Abstract
Background
Abnormally high levels of T-cell activation can persist in HIV-infected subjects despite effective anti-retroviral therapy (ART) and has been associated with negative health outcomes. The nature of the antigenic drivers or other causes of this residual T-cell activation remain uncertain. Anelloviruses are universally acquired soon after birth, resulting in persistent viremia, and considered part of the commensal human virome. Reduced immunocompetence results in increased anellovirus levels.
Objectives
To test whether increased levels of anelloviruses or other viruses in plasma are associated with higher levels of persistent T-cell activation during ART.
Study design
Two amplification methods combined with next generation sequencing were used to detect all viruses and estimate relative anellovirus levels in plasma from 19 adults on effective ART who exhibited a wide range of T-cell activation levels.
Results
Nucleic acids from HBV and HCV were detected in one patient each while pegivirus A (GBV-C) was found in three patients. Anellovirus DNA was detected in all patients with some individuals carrying up to eight different genotypes. Specific anellovirus genotypes or higher level of co-infections were not detected in subjects with higher levels of T-cell activation. No association was detected between relative plasma anellovirus DNA levels and the percentage of activated CD4 or CD8 T cells.
Conclusions
Human anelloviruses were detected in all HIV suppressed subjects, exhibited a wide range of viremia levels, and were genetically highly diverse. The level of persistent T-cell activation was not correlated with the level of viremia or genotypes present indicating that anellovirus antigens are unlikely to be a dominant source of antigens driving chronic T-cell activation.
Keywords: Anellovirus, Torque teno virus, Virome, HLA-DR+CD38+, T-cell activation, HIV
1. Background
Activated T cells, identified by expression of CD38 and HLA-DR, are strongly implicated in the pathogenesis and disease progression of HIV infection [1–5]. The mechanisms underlying the persistent T-cell activation among virally suppressed HIV-infected subjects are not known but immune stimulatory microbial products from the microbiome in the gut are thought to be involved [6,7]. Chronic CMV infection has also been implicated [5]. Here we tested whether the presence or concentration of other human plasma viruses could be correlated with the highly variable levels of T-cell activation seen in HIV suppressed patients.
The most common human viruses detected in human plasma are anelloviruses, which are non-enveloped, small icosahedral viruses with a single-stranded negative-sense circular DNA genome of ~2–4 kb [8]. The Anelloviridae family is composed of nine (Alpha-Iotatorquevirus) and three putative genera (Kappa-Mutorquevirus) infecting a wide range of mammals including humans, chimpanzees, rhesus macaques and African green monkeys [9–15]. Anelloviruses are generally considered non-pathogenic, commensal, infections with the possible exemption of a porcine anellovirus when co-infecting with other viruses [16,17]. A sea mammal anellovirus was also associated with an unexplained mortality event in California sea lions [18]. In humans anellovirus infections occurs early after birth resulting in chronic viremia, fecal shedding, and frequent co-infections with different variants [9,19–26]. Three out of the nine genera are known to infect humans: Alphatorquevirus also known as torque teno virus (TTV) or Sen virus; Betatorquevirus, also known as torque teno mini virus (TTMV); and Gammatorquevirus also known as torque teno midi virus (TTMDV). Each of these human anellovirus genera are highly diverse and can be further divided into groups made up of multiple species [8,27,28]. Alphatorquevirus (TTVs) are currently classified into five major phylogenetic groups subdivided into a total of 29 species, based on the cutoff value of 35% nucleotide sequence identity of the major open reading frame 1(ORF1) as determined by ICTV [8,29].
Plasma levels of anelloviruses appear to be negatively correlated with the level of host immune competency [30–32]. HIV-infected individuals showed higher detection rate and estimated titer of human anelloviruses than healthy blood donors [33–35]. Anellovirus loads were higher in HIV-infected individuals with low versus high CD4+ T-cell counts [31,36–38]. For HIV-infected individuals on antiretroviral therapy (ART), the improved immune status led to a decrease of the TTV viral load and number of TTV genogroups in the serum [39,40]. Anellovirus levels increases in both plasma and bronchoalveolar lavage with the level of immune suppression in transplant patients [30,32]. Critically ill patients with sepsis also show an increase on anellovirus viremia [41].
2. Objectives
Although ART is able to restore CD4+ T-lymphocyte levels, high level of both CD4+ and CD8+ T-cell activation (measured by expression of HLA-DR and CD38 on T-cells) persist despite sustained HIV suppression. Persistent T-cell activation is associated with higher risk of morbid non-AIDS events [1,4,42]. The nature of the antigenic or other stimuli thought responsible for such T-cell activation during untreated or treated HIV disease remains uncertain although increased microbial translocation of bacterial products into the plasma of HIV positive subjects have been reported as T-cell pro-inflammatory signals [42–45]. As the mechanisms of persistent T-cell activation may give insights into a clinically important feature of HIV disease we tested whether increased levels of replication by other viruses could also be associated with higher levels of T-cell activation following successful HIV viremia control using ART.
3. Study design
3.1. Study subjects
HIV-infected subjects were sampled retrospectively from the UCSF-based SCOPE Cohort (Table 1). To be included in this study, the subjects had to have ART-mediated viral suppression with plasma HIV RNA levels below the level of detection (<50 copies/ml). Nineteen subjects were further selected according to the percentages of activated T cells (HLA-DR+ CD38+ CD3+ CD4+ or CD8+) in their peripheral blood mononuclear cells, identified by four-color flow cytometry (Epics XL flow cytometer, Beckman Coulter). The percentage or expression level of other surface markers on T cells were also measured including CCR5, PD-1, CD45RA and CCR7 (Table S1). The age range was 37–57 years (mean 47.8 ± 5.1 years), with a male/female ratio of 17/2. The plasma samples were stored at −80 ºC.
Table 1.
Profiles of selected HIV infected subjects.
| ID | Sex | Ethnic group | Age (year) | Therapy | HIV viral load (copies/ml) | CD4+ (cells/μl) | CD8+ (cells/μl) | CD4+CD38+ T-cell (%) | CD8+CD38+ T-cell (%) |
|---|---|---|---|---|---|---|---|---|---|
| P1 | M | White | 50 | ABC/3TC, NVP | <50 | 612 | 501 | 2.29 | 10.12 |
| P2 | M | White | 44 | TDF, AZT/3TC, LPV/r | <50 | 856 | 679 | 3.01 | 15.49 |
| P3 | M | White | 55 | FTC/TDF, NVP | <50 | 256 | 933 | 3.40 | 11.20 |
| P4 | F | African American | 50 | AZT/3TC, LPV/r | <50 | 569 | 1004 | 4.38 | 17.34 |
| P5 | M | White | 44 | FTC, TDF, LPV/r | <50 | 478 | 1704 | 4.45 | 19.78 |
| P6 | M | Pacific Islander | 44 | FTC/TDF, ATV, RTV | <50 | 607 | 778 | 4.93 | 11.74 |
| P7 | M | White | 50 | FTC/TDF, ATV, RTV | <50 | 396 | 700 | 6.73 | 26.50 |
| P8 | M | African American | 49 | EFV/TDF/FTC | <50 | 293 | 1199 | 6.84 | 33.23 |
| P9 | M | White | 54 | FTC/TDF, EFV | <50 | 236 | 356 | 7.76 | 16.30 |
| P10 | M | White | 48 | ABC/3TC, EFV, LPV/r | <50 | 354 | 1317 | 7.81 | 33.92 |
| P11 | M | White | 50 | ABC/3TC, NVP | <50 | 279 | 1585 | 9.27 | 31.91 |
| P12 | M | White | 43 | ABC/3TC, NVP | <50 | 292 | 685 | 10.15 | 23.51 |
| P13 | M | White | 39 | FTC/TDF, NVP | <50 | 483 | 835 | 11.00 | 32.60 |
| P14 | M | African American | 48 | EFV/TDF/FTC | <50 | 154 | 1039 | 11.30 | 34.10 |
| P15 | F | White | 37 | ABC/3TC, ATV, RTV | <50 | 394 | 970 | 11.90 | 26.20 |
| P16 | M | Latino | 44 | 3TC, TDF, LPV/r | <50 | 370 | 926 | 13.52 | 33.47 |
| P17 | M | White | 50 | ABC, D4T, LPV/r | <50 | 314 | 1088 | 15.57 | 26.81 |
| P18 | M | White | 53 | LPV/r, EFV/TDF/FTC | <50 | 635 | 943 | 17.00 | 53.70 |
| P19 | M | White | 57 | AZT/3TC/ABC, LPV/r | <50 | 811 | 683 | 20.70 | 22.90 |
3.2. Nucleic acid extraction and M13 phage spike
Plasma samples collected in EDTA were processed as previously described [46]. Briefly, plasma sample was clarified by centrifugation at 13,000 × g for 5 min. Then 3 × 106 pfu of M13-KE phage (diluted from a 1.0 × 1013 pfu/ml stock from New England Biolabs) was added to 250 μl of plasma supernatant. The plasma was then filtered through a 0.45 μm filter (Millipore) to remove any remaining large cellular debris. The viral-particle containing filtrate was then digested with a mixture of DNases and RNases to remove unprotected nucleic acids [46]. Viral nucleic acids were then extracted using the QIAamp viral RNA Mini kit (Qiagen) and re-suspended in 60 μl water plus 40U of RNase inhibitor (Fermentas) and stored at −80 ºC.
3.3. NGS Library preparation and sequencing
Viral DNA was amplified by rolling circle amplification (RCA) using the TempliPhi 100 Amplification Kit (GE Healthcare). Briefly, 1 μl of Qiagen nucleic acid extract plus 5 μl of the kit sample buffer was heated to 95 ºC for 3 min, cooled on ice, and 5 μl of the kit reaction buffer and 0.2 μl of Enzyme mix (Phi29 DNA polymerase) added. The RCA reaction was incubated at 30 ºC for 16 hrs, followed by heat-inactivation at 65 ºC for 10 min. Random RT-PCR amplification (RA) was also employed to amplify viral RNA and DNA as previously described [46]. Viral cDNA synthesis was performed by incubation of 10 μl extracted viral nucleic acids with 100 pmol of a primer containing a fixed 18 bp sequence plus a random nonamer at the 3′ end (GCCGACTAATGCGTAGTCNNNNNNNNN) at 85 ºC for 2 min. Then, 200U SuperScript III reverse transcriptase (Invitrogen), 0.5 mM of each deoxynucloside triphosphate (dNTP), 10 mM dithiothreitol, and 1 × first-strand extension buffer were added to the mixture and incubated at 25 ºC for 10 min, followed by 50 ºC incubation for 1 h. The 2nd strand DNA synthesis was performed by incubation with 50 pmol of random primer at 95 ºC for 2 min, 4 ºC for 2 min, and then with 5U Klenow Fragment (New England Biolabs) at 37 ºC for 1 h. The resulting products were PCR amplified by using 5 μl of the RT-Klenow DNA products and 2.5 μM primer consisting of the fixed 18 bp portion of the random primer (GCCGACTAATGCGTAGTC) with 1U AmpliTaq Gold DNA polymerase (Life Technologies), 2.5 mM MgCl2, 0.2 mM dNTPs, and 1 × PCR Gold buffer in a reaction volume of 50 μl. Temperature cycling was performed as follows: 1 cycle of 95 ºC for 5 min, 30 cycles of denaturing at 95 ºC for 30 s, 55 ºC for 30 s, 72 ºC for 1.5 min. An additional extension for 10 min at 72 ºC was added to the end of the run. Both RCA and RA products were then purified by QIAquick PCR Purification Kit (QIAGEN) and eluted in 30 μl of water. For each sample, ~5 ng (determined by Nanodrop) of the RCA or RA DNA product was used as input for the construction of DNA libraries using Nextera XT DNA sample preparation kit (Illumina). The quality of the libraries was assessed by 2100 Bioanalyzer (Agilent Technologies) and quantified by KAPA Library Quant Kit (Kapa Biosystems) following the manufacturer’s instructions. The resulting libraries were then sequenced by two runs using the MiSeq Illumina platform (2 × 250 cycle and 2 × 300 Illumina MiSeq Reagent Kit v2 for the RCA and RA generated DNA respectively). Each of the 19 individual samples in each library was labeled with unique dual barcodes.
3.4. Sequence analysis
Sequence reads were processed by our viral identification pipeline [47,48]. Human host reads and bacterial reads were first subtracted by mapping the reads to human reference genome hg19 and bacterial RefSeq genomes release 66 using bowtie2 [49]. Reads were considered duplicates if 5 bp to 55 bp from 5 prime end is identical and one random copy of duplicates was kept. Low sequencing quality tails were trimmed using Phred quality score 10 as the threshold. Adaptor and primer sequences were trimmed using the default parameters of VecScreen [50]. These cleaned reads were de-novo assembled using EnsembleAssembler [47]. The assembled contigs, along with singlets were aligned to an in-house viral proteome database using BLASTx using E-value cutoff 10−5. The significant hits to virus were then aligned to an in-house nonvirus- non-redundant (NVNR) universal proteome database using BLASTx. Hits with more significant adjusted E-value to NVNR than to virus were removed. The anellovirus reads identified in the RCA run were further assembled by using CLC Genomics Workbench (CLC bio) and SPAdes Genome Assembler [51]. Contigs longer than >1500 bp were examined by the open reading frame (ORF) finder (NCBI), searching for the putative >500 aa anellovirus ORF1. The encoded protein sequences (>500 aa) were aligned by ClustalW with the default settings. Phylogenetic analyses based on aligned aa sequences were generated by the neighbor-joining method in MEGA, using aa p-distances, with 1000 bootstrap replicates.
3.5. Statistical analysis
The statistical analysis was performed using the non-parametric Spearman correlation coefficient and Wilcoxon signed-rank significant tests by Stata/MP software (version 14.0, StataCorp, College Station, TX). All a priori significance levels were set at p < 0.05.
4. Results
4.1. Deep sequencing with rolling circle amplification versus random amplification
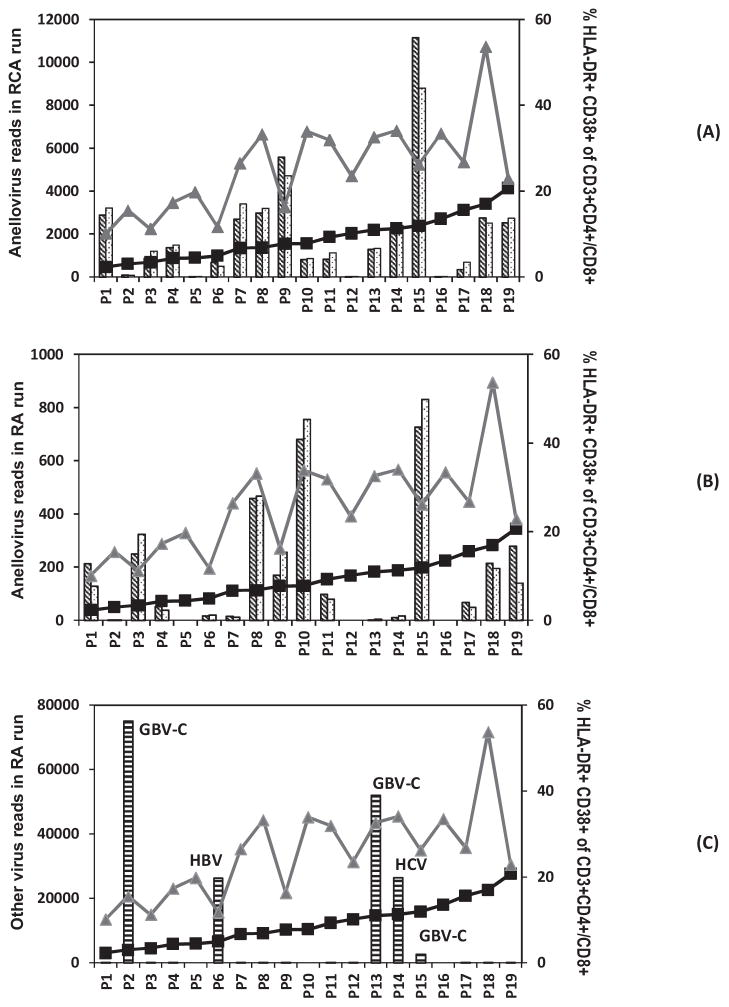
The 19 plasma samples pre-amplified by RCA and the RA method were deep sequenced using two MiSeq runs. The RCA run generated a total of ~24 million reads (mean 1.3 ± 0.3 million/sample), while the RA run produced ~23 million reads (mean 1.2 ± 0.2 million/sample). The resulting sequence contigs and singlets were compared to the viral reference database using BLASTx search with an E-value cutoff of 10−5. M13 bacteriophage with a circular ssDNA genome of ~7 kb was spiked into each plasma sample before extraction to function as an indicator of the amplification of circular ssDNA genomes of anelloviruses. Each sample in the RCA run had an average of 43,000 ± 9000 M13 reads, and the samples in the RA run had an average of 68,000 ± 26,000 M13 reads. The RCA and RA MiSeq raw sequence data including barcodes for each nineteen subjects were deposited in GenBank Sequence Read Archive under accession number SRP059897. In the RA run, the anellovirus sequences were identified in 16/19 subjects, with an average of 172 reads (range: 0–727 reads), while in the RCA run, the anellovirus sequences were identified in all 19 subjects, with an average of 2058 reads (range: 2–11,158 reads) (Fig. 1A, B). The numbers of anellovirus reads obtained by RCA run were significantly higher than those by RA run (Wilcoxon signed-rank test, p = 0.0001), and there was a moderate correlation between the numbers of anellovirus reads obtained by RCA run and those by RA run (Spearman rho = 0.68, p = 0.0014, Fig. S1).
Fig. 1.
Influence of activated T-cells (HLA-DR+CD38+ CD3+ CD4+/CD8+) on the abundance of anellovirus and other viruses in the plasma viromes of 19 patients. (A) The percentage of activated T-cells and the original/normalized anellovirus reads in RCA run (B) the percentage of activated T-cells and the original/normalized anellovirus reads in RA run (C) the percentage of activated T-cells and the other virus reads in RA run (black square: % HLA-DR+ CD38+ of CD3+CD4+; gray triangle: % HLA-DR+ CD38+ of CD3+CD8+; columns with diagonal lines: raw anellovirus read numbers; columns with dots: normalized anellovirus reads; columns with horizontal lines: other virus reads).
4.2. Other viruses detected
By comparing translated sequences with proteins of known eukaryotic viruses, the RCA run detected viral sequences related to anelloviruses and human hepatitis B virus (HBV), both viruses with circular DNA genomes preferentially amplified by RCA. The RA run revealed DNA and RNA viral sequences including anellovirus, human hepatitis B virus (HBV) as well as single stranded RNA genomes in the Flaviviridae family from human hepatitis C (HCV) virus and GB virus C (GBV-C) with single stranded RNA genomes. Of the 19 selected subjects, three had moderate to high level of GBV-C infections (2000–70,000 reads), one had high titer HCV infection (>20,000 reads), and one had high titer HBV infection (>20,000 reads) (Fig. 1C). No HIV reads were detected as expected from their undetectable HIV viremia (Table 1).
4.3. Relative quantitation of anelloviruses and association with T-cell activation
The anellovirus read numbers obtained by RCA and RA methods were normalized according to total raw reads and M13 phage reads (Table S2 & S3). The anellovirus reads obtained by both RCA and RA run (both original and normalized) showed no significant association with either CD4+ or CD8+ T-cell counts and the percentage of activated CD4+ or CD8+ T cells (HLA-DR+ CD38+ CD3+) (Spearman correlation, p > 0.05). The anellovirus reads numbers also showed no significant correlation with the percentage or expression intensity of other surface markers on T cells including CCR5, PD-1, CD45RA and CCR7 (Spearman correlation, p > 0.05) (data not shown).
4.4. Genetic diversity of anelloviruses
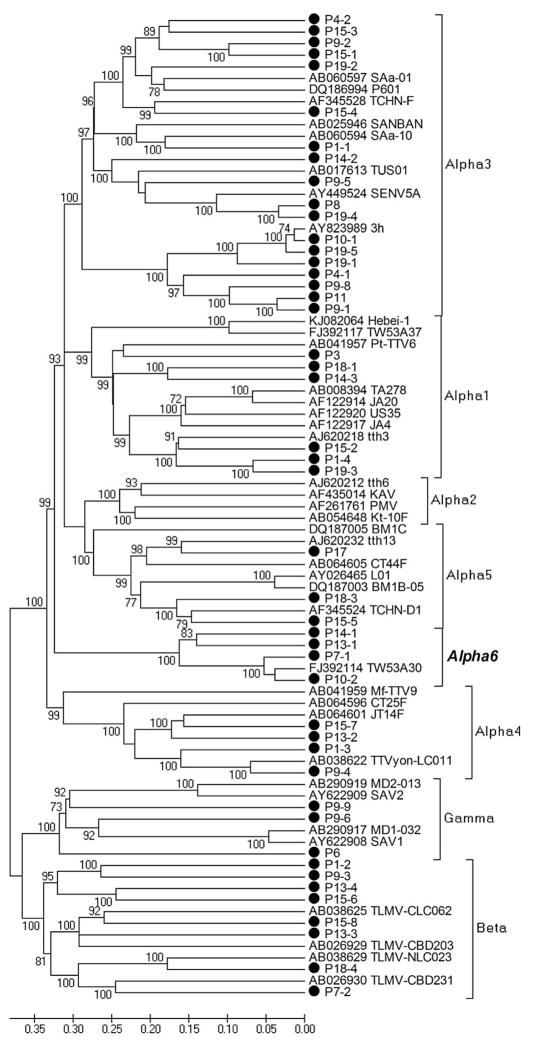
We used reads from the RCA run to generate contigs covering a large fraction of the anellovirus genomes in different subjects. Forty-six contigs could be generated with translated ORF1 protein > 500 aa (covering >70% of the complete anellovirus ORF1) from 15/19 subjects. Phylogenetic analysis of human anellovirus ORF1 protein sequences showed these 46 sequences to be distributed among the three human anellovirus genera (Alpha, Beta and Gamma), including 4 out of 5 of the major phylogenetic groups of Alphatorquevirus and a recently proposed Alphatorquevirus 6 group (Fig. 2).
Fig. 2.
Phylogenetic analysis of human anellovirus sequences, generated with the protein encoded by ORF1 (>500 aa) using neighbor-joining method with p-distance and 1000 bootstrap replications. The scale indicates amino acid substitutions per position. Bootstrap values for each node were shown if >70%. The three human anellovirus genera, Alphatorquevirus, Betatorquevirus and Gammatorquevirus, and 6 phylogenetic genetic groups of Alphatorquevirus 1–6 are marked on the tree.
The Alphatorquevirus group 3 was the most prevalent genetic group with 18 strains detected in 9 subjects, followed by Betatorquevirus with 8 strains detected in 6 subjects and Alphatorquevirus group 1 with 6 strains found in 6 subjects. No sequences were phylogenetically assigned to Alphatorquevirus group 2. The subjects P15, P9, P1 and P19 had high degree of anellovirus heterogeneity with 8, 8, 4, 5 strains from 5, 4, 4, 2 genetic groups, respectively. Neither the number of strains nor the number of genetic groups detected in each subjects showed significant association with CD4+/CD8+ T-cell counts nor the percentage of activated T cells (Spearman correlation, p > 0.05).
5. Discussion
Four different viruses were detected including the highly variable anelloviruses. HBV and HCV were detected in one patient each and GBV-C in three patients. The small number of patients infected with HBV, HCV, and GBV-C precluded testing their association with T-cell activation levels. Consistent with their well-documented high prevalence in humans, anelloviruses were detected in all patients [9,19–26].
Human anellovirus infections are typically detected by PCR targeting conserved regions in 3′-UTR, ORF1 and ORF2 of the viral DNA genomes [33,39,52,53]. Because of the extensive genetic variability among human anelloviruses the efficiency of PCR based methods across different genera and species is uncertain. Recently, deep sequencing has been used to get an overview of the anellovirus population in the plasma of humans or non-human primates [9,30–32]. We used two distinct unbiased amplification methods in combination with deep sequencing to estimate the relative levels and genetic composition of anelloviruses in the plasma of HIV-infected subjects on suppressive ART. RCA is an isothermal DNA amplification technique using bacteriophage phi29 DNA polymerase and random hexamer primers to preferentially amplify circular single-or double-stranded DNA molecules with high fidelity and high amplicon yields and has been used in the identification and characterization of many circular DNA genomes [54–56]. The RA method has been routinely used to non-specifically amplify both DNA and RNA viral genomes (following randomly primed reverse transcription) to generate DNA for input into deep sequencing platforms [46]. Our results showed that the RCA method was more efficient for the identification of anelloviruses, generating ~10 times more anellovirus reads on average than the RA method. The RA methods also detected RNA viruses that were not amplified by RCA.
We tested whether anelloviruses may play a role in the abnormal T-cell activation in HIV-infected subjects who controlled HIV following effective ART. In the 19 selected subjects, neither the relative plasma anellovirus load (as reflected by viral read numbers from RCA and RA) nor anellovirus diversity (as reflected by number of different genetic variants) showed a significant relationship with the generally high CD4+ (average 441) and CD8+ T-cell counts or the percentage of activated T cells (HLA-DR+ CD38+ CD3+ CD4+ or CD8+). Our results indicated that plasma levels of anelloviruses, a nearly universally common chronic human viral infection, do not appear to be a major driving force in the stimulation of persistently activated T-cells in these treated HIV positive subjects.
On average a minimum of 2.4 distinct anellovirus were detected (range 0–8) per patient including only ORF1 contigs >1500 bases. Neither the numbers of co-infecting anelloviruses nor specific phylogenetic groups were associated with higher levels of T-cell activation.
The pegivirus A (GBV-C) in the Flaviviridae family has been classified into at least 7 genotypes with distinct geographical distribution [57]. Here, all three GBV-C detected belonged to genotype 2, mostly reported in Europe and North America [57,58]. HCV has at least 6 major genotypes consisting of more than 70 subtypes. The HCV we obtained was genotype/subtype 1a [59,60] common in North America. HBV has been classified into 10 genotypes A–J. The HBV identified in this study was genotype G, which has been reported in the US [61–63]. The numbers of HBV, HCV, and GBV-C infections detected were too small to test possible association with T-cell activation levels.
Treatment of HIV-infected CMV-seropositive patients with valganciclovir has been shown to abolish CMV DNA detection and reduce CD8 activation [5]. Despite the frequent detection, using PCR, of diverse herpes virus (CMV, EBV, HHV-6, HHV-6) DNA in seminal plasma and other biological samples from patients successfully treated for HIV [5], we did not detect sequence reads from any viruses in the Herpesviridae family using deep sequencing. Despite the ability of our deep sequencing method to detect herpes viruses in a positive control reagent [46], the absence of detection seen here may indicates a lower level of sensitivity relative to PCR. The preferential association of herpes viruses with cells, and the generally low level of immunodeficiency (average CD4 count of 441) of these successfully treated (HIV plasma levels <50 RNA copies/ml) patients may also contribute to our lack of herpes virus DNA detection in plasma by deep sequencing.
In conclusion this study has demonstrated that human anelloviruses were ubiquitous and highly diverse in a cohort of HIV-infected adults on effective ART. High levels of anellovirus co-infections were detected. No association was found between estimated anellovirus levels in plasma and T-cell activation levels. The higher levels of persistent T-cell activation in some subjects may therefore not be attributed simply to higher levels of anellovirus replication. It remains possible that anellovirus replication plays only a minor role in persistent T-cell activation or that replication in specific tissues such as the liver or lymph nodes, which may not be reflected in plasma levels, is more effective at stimulating T cell. The faction of T cells specific to these chronic anelloviral infections remains unknown, but like the high number of CD8 cells specific to human cytomegalovirus [64–67] could make up a significant percentage of the T-cell population.
Supplementary Material
Acknowledgments
The work was supported by the Blood Systems Research Institute and NIH R01HL105770 to Dr. Delwart. The SCOPE cohort was supported by the UCSF/Gladstone Institute of Virology & Immunology CFAR (P30 AI027763), the UCSF Clinical and Translational Research Institute Clinical Research Center (UL1 RR024131), the CFAR Network of Integrated Systems (R24 AI067039) and NIAID (AI096109; AI069994).
Appendix A. Supplementary data
Supplementary data associated with this article can be found, in the online version, at http://dx.doi.org/10.1016/j.jcv.2015.09.004.
Footnotes
Competing interests
The authors have no competing interests to report.
Ethical approval
No ethical approval was required for this study.
References
- 1.Hazenberg MD, Otto SA, van Benthem BH, Roos MT, Coutinho RA, Lange JM, et al. Persistent immune activation in HIV-1 infection is associated with progression to AIDS. AIDS. 2003;17:1881–1888. doi: 10.1097/00002030-200309050-00006. [DOI] [PubMed] [Google Scholar]
- 2.Rosso R, Fenoglio D, Terranova MP, Lantieri F, Risso D, Pontali E, et al. Relevance of CD38 expression on CD8 T cells to evaluate antiretroviral therapy response in HIV-1-infected youths. Scand J Immunol. 2010;71:45–51. doi: 10.1111/j.1365-3083.2009.02345.x. [DOI] [PubMed] [Google Scholar]
- 3.Savarino A, Bottarel F, Malavasi F, Dianzani U. Role of CD38 in HIV-1 infection: an epiphenomenon of T-cell activation or an active player in virus/host interactions? AIDS. 2000;14:1079–1089. doi: 10.1097/00002030-200006160-00004. [DOI] [PubMed] [Google Scholar]
- 4.Vigano A, Saresella M, Villa ML, Ferrante P, Clerici M. CD38+CD8+ T cells as a marker of poor response to therapy in HIV-infected individuals. Chem Immunol. 2000;75:207–217. doi: 10.1159/000058770. [DOI] [PubMed] [Google Scholar]
- 5.Hunt PW, Martin JN, Sinclair E, Epling L, Teague J, Jacobson MA, et al. Valganciclovir reduces T cell activation in HIV-infected individuals with incomplete CD4+ T cell recovery on antiretroviral therapy. J Infect Dis. 2011;203:1474–1483. doi: 10.1093/infdis/jir060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Handley SA, Thackray LB, Zhao G, Presti R, Miller AD, Droit L, et al. Pathogenic simian immunodeficiency virus infection is associated with expansion of the enteric virome. Cell. 2012;151:253–266. doi: 10.1016/j.cell.2012.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li SK, Leung RK, Guo HX, Wei JF, Wang JH, Kwong KT, et al. Detection and identification of plasma bacterial and viral elements in HIV/AIDS patients in comparison to healthy adults. Clin Microbiol Infect. 2012;18:1126–1133. doi: 10.1111/j.1469-0691.2011.03690.x. [DOI] [PubMed] [Google Scholar]
- 8.International Committee on Taxonomy of Viruses, King AMQ, International Union of Microbiological Societies. Virus taxonomy classification and nomenclature of viruses: ninth report of the International Committee on Taxonomy of Viruses. Elsevier; London: 2012. Virology Division; p. 1. online resource (x, 1327 p.) ill. (some col) [Google Scholar]
- 9.Kapusinszky B, Mulvaney U, Jasinska AJ, Deng X, Freimer N, Delwart E. Local virus extinctions following a host population bottleneck. J Virol. 2015 doi: 10.1128/JVI.00671-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li L, Giannitti F, Low J, Keyes C, Ullmann LS, Deng X, et al. Exploring the virome of diseased horses. J Gen Virol. 2015 doi: 10.1099/vir.0.000199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Parras-Molto M, Suarez-Rodriguez P, Eguia A, Aguirre-Urizar JM, Lopez-Bueno A. Genome sequence of two novel species of torque teno minivirus from the human oral cavity. Genome Announc. 2014:2. doi: 10.1128/genomeA.00868-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jones MS, Kapoor A, Lukashov VV, Simmonds P, Hecht F, Delwart E. New DNA viruses identified in patients with acute viral infection syndrome. J Virol. 2005;79:8230–8236. doi: 10.1128/JVI.79.13.8230-8236.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nishiyama S, Dutia BM, Stewart JP, Meredith AL, Shaw DJ, Simmonds P, et al. Identification of novel anelloviruses with broad diversity in UK rodents. J Gen Virol. 2014;95:1544–1553. doi: 10.1099/vir.0.065219-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Okamoto H, Nishizawa T, Tawara A, Peng Y, Takahashi M, Kishimoto J, et al. Species-specific TT viruses in humans and nonhuman primates and their phylogenetic relatedness. Virology. 2000;277:368–378. doi: 10.1006/viro.2000.0588. [DOI] [PubMed] [Google Scholar]
- 15.Thom K, Morrison C, Lewis JC, Simmonds P. Distribution of TT virus (TTV), TTV-like minivirus, and related viruses in humans and nonhuman primates. Virology. 2003;306:324–333. doi: 10.1016/s0042-6822(02)00049-1. [DOI] [PubMed] [Google Scholar]
- 16.Aramouni M, Kekarainen T, Ganges L, Tarradas J, Segales J. Increased viral load and prevalence of Torque teno sus virus 2 (TTSuV2) in pigs experimentally infected with classical swine fever virus (CSFV) Virus Res. 2013;172:81–84. doi: 10.1016/j.virusres.2012.12.010. [DOI] [PubMed] [Google Scholar]
- 17.Kekarainen T, Segales J. Torque teno sus virus in pigs: an emerging pathogen? Transbound Emerg Dis. 2012;59(Suppl 1):103–108. doi: 10.1111/j.1865-1682.2011.01289.x. [DOI] [PubMed] [Google Scholar]
- 18.Ng TF, Suedmeyer WK, Wheeler E, Gulland F, Breitbart M. Novel anellovirus discovered from a mortality event of captive California sea lions. J Gen Virol. 2009;90:1256–1261. doi: 10.1099/vir.0.008987-0. [DOI] [PubMed] [Google Scholar]
- 19.Bagaglio S, Sitia G, Prati D, Cella D, Hasson H, Novati R, et al. Mother-to-child transmission of TT virus: sequence analysis of non-coding region of TT virus in infected mother-infant pairs. Arch Virol. 2002;147:803–812. doi: 10.1007/s007050200027. [DOI] [PubMed] [Google Scholar]
- 20.Chan PK, Tam WH, Yeo W, Cheung JL, Zhong S, Cheng AF. High carriage rate of TT virus in the cervices of pregnant women. Clin Infect Dis. 2001;32:1376–1377. doi: 10.1086/319983. [DOI] [PubMed] [Google Scholar]
- 21.Kapusinszky B, Minor P, Delwart E. Nearly constant shedding of diverse enteric viruses by two healthy infants. J Clin Microbiol. 2012;50:3427–3434. doi: 10.1128/JCM.01589-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gerner P, Oettinger R, Gerner W, Falbrede J, Wirth S. Mother-to-infant transmission of TT virus: prevalence, extent and mechanism of vertical transmission. Pediatr Infect Dis J. 2000;19:1074–1078. doi: 10.1097/00006454-200011000-00009. [DOI] [PubMed] [Google Scholar]
- 23.Ohto H, Ujiie N, Takeuchi C, Sato A, Hayashi A, Ishiko H, et al. TT virus infection during childhood. Transfusion. 2002;42:892–898. doi: 10.1046/j.1537-2995.2002.00150.x. [DOI] [PubMed] [Google Scholar]
- 24.Okamoto H, Takahashi M, Nishizawa T, Ukita M, Fukuda M, Tsuda F, et al. Marked genomic heterogeneity and frequent mixed infection of TT virus demonstrated by PCR with primers from coding and noncoding regions. Virology. 1999;259:428–436. doi: 10.1006/viro.1999.9770. [DOI] [PubMed] [Google Scholar]
- 25.Vasconcelos HC, Cataldo M, Niel C. Mixed infections of adults and children with multiple TTV-like mini virus isolates. J Med Virol. 2002;68:291–298. doi: 10.1002/jmv.10187. [DOI] [PubMed] [Google Scholar]
- 26.Ninomiya M, Takahashi M, Nishizawa T, Shimosegawa T, Okamoto H. Development of PCR assays with nested primers specific for differential detection of three human anelloviruses and early acquisition of dual or triple infection during infancy. J Clin Microbiol. 2008;46:507–514. doi: 10.1128/JCM.01703-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Biagini P. Classification of TTV and related viruses (anelloviruses) Curr Top Microbiol Immunol. 2009;331:21–33. doi: 10.1007/978-3-540-70972-5_2. [DOI] [PubMed] [Google Scholar]
- 28.Spandole S, Cimponeriu D, Berca LM, Mihaescu G. Human anelloviruses: an update of molecular, epidemiological and clinical aspects. Arch Virol. 2015;160:893–908. doi: 10.1007/s00705-015-2363-9. [DOI] [PubMed] [Google Scholar]
- 29.Peng YH, Nishizawa T, Takahashi M, Ishikawa T, Yoshikawa A, Okamoto H. Analysis of the entire genomes of thirteen TT virus variants classifiable into the fourth and fifth genetic groups, isolated from viremic infants. Arch Virol. 2002;147:21–41. doi: 10.1007/s705-002-8301-7. [DOI] [PubMed] [Google Scholar]
- 30.De Vlaminck I, Khush KK, Strehl C, Kohli B, Luikart H, Neff NF, et al. Temporal response of the human virome to immunosuppression and antiviral therapy. Cell. 2013;155:1178–1187. doi: 10.1016/j.cell.2013.10.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li L, Deng X, Linsuwanon P, Bangsberg D, Bwana MB, Hunt P, et al. AIDS alters the commensal plasma virome. J Virol. 2013;87:10912–10915. doi: 10.1128/JVI.01839-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Young JC, Chehoud C, Bittinger K, Bailey A, Diamond JM, Cantu E, et al. Viral metagenomics reveal blooms of anelloviruses in the respiratory tract of lung transplant recipients. Am J Transplant. 2015;15:200–209. doi: 10.1111/ajt.13031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pirouzi A, Bahmani M, Feizabadi MM, Afkari R. Molecular characterization of Torque teno virus and SEN virus co-infection with HIV in patients from Southern Iran. Rev Soc Bras Med Trop. 2014;47:275–279. doi: 10.1590/0037-8682-0073-2014. [DOI] [PubMed] [Google Scholar]
- 34.Sherman KE, Rouster SD, Feinberg J. Prevalence and genotypic variability of TTV in HIV-infected patients. Dig Dis Sci. 2001;46:2401–2407. doi: 10.1023/a:1012307416122. [DOI] [PubMed] [Google Scholar]
- 35.Touinssi M, Gallian P, Biagini P, Attoui H, Vialettes B, Berland Y, et al. TT virus infection: prevalence of elevated viraemia and arguments for the immune control of viral load. J Clin Virol. 2001;21:135–141. doi: 10.1016/s1386-6532(01)00157-3. [DOI] [PubMed] [Google Scholar]
- 36.Shibayama T, Masuda G, Ajisawa A, Takahashi M, Nishizawa T, Tsuda F, et al. Inverse relationship between the titre of TT virus DNA and the CD4 cell count in patients infected with HIV. AIDS. 2001;15:563–570. doi: 10.1097/00002030-200103300-00004. [DOI] [PubMed] [Google Scholar]
- 37.Thom K, Petrik J. Progression towards AIDS leads to increased Torque teno virus and Torque teno minivirus titers in tissues of HIV infected individuals. J Med Virol. 2007;79:1–7. doi: 10.1002/jmv.20756. [DOI] [PubMed] [Google Scholar]
- 38.Christensen JK, Eugen-Olsen J, SŁrensen M, Ullum H, Gjedde SB, Pedersen BK, et al. Prevalence and prognostic significance of infection with TT virus in patients infected with human immunodeficiency virus. J Infect Dis. 2000;181:1796–1799. doi: 10.1086/315440. [DOI] [PubMed] [Google Scholar]
- 39.Devalle S, Rua F, Morgado MG, Niel C. Variations in the frequencies of torque teno virus subpopulations during HAART treatment in HIV-1-coinfected patients. Arch Virol. 2009;154:1285–1291. doi: 10.1007/s00705-009-0440-7. [DOI] [PubMed] [Google Scholar]
- 40.Madsen CD, Eugen-Olsen J, Kirk O, Parner J, Kaae Christensen J, Brasholt MS, et al. TTV viral load as a marker for immune reconstitution after initiation of HAART in HIV-infected patients. HIV Clin Trials. 2002;3:287–295. doi: 10.1310/8c94-vypq-ng1h-4cnw. [DOI] [PubMed] [Google Scholar]
- 41.Walton AH, Muenzer JT, Rasche D, Boomer JS, Sato B, Brownstein BH, et al. Reactivation of multiple viruses in patients with sepsis. PLoS One. 2014;9:e98819. doi: 10.1371/journal.pone.0098819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hunt PW, Brenchley J, Sinclair E, McCune JM, Roland M, Page-Shafer K, et al. Relationship between T cell activation and CD4+ T cell count in HIV-seropositive individuals with undetectable plasma HIV RNA levels in the absence of therapy. J Infect Dis. 2008;197:26–133. doi: 10.1086/524143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Benito JM, Lopez M, Martin JC, Lozano S, Martinez P, Gonzalez-Lahoz J, et al. Differences in cellular activation and apoptosis in HIV-infected patients receiving protease inhibitors or nonnucleoside reverse transcriptase inhibitors. AIDS Res Hum Retrovir. 2002;18:1379–1388. doi: 10.1089/088922202320935456. [DOI] [PubMed] [Google Scholar]
- 44.Massanella M, Negredo E, Perez-Alvarez N, Puig J, Ruiz-Hernandez R, Bofill M, et al. CD4 T-cell hyperactivation and susceptibility to cell death determine poor CD4 T-cell recovery during suppressive HAART. AIDS. 2010;24:959–968. doi: 10.1097/QAD.0b013e328337b957. [DOI] [PubMed] [Google Scholar]
- 45.Brenchley JM, Douek DC. HIV infection and the gastrointestinal immune system. Mucosal Immunol. 2008;1:23–30. doi: 10.1038/mi.2007.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li L, Deng X, Mee ET, Collot-Teixeira S, Anderson R, Schepelmann S, et al. Comparing viral metagenomics methods using a highly multiplexed human viral pathogens reagent. J Virol Methods. 2015;213:139–146. doi: 10.1016/j.jviromet.2014.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Deng X, Naccache SN, Ng T, Federman S, Li L, Chiu CY, et al. An ensemble strategy that significantly improves de novo assembly of microbial genomes from metagenomic next-generation sequencing data. Nucleic Acids Res. 2015;43:e46. doi: 10.1093/nar/gkv002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li L, Diab S, McGraw S, Barr B, Traslavina R, Higgins R, et al. Divergent astrovirus associated with neurologic disease in cattle. Emerg Infect Dis. 2013;19:1385–1392. doi: 10.3201/eid1909.130682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.McGinnis S, Madden TL. BLAST: at the core of a powerful and diverse set of sequence analysis tools. Nucleic Acids Res. 2004;32:W20–W25. doi: 10.1093/nar/gkh435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 2012;19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Garcia-Alvarez M, Berenguer J, Alvarez E, Guzman-Fulgencio M, Cosin J, Miralles P, et al. Association of torque teno virus (TTV) and torque teno mini virus (TTMV) with liver disease among patients coinfected with human immunodeficiency virus and hepatitis C virus. Eur J Clin Microbiol Infect Dis. 2013;32:289–297. doi: 10.1007/s10096-012-1744-1. [DOI] [PubMed] [Google Scholar]
- 53.Nasser TF, Brajade Oliveira K, Reiche EM, Amarante MK, Pelegrinelli Fungaro MH, Watanabe MA. Detection of TT virus in HIV-1 exposed but uninfected individuals and in HIV-1 infected patients and its influence on CD4+ lymphocytes and viral load. Microb Pathog. 2009;47:33–37. doi: 10.1016/j.micpath.2009.04.010. [DOI] [PubMed] [Google Scholar]
- 54.Kluba J, Linnenweber-Held S, Heim A, Ang AM, Raggub L, Broecker V, et al. A rolling circle amplification screen for polyomaviruses other than BKPyV in renal transplant recipients confirms high prevalence of urinary JCPyV shedding. Intervirology. 2015;58:88–94. doi: 10.1159/000369210. [DOI] [PubMed] [Google Scholar]
- 55.Kocjan BJ, Bzhalava D, Forslund O, Dillner J, Poljak M. Molecular methods for identification and characterization of novel papillomaviruses. Clin Microbiol Infect. 2015 doi: 10.1016/j.cmi.2015.05.011. [DOI] [PubMed] [Google Scholar]
- 56.Peretti A, FitzGerald PC, Bliskovsky V, Buck CB, Pastrana DV. Hamburger polyomaviruses. J Gen Virol. 2015;96:833–839. doi: 10.1099/vir.0.000033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Feng Y, Zhao W, Dai J, Li Z, Zhang X, Liu L, et al. A novel genotype of GB virus C: its identification and predominance among injecting drug users in Yunnan, China. PLoS One. 2011;6:e21151. doi: 10.1371/journal.pone.0021151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Muerhoff AS, Simons JN, Leary TP, Erker JC, Chalmers ML, Pilot-Matias TJ, et al. Sequence heterogeneity within the 5′-terminal region of the hepatitis GB virus C genome and evidence for genotypes. J Hepatol. 1996;25:379–384. doi: 10.1016/s0168-8278(96)80125-5. [DOI] [PubMed] [Google Scholar]
- 59.McOmish F, Yap PL, Dow BC, Follett EA, Seed C, Keller AJ, et al. Geographical distribution of hepatitis C virus genotypes in blood donors: an international collaborative survey. J Clin Microbiol. 1994;32:884–892. doi: 10.1128/jcm.32.4.884-892.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Robertson B, Myers G, Howard C, Brettin T, Bukh J, Gaschen B, et al. Classification, nomenclature, and database development for hepatitis C virus (HCV) and related viruses: proposals for standardization. International Committee on Virus Taxonomy. Arch Virol. 1998;143:2493–2503. doi: 10.1007/s007050050479. [DOI] [PubMed] [Google Scholar]
- 61.Guirgis BS, Abbas RO, Azzazy HM. Hepatitis B virus genotyping: current methods and clinical implications. Int J Infect Dis. 2010;14:e941–e953. doi: 10.1016/j.ijid.2010.03.020. [DOI] [PubMed] [Google Scholar]
- 62.van der Kuyl AC, Zorgdrager F, Hogema B, Bakker M, Jurriaans S, Back NK, et al. High prevalence of hepatitis B virus dual infection with genotypes A and G in HIV-1 infected men in Amsterdam, the Netherlands, during 2000–2011. BMC Infect Dis. 2013;13:540. doi: 10.1186/1471-2334-13-540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sunbul M. Hepatitis B virus genotypes: global distribution and clinical importance. World J Gastroenterol. 2014;20:5427–5434. doi: 10.3748/wjg.v20.i18.5427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sylwester AW, Mitchell BL, Edgar JB, Taormina C, Pelte C, Ruchti F, et al. Broadly targeted human cytomegalovirus-specific CD4+ and CD8+ T cells dominate the memory compartments of exposed subjects. J Exp Med. 2005;202:673–685. doi: 10.1084/jem.20050882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Jin X, Demoitie MA, Donahoe SM, Ogg GS, Bonhoeffer S, Kakimoto WM, et al. High frequency of cytomegalovirus-specific cytotoxic T-effector cells in HLA-A*0201-positive subjects during multiple viral coinfections. J Infect Dis. 2000;181:165–175. doi: 10.1086/315201. [DOI] [PubMed] [Google Scholar]
- 66.Komanduri KV, Donahoe SM, Moretto WJ, Schmidt DK, Gillespie G, Ogg GS, et al. Direct measurement of CD4+ and CD8+ T-cell responses to CMV in HIV-1-infected subjects. Virology. 2001;279:459–470. doi: 10.1006/viro.2000.0697. [DOI] [PubMed] [Google Scholar]
- 67.Naeger DM, Martin JN, Sinclair E, Hunt PW, Bangsberg DR, Hecht F, et al. Cytomegalovirus-specific T. cells persist at very high levels during long-term antiretroviral treatment of HIV disease. PLoS One. 2010;5:e8886. doi: 10.1371/journal.pone.0008886. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.