Abstract
Parthenolide (PTL) is a sesquiterpene lactone natural product with anti-proliferative activity to cancer cells. Selective eradication of leukemic stem cells (LSCs) over healthy hematopoietic stem cells (HSCs) by PTL has been demonstrated in previous studies, which suggests PTL and related molecules may be useful for targeting LSCs. Eradication of LSCs is required for curative therapy. Chemical optimizations of PTL to improve potency and pharmacokinetic parameters have focused largely on the α-methylene-γ-butyrolactone, which is essential for activity. Conversely, we evaluated modifications to the C1-C10 olefin and benchmarked new inhibitors to PTL with respect to inhibitory potency across a panel of cancer cell lines, ability to target drug-resistant acute myeloid leukemia (AML) cells, efficacy for inhibiting clonal growth of AML cells, toxicity to healthy bone marrow cells, and efficiency for promoting intracellular reactive oxygen species (ROS) levels. Cyclopropane 4 was found to possess less toxicity to healthy bone marrow cells, enhanced potency for the induction of cellular ROS, and similar broad-spectrum anti-proliferative activity to cancer cells in comparison to PTL.
Keywords: Bone marrow toxicity, Cyclopropanation, Acute myeloid leukemia, Parthenolide, Sesquiterpene lactone
Graphical abstract
1. Introduction
Sesquiterpene lactones (SL) are a diverse family of plant-derived natural products with utilities in treating inflammatory diseases and cancer.1, 2 Parthenolide (PTL) is a well-studied SL derived from the feverfew plant Tanacetum parthenium,3 bearing broad-spectrum anti-proliferative activities to a variety of cancer types through multiple mechanisms of inhibition.4, 5 The seminal discovery that PTL induces apoptosis in acute myeloid leukemia (AML) stem and progenitor cells without exhibiting comparable toxicity to healthy hematopoietic stem cells (HSCs) has anointed PTL as the prototypical member of next-generation therapies for eradicating leukemic stem cells (LSCs).6 AML growth is hierarchical and originates from LSCs.7, 8 Therefore, small molecules that eliminate LSCs are expected to confer more durable and potentially curative therapies.9–13 In addition to its anti-leukemic activity, PTL has been explored as a potential therapeutic for a spectrum of indications.14
Chemical optimizations of PTL have been required to optimize the natural product for in vivo applications. A Phase I dose escalation trial of feverfew extract failed to achieve measurable levels of PTL in serum and oral dosing (40 mg/kg) of PTL in mice yielded approximately 200 nM concentrations in serum, which is not sufficient to confer anti-proliferative activity.15, 16 Conversion of PTL to prodrug dimethylamino-parthenolide fumarate, DMAPT (or LC-1), increased water solubility by ~1000-fold and yielded an analogue with substantially improved pharmacokinetic parameters (mice: Cmax = 25 µM, t1/2 = 0.6 hr; canine: Cmax = 61 µM, t1/2 = 1.9 hr) with oral dosing (100 mg/kg).17, 18 Hetero-Michael addition of aliphatic amines to natural products and synthetic analogues bearing -methylene- -butyrolactones has constituted a modular strategy to enhance water solubilities through prodrug formation.19–25 Semisynthetic modifications to PTL outside of the -methylene- -butyrolactone warhead, however, are significantly less developed. Acid-catalyzed conversion of PTL to 5–7-5 guaianolide, Micheliolide (1), has been achieved, yielding a derivative with anti-proliferative activity comparable to its predecessor.21, 26, 27 Photochemical isomerization of the C1-C10 olefin of PTL has also been reported, yielding cis-olefin 2.28–31 Allylic oxidation of the C1-C10 vinyl methyl group of PTL results in formation of another natural product, Melampomagnolide B (MelB), which exhibits comparable anti-proliferative activity as PTL, but contains an allylic alcohol, which is useful for further transformations (e.g., synthesis of affinity pulldown reagents and O-functionalized analogues).32–36 Recently, the Fasan laboratory has utilized P450 enzymes to oxygenate proximal to the C1-C10 olefin of PTL, yielding alcohols at C9 and C14, which were subsequently esterified with substituted benzoic acids to yield PTL analogues with anti-leukemic activities.37
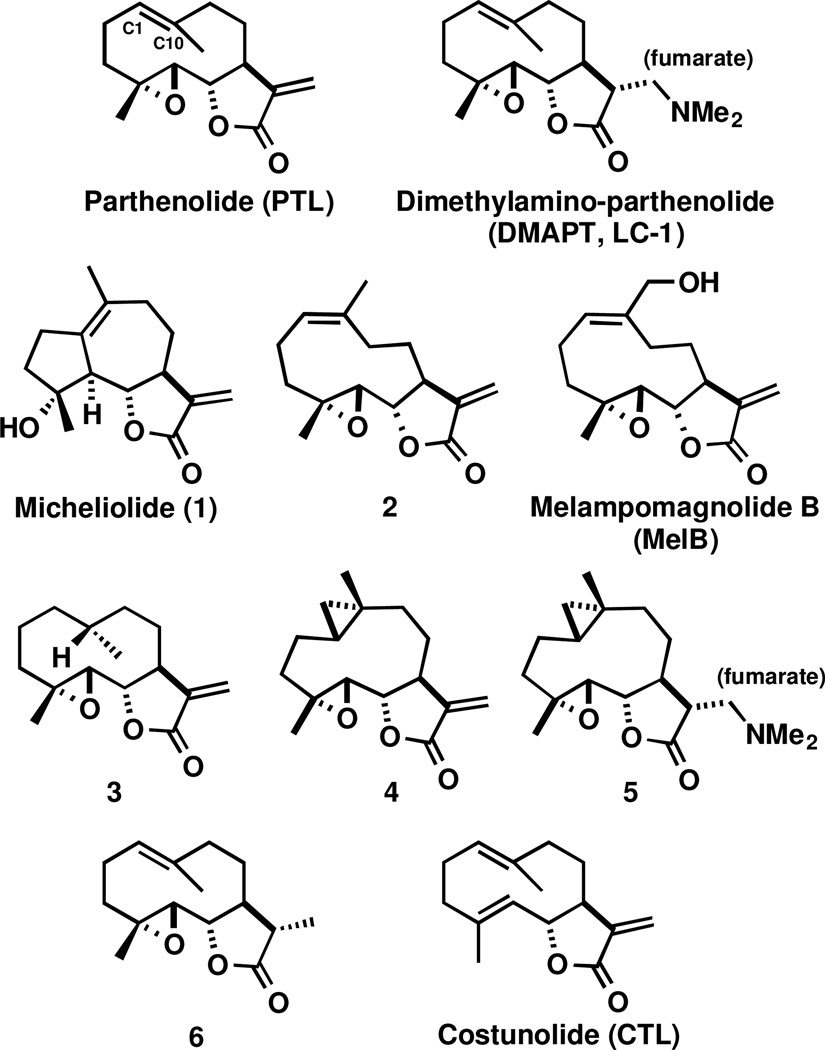
In this study, we examined the necessity of the PTL C1-C10 olefin and its tolerance to structural modification with respect to sustained anti-proliferative activities to cancer cells through the synthesis and biochemical screening of C1-C10 modified PTL analogues. Included among our small library of compounds are established PTL analogues, such as Micheliolide (1), cis-PTL (2), and MelB, as well as additional mechanistic probes, such as 3 (reduced C1-C10 olefin) and 4 (cyclopropanated C1-C10 olefin). Interestingly, cyclopropanated analogue 4 was found to exhibit similar anti-proliferative activity to cancer cells as PTL, but conferred less toxicity to healthy bone marrow and more potently induced cellular reactive oxygen species (ROS), which is known to promote cell death to AML stem cells and other cancer cells.38–47
2. Results and discussion
2.1 Design and synthesis of PTL analogues
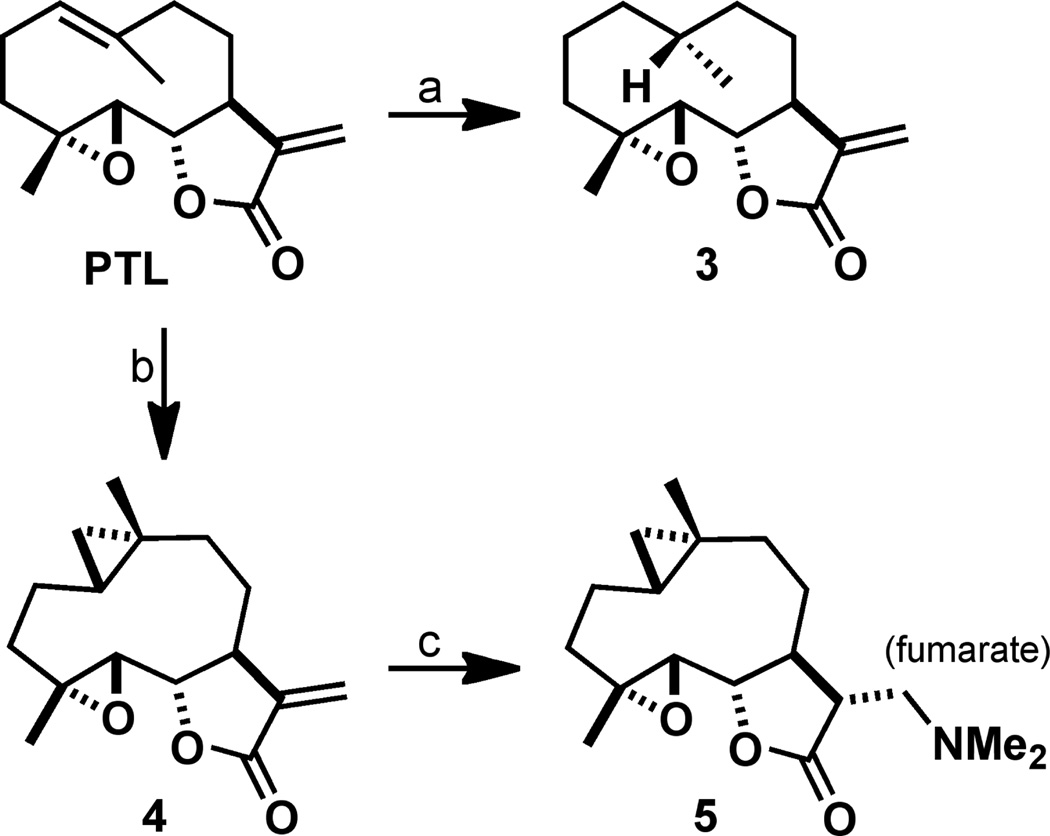
A small library of C1-C10 olefin modified PTL analogues and control compounds were synthesized or purchased commercially (Figure 1). Previous studies have found that the C1-C10 olefin of PTL can participate in electrophilic transannular cyclizations with the C4-C5 epoxide under Brønsted or Lewis acid conditions to yield guaianolide analogues.28, 29 Therefore, we synthesized reduced analogue 3 to eliminate any potential for acid instability. The exocyclic methylene on the -methylene- -butyrolactone of PTL was transiently protected by dimethylamine addition to yield dimethylamino-PTL, which was not converted to the fumarate salt for this application (Scheme 1). Hydrogenation of dimethylamino-PTL free base with PtO2 catalyst at 50 psi H2 resulted in facial selective reduction (~15:1), yielding 10R-diastereomer 3. The exocyclic methylene was deprotected under Hofmann elimination conditions (excess MeI, THF, H2O)20, 48, 49 to yield 3 in 32% yield (over three steps). The stereochemistry of the methyl group was assigned by X-ray crystallography (SI).
Figure 1.
Structures of PTL and DMAPT, C1-C10 modified PTL analogues (1-5 and MelB), and control compounds for biochemical assays (6 and CTL).
Scheme 1.
a) NHMe2, MeOH; Pt2O, H2 (50 psi), EtOAc; MeI, THF, H2O, 45 °C, 32% (3 steps); b) Zn(CH2I)2, DME, CH2Cl2, 41%; c) NHMe2, MeOH; fumaric acid, 85%.
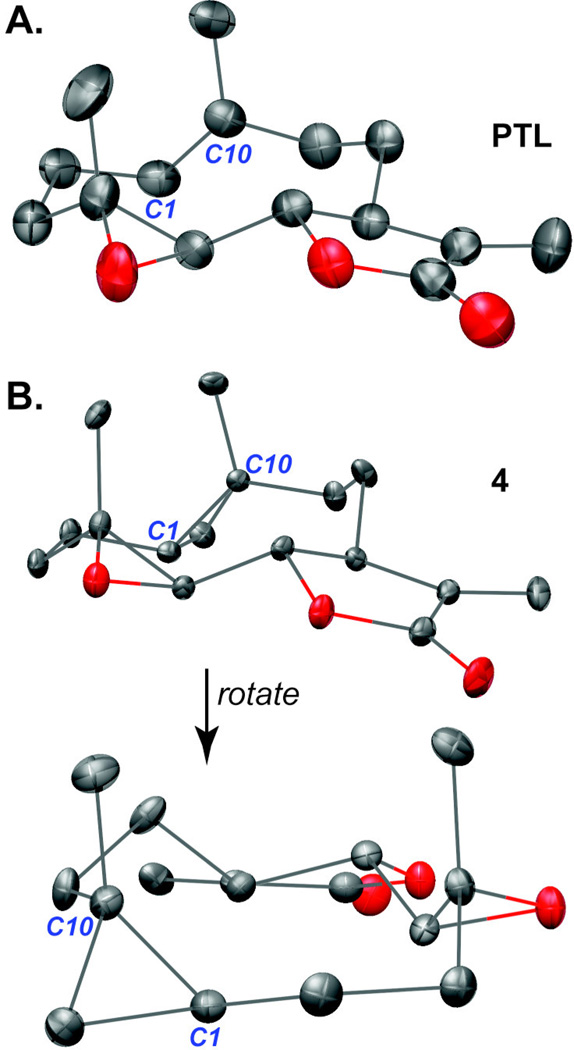
Cyclopropanes are unique ring systems with significant sp2-character, thereby mimicking the electronics of double bonds.50 Such modifications can be valuable for increasing the stability of a drug candidate. In the case of PTL, replacement of the C4-C5 epoxide with a cyclopropane significantly enhanced plasma half-life (t1/2 = 13.9 hr versus 1.6 hr for PTL; testing in mouse plasma).51 To further probe the role of the C1-C10 olefin in PTL with a structurally analogous mimetic, we synthesized C1-C10-cyclopropanated 4. Utilizing the Furukawa modification (ZnEt2) to the classical Simmons-Smith reaction,52–54 PTL was treated with pre-formed Zn(CH2I)2 in a solution of DME and CH2Cl2, which yielded (1S,10R) 4 in 41% yield following silica gel chromatography (Scheme 1). Interestingly, no attack to the exocyclic olefin was observed, with the remaining mass balance consisting of mostly unreacted PTL. The structure of 4 was assigned by X-ray crystallography (Figure 2 and SI). As expected, the solid-state structure of cyclopropane 4 was highly similar to a recently reported PTL X-ray structure51 with a root-mean-square deviation of 0.167 Å (Figure 2; alignment of structures provided in the SI). Recognizing that 4 may suffer from poor aqueous solubility akin to PTL, we synthesized dimethylamine congener 5, which was converted to the fumarate salt for consistency with DMAPT.18 PTL and Costunolide (CTL) were purchased from commercial vendors and the remaining analogues in our library (1, 2, MelB, and 6) were synthesized as previously reported.18, 21–26, 28, 29, 32–35 The structure of synthesized Micheliolide (1) was verified by X-ray crystallography (SI) and compared to a previous report.55
Figure 2.
X-ray crystal structures. (A) PTL18 and (B) cyclopropane 4 adopt similar conformations in the solid-state. Root-mean-square deviation between the two structures is 0.167 Å.
2.2 Lipophilicity Analyses
The distribution coefficients (LogD) of select C1-C10 PTL analogues were assessed through calculated and measured analyses (Table 1). Calculated LogD values were less predictive in comparison to experimentally derived measurements for PTL (cLogD7.4 = 3.07 versus LogD7.4 = 1.79) and cis-PTL isomer 2 (cLogD7.4 = 3.07 versus LogD7.4 = 2.00), whereas calculated and measured values were generally consistent for the remaining analogues (Table 1). Both reduction and cyclopropanation of the C1-C10 olefin increased the overall lipophilicity of the PTL skeleton (LogD7.4 = 2.30 for 3 and LogD7.4 = 2.29 for 4). CTL, which contains a C4-C5 olefin in place of the epoxide in PTL, was substantially more lipophilic (LogD7.4 = 2.90 for CTL versus LogD7.4 = 1.79 for PTL). The distribution coefficients of dimethylamine fumarate salts of PTL and 4, analogues DMAPT and 5, respectively, were not measured because their calculated LogD values (cLogD7.4 = 0.50, DMAPT; cLogD7.4 = 0.56, 5) were outside the measurable range of the assay (1–5 units).
Table 1.
Calculated LogD values (cLogD7.4; MarvinSketch) and measured LogD values (LogD7.4; Sirius Analytical, average of two measurements) at pH 7.4 for select C1-C10 PTL analogues.
| Compound | cLogD7.4 | LogD7.4 |
|---|---|---|
| PTL | 3.07 | 1.79 |
| 1 | 1.97 | 2.18 |
| 2 | 3.07 | 2.00 |
| 3 | 3.48 | 2.30 |
| 4 | 3.16 | 2.29 |
| CTL | 4.22 | 2.90 |
2.3 Cellular cytotoxicity screening
All compounds were screened for anti-proliferative activity against 7 cell lines representing blood lineage cancers (HL-60 and CCRF-CEM) and solid tumors (U-87 MG, GBM6, MCF-7, DU-145, and NCI/ADR-RES). An established drug-resistant tumor cell line, NCI/ADR-RES, was included in our screen to determine if PTL and related analogues possess activity against cancer cell lines with high P-glycoprotein expression.56, 57 Clinically used chemotherapeutic drugs gemcitabine and doxorubicin are inactive (IC50 > 500 µM) against NCI/ADR-RES cells,58 and cancer stem cells (CSCs) are known to have high expression levels of drug efflux machinery.59–61 Additionally, we included the GBM6 glioblastoma multiforme (GBM) cell line in our primary screen because it possesses a CD133+ population of cells,62 which is a frequently used marker of GBM stem cells.63
Screening of PTL and related analogues revealed broad-spectrum, low-micromolar IC50 growth inhibitory activity to all cancer cell lines regardless of modification to the C1-C10 olefin (Table 2). In contrast, 6, which bears a reduced exocyclic methylene on the -methylene- -butyrolactone, was found to be completely inactive against all cell lines examined (IC50 > 500 µM). These data are consistent with previous reports,18, 32, 64 and reinforce the necessity of the -methylene- -butyrolactone for anti-proliferative activity of molecules of this class.1, 20, 23, 65 CTL was found to be equipotent to the C1-C10 modified analogues, suggesting the C4-C5 epoxide is non-essential for activity, which is also consistent with a previous report.51 All compounds except for exomethylene-reduced 6 were active against drug-resistant NCI/ADR-RES cells (IC50 range: 9.4 – 22.0 µM).
Table 2.
Growth inhibitory activities (IC50) of PTL, C1-C10-modified PTL analogues, and related probes to cell lines: HL-60 (acute promyelocytic leukemia), CCRF-CEM (acute lymphoblastic leukemia), U-87 MG (glioblastoma multiforme), GBM6 (glioblastoma multiforme), MCF-7 (breast adenocarcinoma), DU-145 (prostate cancer), and NCI/ADR-RES (ovarian cancer; adriamycin-resistant), IC50 values are mean ± S.D. in µM (n • 3 analyses).
| Compound | HL-60 | CCRF-CEM | U-87 MG | GBM6 | MCF-7 | DU-145 | NCI/ADR-RES |
|---|---|---|---|---|---|---|---|
| PTL | 9.3 ± 3.8a | 4.7 ± 1.6 | 8.8 ± 2.1a | 3.4 ± 1.1a | 9.7 ± 2.8 | 8.9 ± 4.6a | 11.4 ± 2.4b |
| LC-1 | 7.1 ± 0.4 | 1.9 ± 0.4 | 8.8 ± 1.9a | 3.5 ± 1.1a | 10.4 ± 1.2 | 8.4 ± 4.5 | 12.3 ± 2.7 |
| 1 | 9.2 ± 2.2 | 2.7 ± 1.1 | 17.8 ± 5.0 | 8.5 ± 0.7 | 9.6 ± 1.0 | 14.8 ± 4.0 | 22.0 ± 5.3 |
| 2 | 8.0 ± 1.1 | 2.2 ± 1.0 | 9.1 ± 4.4 | 3.3 ± 0.8 | 7.5 ± 0.8 | 6.0 ± 4.2 | 9.4 ± 2.1 |
| MelB | 7.5 ± 3.0 | 5.5 ± 1.2 | 16.3 ± 6.8 | 4.5 ± 1.9 | 9.5 ± 1.9 | 14.3 ± 5.9 | 15.0 ± 4.9 |
| 3 | 9.0 ± 2.1 | 2.5 ± 2.1 | 7.5 ± 0.4 | 2.0 ± 0.3 | 20.1 ± 1.3 | 5.5 ± 1.5 | 15.6 ± 3.1 |
| 4 | 4.4 ± 1.3 | 2.0 ± 0.6 | 11.6 ± 1.4 | 2.3 ± 0.5 | 14.1 ± 1.0 | 14.8 ± 6.7 | 12.0 ± 2.5 |
| 5 | 6.5 ± 2.7 | 2.9 ± 0.6 | 10.5 ± 0.9 | 3.2 ± 0.7 | 16.9 ± 2.0 | 13.3 ± 2.2 | 11.7 ± 3.1 |
| 6 | >500 | >500 | >500 | >500 | >500 | >500 | >500 |
| CTL | 13.0 ± 0.2 | 2.3 ± 0.2 | 9.6 ± 0.8 | 7.0 ± 1.8 | 17.5 ± 3.7 | 7.7 ± 2.8 | 17.1 ± 0.9 |
2.4 Bone marrow toxicity studies
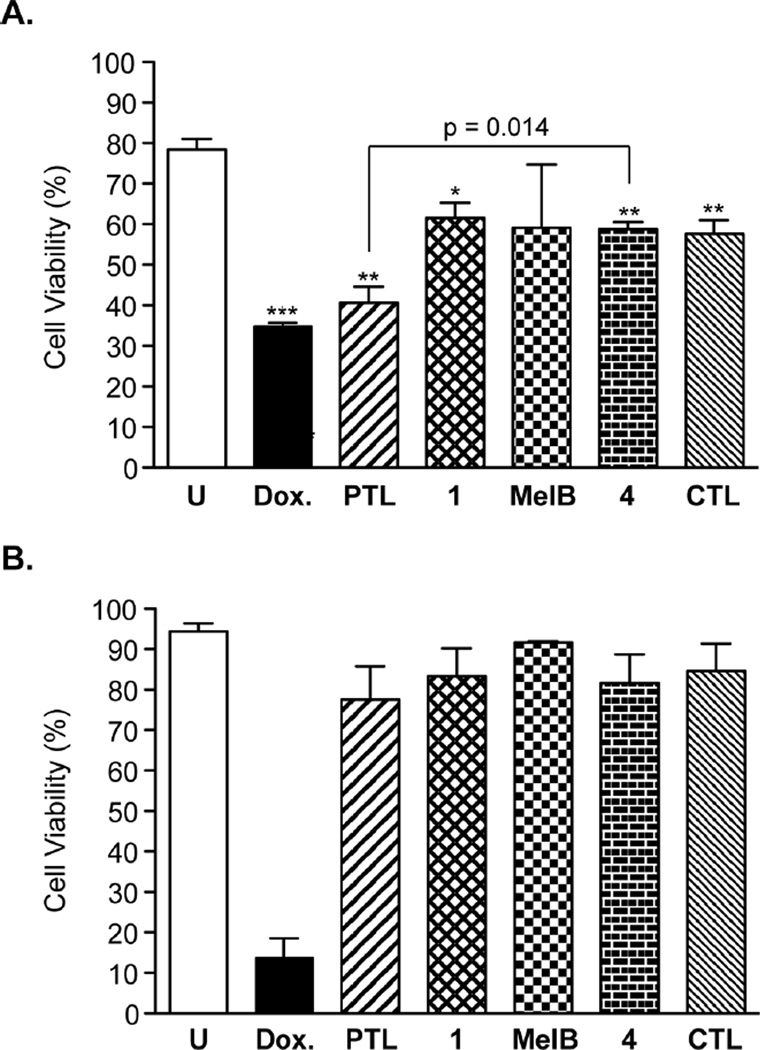
The CD34+CD38− bone marrow (BM) immunophenotype is enriched for self-renewing stem cells.66, 67 Previous studies have demonstrated that PTL is non-toxic to total BM and CD34+CD38− BM cells when dosed at 5 µM for 18 hrs.6 To assess BM toxicity of the synthesized C1-C10 PTL analogues in comparison to PTL, we performed flow cytometry assays with human BM cells and measured cellular viability by flow cytometry using markers for apoptosis (Annexin V) and necrosis (7-AAD). Since PTL has been shown to elicit some overall BM toxicity at a dose of 7.5 µM for 18 hrs,6 we elected to utilize a slightly higher dose to exacerbate the toxicity of PTL so that analogues with less toxicity to BM in comparison to PTL could be measured. Doxorubicin (DOX) was included as a positive control since it is known to elicit BM toxicity.68 The mean overall viability of the BM specimen utilized in our study was 78% (Figure 3A) and 94% for the CD34+CD38− population (Figure 3B). Treatment with 0.5 µM DOX for 12 hr resulted in a 56% reduction in total BM viability and an 85% reduction in the primitive CD34+CD38− BM population (Figure 3). PTL treatment at 25 µM resulted in a 48% reduction in total BM viability, whereas C1-C10 modified PTL analogues 1, MelB, and 4, as well as control analogue, CTL, were less toxic (range: 22–26% average reduction of total BM viable cells). PTL was found to elicit no significant toxicity to primitive CD34+CD38− BM cells at 25 µM dose and the C1-C10 PTL analogues were similarly non-toxic at the same concentration (Figure 3B). Therefore, modification to the C1-C10 olefin of PTL significantly lowers its overall toxicity to BM cells. However, the observed toxicity of PTL to total BM would be expected to be transient since little cell death was measured in the CD34+CD38− BM population upon PTL treatment, which is responsible for BM clonal growth.66, 67 Studies in our group using PTL prodrug, DMAPT, have revealed no measurable toxicity to mice upon continuous oral dosing (100 mg/kg, daily) for over four weeks.64
Figure 3.
Toxicity of PTL and analogues to human bone marrow cells. BM was dosed with DOX (0.5 µM), and PTL, 1, MelB, 4, and CTL (25 µM) for 12 h. Cellular viability was then measured by flow cytometry and viable cells (%) were those not stained by Annexin V (apoptosis) and 7-AAD (necrosis) reagents. (A) Viability of the total bone marrow cell population and (B) viability of the CD34+CD38− population. Data is the mean (n ≥ 3 analyses) ± S.D. *p = 0.05, **p ≤ 0.01, ***p ≤ 0.001 in comparison to untreated control.
2.5 Inhibition of drug-resistant AML and toxicity to LSCs
Given the selectivity of our compounds for inhibiting growth of blood lineage cancer cells (e.g., HL-60 and CCRF-CEM, Table 2) and their lack of toxicity to healthy BM (Figure 3), we focused subsequent efforts on characterizing the anti-leukemic activities of our molecules. Four murine AML cell lines were utilized for our initial screen (Table 3). B117P and B140P are murine cell lines isolated from the BXH-2 mice strain that spontaneously develops AML due to the presence of a murine leukemia virus.69 These cells are sensitive to cytarabine (AraC), which is used in standard-of-care AML therapy. Continual low dosing of B117P and B140P with cytarabine resulted in cytarabine-resistant cell lines B117H and B140H, respectively.70, 71 Cytarabine resistance is conferred in B117H and B140H by loss-of-function mutations in the deoxycytidine kinase gene Dck, which inhibits intracellular metabolism of cytarabine to its 5’-monophosphate.72 PTL and C1-C10-modified analogues 1, MelB, and 4, and control analogue CTL all inhibited the growth of this panel of cell lines with low micromolar activity (IC50 range: 1.1 – 13.5 µM). No loss in potency was observed between cytarabine-sensitive (parental) cell lines B117P and B140P and cytarabine-resistant lines B117H and B140H for any of the molecules tested. These data suggest that PTL analogues have the potential to sustain anti-proliferative activities to AML cell lines that become sensitive to cytarabine.
Table 3.
Growth inhibitory activities (IC50) of PTL, C1-C10-modified PTL analogues, and related probes to murine AML cell lines B117P, B117H, B140P, and B140H. IC50 values are mean ± S.D. in µM (n • 3 analyses).
| Compound | B117P | B117H | B140P | B140H |
|---|---|---|---|---|
| PTL | 1.1 ± 0.2 | 4.7 ± 1.6 | 5.8 ± 2.3 | 3.4 ± 1.1 |
| 1 | 6.4 ± 1.0 | 8.8 ± 1.7 | 10.0 ± 0.4 | 9.3 ± 1.5 |
| MelB | 2.1 ± 0.8 | 2.5 ± 1.0 | 2.9 ± 1.0 | 2.2 ± 0.7 |
| 4 | 2.9 ± 0.6 | 6.4 ± 2.1 | 13.5 ± 2.3 | 5.9 ± 2.4 |
| CTL | 3.5 ± 0.4 | 4.8 ± 0.7 | 3.6 ± 1.2 | 2.8 ± 0.7 |
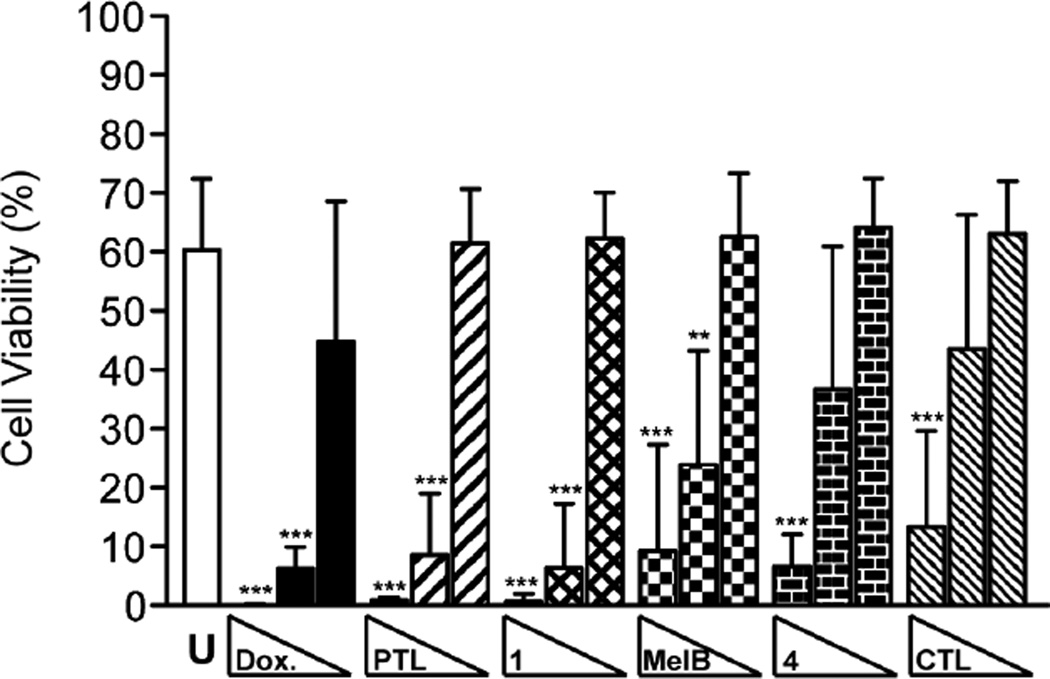
PTL is known to eradicate LSCs,6 and therefore, we investigated if the C1-C10-modified PTL analogues could inhibit LSCs with similar potency as the parent natural product. We utilized an engineered leukemia cell line, TEX, for these assays. TEX cells are derived from lineage depleted (Lin−) human cord blood cells transduced with the fusion gene TLS-ERG. TEX cells effectively mimic human AML by maintaining the potential for multi-lineage differentiation through their heterogeneous population of cells with hierarchical growth properties. A large population of primitive CD34+ cells are present in the TEX model system, which has also been utilized in high-throughput screening for small molecule inhibitors of LSCs.73, 74 Screening of PTL and related analogues (1, MelB, 4, and CTL) revealed broad-spectrum inhibitory activity (IC50 range: 2.7 – 6.8 µM) by metabolic viability staining following 48 hr treatment (Table 4). Analysis of the LSC-enriched CD34+CD38− population of TEX cells treated with 25 µM PTL, 1, MelB, and CTL for 12 hr revealed a nearly complete reduction in cellular viability by flow cytometry analysis (cell viability range: 1–10%, Figure 4), with the majority of cells staining positive for 7-AAD, indicating necrotic cell death. Treatment of cells with 15 µM PTL analogues yielded slightly higher amounts of viable cells (cell viability range: 10–25%) with no statistical significant differences in potencies between the analogues tested. A relatively low dose of DOX (0.5 µM) was sufficient to reduce the viability of CD34+CD38− TEX cells to 6%. Consequently, all of the PTL analogues tested were able to induce cell death in the LSC-enriched CD34+CD38− population of TEX cells. To corroborate that the observed inhibition of primitive CD34+CD38− TEX cells also affects the LSC population, we performed methylcellulose clonal growth assays of TEX cells in the presence of our compounds. As previously mentioned, AML is characterized by hierarchical growth properties that originate from LSCs,7, 8 and therefore, small molecule inhibition of LSCs will prevent clonal growth of cells in methylcellulose. Control (DMSO) treated TEX cells yield on average 20.8 clones per assay (Table 5). Treatment with PTL or analogues 1, MelB, 4, or CTL all potently inhibit clonal outgrowth of TEX cells at 15 µM dose, with PTL and 1 yielding no measurable clones. Decreasing the dosage to 2.5 µM with the same compounds also elicits inhibition of TEX clonal growth (range: 2.6 – 7.0 clone average). DOX and AraC were both able to inhibit TEX clonal growth in our assay, with 0.5 µM treatment of both compounds completely inhibiting cell growth. Taken together, these data demonstrate PTL and related analogues are proficient at inhibiting LSCs, which is likely mediated through their common pharmacophore, the -methylene- -butyrolactone.
Table 4.
Growth inhibitory activities (IC50) of PTL, C1-C10-modified PTL analogues, and related probes to TEX cells. IC50 values are mean ± S.D. in µM (n • 3 analyses).
| Compound | TEX |
|---|---|
| PTL | 2.8 ± 0.2 |
| 1 | 2.7 ± 0.4 |
| MelB | 6.8 ± 2.3 |
| 4 | 3.8 ± 0.2 |
| CTL | 4.9 ± 0.4 |
Figure 4.
Cellular viability (%) of CD34+CD38− TEX cells treated with DOX (2.5, 0.5, 0.05 µM) and PTL, 1, MelB, 4, CTL (25, 15, 2.5 µM). Viable cells were those not stained by Annexin V (apoptosis) and 7-AAD (necrosis) markers. Values are mean ± S.D. (n • 5 analyses). * p = 0.05, ** p ≤ 0.01, *** p ≤ 0.001 in comparison to untreated control.
Table 5.
Clonal growth assay with TEX cells. TEX cells were plated 6,000 cells/well and dosed with DMSO (0.05%, control wells) or compounds at the concentrations noted. Values are the mean number of colonies ± S.D. (3 biological replicates) observed after 11 d growth on methylcellulose. N.D., clonal growth not detected. p ≤ 0.001 in comparison to DMSO control for all samples, except 4 at 2.5 µM (p ≤ 0.01).
| Colonies | ||
|---|---|---|
| Compound | 2.5 µM | 15 µM |
| PTL | 2.6 ± 2.7 | N.D. |
| 1 | 4.2 ± 2.5 | N.D. |
| MelB | 5.3 ± 1.6 | 0.1 ± 0.3 |
| 4 | 7.0 ± 5.1 | 0.6 ± 0.9 |
| CTL | 6.9 ± 3.6 | 5.6 ± 4.7 |
| 0.1 µM | 0.5 µM | |
| DOX | 0.7 ± 1.3 | N.D. |
| AraC | 2.6 ± 1.0 | N.D. |
| DMSO | 20.8 ± 9.0 | |
2.6 Induction of reactive oxygen species
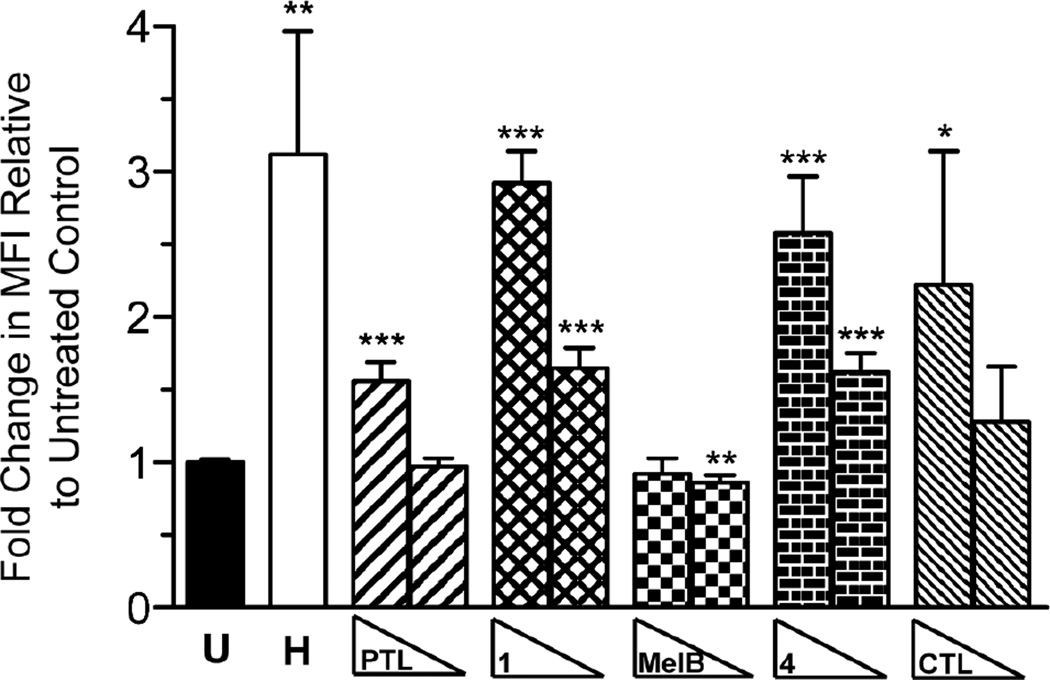
The mechanism by which PTL eradicates cancer cell viability is an area of substantial debate. PTL has been shown to affect a variety of cellular processes, including (among many others) inhibition of NF-κB signaling and microtubule detyrosination, reduction in DNA methylation, and induction of cellular ROS (reviewed in 1, 2, 4, 5, 75, 76). Multiple studies have implicated ROS induction as a mechanism of PTL-mediated cancer cell death.40, 43, 46, 47 Consequently, we measured changes in intracellular ROS in TEX cells resulting from treatment with PTL, 1, MelB, 4, and CTL to rank order the pharmacological utility of our compounds. Treatment of TEX cells with hydrogen peroxide results in a 3.1-fold induction of intracellular ROS after 30 min, which is consistent with a previous study where a similar induction of ROS was measured by flow cytometry in HL-60 cells.77 Dosage of TEX cells with 1 (2.9-fold), 4 (2.6-fold), and CTL (2.2-fold) all substantially induced ROS levels at 100 µM dose in comparison to untreated control (Figure 5). PTL induced ROS as well, but to a lower level (1.6-fold). No observable induction of ROS was detected with MelB at either concentration tested. Decreasing the concentration of compounds to 25 µM also resulted in a significant induction of ROS levels for 1 (1.7-fold) and 4 (1.6-fold). Consequently, these data suggest that 1 and 4 more potently induce ROS than parent natural product, PTL.
Figure 5.
Intracellular ROS induction by PTL and analogues. TEX cells were treated with hydrogen peroxide H (100 µM) and PTL, 1, MelB, 4, CTL (100, 25 µM) and ROS activity was measured by flow cytometry using CellROX Green reagent. The median fluorescence intensity (MFI) of each sample was normalized to the untreated control and averaged. Values are mean MFI ± S.D. (n • 3 analyses). * p = 0.05, ** p ≤ 0.01, *** p ≤ 0.001 in comparison to untreated control.
3. Conclusions
A small library of C1-C10 PTL analogues was synthesized and evaluated for anti-proliferative activity to cancer cells, toxicity to healthy BM, ability to inhibit drug-resistant AML and target LSCs, and proficiency at inducing intracellular ROS. All compounds with the exception of 6 were capable of inducing cancer cell death with low micromolar potency. However, Micheliolide (1) and cyclopropane 4 were found to inhibit the growth of drug-resistant AML and eliminate LSCs similarly to PTL, but offer the advantages of being less toxic to healthy BM and more potently activating ROS in AML cells than PTL. Additionally, elaboration of 4 to its dimethylamine congener 5 provided an analogous prodrug to DMAPT (LC-1). Given the continued interest in PTL, highlighted by its first total synthesis,14 and the rekindled popularity of covalent drugs in general,78, 79 C1-C10 modifications such as cyclopropanation may be useful for optimizing PTL and related germacranolides for therapeutic applications.
4. Material and methods
4.1 LogD measurements
Calculated LogD values were obtained using MarvinSketch (ChemAxon). cLogD7.4 was calculated using 0.1 mol/dm3 electrolyte concentrations (Cl−, Na+, K+) at pH 7.4. Experimental LogD7.4 measurements were performed by Sirius Analytical. The LogD7.4 of each sample was determined using the LDA (liquid-liquid distribution chromatography) method. The data is the average of two measurements.
4.2 Preparation of stock solutions
Compound stock solutions were prepared in DMSO (20–100 mM concentrations) and stored at −20 °C when not in use. Compound purities were assessed frequently by analytical reverse-phase HPLC analysis and fresh solutions were prepared as needed.
4.3 Cell culture
All cell lines were maintained in a humidified 5% CO2 environment at 37 °C in tissue culture flasks (Corning) under normoxic conditions. Adherent cells were dissociated using either Trypsin-EDTA solution (0.25%, Gibco) or TrypLE Express solution (Invitrogen). HL-60, CCRF-CEM, U-87 MG, GBM6, DU-145, and NCI/ADR-RES cells were cultured as described previously.58, 64, 80 MCF-7 cells (ATCC, HTB-22) were cultured in MEM media (Cellgro) supplemented with 10% FBS (Gibco), bovine insulin (0.01 mg/mL, Sigma), penicillin (100 I.U./mL, ATCC), and streptomycin (100 µg/mL, ATCC). TEX cells73, 74 were cultured in IMDM containing L-glutamine (Cellgro) supplemented with 15% FBS (Gibco), Stem Cell Factor (20 ng/mL, PeproTech), Interleukin-3 (2 ng/mL, PeproTech), penicillin (100 I.U./mL, ATCC), and streptomycin (100 µg/mL, ATCC).
4.4 Human cancer cell line cytotoxicity assays
Alamar blue cellular cytotoxicity assays and data analyses were performed as previously described.58, 64, 80 Suspension cell lines (HL-60, CCRF-CEM and TEX73, 74) were seeded at a density of 10,000 cells/well in media (50 µL) and adherent cell lines (U-87 MG, GBM6, MCF-7, DU-145, and NCI/ADR-RES) were seeded at a density of 5,000 cells/well in media (50 µL) 24 h prior to treatment with compounds in 96-well plates (Costar 3595, Corning, Inc.). IC50 values (n • 3 biological replicates) are the mean ± S.D.
4.5 Murine cytotoxicity assay
Cell culture and cytotoxicity assays with murine cell lines B117P, B117H, B140P, and B140H were performed as previously described.71,72 Cells were seeded at a density of 25,000, 28,000, 36,000 and 44,000 cells/well for B117P, B117H, B140P, and B140H cell lines, respectively, in media (200 µL) in 96-well plates (Costar 3596, Corning, Inc.). Assays were conducted in biological triplicate and IC50 values are the mean ± S.D.
4.6 Bone marrow cell culture
Frozen human mononuclear bone marrow cells were purchased from AllCells (Cat. #ABM011F). These bone marrow cells were from two donors (#5630 [Lot #BM4565] and #4887 [Lot #BM4118]). The cells were thawed according to vendor instructions and then cultured in StemSpan SFEM (STEMCELL Technologies, Inc.) media supplemented with StemSpan CC100 cytokine cocktail (STEMCELL Technologies, Inc.) in a humidified 5% CO2 environment at 37 °C in tissue culture flasks (Corning) under normoxic conditions.
4.7 Flow cytometry analysis of cytotoxicity in bone marrow and TEX cells
Human bone marrow or TEX cells were plated in their respective media at 1 × 106 cells/mL (1 mL/well) in a 24-well plate format (Corning). Cells were dosed with compounds or 1% DMSO/media and incubated for 12 h at 37 °C, 20% O2, and 5% CO2. The final DMSO concentration was 0.03% (v/v) per well. After 12 h of incubation, each sample was transferred into FACS tubes and centrifuged for 5 min at 800 rpm. The supernatant was decanted and each sample was washed with cold 1X PBS (1 mL) and centrifuged again. After centrifugation, the supernatant was decanted and the samples were stained with Brilliant Violet 421 mouse anti-human CD34 (BD Biosciences [Cat. #562577]; 5 µL/sample) and APC mouse anti-human CD38 (BD Biosciences [Cat. #555462]; 20 µL/sample) antibodies in FACS buffer (1X PBS, 2% FBS, 0.1% sodium azide; 100 µL total volume/sample) for 10 minutes at 4 °C. The cells were then diluted with FACS buffer (1 mL) and centrifuged. The supernatant was decanted and stained with Annexin V-FITC (BD Biosciences [Cat. #556420]; 5 µL/sample) and 7-AAD (eBioscience [Cat. #00-6993-50]; 5 µL/sample) in FACS buffer (100 µL total volume/sample) for 10 minutes at room temperature in the dark. The samples were diluted with FACS buffer (300 µL), and kept on ice during analysis by flow cytometry using a BD Biosciences LSR II flow cytometer. Greater than 5 × 104 events were measured for each sample during analysis. All antibodies and stains were stored at 4 °C in the dark. After data collection, each sample was processed using FlowJo (Tree Star; v 7.6.5). The cell viability is expressed as a mean of 3–5 biological replicates ± S.D. Statistical significance was determined using unpaired t-tests (GraphPad Prism v. 5.0). An example of the data processing is shown in the Supporting Information.
4.8 Colony growth assay
TEX cells were added to Methocult H4230 (STEMCELL Technologies Inc.) supplemented with penicillin (100 I.U./mL, ATCC) and streptomycin (100 µg/mL, ATCC) at a final cell density of 1.2 × 104 cells/mL. Compounds were diluted in TEX cell media and dosed to each cell suspension to obtain the respective concentration. Each sample was vortexed vigorously to evenly distribute the cells before and after compound dosing. Each sample (1.5 mL) was plated into three wells (0.5 mL/well, three technical replicates) of a 24-well plate (Corning) and incubated under normoxic conditions at 37 °C, 5% CO2 for 11 days before scoring colonies. Colonies were counted for each well at 10X magnification with a light microscope by two people independently and averaged for each sample. The data is the mean number of colonies for three biological replicate ± S.D. Statistical significance was determined using unpaired t-tests (GraphPad Prism v. 5.0).
4.9 ROS Assay
TEX cells were seeded in 24-well plates at 5 × 105 cells/mL/well and incubated overnight at 37°C and 5% CO2. The cells were then treated with compounds (25 and 100 µM), including H2O2 (100 µM; positive control). Immediately after treatment with compounds, CellROX Green (Invitrogen) reagent was added to the appropriate samples at a final concentration of 5 µM. The cells were then incubated for 30 minutes at 37°C and 5% CO2. Following incubation, the samples were transferred to 5 mL FACS tubes and washed with FACS buffer (3 mL, 2X). The samples were run using a BD Biosciences LSR II flow cytometer and 5 × 104 events were recorded for each sample. Flow cytometry data was analyzed using FlowJo software (Tree Star; version 7.6.5). Samples were run in quadruplicate with the exception of H2O2 (triplicate data). Median fluorescence intensity (MFI) values were obtained for each sample and were normalized to the untreated control. Data are shown as mean MFI value ± S.D. Statistical significance was determined using unpaired t-tests (GraphPad Prism v. 5.0).
4.10 General synthesis information
Chemical reagents were typically purchased from Sigma-Aldrich and used without additional purification unless noted. Bulk solvents were from Fisher Scientific. PTL was purchased from Enzo Life Sciences and CTL was purchased from Santa Cruz Biotechnology. Previously reported analogues DMAPT, MelB, 1, 2 and 6 were synthesized as described.18, 21–26, 28, 29, 32–35 The structure of 1 was further confirmed by small molecule X-ray crystallography (SI; CCDC 1033012) and compared to the previous report.55 Tetrahydrofuran (THF) was rendered anhydrous by passing through the resin column of a solvent purification system (MBraun). Reactions were performed under an atmosphere of dry N2 unless noted. Silica gel chromatography was performed on a Teledyne-Isco Combiflash Rf-200 instrument utilizing Redisep Rf Gold High Performance silica gel columns (Teledyne-Isco). Analytical HPLC analysis was performed on an Agilent 1200 series instrument equipped with a diode array detector and a Zorbax SB-C18 column (4.6 × 150 mm, 3.5 µm, Agilent Technologies). The method started with 10% CH3CN (with 0.1% trifluoroacetic acid (TFA)) in H2O (0.1% TFA). The 10% CH3CN (with 0.1% TFA) was increased to 85% over 22 minutes, and then increased to 95% CH3CN (with 0.1% TFA) over 2 more minutes. Nuclear magnetic resonance (NMR) spectroscopy was employed by using either a Bruker Avance (400 MHz for 1H; 100 MHz for 13C) or Bruker Ascend (500 MHz for 1H; 125 MHz for 13C) NMR operating at ambient temperature. Chemical shifts are reported in parts per million and normalized to internal solvent peaks or tetramethylsilane. High-resolution masses were obtained from the University of Minnesota Department of Chemistry Mass Spectrometry lab, employing a Bruker BioTOF II instrument.
C1-C10 Reduced 3
To a stirred solution of PTL (0.050 g, 0.201 mmol) in MeOH (2 mL) was added dimethylamine (2.0 M in MeOH, 1 mL). The reaction was allowed to stir at RT overnight and then concentrated in vacuo. The crude material was used without further purification. The residual material was dissolved in EtOAc (3 mL) and PtO2 (0.005 g, 0.022 mmol) was added. The reaction mixture was degassed, then shaken for 8 hr in a Parr shaker under an atmosphere of H2 (50 psi). The mixture was then degassed, filtered through celite, and concentrated in vacuo. The crude material was taken on to the next step without further purification. The reaction mixture was dissolved in THF (3 mL) and iodomethane was added in excess (0.10 mL, 1.60 mmol). The reaction was allowed to stir at RT for 2 h. The solvent and excess iodomethane were removed in vacuo resulting in a white solid. Water (10 mL) was added and the reaction was heated to 45 °C. Complete solvation of the yellowish material resulted within minutes of heating. The reaction was allowed to stir with heating for 3 h, and then solvent was removed in vacuo. Aqueous NaHCO3 (sat’d, 5 mL) was added to the reaction mixture, and the product was extracted with DCM (3 × 20 mL). The combined organic layers were washed with brine (20 mL), and dried with Na2SO4. The reaction was purified by flash chromatography over SiO2 (10%–50% ethyl acetate in hexanes gradient) to yield 3 as a white solid (0.014 g, 32 %). 1H NMR (CDCl3, 500 MHz): δ 6.24 (d, J = 2.8 Hz 1H), 5.53 (d, J = 2.4 Hz, 1H), 3.84 (t, J = 7.6 Hz, 1H), 3.10 (d, J = 7.6 Hz, 1H), 2.99-2.94 (m, 1H), 2.20-2.14 (m, 2H), 1.81-1.75 (m, 2H), 1.75-1.56 (m, 2H), 1.51 (s, 3H), 1.51-1.40 (m, 2H), 1.26 (m, 2H), 1.17-1.14 (m, 2H), 0.93 (d, J = 4.8 Hz, 3H). 13C NMR (CDCl3, 125 MHz): 169.7, 139.5, 119.7, 81.0, 66.4, 61.3, 43.9, 36.7, 36.1, 30.1, 27.9, 24.7, 21.3, 20.6, 19.2. HRMS (ESI+) m/z calc’d for [C18H22O3+Na]+ 273.1467; found 273.1470. The structure of 3 was further confirmed by small molecule X-ray crystallography (SI; CCDC 1033013).
Cyclopropane 4
A 0.20 M solution of Zn(CH2I)2·DME complex was made in the following manner: To a stirred solution of diethyl zinc (1.0 M solution in hexanes, 4.0 mL, 4.00 mmol) in CH2Cl2 (20 mL) and DME (0.50 mL) at 0 °C was added diiodomethane (0.80 mL, 9.92 mmol) under N2. The mixture was stirred for 10 minutes. PTL (0.090 g, 0.36 mmol) in CH2Cl2 (3 mL) was added dropwise over 10 min to the (CH2I)2·DME complex at 0 °C. The reaction was allowed to warm to rt over 12 h. The reaction was quenched with aqueous NH4Cl (sat’d, 5 mL) and extracted with CH2Cl2 (3 × 20 mL). The combined organic layers were washed with aqueous NaHCO3 (sat’d, 20 mL), brine (20 mL), dried over Na2SO4 and concentrated in vacuo. The crude mixture was purified using silica gel chromatography (gradient 10–30% EtOAc in hexanes over 15 min) to yield 4 (0.036 g, 40%) as a colorless oil and recovered PTL (0.037 g, 41%). 1H NMR (400 MHz, CDCl3) δ: 6.28 (d, J = 3.7 Hz, 1H), 5.57 (d, J = 3.3 Hz, 1H), 3.96 (t, J = 9.1 Hz, 1H), 2.98 (d, J = 9.0 Hz, 1H), 2.67 (m, 1H), 2.39 (dd, J = 8.0 Hz, J = 14.7 Hz, 1H), 2.19 (dd, J = 2.3 Hz, J = 8.3 Hz, 1H), 1.95 (m, 2H), 1.70 (m, 1H), 1.40 (s, 3H), 1.28 (m, 2H), 1.09 (s, 3H), 0.85 (dd, J = 11.1 Hz, J = 14.7, 1H), 0.64 (td, J = 6.0 Hz, J = 9.5 Hz, 1H), 0.39 (dd, J = 4.3 Hz, J = 9.4 Hz, 1H), −0.08 (dd, J = 4.6 Hz, J = 5.6 Hz, 1H). 13C NMR (100 MHz, CDCl3) δ: 169.4, 139.9, 120.5, 82.7, 65.5, 60.6, 48.0, 42.3, 38.4, 25.7, 24.5, 22.3, 20.4, 18.8, 18.5, 17.1. HRMS (ESI+) m/z calc’d for [C16H22O3+Na]+ 285.1467; found 285.1470. The structure of 4 was further confirmed by small molecule X-ray crystallography (SI; CCDC 1033014).
Cyclopropyl-PTL Dimethylamine Fumarate 5
To a stirred solution of 4 (0.009 g, 0.034 mmol) in MeOH (2 mL) was added dimethylamine (2.0 M in MeOH, 1.5 mL). The reaction was stirred for 12 h at rt. The reaction mixture was concentrated in vacuo purified by silica gel chromatography (gradient 0% – 50% EtOAc in hexanes over 10 min, then gradient 0%–25% MeOH in CH2Cl2 over 10 min) to yield the dimethylamino product as a white solid (0.009 g). To a stirred solution of this product in THF (5 mL) was added fumaric acid (0.0034 g, 0.029 mmol). A white precipitate was observed after stirring overnight at rt. The reaction mixture was concentrated in vacuo to give the fumarate salt 5 as white solid (0.0124 g, 85%). 1H NMR (DMSO-d6, 400 MHz): δ 6.61 (s, 2H), 4.09 (t, J = 9.5 Hz, 1H), 3.04 (d, J = 9.2 Hz, 1H), 2.64 (m, 3H), 2.24 (s, 6H), 2.14 (m, 2H), 2.05 (m, 1H), 1.78 (m, 2H), 1.59 (m, 1H), 1.31 (s, 3H), 1.19 (m, 2H), 1.02 (s, 3H), 0.74 (m, 2H), 0.28 (dd, J = 3.8 Hz, J = 9.2 Hz, 1H), −0.18 (t, J = 4.8 Hz, 1H). 13C NMR (DMSO-d6, 100 MHz): 176.6, 166.1, 134.1, 81.8, 64.3, 60.3, 57.2, 47.4, 45.6, 45.3, 41.7, 38.0, 24.2, 24.0, 21.6, 20.1, 18.4, 18.3, 16.7. HRMS (ESI+) m/z calc’d for [C18H30NO3+H]+ 308.2226; found 308.2216.
4.11 Analysis of X-ray structures
The X-ray structures of 4 and PTL51 were analyzed for root-mean-square deviation (RMSD) and graphics were rendered using UCSF Chimera.81, 82 Thermal ellipsoids were drawn at the 50% probability level. RMSD was calculated for all non-hydrogen atoms.
Supplementary Material
Acknowledgments
We thank Ezra Menon and Margaret Olson (University of Minnesota, UMN) for assistance with anti-proliferative activity experiments and Victor G. Young, Jr. of the Department of Chemistry, X-Ray Crystallographic Laboratory (UMN) for solving the X-ray structures of 1, 3, and 4. We thank Professor Michael Verneris (UMN) for assistance with flow cytometry analysis and NIH P30 CA77598, which supports the Flow Cytometry shared resource of the Masonic Cancer Center (UMN). Professor John Dick (University of Toronto) is gratefully acknowledged for the gift of the TEX cell line. GBM6 cells were a gift from the late Professor John Ohlfest (UMN). This research was supported by grants from the UMN, Academic Health Center (Seed Grant 2010.01); UMN, Leukemia Research Fund – Danny Thompson Memorial Golf Tournament; Children’s Cancer Research Fund, Minneapolis, MN; American Cancer Society (IRG-58-001-52-IRG68); Hyundai Hope on Wheels, Hope Grant Award; The V Foundation for Cancer Research, V Scholar Award to D.A.H.; and startup funds to D.A.H. from the UMN. J.C.W. thanks the UMN, College of Pharmacy for a Bighley Graduate Fellowship. D.W. acknowledges the American Heart Association for a predoctoral fellowship (11PRE7240035).
Abbreviations
- PTL
parthenolide
- LSC
leukemic stem cell
- AML
acute myeloid leukemia
- SL
sesquiterpene lactone
- HSC
hematopoietic stem cell
- DMAPT
dimethylamino-parthenolide
- MelB
melampomagnolide B
- ROS
reactive oxygen species
- PtO2
platinum (IV) oxide
- DME
dimethoxyethane
- logD
distribution coefficient
- CSC
cancer stem cell
- CTL
costunolide
- BM
bone marrow
- DOX
doxorubicin
- AraC
cytarabine
- MEM
minimum essential medium
- FBS
fetal bovine serum
- DMEM
Dulbecco’s modification of Eagle’s medium
- HEPES
(4-2-hydroxyethyl)-1-piperazineethanesulfonic acid)
- IMDM
Iscove’s modified Dulbecco’s medium
- MTS
3-(4,5-dimethylthiaol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium
- SFEM
serum-free expansion medium
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Supplementary data
NMR spectra, HPLC characterization data, and additional details of biochemical assays and X-ray crystallographic structures are available. CCDC (1033012-1033014) contains the supplementary crystallographic data for compounds 1, 3, and 4 in this paper. These data can be obtained free of charge from The Cambridge Crystallographic Data Centre via www.ccdc.cam.ac.uk/data_request/cif.
References and notes
- 1.Merfort I. Curr. Drug Targets. 2011;12:1560. doi: 10.2174/138945011798109437. [DOI] [PubMed] [Google Scholar]
- 2.Ghantous A, Gali-Muhtasiv H, Vuorela H, Saliba NA, Darwiche N. Drug Discovery Today. 2010;15:668. doi: 10.1016/j.drudis.2010.06.002. [DOI] [PubMed] [Google Scholar]
- 3.Knight DW. Nat. Prod. Rep. 1995;12:271. doi: 10.1039/np9951200271. [DOI] [PubMed] [Google Scholar]
- 4.Ghantous A, Sinjab A, Herceg Z, Darwiche N. Drug Discovery Today. 2013;18:894. doi: 10.1016/j.drudis.2013.05.005. [DOI] [PubMed] [Google Scholar]
- 5.Kreuger MR, Grootjans S, Biavatti MW, Vandenabeele P, D'Herde K. Anti-Cancer Drugs. 2012;23:883. doi: 10.1097/CAD.0b013e328356cad9. [DOI] [PubMed] [Google Scholar]
- 6.Guzman ML, Rossi RM, Karnischky L, Li XJ, Peterson DR, Howard DS, Jordan CT. Blood. 2005;105:4163. doi: 10.1182/blood-2004-10-4135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lapidot T, Sirard C, Vormoor J, Murdoch B, Hoang T, Caceres-Cortes J, Minden M, Paterson B, Caligiuri MA, Dick JE. Nature. 1994;367:645. doi: 10.1038/367645a0. [DOI] [PubMed] [Google Scholar]
- 8.Hope KJ, Jin L, Dick JE. Nat. Immunol. 2004;5:738. doi: 10.1038/ni1080. [DOI] [PubMed] [Google Scholar]
- 9.Crews LA, Jamieson CH. Cancer Lett. 2013;338:15. doi: 10.1016/j.canlet.2012.08.006. [DOI] [PubMed] [Google Scholar]
- 10.Jordan CT. Sci. Transl. Med. 2010;2:31ps21. doi: 10.1126/scitranslmed.3000914. [DOI] [PubMed] [Google Scholar]
- 11.Dick JE. Blood. 2008;112:4793. doi: 10.1182/blood-2008-08-077941. [DOI] [PubMed] [Google Scholar]
- 12.Horton SJ, Huntly BJ. Haematologica. 2012;97:966. doi: 10.3324/haematol.2011.054734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guzman ML, Allan JN. Stem Cells. 2014;32:844. doi: 10.1002/stem.1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Foo K, Usui I, Gotz DCG, Werner EW, Holte D, Baran PS. Angew. Chem. Int. Ed. Engl. 2012;51:11491. doi: 10.1002/anie.201206904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Curry III EA, Murry DJ, Yoder C, Fife K, Armstrong V, Nakshatri H, O'Connell M, Sweeney CJ. Invest. New Drugs. 2004;22:299. doi: 10.1023/B:DRUG.0000026256.38560.be. [DOI] [PubMed] [Google Scholar]
- 16.Sweeney CJ, Mehrotra S, Sandaria MR, Kumar S, Shortle NH, Roman Y, Sheridan C, Campbell RA, Murry DJ, Badve S, Nakshatri H. Mol. Cancer Ther. 2005;4:1004. doi: 10.1158/1535-7163.MCT-05-0030. [DOI] [PubMed] [Google Scholar]
- 17.Guzman ML, Rossi R, Neelakantan S, Li XJ, Corbett CA, Hassane DC, Becker MW, Bennett JM, Sullivan E, Lachowicz JL, Vaughan A, Sweeney CJ, Matthews W, Carroll M, Liesveld JL, Crooks PA, Jordan CT. Blood. 2007;110:4427. doi: 10.1182/blood-2007-05-090621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Neelakantan S, Nasim S, Guzman ML, Jordan CT, Crooks PA. Bioorg. Med. Chem. Lett. 2009;19:4346. doi: 10.1016/j.bmcl.2009.05.092. [DOI] [PubMed] [Google Scholar]
- 19.Hejchman E, Haugwitz RD, Cushman M. J. Med. Chem. 1995;38:3407. doi: 10.1021/jm00017a025. [DOI] [PubMed] [Google Scholar]
- 20.Woods JR, Mo H, Bieberich AA, Alavanja T, Colby DA. MedChemComm. 2013;4:27. [Google Scholar]
- 21.Zhang Q, Lu Y, Ding Y, Zhai J, Ji Q, Ma W, Yang M, Fan H, Long J, Tong Z, Shi Y, Jia Y, Han B, Zhang W, Qiu C, Ma X, Li Q, Shi Q, Zhang H, Li D, Zhang J, Lin J, Li L-Y, Gao Y, Chen Y. J. Med. Chem. 2012;55:8757. doi: 10.1021/jm301064b. [DOI] [PubMed] [Google Scholar]
- 22.Nasim S, Crooks PA. Bioorg. Med. Chem. Lett. 2008;18:3870. doi: 10.1016/j.bmcl.2008.06.050. [DOI] [PubMed] [Google Scholar]
- 23.Hwang D-R, Wu Y-S, Chang C-W, Lien T-W, Chen W-C, Tan U-K, Hsu JTA, Hsieh H-P. Bioorg. Med. Chem. 2006;14:83. doi: 10.1016/j.bmc.2005.07.055. [DOI] [PubMed] [Google Scholar]
- 24.Matsuda H, Toguchida K, Ninomiya K, Kageura T, Morikawa T, Yoshikawa M. Bioorg. Med. Chem. 2003;11:709. doi: 10.1016/s0968-0896(02)00471-6. [DOI] [PubMed] [Google Scholar]
- 25.Srivastava SK, Abraham A, Bhat B, Jaggi M, Singh AT, Sanna VK, Singh G, Agarwal SK, Mukherjee R, Burman AC. Bioorg. Med. Chem. Lett. 2006;16:4195. doi: 10.1016/j.bmcl.2006.05.083. [DOI] [PubMed] [Google Scholar]
- 26.Castaneda-Acosta J, Fischer NH, Vergas D. J. Nat. Prod. 1993;56:90. doi: 10.1021/np50091a013. [DOI] [PubMed] [Google Scholar]
- 27.An Y, Guo W, Li L, Xu C, Yang D, Wang S, Lu Y, Zhang Q, Zhai J, Fan H, Qiu C, Qi J, Chen Y, Yuan S. PLoS One. 2015;10:e0116202. doi: 10.1371/journal.pone.0116202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Neukirch H, Guerriero A, D'Ambrosio M. Eur. J. Org. Chem. 2003:3696. [Google Scholar]
- 29.Neukirch H, Kaneider NC, Wiedermann CJ, Guerriero A, D'Ambrosio M. Bioorg. Med. Chem. 2003;11:1503. doi: 10.1016/s0968-0896(02)00553-9. [DOI] [PubMed] [Google Scholar]
- 30.Long J, Zhang S-F, Wang P-P, Zhang X-M, Yang Z-J, Zhang Q, Chen Y. J. Med. Chem. 2014;57:7098. doi: 10.1021/jm5009456. [DOI] [PubMed] [Google Scholar]
- 31.Dell'Agli M, Galli GV, Bosisio E, D'Ambrosio M. Bioorg. Med. Chem. Lett. 2009;19:1858. doi: 10.1016/j.bmcl.2009.02.080. [DOI] [PubMed] [Google Scholar]
- 32.Kwok BH, Koh B, Ndubuisi MI, Elofsson M, Crews CM. Chem. Biol. 2001;8:759. doi: 10.1016/s1074-5521(01)00049-7. [DOI] [PubMed] [Google Scholar]
- 33.Macias FA, Velasco RF, Alvarez JA, Castellano D, Galindo JCG. Tetrahedron. 2004;60:8477. [Google Scholar]
- 34.Nasim S, Pei S, Hagen FK, Jordan CT, Crooks PA. Bioorg. Med. Chem. 2011;19:1515. doi: 10.1016/j.bmc.2010.12.045. [DOI] [PubMed] [Google Scholar]
- 35.El-Feraly FS. Phytochemistry. 1984;23:2372. [Google Scholar]
- 36.Janganati V, Penthala NR, Madadi NR, Chen Z, Crooks PA. Bioorg. Med. Chem. Lett. 2014;24:3499. doi: 10.1016/j.bmcl.2014.05.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kolev JN, O'Dwyer KM, Jordan CT, Fasan R. ACS Chem. Biol. 2014;9:164. doi: 10.1021/cb400626w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Raj L, Ide T, Gurkar AU, Foley M, Schenone M, Li X, Tolliday NJ, Golub TR, Carr SA, Shamji AF, Stern AM, Mandinova A, Schreiber SL, Lee SW. Nature. 2011;475:231. doi: 10.1038/nature10167. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 39.Schumacker PT. Cancer Cell. 2006;10:175. doi: 10.1016/j.ccr.2006.08.015. [DOI] [PubMed] [Google Scholar]
- 40.Skalska J, Brookes PS, Nadtochiy SM, Hilchey SP, Jordan CT, Guzman ML, Maggirwar SB, Briehl MM, Bernstein SH. PLoS One. 2009;4:e8115. doi: 10.1371/journal.pone.0008115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lagadinou ED, Sach A, Callahan K, Rossi RM, Neering SJ, Minhajuddin M, Ashton JM, Pei S, Grose V, O'Dwyer KM, Liesveld JL, Brookes PS, Becker MW, Jordan CT. Cell Stem Cell. 2013;12:329. doi: 10.1016/j.stem.2012.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Diehn M, Cho RW, Lobo NA, Kalisky T, Dorie MJ, Kulp AN, Qian D, Lam JS, Ailles LE, Wong M, Joshua B, Kaplan MJ, Wapnir I, Dirbas FM, Somlo G, Garberoglio C, Paz B, Shen J, Lau SK, Quake SR, Brown JM, Weissman IL, Clarke MF. Nature. 2009;458:780. doi: 10.1038/nature07733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shanmugam R, Kusumanchi P, Cheng L, Crooks PA, Neelakantan S, Matthews W, Nakshatri H, Sweeney CJ. Prostate. 2010;70:1074. doi: 10.1002/pros.21141. [DOI] [PubMed] [Google Scholar]
- 44.Trachootham D, Alexandre J, Huang P. Nat. Rev. Drug Discov. 2009;8:579. doi: 10.1038/nrd2803. [DOI] [PubMed] [Google Scholar]
- 45.Adams DJ, Boskovic ZV, Theriault JR, Wang AJ, Stern AM, Wagner BK, Shamji AF, Schreiber SL. ACS Chem. Biol. 2013;8:923. doi: 10.1021/cb300653v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pei S, Minhajuddin M, Callahan KP, Balys M, Ashton JM, Neering SJ, Lagadinou ED, Corbett C, Ye H, Liesveld JL, O'Dwyer KM, Li Z, Shi L, Greninger P, Settleman J, Benes C, Hagen FK, Munger J, Crooks PA, Becker MW, Jordan CT. J. Biol. Chem. 2013;288:33542. doi: 10.1074/jbc.M113.511170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wen J, You KR, Lee SY, Song CH, Kim DG. J. Biol. Chem. 2002;277:38954. doi: 10.1074/jbc.M203842200. [DOI] [PubMed] [Google Scholar]
- 48.Jain TC, Banks CM, McCloskey JE. Tetrahedron. 1976;32:765. [Google Scholar]
- 49.Lee KH, Furukawa H. J. Med. Chem. 1972;15:609. doi: 10.1021/jm00276a010. [DOI] [PubMed] [Google Scholar]
- 50.de Meijere A. Angew. Chem. Int. Ed. Engl. 1979;18:809. [Google Scholar]
- 51.Long J, Ding Y-H, Wang P-P, Zhang Q, Chen Y. J. Org. Chem. 2013;78:10512. doi: 10.1021/jo401606q. [DOI] [PubMed] [Google Scholar]
- 52.Furukawa J, Kawabata N, Nishimura J. Tetrahedron Lett. 1968;31:3495. [Google Scholar]
- 53.Simmons HE, Smith RD. J. Am. Chem. Soc. 1958;80:5323. [Google Scholar]
- 54.Simmons HE, Smith RD. J. Am. Chem. Soc. 1959;81:4256. [Google Scholar]
- 55.Acosta JC, Fronczek FR, Fischer NH. Acta Crystallogr., Sect. C: Cryst. Struct. Commun. 1991;47:2702. [Google Scholar]
- 56.Batist G, Tulpule A, Sinha BK, Katki AG, Myers CE, Cowan KH. J. Biol. Chem. 1986;261:15544. [PubMed] [Google Scholar]
- 57.Ke W, Yu P, Wang J, Wang R, Guo C, Zhou L, Li C, Li K. Med. Oncol. 2011;28(Suppl 1):S135. doi: 10.1007/s12032-010-9747-1. [DOI] [PubMed] [Google Scholar]
- 58.Wen B, Hexum JK, Widen JC, Harki DA, Brummond KM. Org. Lett. 2013;15:2644. doi: 10.1021/ol400904y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hirschmann-Jax C, Foster AE, Wulf GG, Nuchtern JG, Jax TW, Gobel U, Goodell MA, Brenner MK. Proc. Natl. Acad. Sci. U. S. A. 2004;101:14228. doi: 10.1073/pnas.0400067101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Holohan C, Van Schaeybroeck S, Longley DB, Johnston PG. Nat. Rev. Cancer. 2013;13:714. doi: 10.1038/nrc3599. [DOI] [PubMed] [Google Scholar]
- 61.Sotiropoulou PA, Christodoulou MS, Silvani A, Herold-Mende C, Passarella D. Drug Discovery Today. 2014 doi: 10.1016/j.drudis.2014.05.002. [DOI] [PubMed] [Google Scholar]
- 62.Swaminathan SK, Olin MR, Forster CL, Cruz KS, Panyam J, Ohlfest JR. J. Immunol. Methods. 2010;361:110. doi: 10.1016/j.jim.2010.07.007. [DOI] [PubMed] [Google Scholar]
- 63.Andrews TE, Wang D, Harki DA. Drug. Deliv. Transl. Res. 2013;3:121. doi: 10.1007/s13346-012-0075-1. [DOI] [PubMed] [Google Scholar]
- 64.Hexum JK, Becker CM, Kempema AM, Ohlfest JR, Largaespada DA, Harki DA. Bioorg. Med. Chem. Lett. 2015 doi: 10.1016/j.bmcl.2015.04.058. http://dx.doi.org/10.1016/j.bmcl.2015.04.058. [DOI] [PubMed] [Google Scholar]
- 65.Kitson RRA. Angew. Chem. Int. Ed. Engl. 2009;48:9426. doi: 10.1002/anie.200903108. [DOI] [PubMed] [Google Scholar]
- 66.Bhatia M, Wang JC, Kapp U, Bonnet D, Dick JE. Proc. Natl. Acad. Sci. U. S. A. 1997;94:5320. doi: 10.1073/pnas.94.10.5320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Larochelle A, Vormoor J, Hanenberg H, Wang JC, Bhatia M, Lapidot T, Moritz T, Murdoch B, Xiao XL, Kato I, Williams DA, Dick JE. Nat. Med. 1996;2:1329. doi: 10.1038/nm1296-1329. [DOI] [PubMed] [Google Scholar]
- 68.Sundman-Engberg B, Tidefelt U, Paul C. Cancer Chemother. Pharmacol. 1998;42:17. doi: 10.1007/s002800050779. [DOI] [PubMed] [Google Scholar]
- 69.Bedigian HG, Taylor BA, Meier H. J. Virol. 1981;39:632. doi: 10.1128/jvi.39.2.632-640.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Largaespada DA, Shaughnessy JD, Jenkins NA, Copeland NG. J. Virol. 1995;69:5095. doi: 10.1128/jvi.69.8.5095-5102.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yin B, Kogan SC, Dickins RA, Lowe SW, Largaespada DA. Exp. Hematol. 2006;34:631. doi: 10.1016/j.exphem.2006.01.015. [DOI] [PubMed] [Google Scholar]
- 72.Rathe SK, Moriarity BS, Stoltenberg CB, Kurata M, Aumann NK, Rahrmann EP, Bailey NJ, Melrose EG, Beckmann DA, Liska CR, Largaespada DA. Sci. Rep. 2014;4:6048. doi: 10.1038/srep06048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.McDermott SP, Eppert K, Notta F, Isaac M, Datti A, Al-awar R, Wrana J, Minden M, Dick JE. Blood. 2012;119:1200. doi: 10.1182/blood-2011-01-330019. [DOI] [PubMed] [Google Scholar]
- 74.Warner JK, Wang JCY, Takenaka K, Doulatov S, McKenzie JL, Harrington L, Dick JE. Leukemia. 2005;19:1794. doi: 10.1038/sj.leu.2403917. [DOI] [PubMed] [Google Scholar]
- 75.Mathema VB, Koh YS, Thakuri BC, Sillanpaa M. Inflammation. 2012;35:560. doi: 10.1007/s10753-011-9346-0. [DOI] [PubMed] [Google Scholar]
- 76.Pajak B, Gajkowska B, Orzechowski A. Folia Histochem. Cytobiol. 2008;46:129. doi: 10.2478/v10042-008-0019-2. [DOI] [PubMed] [Google Scholar]
- 77.Tartier L, McCarey YL, Biaglow JE, Kochevar IE, Held KD. Cell Death Differ. 2000;7:1002. doi: 10.1038/sj.cdd.4400726. [DOI] [PubMed] [Google Scholar]
- 78.Singh J, Petter RC, Baillie TA, Whitty A. Nat. Rev. Drug Discov. 2011;10:307. doi: 10.1038/nrd3410. [DOI] [PubMed] [Google Scholar]
- 79.Potashman MH, Duggan ME. J. Med. Chem. 2009;52:1231. doi: 10.1021/jm8008597. [DOI] [PubMed] [Google Scholar]
- 80.Hexum JK, Tello-Aburto R, Struntz NB, Harned AM, Harki DA. ACS Med. Chem. Lett. 2012;3:459. doi: 10.1021/ml300034a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Meng EC, Pettersen EF, Couch GS, Huang CC, Ferrin TE. BMC Bioinform. 2006;7:339. doi: 10.1186/1471-2105-7-339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. J. Comput. Chem. 2004;25:1605. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.