Abstract
The main pathological feature in isolated hereditary dentin disorders is the abnormality of dentin mineralization. Dentin sialophosphoprotein (DSPP) gene is the only identified causative gene for the disorders. The present study aims to explore the molecular association between Dspp mutations and the disrupted mineralization homeostasis during odontoblast differentiation. We generated lentivirus constructs with the mouse full-length wild type Dspp cDNA and 3 Dspp mutants and transfected them into mouse odontoblast-lineage cells (OLCs) which were then performed 21-day mineralization inducing differentiation. The formation of mineralized nodules was obviously fewer in mutants. Digital Gene Expression (DGE) showed that Dspp mutation affected the OLC differentiation in a degree. Further examination validated that Dspp (LV-Dspp) overexpressing OLCs possessed the ability to strictly orchestrate framework for mineralization inductors like Bmp2, Col1 and Runx2, and proliferative markers for mineralization like Alp and Ocn, as well as mineral homeostasis feedback regulators Mgp and Htra1. However, the missense mutation in Dspp signal peptide region (LV-M2) and the nonsense mutation (LV-M5) broke this orchestration. The results suggested that the mutant Dspp disrupt the dynamic homeostasis of mineralization during OLC differentiation. We are the first to use full-length mouse Dspp gene expression system to explore the mineralization mechanism by which inductors and inhibitors adjust each other during odontoblast differentiation. Our findings shed new light on association between Dspp and the dynamic homeostasis of mineralization inductors and inhibitors, and indicate the disruption of mineralization homeostasis might be a crucial reason for Dspp mutations resulting in dentin disorders.
Keywords: Dentin sialophosphoprotein, odontoblast-lineage cells (OLCs), mutation, dentin, mineralization
Introduction
Dentinogenesis Imperfecta type II and type III (DGI-II and DGI-III) and Dentin Dysplasia type II (DD-II) are isolated autosomal dominant dentin disorders. Dentin sialophosphoprotein gene (DSPP) is the only identified causative gene for these disorders. The gene encodes a single transcript which cleavages into two main protein products: dentin sialoprotein (DSP) and dentin phosphoprotein (DPP) [1]. DSP is a glycoprotein with a relatively high sialic-acid content and accounts for 5-8% of the dentin extracellular matrix (DECM) excluding collagen [2]. Found as the major noncollagenous DECM protein, DPP is rich in aspartic acid and phosphorylated serine, and might regulate biomineralization processes by binding to the matrix of structural proteins, nucleating mineralization and controlling crystal growth [3,4]. The tooth DECM is a structurally dynamic scaffold that strictly regulates framework for mineralization and orchestrates many cellular processes required for maintaining tooth integrity. Previous findings suggested that DSPP function to stimulate progression of the osteogenic pathway and could involve in multiple signaling pathways, other than functioning as secretory protein to regulate mineral deposition and crystal growth [5,6].
The main pathological feature in DGI-II, DGI-III and DD-II is the abnormality of dentin mineralization. Many mutations in DSPP gene had been identified in families with the above disorders [7-14], however, the molecular mechanisms involving in bridging the mutations and the resulting abnormal dentin mineralization have not been elucidated nowadays. Mineralization is a homeostasis which involves both the enhancement of mineralization-inducing molecules and the induction of mineralization inhibitors. Abnormal mineralization usually means the disrupted homeostasis. The molecular association between DSPP mutations and the disrupted mineralization homeostasis in human dentin disorders has not been established. In the present study, we generated 3 Dspp mutants (Figure 1) which were consistent with the previous identified DSPP mutations in human dentin disorders [7-10]. The mutants were transfected into mouse Odontoblast-lineage cells (OLCs) [15,16], which were then performed mineralization inducing differentiation.
Figure 1.
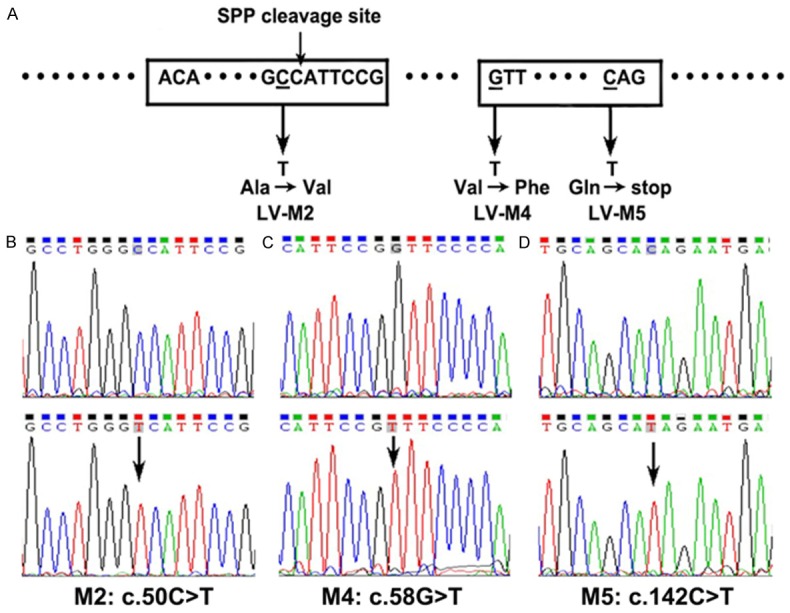
Sketch for 3 Dspp mutations and the results of sequencing the mutants (NM_010080). A. Sketch for 3 Dspp mutations (SPP cleavage site: signal peptide peptidase cleavage site). B-D. The results of sequencing the mutants. The upper pictures present the normal sequences, and the arrows indicate the location of mutations.
The digital gene expression (DGE) [17] is a sequence-based approach for gene expression analysis, which generates such extensive sequence data and depth-of-coverage that even the rare transcripts can be detected and quantified. The expression level of virtually all genes in the sample is defined by the quantity of mRNA for each gene. By phenotype detection, DGE analysis, Quantitative real-time PCR and Western blot verification, we aim to identify the regulating factors involving in the mutants causing abnormal mineralization and try to explore the molecular association between Dspp mutations and the disrupted mineralization homeostasis during dentin formation.
Materials and methods
Generation of lentivirus constructs expressing normal and mutant Dspp
The full-length mouse Dspp construct pcDNA3.0-Dspp (a donation from Professor Chunlin Qin) [18] was digested using BamHI and EcoRV restriction enzymes and subcloned into the lentivirus vector LV-PURO-GFP (GenePharma JiangSu, China), to generate the recombinant shuttle plasmid LV-Dspp-PURO-GFP.
Then we generated 3 mutants according to the identified DSPP mutations (Figure 1A): Mutation 2 (M2) [9] is a missense mutation located in the signal peptide region; Mutation 4 (M4) [7,10] is a proposed mutational “hotspot” occurring at the first nucleotide of exon 3; Mutation 5 (M5) [8,10] is a nonsense mutation at codon 45. According to the manufacturer’s instruction, the constructs were generated with a QuikChange Lightning Site-Directed Mutagenesis Kit (Stratagene, catalog#210518, USA) and verified by sequencing the PCR products (Figure 1B-D). Table 1 showed the mutation points and primers.
Table 1.
The mutation points and primer sequences
| Human NM_014208 | Mouse NM_010080 | Primer | |
|---|---|---|---|
| LV-M2 | c.44C>T | c.50C>T | 5-CAACTGCCTGGGTCATTCCGGTTCCCCAGTTA-3 |
| 5-TGTCATTGTGTTCTTCAGCAGTGTTCCCCTGTTCG-3 | |||
| LV-M4 | c.52G>T | c.58G>T | 5-CCTGGGCCATTCCGTTTCCCCAGTTAGTAC-3 |
| 5-TTTCTATGTCATTGTGTTCTTCAGCAGTGTTCCCC-3 | |||
| LV-M5 | c.133C>T | c.142C>T | 5-TCCAGGAACTGCAGCATAGAATGAGTTATCTATCA-3 |
| 5-GCATGTACCCCATCATGACCGTATGTTTCTATGTC-3 |
The bacterial host DH5α (Invitrogen, USA) were transfected with LV-Dspp/M2/M4/M5-PURO-GFP respectively, and then cultured in Luria-Bertani medium with ampicillin at 25°C. The target recombinant lentivirus plasmids and Packaging plasmids (pGag/Pol, pRev, pVSV-G) were amplified in 293T cells and packaged into virus particles (LV-Dspp/M2/M4/M5). The empty lentivirus vector LV-PURO-GFP was also packaged as a control (LV-GFP). Viral titers were estimated by optical density (OD) and a standard plaque assay.
Transfection of virus particles into OLC cells and selection of stable transfectants
OLCs (donation from Professor Toshihiro Sugiyama) were transfected with lentivirus particles. Following 96 hours transfection, puromycin (3.5 ng/ul Invitrogen, USA) was used to select and maintain stable eukaryotic cell lines. After 2 weeks screening, we got the OLCs stably expressing the wild-type Dspp, mutant Dspp and the control. Then the OLCs were divided into the following groups: LV-Dspp, LV-M2, LV-M4, LV-M5 and the control LV-GFP group.
Cell differentiation
All groups with stably transfected OLCs were cultured in alpha-minimum essential medium with 10% fetal bovine serum (Hyclone, USA) and 1% penicillin/streptomycin on type I collagen-coated culture plates, at 37°C in a humidified atmosphere of 5% CO2 in air. When cells reached 70% confluence, they were transferred to mineralization inducing α-MEM containing 10% FBS, 1% penicillin/streptomycin, 10 nmol/L dexamethasone (Sigma, USA), 50 mg/L ascorbic acid (Sigma, USA), and 10 mmol/L β-sodium glycerophosphate (Sigma, USA), and incubated with 2 ng/ul puromycin. Day 1 was defined as the day of mineralization medium supplementation to the cell culture. The medium was changed every 3 days.
Alizarin red S staining for mineralization assay
The mineralization was assessed by staining with Alizarin red S on day 7 and 21 for all groups. The cells were fixed in 4% paraformaldehyde for 15 min, and washed with PBS. 1% Alizarin red S (Sigma-Aldrich, USA) solution in 0.1% NH4OH at PH 4.1-4.3 was added to each well, and incubated for 30 min at 37°C. Then they were rinsed several times with PBS to remove all unincorporated staining.
ALP activity
Cultures of the stably transfected OLCs were washed 3 times with PBS. The activity of the cells in all groups was measured on day 0, 7, 14 and 21 in freshly prepared colorimetric substrate 0.1 M NaHCO3-Na2CO3 buffer (pH 10) containing 0.1% Triton X-100, 2 mM MgSO4, and 5 mM p-nitrophenyl phosphate. ALP activity was calculated as moles of p-nitrophenol per mg protein. The optical density was measured at 405 nm by a HTS 7000 Plus Bio Assay reader (PE, USA), and normalized to total protein.
Screening of differentially expressed genes by DGE
Total RNA was extracted from cultured stably transfected OLCs of LV-Dspp, LV-M5 and LV-GFP groups using HP Total RNA Kit (OMEGA, USA) and was quantitated into 1 μg. Sequence tag preparation was in the charge of Huada Gene technology company (Shenzhen, China). The expression pattern of these differentially expressed genes (DEGs) was visualized using the heat-map function, and its log2 Ratio (group A/group B) will be clustered. In the pairwise comparison, the former one (group A) is considered as the control, and the latter one (group B) is considered as the treatment.
Based on DGE profiles [19], Gene Ontology (GO) functional enrichment analysis and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis were performed. GO enrichment analysis firstly maps all DEGs to GO terms in the database (http://www.geneontology.org/), calculating gene numbers for every term, then using hypergeometric test to find significantly enriched GO terms in DEGs comparing to the genome background. The calculated P-value goes through Bonferroni Correction, taking corrected P-value≤0.05 as a threshold. With nr annotation, we used Blast2GO program to get GO annotation of DEGs, then we used WEGO software [20] to do GO functional classification for DEGs and to understand the distribution of gene functions of the species from the macro level.
KEGG is the major public pathway-related database [21]. Pathway enrichment analysis identifies significantly enriched metabolic pathways or signal transduction pathways in DEGs comparing with the whole genome background. The calculating formula is the same as that in GO analysis, and pathways with Q-value≤0.05 as significantly enriched in DEGs.
Based on the results of the DGE analysis, some significant molecules were selected to be examined and verified by Quantitative real-time PCR and Western-blot examination in all groups including LV-Dspp, LV-M2, LV-M4, LV-M5 and the control LV-GFP group on day 0, 7, 14 and 21 separately.
Quantitative real-time PCR
Total RNA was extracted from cultured stably transfected OLCs using HP Total RNA Kit (OMEGA, USA). RNA was reverse-transcribed into cDNA with the RevertAid First Strand cDNA Synthesis Kit (Takara, China). Quantitative real-time PCR was performed with 50 ng of cDNA and FastStart Universal SYBR Green Mastor (Rox) (Roche, USA) using the 7900HT Fast Real Time PCR System (Applied Biosystems, USA). The following temperature profile was used: 95°C for 10 minutes, and subjected to 40 cycles of denaturation at 95°C for 15 s, annealing at 60°C for 60 s, and melt curve from 60°C to 90°C. Expressions of the target genes were normalized to Puromycin (Puro) levels and the 2-ΔΔCt method was used to calculate relative expression levels. Table 2 showed the primers sequences of the examined genes.
Table 2.
Oligonucleotide primers for real-time RCR with expected fragment size
| Gene | Primer | bp |
|---|---|---|
| Dsp | F: 5-TGAAAACTCTGTGGCTGTGC-3 | 248 |
| R: 5-TGTGTTCTTCAGCAGTGTTCC-3 | ||
| Bmp2 | F: 5-TGACTGGATCGTGGCACCTC-3 | 112 |
| R: 5-CAGAGTCTGCACTATGGCATGGTTA-3 | ||
| Col1 | F: 5-TGCTTGCAGTAACTTCGTGCCTA-3 | 143 |
| R: 5-CATGGGACCATCAACACCATC-3 | ||
| Runx2 | F: 5-ATGATGACACTGCCACCTCTGAC-3 | 106 |
| R: 5-AACTGCCTGGGGTCTGAAAAAGG-3 | ||
| Ocn | F: 5-CCGGGAGCAGTGTGAGCTTA-3 | 54 |
| R: 5-CCATACTGGTCTGATAGCTC-3 | ||
| Mgp | F: 5-GTCCTATGAAATCAGTCCCTTCA-3 | 108 |
| R: 5-TTGTTGCGTTCCTGGACTCT-3 | ||
| Htra1 | F: 5-CATCTCCTTCGCAATTCCAT-3 | 155 |
| R: 5-GACGGTCCTTCAGCTCTTTG-3 | ||
| Puro | F: 5-CAAGGAGCCCGCGTGGTT-3 | 190 |
| R: 5-GTCGGCGGTGACGGTGAA-3 |
F=Forward; R=Reverse. All primers were designed with the software Primer 3.0 based on the published sequence.
Western blot analysis
Cell layers were washed 3 times with PBS before cell dissolving, then total proteins were extracted by RIPA lysis and extraction buffer (Beyotime, China), followed by centrifugation (13,000 g for 5 min) to remove cellular debris. Protein quantification was performed with the Bio-Rad DC Protein Assay (Bio-Rad, USA). Equal amounts of protein were subjected to SDS-PAGE gel electrophoresis on 10%-20% (w/v) acrylamide gel and then transferred onto PVDF membrane (Millipore, USA). After blocking with TBST containing 5% nonfat dry milk the blots were detected by specific commercial antibodies diluted in TBST overnight at 4°C: DSP (Santa Cruz Biotechnology, sc-33587, USA), Bmp2 (Abcam, ab82511, UK), Col1 (Abcam, ab88147, UK), Runx2 (Abcam, ab76956, UK), Ocn (Santa Cruz Biotechnology, sc-376835, USA), Mgp (Santa Cruz Biotechnology, sc-66965, USA), Htra1 (Abcam, ab38611, UK), and β-actin (Epitomics, USA). There after, membranes were incubated with horseradish peroxidase-conjugated secondary antibody (Antgene, China) for 1 h at room temperature. Immunocomplexes were visualized using chemiluminescence, and then exposed. The relative integrated density of protein bands were analyzed by NIH ImageJ 1.48 v, and then normalized to that of the β-actin.
Statistical analysis
The data for Q-PCR, Western-blot and ALP activity were presented as mean ± SD from three independent experiments and the results were compared by one-way ANOVA using Statview software (SAS Institute Inc., Lary, NC). The differences between each test group and the control LV-GFP group at each time-point were presented and the statistical significance was P < 0.05.
Results
Stable transfectants
The expression of the GFP reporter gene in the infected cells was used to determine the infection efficiency by fluorescent microscopy. OLCs grew well and showed strong green fluorescence after 2 weeks’ puromycin selection, with transfer efficiency of 99% (data not shown).
Alizarin red S staining for mineralization assay
More and larger mineralized nodules were observed in LV-Dspp/M4/GFP groups on day 7 and day 21, and very few mineral deposits were visualized in LV-M2 and LV-M5 groups (Figure 2A).
Figure 2.
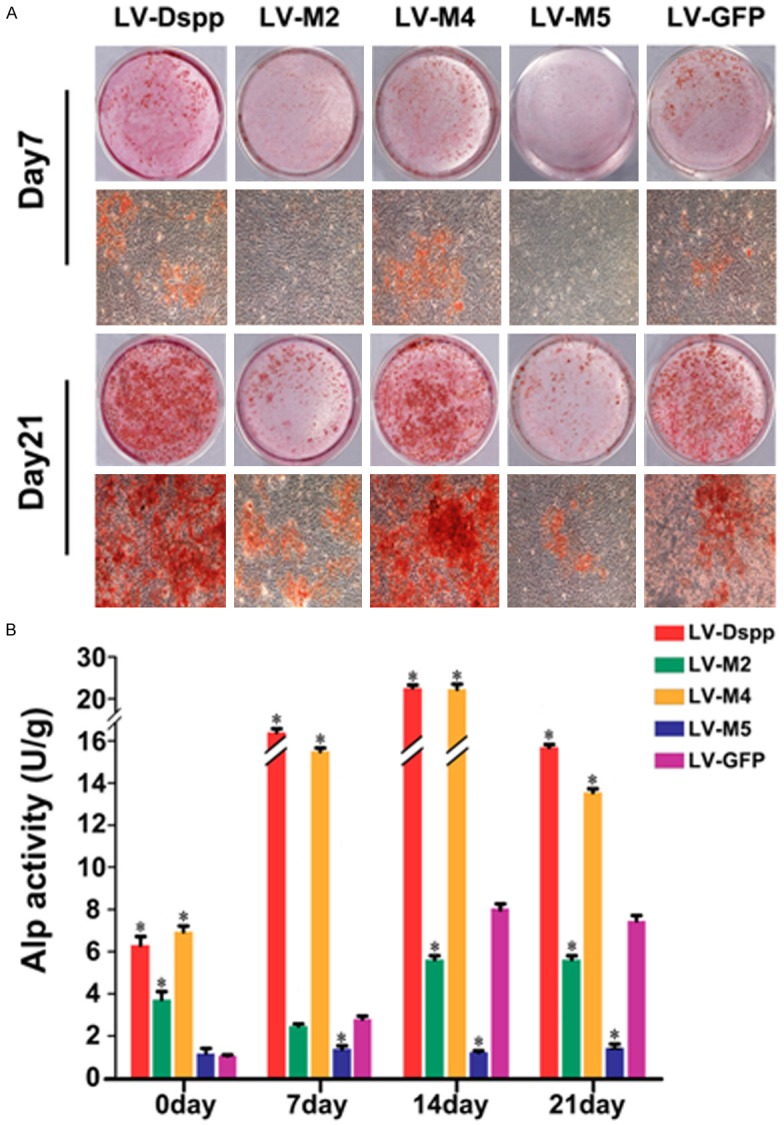
Formation of mineralization nodules and ALP activity assay. A. Alizarin red staining of OLCs transfected with LV-Dspp/M2/M4/M5 and LV-GFP on day 7 and 21. B. ALP activity in each group during the differentiation of transfected OLCs. *Indicates statistical difference between the test group and the control, P<0.05.
ALP activity assay
Alp activity was much higher in LV-Dspp/M4 groups than in other groups at all time and peaked on day 14. LV-M2/M5 groups showed obviously lower Alp activity on day 14 and 21, and LV-M5 group always showed the lowest level (Figure 2B).
Analysis of DGE libraries
On the basis of the Hiseq of Illumina 3’-tag DGE protocol, we generated raw tags between 11.7 and 12.5 million for each of the 12 samples. After removing low-quality reads, the total number of clean tags per library ranged from 11.6 to 12.4 million and the number of tag entities with unique match ranged from 8.6 to 9.3 million (Supplemental Table 1). The number of DEGs with expression difference was less in LV-GFP-VS-LV-M5 than in LV-GFP-VS-LV-Dspp (Supplemental Figure 1).
Clustering of DEGs
In this study, we focused our attention on the genes connected to mineralization. Among these genes (Figure 3), Dspp, Bmp2, Mmps, Mgp, Htra1 and some Wnts were up-regulated, Col1a1, Col1a2, Col2a1 and some other Wnts were down-regulated, but each group showed different expression pattern. Runx2 had the expression defect in LV-M5 group, Ocn (Bglap), Bglap2 and Wnt10a were detected only in LV-Dspp group. The expression level of Mmps was similar in the three groups.
Figure 3.
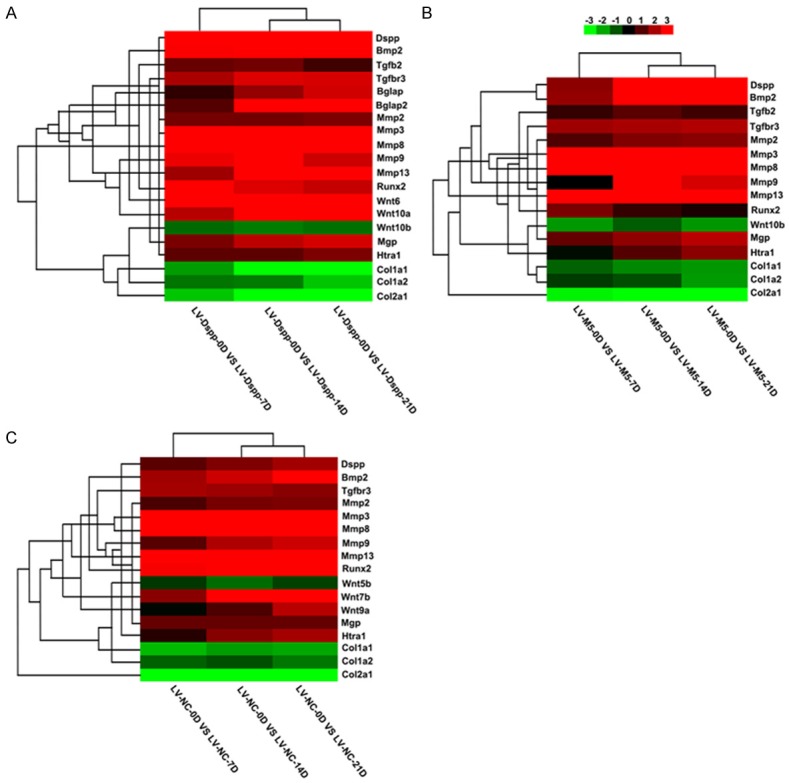
Heat map of selected identified genes P≤0.05. Hierarchical clustering of differentially expressed genes in LV-Dspp (A), LV-M5 (B) and LV-GFP (C) group. Each column represents an experimental condition, each row represents a gene, red indicates up-regulation, and green indicates down-regulation.
GO analysis and KEGG pathway
A total of 15584 differentially expressed genes were detected and mapped to terms in GO database and compared with the whole genome background. The GO comparison among LV-DSPP, LV-M5 and LV-GFP reflects most genes are enriched in biological process including cell proliferation, locomotion, response to stimulus, signaling, et al, cellular component including extracellular region and macromolecular complex, et al, and molecular function including molecular transducer activity, et al (Figure 4). Comparison between the results of LV-GFP-VS-LV-Dspp and LV-GFP-VS-LV-M5 showed that the DEGs for cell proliferation, locomotion, reproduction, reproduction process were less in LV-GFP-VS-LV-M5 (Figure 4B), and that the DEGs for macromolecular complex and transporter activity pass from a greater to a smaller number in LV-GFP-VS-LV-Dspp (Figure 4A), while in LV-GFP-VS-LV-M5 the number of DEGs for macromolecular complex had no significant change and the DEGs for transporter activity increased (Figure 4B).
Figure 4.
Categories of DEGs based on GO function in LV-GFP-VS-LV-Dspp (A) and LV-GFP-VS-LV-M5 (B). The results are summarized in three major categories: biological process, cellular component and molecular function. P≤0.05 as a threshold.
KEGG analysis showed the number of genes in pathways including ECM-receptor interaction pathway, cell adhesion molecules pathway, focal adhesion pathway and some amino acids pathway associated with mineralization metabolism was obviously less in LV-GFP-VS-LV-M5 than in LV-GFP-VS-LV-DSPP (Figure 5). The RichFactor of top 20 pathways in the KEGG database of LV-GFP-VS-LV-M5 gradually decreased, and the pathways with statistical significance only showed on day 0 in LV-GFP-VS-LV-M5 (Figure 5B). The RichFactor and the gene numbers of ECM-receptor interaction pathway in LV-GFP-VS-LV-DSPP behave like a parabola (Figure 5A), whereas the expression of this pathway gradually decreased, even disappeared from day 14 in LV-GFP-VS-LV-M5 (Figure 5B).
Figure 5.
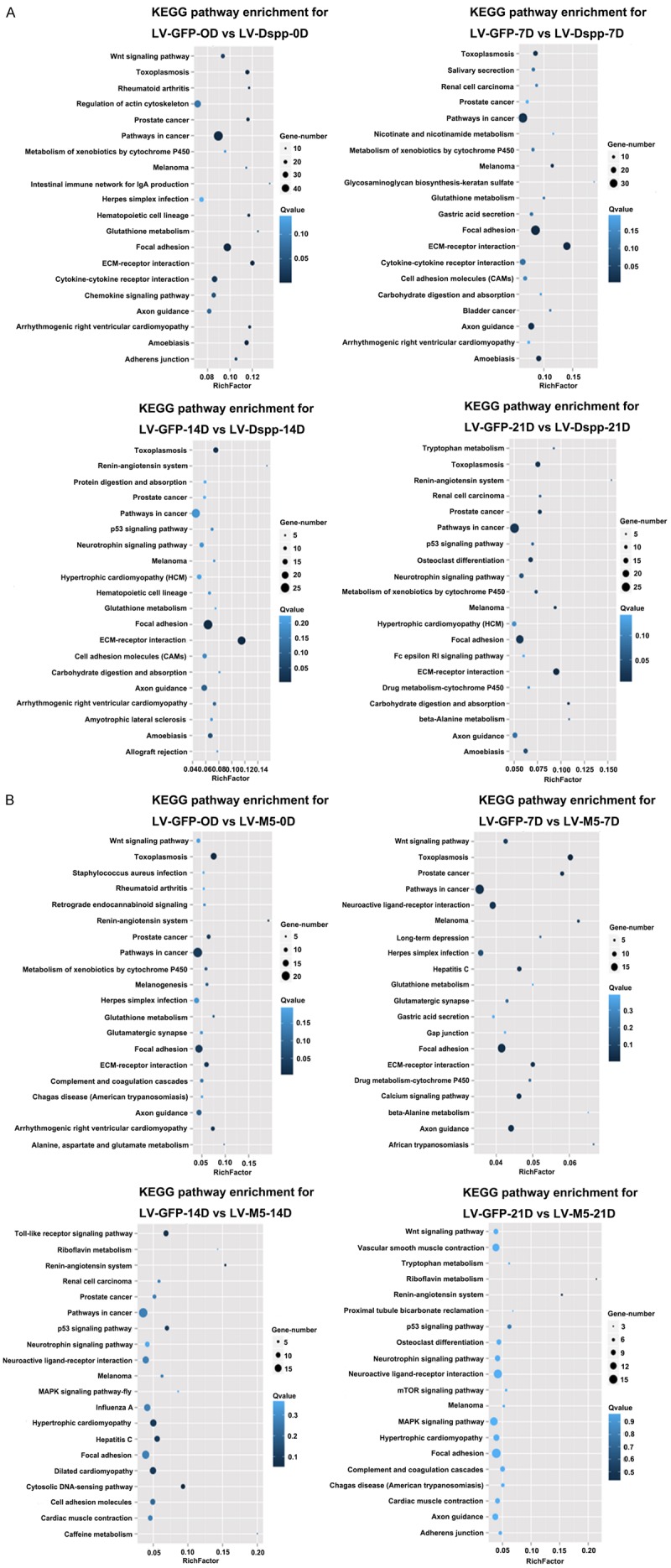
The scatter plot for KEGG enrichment result. Top 20 Statistics of KEGG pathway enrichment for LV-GFP-VS-LV-Dspp (A) and LV-GFP-VS-LV-M5 (B). Rich Factor is the ratio of differentially expressed gene numbers annoted in this pathway terms to all gene numbers annoted in this pathway term. Q≤0.05 as significantly enriched.
Identification and verification of examined molecules during OLC mineralizing induction
Based on the results of the DGE analysis, 7 significant molecules (Dspp, Bmp2, Col1, Runx2, Ocn, Mgp and Htra1) were selected to be examined and verified by Quantitative real-time PCR and Western-blot examination.
The integration of exogenous gene induced the extremely high expression of Dsp in all transfected test groups. Dsp expression in LV-M2/M5 groups was obviously lower than that in LV-Dspp/M4 groups, and was significantly higher than that in the control group at transcription level (Figure 6A), while the two groups showed lower protein expression than other three groups at translational level especially on day 14 and day 21 (Figure 6B and 6C).
Figure 6.
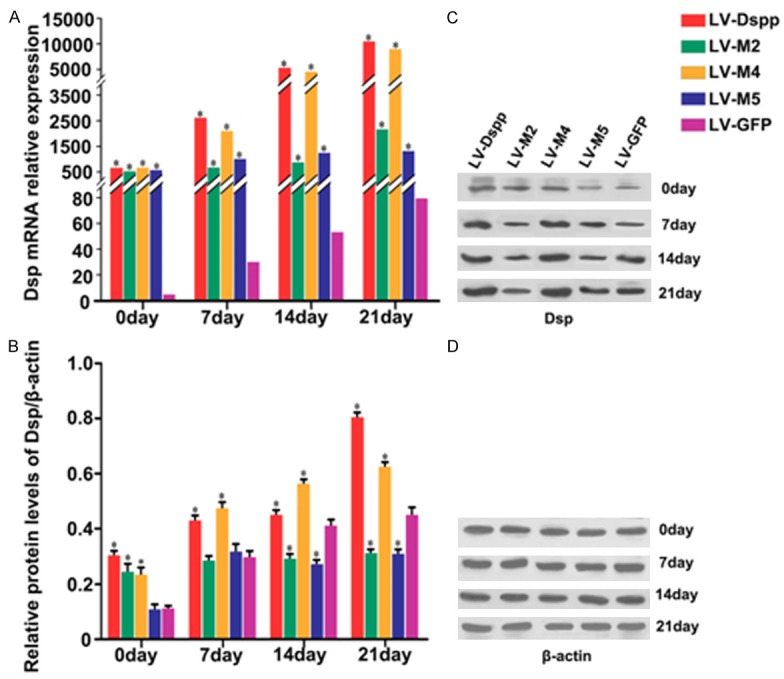
Expression of Dsp in each group during the differentiation of transfected OLCs. A. mRNA expression of Dsp was investigated by Real-time PCR analysis. B, C. Protein expression of Dsp was detected by Western-Blot. D. β-actin served as an internal control. *Indicates statistical difference between the test group and the control P < 0.05. It is worth mentioning that the levels of Dsp were thousands times higher in the test groups than in the control, resulting in the large margin of error bars, so the error bars in the graph were not indicated.
Relative mRNA and protein expressions of specific mineralization inductors including Bmp2, Col1 and Runx2, proliferative marker Ocn, mineralization negative regulator Mgp and Htra1 are as follows: Expression of Bmp2/Bmp2 was obviously up-regulated in all groups during cell differentiation, whereas in LV-M2/M5 groups it showed a smooth rising-trend and exhibited significantly lower expression than in LV-Dspp/M4/GFP groups on day 21 (Figure 7A, 7B and 7B’); During cell maturation, Col1 mRNA and protein levels were significantly declined in all groups although the control group has little variation in mRNA level: Col1/Col1 expression in LV-M5 group showed the highest level during the whole process, and in LV-M2 group it was higher than in LV-DSPP/M4 groups during the late stage of mineralization (Figure 7C, 7D and 7D’); Runx2/Runx2 expression in LV-M2/M5 groups maintained lasted low level and was much lower than in other groups (Figure 8A, 8B and 8B’); Ocn/Ocn expression was obviously up-regulated and peaked on day 21, but it showed extremely lower level in LV-M2/M5 groups than in LV-Dspp/M4/GFP groups on day 14 and 21 (Figure 8C, 8C and 8D’); Mgp/Mgp expression kept high level and was obviously higher in LV-M2/M5 groups than in LV-Dspp/M4/GFP groups on day 14 and 21 (Figure 9A, 9B and 9B’); Htra1/Htra1 expression in LV-M2/M5 groups continuously up-regulated and peaked on day 21, and exhibited obviously higher level than in LV-Dspp/M4/GFP groups on day 21 (Figure 9C, 9C and 9D’).
Figure 7.
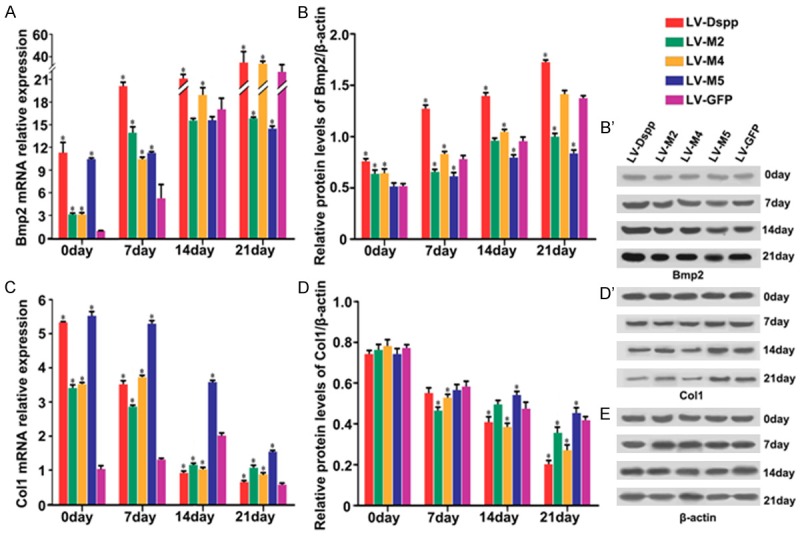
Expression of Bmp2 and Col1 in each group during the differentiation of transfected OLCs. mRNA expression of Bmp2 (A) and Col1 (C) were investigated by Real-time PCR analysis. Protein expression of Bmp2 (B, B’) and Col1 (D, D’) were detected by Western-Blot. (E) β-actin served as an internal control. *Indicates statistical difference between the test group and the control P < 0.05.
Figure 8.
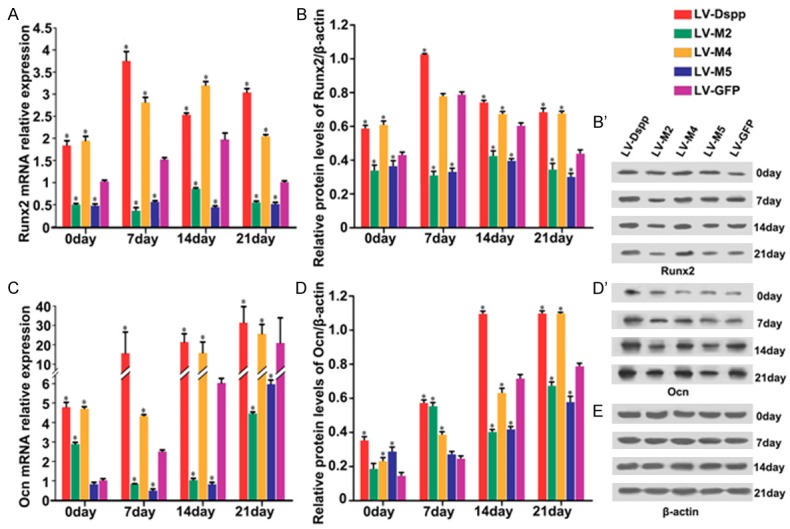
Expression of Runx2 and Ocn in each group during the differentiation of transfected OLCs. mRNA expression of Runx2 (A) and Ocn (C) were investigated by Real-time PCR analysis. Protein expression of Runx2 (B, B’) and Ocn (D, D’) were detected by Western-Blot. (E) β-actin served as an internal control. *indicates statistical difference between the test group and the control P < 0.05.
Figure 9.
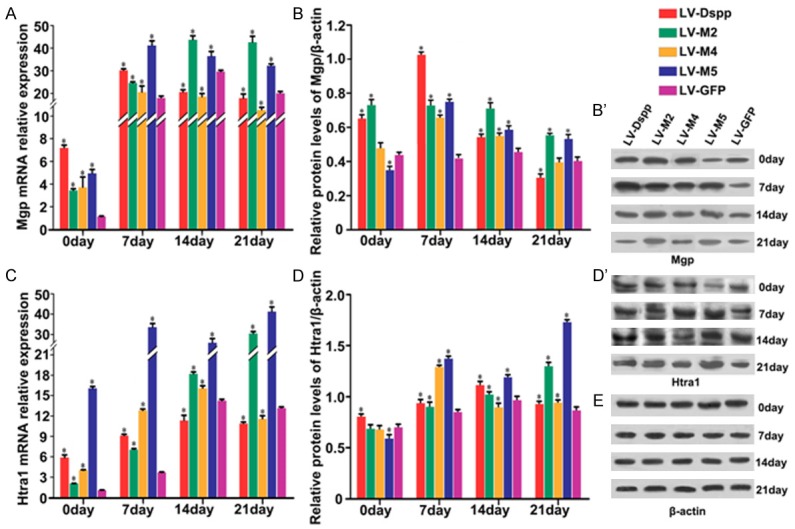
Expression of Mgp and Htra1 in each group during the differentiation of transfected OLCs. mRNA expression of Mgp (A) and Htra1 (C) were investigated by Real-time PCR analysis. Protein expression of Mgp (B, B’) and Htra1 (D, D’) were detected by Western-Blot. (E) β-actin served as an internal control. *indicates statistical difference between the test group and the control P < 0.05.
Discussion
To date, no full-length DSPP protein has been identified or isolated from dentin. We firstly reported the differentiation and mineralization of OLCs which were transfected with the mouse full-length Dspp gene and the full-length mutant Dspp. Because of the highly repetitive sequence in DPP region, Dspp gene appeared to be cytotoxic to many kinds of cells. It was a great challenge to obtain the cells stably expressing the full-length Dspp [22], which becomes an obstacle for researchers to further study the precise function of DSPP during dentin formation. After failure of transfecting a variety of cell types, we fortunately received the OLCs, and eventually obtained the stable OLCs transfected with the full-length Dspp and the full-length mutant Dspp.
Mutant DSPP protein is expected to have a dominant negative effect on cell function and/or dentin matrix mineralization [14]. The genotype-phenotype correlation appears to be affected not only by the degree of reduction in the amount of wild-type (WT) DSPP but also by the quantity of mutant protein [14]. Secretion of WT DSPP is reduced when coexpressed with mutant proteins, and the decline level is in a dose-dependent manner. Compared to WT DSPP alone, co-expression of equal amount of the mutant DSPP protein caused a 40% loss in secreted WT and this increased to a>70% loss at the modest dose of 4:1 (mutant:WT) [23], which would result in the normal DSPP protein not being properly processed for rapid accumulating dentin matrix. The OLC is a kind of odontoblast-lineage cell able to express the endogenous and exogenous Dspp. Our results showed that the OLCs transfected with different types of mutant Dspp suffered diverse degree of impairment on mineralization differentiation, which is like the different DSPP mutations resulting in the phenotype variation in the dentin disorders.
LV-M2 in the present study was a missense mutation which located in the last amino acid of signal peptide region of mouse Dspp. The mutation would severely compromise the ability of translocating the primary translation product into endoplasmic reticulum, and then cause the decrease of the Dsp and Dpp dramatically. The nonsense mutation (c.133C>T) of DSPP, corresponding to LV-M5 in the present study, was associated with the DGI-II phenotype, which could affect the coding proteins by the nonsense-mediated mRNA decay (NMD) [24]. The nonsense mutation may exert its effect by greatly reducing the quantity of Dsp and Dpp. The two mutations impair the function of Dpp or/and Dsp and induce defective dentin mineralization.
A complex molecular network of regulators including mineralization inductors and inhibitors governs the odontogenesis of odontoblast [25]. Mutations in DSPP resulted in hereditary human dentin disorders which characterized as abnormal mineralization. The molecular regulation mechanism between mutational DSPP and dynamic homeostasis of mineralization regulators would be the important etiology to develop the abnormal dentin phenotype. The findings in the present study indicated that mutant Dspp could disrupt the strictly orchestrated molecular framework for odontoblast differentiation.
In the present study, the formation of mineralized nodules was obviously fewer in LV-M2/M5 groups which indicated the mutant Dspp disrupted the mineralization homeostasis. Alp is recognized as an early marker of osteogenic differentiation and mineralization [5]. The different pattern of Alp activity in LV-M2 and LV-M5 groups suggested that the two types of mutations could exert diverse impacts on the dynamic homeostasis of dentin mineralization. To explore the molecular association between Dspp and the homeostasis of mineralization, we performed a large-scale analysis of gene expressions in LV-Dspp, LV-M5 and LV-GFP groups using DGE approach. A number of genes were involved in biological process, cellular component and molecular function, suggesting that these function may be associated with cellular metabolism, response to mineralization stimulus and the activity of signaling pathways.
Compared with the results of LV-GFP-VS-LV-Dspp, the number of DEGs for cell proliferation, locomotion, reproduction, reproduction process was obviously less in results of LV-GFP-VS-LV-M5, which could indicate that these cell activities associate with metabolism of ECM and cell accelerated growth be inactive in LV-M5 group. The DEGs for macromolecular complex and transporter activity passed from a greater to a smaller number in LV-GFP-VS-LV-Dspp during mineralization, suggesting the formation of ECM compound and the activity of transporter started earlier and actively in LV-Dspp group, while in LV-GFP-VS-LV-M5 the number of DEGs for this process displayed in irregular manner, which indicated that activity of ECM compound and cellular delivery be different from those in normal mineralization process.
KEGG analysis suggested that several pathways are affected, including ECM-receptor interaction pathway, cell adhesion molecules pathway, focal adhesion pathway and some amino acids pathway associated with mineralization metabolism. Less expression of these pathways in LV-GFP-VS-LV-M5 implied that the intercellular and extracellular communications were passive and the metabolism of ECM responding to mineralize stimulus was inactive. Meanwhile the top 20 pathways in the KEGG database in LV-GFP-VS-LV-M5 were disorderly and the number of pathways with statistical significance was very few, indicating the pathways strictly orchestrating framework for mineralization homeostasis was disrupted in LV-M5 group.
The canonical Smad/BMP2 plays important role in the interaction between DSPP and odontoblast differentiation [6,26]. In our study, Bmp2 expression was up-regulated during cell maturation although the rising trend in LV-M2/M5 groups exhibited smooth profile. It can be speculated that BMP2 is an indispensable factor in dentin development and is not easily manipulated by mutant Dspp. LV-M2 and LV-M5 may exert effect by greatly reducing the quantity of Dpp and/or Dsp, which would impair the cooperation between Dspp and Bmp2 to regulate odontoblast differentiation and mineral deposition. Previous studies [27] demonstrated that DPP acted as an accelerator of recombinant human BMP2 induction of orthotopic hard tissue formation, when DPP was covalently cross-linked to type I collagen. The potential of the DPP immobilized to type I collagen for apatite induction is similar to that of crystal growth on seeded hydroxyapatite [28]. Col1 serves as a mineralization scaffold rarely appears or absents in the peritubular dentin that is a hypermineralized zone surrounding tubules [29]. However, our previous study found that amounts of collagen fibers were around dentin tubules in peritubular dentin which was hypomineralized in DGI specimen [10]. In the present study, Col1 were significantly declined in all groups, and showed the highest expression in LV-M5 group. Although Col1 expression in LV-M2 group was not noticeable as that in LV-M5 group, it remained higher than that in LV-Dspp/M4 groups during the late stage of mineralization. Our findings indicated that LV-M2/M5 mutants might compromise Dspp synthesis and then interfere in Dpp binding to collagen type I for apatite induction and mineral deposition.
Because Wnt/β-catenin binds the Runx2 promoter and controls its transcription [30], meanwhile Ocn is a known target gene for Runx2 that is directly or indirectly regulated by the interaction between Bmp2 and Dspp [26,31], we chose the two factors to further validate. Runx2 and Ocn involve in dentin formation likely dependent on the stage of cytodifferentiation. Runx2 expresses in preodontoblasts, and then strongly expresses in early mature odontoblast, but downregulates during mineralizing maturation [32,33]. Ocn is a marker of late stage osteoblast differentiation and strongly expresses in maturated osteoblasts [34]. In the present study, Runx2/Runx2 expression in LV-M2/M5 groups was always in a significantly low level at all stages of mineralization, as well as DGE showed Runx2 had the expression defect in LV-M5 group. Although Ocn/Ocn expression in LV-M2/M5 groups showed a similar up-regulated trend as other groups during the progression of mineralization, it was still lower than that in other groups on day 14 and 21, as DGE showed it was absent in LV-M5 group. The results suggested that as a signaling molecule in cell differentiation, abnormal Dspp would cause abnormal Runx2 regulation which disturb odontoblast differentiation and/or lead Ocn unable to properly remodel the differentiation of odontoblasts.
Mineralization is a process of dynamic homeostasis requiring a feedback mechanism to prevent further calcification. MGP and Htra1 play key roles during this well orchestration, and they might regulate mineralization in an interdependent manner [35]. Previous studies demonstrated that Htra1 interacting with Mgp played a role in reparative dentin formation [36]. Htra1-mediated cleavage of Mgp at the C-terminus may enhance the ability of Mgp to bind to the ECM, and next may inhibit Bmp2 signaling and mineralization, which suggested that Htra1 regulate matrix calcification via the inhibition of Bmp2 signaling [37,38]. In the present study, the expression profile of Htra1 was in accordance with that of Mgp, and their expression were always inversely proportional to Bmp2. While the expression level in LV-M2/M5 groups was significantly higher for Mgp/Htra1 and obviously lower for Bmp2 than that in other groups especially at late stage. The results indicated that the mutant Dspp altered the expression pattern of Mgp and Htra1 and then could inhibit Bmp2 signaling, which would consequently cause an imbalance for mineralization homeostasis. How Mgp and Htra1 regulate dentin mineralization in an interdependent manner, and how Dspp participate in or converge BMP2 to mediate this process need to be further elucidated.
In our study, lentivirus was subcloned with Dspp cDNA, no splicing process existed. So LV-M4 mutant is predicted to result in an amino acid substitution. However, Mutations located at or near exon-intron junctions could affect the recognition of splice sites [11,14]. Our findings showed the expression of most examined molecules and mineral deposits in LV-M4 group were significantly different from those in LV-M2 and LV-M5 groups and exhibited manifestation similar to those in LV-Dspp group. So it should be the disturbance of the splice process rather than the substitution of amino acid residue for LV-M4 mutation affecting the structural integrity and function of DSPP in human dentin disorders.
Although little is known about the mechanisms involving in regulating such hallmark cellular responses by Dspp, it can be proposed that Dspp could signal odontoblast precursor cells differentiate into mature odontoblasts during the early stages of tooth development, and that Dspp might contribute to positive signaling related to the regulation of mineral deposition, crystal formation and growth in the later stage [3,5,6,39]. Furthermore, Dspp also might regulate the subsequent feedback inhibition of signaling in order to maintain tissue homeostasis as well as morphogenesis [6,31,40]. Overall, our findings shed new light on association between Dspp and the dynamic homeostasis of mineralization inductors and inhibitors, and indicated that the disruption of the homeostasis might be the crucial reason for Dspp mutations resulting in the dentin disorders. Further studies need to reveal whether Dspp regulates these mineralization inductors and inhibitors directly or as upstream signal factors playing key roles in maintaining dynamic homeostasis indirectly.
Acknowledgements
The study was supported by grants from the National Natural Science Foundation of China (No. 81120108010, 81170957 and 81470727), and by the grant from the Bureau of Science and Technology of Wuhan ([2014]160). We thank Professor Chunlin Qin for providing the full-length mouse Dspp construct pcDNA3.0-Dspp and Dr Arany (Department of Biochemistry, Akita University School of Medicine, Akita, Japan) for donation of OLCs.
Disclosure of conflict of interest
None.
Supporting Information
References
- 1.MacDougall M, Simmons D, Luan X, Nydegger J, Feng J, Gu TT. Dentin phosphoprotein and dentin sialoprotein are cleavage products expressed from a single transcript coded by a gene on human chromosome 4. Dentin phosphoprotein DNA sequence determination. J Biol Chem. 1997;272:835–842. doi: 10.1074/jbc.272.2.835. [DOI] [PubMed] [Google Scholar]
- 2.Butler WT, Bhown M, Brunn JC, D’Souza RN, Farach-Carson MC, Happonen RP, Schrohenloher RE, Seyer JM, Somerman MJ, Foster RA, et al. Isolation, characterization and immunolocalization of a 53-kDal dentin sialoprotein (DSP) Matrix. 1992;12:343–351. doi: 10.1016/s0934-8832(11)80030-2. [DOI] [PubMed] [Google Scholar]
- 3.George A, Bannon L, Sabsay B, Dillon JW, Malone J, Veis A, Jenkins NA, Gilbert DJ, Copeland NG. The carboxyl-terminal domain of phosphophoryn contains unique extended triplet amino acid repeat sequences forming ordered carboxyl-phosphate interaction ridges that may be essential in the biomineralization process. J Biol Chem. 1996;271:32869–32873. doi: 10.1074/jbc.271.51.32869. [DOI] [PubMed] [Google Scholar]
- 4.George A, Hao J. Role of phosphophoryn in dentin mineralization. Cells Tissues Organs. 2005;181:232–240. doi: 10.1159/000091384. [DOI] [PubMed] [Google Scholar]
- 5.Wu L, Zhu F, Wu Y, Lin Y, Nie X, Jing W, Qiao J, Liu L, Tang W, Zheng X, Tian W. Dentin sialophosphoprotein-promoted mineralization and expression of odontogenic genes in adipose-derived stromal cells. Cells Tissues Organs. 2008;187:103–112. doi: 10.1159/000110079. [DOI] [PubMed] [Google Scholar]
- 6.Lee SY, Kim SY, Park SH, Kim JJ, Jang JH, Kim EC. Effects of recombinant dentin sialoprotein in dental pulp cells. J Dent Res. 2012;91:407–412. doi: 10.1177/0022034511436113. [DOI] [PubMed] [Google Scholar]
- 7.Xiao S, Yu C, Chou X, Yuan W, Wang Y, Bu L, Fu G, Qian M, Yang J, Shi Y, Hu L, Han B, Wang Z, Huang W, Liu J, Chen Z, Zhao G, Kong X. Dentinogenesis imperfecta 1 with or without progressive hearing loss is associated with distinct mutations in DSPP. Nat Genet. 2001;27:201–204. doi: 10.1038/84848. [DOI] [PubMed] [Google Scholar]
- 8.Zhang X, Zhao J, Li C, Gao S, Qiu C, Liu P, Wu G, Qiang B, Lo WH, Shen Y. DSPP mutation in dentinogenesis imperfecta Shields type II. Nat Genet. 2001;27:151–152. doi: 10.1038/84765. [DOI] [PubMed] [Google Scholar]
- 9.Malmgren B, Lindskog S, Elgadi A, Norgren S. Clinical, histopathologic, and genetic investigation in two large families with dentinogenesis imperfecta type II. Hum Genet. 2004;114:491–498. doi: 10.1007/s00439-004-1084-z. [DOI] [PubMed] [Google Scholar]
- 10.Song Y, Wang C, Peng B, Ye X, Zhao G, Fan M, Fu Q, Bian Z. Phenotypes and genotypes in 2 DGI families with different DSPP mutations. Oral Surg Oral Med Oral Pathol Oral Radiol Endod. 2006;102:360–374. doi: 10.1016/j.tripleo.2005.06.020. [DOI] [PubMed] [Google Scholar]
- 11.Lee SK, Hu JC, Lee KE, Simmer JP, Kim JW. A dentin sialophosphoprotein mutation that partially disrupts a splice acceptor site causes type II dentin dysplasia. J Endod. 2008;34:1470–1473. doi: 10.1016/j.joen.2008.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McKnight DA, Suzanne Hart P, Hart TC, Hartsfield JK, Wilson A, Wright JT, Fisher LW. A comprehensive analysis of normal variation and disease-causing mutations in the human DSPP gene. Hum Mutat. 2008;29:1392–1404. doi: 10.1002/humu.20783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Song YL, Wang CN, Fan MW, Su B, Bian Z. Dentin phosphoprotein frameshift mutations in hereditary dentin disorders and their variation patterns in normal human population. J Med Genet. 2008;45:457–464. doi: 10.1136/jmg.2007.056911. [DOI] [PubMed] [Google Scholar]
- 14.Lee KE, Lee SK, Jung SE, Lee Z, Kim JW. Functional splicing assay of DSPP mutations in hereditary dentin defects. Oral Dis. 2011;17:690–695. doi: 10.1111/j.1601-0825.2011.01825.x. [DOI] [PubMed] [Google Scholar]
- 15.Arany S, Nakata A, Kameda T, Koyota S, Ueno Y, Sugiyama T. Phenotype properties of a novel spontaneously immortalized odontoblast-lineage cell line. Biochem Biophys Res Commun. 2006;342:718–724. doi: 10.1016/j.bbrc.2006.02.020. [DOI] [PubMed] [Google Scholar]
- 16.He W, Qu T, Yu Q, Wang Z, Wang H, Zhang J, Smith AJ. Lipopolysaccharide enhances decorin expression through the Toll-like receptor 4, myeloid differentiating factor 88, nuclear factor-kappa B, and mitogen-activated protein kinase pathways in odontoblast cells. J Endod. 2012;38:464–469. doi: 10.1016/j.joen.2011.12.021. [DOI] [PubMed] [Google Scholar]
- 17.Hanriot L, Keime C, Gay N, Faure C, Dossat C, Wincker P, Scote-Blachon C, Peyron C, Gandrillon O. A combination of LongSAGE with Solexa sequencing is well suited to explore the depth and the complexity of transcriptome. BMC Genomics. 2008;9:418. doi: 10.1186/1471-2164-9-418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sun Y, Lu Y, Chen S, Prasad M, Wang X, Zhu Q, Zhang J, Ball H, Feng J, Butler WT, Qin C. Key proteolytic cleavage site and full-length form of DSPP. J Dent Res. 2010;89:498–503. doi: 10.1177/0022034510363109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Audic S, Claverie JM. The significance of digital gene expression profiles. Genome Res. 1997;7:986–995. doi: 10.1101/gr.7.10.986. [DOI] [PubMed] [Google Scholar]
- 20.Kanehisa M, Araki M, Goto S, Hattori M, Hirakawa M, Itoh M, Katayama T, Kawashima S, Okuda S, Tokimatsu T, Yamanishi Y. KEGG for linking genomes to life and the environment. Nucleic Acids Res. 2008;36:D480–484. doi: 10.1093/nar/gkm882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hooper SD, Bork P. Medusa: a simple tool for interaction graph analysis. Bioinformatics. 2005;21:4432–4433. doi: 10.1093/bioinformatics/bti696. [DOI] [PubMed] [Google Scholar]
- 22.von Marschall Z, Fisher LW. Dentin sialophosphoprotein (DSPP) is cleaved into its two natural dentin matrix products by three isoforms of bone morphogenetic protein-1 (BMP1) Matrix Biol. 2010;29:295–303. doi: 10.1016/j.matbio.2010.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.von Marschall Z, Mok S, Phillips MD, McKnight DA, Fisher LW. Rough endoplasmic reticulum trafficking errors by different classes of mutant dentin sialophosphoprotein (DSPP) cause dominant negative effects in both dentinogenesis imperfecta and dentin dysplasia by entrapping normal DSPP. J Bone Miner Res. 2012;27:1309–1321. doi: 10.1002/jbmr.1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shyu AB, Wilkinson MF, van Hoof A. Messenger RNA regulation: to translate or to degrade. EMBO J. 2008;27:471–481. doi: 10.1038/sj.emboj.7601977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Staines KA, MacRae VE, Farquharson C. The importance of the SIBLING family of proteins on skeletal mineralisation and bone remodelling. J Endocrinol. 2012;214:241–255. doi: 10.1530/JOE-12-0143. [DOI] [PubMed] [Google Scholar]
- 26.Ducy P, Karsenty G. Two distinct osteoblast-specific cis-acting elements control expression of a mouse osteocalcin gene. Mol Cell Biol. 1995;15:1858–1869. doi: 10.1128/mcb.15.4.1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Saito T, Kobayashi F, Fujii T, Bessho K. Effect of phosphophoryn on rhBMP-2-induced bone formation. Arch Oral Biol. 2004;49:239–243. doi: 10.1016/j.archoralbio.2003.08.003. [DOI] [PubMed] [Google Scholar]
- 28.Saito T, Yamauchi M, Abiko Y, Matsuda K, Crenshaw MA. In vitro apatite induction by phosphophoryn immobilized on modified collagen fibrils. J Bone Miner Res. 2000;15:1615–1619. doi: 10.1359/jbmr.2000.15.8.1615. [DOI] [PubMed] [Google Scholar]
- 29.Weiner S, Veis A, Beniash E, Arad T, Dillon JW, Sabsay B, Siddiqui F. Peritubular dentin formation: crystal organization and the macromolecular constituents in human teeth. J Struct Biol. 1999;126:27–41. doi: 10.1006/jsbi.1999.4096. [DOI] [PubMed] [Google Scholar]
- 30.Gaur T, Lengner CJ, Hovhannisyan H, Bhat RA, Bodine PV, Komm BS, Javed A, van Wijnen AJ, Stein JL, Stein GS, Lian JB. Canonical WNT signaling promotes osteogenesis by directly stimulating Runx2 gene expression. J Biol Chem. 2005;280:33132–33140. doi: 10.1074/jbc.M500608200. [DOI] [PubMed] [Google Scholar]
- 31.Jadlowiec JA, Zhang X, Li J, Campbell PG, Sfeir C. Extracellular matrix-mediated signaling by dentin phosphophoryn involves activation of the Smad pathway independent of bone morphogenetic protein. J Biol Chem. 2006;281:5341–5347. doi: 10.1074/jbc.M506158200. [DOI] [PubMed] [Google Scholar]
- 32.Chen S, Rani S, Wu Y, Unterbrink A, Gu TT, Gluhak-Heinrich J, Chuang HH, Macdougall M. Differential regulation of dentin sialophosphoprotein expression by Runx2 during odontoblast cytodifferentiation. J Biol Chem. 2005;280:29717–29727. doi: 10.1074/jbc.M502929200. [DOI] [PubMed] [Google Scholar]
- 33.Maruyama Z, Yoshida CA, Furuichi T, Amizuka N, Ito M, Fukuyama R, Miyazaki T, Kitaura H, Nakamura K, Fujita T, Kanatani N, Moriishi T, Yamana K, Liu W, Kawaguchi H, Nakamura K, Komori T. Runx2 determines bone maturity and turnover rate in postnatal bone development and is involved in bone loss in estrogen deficiency. Dev Dyn. 2007;236:1876–1890. doi: 10.1002/dvdy.21187. [DOI] [PubMed] [Google Scholar]
- 34.Qian H, Zhao Y, Peng Y, Han C, Li S, Huo N, Ding Y, Duan Y, Xiong L, Sang H. Activation of cannabinoid receptor CB2 regulates osteogenic and osteoclastogenic gene expression in human periodontal ligament cells. J Periodontal Res. 2010;45:504–511. doi: 10.1111/j.1600-0765.2009.01265.x. [DOI] [PubMed] [Google Scholar]
- 35.Wajih N, Borras T, Xue W, Hutson SM, Wallin R. Processing and transport of matrix gamma-carboxyglutamic acid protein and bone morphogenetic protein-2 in cultured human vascular smooth muscle cells: evidence for an uptake mechanism for serum fetuin. J Biol Chem. 2004;279:43052–43060. doi: 10.1074/jbc.M407180200. [DOI] [PubMed] [Google Scholar]
- 36.Li X, Zhou M, Wang X, Li R, Han N, Zhang Q. Quantitative determination of high-temperature requirement protein A1 and its possible associated molecules during induced reparative dentin formation. J Endod. 2012;38:814–820. doi: 10.1016/j.joen.2012.03.009. [DOI] [PubMed] [Google Scholar]
- 37.Oka C, Tsujimoto R, Kajikawa M, Koshiba-Takeuchi K, Ina J, Yano M, Tsuchiya A, Ueta Y, Soma A, Kanda H, Matsumoto M, Kawaichi M. HtrA1 serine protease inhibits signaling mediated by Tgfbeta family proteins. Development. 2004;131:1041–1053. doi: 10.1242/dev.00999. [DOI] [PubMed] [Google Scholar]
- 38.Hadfield KD, Rock CF, Inkson CA, Dallas SL, Sudre L, Wallis GA, Boot-Handford RP, Canfield AE. HtrA1 inhibits mineral deposition by osteoblasts: requirement for the protease and PDZ domains. J Biol Chem. 2008;283:5928–5938. doi: 10.1074/jbc.M709299200. [DOI] [PubMed] [Google Scholar]
- 39.Zurick KM, Qin C, Bernards MT. Adhesion of MC3T3-E1 cells bound to dentin phosphoprotein specifically bound to collagen type I. J Biomed Mater Res A. 2012;100:2492–2498. doi: 10.1002/jbm.a.34159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Suzuki S, Sreenath T, Haruyama N, Honeycutt C, Terse A, Cho A, Kohler T, Muller R, Goldberg M, Kulkarni AB. Dentin sialoprotein and dentin phosphoprotein have distinct roles in dentin mineralization. Matrix Biol. 2009;28:221–229. doi: 10.1016/j.matbio.2009.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.