CONSPECTUS
Protein kinases are ubiquitous enzymes with critical roles in cellular processes and pathology. As a result, researchers have studied their activity and regulatory mechanisms extensively. Thousands of X-ray structures give snapshots of the architectures of protein kinases in various states of activation and ligand binding. However, the extent of and manner by which protein motions and conformational dynamics underlie the function and regulation of these important enzymes is not well understood. Nuclear magnetic resonance (NMR) methods provide complementary information about protein conformation and dynamics in solution. However, until recently, the large size of these enzymes prevented researchers from using these methods with kinases. Developments in transverse relaxation-optimized spectroscopy (TROSY)-based techniques and more efficient isotope labeling strategies are now allowing researchers to carry out NMR studies on full-length protein kinases.
In this Account, we describe recent insights into the role of dynamics in protein kinase regulation and catalysis that have been gained from NMR measurements of chemical shift changes and line broadening, residual dipolar couplings, and relaxation. These findings show strong associations between protein motion and events that control kinase activity. Dynamic and conformational changes occurring at ligand binding sites and other regulatory domains of these proteins propagate to conserved kinase core regions that mediate catalytic function. NMR measurements of slow time scale (microsecond to millisecond) motions also reveal that kinases carry out global exchange processes that synchronize multiple residues and allosteric interconversion between conformational states. Activating covalent modifications or ligand binding to form the Michaelis complex can induce these global processes. Inhibitors can also exploit the exchange properties of kinases by using conformational selection to form dynamically quenched states.
These investigations have revealed that kinases are highly dynamic enzymes, whose regulation by interdomain interactions, ligand binding, and covalent modifications involve changes in motion and conformational equilibrium in a manner that can be correlated with function. Thus, NMR provides a unique window into the role of protein dynamics in kinase regulation and catalysis with important implications for drug design.
The involvement of eukaryotic protein kinases in nearly all intracellular processes has prompted extensive structural studies on this important class of enzymes, beginning with the first X-ray structure of a protein kinase more than 20 years ago.1,2 Since then, more than 6000 kinase structures have been added to the PDB database, yielding deep insights into the mechanisms underlying kinase regulation. The static views obtained by X-ray crystallography are greatly enhanced by complementary solution studies that probe conformational dynamics. NMR spectroscopy is a powerful technique to study the dynamics of proteins in solution, but until recently there have been only limited applications of NMR to studies of protein kinases due to their relatively large size, which leads to fast relaxation of the NMR signals. NMR techniques that increase the signal-to-noise for larger proteins include transverse relaxation-optimized spectroscopy (TROSY) methods,3,4 which select slow relaxation signals, and protein labeling methods5,6 such as perdeuteration, which reduces the effect of surrounding protons on relaxation. These now allow glimpses into solution structures and dynamics of protein kinases. This Account highlights recent studies that use NMR to examine the contributions of dynamics to regulation of protein kinases, yielding fundamental insights into their mechanisms for activation, inhibition, and catalytic function.
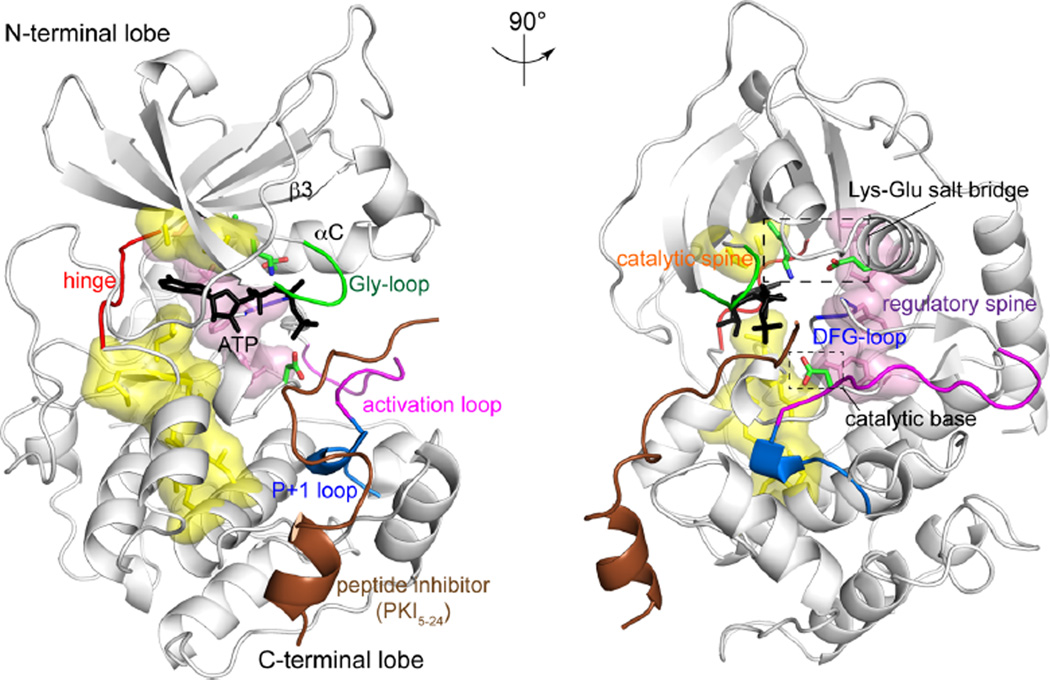
Eukaryotic protein kinases share a conserved catalytic domain, comprised of N-terminal and C-terminal lobes connected by a hinge (Figure 1).2,7,8 ATP binds the active site cleft between the lobes, forming critical contacts with residues and motifs that are conserved among kinases. These contacts include a conserved lysine residue and backbone amides in a glycine-rich motif (usually referred to as “Gly-loop” in protein kinases and “P-loop” in other kinases, dehydrogenases, and ATPases) in the N-terminal lobe, which form hydrogen bonds to the ATP phosphoryl oxygens, backbone atoms in the hinge, which hydrogen bond with the adenine ring, and the aspartate side chain in a conserved Asp-Phe-Gly motif (DFG-loop) in the C-terminal lobe, which coordinates Mg2+. The activation loop and peptide recognition segment (P + 1 loop) in the C-terminal lobe of the kinase form contacts with substrate, conferring sequence specificity and positioning of the substrate hydroxyl acceptor. A conserved aspartate residue in the active site serves as the catalytic base for phosphoryl transfer from ATP to substrate.
Figure 1.
The architecture of protein kinases. The X-ray structure of the PKA catalytic subunit bound to ATP (black) and peptide inhibitor, PKI5–24 (brown) (PDB 1ATP43). Elements conserved among protein kinases that are needed for catalytic function are labeled, including the Gly-loop, Lys–Glu salt bridge, DFG-loop, hinge, activation loop, P + 1 loop, and catalytic base. Space filled segments indicate internal hydrophobic structural motifs, named regulatory (pink) and catalytic (yellow) “spines”.
Key structural arrangements accompany kinase activation by covalent modifications or by interactions with regulatory subunits and domains.9–11 The activation loop is a flexible segment that can rearrange substantially and is often the location for regulatory phosphorylation.10 Phosphorylation at the activation loop allows ion pair interactions with an active site arginine residue to orient the catalytic base. The conserved N-terminal lysine, located in strand β3, interacts with a glutamate residue in helix αC to form a Lys–Glu salt bridge, often coupled with lateral movement of helix αC.12 Catalytic activity requires the proper alignment of internal hydrophobic residues, which form structural motifs, termed regulatory and catalytic “spines”, which stabilize the position of active site residues.13 In particular, an assembled regulatory spine often indicates an active kinase structure.13 Together, these represent common locations for conformational changes that contribute to kinase activation.
Recent reports illustrate the use of TROSY-based NMR correlation and relaxation experiments to address the essential role of dynamics in kinase regulation.14–20 Studies of the tyrosine kinases Eph and c-Abl, the MAP kinases p38α and ERK2, and the cAMP-dependent kinase reveal regulation of solution conformational equilibria and active site dynamics by interdomain interactions, regulatory covalent modifications, and binding of nucleotide, substrates, and inhibitors. These studies expand our understanding of protein kinases as dynamic enzymes, whose motions underlie mechanisms for regulation.
AUTOINHIBITORY DOMAIN INTERACTIONS IN THE Eph RECEPTOR TYROSINE KINASE
NMR analysis of the ephrin receptor tyrosine kinase (Eph RTK) reveals that dynamics in the active site can be controlled through autoinhibitory domain interactions.14 Eph RTK is a transmembrane receptor involved in promoting axon guidance and cell migration, which is activated by dimerization upon binding the ephrin ligand to its extracellular domain.21,22 X-ray structures of the ephrin type-B receptor 2 (EphB2) show that an autoinhibitory α-helical juxtamembrane segment (JMS) contacts the N-terminal lobe of the kinase domain (Figure 2a), resulting in distortion of helix αC into an inactive, kinked conformation and disorder of the activation loop.14,22 Ephrin binding induces autophosphorylation at two tyrosine residues in the JMS, which disrupts intramolecular contacts and activates the kinase.23 However, because the X-ray structures of activated Eph RTKs retain the “inactive” helix αC conformation and disordered activation loop,14,24 the mechanism of activation was uncertain. Thus, it is necessary to employ other tools, such as NMR, to elucidate how the active state could be achieved.
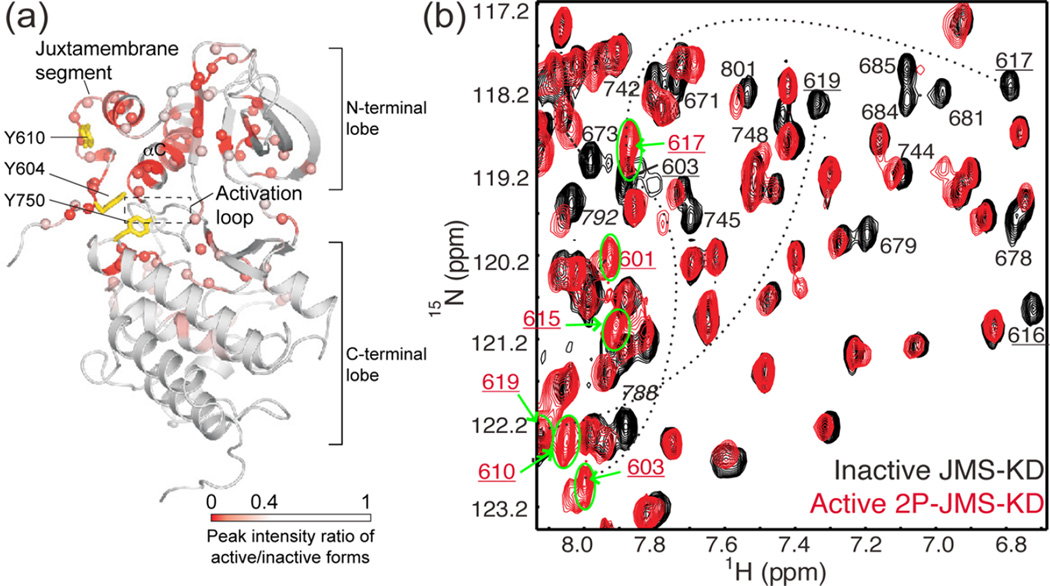
Figure 2.
Activation of Eph alters conformational dynamics in the kinase domain and juxtamembrane segment (JMS). (a) X-ray structure of the autoinhibited JMS-KD of EphB2 (PDB 1JPA14), mapping residues with spectral perturbations (red/pink) accompanying activation by phosphorylation. NMR line broadening and chemical shift perturbations reflect conformational and dynamic changes from the JMS contact site propagating to the active site cleft. Sites for activating tyrosine phosphorylation (Y604, Y610 for murine EphB2) and an activating mutation (Y750) are represented with yellow sticks. (b) 15N-HSQC spectra overlaying autoinhibited JMS-KD (black) and activated EphB2 2P-JMS-KD (red), where resonances showing significant spectral perturbations are labeled. In 2P-JMS-KD, peaks corresponding to JMS residues (numbers underlined) shifted to ~8 ppm (green circle and bold arrows), revealing an order-to-disorder transition in this region upon activation. Adapted with permission from ref 14. Copyright 2006 EMBO Press.
Autoinhibited and activated forms of EphB2 were examined using two-dimensional (2D) 1H,15N correlation experiments, which correlate the chemical shifts of backbone amide groups.14 These chemical shifts reflect the local chemical environment and therefore serve as probes for conformational changes. The autoinhibited state, modeled by a JMS-kinase domain (JMS-KD) construct, was compared with three active kinase forms, corresponding to JMS-KD phosphorylated at both tyrosine residues (2P-JMS-KD), JMS-KD with an activating mutation (JMS-KD-Y750A), and the kinase domain alone. The active kinases, 2P-JMS-KD and JMS-KD-Y750A, each showed spectral perturbations of JMS amides relative to the autoinhibited form. Upon activation, the 1H chemical shifts of JMS amides moved to ~8 ppm, a region typical for unfolded polypeptides (Figure 2b). This signifies a major conformational change in the JMS, involving its transition from an ordered structure to a disordered state upon release of autoinhibition.
Changes in conformational dynamics were also seen within the kinase domain upon activation.14 This was revealed by NMR line shape analysis, where nuclei that interconvert between multiple conformations can adopt varying line widths and numbers of peaks, depending on the ratio of the difference in chemical shift (|Δω|) to the rate constant for exchange between conformers (kex).25 Assuming two-state exchange (e.g., A ⇌ B, where kex = kAB + kBA), slow interconversion (kex ≪ |Δω|) leads to two resolved peaks, each representing a distinct state, whereas fast interconversion (kex ≫ |Δω|) yields only one peak having a chemical shift at the weighted average population of the two states. When interconversion occurs in an intermediate time regime (kex ≈ |Δω|), line broadening leads to reduction of peak height, often below noise. Thus, changes in peak number or line shape can reflect changes in dynamics and information about exchange rate constants.
In all three active states of EphB2, a similar set of amide resonances showed line broadening or chemical shift perturbations from the autoinhibited state.14 These spectral changes occurred in regions throughout the N-terminal lobe and in the active site cleft, including residues in the Gly-loop, the hinge, the activation loop, and the catalytic site (Figure 2a). Many of these regions were too far from the JMS-contacting sites to be explained simply by disrupted JMS interactions. Instead, they revealed allosteric changes in conformation or altered protein dynamics, propagating from the JMS interaction region in helix αC to the active site. The active mutant JMS-KD-Y750A showed multiple amides that had two peaks in slow exchange, whose chemical shifts indicated a major autoinhibited state and a new, lower populated state.14 This suggests a model in which the dynamics of the activated Eph RTK enables it to sample rare active conformers against a larger population of autoinhibited conformers. Thus, the NMR chemical shift and line broadening results reveal correlations between kinase activation and dynamics within the active site and explain why X-ray structures of active Eph RTKs resemble the inactive state.
CONTROL OF DYNAMICS BY SMALL MOLECULE INHIBITORS IN c-Abl AND p38α MAP KINASE
c-Abl
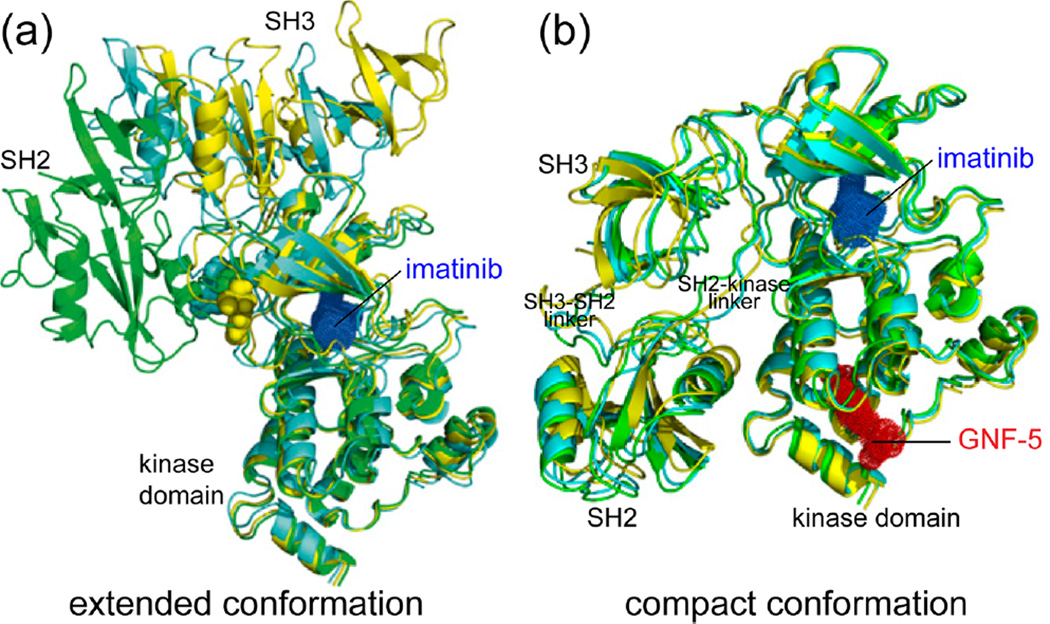
NMR studies of the tyrosine kinase c-Abl show how inhibitor binding can modulate conformational dynamics.15 c-Abl contains SH3, SH2, and kinase domains with flexible linkers between each domain.26 X-ray structures show that the SH3–SH2 domain interacts with the N-terminal and C-terminal lobes of the kinase domain to form a compact conformation, common among members of the Src tyrosine kinase family.27–29 The compact, inhibited conformation in c-Abl is maintained by an N-terminal myristoyl group, which inserts into a hydrophobic pocket in the C-terminal lobe of the kinase domain and stabilizes SH2–kinase interactions.27 X-ray structures of activated tyrosine kinases (e.g., c-Src) show changes in the relative orientation of the SH3–SH2 domain to form an extended conformation with fewer interdomain interactions.30 A similar disruption of the interaction between the SH3–SH2 and kinase domains has been proposed for the active c-Abl.28
Imatinib (Gleevec) is an ATP-competitive inhibitor of c-Abl, whose potency toward the constitutively active Bcr–Abl fusion protein has led to its clinical use for treating chronic myelogenous leukemia (CML).31,32 Several NMR experiments revealed that the binding of imatinib to c-Abl disrupts interactions between the SH3–SH2 and kinase domains, allowing formation of an extended conformation (Figure 3a).15 This was unexpected, given that the extended conformation is usually associated with activated forms of Src family kinases.28,30 It should be noted that the c-Abl protein in this NMR study was not myristoylated and therefore might be more prone to escaping autoinhibition. Nevertheless, 2D-NMR correlation experiments showed significant chemical shift perturbations for backbone amide resonances throughout the kinase and SH3–SH2 domains upon imatinib binding.15 These were supplemented with 15N relaxation experiments, from which the apparent rotational correlation time of molecular tumbling (τc) can be extracted.33 In the c-Abl/imatinib complex, faster tumbling within the SH3 and SH2 domains indicated their independent movement relative to the kinase domain.15 Finally, NMR residual dipolar coupling (RDC) experiments, which yield information on the relative orientations of individual amide bond vectors,34 confirmed that imatinib binding induces a change in the orientation of the SH3 and SH2 domains with respect to the kinase domain.15 By combining RDC constraints with radius of gyration (Rgyr) measurements from small-angle X-ray scattering (SAXS), structural models were calculated showing an extended conformation for the c-Abl/imatinib complex (Figure 3a).
Figure 3.
Differential regulation of interdomain interactions by c-Abl kinase inhibitors. (a) The lowest energy structural models of the c-Abl/imatinib complex calculated from RDC and SAXS measurements are shown in different colors. Each model shows the SH3–SH2 domain oriented in an extended conformation, relative to the kinase domain. (b) The lowest energy models of the ternary c-Abl/imatinib/GNF-5 complex show a compact conformation with multiple contacts between SH3–SH2 and kinase domains. Adapted with permission from ref 15. Copyright 2013 National Academy of Sciences.
NMR was also used to shed light on the role of dynamics in the interaction between imatinib and a new class of allosteric inhibitors.15 Chronic myelogenous leukemia patients treated with imatinib often acquire resistance through mutations in the kinase domain of Bcr–Abl that block inhibitor binding.35 To overcome this, ATP noncompetitive inhibitors have been developed, including GNF-5, which occupies the myristoyl-binding pocket of c-Abl (Figure 3b).36–38 In contrast to imatinib, the binding of GNF-5 to c-Abl induced local chemical shift perturbations that were restricted to the myristoyl-binding pocket and rotational correlation times indicating a rigid structure for the SH3–SH2 and kinase domains.15 These together with RDC constraints and Rgyr measurements showed that c-Abl/GNF-5 adopts the compact conformation.15
Significantly, the NMR measurements also indicated a compact conformation of the c-Abl/imatinib/GNF-5 ternary complex (Figure 3b).15 This means that the two classes of inhibitors have dramatically different effects on the protein conformation, where GNF-5 binding reverses the extended conformation induced by imatinib. Thus, GNF-5 alters the dynamic equilibrium of c-Abl to maintain the compact, autoinhibited state. Such behavior may provide inhibitors of this class with favorable clinical properties, given that GNF-5 or a related analogue, when combined with the ATP-competitive inhibitors imatinib or nilotinib, extend survival in animal models of Bcr–Abl-induced leukemia and delay the emergence of drug resistance mutations in Bcr–Abl.37
p38α MAP Kinase
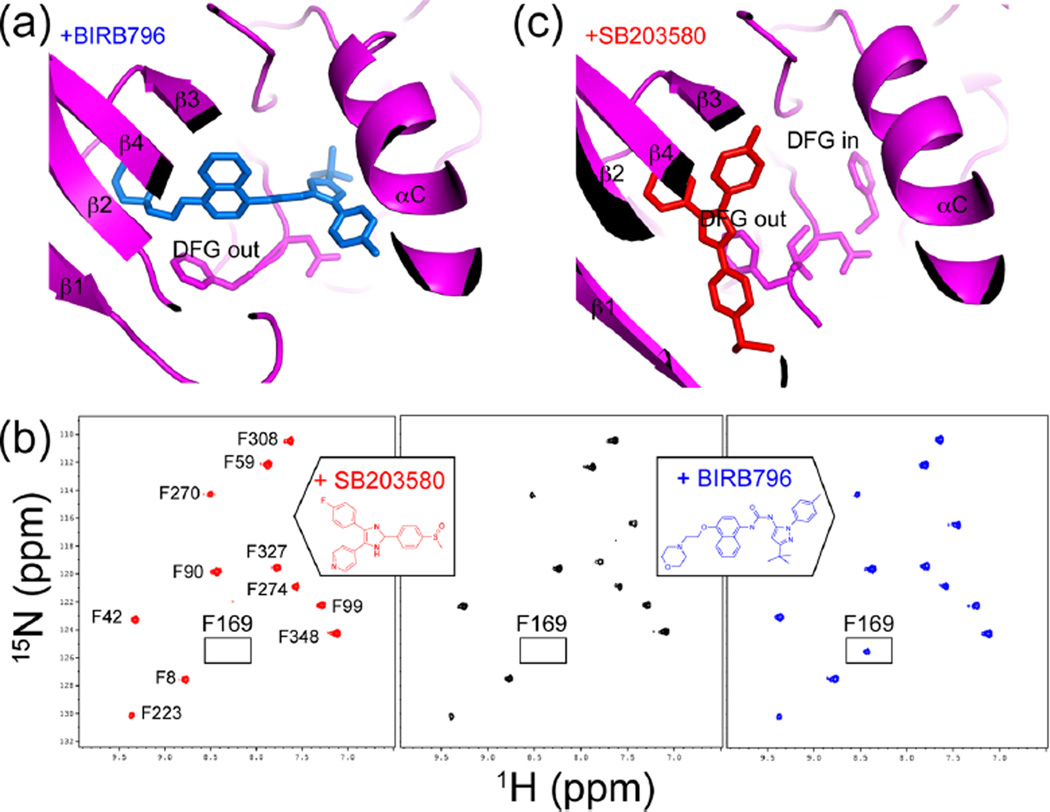
Alterations in kinase dynamics by inhibitor binding were also observed in the p38α MAP kinase,16 which mediates cell stress responses leading to inflammation and apoptosis.39 The X-ray structure of p38α bound to the ATP-competitive inhibitor BIRB796 reveals an allosteric mechanism involving movement of the Mg2+-coordinating aspartate residue located within the conserved DFG motif (Figure 4a).40 Here, aspartate is displaced from its active site conformation (“DFG-in”) through rotation of the Asp–Phe backbone to form an inactive “DFG-out” conformation, which exposes a hydrophobic pocket.40,41 BIRB796, like other so-called “type II” kinase inhibitors that have been identified for many kinases, occupies this new pocket and thus selects for and stabilizes the DFG-out conformation.41
Figure 4.
Conformational selection in binding the p38α inhibitor BIRB796. (a) X-ray structure of p38α (magenta) bound to BIRB796 (blue) (PDB 1KV240) shows selection for the inactive DFG-out conformation. (b) 15N-HSQC spectra show the Phe169 resonance, observed in p38α/BIRB796 (blue) but not observed in apo-p38α (black) or p38α/SB203580 (red) due to line broadening. Line broadening reflects altered dynamics leading to conformational exchange in the intermediate time regime. (c) X-ray structure of p38α (magenta) bound to SB203580 (red) (PDB 2EWA16) shows that both DFG-in and DFG-out conformers can form. Adapted with permission from ref 16. Copyright 2006 John Wiley and Sons.
NMR has been used to show how the dynamics of p38α enables conformation selection by type II inhibitors.16 Line shape analysis of the amides of phenylalanine residues in p38α, followed using selective labeling with 1H,15N-phenylalanine, showed that the DFG-in and DFG-out conformations exist in equilibrium. In apo-p38α, the resonance for Phe169 within the DFG motif was not observed due to line broadening (Figure 4b), indicating its conformational exchange in the intermediate (microsecond to millisecond) regime. Therefore, the apoenzyme interconverts between conformers on a microsecond to millisecond time scale, transiently exposing the allosteric pocket in the DFG-out state. In contrast, in the p38α/BIRB796 complex, Phe169 appeared as a single amide resonance (Figure 4b), which could be associated with the DFG-out conformer based on the X-ray structure (Figure 4a). Thus, BIRB796 alters the motions in p38α by binding with higher affinity to the DFG-out conformer and trapping this inactive state. Interestingly, line broadening of Phe169 was also seen in p38α bound to the inhibitor SB203580, which does not occupy this pocket (Figure 4c). Thus, the dynamics of p38α provide properties of conformation selection that can be exploited specifically by type II inhibitors.
LINKAGE BETWEEN KINASE DYNAMICS AND CATALYSIS IN cAMP-DEPENDENT PROTEIN KINASE (PKA)
Elegant NMR experiments on the catalytic subunit of PKA have linked internal dynamics to its catalytic function.17–19 Two-dimensional NMR correlation experiments were used to examine binary and ternary intermediates of PKA in complex with the ATP analog β,γ-imidoadenosine 5′-triphosphate (AMP-PNP) and a peptide substrate.17 The binding of AMP-PNP induced chemical shift perturbations or line broadening of amide resonances for residues throughout the active site that are needed for catalytic function, including residues in the Gly-loop, the Lys–Glu salt bridge, the DFG-loop, the substrate-positioning (P + 1) loop, and comprising the hydrophobic spines. In contrast, an inactivating point mutation in the P + 1 loop attenuated the perturbations induced upon nucleotide binding, while also eliminating positive cooperativity in binding affinities between nucleotide and peptide substrate.17 Thus, a network of spectral changes induced by AMP-PNP reflect propagation of conformational and dynamic responses to the active site and correlate with ligand binding cooperativity.
NMR relaxation studies comparing apo- vs ligand-bound PKA demonstrated the relevance of perturbations in dynamics to enzyme turnover.18 Fast dynamics (picosecond to nanosecond) of amide bonds can be monitored by heteronuclear nuclear Overhauser effect (H–X NOE) experiments, which measure the relaxation of 15N nuclei via dipole–dipole interactions with 1H.33 Both the binary PKA/AMP-PNP and ternary PKA/AMP-PNP/peptide substrate complexes showed increased numbers of residues with fast dynamics, including residues in the catalytically important active site regions.18 Strikingly, binding of a high-affinity peptide inhibitor (PKI) to PKA/AMP-PNP reduced the fast time scale dynamics to levels matching that of apoenzyme.19 Thus, nucleotide binding increases dynamics within the active site, whereas inhibitor binding creates a conformationally “quenched” state.
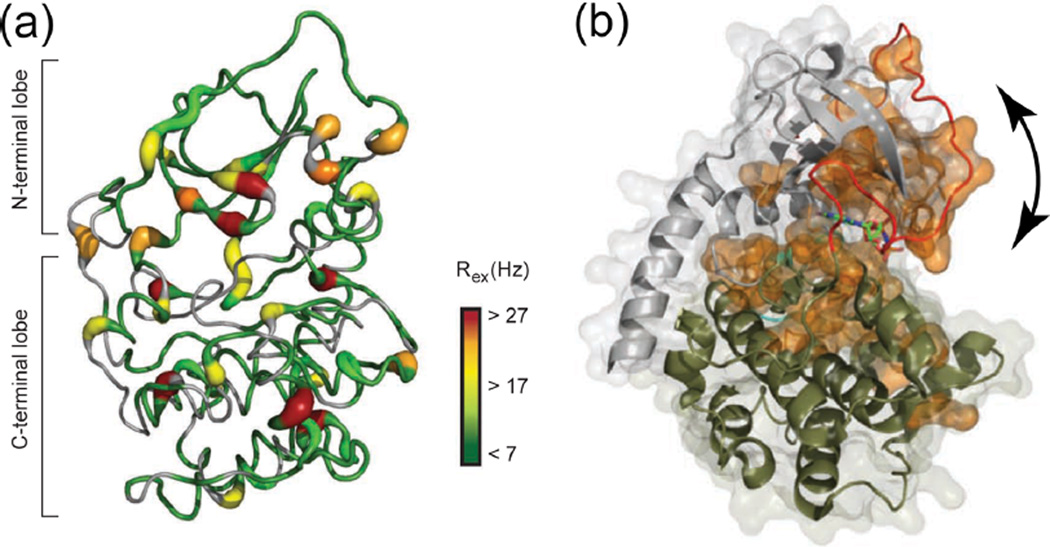
Slower motions were monitored by 15N relaxation experiments that measure Rex, the apparent rate constant for relaxation arising from chemical exchange between two states (e.g., A ⇌ B).42 In this experiment, Rex was estimated by a simplified form of the Hahn-echo experiment, where the transverse relaxation rate (R2) was measured at only one frequency, and chemical exchange-dependent processes were isolated by removing contributions from exchange-independent processes. Binding of AMP-PNP and peptide to mimic the Michaelis complex led to an increased number of residues that showed millisecond dynamics, again including those in the Gly, DFG, activation, and P + 1 loops (Figure 5a).18 Significantly, many residues showed synchrony with a global exchange process (Figure 5b). This was indicated by linear correlations between the square of the chemical shift difference (Δω2) and the Rex, suggesting that their exchange rate constants are the same.18 Thus, ligand binding to PKA induces a slow global motion described by two-state exchange. Based on X-ray structures of apoenzyme and ternary complex,1,18,43 this was interpreted as interdomain movements between N- and C-terminal lobes, leading to interconversion between open and closed conformations (Figure 5b). Assuming that the closed conformation is dominant in the ternary PKA/AMP-PNP/peptide complex, the rate constant for opening (kopen) could be estimated as 31 s−1.18 This value is similar to the kcat of 20 s−1, which is rate-limited by product dissociation (koff = 23 s−1).8 Thus, NMR reveals substantial effects of nucleotide and substrate binding on backbone dynamics at both fast and slow time scales. The slower dynamics in particular reveal global motions indicating ligand-induced exchange between open and closed conformations, where interdomain opening correlates with the rate-limiting step in catalytic turnover.
Figure 5.
Ligand binding regulates conformational dynamics of PKA. (a) PKA backbone structure showing slow (microsecond to millisecond) dynamics of amides in the ternary PKA/AMP-PNP/peptide complex, reflected by the magnitude of relaxation caused by chemical exchange (Rex). (b) Space filled residues (orange) localized around the active site reflect amides that synchronize to the same exchange process, modeled as conformational interconversion between open and closed states (arrow). Adapted with permission from ref 18. Copyright 2010 Nature Publishing Group.
GLOBAL MOTIONS ARE INDUCED BY PHOSPHORYLATION IN ERK2
The MAP kinase extracellular regulated kinase-2 (ERK2) is activated by dual phosphorylation at threonine and tyrosine residues in the activation loop,44 increasing the rate of phosphoryltransfer by 60000-fold.45 X-ray structures of the active, phosphorylated kinase (2P-ERK2) and the inactive, unphosphorylated kinase (0P-ERK2) show that phosphorylation rearranges the activation loop to accommodate substrate binding, reorganizes active site residues, and exposes a pocket for substrate docking (Figure 6a,b).46–48 NMR revealed global dynamics induced upon kinase activation, which were not apparent from the X-ray structures.20 Here, NMR relaxation experiments and line shape analysis were used to monitor motions of 1H,13C methyls on isoleucine, leucine, and valine side chains, in protein prepared using isotopically labeled precursors. The fast rotation of methyls offers favorable relaxation properties, yielding high signal-to-noise and sharp peaks.6
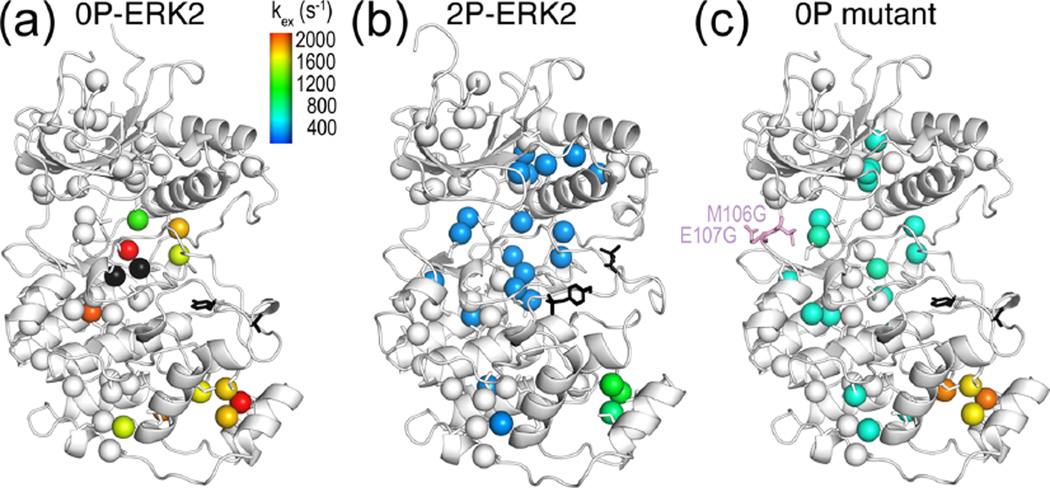
Figure 6.
Global motion accompanies activation of ERK2 by phosphorylation. (a) Isoleucine, leucine, and valine methyls in the kinase core of 0P-ERK2 show large variations in kex, reflecting uncoupled, local motions. Each methyl could be fitted individually, except those in black spheres, which show microsecond to millisecond dynamics but high errors in kex. (b) In contrast, methyls throughout the core of 2P-ERK2 (blue) could be fit together to a two-state exchange process with global kex ≈ 300 s−1. Black sticks show the phosphorylation sites in the activation loop. (c) A mutation at the hinge (M106G, E107G) induces global exchange, even in the absence of phosphorylation. Adapted with permission from ref 20. Copyright 2014 National Academy of Sciences.
The Carr–Purcell–Meiboom–Gill (CPMG) relaxation dispersion experiment is used to monitor dynamics on the microsecond to millisecond time scale.25,49 It applies a Hahn-echo experiment at varying refocusing pulse frequencies, measuring exchange by analysis of transverse relaxation rates as a function of the interval between pulses. Under optimal conditions, each methyl can be fit to a two-state model to extract the rate constant for exchange (kex) and the populations of the two states (pA, pB).50 The CPMG experiments revealed striking changes in dynamics upon ERK2 activation.20 Fitted individually, methyls within the catalytic core of 0P-ERK2 showed millisecond dynamics with large variations in kex (Figure 6a). By contrast, the methyls in 2P-ERK2 converged to a narrow range of rates, allowing most to be fit globally to a single two-state model with kex = 300 s−1 and pA/pB = 80%:20% (Figure 6b). Thus, motions in 0P-ERK2 appear uncoupled, whereas motions in 2P-ERK2 are dominated by a global exchange process.
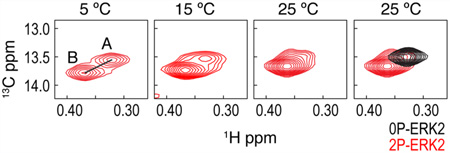
Multiple methyl resonances in 2P-ERK2 appeared as two peaks in slow exchange, yielding populations from their relative intensities that agreed well with those measured by CPMG.20 By quantifying changes in population as a function of temperature, enthalpies for exchange were found to be similar at different methyls, confirming the same underlying process. By comparison of chemical shifts, the single peaks in 0P-ERK2 could be matched to the peaks with lower populations in 2P-ERK2.20 This means that the unphosphorylated kinase is constrained into one conformer (e.g., A), whereas the phosphorylated kinase forms two separate conformers that exchange on a millisecond time scale (e.g., A ⇌ B). Thus, phosphorylation alters the dynamics of ERK2 to allow interconversion with a new conformer. Insight into the nature of the constraint in 0P-ERK2 was provided by a mutant that modified hinge residues to increase flexibility (M106G, E107G). This mutant altered the conformational exchange such that many residues showed global exchange, even in the absence of phosphorylation (Figure 6c).20 Kinetic studies showed that this mutant activated the kinase in a manner that bypassed the need for tyrosine phosphorylation.51 This implies that the global motions induced by phosphorylation involve perturbations that propagate over long distances, from the activation loop to the hinge.
Additional studies showed that the equilibrium between the two states in 2P-ERK2 can be shifted by inhibitor binding, revealing a new mechanism for conformation selection. Chemical shifts of 2P-ERK2 in complex with an ATP-competitive inhibitor, Vertex-11e, showed complete conversion to the new conformer.52 This means that the effect of phosphorylation on dynamics exposes sites that can be selectively recognized by Vertex-11e. This is analogous to the recognition of DFG-out conformations by type II inhibitors; however, the DFG-in/DFG-out mechanism does not occur in ERK2.46,47 Instead, the global motions in 2P-ERK2 reflect a different allosteric mechanism, which can be exploited by a new class of conformation selective inhibitors.52 Structural analyses of this mechanism should yield new models of kinase activation and inhibition.
PERSPECTIVES AND OUTLOOK
There are only a handful of examples where NMR has been used to study functional protein kinases. Nevertheless, these studies have revealed a central role for solution conformational changes and dynamics in kinase regulation and function. NMR chemical shift perturbation and line shape analyses reveal that changes in conformation and dynamics are strongly affected by enzyme activation and ligand binding. NMR relaxation experiments provide a powerful approach to quantify changes in the kinetics of conformational exchange, and to distinguish uncoupled motions between individual residues from global processes. The NMR data do not define the conformational changes undergoing dynamics (e.g., DFG in/out, conformational selection); the interpretation of which requires structural information from X-ray crystallography or other experiments. Nevertheless, the ability of NMR to measure populations of conformations in solution and provide a kinetic view of their interconversion, is a singular strength of this method that is difficult to achieve by other ensemble techniques.
Certain common findings emerge from these studies. First, as seen in Eph and PKA,14,17 common patterns of regulated dynamics occur around the kinase active site, involving conserved motifs that participate in enzyme turnover. These are often distant from interaction sites with regulatory domains or ligands, suggesting that the kinase architecture enables propagation of motions from distal sites to catalytically important regions. Second, global motions can be regulated in kinases, induced by ligand binding to form the Michaelis complex in PKA,18 or regulatory phosphorylation to activate ERK2.20 These demonstrate the importance of motions involving conformational interconversion on microsecond to millisecond time scales for catalytic function. Most conclusions so far are based on correlative results. Therefore, a goal for the future will be to prove that these patterns of regulated dynamics and global motions are functionally important for kinase activity, which will require deeper comparisons between NMR relaxation measurements and enzyme kinetics.
Finally, NMR studies reveal a profound influence of inhibitor binding on kinase dynamics. Most inhibitors restrict dynamics or impede allosteric mechanisms through conformational selection, as in the examples of PKI binding to PKA,19 BIRB796 binding to p38α,16 GNF-5 binding to c-Abl,15 and Vertex-11e binding to ERK2.52 Other inhibitors unexpectedly show the opposite effect of releasing constraints to conformational interconversion, as in the example of imatinib binding to c-Abl.15 Thus, NMR yields novel insight into the solution behavior of kinases, by showing that different inhibitors elicit distinct effects on dynamics.
ACKNOWLEDGMENTS
This work was supported by NIH Grants R01GM074134 (N.G.A.) and T32GM008759 (J.C.L.). We are grateful to Dr. Johannes Rudolph for insightful discussions and review of the manuscript and to Drs. Julie Forman-Kay, Silke Wiesner, Stephan Grzesiek, Harald Schwalbe, and Gianluigi Veglia for permission to reproduce figures and data.
Biographies
Yao Xiao received her B.S. in Biological Sciences from Fudan University in 2008 and is currently a Ph.D. candidate with Natalie Ahn and Arthur Pardi at U. Colorado, Boulder. Her research focuses on NMR studies of conformational dynamics in ERK2.
Jennifer Liddle received her B.S. in Biochemistry from Western Washington University in 2011 and is currently a Ph.D. candidate with Natalie Ahn at U. Colorado, Boulder. Her research focuses on characterizing conformational mobility in MAP kinases using hydrogen exchange mass spectrometry.
Arthur Pardi obtained his A.B. in Chemistry from U. California, San Diego, and his Ph.D. in Chemistry from U. California, Berkeley, working with I. Tinoco, Jr. He was a NSF postdoctoral fellow with K. Wüthrich at E.T.H., Zurich. He was Assistant Professor at Rutgers University before moving to U. Colorado, Boulder, where he is currently Professor of Chemistry and Biochemistry. His research focuses on studies of the structure, function, and dynamics of proteins and RNA using NMR and other biophysical approaches.
Natalie Ahn received her B.S. in Chemistry at U. Washington and her Ph.D. in Chemistry at U. California, Berkeley, under Judith Klinman. She carried out postdoctoral studies at U. Washington under Edwin G. Krebs and Christoph de Haën. She is currently Professor of Distinction of Chemistry and Biochemistry at U. Colorado, Boulder, where her research integrates proteomics, cell biology, and biophysics to discover new signal transduction mechanisms.
Footnotes
The authors declare no competing financial interest.
REFERENCES
- 1.Knighton DR, Zheng JH, Ten Eyck LF, Ashford VA, Xuong NH, Taylor SS, Sowadski JM. Crystal structure of the catalytic subunit of cyclic adenosine monophosphate-dependent protein kinase. Science. 1991;253:407–414. doi: 10.1126/science.1862342. [DOI] [PubMed] [Google Scholar]
- 2.Taylor SS, Kornev AP. Protein kinases: Evolution of dynamic regulatory proteins. Trends Biochem. Sci. 2011;36:65–77. doi: 10.1016/j.tibs.2010.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Venters RA, Thompson R, Cavanagh J. Current approaches for the study of large proteins by NMR. J. Mol. Struct. 2002;602:275–292. [Google Scholar]
- 4.Wider G, Wuthrich K. NMR spectroscopy of large molecules and multimolecular assemblies in solution. Curr. Opin. Struct. Biol. 1999;9:594–601. doi: 10.1016/s0959-440x(99)00011-1. [DOI] [PubMed] [Google Scholar]
- 5.Grzesiek S, Anglister J, Ren H, Bax A. C-13 Line Narrowing by H-2 Decoupling in H-2/C-13/N-15-Enriched Proteins - Application to Triple-Resonance 4d J-Connectivity of Sequential Amides. J. Am. Chem. Soc. 1993;115:4369–4370. [Google Scholar]
- 6.Ruschak AM, Kay LE. Methyl groups as probes of supra-molecular structure, dynamics and function. J. Biomol. NMR. 2010;46:75–87. doi: 10.1007/s10858-009-9376-1. [DOI] [PubMed] [Google Scholar]
- 7.Johnson DA, Akamine P, Radzio-Andzelm E, Madhusudan M, Taylor SS. Dynamics of cAMP-dependent protein kinase. Chem. Rev. 2001;101:2243–2270. doi: 10.1021/cr000226k. [DOI] [PubMed] [Google Scholar]
- 8.Adams JA. Kinetic and catalytic mechanisms of protein kinases. Chem. Rev. 2001;101:2271–2290. doi: 10.1021/cr000230w. [DOI] [PubMed] [Google Scholar]
- 9.Huse M, Kuriyan J. The conformational plasticity of protein kinases. Cell. 2002;109:275–282. doi: 10.1016/s0092-8674(02)00741-9. [DOI] [PubMed] [Google Scholar]
- 10.Nolen B, Taylor S, Ghosh G. Regulation of protein kinases; controlling activity through activation segment conformation. Mol. Cell. 2004;15:661–675. doi: 10.1016/j.molcel.2004.08.024. [DOI] [PubMed] [Google Scholar]
- 11.Endicott JA, Noble ME, Johnson LN. The structural basis for control of eukaryotic protein kinases. Annu. Rev. Biochem. 2012;81:587–613. doi: 10.1146/annurev-biochem-052410-090317. [DOI] [PubMed] [Google Scholar]
- 12.Palmieri L, Rastelli G. alpha C helix displacement as a general approach for allosteric modulation of protein kinases. Drug Discovery Today. 2013;18:407–414. doi: 10.1016/j.drudis.2012.11.009. [DOI] [PubMed] [Google Scholar]
- 13.Kornev AP, Haste NM, Taylor SS, Eyck LF. Surface comparison of active and inactive protein kinases identifies a conserved activation mechanism. Proc. Natl. Acad. Sci. U. S. A. 2006;103:17783–17788. doi: 10.1073/pnas.0607656103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wiesner S, Wybenga-Groot LE, Warner N, Lin H, Pawson T, Forman-Kay JD, Sicheri F. A change in conformational dynamics underlies the activation of Eph receptor tyrosine kinases. EMBO J. 2006;25:4686–4696. doi: 10.1038/sj.emboj.7601315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Skora L, Mestan J, Fabbro D, Jahnke W, Grzesiek S. NMR reveals the allosteric opening and closing of Abelson tyrosine kinase by ATP-site and myristoyl pocket inhibitors. Proc. Natl. Acad. Sci. U. S. A. 2013;110:E4437–E4445. doi: 10.1073/pnas.1314712110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vogtherr M, Saxena K, Hoelder S, Grimme S, Betz M, Schieborr U, Pescatore B, Robin M, Delarbre L, Langer T, Wendt KU, Schwalbe H. NMR characterization of kinase p38 dynamics in free and ligand-bound forms. Angew. Chem., Int. Ed. 2006;45:993–997. doi: 10.1002/anie.200502770. [DOI] [PubMed] [Google Scholar]
- 17.Masterson LR, Mascioni A, Traaseth NJ, Taylor SS, Veglia G. Allosteric cooperativity in protein kinase A. Proc. Natl. Acad. Sci. U. S. A. 2008;105:506–511. doi: 10.1073/pnas.0709214104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Masterson LR, Cheng C, Yu T, Tonelli M, Kornev A, Taylor SS, Veglia G. Dynamics connect substrate recognition to catalysis in protein kinase A. Nat. Chem. Biol. 2010;6:821–828. doi: 10.1038/nchembio.452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Masterson LR, Shi L, Metcalfe E, Gao J, Taylor SS, Veglia G. Dynamically committed, uncommitted, and quenched states encoded in protein kinase A revealed by NMR spectroscopy. Proc. Natl. Acad. Sci. U. S. A. 2011;108:6969–6974. doi: 10.1073/pnas.1102701108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xiao Y, Lee T, Latham MP, Warner LR, Tanimoto A, Pardi A, Ahn NG. Phosphorylation releases constraints to domain motion in ERK2. Proc. Natl. Acad. Sci. U. S. A. 2014;111:2506–2511. doi: 10.1073/pnas.1318899111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hubbard SR. Juxtamembrane autoinhibition in receptor tyrosine kinases. Nat. Rev. Mol. Cell Biol. 2004;5:464–471. doi: 10.1038/nrm1399. [DOI] [PubMed] [Google Scholar]
- 22.Egea J, Klein R. Bidirectional Eph-ephrin signaling during axon guidance. Trends Cell Biol. 2007;17:230–238. doi: 10.1016/j.tcb.2007.03.004. [DOI] [PubMed] [Google Scholar]
- 23.Binns KL, Taylor PP, Sicheri F, Pawson T, Holland SJ. Phosphorylation of tyrosine residues in the kinase domain and juxtamembrane region regulates the biological and catalytic activities of Eph receptors. Mol. Cell. Biol. 2000;20:4791–4805. doi: 10.1128/mcb.20.13.4791-4805.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nowakowski J, Cronin CN, McRee DE, Knuth MW, Nelson CG, Pavletich NP, Rogers J, Sang BC, Scheibe DN, Swanson RV, Thompson DA. Structures of the cancer-related Aurora-A, FAK, EphA2 protein kinases from nanovolume crystallography. Structure. 2002;10:1659–1667. doi: 10.1016/s0969-2126(02)00907-3. [DOI] [PubMed] [Google Scholar]
- 25.Palmer AG., 3rd NMR characterization of the dynamics of biomacromolecules. Chem. Rev. 2004;104:3623–3640. doi: 10.1021/cr030413t. [DOI] [PubMed] [Google Scholar]
- 26.Hantschel O, Superti-Furga G. Regulation of the c-Abl and Bcr-Abl tyrosine kinases. Nat. Rev. Mol. Cell Biol. 2004;5:33–44. doi: 10.1038/nrm1280. [DOI] [PubMed] [Google Scholar]
- 27.Nagar B, Hantschel O, Young MA, Scheffzek K, Veach D, Bornmann W, Clarkson B, Superti-Furga G, Kuriyan J. Structural basis for the autoinhibition of c-Abl tyrosine kinase. Cell. 2003;112:859–871. doi: 10.1016/s0092-8674(03)00194-6. [DOI] [PubMed] [Google Scholar]
- 28.Nagar B, Hantschel O, Seeliger M, Davies JM, Weis WI, Superti-Furga G, Kuriyan J. Organization of the SH3-SH2 unit in active and inactive forms of the c-Abl tyrosine kinase. Mol. Cell. 2006;21:787–798. doi: 10.1016/j.molcel.2006.01.035. [DOI] [PubMed] [Google Scholar]
- 29.Xu W, Doshi A, Lei M, Eck MJ, Harrison SC. Crystal structures of c-Src reveal features of its autoinhibitory mechanism. Mol. Cell. 1999;3:629–638. doi: 10.1016/s1097-2765(00)80356-1. [DOI] [PubMed] [Google Scholar]
- 30.Cowan-Jacob SW, Fendrich G, Manley PW, Jahnke W, Fabbro D, Liebetanz J, Meyer T. The crystal structure of a c-Src complex in an active conformation suggests possible steps in c-Src activation. Structure. 2005;13:861–871. doi: 10.1016/j.str.2005.03.012. [DOI] [PubMed] [Google Scholar]
- 31.Hosfield DJ, Mol CD. Cancer Drug Design and Discovery. New York: Academic Press; 2008. Targeting inactive kinases: Structure as a foundation for cancer drug discovery; pp. 229–252. [Google Scholar]
- 32.Capdeville R, Buchdunger E, Zimmermann J, Matter A. Glivec (STI571, imatinib), a rationally developed, targeted anticancer drug. Nat. Rev. Drug Discovery. 2002;1:493–502. doi: 10.1038/nrd839. [DOI] [PubMed] [Google Scholar]
- 33.Kay LE, Torchia DA, Bax A. Backbone dynamics of proteins as studied by N-15 inverse detected heteronuclear NMR-spectroscopy - application to staphylococcal nuclease. Biochemistry. 1989;28:8972–8979. doi: 10.1021/bi00449a003. [DOI] [PubMed] [Google Scholar]
- 34.Fitzkee NC, Bax A. Facile measurement of H-1-N-15 residual dipolar couplings in larger perdeuterated proteins. J. Biomol. NMR. 2010;48:65–70. doi: 10.1007/s10858-010-9441-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gorre ME, Mohammed M, Ellwood K, Hsu N, Paquette R, Rao PN, Sawyers CL. Clinical resistance to STI-571 cancer therapy caused by BCR-ABL gene mutation or amplification. Science. 2001;293:876–880. doi: 10.1126/science.1062538. [DOI] [PubMed] [Google Scholar]
- 36.Adrian FJ, Ding Q, Sim T, Velentza A, Sloan C, Liu Y, Zhang G, Hur W, Ding S, Manley P, Mestan J, Fabbro D, Gray NS. Allosteric inhibitors of Bcr-abl-dependent cell proliferation. Nat. Chem. Biol. 2006;2:95–102. doi: 10.1038/nchembio760. [DOI] [PubMed] [Google Scholar]
- 37.Zhang J, Adrian FJ, Jahnke W, Cowan-Jacob SW, Li AG, Iacob RE, Sim T, Powers J, Dierks C, Sun F, Guo GR, Ding Q, Okram B, Choi Y, Wojciechowski A, Deng X, Liu G, Fendrich G, Strauss A, Vajpai N, Grzesiek S, Tuntland T, Liu Y, Bursulaya B, Azam M, Manley PW, Engen JR, Daley GQ, Warmuth M, Gray NS. Targeting Bcr-Abl by combining allosteric with ATP-binding-site inhibitors. Nature. 2010;463:501–506. doi: 10.1038/nature08675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Iacob RE, Zhang J, Gray NS, Engen JR. Allosteric interactions between the myristate- and ATP-site of the Abl kinase. PLoS One. 2011;6(e15929) doi: 10.1371/journal.pone.0015929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zarubin T, Han J. Activation and signaling of the p38 MAP kinase pathway. Cell Res. 2005;15:11–18. doi: 10.1038/sj.cr.7290257. [DOI] [PubMed] [Google Scholar]
- 40.Pargellis C, Tong L, Churchill L, Cirillo PF, Gilmore T, Graham AG, Grob PM, Hickey ER, Moss N, Pav S, Regan J. Inhibition of p38 MAP kinase by utilizing a novel allosteric binding site. Nat. Struct. Biol. 2002;9:268–272. doi: 10.1038/nsb770. [DOI] [PubMed] [Google Scholar]
- 41.Zhao Z, Wu H, Wang L, Liu Y, Knapp S, Liu Q, Gray NS. Exploration of type II binding mode: A privileged approach for kinase inhibitor focused drug discovery? ACS Chem. Biol. 2014;9:1230–1241. doi: 10.1021/cb500129t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang C, Rance M, Palmer AG., 3rd Mapping chemical exchange in proteins with MW > 50 kD. J. Am. Chem. Soc. 2003;125:8968–8969. doi: 10.1021/ja035139z. [DOI] [PubMed] [Google Scholar]
- 43.Zheng J, Trafny EA, Knighton DR, Xuong NH, Taylor SS, Ten Eyck LF, Sowadski JM. 2.2 Å refined crystal structure of the catalytic subunit of cAMP-dependent protein kinase complexed with MnATP and a peptide inhibitor. Acta Crystallogr., Sect. D: Biol. Crystallogr. 1993;49:362–365. doi: 10.1107/S0907444993000423. [DOI] [PubMed] [Google Scholar]
- 44.Payne DM, Rossomando AJ, Martino P, Erickson AK, Her JH, Shabanowitz J, Hunt DF, Weber MJ, Sturgill TW. Identification of the regulatory phosphorylation sites in pp42/mitogen-activated protein kinase (MAP kinase) EMBO J. 1991;10:885–892. doi: 10.1002/j.1460-2075.1991.tb08021.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Prowse CN, Lew J. Mechanism of activation of ERK2 by dual phosphorylation. J. Biol. Chem. 2001;276:99–103. doi: 10.1074/jbc.M008137200. [DOI] [PubMed] [Google Scholar]
- 46.Zhang F, Strand A, Robbins D, Cobb MH, Goldsmith EJ. Atomic structure of the MAP kinase ERK2 at 2.3 A resolution. Nature. 1994;367:704–711. doi: 10.1038/367704a0. [DOI] [PubMed] [Google Scholar]
- 47.Canagarajah BJ, Khokhlatchev A, Cobb MH, Goldsmith EJ. Activation mechanism of the MAP kinase ERK2 by dual phosphorylation. Cell. 1997;90:859–869. doi: 10.1016/s0092-8674(00)80351-7. [DOI] [PubMed] [Google Scholar]
- 48.Lee T, Hoofnagle AN, Kabuyama Y, Stroud J, Min X, Goldsmith EJ, Chen L, Resing KA, Ahn NG. Docking motif interactions in MAP kinases revealed by hydrogen exchange mass spectrometry. Mol. Cell. 2004;14:43–55. doi: 10.1016/s1097-2765(04)00161-3. [DOI] [PubMed] [Google Scholar]
- 49.Mittermaier AK, Kay LE. Observing biological dynamics at atomic resolution using NMR. Trends Biochem. Sci. 2009;34:601–611. doi: 10.1016/j.tibs.2009.07.004. [DOI] [PubMed] [Google Scholar]
- 50.Korzhnev DM, Kloiber K, Kanelis V, Tugarinov V, Kay LE. Probing slow dynamics in high molecular weight proteins by methyl-TROSY NMR spectroscopy: application to a 723-residue enzyme. J. Am. Chem. Soc. 2004;126:3964–3973. doi: 10.1021/ja039587i. [DOI] [PubMed] [Google Scholar]
- 51.Sours KM, Xiao Y, Ahn NG. Extracellular-regulated kinase 2 is activated by the enhancement of hinge flexibility. J. Mol. Biol. 2014;426:1925–1935. doi: 10.1016/j.jmb.2014.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rudolph J, Xiao Y, Pardi A, Ahn NG. Slow inhibition and conformation selective properties of extracellular signal-regulated kinase 1 and 2 inhibitors. Biochemistry. 2015;54:22–31. doi: 10.1021/bi501101v. [DOI] [PMC free article] [PubMed] [Google Scholar]