Abstract
Loss of hyaline chondrocyte phenotype during the monolayer culture in vitro is a major obstacle for cell-based articular cartilage repair. Increasing evidence implicates an important role of the epigenetic regulation in maintaining the chondrocyte phenotype. DNA methylation, histone modifications and microRNAs have all been shown to contribute to chondrocyte dedifferentiation and hypertrophy. Moreover, the interplay among epigenetic regulators forms a complicated epigenetic network in regulating chondrocyte dedifferentiation. This review provides a detailed overview of the epigenetic regulation in maintaining the chondrocyte phenotype for chondrocyte-based cartilage repair.
Keywords: DNA methylation, histone acetylation, microRNAs, chondrocyte phenotype, cartilage repair
Introduction
Articular cartilage has very poor regeneration ability. Once damaged, it is very difficult for the cartilage to get repaired by itself. Traditional therapies (e.g. micro-fracture) have limited capacity to treat cartilage defects and the long-term outcome is often unsatisfactory [1]. Since 1994, autologous chondrocyte implantation (ACI) was established to treat cartilage defects [2], it has attracted much attention and become a golden therapy to repair focal defects except in the case of osteoarthritis (OA) or other complications [3]. However, the efficacy of ACI in repairing cartilage defects is unsatisfactory due to its variability in forming the desirable hyaline cartilage [4,5]. Monolayer expansion in vitro is a crucial step for the treatment of cartilage defects with ACI. One of the major obstacles accompanying with the monolayer culture is the loss of hyaline chondrocyte phenotype, leading to the chondrocyte dedifferentiation or chondrocyte hypertrophy, and the production of inferior matrix unsuitable for ACI [3]. Epigenetics plays an essential role in the maintenance of articular cartilage and has been implicated in the degenerative articular cartilage of OA that is characterized by the excessive chondrocyte dedifferentiation and hypertrophy [6-9]. Several lines of evidence have suggested the role of epigenetic modification in regulating chondrocyte phenotype. A detailed overview of the epigenetic regulation in gene expression during the chondrocyte dedifferentiation and hypertrophy may present a new perspective on the maintenance of chondrocyte phenotype.
In this review, we first summarize several key genes during the chondrocyte differentiation and cartilage matrix homeostasis. Secondly, we review several epigenetic mechanisms including DNA methylation, histone modifications and microRNAs (miRNAs) involved in the chondrocyte dedifferentiation and hypertrophy during in vitro chondrocyte expansion and passage. Thirdly, we also discuss recent advances in epigenetic research on cartilage defect repair including 5-hydroxymethylcytosine (5-hmC) and long non-coding RNAs (lncRNAs). Finally, we highlight the interactions between epigenetic regulators in the maintenance of chondrocyte phenotype.
Key genes involved in the chondrocyte differentiation and cartilage homeostasis
Accumulating evidence has shown that epigenetics regulates the gene expression during the chondrocyte proliferation, differentiation and function. Derived from mesenchymal stem cells (MSCs), chondrocytes migrate and are sparsely distributed in the articular cartilage. The chondrocyte is the only cell type responsible for the remodeling of the cartilage extracellular matrix (ECM). The cartilage ECM contains collagen proteins such as type II, XI, IX collagen (COL-2, COL-11, COL-9), and non-collagen proteins such as hyaluronan, aggrecan (ACAN), cartilage link protein 1 (CRTL-1) (reviewed in Table 1). Catabolic genes such as matrix metalloproteinase-13 (MMP-13) and a disintegrin and metalloproteinase with thrombospondin motifs (ADAMTs) maintain the low turnover of cartilage matrix. In turn, cartilage ECM influences the nature of chondrocyte phenotype during the cartilage matrix remodeling [10]. Once the cartilage remodeling is disturbed, hyaline chondrocytes will dedifferentiate into fibroblast-like cells or undergo hypertrophy-like changes, and eventually result in apoptosis.
Table 1.
Related genes in the chondrocyte differentiation
| Marker | Function | References |
|---|---|---|
| COL-2A1, COL-11A1 | Cartilage-specific ECM | [22] |
| COL-9A1 | Regulating the integrity and stability of articular cartilage | [36,76] |
| COL-12A1 | Providing a microenvironment that supports hyaline cartilage formation | [77] |
| COL-1A1, COL-3A1 | Collagens associated with dedifferentiation | [22] |
| COL-10A1 | A hypertrophic marker | [78] |
| ALPL | A transcriptional factor regulating hypertrophy | [78] |
| RUNX-2 | A transcriptional factor promoting chondrocyte hypertrophy | [19,78,79] |
| IHH | A transcriptional factor regulating chondrocyte hypertrophy | |
| COX-2 | A transcriptional factor promoting chondrocyte hypertrophy | |
| PTHR | Inhibiting chondrocyte hypertrophy | [80-82] |
| SOX-9 | A critical factor for chondrocyte differentiation | [83,84] |
| CRTL-1, MAGP2 | The most differentially expressed transcripts between chondrocyte and synovial cell cultures | [85] |
| CD44 | Hyaluronan receptor | [86,87] |
| ACAN | A chondrogenic marker | [22] |
| VCAN | A large droitin sulfate proteoglycan expressed by fibroblasts | [23,24] |
| MMP-13 | COL-2-degrading enzyme | [79] |
| ADAMTs | Aggrecan-degrading enzymes | [88,89] |
| Matrilin-3 | Inhibiting chondrocyte hypertrophy; Essential for cartilage matrix stability | [76,90-92] |
| COMP | A non-collagenous glycoprotein of the extracellular matrix, which enhancing cartilage ECM organization and assembly | [83,93-95] |
| MGP | Mineralization inhibitory protein | [96-98] |
| HDAC | Inhibiting chondrocyte dedifferentiation and hypertrophy | [42,43] |
Along with the articular chondrocyte expansion in the monolayer culture, the expression of the sex determining region Y box gene 9 (SOX-9) and the regenerative ability of articular chondrocytes to cartilage tissue are decreased [11,12]. SOX-9 is one of the key transcriptional factors that maintain the chondrocyte phenotype and cartilage homeostasis [13,14]. SOX-9 activates the transcription of COL-9A1, which is important for the integrity and stability of articular cartilage [15]. However, the SOX-9 activity negatively regulates the MMP-13 expression [16-18]. MMP-13 is the specific collagenase that is responsible for the degradation of COL-2 and the increased expression of MMP-13 is a notable character of chondrocyte hypertrophy [19]. Interestingly, the impact of SOX-9 on the promoter activity of COL-2A1 depends on the phenotype of chondrocytes and culture conditions. In a 2-dimensional (2-D) culture system, low-level of the SOX-9 expression promotes the COL-2A1 gene transcription, while the elevated SOX9 expression inhibits the COL-2A1 gene expression in both fully differentiated and slightly phenotypically altered chondrocytes. By contrast, in the advanced stages of dedifferentiation, SOX-9, independently of its expression level, depresses the COL-2A1 transcriptional activity [20]. Under 3-D culture conditions, the overexpression of SOX-9 promotes the dedifferentiated articular chondrocytes to regain a chondrocyte phenotype and the COL-2A1 gene expression to form a cartilaginous matrix [11]. Kim et al. reported that increased DNA methylation and decreased acetylation in the SOX-9 gene promoter region resulted in the loss of phenotypes [21]. Therefore, the epigenetic modification may be involved in the differential roles of SOX-9 in the COL-2A1 transcription.
During the monolayer culture in vitro, hyaline chondrocytes are prone to lose their phenotype and dedifferentiate into fibro-chondrocytes, which are characterized by the increased expressions of COL-1, COL-3, and versican [22-24].
During the prolonged in vitro culture, a portion of articular chondrocytes may undergo the hypertrophic transformation [25,26]. A negative feedback loop between IHH and PTHrP has been reported to be responsible for the early chondrocyte differentiation and the initiation of the hypertrophic differentiation [27]. Indian hedgehog (IHH) contributes to the chondrocyte hypertrophy [19], while the parathyroid hormone (PTH)-related peptide (PTHrP) signaling pathway can inhibit this process [28,29]. A very recent study has reported that IHH can induce the runt-related transcription factor 2 (RUNX-2) and COL-10 expression in chondrocytes [30]. Bone morphogenetic protein 2 (BMP-2) is shown to be a potent inducer of chondrogenesis. BMP-2 in combination with IHH could induce the chondrogenesis from human primary MSCs without the chondrocyte hypertrophy induction. However, BMP-2 treatment alone could induce the chondrocyte hypertrophy in vitro and in vivo [31]. Moreover, IHH gene transfer is sufficient to improve the cartilage repair quality in vivo, whereas BMP-2 treatment may increase the risk of the intra-lesional bone formation [32]. Therefore, the combinational and individual application of BMP-2 and IHH lead to different phenotype changes of chondrocytes. However, the mechanisms underlying these dramatic changes need further investigation.
Recently, the role of nuclear factor of activated T-cells (NFAT) in chondrocyte phenotype has been studied. NFAT-1 inhibits the chondrocyte hypertrophy and maintains the metabolic balance in articular cartilage [33]. NFAT-1 deficiency in mice leads to the loss of COL-2 and ACAN, meanwhile, the up-regulation of specific matrix-degrading proteinases in young adult articular cartilage of load-bearing joints [34]. NFAT-1 has been shown to affect the function of adult articular chondrocyte via the induction of histone methylation [35].
Epigenetic regulation in the chondrocyte differentiation and the cartilage homeostasis
DNA methylation
DNA methylation is the most stable epigenetic modification that involves the addition of a methyl group to the 5′ carbon of a cytosine base pair and occurs most often in a CG dinucleotide (CG site). DNA methyltransferase (DNA MTase) family including DNMT1, DNMT2, DNMT3A, DNMT3B, and DNMT3L are involved in the DNA methylation modification. Pathological loss of DNA methylation may result in aberrant gene induction, whereas increased methylation may silence normally expressed genes [3]. Results of our study showed that DNMT3A expression level was increased in chondrocytes undergoing dedifferentiation following serial passage in monolayer culture. DNA methylation inhibitor 5-azacytidine (5-AzaC) efficiently inhibited the chondrocyte dedifferentiation and decreased the DNMT3A expression level (Figure 1). These data suggest that DNMT3A, but not DNMT3B, may be involved in regulating the chondrocyte dedifferentiation. We are currently investigating the differential role of DNMT3A and DNMT3B in regulating chondrocyte phenotype.
Figure 1.
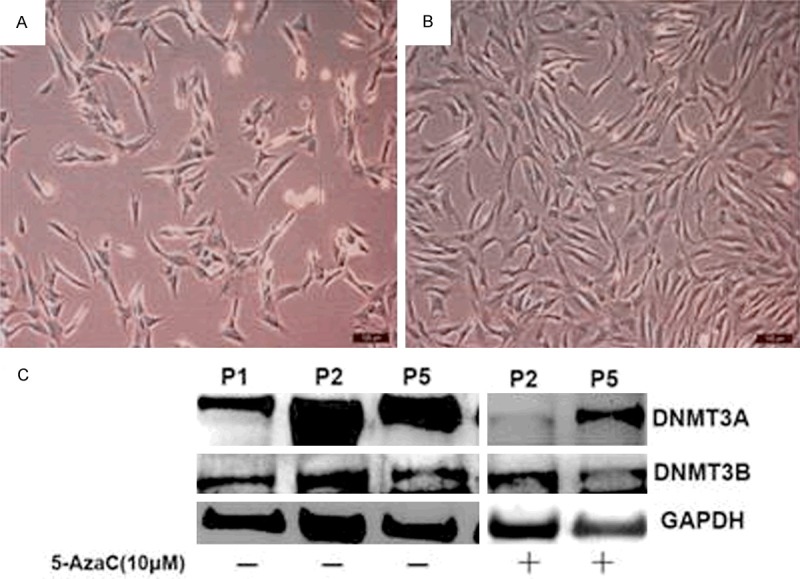
A crucial role of DNMT3A in the chondrocyte dedifferentiation. Chondrocyte dedifferentiation always accompanied with the monolayer chondrocyte expansion in vitro. After the treatment of 5-AzaC, the cells show a typical polygonal shape (A). The dedifferentiated chondrocytes have a fibroblast-like phenotype (B). With chondrocyte passaging (from P1 to P5), the DNMT3A expression level was increased. With 5-AzaC treatment, the DNMT3A expression level was decreased while no changes of DNMT3B were observed (C).
In vitro and in vivo experiments have demonstrated that COL-9A1 gene expression levels are modulated by DNA methylation in chondrocytes. Six CpG sites of the COL-9A1 promoter have been identified to be significantly hypermethylated in chondrocytes isolated from diseased OA cartilage, leading to a significant decrease (6,000-fold) in the COL-9A1 mRNA expression. The presence of 5-AzaC prevents passaging-induced hypermethylation and up-regulates COL-9A1 gene expression. A further mechanism study indicates that DNA methylation inhibits the activity of COL-9A1 promoter via attenuating the binding of transcriptional factor SOX-9 to the COL-9A1 promoter [36].
MMP-13, a catabolic gene of cartilage matrix, has been shown to be epigenetically regulated by DNA methylation [37,38]. The depletion of DNMT1 and DNMT3A triggers a significant increase in the MMP-13 expression. On the contrary, DNMT3B depletion decreases MMP-13 mRNA in human articular chondrocytes. The -104 CpG site is found to be critical for the regulation of MMP-13 by DNA methylation because its inhibition by 5-AzaC significantly increases MMP-13 expression in human articular chondrocytes. However, CpG methylation is not engaged in the regulation of ACAN promoter activity [39].
The BMP-2 expression is suppressed during the process of dedifferentiation in human articular chondrocytes. When treated with 5-AzaC, the BMP-2 expression is up-regulated and the process of chondrocyte dedifferentiation is decelerated [3], implicating an important role of DNA methylation in the BMP-2 expression in maintaining chondrocyte phenotype. However, the underlying molecular mechanisms remain to be investigated. In a previous study, DNA methylation was shown to have no effect on the COL-2 gene expression during the chicken chondrocyte differentiation and dedifferentiation [40]. Ma et al. demonstrated that 5-AzaC had no influence on the COL-2A1 expression in human chondrocytes, suggesting that DNA methylation is unlikely to be implicated in the COL-2A1 down-regulation in chondrocyte dedifferentiation [3].
Histone deacetylation
Acetylation and deacetylation are two major modifications of nucleosomal histones. In comparison to acetylation, deacetylation has been better characterized. There are two types of deacetylases involved in histone deacetylation: histones deacetylases (HDACs) and sirtuins deacetylases. HDACs function through a zinc-catalyzed mechanism of deacetylation, while sirtuins deacetylases are nicotinamide adenine dinucleotide (NAD+) dependent [41]. The major function of HDACs is to remove acetyl groups from histones, which causes condensation of the chromatins and ultimately leads to transcriptional repression. HDAC activity in primary articular chondrocytes decreases during dedifferentiation induced by a serial monolayer culture of chondrocytes and the activity recovers during re-differentiation when cultured in a 3-D system. Inhibition of HDAC with trichostatin A or PXD101 is sufficient to block the COL-2 expression in primary chondrocytes. Also, HDAC inhibitor blocks the re-differentiation of dedifferentiated chondrocytes by suppressing the synthesis and accumulation of COL-2. The detailed mechanism behind the inhibitory effect is that HDAC inhibitor could increase the expression of WNT-5A, which is known to inhibit the COL-2 expression [42]. In addition, the induction of WNT-5A expression by HDAC inhibitors is associated with acetylation of the WNT-5A promoter. These results indicate that HDACs promote the COL-2 expression at least partially by suppressing the transcription of WNT-5A [43]. Recently, HDAC4 is reported to play a chondroprotective role under inflammatory conditions. In addition to decrease the expression of interleukin 1 beta (IL-1β), overexpression of HDAC4 also blocks IL-1β-induced activation of catabolic events in human OA chondrocytes. Moreover, HDAC4 inhibits the promoter activity of RUNX-2 and MMP-13 in a dose-dependent manner, and its down-regulation is associated with the up-regulation of RUNX-2 and other OA-related genes (MMP-13, IHH, and COL-10) in human OA cartilage [44].
Silent information regulator 2 (SIR2) is an NAD-dependent deacetylase involved in chromatin silencing, which comprises seven homologues of SIRT1-7. Down-regulation of SIRT1 expression induces cellular senescence in rabbit articular chondrocytes, which in turn promotes the OA progression. Using human chondrocytes derived from OA patients, SIRT1 is found to promote target gene expression by the recruitment of SOX-9 transcription factor to cartilage-specific gene promoters. Therefore, SIRT1 may be critical in maintaining chondrocyte phenotype. Additionally, SIRT1 signaling is involved in IL-1β-mediated chondrocyte dedifferentiation, in which both p38 kinase and extracellular regulated protein kinases (ERK) are activated to initiate SIRT1-ERK signaling [6].
Histone methylation
Besides histone acetylation, histone methylation has been extensively studied and is the best characterized modifications of nucleosomal histones. Methylation occurs on both lysine (K) and arginine (E) residues. In histone H3, different lysine residues (K4, K9, K27, K36 and K79) can be methylated. Unlike acetylation that is generally associated with transcriptional activation, histone lysine methylation is associated with either gene activation or repression, depending on the specific residue modified. Methylation of histone H3 lysine 4 (H3K4), H3K36 and H3K79 is generally associated with transcriptional gene activation, whereas methylation of H3K9 and H3K20 is associated with transcriptional gene silencing. For example, in human osteoarthritic chondrocytes, the H3K4-Methyl epigenome regulates IL-1-induced cyclooxygenase 2 (COX-2) and inducible nitric oxide synthase (iNOS) expression [45]. NFAT1 is regulated by histone methylation, in which H3K4me2 is found to promote the NFAT1 expression, whereas H3K9me2 inhibits the NFAT1 expression in chondrocytes from healthy human articular cartilage. Lysine-specific demethylase-1 (Lsd1) has an inhibitory effect on the H3K4me2 activity at the NFAT1 promoter in embryonic articular chondrocytes. In 6-month articular chondrocytes, Jumonji C (Jmjc)-containing histone demethylase-2a (Jhdm2a) inhibits H3K9me2 at the NFAT1 promoter [35]. The SOX-9 transcriptional activity is also regulated by histone demethylation. PHF2, a histone lysine demethylase, positively regulates the SOX9-mediated chondrocyte differentiation. AT-rich interactive domain 5b (Arid5b) inhibits the H3K9me2 expression and thus demethylates the target genes of SOX-9. Arid5b recruits PHF2 to the promoter region of SOX-9 target genes and stimulates the H3K9me2 activation [46].
ERG-associated protein with SET domain (ESET), a histone methyltransferase enzyme, plays a vital role in the chondrocyte hypertrophy. Deletion of the ESET gene accelerates the chondrocyte hypertrophy in both embryos and young animals. ESET interacts with HDAC4 could inhibit the activity of RUNX-2, a hypertrophy-promoting transcription factor. The underlying mechanism by which ESET represses RUNX-2-mediated gene transactivation is dependent on its H3K9 methyltransferase activity and histone deacetylase activity. In addition, knockout of ESET is associated with the repression of IHH gene in pre- and early hypertrophic chondrocytes [8,47]. Another research found that histone methyltransferase Set7/9 could inhibit SIRT1 and potentiate the euchromatin formation on the promoter site of COL-2A1, resulting in the morphology-dependent COL-2A1 gene transactivation [48].
MiRNAs
MiRNAs are small single-stranded non-coding RNA molecules (~21-22nt) that affect both mRNA expression and protein synthesis. It is estimated that one-third of all human genes are regulated by miRNAs [49]. It is generally accepted that the main function of miRNAs is to knock down gene expression, though it has been shown to activate transcription too [50].
MiRNAs emerges as a new class of gene expression regulators in chondrogenesis. Recent miRNA expression profiling studies have identified subsets of miRNAs that are up-regulated or down-regulated during the process of human articular chondrocyte dedifferentiation. Several miRNAs, including miR-221, miR-222, miR-143, miR-145, miR-146a, miR-548e, miR-342-5p and miR-1248 are up-regulated in dedifferentiated articular chondrocytes [51,52]. Potential targets of of miR-145 include SOX-9 and COL-2A1 [51]. MiR-222 expression is higher in the weight-bearing anterior medial condyle as compared with the posterior non-weight bearing medial condyle. Therefore, the loss of mechanical stimulating factors may be one of the reasons accounting for the dedifferentiation of chondrocytes when cultured in monolayer in vitro [52]. On the other hand, miR-140, miR-491-3p, let-7d and miR-26a are down-regulated during the chondrocyte dedifferentiation [52]. MiR-140 is abundantly and relatively specifically expressed in chondrocytes, and its expression is positively correlated to the expression of chondrocyte markers [53]. The small-GTPase RalA and ADAMTS-5 have been identified as the target genes of miR-140 [54,55]. Loss of miR-140 moderately accelerates the hypertrophic differentiation of chondrocytes, suggesting miR-140 as an inhibitor of chondrocyte hypertrophy.
MiR-125b is down-regulated in OA chondrocytes compared to normal chondrocytes. Overexpression of miR-125b in human OA chondrocytes reverses the effect of IL-1β on ADAMTS-4 [56]. Thus, miR-125b may play an important role in inhibiting the cartilage matrix degradation. Another miRNA, such as miR-21 is significantly elevated in OA patients, and its overexpression could attenuate the process of chondrogenesis. The levels of MMP-1, MMP-2, MMP-3, and MMP-9 are significantly increased when CH8 cells (a human articular chondrocyte cell line) are transfected with synthetic miR-21 precursor, and the inhibition of miR-21 increases levels of MMPs [57]. Moreover, miR-21 silences the expression of growth differentiation factor 5 (GDF5) target gene through promoting its mRNA decay.
Since the identification of differentially expressed microRNA in differentiated and dedifferentiated chondrocytes, it is an attractive possibility that manipulate the miRNA expression could provide new strategies for improving cartilage tissue engineering. A thorough understanding of miRNA expression profile in chondrocytes and the manipulation of individual miRNA via introducing miRNA precursor molecules or antagonists, may open a new window for developing therapeutic strategies to inhibit the chondrocyte dedifferentiation and enhance the ECM synthesis.
Recent findings in epigenetics involved in chondrocyte dedifferentiation
Recent studies have identified 5-hydroxymethylcytosine (5-hmC) as a new epigenetic marker widely distributed in all types of tissues with varying degrees of abundance [58]. 5-hmC is an intermediate derived from the ten-eleven translocation (TET) enzyme-mediated DNA demethylation [59]. The expression levels of 5-hmC and TET-1 activity in human chondrocytes are suppressed by IL-1β and tumor necrosis factor alpha (TNF-α). Increased 5-hmC expression levels or its activities have been shown to counteract the pro-inflammatory effect to regulate the chondrocyte dedifferentiation [60].
Emerging as a new field, the lncRNAs have attracted much attention. The lncRNAs have been tentatively defined as noncoding RNAs of more than 200 nucleotides in length and are characterized by their complexity and diversity of sequences as well as mechanisms of action [61]. Accumulating evidence has shown that altered lncRNAs expression can result in aberrant gene expression, which contributes to a variety of disease states and biological functions [62-64]. OA cartilage and OA chondrocytes express higher levels of cartilage injury-related lncRNA (lncRNA-CIR). LncRNA-CIR is a negative regulator for collagen and ACAN while server as a positive regulator for matrix-degrading enzymes, such as MMP-13 and ADAMTS-5 [65]. HOTTIP is a novel lncRNA that is significantly up-regulated and suppresses the cartilage integrity factor integrin-α1 expression in OA progression [66]. Another lncRNA, growth arrest-specific 5 (GAS5) negatively regulates the miR-21 expression and thus contributes to the pathogenesis of OA through regulating the cell survival [67]. Since lncRNAs exert their functions in chondrocytes, it will be interesting to identify more IncRNAs related to the chondrocyte phenotype, which may provide new targets for intervention in treating OA patients.
Interactions between different epigenetic regulations
Recent studies have suggested that epigenetic regulators interact with each other to regulate chondrocyte phenotype (Figures 2 and 3). In this review, the interactions among miRNAs, DNA methylation and histone deacetylation will be discussed.
Figure 2.
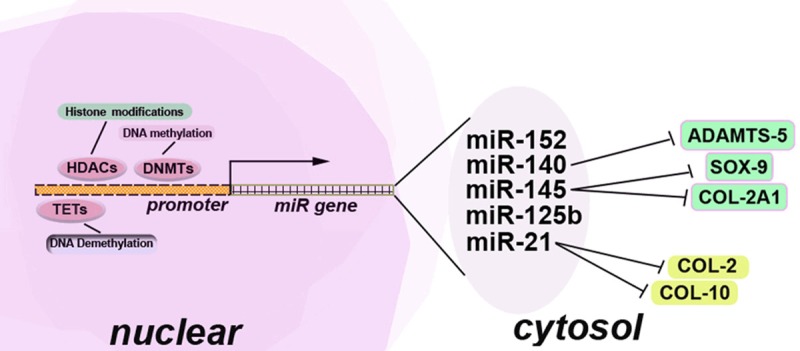
Epigenetics regulation of miRNA expression during the chondrocyte dedifferentiation. CpG islands are located on the binding sites where transcription factors actively regulate promoters of individual miRNAs. HDAC, MBDs and DNMTs compete with transcriptional factors to bind the promoters of miRNAs, which leads to the formation of compact inactive chromatin. In addition, DNA demethylation exerts its regulatory effect on the promoters via TETs. Under the regulation of HDAC, DNMTs, and TETs, miR-140 inhibits the catabolic genes of the cartilage matrix metabolism, such as ADAMTS-5. MiR-145 negatively regulates anabolic genes of the chondrocyte differentiation, including SOX-9 and COL-2A1. MiR-21 degrades the chondrocyte hypertrophy marker COL-10 and hyaline chondrocyte marker COL-2.
Figure 3.
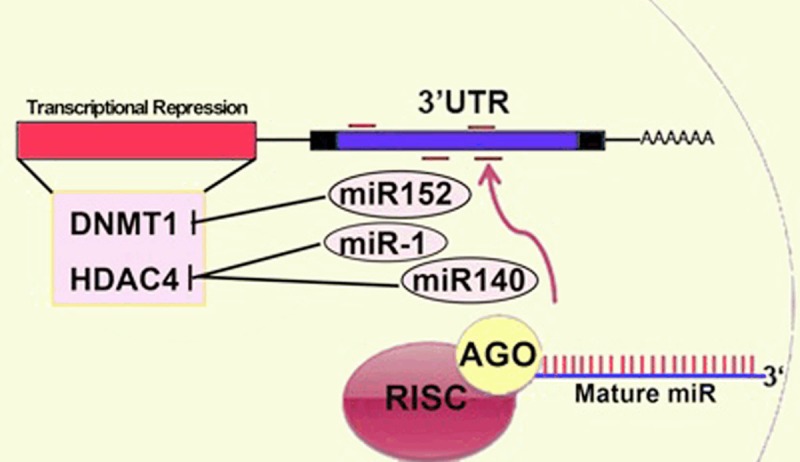
MiRNA in the regulation of DNA methylation and histone deacetylation during the chondrocyte dedifferentiation. Mature miRNAs regulate the DNMT and HDAC expression. MiR-152 directly targets DNMT1 by binding a recognition site located not in the usual 3’UTR but in the coding region. Both miR-1 and miR-140 directly modulate the HDAC4 expression.
The interplay between miRNAs and DNA methylation
Some miRNAs can affect the methylation pattern of the whole genome by direct target DNA methyltransferases to inhibit their activities. For instance, miRNA-152 modulates the canonical WNT pathway via targeting DNMT-1 in an arthritic rat model [68]. Conversely, DNA methylation can inhibit the transcription of miRNAs. The methylation of miRNAs in mammals was demonstrated by Yuan et al., in which they verified a tRNA methyltransferase NSun2 mediated methylation of the primary, precursor and mature miR-125b in vitro and in vivo [69]. NSun2 inhibits MiR-125b expression via inhibiting miR-125b maturation and RISC recruitment. Methyl CpG binding protein 2 (MeCP2) binds to methylated DNA and recruits the HDAC complex, and then regulates gene expression post-transcriptionally by suppressing nuclear miRNA processing [70].
The interactions between miRNAs and histone deacetylation
MiR-1 plays an important role in the regulation of chondrocyte phenotype during the development of growth plate via interacting with HDAC4 [71]. Evidence has demonstrated that miR-1 down-regulates HDAC4 by directly targeting its three prime untranslated region (3’-UTR), while inhibition of miR-1 attenuates its repression effect on HDAC4. Conversely, overexpression of HDAC4 inhibits the effect of miR-1 on the chondrocyte phenotype.
Recently, HDAC4 has been shown to be a target of miR-140. MiR-140 inhibits the mRNA translation via direct binding to the 3’-UTR of HDAC4 mRNA [72]. Another study has shown that, miR-140 reduces the myocyte-specific enhancer factor 2C expression, which results in enhancing the PTHrP-HDAC4 pathway to suppress the chondrocyte hypertrophy [73]. These results support an important role of the interactions between miRNAs and HDAC in maintaining the chondrocyte phenotype.
Future perspectives
The interactions between DNMT and histone deacetylase in many cell types suggest a possible crosstalk between these epigenetic regulators [74]. Given the important functions of epigenetic regulation, inhibition of DNA methylation could provide a promise approach in maintaining the chondrocyte phenotype, while site-specific promoter methylation or demethylation must be optimized. Epigenetic profiling and subsequent bioinformatics analysis may represent powerful strategies to identify region-specific DNA methylation that is critical for chondrocyte phenotype. In addition, recent studies have shown that RNA methylation is involved in the cell fate decision [75]. It is speculated that RNA methylation may play an important role in the chondrocyte phenotype maintenance. It may also open a new avenue to decelerate the chondrocyte dedifferentiation and hypertrophy. To design effective miRNA-based treatment modalities that have minimal or no off-target side effects, future work should be directed toward identifying key miRNA targets that functionally impact on the chondrocyte dedifferentiation and hypertrophy. More information regarding new epigenetic regulators for chondrocyte phenotype are needed to explore. With the increasing knowledge of the molecular mechanisms underlying the chondrocyte dedifferentiation and hypertrophy, better treatment strategies may be on the horizon to maintain the chondrocyte phenotype and simultaneously enhance the hyaline cartilage matrix to improve the outcome of ACI.
Acknowledgements
This study was financially supported by the following grants: Natural Science Foundation of China (No. 81572198; No. 81260161; No. 81000460); Natural Science Foundation of Guangdong Province, China (No. 2015A030313772; No. S2012010008129); China Postdoctoral Science Foundation Funded Project (No. 2013M530385); The Promotion Program for Shenzhen Key Laboratory, Shenzhen, China (No. ZDSY20120614154551201); Shenzhen Science and Technology Project (No. JSGG2014051905550503; No. GJHZ20130412153906739; No. CXZZ20120614160234842; No. JCYJ20140414170821160; No. JCYJ20140414170821200). We wish to thank Dr. Wenlan Liu for his valuable comments in improving the quality of the review article.
Disclosure of conflict of interest
None.
Abbreviations
- ACI
autologous chondrocyte implantation
- ACAN
aggrecan
- ADAMTs
a disintegrin and metalloproteinase with thrombospondin motifs
- Arid5b
AT-rich interactive domain 5b
- BMP-2
bone morphogenetic protein 2
- COL-1
type I collagen
- COL-2
type II collagen
- COX-2
cyclooxygenase 2
- CRTL-1
cartilage link protein 1
- DNMTs
DNA methyltransferases
- ECM
extracellular matrix
- ERK
extracellular regulated protein kinases
- ESET
an ERG-associated protein with a SET domain
- 5-AzaC
5-azacytidine
- 5-hmC
5-hydroxymethylcytosine
- HDACs
histones deacetylases
- H3K4
histone H3 lysine 4
- IHH
Indian hedgehog
- IL-1β
interleukin-1 beta
- iNOS
inducible nitric oxide synthase
- Jmjc
Jumonji C
- Jhdm2a
Jumonji C containing histone demethylase-2a
- LncRNAs
long noncoding RNAs
- LncRNA-CIR
cartilage injury-related lncRNA
- Lsd1
Lysine-specific demethylase-1
- MMP-13
matrix metalloproteinase-13
- MeCP2
methyl CpG binding protein 2
- miRNAs
microRNAs
- MSCs
mesenchymal stem cells
- NAD
nicotinamide adenine dinucleotide
- NFAT
nuclear factor of activated T-cells
- OA
osteoarthritis
- PTHrP
parathyroid hormone related peptide
- RUNX-2
runt-related transcription factor 2
- SIR2
silent information regulator 2
- SOX-9
sex determining region Y box gene 9
- 2-D
2-dimension
- TET
ten-eleven translocation
- 3’-UTR
three prime untranslated region
- TNF-α
tumor necrosis factor alpha
References
- 1.Mithoefer K, McAdams T, Williams RJ, Kreuz PC, Mandelbaum BR. Clinical efficacy of the microfracture technique for articular cartilage repair in the knee: an evidence-based systematic analysis. Am J Sports Med. 2009;37:2053–2063. doi: 10.1177/0363546508328414. [DOI] [PubMed] [Google Scholar]
- 2.Brittberg M, Lindahl A, Nilsson A, Ohlsson C, Isaksson O, Peterson L. Treatment of deep cartilage defects in the knee with autologous chondrocyte transplantation. N Engl J Med. 1994;331:889–895. doi: 10.1056/NEJM199410063311401. [DOI] [PubMed] [Google Scholar]
- 3.Ma B, Leijten JC, Wu L, Kip M, van Blitterswijk CA, Post JN, Karperien M. Gene expression profiling of dedifferentiated human articular chondrocytes in monolayer culture. Osteoarthritis Cartilage. 2013;21:599–603. doi: 10.1016/j.joca.2013.01.014. [DOI] [PubMed] [Google Scholar]
- 4.Bartlett W, Skinner JA, Gooding CR, Carrington RW, Flanagan AM, Briggs TW, Bentley G. Autologous chondrocyte implantation versus matrixinduced autologous chondrocyte implantation for osteochondral defects of the knee: a prospective, randomised study. J Bone Joint Surg Br. 2005;87:640–645. doi: 10.1302/0301-620X.87B5.15905. [DOI] [PubMed] [Google Scholar]
- 5.Hubka KM, Dahlin RL, Meretoja VV, Kasper FK, Mikos AG. Enhancing chondrogenic phenotype for cartilage tissue engineering: monoculture and coculture of articular chondrocytes and mesenchymal stem cells. Tissue Eng Part B Rev. 2014;20:641–654. doi: 10.1089/ten.teb.2014.0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hong EH, Yun HS, Kim J, Um HD, Lee KH, Kang CM, Lee SJ, Chun JS, Hwang SG. Nicotinamide phosphoribosyltransferase is essential for interleukin-1beta-mediated dedifferentiation of articular chondrocytes via SIRT1 and extracellular signal-regulated kinase (ERK) complex signaling. J Biol Chem. 2011;286:28619–28631. doi: 10.1074/jbc.M111.219832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sassi N, Gadgadi N, Laadhar L, Allouche M, Mourali S, Zandieh-Doulabi B, Hamdoun M, Nulend JK, Makni S, Sellami S. Notch signaling is involved in human articular chondrocytes de-differentiation during osteoarthritis. J Recept Signal Transduct Res. 2014;34:48–57. doi: 10.3109/10799893.2013.856920. [DOI] [PubMed] [Google Scholar]
- 8.Lawson KA, Teteak CJ, Zou J, Hacquebord J, Ghatan A, Zielinska-Kwiatkowska A, Fernandes RJ, Chansky HA, Yang L. Mesenchyme-specific knockout of ESET histone methyltransferase causes ectopic hypertrophy and terminal differentiation of articular chondrocytes. J Biol Chem. 2013;288:32119–32125. doi: 10.1074/jbc.M113.473827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tchetina EV, Squires G, Poole AR. Increased type II collagen degradation and very early focal cartilage degeneration is associated with upregulation of chondrocyte differentiation related genes in early human articular cartilage lesions. J Rheumatol. 2005;32:876–886. [PubMed] [Google Scholar]
- 10.Houard X, Goldring MB, Berenbaum F. Homeostatic mechanisms in articular cartilage and role of inflammation in osteoarthritis. Curr Rheumatol Rep. 2013;15:375. doi: 10.1007/s11926-013-0375-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tew SR, Li Y, Pothacharoen P, Tweats LM, Hawkins RE, Hardingham TE. Retroviral transduction with SOX9 enhances re-expression of the chondrocyte phenotype in passaged osteoarthritic human articular chondrocytes. Osteoarthritis Cartilage. 2005;13:80–89. doi: 10.1016/j.joca.2004.10.011. [DOI] [PubMed] [Google Scholar]
- 12.Stokes DG, Liu G, Coimbra IB, Piera-Velazquez S, Crowl RM, Jimenez SA. Assessment of the gene expression profile of differentiated and dedifferentiated human fetal chondrocytes by microarray analysis. Arthritis Rheum. 2002;46:404–419. doi: 10.1002/art.10106. [DOI] [PubMed] [Google Scholar]
- 13.Ikeda T, Kamekura S, Mabuchi A, Kou I, Seki S, Takato T, Nakamura K, Kawaguchi H, Ikegawa S, Chung UI. The combination of SOX5, SOX6, and SOX9 (the SOX trio) provides signals sufficient for induction of permanent cartilage. Arthritis Rheum. 2004;50:3561–3573. doi: 10.1002/art.20611. [DOI] [PubMed] [Google Scholar]
- 14.Cucchiarini M, Thurn T, Weimer A, Kohn D, Terwilliger EF, Madry H. Restoration of the extracellular matrix in human osteoarthritic articular cartilage by overexpression of the transcription factor SOX9. Arthritis Rheum. 2007;56:158–167. doi: 10.1002/art.22299. [DOI] [PubMed] [Google Scholar]
- 15.Genzer MA, Bridgewater LC. A Col9a1 enhancer element activated by two interdependent SOX9 dimers. Nucleic Acids Res. 2007;35:1178–1186. doi: 10.1093/nar/gkm014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grigolo B, De Franceschi L, Roseti L, Cattini L, Facchini A. Down regulation of degenerative cartilage molecules in chondrocytes grown on a hyaluronan-based scaffold. Biomaterials. 2005;26:5668–5676. doi: 10.1016/j.biomaterials.2005.02.030. [DOI] [PubMed] [Google Scholar]
- 17.Orfanidou T, Iliopoulos D, Malizos KN, Tsezou A. Involvement of SOX-9 and FGF-23 in RUNX-2 regulation in osteoarthritic chondrocytes. J Cell Mol Med. 2009;13:3186–3194. doi: 10.1111/j.1582-4934.2009.00678.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Borzi RM, Olivotto E, Pagani S, Vitellozzi R, Neri S, Battistelli M, Falcieri E, Facchini A, Flamigni F, Penzo M, Platano D, Santi S, Facchini A, Marcu KB. Matrix metalloproteinase 13 loss associated with impaired extracellular matrix remodeling disrupts chondrocyte differentiation by concerted effects on multiple regulatory factors. Arthritis Rheum. 2010;62:2370–2381. doi: 10.1002/art.27512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wei F, Zhou J, Wei X, Zhang J, Fleming BC, Terek R, Pei M, Chen Q, Liu T, Wei L. Activation of Indian hedgehog promotes chondrocyte hypertrophy and upregulation of MMP-13 in human osteoarthritic cartilage. Osteoarthritis Cartilage. 2012;20:755–763. doi: 10.1016/j.joca.2012.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kypriotou M, Fossard-Demoor M, Chadjichristos C, Ghayor C, de Crombrugghe B, Pujol JP, Galera P. SOX9 exerts a bifunctional effect on type II collagen gene (COL2A1) expression in chondrocytes depending on the differentiation state. DNA Cell Biol. 2003;22:119–129. doi: 10.1089/104454903321515922. [DOI] [PubMed] [Google Scholar]
- 21.Kim KI, Park YS, Im GI. Changes in the epigenetic status of the SOX-9 promoter in human osteoarthritic cartilage. J Bone Miner Res. 2013;28:1050–1060. doi: 10.1002/jbmr.1843. [DOI] [PubMed] [Google Scholar]
- 22.Dehne T, Schenk R, Perka C, Morawietz L, Pruss A, Sittinger M, Kaps C, Ringe J. Gene expression profiling of primary human articular chondrocytes in high-density micromasses reveals patterns of recovery, maintenance, reand dedifferentiation. Gene. 2010;462:8–17. doi: 10.1016/j.gene.2010.04.006. [DOI] [PubMed] [Google Scholar]
- 23.Albrecht C, Tichy B, Nurnberger S, Zak L, Handl MJ, Marlovits S, Aldrian S. Influence of cryopreservation, cultivation time and patient’s age on gene expression in Hyalograft(R) C cartilage transplants. Int Orthop. 2013;37:2297–2303. doi: 10.1007/s00264-013-2009-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zimmermann DR, Ruoslahti E. Multiple domains of the large fibroblast proteoglycan, versican. EMBO J. 1989;8:2975–2981. doi: 10.1002/j.1460-2075.1989.tb08447.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pacifici M, Golden EB, Adams SL, Shapiro IM. Cell hypertrophy and type X collagen synthesis in cultured articular chondrocytes. Exp Cell Res. 1991;192:266–270. doi: 10.1016/0014-4827(91)90185-w. [DOI] [PubMed] [Google Scholar]
- 26.Wu L, Gonzalez S, Shah S, Kyupelyan L, Petrigliano FA, McAllister DR, Adams JS, Karperien M, Tuan TL, Benya PD, Evseenko D. Extracellular matrix domain formation as an indicator of chondrocyte dedifferentiation and hypertrophy. Tissue Eng Part C Methods. 2014;20:160–168. doi: 10.1089/ten.tec.2013.0056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vortkamp A, Lee K, Lanske B, Segre GV, Kronenberg HM, Tabin CJ. Regulation of rate of cartilage differentiation by Indian hedgehog and PTH-related protein. Science. 1996;273:613–622. doi: 10.1126/science.273.5275.613. [DOI] [PubMed] [Google Scholar]
- 28.Guo J, Chung UI, Yang D, Karsenty G, Bringhurst FR, Kronenberg HM. PTH/PTHrP receptor delays chondrocyte hypertrophy via both Runx2-dependent and -independent pathways. Dev Biol. 2006;292:116–128. doi: 10.1016/j.ydbio.2005.12.044. [DOI] [PubMed] [Google Scholar]
- 29.Harrington EK, Lunsford LE, Svoboda KK. Chondrocyte terminal differentiation, apoptosis, and type X collagen expression are downregulated by parathyroid hormone. Anat Rec A Discov Mol Cell Evol Biol. 2004;281:1286–1295. doi: 10.1002/ar.a.20129. [DOI] [PubMed] [Google Scholar]
- 30.Amano K, Densmore M, Nishimura R, Lanske B. Indian hedgehog signaling regulates transcription and expression of collagen type X via Runx2/Smads interactions. J Biol Chem. 2014;289:24898–24910. doi: 10.1074/jbc.M114.570507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Steinert AF, Weissenberger M, Kunz M, Gilbert F, Ghivizzani SC, Gobel S, Jakob F, Noth U, Rudert M. Indian hedgehog gene transfer is a chondrogenic inducer of human mesenchymal stem cells. Arthritis Res Ther. 2012;14:R168. doi: 10.1186/ar3921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sieker JT, Kunz M, Weissenberger M, Gilbert F, Frey S, Rudert M, Steinert AF. Direct bone morphogenetic protein 2 and Indian hedgehog gene transfer for articular cartilage repair using bone marrow coagulates. Osteoarthritis Cartilage. 2015;23:433–442. doi: 10.1016/j.joca.2014.11.008. [DOI] [PubMed] [Google Scholar]
- 33.Caldwell KL, Wang J. Cell-based articular cartilage repair: the link between development and regeneration. Osteoarthritis Cartilage. 2015;23:351–362. doi: 10.1016/j.joca.2014.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang J, Gardner BM, Lu Q, Rodova M, Woodbury BG, Yost JG, Roby KF, Pinson DM, Tawfik O, Anderson HC. Transcription factor Nfat1 deficiency causes osteoarthritis through dysfunction of adult articular chondrocytes. J Pathol. 2009;219:163–172. doi: 10.1002/path.2578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rodova M, Lu Q, Li Y, Woodbury BG, Crist JD, Gardner BM, Yost JG, Zhong XB, Anderson HC, Wang J. Nfat1 regulates adult articular chondrocyte function through its age-dependent expression mediated by epigenetic histone methylation. J Bone Miner Res. 2011;26:1974–1986. doi: 10.1002/jbmr.397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Imagawa K, de Andres MC, Hashimoto K, Itoi E, Otero M, Roach HI, Goldring MB, Oreffo RO. Association of reduced type IX collagen gene expression in human osteoarthritic chondrocytes with epigenetic silencing by DNA hypermethylation. Arthritis Rheumatol. 2014;66:3040–3051. doi: 10.1002/art.38774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Iliopoulos D, Malizos KN, Tsezou A. Epigenetic regulation of leptin affects MMP-13 expression in osteoarthritic chondrocytes: possible molecular target for osteoarthritis therapeutic intervention. Ann Rheum Dis. 2007;66:1616–1621. doi: 10.1136/ard.2007.069377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Roach HI, Yamada N, Cheung KS, Tilley S, Clarke NM, Oreffo RO, Kokubun S, Bronner F. Association between the abnormal expression of matrix-degrading enzymes by human osteoarthritic chondrocytes and demethylation of specific CpG sites in the promoter regions. Arthritis Rheum. 2005;52:3110–3124. doi: 10.1002/art.21300. [DOI] [PubMed] [Google Scholar]
- 39.Poschl E, Fidler A, Schmidt B, Kallipolitou A, Schmid E, Aigner T. DNA methylation is not likely to be responsible for aggrecan down regulation in aged or osteoarthritic cartilage. Ann Rheum Dis. 2005;64:477–480. doi: 10.1136/ard.2004.022509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fernandez MP, Young MF, Sobel ME. Methylation of type II and type I collagen genes in differentiated and dedifferentiated chondrocytes. J Biol Chem. 1985;260:2374–2378. [PubMed] [Google Scholar]
- 41.Inoue T, Hiratsuka M, Osaki M, Oshimura M. The molecular biology of mammalian SIRT proteins: SIRT2 in cell cycle regulation. Cell Cycle. 2007;6:1011–1018. doi: 10.4161/cc.6.9.4219. [DOI] [PubMed] [Google Scholar]
- 42.Ryu JH, Chun JS. Opposing roles of WNT-5A and WNT-11 in interleukin-1beta regulation of type II collagen expression in articular chondrocytes. J Biol Chem. 2006;281:22039–22047. doi: 10.1074/jbc.M601804200. [DOI] [PubMed] [Google Scholar]
- 43.Huh YH, Ryu JH, Chun JS. Regulation of type II collagen expression by histone deacetylase in articular chondrocytes. J Biol Chem. 2007;282:17123–17131. doi: 10.1074/jbc.M700599200. [DOI] [PubMed] [Google Scholar]
- 44.Cao K, Wei L, Zhang Z, Guo L, Zhang C, Li Y, Sun C, Sun X, Wang S, Li P, Wei X. Decreased histone deacetylase 4 is associated with human osteoarthritis cartilage degeneration by releasing histone deacetylase 4 inhibition of runt-related transcription factor-2 and increasing osteoarthritis-related genes: a novel mechanism of human osteoarthritis cartilage degeneration. Arthritis Res Ther. 2014;16:491. doi: 10.1186/s13075-014-0491-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.El Mansouri FE, Chabane N, Zayed N, Kapoor M, Benderdour M, Martel-Pelletier J, Pelletier JP, Duval N, Fahmi H. Contribution of H3K4 methylation by SET-1A to interleukin-1-induced cyclooxygenase 2 and inducible nitric oxide synthase expression in human osteoarthritis chondrocytes. Arthritis Rheum. 2011;63:168–179. doi: 10.1002/art.27762. [DOI] [PubMed] [Google Scholar]
- 46.Hata K, Takashima R, Amano K, Ono K, Nakanishi M, Yoshida M, Wakabayashi M, Matsuda A, Maeda Y, Suzuki Y, Sugano S, Whitson RH, Nishimura R, Yoneda T. Arid5b facilitates chondrogenesis by recruiting the histone demethylase Phf2 to Sox9-regulated genes. Nat Commun. 2013;4:2850. doi: 10.1038/ncomms3850. [DOI] [PubMed] [Google Scholar]
- 47.Yang L, Lawson KA, Teteak CJ, Zou J, Hacquebord J, Patterson D, Ghatan AC, Mei Q, Zielinska-Kwiatkowska A, Bain SD, Fernandes RJ, Chansky HA. ESET histone methyltransferase is essential to hypertrophic differentiation of growth plate chondrocytes and formation of epiphyseal plates. Dev Biol. 2013;380:99–110. doi: 10.1016/j.ydbio.2013.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Oppenheimer H, Kumar A, Meir H, Schwartz I, Zini A, Haze A, Kandel L, Mattan Y, Liebergall M, Dvir-Ginzberg M. Set7/9 impacts COL2A1 expression through binding and repression of SirT1 histone deacetylation. J Bone Miner Res. 2014;29:348–360. doi: 10.1002/jbmr.2052. [DOI] [PubMed] [Google Scholar]
- 49.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 50.Vasudevan S, Tong Y, Steitz JA. Switching from repression to activation: microRNAs can up-regulate translation. Science. 2007;318:1931–1934. doi: 10.1126/science.1149460. [DOI] [PubMed] [Google Scholar]
- 51.Karlsen TA, Shahdadfar A, Brinchmann JE. Human primary articular chondrocytes, chondroblasts-like cells, and dedifferentiated chondrocytes: differences in gene, microRNA, and protein expression and phenotype. Tissue Eng Part C Methods. 2011;17:219–227. doi: 10.1089/ten.TEC.2010.0200. [DOI] [PubMed] [Google Scholar]
- 52.Lin L, Shen Q, Zhang C, Chen L, Yu C. Assessment of the profiling microRNA expression of differentiated and dedifferentiated human adult articular chondrocytes. J Orthop Res. 2011;29:1578–1584. doi: 10.1002/jor.21423. [DOI] [PubMed] [Google Scholar]
- 53.Hong E, Reddi AH. Dedifferentiation and redifferentiation of articular chondrocytes from surface and middle zones: changes in microRNAs-221/-222, -140, and -143/145 expression. Tissue Eng Part A. 2013;19:1015–1022. doi: 10.1089/ten.TEA.2012.0055. [DOI] [PubMed] [Google Scholar]
- 54.Miyaki S, Sato T, Inoue A, Otsuki S, Ito Y, Yokoyama S, Kato Y, Takemoto F, Nakasa T, Yamashita S, Takada S, Lotz MK, Ueno-Kudo H, Asahara H. MicroRNA-140 plays dual roles in both cartilage development and homeostasis. Genes Dev. 2010;24:1173–1185. doi: 10.1101/gad.1915510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Karlsen TA, Jakobsen RB, Mikkelsen TS, Brinchmann JE. microRNA-140 targets RALA and regulates chondrogenic differentiation of human mesenchymal stem cells by translational enhancement of SOX9 and ACAN. Stem Cells Dev. 2014;23:290–304. doi: 10.1089/scd.2013.0209. [DOI] [PubMed] [Google Scholar]
- 56.Matsukawa T, Sakai T, Yonezawa T, Hiraiwa H, Hamada T, Nakashima M, Ono Y, Ishizuka S, Nakahara H, Lotz MK, Asahara H, Ishiguro N. MicroRNA-125b regulates the expression of aggrecanase-1 (ADAMTS-4) in human osteoarthritic chondrocytes. Arthritis Res Ther. 2013;15:R28. doi: 10.1186/ar4164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang Y, Jia J, Yang S, Liu X, Ye S, Tian H. MicroRNA-21 controls the development of osteoarthritis by targeting GDF-5 in chondrocytes. Exp Mol Med. 2014;46:e79. doi: 10.1038/emm.2013.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Li W, Liu M. Distribution of 5-hydroxymethylcytosine in different human tissues. J Nucleic Acids. 2011;2011:870726. doi: 10.4061/2011/870726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, Agarwal S, Iyer LM, Liu DR, Aravind L, Rao A. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324:930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Haseeb A, Makki MS, Haqqi TM. Modulation of ten-eleven translocation 1 (TET1), Isocitrate Dehydrogenase (IDH) expression, alpha-Ketoglutarate (alpha-KG), and DNA hydroxymethylation levels by interleukin-1beta in primary human chondrocytes. J Biol Chem. 2014;289:6877–6885. doi: 10.1074/jbc.M113.512269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yang F, Zhang L, Huo XS, Yuan JH, Xu D, Yuan SX, Zhu N, Zhou WP, Yang GS, Wang YZ, Shang JL, Gao CF, Zhang FR, Wang F, Sun SH. Long noncoding RNA high expression in hepatocellular carcinoma facilitates tumor growth through enhancer of zeste homolog 2 in humans. Hepatology. 2011;54:1679–1689. doi: 10.1002/hep.24563. [DOI] [PubMed] [Google Scholar]
- 62.Faghihi MA, Modarresi F, Khalil AM, Wood DE, Sahagan BG, Morgan TE, Finch CE, St Laurent G 3rd, Kenny PJ, Wahlestedt C. Expression of a noncoding RNA is elevated in Alzheimer’s disease and drives rapid feed-forward regulation of beta-secretase. Nat Med. 2008;14:723–730. doi: 10.1038/nm1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cui Z, Ren S, Lu J, Wang F, Xu W, Sun Y, Wei M, Chen J, Gao X, Xu C, Mao JH, Sun Y. The prostate cancer-up-regulated long noncoding RNA PlncRNA-1 modulates apoptosis and proliferation through reciprocal regulation of androgen receptor. Urol Oncol. 2013;31:1117–1123. doi: 10.1016/j.urolonc.2011.11.030. [DOI] [PubMed] [Google Scholar]
- 64.Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, Tsai MC, Hung T, Argani P, Rinn JL, Wang Y, Brzoska P, Kong B, Li R, West RB, van de Vijver MJ, Sukumar S, Chang HY. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464:1071–1076. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Liu Q, Zhang X, Dai L, Hu X, Zhu J, Li L, Zhou C, Ao Y. Long noncoding RNA related to cartilage injury promotes chondrocyte extracellular matrix degradation in osteoarthritis. Arthritis Rheumatol. 2014;66:969–978. doi: 10.1002/art.38309. [DOI] [PubMed] [Google Scholar]
- 66.Kim D, Song J, Han J, Kim Y, Chun CH, Jin EJ. Two non-coding RNAs, MicroRNA-101 and HOTTIP contribute cartilage integrity by epigenetic and homeotic regulation of integrin-alpha1. Cell Signal. 2013;25:2878–2887. doi: 10.1016/j.cellsig.2013.08.034. [DOI] [PubMed] [Google Scholar]
- 67.Song J, Ahn C, Chun CH, Jin EJ. A long non-coding RNA, GAS5, plays a critical role in the regulation of miR-21 during osteoarthritis. J Orthop Res. 2014;32:1628–1635. doi: 10.1002/jor.22718. [DOI] [PubMed] [Google Scholar]
- 68.Miao CG, Yang YY, He X, Huang C, Huang Y, Qin D, Du CL, Li J. MicroRNA-152 modulates the canonical Wnt pathway activation by targeting DNA methyltransferase 1 in arthritic rat model. Biochimie. 2014;106:149–156. doi: 10.1016/j.biochi.2014.08.016. [DOI] [PubMed] [Google Scholar]
- 69.Yuan S, Tang H, Xing J, Fan X, Cai X, Li Q, Han P, Luo Y, Zhang Z, Jiang B, Dou Y, Gorospe M, Wang W. Methylation by NSun2 represses the levels and function of microRNA 125b. Mol Cell Biol. 2014;34:3630–3641. doi: 10.1128/MCB.00243-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Cheng TL, Wang Z, Liao Q, Zhu Y, Zhou WH, Xu W, Qiu Z. MeCP2 suppresses nuclear microRNA processing and dendritic growth by regulating the DGCR8/Drosha complex. Dev Cell. 2014;28:547–560. doi: 10.1016/j.devcel.2014.01.032. [DOI] [PubMed] [Google Scholar]
- 71.Li P, Wei X, Guan Y, Chen Q, Zhao T, Sun C, Wei L. MicroRNA-1 regulates chondrocyte phenotype by repressing histone deacetylase 4 during growth plate development. FASEB J. 2014;28:3930–3941. doi: 10.1096/fj.13-249318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tuddenham L, Wheeler G, Ntounia-Fousara S, Waters J, Hajihosseini MK, Clark I, Dalmay T. The cartilage specific microRNA-140 targets histone deacetylase 4 in mouse cells. FEBS Lett. 2006;580:4214–4217. doi: 10.1016/j.febslet.2006.06.080. [DOI] [PubMed] [Google Scholar]
- 73.Papaioannou G, Mirzamohammadi F, Lisse TS, Nishimori S, Wein MN, Kobayashi T. MicroRNA-140 Provides Robustness to the Regulation of Hypertrophic Chondrocyte Differentiation by the PTHrP-HDAC4 Pathway. J Bone Miner Res. 2015;30:1044–1052. doi: 10.1002/jbmr.2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bachman KE, Rountree MR, Baylin SB. Dnmt3a and Dnmt3b are transcriptional repressors that exhibit unique localization properties to heterochromatin. J Biol Chem. 2001;276:32282–32287. doi: 10.1074/jbc.M104661200. [DOI] [PubMed] [Google Scholar]
- 75.Geula S, Moshitch-Moshkovitz S, Dominissini D, Mansour AA, Kol N, Salmon-Divon M, Hershkovitz V, Peer E, Mor N, Manor YS, Ben-Haim MS, Eyal E, Yunger S, Pinto Y, Jaitin DA, Viukov S, Rais Y, Krupalnik V, Chomsky E, Zerbib M, Maza I, Rechavi Y, Massarwa R, Hanna S, Amit I, Levanon EY, Amariglio N, Stern-Ginossar N, Novershtern N, Rechavi G, Hanna JH. Stem cells. m6A mRNA methylation facilitates resolution of naive pluripotency toward differentiation. Science. 2015;347:1002–1006. doi: 10.1126/science.1261417. [DOI] [PubMed] [Google Scholar]
- 76.Brachvogel B, Zaucke F, Dave K, Norris EL, Stermann J, Dayakli M, Koch M, Gorman JJ, Bateman JF, Wilson R. Comparative proteomic analysis of normal and collagen IX null mouse cartilage reveals altered extracellular matrix composition and novel components of the collagen IX interactome. J Biol Chem. 2013;288:13481–13492. doi: 10.1074/jbc.M112.444810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Taylor DW, Ahmed N, Parreno J, Lunstrum GP, Gross AE, Diamandis EP, Kandel RA. Collagen type XII and versican are present in the early stages of cartilage tissue formation by both redifferentating passaged and primary chondrocytes. Tissue Eng Part A. 2015;21:683–693. doi: 10.1089/ten.TEA.2014.0103. [DOI] [PubMed] [Google Scholar]
- 78.Gu J, Lu Y, Li F, Qiao L, Wang Q, Li N, Borgia JA, Deng Y, Lei G, Zheng Q. Identification and characterization of the novel Col10a1 regulatory mechanism during chondrocyte hypertrophic differentiation. Cell Death Dis. 2014;5:e1469. doi: 10.1038/cddis.2014.444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Tsukamoto I, Inoue S, Teramura T, Takehara T, Ohtani K, Akagi M. Activating types 1 and 2 angiotensin II receptors modulate the hypertrophic differentiation of chondrocytes. FEBS Open Bio. 2013;3:279–284. doi: 10.1016/j.fob.2013.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Razzaque MS, Soegiarto DW, Chang D, Long F, Lanske B. Conditional deletion of Indian hedgehog from collagen type 2alpha1-expressing cells results in abnormal endochondral bone formation. J Pathol. 2005;207:453–461. doi: 10.1002/path.1870. [DOI] [PubMed] [Google Scholar]
- 81.Bach FC, Rutten K, Hendriks K, Riemers FM, Cornelissen P, de Bruin A, Arkesteijn GJ, Wubbolts R, Horton WA, Penning LC, Tryfonidou MA. The paracrine feedback loop between vitamin D(3) (1,25(OH)(2)D(3)) and PTHrP in prehypertrophic chondrocytes. J Cell Physiol. 2014;229:1999–2014. doi: 10.1002/jcp.24658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Mak KK, Kronenberg HM, Chuang PT, Mackem S, Yang Y. Indian hedgehog signals independently of PTHrP to promote chondrocyte hypertrophy. Development. 2008;135:1947–1956. doi: 10.1242/dev.018044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Haleem-Smith H, Calderon R, Song Y, Tuan RS, Chen FH. Cartilage oligomeric matrix protein enhances matrix assembly during chondrogenesis of human mesenchymal stem cells. J Cell Biochem. 2012;113:1245–1252. doi: 10.1002/jcb.23455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Hino K, Saito A, Kido M, Kanemoto S, Asada R, Takai T, Cui M, Cui X, Imaizumi K. Master regulator for chondrogenesis, Sox9, regulates transcriptional activation of the endoplasmic reticulum stress transducer BBF2H7/CREB3L2 in chondrocytes. J Biol Chem. 2014;289:13810–13820. doi: 10.1074/jbc.M113.543322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Rapko S, Zhang M, Richards B, Hutto E, Dethlefsen S, Duguay S. Identification of the chondrocyte lineage using microfibril-associated glycoprotein-2, a novel marker that distinguishes chondrocytes from synovial cells. Tissue Eng Part C Methods. 2010;16:1367–1375. doi: 10.1089/ten.tec.2009.0772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ishida O, Tanaka Y, Morimoto I, Takigawa M, Eto S. Chondrocytes are regulated by cellular adhesion through CD44 and hyaluronic acid pathway. J Bone Miner Res. 1997;12:1657–1663. doi: 10.1359/jbmr.1997.12.10.1657. [DOI] [PubMed] [Google Scholar]
- 87.Knudson CB, Knudson W. Hyaluronan and CD44: modulators of chondrocyte metabolism. Clin Orthop Relat Res. 2004:S152–162. [PubMed] [Google Scholar]
- 88.Huang K, Wu LD. Suppression of aggrecanase: a novel protective mechanism of dehydroepiandrosterone in osteoarthritis? Mol Biol Rep. 2010;37:1241–1245. doi: 10.1007/s11033-009-9495-5. [DOI] [PubMed] [Google Scholar]
- 89.Xue J, Wang J, Liu Q, Luo A. Tumor necrosis factor-alpha induces ADAMTS-4 expression in human osteoarthritis chondrocytes. Mol Med Rep. 2013;8:1755–1760. doi: 10.3892/mmr.2013.1729. [DOI] [PubMed] [Google Scholar]
- 90.Pullig O, Weseloh G, Klatt AR, Wagener R, Swoboda B. Matrilin-3 in human articular cartilage: increased expression in osteoarthritis. Osteoarthritis Cartilage. 2002;10:253–263. doi: 10.1053/joca.2001.0508. [DOI] [PubMed] [Google Scholar]
- 91.Yang X, Trehan SK, Guan Y, Sun C, Moore DC, Jayasuriya CT, Chen Q. Matrilin-3 inhibits chondrocyte hypertrophy as a bone morphogenetic protein-2 antagonist. J Biol Chem. 2014;289:34768–34779. doi: 10.1074/jbc.M114.583104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Leighton MP, Nundlall S, Starborg T, Meadows RS, Suleman F, Knowles L, Wagener R, Thornton DJ, Kadler KE, Boot-Handford RP, Briggs MD. Decreased chondrocyte proliferation and dysregulated apoptosis in the cartilage growth plate are key features of a murine model of epiphyseal dysplasia caused by a matn3 mutation. Hum Mol Genet. 2007;16:1728–1741. doi: 10.1093/hmg/ddm121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Di Cesare PE, Carlson CS, Stolerman ES, Hauser N, Tulli H, Paulsson M. Increased degradation and altered tissue distribution of cartilage oligomeric matrix protein in human rheumatoid and osteoarthritic cartilage. J Orthop Res. 1996;14:946–955. doi: 10.1002/jor.1100140615. [DOI] [PubMed] [Google Scholar]
- 94.Di Cesare PE, Chen FS, Moergelin M, Carlson CS, Leslie MP, Perris R, Fang C. Matrix-matrix interaction of cartilage oligomeric matrix protein and fibronectin. Matrix Biol. 2002;21:461–470. doi: 10.1016/s0945-053x(02)00015-x. [DOI] [PubMed] [Google Scholar]
- 95.Pirog-Garcia KA, Meadows RS, Knowles L, Heinegard D, Thornton DJ, Kadler KE, Boot-Handford RP, Briggs MD. Reduced cell proliferation and increased apoptosis are significant pathological mechanisms in a murine model of mild pseudoachondroplasia resulting from a mutation in the C-terminal domain of COMP. Hum Mol Genet. 2007;16:2072–2088. doi: 10.1093/hmg/ddm155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Hale JE, Fraser JD, Price PA. The identification of matrix Gla protein in cartilage. J Biol Chem. 1988;263:5820–5824. [PubMed] [Google Scholar]
- 97.Wallin R, Schurgers LJ, Loeser RF. Biosynthesis of the vitamin K-dependent matrix Gla protein (MGP) in chondrocytes: a fetuin-MGP protein complex is assembled in vesicles shed from normal but not from osteoarthritic chondrocytes. Osteoarthritis Cartilage. 2010;18:1096–1103. doi: 10.1016/j.joca.2010.05.013. [DOI] [PubMed] [Google Scholar]
- 98.Yagami K, Suh JY, Enomoto-Iwamoto M, Koyama E, Abrams WR, Shapiro IM, Pacifici M, Iwamoto M. Matrix GLA protein is a developmental regulator of chondrocyte mineralization and, when constitutively expressed, blocks endochondral and intramembranous ossification in the limb. J Cell Biol. 1999;147:1097–1108. doi: 10.1083/jcb.147.5.1097. [DOI] [PMC free article] [PubMed] [Google Scholar]


