Abstract
Background
Studies conflict regarding the importance of the fluoroquinolone-resistant North American pulsed-field gel electrophoresis type 1 (NAP1) strain in Clostridium difficile infection (CDI) outcome. We describe strain types causing CDI and evaluate their association with patient outcomes.
Methods
CDI cases were identified from population-based surveillance. Multivariate regression models were used to evaluate the associations of strain type with severe disease (ileus, toxic megacolon, or pseudomembranous colitis within 5 days; or white blood cell count ≥15,000/mm3 within one day of positive test), severe outcome (intensive care unit admission after positive test, colectomy for C. difficile infection, or death within 30 days of positive test), and death within 14 days of positive test.
Results
Strain typing results were available for 2,057 cases. Severe disease occurred in 363 (17.7%) cases, severe outcome in 100 (4.9%), and death within 14 days in 56 (2.7%). The most common strain types were NAP1 (28.4%), NAP4 (10.2%) and NAP11 (9.1%). In unadjusted analysis, NAP1 was associated with greater odds of severe disease than other strains. After controlling for patient risk factors, healthcare exposure, and antibiotic use, NAP1 was associated with severe disease (adjusted odds ratio [aOR] 1.74, 95% confidence interval [CI], 1.36–2.22), severe outcome (aOR 1.66, 95% CI, 1.09–2.54), and death within 14 days (aOR 2.12, 95% CI, 1.22–3.68).
Conclusion
NAP1 was the most prevalent strain and a predictor of severe disease, severe outcome, and death. Strategies to reduce NAP1 prevalence, such as antibiotic stewardship to reduce fluoroquinolone use, might reduce CDI morbidity.
Keywords: Clostridium difficile, clinical outcomes, strain typing, epidemiology
INTRODUCTION
Increases in incidence and severity of Clostridium difficile infection (CDI) have been reported in the past decade and were initially attributed to the emergence of a “hypervirulent” strain, the North American pulsed field gel electrophoresis type 1 (NAP1) strain, also described as PCR ribotype 027 and restriction endonuclease analysis (REA) group BI [1–3]. This strain demonstrates increased toxin production in vitro and increased fluoroquinolone resistance compared to previously described strains.
Subsequent reports of the relationship between the NAP1 strain and patient outcomes have conflicted. Although some suggest that infection with the NAP1 strain is associated with more severe disease [4–6], others reported no association [7–11]. These studies used different outcome measures (clinical severity [10] as defined by clinical practice guidelines [12]; a composite of intensive care unit admission, colectomy, and death [5, 7, 9]; and 14 or 30-day mortality [4, 6, 8, 11]), involved small sample sizes, or have focused on cases of infection from a single institution or community.
We sought to clarify the role of C. difficile strain type by determining the relationship between strain type and disease outcomes using a geographically diverse dataset from the United States (US). We used outcome measures similar to those in other studies to facilitate comparisons with prior reports.
METHODS
CDI Surveillance and Study Population
Data were obtained from the Centers for Disease Control and Prevention (CDC)’s Emerging Infections Program (EIP) C. difficile surveillance, which has been described elsewhere [13, 14]. The EIP CDI surveillance system is an active population-based and laboratory-based surveillance system that began in 2009 in selected counties of 6 US states (California, Colorado, Connecticut, Georgia, Minnesota, New York), expanded to 2 additional US states in 2010 (Tennessee and Oregon), and in 2011 expanded to an additional 2 states (Maryland and New Mexico). At each EIP site, trained surveillance officers investigate all positive C. difficile toxin assay or molecular assay reports from clinical, reference, and commercial laboratories for residents of surveillance catchment areas. A CDI case is defined as a positive C. difficile stool specimen in a surveillance area resident aged one year or older who did not have a positive test in the previous 8 weeks.
Cases are classified as community-associated if a positive specimen was collected as an outpatient or within 3 days of an acute care admission, without documentation of an overnight stay in a healthcare facility during the 12 weeks prior to stool collection; otherwise, cases are classified as healthcare-associated. Healthcare-associated cases are further classified as healthcare facility-onset if they occurred during a long-term care facility/nursing home stay or > 3 calendar days after hospital admission; otherwise they are classified as community-onset healthcare facility-associated [15]. All CDI cases classified as either community-associated or community-onset healthcare-facility associated underwent a full medical record review to collect information on symptoms, co-infections, clinical comorbidities (Charlson index) [16], and outcomes, and a 10% sample of the healthcare facility-onset cases were fully reviewed.
A convenience sample of clinical laboratories in each catchment area (n=37 laboratories) submitted all stool specimens from CDI cases with full medical record review to three reference laboratories (Edwards Hines Jr. Veterans Affairs, New York State Department of Health, and Minnesota Department of Health Public Health Laboratory) for culture of C. difficile [17]. Recovered isolates were sent to CDC for molecular typing by pulsed-field gel electrophoresis (PFGE). PFGE patterns were analyzed using BioNumerics v.5.10 (Applied Maths, Austin, TX) and grouped into pulsed-field types using Dice/UPGMA clustering. An 80% similarity threshold was used to assign North American PFGE (NAP) types [18]. Isolates also underwent PCR to detect the presence of tcdA, tcdB and binary toxin (cdtA and ctdB) genes [19].
For this analysis, we limited the data to the CDI cases with stool specimens collected between January 1, 2009–December 31, 2011. Only cases with full medical record review and strain typing results available were included. Cases whose isolates were negative for both tcdA and tcdB (89 cases) were excluded. During 2009–2011, only 8 EIP sites collected stool specimens (CA, CO, CT, GA, MN, NY, OR, and TN). These 8 EIP sites represented a surveillance catchment area of 9,667,103 persons in 2011.
Outcomes of Interest
Three separate outcome measures were evaluated: severe CDI disease, severe CDI outcome, and death within 14 days of infection. The definition of severe disease, adapted from current clinical practice guidelines [12], was development of ileus, toxic megacolon, or pseudomembranous colitis within 5 days of the positive C. difficile stool specimen or serum white blood count ≥15,000 cells/mm3 within one calendar day of collection of the stool specimen. Severe outcome was defined as intensive care unit (ICU) admission within 7 days after stool collection, colectomy for CDI, or death within 30 days of stool collection, in accordance with a recent study from Walk et al [7]. Death within 14 days was also evaluated based on the recent study from Walker et al [6].
Statistical Analysis
Multiple imputation was used to impute missing race (12.6% of cases) based on the distribution of known race by age, sex and surveillance site. Analysis of imputed datasets was performed using PROC MIANALYZE (SAS Institute, Cary, NC) to account for the uncertainty associated with imputation. Baseline differences between groups were evaluated using chi-square or Fisher exact tests for categorical variables, as appropriate, and Wilcoxon rank sum tests for continuous variables. Because a linear relationship between increasing Charlson index and outcome variables was seen (up to a Charlson comorbidity index of 3), the Charlson index was treated as an ordinal variable with levels 0, 1, 2, and ≥3.
For the outcomes studied, initial analyses of the association between individual variables and the outcome of interest were first performed with a univariate logistic regression model. Then a separate multivariate logistic regression model was constructed for each of three outcome measures of interest using stepwise backwards selection. Variables with P ≤ 0.25 in univariate analysis were eligible for inclusion in the corresponding multivariate model. Possible confounding variables (i.e., change of ≥ 10% to the estimated odds ratio for NAP 1 strain) were added to respective multivariate models regardless of P values. Charlson index was also included in all models regardless of P value. To confirm the results found in multivariate models, analyses stratifying the data by age (patients ≤50 versus >50 years of age) and by epidemiologic classification (community-associated versus healthcare-associated) were performed. A sensitivity analysis was also performed excluding the EIP site contributing the largest number of NAP1 cases from models. A 2-tailed P value of < 0.05 was considered statistically significant. All analyses were performed with SAS 9.3 (SAS Institute, Cary, NC).
Human Subjects
CDC and local institutional review boards approved the study. A waiver of informed consent was granted because the study posed no greater than minimal risk to participants.
RESULTS
Description of Clostridium difficile Infection Cases
During 2009–2011, strain typing results were available for 2,057 of the 14,091 total CDI cases identified. Two EIP sites (New York and Minnesota) contributed >50% of cases with strain typing results (Table 1). Compared to CDI cases without strain typing results but with clinical data available (N=5,324), those with strain typing results were similar with respect to age (P=0.41) and the three clinical outcomes of interest: severe disease (P=0.05), severe outcome (P=0.90), and death within 14 days (P=0.18). Although differences in sex (P=0.04) and race (P<0.0001) between CDI cases with strain typing and those without strain typing results reached statistical significance, the relative differences were <6% between groups.
Table 1.
Clostridium difficile Infection (CDI) Cases with Strain Typing Results by Emerging Infections Program (EIP) site, 2009–2011
| CDI Cases, All Strains (n=2,057) |
NAP1 Strain (n=585) | |||
|---|---|---|---|---|
|
|
|
|||
| EIP Site | No. of Cases | % of Total | No. of Cases | % of Total |
| California | 114 | 5.5 | 29 | 5.0 |
| Colorado | 346 | 16.8 | 103 | 17.6 |
| Connecticut | 184 | 9.0 | 82 | 14.0 |
| Georgia | 105 | 5.1 | 32 | 5.5 |
| Minnesota | 469 | 22.8 | 56 | 9.6 |
| New York | 689 | 33.5 | 242 | 41.4 |
| Oregon | 39 | 1.9 | 4 | 0.7 |
| Tennessee | 111 | 5.4 | 37 | 6.3 |
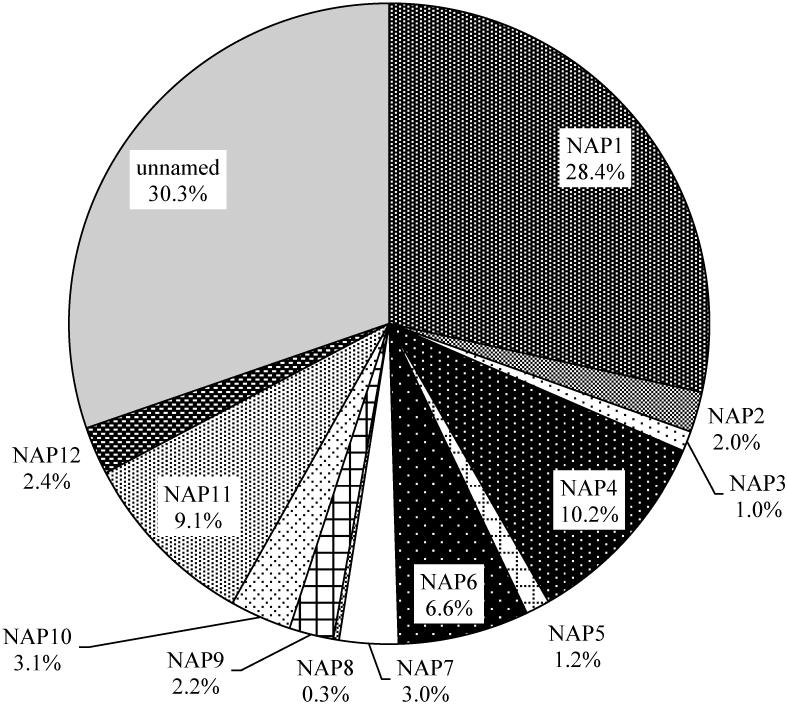
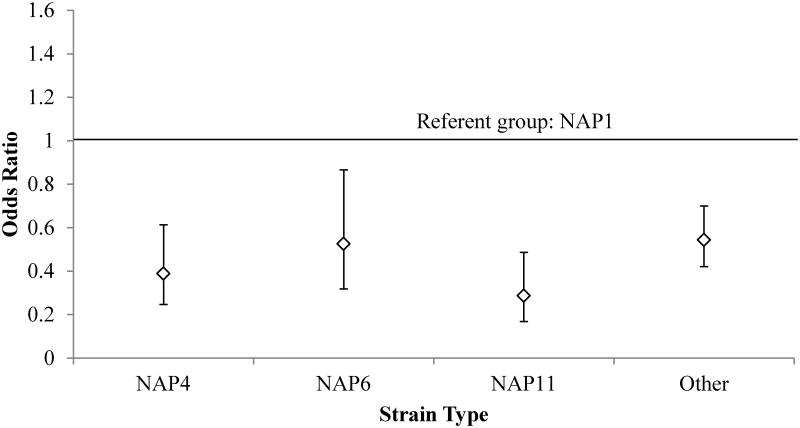
Among the 2,057 CDI cases included in the analysis, the majority (52.8%) were community-associated by design. Overall, the most common strain types were: NAP1 (28.4% of cases), NAP4 (10.2%), NAP11 (9.1%), and NAP6 (6.6%) (Figure 1). Of the 585 NAP1 cases, 17 (2.9%) were negative for binary toxin. Compared to these three most common NAP types, and compared to all others, the NAP1 type was associated with a greater odds of severe disease in unadjusted analysis. (Figure 2). Therefore, remaining analyses compare the NAP1 strain to all other strain types (i.e., non-NAP1 strains).
Figure 1.
Distribution of North American Pulsed-Field Gel Electrophoresis (PFGE) types among Clostridium difficile cases with strain typing results (n=2,057). Note that the “unnamed” strain type consists of many (>200) unrelated PFGE patterns.
Figure 2.
Univariate odds ratios for severe disease by strain type. NAP1 is the referent group, as indicated by the horizontal line at the odds ratio of 1.0. P values for all odds ratios depicted are < 0.05.
NAP1 strain was associated with older age (P<0.0001), healthcare-associated epidemiologic classification (P<0.0001), emergency room visits (P=0.003) in the prior 12 weeks, Charlson index (P<0.0001), and prior receipt of antibiotics (P<0.0001) in univariate analysis (Table 2). Inflammatory bowel disease (P=0.0003) and prior immunosuppressive treatment (P=0.04) were associated with non-NAP1 strains.
Table 2.
Univariate Comparison of Baseline Characteristics of Clostridium difficile Infection Due to the NAP1 Strain Compared to Non-NAP1 Strains
| Characteristic | NAP1 Strain, no. (%) (n=585) |
Non-NAP1 Strain, no. (%) (n=1,472) |
P Value |
|---|---|---|---|
| Demographic variables | |||
| Age > 65 years | 305 (52.1) | 586 (39.8) | <0.0001 |
| Female sex | 346 (59.2) | 909 (61.8) | 0.27 |
| White race | 505 (86.4) | 1262 (85.8) | 0.74 |
| Epidemiologic classification | <0.0001 | ||
| Healthcare facility onset | 111 (19.0) | 211 (14.3) | |
| Community onset, healthcare facility-associated | 232 (39.7) | 416 (28.3) | |
| Community-associated | 242 (41.4) | 845 (57.4) | |
| Healthcare exposures | |||
| Chronic hemodialysis | 39 (6.7) | 68 (4.6) | 0.06 |
| Surgery during 12 weeks prior to infection | 130 (22.2) | 285 (19.4) | 0.15 |
| ER visits during 12 weeks prior to infection | 224 (38.3) | 462 (31.4) | 0.003 |
| Charlson comorbidity index | <0.0001 | ||
| 0 | 206 (35.2) | 689 (46.8) | |
| 1 | 81 (13.9) | 169 (11.5) | |
| 2 | 85 (14.5) | 161 (10.9) | |
| ≥3 | 213 (36.4) | 453 (30.8) | |
| Other underlying conditions | |||
| Diverticular disease | 51 (8.7) | 121 (8.2) | 0.71 |
| Inflammatory bowel disease | 11 (1.9) | 90 (6.1) | 0.0002 |
| Medications during 14 days prior to infection | |||
| Proton pump inhibitor | 192 (32.8) | 466 (31.7) | 0.61 |
| H2 blocker | 52 (8.9) | 146 (9.9) | 0.48 |
| Any immunosuppressive treatment | 82 (14.0) | 261 (17.7) | 0.04 |
| Any antibiotic | 345 (59.0) | 691 (46.9) | <0.0001 |
Abbreviations: ER, emergency room
Outcomes
Severe disease
Criteria for severe disease were met for 363 cases (17.7%) with strain typing results. The majority of these cases (86.0%) met criteria for severe disease because of elevated white blood cell count alone. Infection with the NAP1 strain was significantly associated with severe disease in multivariate analysis (adjusted odds ratio [aOR] 1.74, 95% CI 1.36–2.22) after controlling for age, epidemiologic classification, prior emergency room visits and hospitalizations, Charlson index, prior immunosuppressive treatment, and prior antibiotic use (Table 3).
Table 3.
Multivariate Analysis for Severe Clostridium difficile Disease Among Cases With Strain Typing Results, 2009–2011
| Cases With Strain Typing Results (n=2,057) | ||
|---|---|---|
|
| ||
| Risk Factorsa | aOR (95% CI) | P Value |
| Age > 65 years | 1.69 (1.31–2.18) | <0.0001 |
| Healthcare-associated epidemiologic classificationb | 1.75 (1.32–2.34) | 0.0001 |
| Emergency room visit during 12 weeks prior to infection | 1.31 (1.01-1.69) | 0.04 |
| Charlson index | 1.08 (0.98-1.20) | 0.14 |
| Medications during 14 days prior to infection | ||
| Immunosuppressive treatment | 1.42 (1.05-1.92) | 0.02 |
| Any antibiotic | 1.38 (1.08-1.76) | 0.01 |
| NAP1 strain | 1.74 (1.36-2.22) | <0.0001 |
Abbreviations: aOR, adjusted odds ratio; CI, confidence interval
Candidate variables included in the severe disease model: age; epidemiologic classification; surgery or emergency room visit during 12 weeks prior to infection; diverticular disease; Charlson index; proton pump inhibitor use, immunosuppressive treatment, or antibiotic use during 14 days prior to infection; strain type.
Healthcare facility-onset and community-onset healthcare facility-associated epidemiologic classes did not have significantly different odds of severe disease and were collapsed into a single “healthcare-associated” category.
Severe outcome
Severe outcomes occurred for 100 cases (4.9%). Of these cases, 41 were admitted to an ICU within 7 days after infection, 6 underwent colectomy, and 70 died within 30 days of infection. Cases developing severe outcomes were more likely to be infected with a NAP1 strain compared to those who did not develop severe outcomes (46.0% vs. 27.5%, P<0.0001). NAP 1 strain remained a predictor of severe outcomes in multivariate analysis (aOR 1.66, 95% CI 1.09–2.54) after controlling for older age, white race, healthcare-associated epidemiologic classification, Charlson index, and prior antibiotic use (Table 4).
Table 4.
Multivariate Analysis for Severe Outcome of Clostridium difficile Infection Among Cases With Strain Typing Results, 2009–2011
| Cases With Strain Typing Results (n=2,057) | ||
|---|---|---|
|
| ||
| Risk Factorsa | aOR (95% CI) | P Value |
| Age > 65 years | 1.71 (1.06–2.76) | 0.03 |
| White race | 0.49 (0.29–0.85) | 0.01 |
| Healthcare-associated epidemiologic classificationb | 2.90 (1.63–5.19) | 0.0003 |
| Charlson index | 1.71 (1.38–2.13) | <0.0001 |
| Any antibiotic during 14 days prior to infection | 1.63 (1.04–2.56) | 0.03 |
| NAP1 strain | 1.66 (1.09–2.54) | 0.02 |
Abbreviations: aOR, adjusted odds ratio; CI, confidence interval
Candidate variables included in the severe outcome model: age; race; epidemiologic classification; chronic hemodialysis; emergency room visit during 12 weeks prior to infection; Charlson index; proton pump inhibitor use, H2 blocker use, or antibiotic use during prior 14 days prior to infection; strain type.
Healthcare facility-onset and community-onset healthcare facility-associated epidemiologic classes did not have significantly different odds of severe outcome and were collapsed into a single “healthcare-associated” category.
Death within 14 days
Fifty-six deaths occurred within 14 days of stool collection. The NAP1 strain was more common among those who died within 14 days than among those who survived (51.8% vs. 27.8%, P<0.0001). The 14-day mortality of NAP1 cases was 5.0% and for non-NAP1 cases was 1.8%. In particular, for NAP7/8 cases, which correspond to ribotype 078, the 14-day mortality was 1.5% (1 of 66 cases, 95% CI, 0.04%–8.0%). In univariate analyses, older age (OR 5.6, 95% CI 2.87–10.86), healthcare-associated epidemiologic classification (OR 5.41, 95% CI 2.43–12.07 for community-onset healthcare facility-associated vs community-associated; OR 10.38, 95% CI 4.59–23.43 for healthcare facility-onset vs community-associated), Charlson index (OR 2.68, 95% CI 1.94–3.70), prior proton pump inhibitor use (OR 2.18, 95% CI 1.28–3.71), and prior antibiotic use (OR 2.31, 95% CI 1.30–4.11) were also associated with death within 14 days. In multivariate analysis, age, race, epidemiologic classification, and Charlson index were retained in the final model. After controlling for these risk factors, NAP1 strain remained a significant predictor of 14-day mortality (aOR 2.12, 95% CI 1.22–3.68) (Table 5).
Table 5.
Multivariate Analysis for 14-Day Mortality After Clostridium difficile Infection Among Cases With Strain Typing Results, 2009–2011
| Cases With Strain Typing Results (n=2,057) | ||
|---|---|---|
|
| ||
| Risk Factorsa | aOR (95% CI) | P Value |
| Age > 65 years | 2.98 (1.45–6.11) | 0.003 |
| White race | 0.46 (0.23–0.95) | 0.04 |
| Epidemiologic classification | ||
| Healthcare facility onset | 3.80 (1.62–8.94) | 0.002 |
| Community-onset healthcare facility-associated | 2.33 (1.01–5.36) | 0.05 |
| Community-associated (reference) | ||
| Charlson score | 2.03 1.44–2.86) | <0.0001 |
| NAP1 strain | 2.12 (1.22–3.68) | 0.008 |
Candidate variables included in the model: age; race, epidemiologic classification; chronic hemodialysis; emergency room visit during 12 weeks prior to infection; Charlson index; proton pump inhibitor use, H2 blocker use, or antibiotic use during 14 days prior to infection; strain type.
Sensitivity analyses
In analyses stratified by age group, NAP1 infection remained a predictor of poor outcomes in both younger (≤50 years) and older patients (>50 years) (data not shown). When stratifying by epidemiologic class (healthcare- vs. community-associated), NAP1 remained significantly associated with the three outcomes of interest for healthcare-associated cases, while for community-associated cases, all NAP1 odds ratios were above 1.0 but P values were >0.05. Finally, the associations between the NAP1 strain and severe disease, severe outcome, and 14-day mortality remained significant after excluding cases from the EIP site (New York) that contributed the largest number of NAP1 cases.
DISCUSSION
We found NAP1 to be the most common strain, accounting for over one-quarter of cases in our dataset. NAP1 was associated with greater odds of severe disease than other NAP types in unadjusted analysis and was also associated with older age and a variety of healthcare exposures. After controlling for potential confounders, the NAP1 strain remained a significant predictor of severe disease, severe outcome, and 14-day mortality. Our results represent the largest study to date to examine the association between strain type and disease outcomes. Furthermore, inclusion of cases from a wide geographic spread reduces bias due to regional variation in C. difficile strain type or patient characteristics.
We chose the outcome measures of severe outcome and 14-day mortality to facilitate comparisons with two recent studies on CDI outcome related to the NAP1 strain. Our findings agree with those in a recent report from the United Kingdom showing that the NAP1/027 strain is associated with increased 14-day mortality [5]. However, our results differ from a recent study from the United States by Walk et al that found no association between the NAP1/027 strain and severe outcome [7]. In the US study, the number of patients with NAP1 was small (~40) and fewer than 50 patients met the outcome measure. Though the US study reported a lack of association, the odds for severe outcome were increased among NAP1/027 cases, albeit not achieving statistical significance. Thus, as suggested by others [20, 21], lack of association between the NAP1/027 strain and severe outcome reported in the US study might be largely related to differences in sample size.
The proportion of cases who died in our study (3.8%) is lower than that reported from other studies of CDI outcomes [6, 22]. This discrepancy is likely a result of a larger proportion of community-associated CDI cases in our study, which are associated with better outcomes than healthcare-associated cases [14]. This is unlikely to bias our study towards detection of association between strain type and outcomes, given that we adjusted for epidemiologic classification as a confounder in our analyses.
We found baseline differences between the patient populations infected by C. difficile NAP1 versus non-NAP1 strains. Though we adjusted for patient comorbidities, unmeasured patient-level confounders might still account for some of the relationship we found between the NAP1 strain and patient outcomes. However, our findings persisted in analyses stratified by age, supporting that C. difficile strain is an important predictor of patient outcome independent of patient age. Associations between NAP1 strain and patient outcomes did not remain significant for community-associated cases when stratifying the data by epidemiologic classification. This lack of significance is likely related to low statistical power, as outcomes of interest were uncommon among community-associated cases (10.8% for severe disease, 1.5% for severe outcomes and 0.7% for death within 14 days), and the NAP1 strain was less prevalent in the community.
Our analysis therefore provides additional support for the conclusion that infection by the NAP1 strain adversely affects patient outcomes. The practical implications of this finding remain to be determined. Host-related factors play an important role in the development of CDI, and other studies have suggested that clinical scores or biomarkers based on the immune response of the host (e.g., albumin, serum white blood count, c-reactive protein), rather than strain type, should be the basis for decisions about severity of disease for treatment [12, 23, 24].
Nevertheless, strategies that account for strain-specific factors might complement treatment strategies based on host response and further reduce morbidity due to CDI. The specific virulence factors possessed by the NAP1/027 strain that lead to worsened outcomes still remain to be more clearly elucidated. As such research progresses, vaccines being developed for C. difficile might target such factors.
In addition, antimicrobial stewardship might further aid in preventing infections from the NAP1 strain. Although more judicious antimicrobial use would likely reduce C. difficile infections in general, including those caused by NAP1, stewardship efforts might also be leveraged to have a greater impact on NAP1 prevalence. For example, given that the NAP1/027 strain is more resistant than other strains to the fluoroquinolones [1, 3, 5, 25, 26], antimicrobial stewardship aimed at reducing the overall use of fluoroquinolones might also reduce the prevalence of infections from NAP1 and decrease patient morbidity from CDI. Indeed, fluoroquinolone use has been found to be a risk factor for infection by the NAP1/027 strain [27–29] and the development of fluoroquinolone resistance by the NAP1/027 strain has been suggested to be the primary genetic factor facilitating its spread [29]. Fluoroquinolone restriction has also been reported to be an important component of efforts to control outbreaks of C. difficile from the NAP1/027 strain [30, 31], but further research is needed to determine the utility of the application of this strategy to non-outbreak settings.
We should note the following limitations of our study. First, all clinical data collected were obtained by retrospective review of medical charts. This potentially could have underestimated mortality rates if patients died soon after discharge. Second, we were not able to control for differences in treatment, which might affect patient outcomes. However, as noted earlier, because our data encompass a diverse geographic area, it is unlikely that our findings are driven by individual institutional treatment practices. Third, we could not fully evaluate the potential role of the NAP7/8/ribotype 078 strain, which has also been reported to have increased virulence [5, 32], on outcomes due to limited numbers of these cases in our dataset. Fourth, our analysis might not be representative of all C. difficile infections. For example, only a sample of healthcare facility-onset CDI cases are fully reviewed. However, even though cases with strain typing results represent a convenience sample of the total, comparison to cases without strain typing results suggests that our sample is representative of those cases with full medical record review. Fifth, we do not have data on all cases about the type of diagnostic test that was used to identify each CDI case (e.g., toxin assay versus nucleic acid amplification test [NAAT]). As CDI cases detected by toxin assays have been reported to have higher mortality than cases detected solely by NAAT [33], we could not account for this potential effect modifier of CDI outcomes.
In conclusion, analysis of a large, geographically diverse set of CDI cases from the United States corroborates that the C. difficile NAP1 strain type is an important determinant of patient outcomes. Disease from C. difficile results from a complex interplay between host-related factors and pathogen-specific factors. Efforts to reduce the burden of CDI likely will need to consider both.
Summary: After controlling for patient risk factors and healthcare exposures, NAP1 Clostridium difficile strain type was associated with severe disease, severe outcome, and 14-day mortality in multivariate models when compared to other strains types.
Acknowledgements
We acknowledge the following contributors: Joelle Nadle, Erin Garcia, Erin Parker, Ashley Williamson, California Emerging Infections Program; Wendy Bamberg, Colorado Emerging Infections Program; Jim Meek, Danyel Olson, Connecticut Emerging Infections Program; Wendy Baughman, Leigh Ann Clark, Andrew Revis, Zirka Thompson, Olivia Almendares, Georgia Emerging Infections Program; Lucy Wilson, Malorie Givan, Maryland Emerging Infections Program; Ruth Lynfield, Minnesota Emerging Infections Program; Nathan Blacker, New Mexico Emerging Infections Program, Rebecca Tsay, Deborah Nelson, New York Emerging Infections Program; New York State Department of Health laboratory personnel; Valerie Ocampo, Oregon Emerging Infections Program; Samir Hanna, L. Amanda Ingram, Brenda Rue, Tennessee Emerging Infections Program; Susan Sambol, Laurica Petrella, Hines VA Hospital; L. Clifford McDonald, Brandi Limbago, Duncan MacCannell, Centers for Disease Control and Prevention.
Funding
This work was supported by the Center for Disease Control’s Emerging Infections Program Cooperative Agreement.
Footnotes
Disclaimer
The findings and conclusions in this report are those of the authors and do not necessarily represent the official position of the CDC.
Potential conflicts of interest:
Dr. Gerding is a board member of Merck, Rebiotix, Summit, and Actelion, and consults for Roche, Novartis, Sanofi Pasteur, and Cubist, all of which perform research on potential C. difficile products. Dr. Gerding is a consultant for and has patents licensed to Viropharma, which makes vancomycin used to treat C. difficile infection. The other authors have no potential conflicts to disclose.
Presented in part: IDWeek 2013, October 2–6, 2013, San Francisco, CA. Oral abstract 1218.
References
- 1.Loo VG, Poirier L, Miller MA, et al. A predominantly clonal multi-institutional outbreak of Clostridium difficile-associated diarrhea with high morbidity and mortality. N Engl J Med. 2005;353:2442–9. doi: 10.1056/NEJMoa051639. [DOI] [PubMed] [Google Scholar]
- 2.Warny M, Pepin J, Fang A, et al. Toxin production by an emerging strain of Clostridium difficile associated with outbreaks of severe disease in North America and Europe. Lancet. 2005;366:1079–84. doi: 10.1016/S0140-6736(05)67420-X. [DOI] [PubMed] [Google Scholar]
- 3.McDonald LC, Killgore GE, Thompson A, et al. An epidemic, toxin gene-variant strain of Clostridium difficile. N Engl J Med. 2005;353:2433–41. doi: 10.1056/NEJMoa051590. [DOI] [PubMed] [Google Scholar]
- 4.Labbé AC, Poirier L, MacCannell D, et al. Clostridium difficile infections in a Canadian tertiary care hospital before and during a regional epidemic associated with the BI/NAP1/027 strain. Antimicrob Agents Chemother. 2008;59:3180–7. doi: 10.1128/AAC.00146-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Walker AS, Eyre DW, Wyllie DH, et al. Relationship between bacterial strain type, host biomarkers and mortality in Clostridium difficile infection. Clin Infect Dis. 2013;56:1589–600. doi: 10.1093/cid/cit127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Miller M, Gravel D, Mulvey M, et al. Health care-associated Clostridium difficile infection in Canada: patient age and infecting strain type are highly predictive of severe outcome and mortality. Clin Infect Dis. 2010;50:194–201. doi: 10.1086/649213. [DOI] [PubMed] [Google Scholar]
- 7.Walk ST, Micic D, Jain R, et al. Clostridium difficile ribotype does not predict severe infection. Clin Infect Dis. 2012;55:1661–8. doi: 10.1093/cid/cis786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Goldenberg SD, French GL. Lack of association of tcdC type and binary toxin status with disease severeity and outcome in toxigenic Clostridium difficile. J Infect. 2011;62:355–62. doi: 10.1016/j.jinf.2011.03.001. [DOI] [PubMed] [Google Scholar]
- 9.Cloud J, Noddin L, Pressman A, Hu M, Kelly C. Clostridium difficile strain NAP-1 is not associated with severe disease in a nonepidemic setting. Clin Gastroenterol Hepatol. 2009;7:868–73. doi: 10.1016/j.cgh.2009.05.018. [DOI] [PubMed] [Google Scholar]
- 10.Sirard S, Valiquette L, Fortier L. Lack of association between clinical outcome of Clostridium difficile infections, strain type, and virulence-associated phenotypes. J Clin Microbiol. 2011;49:4040–6. doi: 10.1128/JCM.05053-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wilson V, Cheek L, Satta G, et al. Predictors of death after Clostridium difficile infection: a report on 128 strain-typed cases from a teaching hospital in the United Kingdom. Clin Infect Dis. 2010;50:e77–81. doi: 10.1086/653012. [DOI] [PubMed] [Google Scholar]
- 12.Cohen SH, Gerding DN, Johnson S, et al. Clinical practice guidelines for Clostridium difficile infection in adults: 2010 update by the Society for Healthcare Epidemiology of America (SHEA) and the Infectious Diseases Society of America (IDSA) Infect Control Hosp Epidemiol. 2010;31:431–55. doi: 10.1086/651706. [DOI] [PubMed] [Google Scholar]
- 13.Centers for Disease Control and Prevention Measuring the scope of Clostridium difficile infection in the United States. Available at: http://www.cdc.gov/hai/eip/clostridium-difficile.html. Accessed July 2, 2013.
- 14.Lessa FC. Community-associated Clostridium difficile infection: how real is it? Anaerobe. 2013 doi: 10.1016/j.anaerobe.2013.01.006. [epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McDonald LC, Coignard B, Dubberke E, Song X, Horan T, Kutty PK. Recommendations for surveillance of Clostridium difficile-associated disease. Infect Control Hosp Epidemiol. 2007;28:140–5. doi: 10.1086/511798. [DOI] [PubMed] [Google Scholar]
- 16.Charlson ME, Pompei P, Ales KL, MacKenzie CR. A new method of classifying prognostic comorbidity in longitudinal studies: development and validation. J Chron Dis. 1987;40:373–83. doi: 10.1016/0021-9681(87)90171-8. [DOI] [PubMed] [Google Scholar]
- 17.Lowy I, Molrin DC, Leav BA, et al. Treatment with monoclonal antibodies to Clostridium difficile toxins A and B prevent recurrent infection. New Engl J Med. 2010;362:1–9. doi: 10.1056/NEJMoa0907635. [DOI] [PubMed] [Google Scholar]
- 18.Killgore G, Thompson A, Johnson S, et al. Comparison of seven techniques for typing international epidemic strains of Clostridium difficile: restriction endonuclease analysis, pulsed-field gel electrophoresis, PCR-ribotyping, multilocus sequence typing, multilocus variable-number tandem-repeat analysis, amplified fragment length polymorphism, and surface layer protein A gene sequence typing. J Clin Microbiol. 2008;46:431–7. doi: 10.1128/JCM.01484-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Persson S, Torpdahl M, Olsen KE. New multiple PCR method for the detection of Clostridium difficile toxin A (tcdA) and toxin B (tcdB) and the binary toxin (cdtA/cdtB) genes applied to a Danish strain collection. Clin Microbiol Infect. 2008;2009;1115:1057–64. 296. doi: 10.1111/j.1469-0691.2008.02092.x. Erratum: Clin Microbiol Infect. [DOI] [PubMed] [Google Scholar]
- 20.Gerding DN, Johnson S. Does infection with specific Clostridium difficile strains or clades influence clinical outcome? Clin Infect Dis. 2013;56:1601–3. doi: 10.1093/cid/cit133. [DOI] [PubMed] [Google Scholar]
- 21.Walker AS, Eyre DW, Crook DW, Wilcox MH, Peto TE. Regarding “Clostridium difficile ribotype does not predict severe infection”. Clin Infect Dis. 2013;56:1845–6. doi: 10.1093/cid/cit098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hensgens MP, Goorhuis A, Dekkers OM, van Benthem BH, Kuijper E. All-cause and disease-specific mortality in hospitalized patients with Clostridium difficile infection: a multicenter cohort study. Clin Infect Dis. 2013;56:1108–16. doi: 10.1093/cid/cis1209. [DOI] [PubMed] [Google Scholar]
- 23.Fujitani S, George WL, Murthy AR. Comparison of clinical severity score indices for Clostridium difficile infection. Infect Control Hosp Epidemiol. 2011;32:220–8. doi: 10.1086/658336. [DOI] [PubMed] [Google Scholar]
- 24.Bauer MP, Hensgens MP, Miller MA, et al. Renal failure and leukocytosis are predictors of a complicated course of Clostridium difficile infection if measured on day of diagnosis. Clin Infect Dis. 2012;55(suppl 2):S149–53. doi: 10.1093/cid/cis340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Walkty A, Boyd DA, Gravel D, et al. Molecular characterization of moxifloxacin resistance from Canadian Clostridium difficile clinical isolates. Diagn Microbiol Infect Dis. 2010;66:419–24. doi: 10.1016/j.diagmicrobio.2009.12.002. [DOI] [PubMed] [Google Scholar]
- 26.Vardakas KZ, Konstantelias AA, Loizidis G, Rafailidis PI, Falagas ME. Risk factors for development of Clostridium difficile infection due to BI/NAP1/027 strain: a meta-analysis. Int J Infect Dis. 2012;16:e768–73. doi: 10.1016/j.ijid.2012.07.010. [DOI] [PubMed] [Google Scholar]
- 27.Pépin J, Saheb N, Coulombe M-A, et al. Emergence of fluoroquinolones as the predominant risk factor for Clostridium difficile-associated diarrhea: a cohort study during an epidemic in Quebec. Clin Infect Dis. 2005;49:1254–60. doi: 10.1086/496986. [DOI] [PubMed] [Google Scholar]
- 28.Kazakova SV, Ware K, Baughman B, et al. A hospital outbreak of diarrhea due to an emerging epidemic strain of Clostridium difficile. Arch Int Med. 2006;166:2518–24. doi: 10.1001/archinte.166.22.2518. [DOI] [PubMed] [Google Scholar]
- 29.He M, Miyajima F, Roberts P, et al. Emergence and global spread of epidemic healthcare-associated Clostridium difficile. Nat Genet. 2013;45:109–13. doi: 10.1038/ng.2478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kallen AJ, Thompson A, Ristaino P, et al. Complete restriction of fluoroquinolone use to control and outbreak of Clostridium difficile infection at a community hospital. Infect Control and Hosp Epidemiol. 2009;30:264–72. doi: 10.1086/595694. [DOI] [PubMed] [Google Scholar]
- 31.Aldeyab MA, Devine MJ, Flanagan P, et al. Multihospital outbreak of Clostridium difficile ribotype 027 infection: epidemiology and analysis of control measures. Infect Control Hosp Epidemiol. 2011;32:210–9. doi: 10.1086/658333. [DOI] [PubMed] [Google Scholar]
- 32.Goorhuis A, Bakker D, Corver J, et al. Emergence of Clostridium difficile infection due to a new hypervirulent strain, Polymerase Chain Reaction ribotype 078. Clin Infect Dis. 2008;47:1162–70. doi: 10.1086/592257. [DOI] [PubMed] [Google Scholar]
- 33.Planche TD, Davies KA, Coen PG, et al. Differences in outcome according to Clostridium difficile testing method: a prospective multicenter diagnostic validation study of C. difficile infection. Lancet Infect Dis. 2013;13:936–45. doi: 10.1016/S1473-3099(13)70200-7. [DOI] [PMC free article] [PubMed] [Google Scholar]