Abstract
Many genes in budding yeast Saccharomyces cerevisiae associate with the nuclear pore complex (NPC), which impacts their location within the nucleus and their transcriptional regulation. To understand how eukaryotic genomes are spatially organized, we have used multiple approaches for analyzing the localization and transcription of genes. We have used these approaches to study the role of DNA elements in targeting genomic loci to the NPC and how these interactions regulate transcription, chromatin structure and the spatial organization of the yeast genome. These studies combine yeast molecular genetics with live-cell microscopy and biochemistry. Here, we present detailed protocols for these cytological and molecular approaches.
INTRODUCTION
Eukaryotic genomes are spatially organized within the nucleus. Chromosomes fold back on themselves and are positioned in distinct “territories.” The localization of genes with respect to each other and with respect to nuclear landmarks can be coupled to their expression (Egecioglu & Brickner, 2011). One model for this type of regulation is the movement of genes from the nucleoplasm to the nuclear periphery through interaction with the nuclear pore complex (NPC) upon activation. This phenomenon was discovered in the brewer’s yeast Saccharomyces cerevisiae (Brickner & Walter, 2004; Casolari et al., 2004) and has since been observed in flies, worms, and human cells (Liang & Hetzer, 2011). Genome-wide molecular approaches suggest that hundreds of yeast genes physically associate with the NPC (Casolari, Brown, Drubin, Rando, & Silver, 2005; Casolari et al., 2004). Therefore, the interaction of nuclear pore proteins with genes is both widespread and conserved. We have found that interaction of yeast genes with the NPC is controlled by cis-acting promoter elements (Ahmed et al., 2010; Brickner et al., 2012; Light, Brickner, Brand, & Brickner, 2010). These DNA elements are both necessary and sufficient to confer interaction with the NPC and to target genomic loci to the nuclear periphery. For this reason, we have called them DNA zip codes. These DNA elements may control a more global spatial organization of the yeast nucleus; the targeting of genes to the NPC also results in the interchromosomal clustering of genes that share common zip codes (Brickner et al., 2012). To study these phenomena, we have employed cytological and molecular approaches described in this chapter.
21.1 A QUANTITATIVE ASSAY FOR GENE LOCALIZATION TO THE NUCLEAR PORE COMPLEX IN YEAST
We have used a simple quantitative assay to monitor the targeting of genes to the NPC in yeast. As a proxy for interaction of genomic loci with the NPC, we tag the locus of interest with a protein (or fluorescent protein) and then localize this protein with respect to the nuclear envelope through immunofluorescence and confocal microscopy (Brickner, Light, & Brickner, 2010) (Fig. 21.2A). We then quantify the fraction of the cells in which the locus of interest colocalizes with the nuclear envelope. Here, we describe current methods utilized by our lab to visualize fluorescently tagged genomic loci through confocal microscopy.
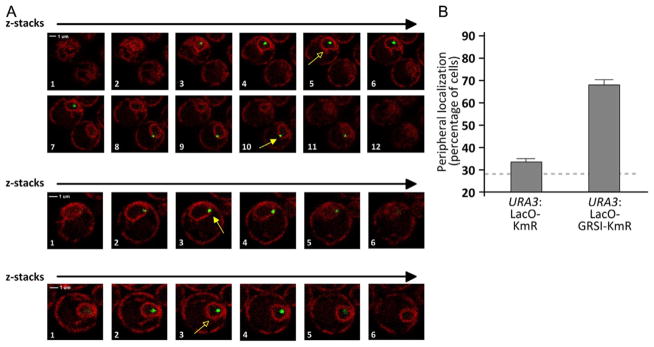
Figure 21.2. Quantifying Gene Localization at the Nuclear Periphery in Live Cells.
(A) Individual z-slices from confocal microscopy of yeast cells having the LacO array integrated at URA3 and expressing both GFP-LacI and mCherry on the endoplasmic reticulum/nuclear envelope. The slice corresponding to the brightest, most focused dot was scored (indicated with yellow arrows). Cells in which the center of the green dot does not colocalize with the nuclear envelope were scored as “OFF” (open arrowheads). Cells in which the center of the green dot colocalizes with the nuclear periphery were scored as “ON” (closed arrowheads). (B) Histogram depicting the localization of the URA3 locus with either the LacO array and KmR cassette inserted in place of the Ampr gene (as in Fig. 21.1B) or with the LacO array and the KmR gene with the GRSI zip code inserted in place of the Ampr gene. Three biological replicates of 30–50 cells each were scored for each strain. For reference, the mean peripheral localization of the URA3 gene from Brickner and Walter (2004) is shown with the dashed line.
21.1.1 Strain construction
We use a strategy similar to that described previously (Brickner et al., 2010) to create yeast strains that allow the determination of the subnuclear localization of genomic loci. However, whereas the strains described previously are only useful for immunofluorescence, the strains described here can be visualized live. Classical and general molecular biology methods used for this section include lithium acetate yeast transformation (Amberg, Burke, & Strathern, 2006a, 2006b) for both plasmids and fragments and standard bacterial plasmid transformation.
All plasmids and strains are listed in the summary table at the end of this chapter. These plasmids, plasmid maps, and strains are available to academic scientists from the Brickner Lab upon request.
To visualize the location of a gene of interest with respect to the nuclear envelope, three elements are required: an array of Lac Operators (LacO array) inserted at the genomic locus of interest, the GFP-tagged Lac repressor (GFP-LacI), and an mCherry-tagged marker for the nuclear envelope/endoplasmic reticulum.
21.1.1.1 Inserting the LacO array into the yeast genome
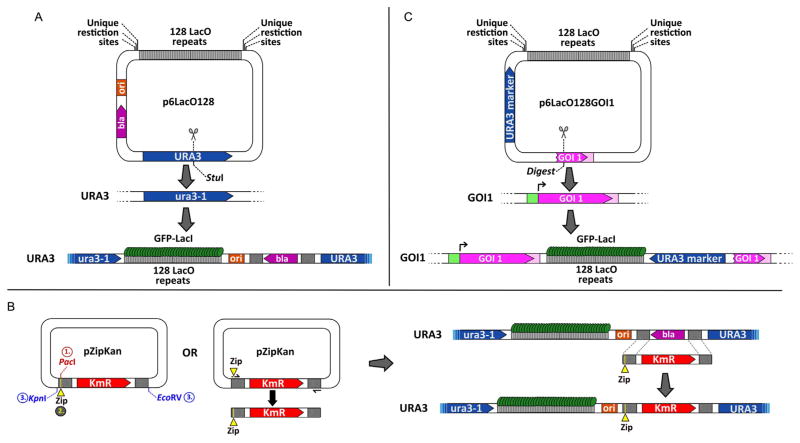
The LacO array we use consists of 128 repeats of the Lac Operator cloned into the yeast-integrating plasmid pRS306 (Sikorski & Hieter, 1989), resulting in the plasmid p6LacO128 (Brickner et al., 2010; Brickner & Walter, 2004) (Fig. 21.1A). This yeast shuttle vector carries the URA3 and Ampr (bla, for β-lactamase in Fig. 21.1) markers for selection in yeast and E. coli, respectively. We have used two different approaches to insert the LacO array into the yeast genome: (1) digesting p6LacO128 with a restriction enzyme that cleaves within URA3 to target integration to the endogenous URA3 locus (Fig. 21.1A) or (2) cloning sequences downstream of a gene of interest into the multiple cloning site in p6LacO128 and digesting the resulting plasmid with a restriction enzyme that cleaves within these sequences to direct integration of the LacO array and URA3 at that locus (Fig. 21.1C). The URA3 locus localizes primarily in the nucleoplasm and colocalizes with the nuclear envelope in only 25–30% of the cells (Brickner & Walter, 2004; Taddei et al., 2006) (e.g., Fig. 21.2B). This represents the fraction of the yeast nuclear volume that cannot be resolved from the nuclear envelope by light microscopy and is expected for an unbiased distribution (Brickner & Walter, 2004). Therefore, URA3 serves as a negative control for targeting to the NPC. For genes that interact with the NPC, we observe between 50% and 75% colocalization with the nuclear envelope (Fig. 21.2B). The fact that this number is lower than 100% reflects the dynamic nature of the association of genes with the NPC; these genes continuously move and occasionally dissociate from the nuclear periphery (Cabal et al., 2006). Furthermore, most experiments represent a snapshot(s) of an asynchronous culture of cells and targeting of active genes to the NPC is regulated through the cell cycle; for 20–30 min after the initiation of S-phase, localization to the nuclear periphery is lost (Brickner & Brickner, 2010). Cells in G1 or G2/M show higher percent colocalization with the nuclear periphery (Brickner & Brickner, 2010).
Figure 21.1. Methodology Used in Strain Construction for Microscopy.
Inserting the LacO array and testing sequences for zip code activity in the yeast genome. (A) The p6LacO128 plasmid is integrated at URA3 by digestion with StuI. Putative DNA zip codes can be cloned bedside the LacO array using the unique restriction sites on either side of the array. (B) Integration of putative zip codes (yellow Zip segments) next to the LacO array already incorporated within yeast genome through Kanr/Ampr exchange. Left: Putative DNA zip codes are either cloned into the PacI site in the pZipKan plasmid and then introduced into yeast by digestion with KpnI+EcoRV or incorporated into the cassette by including the putative zip code in the primer and amplifying the cassette by PCR. Right: The KmR cassette with a zip code can be transformed into a yeast strain having the LacO array integrated at URA3. Transformants are selected on G418 medium and screened by PCR for proper integration. (C) General strategy to incorporate the LacO array next to a gene of interest (GOI1). The 3′-end and 3′-UTR of the gene of interest is cloned into p6LacO128. Digestion within this sequence at a unique site promotes integration of the LacO plasmid downstream of the gene of interest. Ura+ transformants are selected on medium lacking uracil.
21.1.1.2 Inserting DNA zip code variants
Much of our work has focused on deciphering the molecular mechanism(s) by which genes are targeted to the NPC. Many genes are targeted to the NPC by cis-acting promoter elements that function as DNA zip codes. To test the ability of DNA sequences to function as DNA zip codes, we insert them at URA3, along with a LacO array. We define DNA zip codes as DNA elements that are sufficient to cause URA3 to localize at the nuclear periphery. To test elements for zip code activity, DNA sequences can be cloned adjacent to the LacO array in p6LacO128 and the resulting LacO plasmid can be inserted at URA3 (Ahmed et al., 2010). For small DNA elements, we integrate them directly into the backbone of the p6LacO128 plasmid that has already been integrated at URA3 in yeast (Ahmed et al., 2010; Light et al., 2010, 2013) (Fig. 21.1A). Candidate sequences can be either cloned into the PacI site in an integration cassette within the plasmid pZipKan or included in the primers used for PCR amplification of the Kanr marker from this plasmid (KmR in Fig. 21.1B). Yeast transformants that have replaced a portion of the Ampr gene in the p6LacO128 plas-mid at URA3 with the putative zip code and the Kanr gene are selected by plating on G418 medium. The resulting yeast colonies are confirmed through PCR from genomic DNA.
The restriction sites available for cloning a desired fragment of DNA or annealed oligonucleotides encoding zip code variants into p6LacO128 are as follows: Between the LacO array and ori (Fig. 21.1A): SciI, XhoI, AbsI, PspXI, KpnI, and Acc65I.
Between the URA3 gene and the LacO array (Fig. 21.1A): DrdIV, SacII, BstXI, SacI, AleI, EagI, NotI, SpeI, BamHI, XmaI, SmaI, HindIII, SphI, SalI, and Eco53kI.
The primers used to amplify the KmR cassette from the pZipKan plasmid for integration of putative zip codes (Fig. 21.1B) are as listed below. This method is recommended for relatively short inserts (15–20mers). To test longer sequences, cloning annealed oligonucleotides into the PacI site in the pZipKan plasmid followed by digestion of pZipKan with KpnI and EcoRV (Fig. 21.1B) is recommended.
-
Forward primer (without zip code):
5′-AAAAAGGCCGCGTTGCTGGCGTTTTTCCATAGGCTCCGCCCCC CTGCGGATCCCCGGGTTAATTAACATCTTTTACCC-3′
-
Forward primer (with zip code):
5′-AAAAAGGCCGCGTTGCTGGCGTTTTTCCATAGGCTCCGCCCCCCTGCGGATCCCCGGG-[Zip Code]-TTAATTAACATCTTTTACCC-3′
-
Reverse primer:
5′-CGAAATCAAAAAAAAGAATAAAAAAAAAATGATGAATTGAATTGAGAATTCGAGCTCGTTTAAAC-3′
21.1.1.3 Introducing GFP-LacI and a fluorescent ER marker
Before introducing the LacO array, we introduce the GFP-LacI and the fluorescent ER marker. This allows screening for transformants with the best LacO arrays by microscopy. The plasmid pAFS144 expressing GFP-LacI is digested with NheI to target for integration at the HIS3 locus. To mark the endoplasmic reticulum and nuclear envelope, we use mCherry fused to an endoplasmic reticulum membrane protein under the control of the GPD promoter. This plasmid (pmCh-ER04) is digested with either BstXI or AflII to target for integration at the TRP1 locus. This plasmid is derived from pAC08-mCh-L-TM from the Veenhoff lab (Meinema et al., 2011). The GPD promoter from p416-GPD (Mumberg, Muller, & Funk, 1995) was cloned as a SacI–SpeI fragment in place of the GAL1-10 promoter (using SacI–AvrII sites) in the parent plasmid pAC08-mCh-L-TM to create pGPD-mCh-ER16. The entire promoter-fusion protein-3′-UTR was removed from this plasmid as a KpnI–SacI fragment into the integrating plasmid pRS304 (Sikorski & Hieter, 1989).
21.1.2 Microscopy experiments
21.1.2.1 Materials and reagents required for microscopy Experiments
Confocal laser scanning microscope with 488 and 561 nm lasers. In our case, we use the Leica SP5 II LSCM.
Immersion oil for microscopes.
Microscope slides, 25×75×1 mm: Fisher CAT#12-544-4.
Premium cover glass for microscope slides, 24×50 mm: Fisher CAT#12-544-14.
LAS AF or LAS AF Lite software, available from Leica.
21.1.2.2 Microscope settings
We have used several confocal microscopes with 100× 1.44NA objectives with success (Fig. 21.2A). Our lab currently uses a Leica SP5 line scanning confocal microscope with the following settings: the Argon 488 nm and Diode Pumped Solid State 561 nm lasers are used at 10–15% power and images are acquired for a 150 μm × 150 μm field at 2048 × 2048 pixel resolution (without zoom). We collect a z-stack of ~20 × 0.73 μm slices, with a step size of 0.29 μm. This redundancy provides a smoother transition between each slice and enables better visualization of the green dot generated by the GFP-LacI:LacO array interaction (Fig. 21.2A). For each yeast culture, at least two samples are analyzed per slide, and care is taken to ensure that the cells are immobile.
21.1.2.3 Data acquisition through confocal microscopy
Even without deconvolution, laser scanning confocal microscopy produces images that are well resolved in the z dimension and have minimal out-of-focus light. Below is the procedure we use to quantify the localization of genes at the nuclear periphery in the strains described in the previous section:
Because rich medium (YPD) produces significant fluorescence, cells are grown in Synthetic Dextrose Complete (SDC) medium for several generations (usually overnight) at either room temperature (~23°C) or 30°C. Data collection is performed on cultures that are in early to mid-log phase of growth (OD600 ≤ 0.5).
10–50 ml of cells are harvested by centrifugation at 4000 rpm (3041×g) for 1 min, resuspended in fresh SDC media and concentrated 10–20×(final OD600 = 5–10), and stored at room temperature until visualized. Note: Chilling cells on ice or chemically inhibiting metabolism to arrest them has effects on gene targeting to the NPC and is not recommended.
The cells should be promptly visualized under the microscope, although we have not observed any significant difference between cells that were scored immediately after harvesting and cells that were scored 2 h after harvesting.
Immediately before imaging, 1 μl of the concentrated cells is spotted onto a microscope slide and covered with a cover glass. The droplet of cells should spread in the thin space between the slide and the cover glass in an even fashion, without reaching the edges. Using a small volume reduces the movement of cells during microscopy.
21.1.2.4 Data analysis
Each nucleus in which the GFP-LacI dot colocalizes with the nuclear envelope is scored as peripheral (ON); all other nuclei are scored as nucleoplasmic (OFF) (Fig. 21.2A and B). There are a few key considerations when deciding if a dot is ON or OFF:
The z-stack should include the entire nucleus. This ensures that the nucleus is visible and accounted for and that within the total volume of the entire nucleus there is only a single, relatively immobile dot. We do not score dots that move between the nucleoplasm and the nuclear periphery during the scan or nuclei with multiple dots. Dividing nuclei can also add some ambiguity to the scoring of the position of the dot, so it is important to track the entire volume of the nucleus through the z-stacks.
The step size (the distance between z-slices) should be less than half the thickness of a z-slice (i.e., ≤0.34 μm step size for a 0.73-μm slice thickness). This oversampling ensures a smooth transition between each z-stack and a greater chance of finding the z-position at which the dot is brightest and most focused.
The position of the LacI dot should be scored within the z-slice where it is brightest and most focused.
The nuclear envelope should be clearly visible as a bright, continuous ring with a dark nucleoplasm. Do not score green dots that are at the top or the bottom of the nucleus, where the nuclear envelope appears as a sheet or in nuclei in which the membrane is not a continuous ring (Fig. 21.2A).
If the center of the green dot overlaps the nuclear envelope, it is scored as ON (Fig. 21.2A).
We do not score nuclei if the position of the dot is ambiguous because it has moved during the scan, the nuclear envelope is not well resolved or there are two dots within the nucleus.
It is helpful to view each channel (green and red) separately as well as the merged image. This can resolve any ambiguity as to the extent of overlap of the LacI dot (green) with the nuclear envelope (red). The Leica LAS AF or LAS AF Lite software also enables the drawing of lines and arrows for highlighting dots and marking cells.
As cells in a field are scored, we mark them using the basic shape drawing utility to indicate that they have been scored and to identify them as peripheral or nucleoplasmic. This helps avoid scoring the same nucleus more than once and speeds the counting of each class of cells. The .lif file can be saved with the shapes drawn around the cells.
For each experiment, we score at least 30 nuclei per biological replicate and the mean percentage peripheral localization and the standard error of the mean is calculated from at least three biological replicates (Fig. 21.2B). An unpaired t test is applied to determine if two strains or conditions are significantly different.
An alternate and more laborious method for performing this experiment has been used by a number of research groups (Meister, Gehlen, Varela, Kalck, & Gasser, 2010; Taddei et al., 2006). This method divides the nucleus into three concentric “zones” of equal area and the fraction of the population in which the LacI dot localizes within each of these zones is determined. This method can reveal more subtle changes in localization than the method we have described. Because yeast cells do not have perfectly spherical nuclei, computational aid is required to calculate the zones for each nucleus.
21.1.2.5 Conclusion: Analysis of gene localization in live cells through confocal microscopy
The analysis of fluorescently labeled yeast cells through confocal microscopy provides a robust method for studying the localization of a genomic locus. Our studies have focused on quantifying the localization of a gene with respect to the nuclear envelope in a population of cells under specific conditions. In addition to this type of “snapshot” experiment, it is also possible to examine the dynamics of gene positioning within the nucleus over time, preferably using spinning-disc confocal microscopy (Meister et al., 2010).
21.2 MONITORING INTERCHROMOSOMAL CLUSTERING OF GENES AT THE NPC
In addition to causing genes to move from the nucleoplasm to the nuclear periphery, the interaction of DNA zip codes with the NPC can also lead to interchromosomal clustering of genes that share the same zip codes (Brickner et al., 2012). To observe such clustering, we have used two different strategies: (1) tagging loci with different repressor proteins fused to different fluorescent proteins (i.e., RFP-LacI/LacO array and GFP-TetR/TetO array) or (2) integrating LacO arrays of different length that give rise to a larger and a smaller dot. The position of these two loci with respect to each other is determined by measuring the distribution of distances between them in the population (Brickner et al., 2012). Here, we describe both approaches and the important considerations for the microscopy that differ from the method described above.
21.2.1 Strain construction
21.2.1.1 RFP-LacI and GFP-TetR two-dot assay
All plasmids and strains are listed in the summary table at the end of this chapter. These plasmids, plasmid maps and strains can be obtained from the Brickner Lab upon request.
1. RFP-LacI+GFP-TetR two-dot assay
We have used this technique to mark the INO1 gene with GFP-Tet repressor and compare its localization to other genes marked with RFP-Lac repressor (Fig. 21.3A). For introduction of the Lac repressor array into the yeast genome, the methods described above can be used. To integrate the Tet repressor array at INO1, the 3′-end of INO1 was cloned as an XbaI–HindIII fragment into the TetO array plasmid p15816 (Cabal et al., 2006; Grund et al., 2008), producing p15816-INO1.
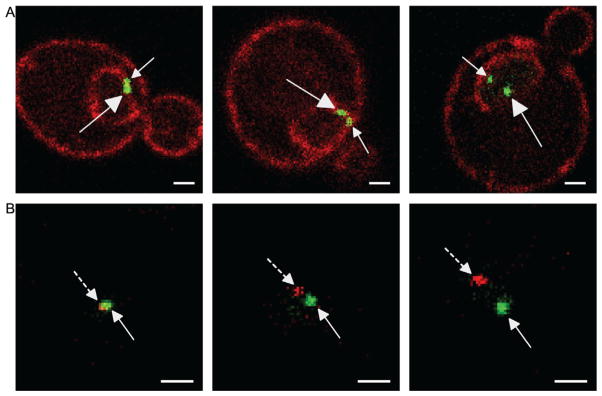
Figure 21.3. Two-Dot Experiments in Live Yeast Cells for Studying Interchromosomal Clustering by Confocal Microscopy.
For both panels, white scale bar is 1 μm and distances between dots increase from left to right. (A) Examples from the GFP-LacI two-dots assay. This is a diploid cell having both copies of the GAL1 gene marked with the LacO array. The dots are indicated with arrows that have closed arrowheads and straight lines. The large arrows indicate the localization of the large dot (LacO256) and the small arrows indicate the localization of the smaller dot (LacO128). (B) Examples of the RFP-LacI+GFP-TetR two-dot assay. The green GFP-TetR dot marks the TetO array integrated at INO1 and the RFP-LacI red dot marks the LacO array and component(s) of the INO1 gene integrated at URA3. The green dots are indicated with arrows that have closed arrowheads and the red dots are indicated with arrows that have closed arrowheads and dashed lines.
Into a strain expressing GFP-TetR (integrated at LEU2) (Grund et al., 2008) and RFP-LacI under the control of the ADH2 promoter (CEN/ARS plasmid pME08; select for by growth in SDC-TRP; Jiang, Frey, Evans, Friel, & Hopper, 2009), plasmid p15816-INO1 is integrated at INO1 (after digestion with MscI) and LacO array plasmids are integrated at various genomic sites using the strategy schematized in Fig. 21.1C. To derepress the ADH2 promoter and allow expression of RFP-LacI, cells were grown in media containing 2% ethanol as a carbon source for at least 6 h prior to harvesting.
21.2.1.2 GFP-LacI two-dots assay
We have also localized two GFP-LacI marked genes with respect to each other (Brickner et al., 2012). For these experiments, we have localized the HSP104 gene with respect to other genes. Plasmid pFS3013, having 256 Lac repressor binding sites beside the HSP104 gene, was integrated at HSP104 by digesting with BsrGI (Dieppois, Iglesias, & Stutz, 2006) in a strain transformed with GFP-LacI plasmid pAFS144 (Robinett et al., 1996) and a plasmid having 128 Lac repressor-binding sites integrated at other sites (i.e., INO1, GAL1, and GAL2). The different sizes of the arrays give rise to dots of distinct size (Brickner et al., 2012). One advantage of this system is that it offers the ability to combine clustering analysis with localization of each gene with respect to the nuclear envelope (Fig. 21.3B) (Brickner et al., 2012). We have also localized the two alleles of the same gene with respect to each other in diploid cells (Fig. 21.3B). In this case, the size of the dots is not distinguishable.
21.2.2 Microscopy settings and methods for clustering experiments
These cells can be either fixed and processed for immunofluorescence (Brickner et al., 2012) or imaged as live cells as described above. We have imaged these strains using both a Zeiss 510 line scanning confocal microscope as well as the Leica SP5.
Cells are selected for measurement based on the appearance of both dots in the same confocal slice. Thus, we have not attempted to measure distances between spots in the z dimension. We measure the distances between the centers of the dots in at least 100 cells using Zeiss LSM or the Leica LAS AF software. The distribution of distances is then plotted for the population. Also, we have defined the fraction of cells in which the two dots are “clustered” as the fraction of cells in which the center of the dots are ≤0.55 μm apart. Dots that are ≤0.5 μm apart are localized within the same 8% of the area of the nucleus (Brickner et al., 2012). These metrics can be used to compare the localization of pairs of loci using either a t-test (for distributions) or a Fisher’s Exact Test (for the fraction of dots that are clustered). Genes that are not clustered show a distribution with a mean distance of ~0.85±0.3 μm and 10–15% clustering. Genes that cluster show a very significant shift to shorter distances, with a mean distance of 0.5±0.3 μm and ~65% clustering (Brickner et al., 2012).
21.2.3 Conclusion: Monitoring interchromosomal clustering of genes at the NPC
We have described the techniques used in the Brickner Lab to visualize multiple genomic loci and their position in relation to each other on the nuclear envelope in the yeast nuclei. With this method, multiple DNA zip codes can be tested for interchromosomal clustering of different genes or genomic loci.
21.3 USING CHROMATIN IMMUNOPRECIPITATION TO PROBE NUCLEAR ORGANIZATION, TRANSCRIPTION, AND CHROMATIN STRUCTURE IN YEAST AND HUMAN CELLS
An orthogonal approach to studying gene localization to the nuclear periphery by microscopy has been to use chromatin immunoprecipitation (ChIP) to monitor the interaction of nuclear pore proteins with chromatin. In fact, this type of experiment led to the initial discovery of this widespread nuclear pore–gene interaction (Casolari et al., 2004). ChIP is a powerful technique that allows the characterization of specific protein/DNA interactions (both direct and indirect) at a given genomic location of interest or genome wide (Hecht, Strahl-Bolsinger, & Grunstein, 1996; Kuo & Allis, 1999). In this procedure, protein and DNA complexes are fixed by crosslinking with formaldehyde, followed by mechanical shearing into small DNA fragments (Orlando, 2000; Orlando, Strutt, & Paro, 1997). Immunoprecipitation of an antigen leads to the coincident recovery of the DNA associated with the target antigen. This DNA can then be recovered, purified and its immunoprecipitation can be quantified by real-time PCR (qPCR). Relative recovery of a locus of interest can then be compared with appropriate control loci (Nelson, Denisenko, & Bomsztyk, 2006).
Using ChIP, our lab has studied the interaction of nuclear pore proteins with chromatin, as well as the downstream events associated with transcription or alterations in chromatin structure. In yeast, we have used Tandem Affinity Purification (TAP)-tagged forms of several nuclear pore proteins, including components of the core channel (i.e., Nup100), the nuclear basket (i.e., Nup2), and associated factors (i.e., Mlp2, SAGA). We have also examined the binding of transcription factors, the recruitment of RNA polymerase II and the preinitiation complex, incorporation of the histone variant H2A.Z, and the methylation of histone H3 using ChIP (Ahmed et al., 2010; Brickner et al., 2012, 2007; Light et al., 2010, 2013). Finally, we have performed ChIP against nuclear pore proteins, RNA polymerase II, and chromatin marks in HeLa cells (Light et al., 2013).
In this section, we will describe our general protocol for ChIP (Fig. 21.4). We include antigen-specific details for ChIP against RNA polymerase II and H3K4me2 or TAP-tagged proteins in yeast and Nup98 in HeLa cells. The immunoprecipitated material is quantified using real-time quantitative PCR and the yield is expressed relative to a sample of the Input.
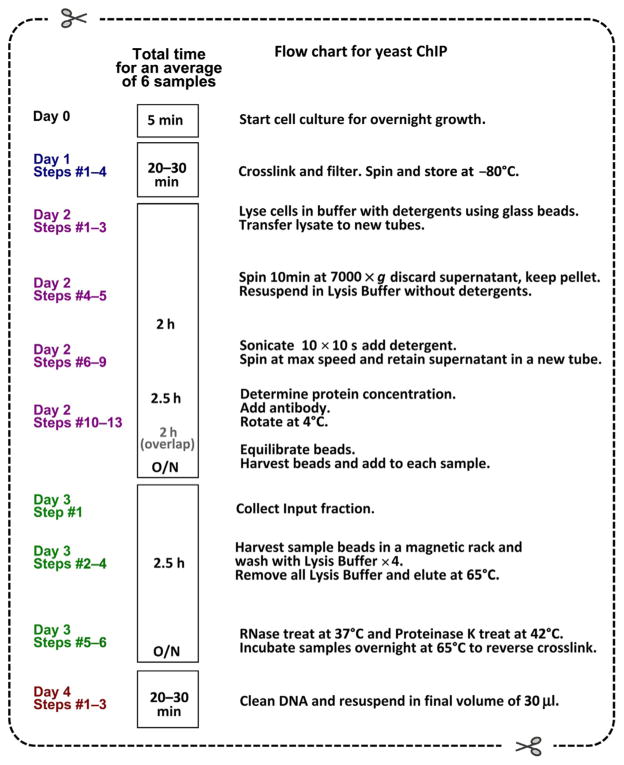
Figure 21.4.
Flow chart for the ChIP protocol described in this chapter.
21.3.1 Equipment, buffers, solutions, and reagents for ChIP
EQUIPMENT
Branson Sonifier 450
8-Tip microtip attachment for sonifier (Cole Parmer catalog# 630-0586)
Magnetic bead harvester (for 1.5–2.0 ml microfuge tubes)
BIO-RAD iCycler iQ5 (Multicolor Real-Time PCR Detection System), Bio-rad Cat# 170-9780
COMMON REAGENTS FOR YEAST AND HUMAN CELL PROTOCOL
TE Buffer
20% SDS
36% Formaldehyde stock: Sigma CAT# F8775-500ML
2.5 M Glycine stock
100 mM Tris–HCl (pH 7.0)
2× ChIP Lysis Buffer Stock (no detergent added): 100 mM HEPES–KOH (pH 7.5), 280 mM NaCl, 2 mM EDTA
1× ChIP-Lysis Buffer: 50 mM HEPES–KOH (pH 7.5), 140 mM NaCl, 1 mM EDTA, 1% Triton X-100, 0.1% NaDOC+protease inhibitors
Elution Buffer: 50 mM Tris (pH 8), 10 mM EDTA, 1% SDS
Qiagen Buffers PB (CAT# 19066), PE (CAT# 19065)
Qiagen PCR cleanup column (QIAquick® Spin Column LOT# 136244976, MAT#1018215). These might be sold separately or provided with the QIAquick PCR Purification Kit CAT#28104)
Pan-Mouse IgG (Pol ll-antibody): (Dynabeads® Pan Mouse IgG, CAT # 110.41)
Protein-G (H3K4me2) magnetic beads: (Dynabeads® Protein G for Immunoprecipitation CAT# 10004D)
RNase A
Proteinase K
Recombinant Taq purified from E. coli (Engelke, Krikos, Bruck, & Ginsburg, 1990)
10× SYBR® Green I Nucleic Acid Gel Stain (Life Technologies/Invitrogen CAT# S-7567. This stock is 10,000×)
FOR YEAST PROTOCOL
Acid-washed 0.5-mm glass beads: BioSpec Products, Inc. CAT#11079105
Nitrocellulose Membrane Filters: Millipore CAT# RAWP09025
Pierce BCA Assay Kit: Pierce™ CAT# 23225 (500 assays) or CAT#23227 (250 assays)
Protease Inhibitor cocktail tablets “complete” from Roche CAT#11 697 498 001 (with EDTA): Use 1 tablet per 50 ml of lysis buffer.
anti-RNA Polymerase ll (AbCam, ab32356 or Covance 8WG16)
anti-H3K4me2 antibody (AbCam, ab5408)
FOR HELA PROTOCOL
TBS Buffer (Tris Buffered Saline, pH 7.5)
MC Lysis Buffer: 3 mM MgCl2, 10 mM Tris (pH 7.5), 10 mM NaCl, and 1%
NP-40
DMEM+10% serum + Pen/Strep
Rabbit Nup98 antibody (AbCam 45584 or Cell Signaling Technologies 2292)
21.3.2 Yeast ChIP
Day 1
Label one set of 50-ml conical tubes and one set of microfuge tubes for the day (for each sample). Obtain ice and an aluminum ice block that fits the sonicator tip spacing. Obtain liquid N2.
-
Grow 100 ml of yeast cells in the appropriate medium to an OD600 of 0.8–1(=80–100 ODs).
Caution: Do not over grow cells as the following crosslinking steps are optimized for the specified OD range. -
Crosslink cells with fresh 1% formaldehyde. Add 2.7 ml of 36% formaldehyde stock to 100 ml of culture, fix for 15 min to 1 h at room temperature with occasional mixing/swirling. Stop fixation by adding 6 ml of 2.5 M glycine to a final concentration of 150 mM, incubate for 5 min.
Crosslinking time must be optimized for each antigen. Harvest cells by vacuum filtration. Wash nitrocellulose membrane filter with 100 ml of 100 mM Tris–HCl (pH 7.0).
Scrape cells by washing gently with buffer (do not use spatulae) off of the filter, resuspend in 1 ml of 100 mM Tris (pH 7.0) and transfer to a microfuge tube. Spin 30 s at top speed (13,000 rpm), remove supernatant, and snap freeze cell pellets in liquid N2. Store cell pellets at −80 °C.
Day 2
Label two new sets of microfuge tubes for all the samples. Label a third set of tubes for the BCA assay.
Resuspend cell pellets in 600 μl ice cold ChIP Lysis Buffer+protease inhibitors. Note: Make fresh from 2×stock buffer.
To lyse the cells, add cells to 600 μl of acid-washed glass beads and vortex at 4°C, six times for 45 s each, with 2 min rests on ice.
-
Puncture the bottom of tubes and caps with an 18G needle, place into a second 1.5-ml tube and collect lysate by gentle centrifugation at 4 °C, 100 RCF for 5 min (do not use lid in the centrifuge). Discard the dry glass beads. The lysate should all be in the new 1.5-ml tube.
Caution: If the lysate did not travel properly to the new tube, check the puncture holes and spin samples again. Spin lysate 10 min at 7000×g at 4 °C.
-
Discard supernatant and resuspend pellet in 1 ml fresh Lysis Buffer.
Note: Most of the chromatin is found in pellet before sonication.
-
With an 8-tip microtip attachment on a Branson Sonifier 450, sonicate at setting #7, 10×10 s*.
All of the tips should be in a tube immersed in either lysate or buffer/water. To prevent foaming, place tip as close to the bottom of tube as possible while sonicating.
*Sonication time (i.e., the number of cycles) should be optimized for each fixation condition because longer fixation usually requires more sonication.
Check the range of DNA sizes after sonication in a 2% agarose gel. They should be centered≤500 bp.
Mix well.
Pellet insoluble material by spinning for 10 min at maximum speed in a microcentrifuge at 4°C.
Transfer supernatant to a new 1.5-ml tube and, if necessary, spin again to remove additional debris and collect the supernatant.
Determine protein concentration in the supernatant fraction using BCA assay (Pierce). Samples should range between 2 and 5 mg/ml. Dilute all samples to equal protein concentrations using Lysis buffer and use 1 ml for subsequent steps.
-
For 5 mg of protein in each sample, add 5 μg of anti-RNA Polymerase ll or anti-H3K4me2 antibody. Rotate for 2 h at 4 °C.
For TAP-tagged proteins, skip this step. -
While step 11 is in progress, equilibrate the magnetic beads. Per 5 μl of antibody, 8 μl of Pan-Mouse IgG (Pol ll-antibody), or Protein-G (H3K4me2) magnetic beads will be required. Harvest beads for 2 min with magnetic bead harvester, remove supernatant (suspension buffer) and resuspend beads in 1-ml Lysis Buffer. Rotate at 4 °C for 2 h.
For TAP-tagged proteins, use 5–10 μl Pan-Mouse IgG beads per 5 mg of chromatin. Harvest IgG beads for 2 min with magnetic bead harvester. Remove supernatant with a pipette and resuspend beads in the initial volume of Lysis Buffer that was removed from the stock bottle. Add the appropriate volume of beads to each experimental sample and rotate at 4 °C overnight.
Day 3
Label one new set of microfuge tubes for the Input and one set of new microfuge tubes for the IP samples.
Before harvesting beads, collect 50 μl of “Input” fraction. Add 200 μl TE+12.5 μl of 20% SDS to Input fractions and set aside.
Pellet beads for 2 min with magnetic bead harvester (Protein G Dynabeads take longer than Pan-mouse IgG Dynabeads), remove supernatant with a pipette and wash 4×1-ml Lysis Buffer, resuspending and harvesting beads for 2 min between each wash.
Elute in 100 μl Elution Buffer (50 mM Tris (pH 8), 10 mM EDTA, 1% SDS), incubate 15 min at 65 °C.
Harvest beads on magnetic rack, transfer eluate to new microfuge tube. Do a second elution by washing beads with 150 μl TE+5μl of 20% SDS. Harvest beads again and pool eluates. Label this tube as your IP samples.
RNAse treat with 50 μg RNAse A for 10 min at 37 °C followed by Proteinase K treatment with 100 μg Proteinase K for 1 h at 42 °C.
Incubate both IP and Input samples at 65 °C for 6–15 h (usually overnight) to reverse crosslinks.
Day 4
Label one set of tubes for each sample (Input or IP) for the Qiagen cleanup step.
Remark: In lieu of the cleanup with the Qiagen column, extraction with phenol:chloroform:isoamyl alcohol (25:24:1) can also be performed—see Note (1).
Add 1.25-ml Qiagen PB Buffer to each sample, mix and load onto Qiagen PCR cleanup column.
Wash column 2×750 μl Qiagen PE.
Spin columns dry for 2 min, top speed in the microcentrifuge, let air dry for 2 min and then elute in 30 μl water.
21.3.3 ChIP in HeLa cells
Harvest ~20 to 50×106 cells by trypsin: Aspirate media and add 6 ml of Trypsin–EDTA to 15-cm plate and incubate at 37 °C until most cells have lifted off the plate (about 5 min or so). Add 19 ml of DMEM+10% serum and pen/strep to stop reaction and pipet up and down several times to resuspend. Transfer to a 50 ml conical flask. Finally, fix with 1% formaldehyde (from a 36% stock, as for the yeast protocol) at room temperature for 15 min, quenching with 125 mM glycine for 5 min.
-
Harvest cells by spinning for 5 min at 1750 rpm. Wash the pellet twice with TBS.
Note: Cell pellet can be frozen in liquid N2 and stored at −80 °C. Suspend cells in 10 ml hypotonic cell lysis with MC Lysis Buffer.
Spin lysate 5 min at 4 °C twice: To do this, pour off supernatant, and add an additional 10 ml of MC Lysis Buffer, spin again at 4 °C 1500 rpm for 5 min. Discard the supernatant and then snap freeze the pellet.
Freeze the pellet fraction in liquid N2 and store at −80 °C.
Suspend cell pellets in 1 ml ChIP Lysis Buffer and sonicate using 8-tip microtip attachment on a Branson 450 sonicator. Use cycles of 15 s on 100% duty cycle, 15 s off on ice for approximately 5 min of total sonication time on tip setting #5.
Spin 10 min at 4 °C at maximum speed in the microcentrifuge.
Collect the supernatant fraction and measure the protein concentration using the BCA assay.
Equalize the chromatin concentration for all samples and proceed with 1 ml of each. Using a minimum 1 mg/ml chromatin in 1 ml, add 5 μg Rabbit Nup98 antibody (AbCam 45584 or Cell Signaling Technologies 2292) or m414/H3K4me3/H3K4me2 antibody and Protein-G DynaBeads (use 5–10 μg/1 ml of chromatin).
-
All subsequent steps are the same as listed from step #12 on Day 2 of the yeast ChIP protocol described above.
Note (1): In lieu of the cleanup with the Qiagen column on Day 4, extraction with phenol:chloroform:isoamyl alcohol (25:24:1) can also be performed: Bring volume of the IP and Input samples to same volume with TE+0.67% SDS (usually about 300 μl) and add an equal volume of phenol:chloroform:isoamyl alcohol. Vortex thoroughly, followed by spinning at room temperature for 5 min at maximum speed in a microcentrifuge. Remove top aqueous layer and add to fresh microfuge tube with equal volume chloroform (300 μl). Vortex to mix and spin for 2.5 min at maximum speed at room temperature. Remove the upper aqueous layer and add to 1 ml ice cold 100% ethanol. Add 1/9th volume of 3 M NaOAc (pH 5.2) and 20 μg glycogen or 2 μl of linear acrylamide (50 mg/ml stock) and mix by inversion several times. Incubate at least 1 h at −20 °C and pellet by centrifugation at maximum speed at 4 °C for 30 min. Remove supernatant and wash pellet with 70% ethanol and spin for 15 min at 4 °C. Remove supernatant and let the pellet air dry until pellet becomes translucent. Resuspend in 30 μl TE and heat for ~10 min at 42 °C to solubilize DNA. Yields using phenol–chloroform extractions are higher than the yields obtained with Qiagen columns. Determining the concentration of DNA is also more straightforward due to the absence of the characteristic background at A260 that comes from Qiagen buffers.
21.3.4 qPCR Reaction and analysis
21.3.4.1 qPCR set Reaction
Dilute Input fractions 1:400 and IP samples 1:10 with water. For standard curves, use six fivefold serial dilutions of genomic DNA (the most concentrated is 20 ng/μl for yeast and 100 ng/μl for HeLa). Set up triplicate 25 μl reactions as follows:
5 μl DNA Template
2.5 μl 10× PCR buffer
0.5 μl 50 μM primer mix
0.25 μl dNTP mix (25 mM stock)
0.25 μl 10× SYBR® Green I Nucleic Acid Gel Stain
0.125 μl Taq
16.4 μl water
25 μl total reaction volume
We use the BIO-RAD iCycler iQ5 (Multicolor Real-Time PCR Detection system) for amplification. The PCR reaction, along with real-time settings, is listed in Table 21.1.
Table 21.1. PCR cycling settings used for the data analysis of ChIP.
| Set points (temperature) (°C) | Dual time (min or s) | Camera | |
|---|---|---|---|
| Cycle 1 | 95 | 3 min | × |
| Cycle 2 | 95 | 10 s | × |
| 50 | 30 s | × | |
| 72 | 45 s Return to Cycle 2, repeat ×40 |
Real-time | |
| Cycle 3 | 95 | 1 min | × |
| Cycle 4 | 55 | 1 min | × |
| Cycle 5 | 55 | 10 s | Melt |
The settings used for the qPCR are shown with their corresponding time frames.
21.3.4.2 Data analysis
To analyze the data, we use the threshold CT qPCR reaction in the linear range of amplification (Brickner et al., 2007). To determine the relationship between DNA concentration and CT value, we use the genomic DNA standard curve. The data are fit to an exponential curve described by: y=ke(−rx), where y is the DNA concentration, k and r are constants, and x is the CT value. This standard curve is performed with every qPCR reaction to confirm that the reagents and the machine are working well. We take the average CT of the three technical replicates of the qPCR for each sample, and DNA concentrations are used to calculate enrichment by dividing the IP/Input for the corresponding samples and plotting. For negative controls in yeast ChIP, we have used primers to amplify repressed loci such as PRM1 and GAL1 genes orthe intergenic region downstream of the RPA34 gene. For HeLa Nup98 ChIP negative control, we have used primers to the CIITA promoter, GAPDH and β-ACTIN locus.
21.4 LIST OF PLASMIDS AND STRAINS
More information on the plasmids and yeast strains described in this chapter is available from the Brickner Lab.
| Plasmids
| |
|---|---|
| Plasmid name | Purpose/description |
| p6LacO128 | Integration of LacO128 at URA3 |
| p6lacO128-GRSIINO1 or p6LacO128-GRSITSA2 | Integration of LacO128-GRSI at URA3 |
| pZipKan | Replacement of Ampr with Kanr |
| pZipKan-GRSIINO1 or pZipKan-GRSITSA2 | Replacement of Ampr with Kanr-GRSI |
| pAFS144 | Integration of GFP-LacI at HIS3 |
| pmCh-ER04 | Integration of mCherry-ER marker at TRP1 |
| p15816-INO1 | Integration of TetO array at INO1 |
| p128-tetR-GFP | Integration of GFP-TetR at LEU2 (Michaelis, Ciosk, & Nasmyth, 1997) |
| pME08 | Nonintegrated RFP-LacI, select for TRP |
| pFS3013 | Integration of LacO256 array at HSP104 |
| Yeast strains
| |
|---|---|
| Yeast strain | Description |
| NDY012 | Live-microscopy acceptor strain for inserts at URA3 using pZipKan |
| DBY434 | NDY012 transformed with Kanr |
| DEY09 or TKY01 | NDY012 transformed with Kanr-GRSI |
| DBY339 | TetO at INO1 (green) and LacO at INO1 integrated at URA3 (red). Strain is transformed with GFP-TetR and RFP-LacI |
| DBY546 | Diploid cell with both copies of GAL1 with the LacO128 integration. Strain is transformed with GFP-LacI |
CONCLUDING REMARKS, POSSIBLE CAVEATS, AND TROUBLESHOOTING
We have presented here the methods used in the Brickner Lab to investigate the spatial organization of the yeast genome and its interactions with nuclear envelope and the NPC. In the sections on microscopy, we have discussed our methods for visualizing the nuclear periphery in yeast cells with one (21.1, pp 463-469) or multiple (21.2, pp 469-472) genomic loci. The position of these genomic loci in relation to the nuclear envelope or in relation to each other on the nuclear periphery reveals a spatial component to regulating gene expression that is mediated by DNA zip codes. Data acquired through confocal microscopy via “snapshots” or over time in live cells can provide important information about the molecular basis of this phenomenon. Movies of over a longer time period can provide important information about the dynamics of this process. An important concern when visualizing live cells is their physiological state. We have found that excess centrifugation, chilling on ice, heating, and overgrowth can alter localization results in live cells.
As a recent study has pointed out, the localization and expression of certain chromosomal sites can be affected by the utilization of biological repressors that target these loci (Dubarry, Loiodice, Chen, Thermes, & Taddei, 2011). These include tagged-LacI or TetR used to target LacO and TetO arrays, which is a frequently used method in the localization experiments we describe. Tight binding by LacI or TetR, in combination with cryptic silencing elements can lead to transcriptional repression. Thus, it is important to compare expression with and without the arrays and to use orthogonal techniques such as ChIP to confirm that the localization and expression of a gene of interest are not perturbed by the arrays. Also, mutant LacI proteins with lower affinity can also avoid some of these problems (Dubarry et al., 2011).
The study of interactions between Nups, PolII, and other markers of gene expression with chromatin as described in detail in Section 21.3 has been indispensable in understanding nuclear and genomic structure and transcriptional regulation. We stress the importance of optimizing the crosslinking and sonication to obtain the best signal-to-noise ratio. Some proteins or complexes are poorly crosslinked, which can lead to discrepancies between proteins, experiments, or laboratories.
Acknowledgments
The authors acknowledge support from NIH grant GM 080484 (J. H. B.), a W.M. Keck Young Scholars in Biomedical Research Award (J. H. B.), Cellular and Molecular Basis of Disease Training Program (institutional predoctoral training grant) 2T32GM008061-31 (A. D.), a Rappaport Award for Research Excellence (W. H. L.), and an American Heart Association Postdoctoral Fellowship 13POST14580066 (D. E. E.). The authors also thank members of the Brickner lab for helpful comments on the chapter.
References
- Ahmed S, Brickner DG, Light WH, Cajigas I, McDonough M, Froyshteter AB, et al. DNA zip codes control an ancient mechanism for gene targeting to the nuclear periphery. Nature Cell Biology. 2010;12(2):111–118. doi: 10.1038/ncb2011. [Research Support, N.I.H., Extramural Research Support, Non-U.S. Gov’t] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amberg DC, Burke DJ, Strathern JN. High-efficiency transformation of yeast. CSH Protocols. 2006a;2006(1) doi: 10.1101/pdb.prot4145. [DOI] [PubMed] [Google Scholar]
- Amberg DC, Burke DJ, Strathern JN. “Quick and dirty” plasmid transformation of yeast colonies. CSH Protocols. 2006b;2006(1) doi: 10.1101/pdb.prot4146. [DOI] [PubMed] [Google Scholar]
- Brickner DG, Ahmed S, Meldi L, Thompson A, Light W, Young M, et al. Transcription factor binding to a DNA zip code controls interchromosomal clustering at the nuclear periphery. Developmental Cell. 2012;22(6):1234–1246. doi: 10.1016/j.devcel.2012.03.012. [Research Support, N.I.H., Extramural Research Support, Non-U.S. Gov’t Research Support, U.S. Gov’t, Non-P.H.S.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brickner DG, Brickner JH. Cdk phosphorylation of a nucleoporin controls localization of active genes through the cell cycle. Molecular Biology of the Cell. 2010;21(19):3421–3432. doi: 10.1091/mbc.E10-01-0065. [Research Support, N.I.H., Extramural] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brickner DG, Cajigas I, Fondufe-Mittendorf Y, Ahmed S, Lee PC, Widom J, et al. H2A.Z-mediated localization of genes at the nuclear periphery confers epigenetic memory of previous transcriptional state. PLoS Biology. 2007;5(4):e81. doi: 10.1371/journal.pbio.0050081. [Research Support, N.I.H., Extramural Research Support, Non-U.S. Gov’t] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brickner DG, Light W, Brickner JH. Quantitative localization of chromosomal loci by immunofluorescence. Methods in Enzymology. 2010;470:569–580. doi: 10.1016/S0076-6879(10)70022-7. [Research Support, N.I.H., Extramural Research Support, Non-U.S. Gov’t] [DOI] [PubMed] [Google Scholar]
- Brickner JH, Walter P. Gene recruitment of the activated INO1 locus to the nuclear membrane. PLoS Biology. 2004;2(11):e342. doi: 10.1371/journal.pbio.0020342. [Research Support, Non-U.S. Gov’t Research Support, U.S. Gov’t, P.H.S.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabal GG, Genovesio A, Rodriguez-Navarro S, Zimmer C, Gadal O, Lesne A, et al. SAGA interacting factors confine sub-diffusion of transcribed genes to the nuclear envelope. Nature. 2006;441(7094):770–773. doi: 10.1038/nature04752. [Research Support, Non-U.S. Gov’t] [DOI] [PubMed] [Google Scholar]
- Casolari JM, Brown CR, Drubin DA, Rando OJ, Silver PA. Developmentally induced changes in transcriptional program alter spatial organization across chromosomes. Genes & Development. 2005;19(10):1188–1198. doi: 10.1101/gad.1307205. [Research Support, Non-U.S. Gov’t Research Support, U.S. Gov’t, P.H.S.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casolari JM, Brown CR, Komili S, West J, Hieronymus H, Silver PA. Genome-wide localization of the nuclear transport machinery couples transcriptional status and nuclear organization. Cell. 2004;117(4):427–439. doi: 10.1016/s0092-8674(04)00448-9. [Research Support, Non-U.S. Gov’t Research Support, U.S. Gov’t, P.H.S.] [DOI] [PubMed] [Google Scholar]
- Dieppois G, Iglesias N, Stutz F. Cotranscriptional recruitment to the mRNA export receptor Mex67p contributes to nuclear pore anchoring of activated genes. Molecular and Cellular Biology. 2006;26(21):7858–7870. doi: 10.1128/MCB.00870-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubarry M, Loiodice I, Chen CL, Thermes C, Taddei A. Tight protein-DNA interactions favor gene silencing. Genes & Development. 2011;25(13):1365–1370. doi: 10.1101/gad.611011. [Research Support, Non-U.S. Gov’t] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egecioglu D, Brickner JH. Gene positioning and expression. Current Opinion in Cell Biology. 2011;23(3):338–345. doi: 10.1016/j.ceb.2011.01.001. [Research Support, N.I.H., Extramural Research Support, Non-U.S. Gov’t Review] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelke DR, Krikos A, Bruck ME, Ginsburg D. Purification of Thermus aquaticus DNA polymerase expressed in Escherichia coli. Analytical Biochemistry. 1990;191(2):396–400. doi: 10.1016/0003-2697(90)90238-5. [Research Support, Non-U.S. Gov’t Research Support, U.S. Gov’t, Non-P.H.S. Research Support, U.S. Gov’t, P.H.S.] [DOI] [PubMed] [Google Scholar]
- Grund SE, Fischer T, Cabal GG, Antunez O, Perez-Ortin JE, Hurt E. The inner nuclear membrane protein Src1 associates with subtelomeric genes and alters their regulated gene expression. The Journal of Cell Biology. 2008;182(5):897–910. doi: 10.1083/jcb.200803098. [Research Support, Non-U.S. Gov’t] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hecht A, Strahl-Bolsinger S, Grunstein M. Spreading of transcriptional repressor SIR3 from telomeric heterochromatin. Nature. 1996;383(6595):92–96. doi: 10.1038/383092a0. [Research Support, Non-U.S. Gov’t Research Support, U.S. Gov’t, P.H.S.] [DOI] [PubMed] [Google Scholar]
- Jiang F, Frey BR, Evans ML, Friel JC, Hopper JE. Gene activation by dissociation of an inhibitor from a transcriptional activation domain. Molecular and Cellular Biology. 2009;29(20):5604–5610. doi: 10.1128/MCB.00632-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo MH, Allis CD. In vivo cross-linking and immunoprecipitation for studying dynamic Protein:DNA associations in a chromatin environment. Methods. 1999;19(3):425–433. doi: 10.1006/meth.1999.0879. [Research Support, U.S. Gov’t, P.H.S. Review] [DOI] [PubMed] [Google Scholar]
- Liang Y, Hetzer MW. Functional interactions between nucleoporins and chromatin. Current Opinion in Cell Biology. 2011;23(1):65–70. doi: 10.1016/j.ceb.2010.09.008. [Research Support, Non-U.S. Gov’t Review] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Light WH, Brickner DG, Brand VR, Brickner JH. Interaction of a DNA zip code with the nuclear pore complex promotes H2A.Z incorporation and INO1 transcriptional memory. Molecular Cell. 2010;40(1):112–125. doi: 10.1016/j.molcel.2010.09.007. [Research Support, N.I.H., Extramural Research Support, Non-U.S. Gov’t] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Light WH, Freaney J, Sood V, Thompson A, D’Urso A, Horvath CM, et al. A conserved role for human Nup98 in altering chromatin structure and promoting epigenetic transcriptional memory. PLoS Biology. 2013;11(3):e1001524. doi: 10.1371/journal.pbio.1001524. [Research Support, N.I.H., Extramural Research Support, Non-U.S. Gov’t] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meinema AC, Laba JK, Hapsari RA, Otten R, Mulder FA, Kralt A, et al. Long unfolded linkers facilitate membrane protein import through the nuclear pore complex. Science. 2011;333(6038):90–93. doi: 10.1126/science.1205741. [Research Support, Non-U.S. Gov’t] [DOI] [PubMed] [Google Scholar]
- Meister P, Gehlen LR, Varela E, Kalck V, Gasser SM. Visualizing yeast chromosomes and nuclear architecture. Methods in Enzymology. 2010;470:535–567. doi: 10.1016/S0076-6879(10)70021-5. [DOI] [PubMed] [Google Scholar]
- Michaelis C, Ciosk R, Nasmyth K. Cohesins: Chromosomal proteins that prevent premature separation of sister chromatids. Cell. 1997;91(1):35–45. doi: 10.1016/s0092-8674(01)80007-6. [Research Support, Non-U.S. Gov’t] [DOI] [PubMed] [Google Scholar]
- Mumberg D, Muller R, Funk M. Yeast vectors for the controlled expression of heterologous proteins in different genetic backgrounds. Gene. 1995;156(1):119–122. doi: 10.1016/0378-1119(95)00037-7. [Research Support, Non-U.S. Gov’t] [DOI] [PubMed] [Google Scholar]
- Nelson JD, Denisenko O, Bomsztyk K. Protocol for the fast chromatin immunoprecipitation (ChIP) method. Nature Protocols. 2006;1(1):179–185. doi: 10.1038/nprot.2006.27. [Research Support, N.I.H., Extramural Research Support, Non-U.S. Gov’t] [DOI] [PubMed] [Google Scholar]
- Orlando V. Mapping chromosomal proteins in vivo by formaldehyde-crosslinked-chromatin immunoprecipitation. Trends in Biochemical Sciences. 2000;25(3):99–104. doi: 10.1016/s0968-0004(99)01535-2. [Research Support, Non-U.S. Gov’t Review] [DOI] [PubMed] [Google Scholar]
- Orlando V, Strutt H, Paro R. Analysis of chromatin structure by in vivo formaldehyde cross-linking. Methods. 1997;11(2):205–214. doi: 10.1006/meth.1996.0407. [DOI] [PubMed] [Google Scholar]
- Robinett CC, Straight A, Li G, Willhelm C, Sudlow G, Murray A, et al. In vivo localization of DNA sequences and visualization of large-scale chromatin organization using lac operator/repressor recognition. The Journal of Cell Biology. 1996;135(6 Pt 2):1685–1700. doi: 10.1083/jcb.135.6.1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski RS, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122(1):19–27. doi: 10.1093/genetics/122.1.19. [Research Support, Non-U.S. Gov’t Research Support, U.S. Gov’t, P.H.S.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taddei A, Van Houwe G, Hediger F, Kalck V, Cubizolles F, Schober H, et al. Nuclear pore association confers optimal expression levels for an inducible yeast gene. Nature. 2006;441(7094):774–778. doi: 10.1038/nature04845. [Research Support, Non-U.S. Gov’t] [DOI] [PubMed] [Google Scholar]