Abstract
Background. West Nile virus (WNV) infection in humans can result in severe, acute encephalitis typically involving subcortical gray matter brain regions. West Nile virus replication within specific human brain regions from a human case of acute encephalitis has not been studied.
Methods. We describe a fatal case of WNV encephalitis in which we obtained tissue from specific brain regions at autopsy to evaluate viral-host interactions using next-generation sequencing and immunohistochemistry analysis.
Results. We found that WNV populations in the injured subcortical brain regions exhibited increased amino acid variation and increased expression of specific interferon genes compared with cortical tissues despite similar viral burden.
Conclusions. These observational, patient-based data suggest that neuronal injury and the strength of viral selection pressure may be associated with the level of the innate immune responses. Further studies in human and animal models evaluating the role of innate immune responses on injury patterns and viral selection pressure are needed.
Keywords: apoptosis, encephalitis, interferon-stimulated genes, next-generation sequencing, West Nile virus
West Nile virus (WNV) is a mosquito-borne infection that can result in infection of the central nervous system (CNS) in humans. Post mortem studies from patients with WNV encephalitis reveal characteristic neuronal loss and glial nodules in the gray matter of the thalamus, medulla, pons, midbrain, basal ganglia, and anterior horn of the spinal cord [1]. However, cortical neurons do not display pathologic injury [1]. In mice, regional neuronal susceptibility is due in part to type I interferon-dependent restriction of WNV infection through the induction of interferon-stimulated genes (ISGs) [2, 3]. In human cases, the underlying distribution of ISG expression, WNV quantitative loads, viral genetic diversity, and the degree to which they associate with CNS injury are not known. Accordingly, we examined these factors in distinct brain regions of a patient with WNV neuroinvasive disease (WNND). Our data suggest that thalamic regions displaying neuropathologic and neuroimaging evidence of injury also exhibit high ISG expression and viral loads associated with increased WNV amino acid diversity. In comparison, cortical neuronal regions exhibit unexpectedly high viral loads but minimal injury, decreased ISG expression, and lower viral amino acid diversity.
METHODS
Central nervous system tissue was placed in RNAlater (Life Technologies) or 10% formalin for downstream analysis, and total RNA was isolated using the High Pure Viral RNA kit (Roche). Fixed brain tissue samples were prepared for immunohistochemistry (IHC) analysis with an additional step of 3 subsequent 7-minute treatments of 0.1% sodium borohydride in phosphate-buffered saline to remove background autofluorescence. Tissues were labeled with rabbit monoclonal antibody against cleaved-caspase 3 ([CC3] Cell Signaling) and mouse monoclonal antibody to WNV envelope (ATCC, clone E18, VR-1611) at a dilution of 1:100 overnight at 4°C. TRITC-conjugated goat anti-immunoglobulin (Ig)G (Jackson ImmunoResearch) was used for secondary staining. Coverslips were mounted with ProLong Gold antifade reagent (Life Technologies), and images were obtained using an Olympus VS120 Virtual Slide system and analyzed using Olympus VS-Desktop software. Secondary-only labeling controls were used to calibrate exposures for each tissue type. All research was reviewed and approved by the University of Colorado Institutional Biosafety Committee and Colorado Multiple Institutional Review Board.
West Nile virus RNA copies per milligram of tissue were determined from total RNA isolated from each indicated CNS region using previously described primers [4]. Ribosomal depleted (RiboMinus, Waltham, MA) total RNA from each brain region was prepared for next-generation sequencing (NGS) using the Ovation RNA-Seq System V2 and Ultralow Library kits (nuGEN, San Carlos, CA), as previously described with sequencing on the NextSeq 500 platform (Illumina; sequenced at Colorado State University IDRC Genomics Core) [5]. The 150 nucleotide paired-end reads were demultiplexed using BaseSpace (Illumina). To determine ISG expression, reads were aligned to human Stat1(GenBank accession no. NM_007315), Rsad2(GenBank accession no. NM_080657), Ifi27(GenBank accession no. NM_001288957), Irg1(GenBank accession no. NM_001258406), Irf1(GenBank accession no. NM_002198), Oas1(GenBank accession no. NM_016816), and Ifit1(GenBank accession no. NM_001548) using MOSAIK [6].
To obtain the consensus WNV sequence from this patient and to define viral genetic diversity, WNV reads from the frontal cortex were assembled using Trinity to create a reference sequence (KT020853) for guided assembly from each region using MOSAIK. Geneious version 7.0.6. was used to compare the assembled WNV sequences to the following WNV strains: NY99(AF196835), 04-214CO(DQ431701), CO5-07(JF957170), BSL6-11(JQ700439), AVA1202600(KC736487), FtC-3699(KR868734), and BSL2-10(JF957185). Intratissue WNV minority nucleotide variants were analyzed using Vphaser2 [7, 8].
RESULTS
A 51-year-old female presented with 2 days of increasing altered mental status and decreasing responsiveness. Illness was preceded by 4 days of fever, nausea, and diarrhea. The patient's past medical history was remarkable for a history of rheumatoid arthritis treated with methotrexate and prednisone, and the last treatment was 1 year before presentation. The patient was evaluated in the emergency room, intubated for airway protection, and lumbar puncture was performed to obtain cerebral spinal fluid (CSF), which exhibited 163 white blood cells/mL with a differential of 99% lymphocytes. Magnetic resonance imaging (MRI) revealed increased signal in the insula, medial temporal lobe, medial left thalamus, and left cerebral peduncle (Figure 1A and B). The CSF was positive for WNV IgG (1.82, normal <1.29 intravenous [IV]) and IgM (8.74, normal <0.89 IV; Focus Diagnostics enzyme-linked immunosorbent assay), but CSF and serum WNV reverse transcription-polymerase chain reaction assays were negative for WNV RNA (ARUP Laboratories, Roche Molecular Systems Inc.). With these data, the patient was diagnosed with WNV encephalitis. During hospitalization in the intensive care unit, the patient was weaned from sedation, remained comatose, and had an electroencephalogram showing diffuse slowing. The patient did exhibit brain stem function but minimal peripheral responses with a physical exam consistent with loss of lower motor neuron function in all 4 extremities. An MRI of the spine exhibited no spinal cord lesions. Despite aggressive supportive care, the patient passed away after a cardiac arrest at day 11 of hospitalization. Autopsy was initiated 22 hours and 30 minutes after death and found no evidence of myocardial infarction or coronary artery disease. Brain tissue was collected at time of autopsy.
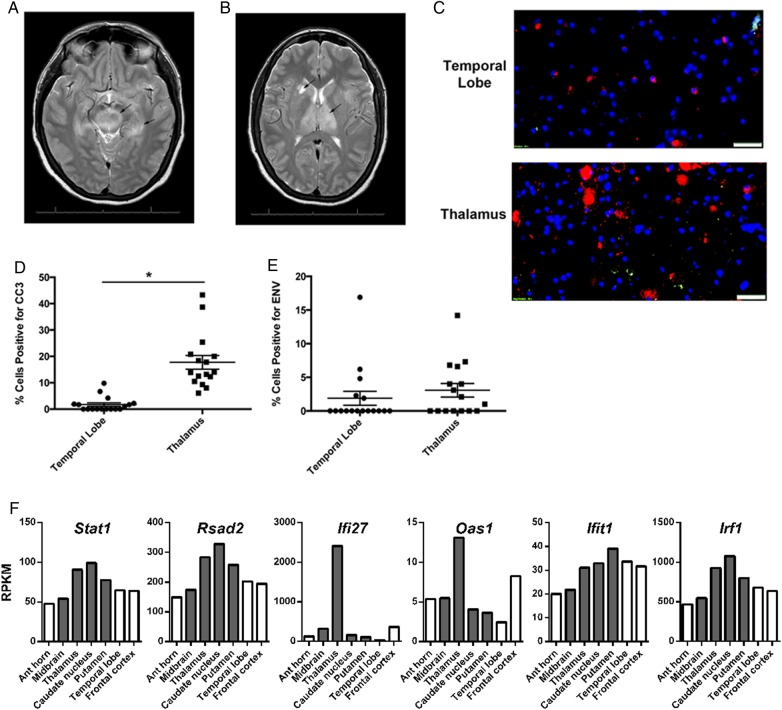
Figure 1.
Regional magnetic resonance imaging injury patterns associated with apoptosis and interferon-stimulated gene (ISG) expression. Magnetic resonance images (T2 sequences) showing increased signal intensity in the (A) midbrain substantia nigra and left mesial temporal lobe (arrows) and (B) the thalamus and right caudate nucleus (arrows). (C) Immunohistochemistry staining for cleaved-caspase 3 ([Cy3] red) and West Nile virus (WNV) envelope (ENV) antigen (TRITC, green) from indicated brain regions. Bar = 50 µm. Percentage of cells per high-power field positive for (D) CC3 and (E) WNV ENV antigen. *P < .0001, unpaired t test. (F) Reads per kilobase per million (RPKM) reads to interferon-stimulated genes from different brain regions were determined by next-generation sequencing. Gray bars indicate brain regions with neuronal injury.
The MRI evidence of injury in the thalamus correlated with a 10-fold increase (P < .0001) in expression of cells positive for a marker of apoptosis (CC3) when compared with an uninjured region in the temporal lobe (17.8 ± 2.6 vs 1.7 ± 0.67, mean positive cells per high-power field ± standard error of the mean; Figure 1C and D). The CC3 expression was not correlated with WNV envelope antigen expression (Figure 1E). Using NGS of whole tissue RNA (RNAseq), we determined the relative expression of ISGs between indicated regions of the CNS during acute WNV encephalitis (Figure 1F). We found that, in general, ISG expression was the highest in the subcortical tissues of the thalamus and basal ganglia (caudate nucleus and putamen) that also exhibit injury.
As detailed above, CSF was positive for WNV IgG and IgM but negative for WNV RNA. However, we used NGS of brain tissue from different brain regions to evaluate WNV population structure in the brain. The frontal cortex, thalamus, and anterior horn of the spinal cord exhibited in excess of 1 × 107 WNV RNA copies per milligram of tissue, whereas the midbrain, caudate nucleus, putamen, and temporal lobe all exhibited <1 × 106 WNV RNA copies per milligram of tissue (Figure 2A). From NGS, each tissue yielded 25–33 million sequencing reads, and 0.04% (frontal cortex) to 0.00004% (temporal lobe) of these aligned to WNV genetic sequences. The WNV population size (WNV RNA copies) directly correlated with the WNV sequencing coverage [5]. The frontal cortex, thalamus, and anterior horn of the spinal cord exhibited in excess of 1 × 107 WNV RNA copies per milligram of tissue, whereas the midbrain, caudate nucleus, putamen, and temporal lobe all exhibited <1 × 106 WNV RNA copies per milligram of tissue (Figure 2A).
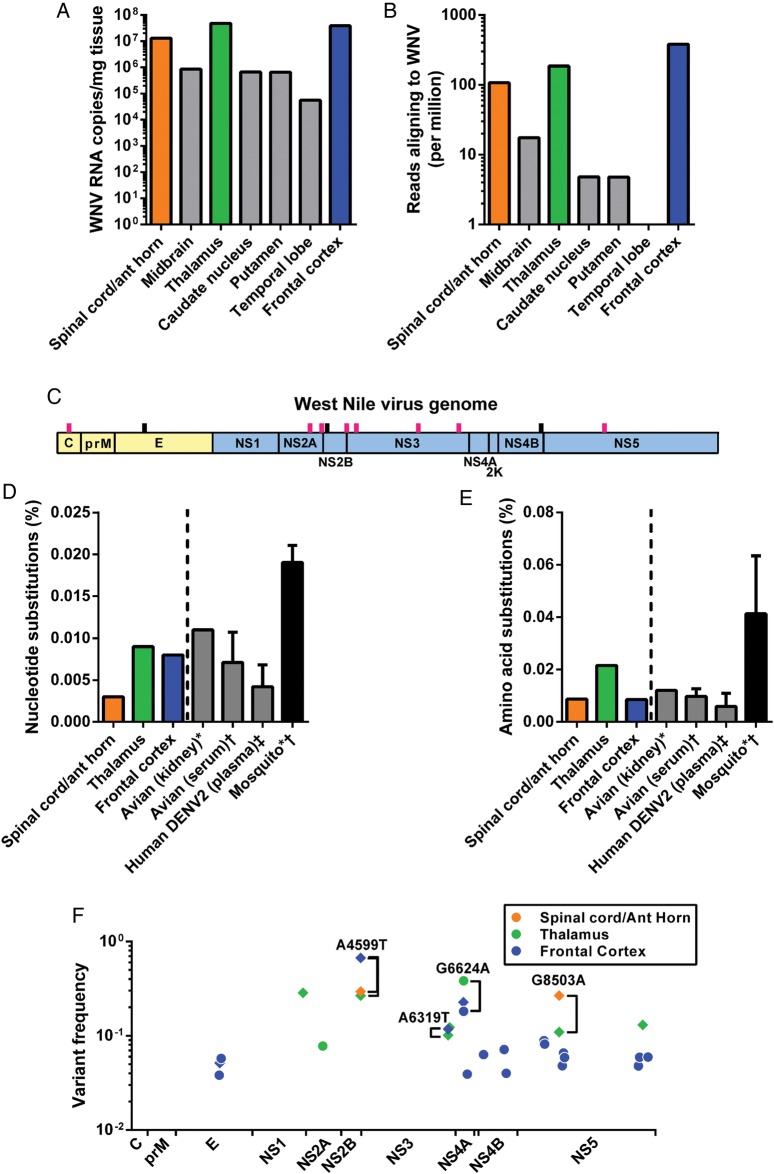
Figure 2.
Variations of West Nile virus (WNV) copies and population structure among brain regions. (A) WNV RNA was quantified from different brain tissues, prepared for next-generation sequencing, and (B) aligned to the WNV genome (displayed as number of aligning reads per million reads sequenced). The colored bars represent tissues with enough WNV coverage for subsequent population genetic analysis. (C) The consensus WNV sequences from each tissue were analyzed, and each line represents an amino acid change compared with the prototype strain NY99 (pink lines represent novel mutations; also see Table 1). The percentage of all sequenced WNV (D) nucleotides and (E) amino acids with substitutions were compared with published reports of WNV and dengue virus in other vertebrate samples and WNV in mosquitoes. (F) Individual intratissue WNV variants were plotted across the genome. Diamonds represent amino acid substitutions and circles represent silent mutations. *Data adapted from Grubaugh ND et al. Experimental evolution of an RNA virus in wild birds: evidence for host-dependent impacts on population structure and competitive fitness. PLoS Pathog. 2015;11:e1004874; †data adapted from Jerzak G et al. Genetic variation in West Nile virus from naturally infected mosquitoes and birds suggests quasispecies structure and strong purifying selection. J Gen Virol. 2005;86:2175; ‡data adapted from Romano et al. Inter- and intra-host viral diversity in a large seasonal DENV2 outbreak. PLoS One. 2013;8:e70318.
The WNV population size (WNV RNA copies) directly correlated with the WNV sequencing coverage, as previously described (Figure 2B) [5]. The consensus WNV genome sequence obtained from this patient belonged to the WN02 genotype; however, the consensus sequence isolate contained several novel amino acid substitutions found mostly within the viral nonstructural proteins (Figure 2C and Table 1).
Table 1.
WNV Amino Acid Substitutions Compared to the Prototype Strain NY99
| Strain (Source, Year, State) | Pairwise % Identity |
Nt Differences | WNV Genome Nt Position |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 268 | 1442 | 4025 | 4208 | 4294 | 4599 | 4749 | 5717 | 6350 | 7635 | 8621 | |||
| NY99 (flamingo, 1999, NY) | G | T | G | A | A | A | C | G | T | A | A | ||
| 04-214CO (human, 2004, CO) | 99.8 | 25 | ● | C | ● | ● | ● | ● | ● | ● | ● | ● | ● |
| CO5-07 (human, 2007, CO) | 99.6 | 41 | ● | C | ● | ● | ● | ● | ● | ● | ● | ● | G |
| BSL6-11 (human, 2011, MS) | 99.4 | 61 | ● | C | ● | ● | G | ● | T | ● | ● | G | ● |
| AVA1202600 (Culex, 2012, TX) | 99.4 | 64 | ● | C | ● | ● | ● | ● | ● | ● | ● | ● | ● |
| FtC-3699 (Culex, 2012, CO) | 99.4 | 64 | ● | C | ● | ● | ● | ● | ● | ● | ● | ● | ● |
| BSL2-10 (human, 2010, AZ) | 99.3 | 68 | ● | C | ● | ● | ● | ● | ● | ● | ● | G | G |
| Spinal cord/ant horn | 99.2 | 85 | A | C | A | ● | G | ●/T | A | ● | ● | G | T |
| Midbraina | A | C | A | ● | G | ●/T | – | T | ● | G | T | ||
| Thalamus | 99.2 | 85 | A | C | A | ● | G | ●/T | A | ● | ● | G | T |
| Caudatea | A | – | A | T | – | ●/T | A | ● | C | G | T | ||
| Putamena | A | – | A | ● | – | – | – | – | ●/C | – | – | ||
| Frontal cortex | 99.2 | 86 | A | C | A | ● | G | T/● | A | ● | ● | G | T |
| Polyprotein aa # | A58T | V449A | R1310K | N1371I | I1400V | Q1501H | F1551L | G1874C | H2985Q | I2513M | K2842M | ||
| Protein aa # | A58T | V159A | R167K | N228I | I26V | Q94H | F46L | G369C | H580Q | I240M | K314M | ||
| Protein | C | E | NS2A | NS2A | NS2B | NS2B | NS3 | NS3 | NS3 | NS4B | NS5 | ||
Abbreviations: aa, amino acid; AZ, Arizona; C, capsid; CO, Colorado; E, envelope; MS, Mississippi; NS, nonstructural; Nt, nucleotide; NY, New York; TX, Texas; WNV, West Nile virus; ●, ancestral nucleotide; -, missing data.
a Could not determine pairwise identity or the number of Nt differences due to incomplete coverage of the coding sequence.
The cortex, thalamus, and anterior horn regions were analyzed for WNV population diversity because these regions had complete coverage of the WNV coding sequence. The percentage of nucleotides and amino acids with substitutions found within these brain regions was comparable to that previously reported for WNV and dengue virus isolated from other vertebrate tissues, but less than from mosquitoes (Figures 2D and E) [5, 9, 10]. The thalamic and cortical tissues contained similar nucleotide diversity, but most of the WNV variants detected in the cortex were silent mutations, which do not result in an amino acid change (Figure 2F). The thalamus contained considerably more WNV amino acid diversity in comparison to the cortex. The intratissue WNV nucleotide variants tended to be concentrated in the nonstructural genome regions, except for 3 envelope variants found in the cortex (Figure 2F). Four nucleotides were maintained as minority variants between at least 2 different regions, but the majority of the nucleotide variants were found only in a single brain region (Figure 2F). We found additional evidence for region-specific differences in WNV populations by comparing the partial consensus sequences from all of the brain tissues obtained (Table 1).
DISCUSSION
West Nile virus is a vector-borne infection most often transmitted to humans by Culex mosquitoes that can result in neuroinvasive disease (WNND) [11]. Presenting clinical symptoms are representative of typical brain regions displaying MRI injury patterns associated with WNV encephalitis and include areas of increased T2 and FLAIR signal and low T1 signal that involve the basal ganglia, thalamus, and brain stem [12, 13].
For the first time, we isolated total RNA from different brain regions in a patient with acute WNV encephalitis and described intratissue viral population diversity using NGS technology. We discovered classic injury patterns, defined by neuroimaging and IHC, in the subcortical gray matter of the thalamus, basal ganglia, and midbrain. To our surprise, we found high viral loads in the frontal cortex despite showing little injury, whereas regions exhibiting injury exhibited highly variable viral loads. These data suggest that in human WNND, neuronal injury may be independent of viral replication and injury in the brain may be due to regional factors.
Post mortem studies have shown that injury patterns correlate with inflammatory infiltrates in the brain, which [1] suggests that inflammation and innate neuronal subtype susceptibility to injury are important determinants of disease. The ISGs Irf1, Irg1, Ifi27, and Rsad2 were shown to inhibit WNV replication in cortical neurons, and they were more highly expressed in the cerebellum compared with the cortex of mice [14]. In this patient, we also show that expression of Irf1, Ifi27, and Rsad2, in addition to Stat1, Oas1, and Ifit1, are regionally heterogeneous, with the high levels generally found in the subcortical regions of the brain. Thus, it may be that regional differences in ISG expression may contribute to the regional patterns of neuronal injury.
Our data on region-specific patterns of WNV genetic diversity further suggest that WNV encounters different selective pressures as it spreads throughout brain. As with injury, the regional differences in immune responses and susceptibility of neuronal subtypes may contribute to the strength of selection as represented by increased amino acid substitutions in the viral genomes. For example, WNV populations in the thalamus exhibited an increased frequency of amino acid substitutions when compared with the cortex, suggesting an association between increased ISG expression and selection in the CNS. Although consensus amino acid differences between the WNV populations of the cortex and the thalamus were minimal, an expanding body of evidence demonstrates that minority variants may directly and significantly influence WNV phenotype [5, 15, 16].
This study has several limitations. This work is from a single patient; therefore, the findings are hypothesis generating and need to be verified in experimental models. However, the confluence of human clinical data and timely acquisition and analysis of human brain tissue is difficult to complete for this sporadic infection. In this 1 host, we used NGS to show evidence of intraregional variation in viral populations and ISG expression. However, other hosts with different ISG patterns may exhibit different regional genetic variation. Thus, further studies of minority WNV variants and immune activation in the human CNS will be needed to identify mechanisms of viral selection that are associated with injury and disease.
CONCLUSIONS
We report the first comprehensive study of WNV variation within a human case of viral encephalitis and describe possible associations between viral nucleotide substitutions, neuroradiographic and pathologic injury patterns, and ISG expression in the human brain. These data should guide future investigations into the role of inflammatory responses in selective viral pressures and injury patterns in the brain following viral infections.
Acknowledgments
We thank Dr. Betty DeMasters and the University of Colorado Hospital Pathology Department for coordination of care and sample acquisition. We also thank Joseph R. Fauver, Erin Petrilli, and Phil Buxton from the Colorado State University for laboratory and sequencing support.
Financial support. This work was funded by grants from the Colorado Clinical and Translational Sciences Institute and Center for Neurosciences at the University of Colorado (to J. D. B.) and the National Institutes of Health (AI067380; to G. D. E.).
Potential conflicts of interest. All authors: No reported conflicts.
All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest.
References
- 1.Armah HB, Wang G, Omalu BI et al. . Systemic distribution of West Nile virus infection: postmortem immunohistochemical study of six cases. Brain Pathol 2007; 17:354–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Samuel MA, Diamond MS. Alpha/beta interferon protects against lethal West Nile virus infection by restricting cellular tropism and enhancing neuronal survival. J Virol 2005; 79:13350–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lazear HM, Pinto AK, Vogt MR et al. . Beta interferon controls West Nile virus infection and pathogenesis in mice. J Virol 2011; 85:7186–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lanciotti RS, Kerst AJ, Nasci RS et al. . Rapid detection of West Nile virus from human clinical specimens, field-collected mosquitoes, and avian samples by a TaqMan reverse transcriptase-PCR assay. J Clin Microbiol 2000; 38:4066–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Grubaugh ND, Smith DR, Brackney DE et al. . Experimental evolution of an RNA virus in wild birds: evidence for host-dependent impacts on population structure and competitive fitness. PLoS Pathog 2015; 11:e1004874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee WP, Stromberg MP, Ward A et al. . MOSAIK: a hash-based algorithm for accurate next-generation sequencing short-read mapping. PLoS One 2014; 9:e90581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Grabherr MG, Haas BJ, Yassour M et al. . Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol 2011; 29:644–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang X, Charlebois P, Macalalad A et al. . V-Phaser 2: variant inference for viral populations. BMC Genomics 2013; 14:674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jerzak G, Bernard KA, Kramer LD, Ebel GD. Genetic variation in West Nile virus from naturally infected mosquitoes and birds suggests quasispecies structure and strong purifying selection. J Gen Virol 2005; 86:2175–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Romano CM, Lauck M, Salvador FS et al. . Inter- and intra-host viral diversity in a large seasonal DENV2 outbreak. PLoS One 2013; 8:e70318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Beckham JD, Tyler KL. Neuro-intensive care of patients with acute CNS infections. Neurotherapeutics 2012; 9:124–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ali M, Safriel Y, Sohi J et al. . West Nile virus infection: MR imaging findings in the nervous system. AJNR Am J Neuroradiol 2005; 26:289–97. [PMC free article] [PubMed] [Google Scholar]
- 13.Petropoulou KA, Gordon SM, Prayson RA, Ruggierri PM. West Nile virus meningoencephalitis: MR imaging findings. AJNR Am J Neuroradiol 2005; 26:1986–95. [PMC free article] [PubMed] [Google Scholar]
- 14.Cho H, Proll SC, Szretter KJ et al. . Differential innate immune response programs in neuronal subtypes determine susceptibility to infection in the brain by positive-stranded RNA viruses. Nat Med 2013; 19:458–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ciota AT, Ngo KA, Lovelace AO et al. . Role of the mutant spectrum in adaptation and replication of West Nile virus. J Gen Virol 2007; 88:865–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jerzak GV, Bernard K, Kramer LD et al. . The West Nile virus mutant spectrum is host-dependant and a determinant of mortality in mice. Virology 2007; 360:469–76. [DOI] [PMC free article] [PubMed] [Google Scholar]