Abstract
MicroRNAs (miRNAs) have been widely reported, which play important roles in cancer development. CXCL2 acts as an oncogene, however, its regulation by miRNAs is not clear in hepatocellular carcinoma (HCC). In our research, it is aimed to study the role of CXCL2 in HCC and the regulation of its expression by miRNAs. Firstly, we found that CXCL2 was up-regulated in the blood of patients with HCC and cell lines compared with the normal controls. CXCL2 could enhance HCC cell proliferation and metastasis. miR-532-5p was predicted as a regulatory miRNA of CXCL2 in HCC, and negatively associated with CXCL2 in HCC samples. It was also verified that miR-532-5p inhibited cell proliferation and metastasis of HCC cells by inhibition CXCL2. Collectively, our findings suggested that miR-532-5p may function as a tumor suppressor in HCC by targeting CXCL2.
Keywords: CXCL2, HCC, miR-532-5p, cell proliferation, metastasis
Introduction
Hepatocellular carcinoma (HCC) is the major type of primary liver cancers and is one of the most deadly malignancies [1]. In the early stage of HCC, the tumor could be removed effectively and has a relative longer survival time, however, in the late stage of HCC, there exsits metastasis and has shorter suvival time [2]. So, it is very important to study the molecular mechanism of HCC and find new therapeutic strategies. CXCL2, belonging to the CXC chemokines, is produced by macrophages, endothelial cells, epithelial cells and tumor cells [3]. CXCL2 plays important roles in various biological progresses such as angiogenesis, inflammation, immune response and cancer biological behaviors [4-10]. Downregulation of CXCL2 expression could induce apoptosis, inhibit proliferation and downregulate several important metastasis-promoting factors like COX2, SPARC and EFEMP in colorectal cancer [11]. A report showed that miR181b down-regulates CXCL2 through a direct binding to its 3’-UTR in breast cancer [12].
MicroRNAs (miRNAs) are small noncoding RNAs that are approximately 20 nucleotides in length [13]. miRNAs can regulate the expression of many target genes and are associated with developmental processes of cancer [14]. Profiling of poorly differentiated solid tumors from multiple organs revealed that miRs are more cancer-specific than mRNAs [13]. miRNAs have been shown to be down-regulated or up-regulated in HCC [14,15]. miR-532-5p is under-expressed in solid tumors including ovarian cancer and cutaneous melanoma [15,16]. However, the role of miR-532-5p in liver cancer has no reports and needs futher research.
To this study, we investigated the potential roles of CXCL2 and miR-532-5p in HCC. CXCL2 was predicted as potential target genes of miR-532-5p and verified in HCC cells. We examined the expression of CXCL2 and miR-532-5p in human HCC cells, tissues and blood from the patients. Cell growth, cell-cycle distribution, colony formation, migration, and invasion were also examined in the HCC cells with miR-532-5p overexpression. The study demonstrates that miR-532-5p is a tumor suppressor by down-regulation of CXCL2 expression in the progression of HCC.
Materials and methods
Cell culture
HCC cell lines including HEL7702, HEL7404, SMMC7721, HepG2 and PG5 MHCC97H and Huh7 cell lines were obtained from ATCC. The cell lines were grown in normal culture conditions. Cells were maintained in the recommended medium such as DMEM or RPMI1640 (Invitrogen, Carlsbad, CA) supplemented with 10% fetal bovine serum (ThermoFisher Scientific, Pittsburgh, PA) and 1% antibiotic-antimycotic (Invitrogen, Carlsbad, CA). Cells were cultured at 37°C in a humidified atmosphere of 5% CO2.
MTT assay
The effect of CXCL2 or miR-532-5p on HCC cell proliferation was measured using an MTT assay. Briefly, 1000 cells from different groups were seeded in 96-well culture plates. At the same time of next day of 0, 1, 3 and 5, 10 ul MTT solution (5 mg/ml) was added to each well, and the cells were incubated for 4 h and then the supernatant was removed from each well. DMSO was added to each well to dissolve the colored formazan crystal and the absorbance value was measured at 490 nm.
Cell cycle assay
Cells were harvested by trypsinization, washed in ice-cold PBS, fixed in ice-cold 80% ethanol in PBS, centrifuged at 4°C and resuspended in chilled PBS. Bovine pancreatic RNAase (Sigma-Aldrich) was added at a final concentration of 2 μg/ml, incubated at 37°C for 30 min, then 20 μg/ml propidium iodide (Sigma-Aldrich) was added and incubated for 20 min at room temperature. In each group, 50,000 cells were analyzed by flow cytometry (FACSCalibur; BD Biosciences, San Jose, CA, USA).
Luciferase reporter assay
The luciferase reporter plasmids with the 3’UTR sequence of CXCL2 or the control were transfected into the liver cancer cells using Fugene HD tansfection reagent (Roche Applied Science, USA) according to the manufacturer’s instruction. After transfection for 48 h, luciferase activity was assayed by luciferase assay kit (Promega, USA) using a luminometer.
Cell migration and invasion assay
HepG2 and PG5 cells were transfected with either miR-532-5p or the control with or without CXCL2 siRNA and cells were collected 24 h later. The cells were washed with PBS and seeded on the up-chamber of a transwell. The membrane of the transwell were coated with matrigel (invasion) and without matrigel (migration). CXCL2 (10 ng/ml) was in the lower chamber as as a chemoattractant. After 12-24 h, migrated or invaded cells were fixed, dyed with 0.1% crystal violet and counted under a microscope.
Tumor samples
Tissue specimens (tumor, adjacent normal mucosa) from 48 patients with HCC were collected after informed written consent from all patients and verification of the samples by a pathologist, and immediately frozen in liquid nitrogen. Tissue screening and documentation process was approved by the ethics committee of Shandong Provincince Hospital Affliated to Shandong Uiniversty.
Statistical analysis
All the analysis of the data was performed using SPSS version 13.0 (SPSS). Spearman correlations among continuous variables were computed. Data were presented as the mean ± standard deviation (SD) from three separate experiments. When two groups were compared, the differences between groups were analyzed using Student’s t-test and when more than two groups were compared, a one-way analysis of variance (ANOVA) was used. In all tests, p values lower than 0.05 were considered significant.
Results
CXCL2 expression is higher in the blood of patients with liver cancer and cell lines
To investigate the role of CXCL2 in liver cancer, in the first, CXCL2 expression in the blood sample of liver cancer was examined by ELISA. The data indicated that CXCL2 was much higher in the samples from the liver cancer than the normal controls (Figure 1A). To confirm the result, we used various liver cancer cell lines to verify and found that CXCL2 protein in the medium from the liver cancer cells including HepG2, BEL-7405, PG5, SMMC7721, MHCC97H and BEL-7404 was much more than in the medium from the normal liver cells 7702 (Figure 1B).
Figure 1.
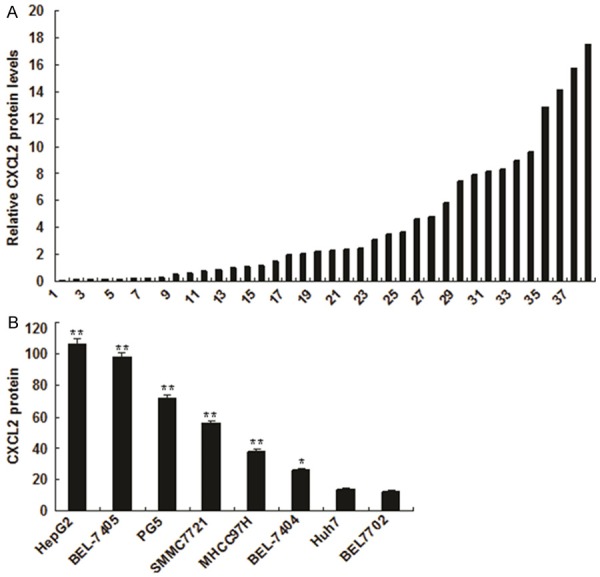
CXCL2 expression increased in liver cancer. A. CXCL2 expression in the blood of patients with liver cancer was higher than in the blood of patients with non-liver cancer. CXCL2 expression was analyzed by ELISA. B. CXCL2 expression in the medium from liver cancer cells was increased comparing with the normal control. CXCL2 expression was analyzed by ELISA. The experiments were done at least for three times. **p<0.01, *p<0.05.
CXCL2 promotes HCC cell proliferation and metastasis
Next, we investigated the celluar function of CXCL2 in liver cancer cells. HepG2 and PG5 cells were treated with CXCL2 (10 ng/ml) for different time, and cell proliferation was assayed by MTT method. The result showed that cell survival rate became higher in HepG2 and PG5 cells with CXCL2 treatment than the control (Figure 2A and 2B). We used cell cycle analysis to confirm the results and found that CXCL2 could lead to G1 arrest in HCC cells (Figure 2C and 2D). We tested whether CXCL2 involves in liver cancer cell metastasis. The results from migration assay showed that liver cancer cells with CXCL2 treatment had higher migration ability (Figure 2E), so did cell invasion (Figure 2F).
Figure 2.

CXCL2 promoted liver cancer cell proliferation and metastasis. A. Cell proliferation was analyzed by MTT assay in the HepG2 cells with CXCL2 treatment. B. Cell proliferation was analyzed by MTT assay in the PG5 cells with CXCL2 treatment. C. Cell cycle was analyzed by flow cytometry in the HepG2 cells with CXCL2 treatment. D. Cell cycle was analyzed by flow cytometry in the PG5 cells with CXCL2 treatment. E. Cell migration was analyzed by transwell system. In the low chamber, there was CXCL2. HepG2 or PG5 cells were seeded in the up chamber. F. Cell invasion was analyzed by transwell system. In the low chamber, there was CXCL2. HepG2 or PG5 cells were seeded in the up chamber. with CXCL2 treatment. The experiments were done at least for three times. **p<0.01, *p<0.05.
CXCL2 is a target gene of miR-532-5p in liver cancer cells
Bioinformatic analysis showed that CXCL2 might be regulated by miR-532-5p (Figure 3A). As shown in Figure 3B and 3C, the luciferase activities of wide typed CXCL2 in HepG2 and PG5 cells were much lower than in control cells. The luciferase activity of mutated CXCL2 was rescued in the two cell lines. We next examined whether miR-532-5p could regulate endogenous CXCL2 expression in the above two cell lines. Compared with the control, endogenous CXCL2 mRNA levels were down-regulated when cells were transfected with miR-532-5p (Figure 3D). The protein levels CXCL2 was reduced in the HepG2 and PG5 cells with miR-532-5p overexpression (Figure 3E).
Figure 3.

miR-532-5p downregulates CXCL2 expression in liver cancer cells. A. Sequence alignment of human CXCL2 3’UTR with miR-532-5p. B and C. miR-532-5p targeted the wild-type but not the mutant 3’UTR of CXCL2. Luciferase activity assay of indicated HepG2 and PG5 cells transfected with the pGL3-CXCL2-3’UTR reporter with miR-532-5p. D. Ectopic expression of miR-532-5p downregulated CXCL2 mRNA expression in HepG2 and PG5 cells as determined by real time RT-PCR. E. miR-532-5p decreased CXCL2 protein level in HepG2 and PG5 cells by ELISA. Error bars represent from three independent experiments. **p< 0.01. *p< 0.05.
Relationship between miR-532-5p and CXCL2 expression in liver cancer
To investigate the possible role of miR-532-5p in liver cancer, we first examined the expression of miR-532-5p in liver cancer specimens by real time RT-PCR. We examined the expression of miR-532-5p in liver cancer samples and their compared normal tissues. As shown in Figure 4A, the expression levels of miR-532-5p in liver cancer samples were lower than those in normal samples, so did in blood samples (Figure 4B). Similarly, miR-532-5p was lower in human liver cancer cells compared with the normal cells (Figure 4C). Figure 1 showed us that CXCL2 was up-regulated in liver cancer. We further analyzed the relationship between CXCL2 and miR-532-5p, and found that CXCL2 expression was negatively related to miR-532-5p expression in liver cancer tissue samples (Figure 4D).
Figure 4.

Relationship between miR-532-5p and CXCL2 expression in liver cancer. A. miR-532-5p expression in the blood of patients with liver cancer was lower than in the blood of patients with non-liver cancer. miR-532-5p expression was analyzed by real time RT-PCR. B. miR-532-5p expression in the samples from liver cancer cells decreased comparing with the normal control. miR-532-5p expression was analyzed by real time RT-PCR. C. miR-532-5p expression in liver cancer cell lines and nomal liver cells. miR-532-5p expression was analyzed by real time RT-PCR. D. Relationship between miR-532-5p and CXCL2 in liver cancer samples. Error bars represent from three independent experiments. **p< 0.01. *p< 0.05.
miR-532-5p inhibits proliferation and metastasis of liver cancer cells by targeting CXCL2
miR-532-5p down-regulates CXCL2 expression in liver cancer cells, which suggests that miR-532-5p may act as a tumor suppressor. To test it, HepG2 and PG5 cells were transfected with miR-532-5p and the tranfected effect was verified by real time RT-PCR (Figure 5A). Overexpression of miR-532-5p in HepG2 and PG5 cells led to increasing of cell proliferation by MTT assay (Figure 5B and 5C). We used another liver cancer cell line PG5 to confirm the results (Figure 5D and 5E). To further know the role of miR-532-5p in liver cancer metastasis, HepG2 and PG5 cells were transfected with miR-532-5p and it was found that miR-532-5p inhibited cell migration and invasiveness of HepG2 and PG5 cells (Figure 5F and 5G).
Figure 5.

miR-532-5p inhibits proliferation and metastasis of liver cancer cells by inhibition of CXCL2. A. HepG2 and PG5 cells were transfected with miR-532-5p or miRNA control and then miR-532-5p expression was analyzed by real time RT-PCR. B. Cell proliferation was analyzed by MTT assay in the HepG2 cells with miR-532-5p transfection and then treated with CXCL2. C. Cell proliferation was analyzed by MTT assay in the PG5 cells transfected with miR-532-5p and CXCL2 treatment. D. Cell migration was analyzed by transwell system. HepG2 or PG5 cells were transfected with miR-532-5p and seeded in the up chamber. E. Cell invasion was analyzed by transwell system. HepG2 or PG5 cells were transfected with miR-532-5p and seeded in the up chamber.
miR-532-5p inhibits liver cancer by suppressing CXCL2 actived signal pathways
We further explored the molecular mechanism of miR-532-5p inhibiting liver cancer by down-regulation of CXCL2. Cell proliferation and metastasis associated markers were detected by western blot and the results showed that the expression of PCNA, MMP9, SNAIL1, Vimentin, α-SMA decreased in the HepG2 and PG5 cells with LV-miR-532-5p. CXCL2 could activate STAT3 signal pathway in cancer, so did in the liver cancer cells HepG2 and PG5 (Figure 6A and 6B). The result indicated that miR-532-5p inhibited CXCL2 activated STAT3 signal pathway. Figure 6B was the summary of our research.
Figure 6.
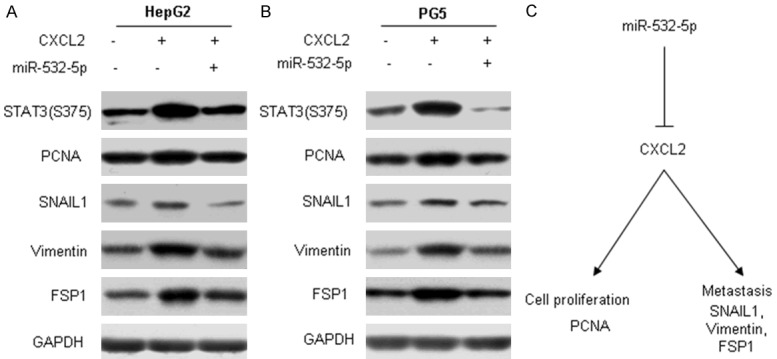
miR-532-5p inhibits liver cancer by suppressing CXCL2 actived signal pathways. A and B. HepG2 or PG5 cells were transfected with miR-532-5p and then treated with CXCL2. Protein was extracted for western blotting analysis. C. Summary of the study.
Discussion
From the member of chemokine family, CXCL2 play important roles like other chemokines. They are ubiquitously expressed and play important roles in a wide variety of cellular processes such as membrane receptor signaling, membrane trafficking, actin cytoskeleton reorganizations, cell adhesion and cell motility [12,13]. Recent reports showed that CXCL2 play important roles in development and progression of cancer [12,13]. CXCL2 was often found overexpressed in breast cancer, colorectal cancer, liver cancer and esophageal squamous cell carcinoma [15-17]. Overexpression of CXCL2 in esophageal squamous cell carcinoma cells could enhance cell proliferation. Knocking down CXCL2 suppressed the proliferation and tumorigenicity of breast cancer cells both in vitro and in vivo [12]. Our study showed the similar results that CXCL2 enhanced liver cancer cell proliferation, migration and invasion.
Reports showed that miR-532-5p expression is lower in tumor tissues as compared with adjacent normal tissues and down-regulated in the macrophage inflammatory response. miR-532-5p was also been detected in the serum. However, in some type of cancer, miR-532-5p expression is up-regulated [15-18]. In our work, we found that miR-532-5p expression in liver cell lines was much lower than it in the normal cells. Overexpression of miR-532-5p led to inhibition of cell growth, migration, invasion and stem-cell like properties in liver cancer cells. Further research demonstrated that miR-532-5p expression was reduced in most of the tested liver cancer tissues and the results was consistent with the cell lines. miR-532-5p is also induced in soft tissue sarcoma cells under the hypoxic environment [11], which suggests that miR-532-5p may be controlled by the tumor microenvironment.
miRNAs play important roles in HCC. There are many potential target genes of miR-532-5p in the cells. According to the results from the prediction, miR-532-5p has many potential target genes. In our research, CXCL2 was selected due to its high score which means there was high possibility to be regulated by miR-532-5p. Using dual luciferase system, CXCL2 was validated as the target gene of miR-532-5p. CXCL2 mRNA and protein were suppressed by miR-532-5p in liver cancer cells. CXCL2 expresssion is negatively correlated to miR-532-5p in liver cancer samples. These results clearly indicated that CXCL2 expression is regulated by miR-532-5p in liver cancer.
In a conclusion, our study found that miR-532-5p was identified as a tumor suppressor miRNA in liver cancer cells that can negatively regulate CXCL2 expression in vitro and in vivo. Taken together with clinical observations, our finding suggests that loss of miR-532-5p expression in liver cancer is a significant biomarker for metastasis and could be targets for the development of antimetastasis strategy in the treatment of liver cancer. But the molecular mechnism of low expression of miR-532-5p in liver cancer needs to further research.
Acknowledgements
The study was supported by the National Natural Science Foundation of China (No. 81000731 and 81401696) and Shandong province science and technology development program of China (No. 2012G0021844).
Disclosure of conflict of interest
None.
References
- 1.Govaere O, Roskams T. Pathogenesis and Prognosis of Hepatocellular Carcinoma at the Cellular and Molecular Levels. Clin Liver Dis. 2015;19:261–276. doi: 10.1016/j.cld.2015.01.002. [DOI] [PubMed] [Google Scholar]
- 2.Pelus LM, Fukuda S. Peripheral blood stem cell mobilization: the CXCR2 ligand GRObeta rapidly mobilizes hematopoietic stem cells with enhanced engraftment properties. Exp Hematol. 2006;34:1010–1020. doi: 10.1016/j.exphem.2006.04.004. [DOI] [PubMed] [Google Scholar]
- 3.Vansaun MN, Mendonsa AM, Lee Gorden D. Hepatocellular proliferation correlates with inflammatory cell and cytokine changes in a murine model of nonalchoholic fatty liver disease. PLoS One. 2013;8:e73054. doi: 10.1371/journal.pone.0073054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jablonska J, Wu CF, Andzinski L, Leschner S, Weiss S. CXCR2-mediated tumor-associated neutrophil recruitment is regulated by IFN-β. Int J Cancer. 2014;134:1346–1358. doi: 10.1002/ijc.28551. [DOI] [PubMed] [Google Scholar]
- 5.Killian PH, Kronski E, Michalik KM, Barbieri O, Astigiano S, Sommerhoff CP, Pfeffer U, Nerlich AG, Bachmeier BE. Curcumin inhibits prostate cancer metastasis in vivo by targeting the inflammatory cytokines CXCL1 and -2. Carcinogenesis. 2012;33:2507–2519. doi: 10.1093/carcin/bgs312. [DOI] [PubMed] [Google Scholar]
- 6.Shen J, Abel EL, Riggs PK, Repass J, Hensley SC, Schroeder LJ, Temple A, Chau A, McClellan SA, Rho O, Kiguchi K, Ward MD, Semmes OJ, Person MD, Angel JM, Digiovanni J. Proteomic and pathway analyses reveal a network of inflammatory genes associated with differences in skin tumor promotion susceptibility in DBA/2 and C57BL/6 mice. Carcinogenesis. 2012;33:2208–2219. doi: 10.1093/carcin/bgs213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Oue E, Lee JW, Sakamoto K, Iimura T, Aoki K, Kayamori K, Michi Y, Yamashiro M, Harada K, Amagasa T, Yamaguchi A. CXCL2 synthesized by oral squamous cell carcinoma is involved in cancer-associated bone destruction. Biochem Biophys Res Commun. 2012;424:456–461. doi: 10.1016/j.bbrc.2012.06.132. [DOI] [PubMed] [Google Scholar]
- 8.Gandhi V, Balakrishnan K. CCL2 in chronic lymphocytic leukemia: a macro in microenvironment? Leuk Lymphoma. 2012;53:1849–1850. doi: 10.3109/10428194.2012.688966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Burgess M, Cheung C, Chambers L, Ravindranath K, Minhas G, Knop L, Mollee P, McMillan NA, Gill D. CCL2 and CXCL2 enhance survival of primary chronic lymphocytic leukemia cells in vitro. Leuk Lymphoma. 2012;53:1988–1998. doi: 10.3109/10428194.2012.672735. [DOI] [PubMed] [Google Scholar]
- 10.Kronski E, Fiori ME, Barbieri O, Astigiano S, Mirisola V, Killian PH, Bruno A, Pagani A, Rovera F, Pfeffer U, Sommerhoff CP, Noonan DM, Nerlich AG, Fontana L, Bachmeier BE. miR181b is induced by the chemopreventive polyphenol curcumin and inhibits breast cancer metastasis via down-regulation of the inflammatory cytokines CXCL1 and -2. Mol Oncol. 2014;8:581–595. doi: 10.1016/j.molonc.2014.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shibata C, Otsuka M, Kishikawa T, Ohno M, Yoshikawa T, Takata A, Koike K. Diagnostic and therapeutic application of noncoding RNAs for hepatocellular carcinoma. World J Hepatol. 2015;7:1–6. doi: 10.4254/wjh.v7.i1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhu Z, Zhang X, Wang G, Zheng H. Role of MicroRNAs in Hepatocellular Carcinoma. Hepat Mon. 2014;14:e18672. doi: 10.5812/hepatmon.18672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.He H, Wang L, Zhou W, Zhang Z, Wang L, Xu S, Wang D, Dong J, Tang C, Tang H, Yi X, Ge J. MicroRNA Expression Profiling in Clear Cell Renal Cell Carcinoma: Identification and Functional Validation of Key miRNAs. PLoS One. 2015;10:e0125672. doi: 10.1371/journal.pone.0125672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee H, Park CS, Deftereos G, Morihara J, Stern JE, Hawes SE, Swisher E, Kiviat NB, Feng Q. MicroRNA expression in ovarian carcinoma and its correlation with clinicopathological features. World J Surg Oncol. 2012;10:174. doi: 10.1186/1477-7819-10-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mosakhani N, Räty R, Tyybäkinoja A, Karjalainen-Lindsberg ML, Elonen E, Knuutila S. MicroRNA profiling in chemoresistant and chemosensitive acute myeloid leukemia. Cytogenet Genome Res. 2013;141:272–6. doi: 10.1159/000351219. [DOI] [PubMed] [Google Scholar]
- 16.Kitago M, Martinez SR, Nakamura T, Sim MS, Hoon DS. Regulation of RUNX3 tumor suppressor gene expression in cutaneous melanoma. Clin Cancer Res. 2009;15:2988–2994. doi: 10.1158/1078-0432.CCR-08-3172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li P, Teng F, Gao F, Zhang M, Wu J, Zhang C. Identification of circulating microRNAs as potential biomarkers for detecting acute ischemic stroke. Cell Mol Neurobiol. 2015;35:433–447. doi: 10.1007/s10571-014-0139-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cheng Y, Kuang W, Hao Y, Zhang D, Lei M, Du L, Jiao H, Zhang X, Wang F. Downregulation of miR-27a* and miR-532-5p and upregulation of miR-146a and miR-155 in LPS-induced RAW264.7 macrophage cells. Inflammation. 2012;35:1308–13. doi: 10.1007/s10753-012-9443-8. [DOI] [PubMed] [Google Scholar]


