–1213. doi: 10.1093/annonc/mdr387. [DOI] [PubMed] [Google Scholar]
- 25.Yoon KA, Gil HJ, Han J, Park J, Lee JS. Genetic polymorphisms in the polycomb group gene EZH2 and the risk of lung cancer. J Thorac Oncol. 2010;5:10–16. doi: 10.1097/JTO.0b013e3181c422d9. [DOI] [PubMed] [Google Scholar]
- 26.Subramaniam D, Thombre R, Dhar A, Anant S. DNA methyltransferases: a novel target for prevention and therapy. Front Oncol. 2014;4:80. doi: 10.3389/fonc.2014.00080. [DOI] [PMC free article] [PubMed] [Google Scholar]