Abstract
Knowledge integration includes knowledge management, synthesis, and translation processes. It aims to maximize the use of collected scientific information and accelerate translation of discoveries into individual and population health benefits. Accumulated evidence in cancer epidemiology constitutes a large share of the 2.7 million articles on cancer in PubMed. We examine the landscape of knowledge integration in cancer epidemiology. Past approaches have mostly used retrospective efforts of knowledge management and traditional systematic reviews and meta-analyses. Systematic searches identify 2,332 meta-analyses, about half of which are on genetics and epigenetics. Meta-analyses represent 1:89-1:1162 of published articles in various cancer subfields. Recently, there are more collaborative meta-analyses with individual-level data, including those with prospective collection of measurements [e.g., genotypes in genome-wide association studies (GWAS)]; this may help increase the reliability of inferences in the field. However, most meta-analyses are still done retrospectively with published information. There is also a flurry of candidate gene meta-analyses with spuriously prevalent "positive" results. Prospective design of large research agendas, registration of datasets, and public availability of data and analyses may improve our ability to identify knowledge gaps, maximize and accelerate translational progress or—at a minimum—recognize dead ends in a more timely fashion.
Introduction
Given the rapid expansion of scientific information, there is a critical need to ensure that maximal use is made of the collected data in a most efficient and unbiased way. "Knowledge integration" describes the processes that aim at effective use of information from many sources for accelerating translation of scientific discoveries into clinical applications, evidence-based recommendations, use in practice, and eventually health benefits for individuals and populations (Fig. 1). The term has been used with different definitions and perspectives (1–3), but here we adopt the definition that includes knowledge management (KM), knowledge synthesis (KS), and knowledge translation (KT; ref. 1). These 3 processes can inform the continuum of translational research from discovery to population health impact (1). The knowledge integration engine can drive research progress, especially in data-intensive fields.
Figure 1.
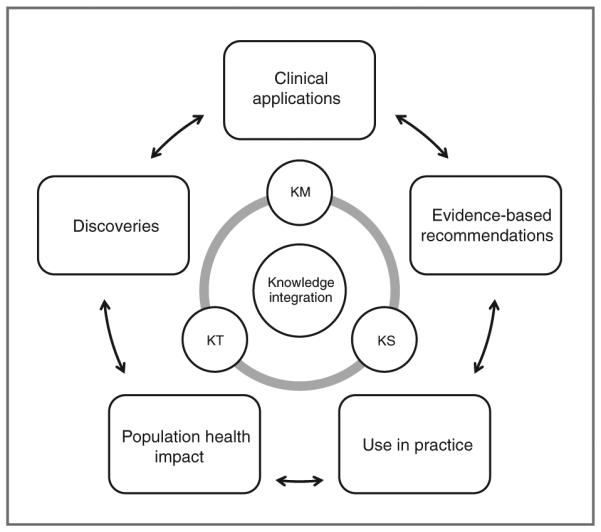
The central role of knowledge integration in driving translational research.
Previously, knowledge integration has depended mostly on retrospective efforts of horizon scanning, traditional systematic reviews and meta-analyses, and nonsystematic knowledge brokering between stakeholders. These efforts may be inadequate in the current era of rapid accumulation of multilevel information—from molecular to the macro-level of environmental exposures and health system attributes. Revamping the current knowledge integration processes may help drive the future of cancer epidemiology across the translational research continuum. Here, we overview the accumulated experience on knowledge integration with emphasis on cancer epidemiology. We discuss what methods have been used over time, their strengths and limitations, and what alternative approaches might lead to more efficient integration of emerging information.
Published information on cancer
PubMed (search August 27, 2012) lists 2,673,926 articles with the search on "cancer," of which more than three quarters (n = 2,146,156) are tagged with "human." Table 1 shows selective subsets that present an overall picture of published evidence for different types of designs relating to cancer epidemiology. More than 50,000 articles are identified with the search word "cohort," and slightly more with "case control," whereas the term "risk" retrieves more than 200,000 articles, and "biomarker" just as many. At the same time, there are also more than 100,000 clinical trial publications, many of which are randomized trials. Efforts to synthesize this information are represented by more than 6,000 meta-analyses, almost 30,000 systematic reviews, and more than 300,000 nonsystematic reviews. Thus, most efforts at evidence integration still use subjective opinion and nonsystematic methods for data overview and interpretation.
Table 1.
Published articles in cancer literature
| PubMed | Treatment | Prevention | Not (trial or treatment) | |
|---|---|---|---|---|
| Cancer | 2,673,926 | 1,360,697 | 208,187 | 1,295,958 |
| +Animal | 481,080 | 206,009 | 40,891 | 269,653 |
| +Cell | 1,118,600 | 510,286 | 66,935 | 604,187 |
| +Cohort | 53,567 | 29,808 | 8,683 | 22,819 |
| +Case–control | 59,248 | 11,255 | 5,848 | 26,973 |
| +Risk | 267,490 | 158,529 | 58,002 | 105,276 |
| +Biomarker | 204,419 | 91,298 | 15,675 | 111,025 |
| +Clinical trial, type | 105,939 | 93,172 | 14,807 | 4,535 |
| +RCT, type | 34,449 | 31,862 | 8,047 | 0 |
| +Meta-analysis, type | 6,406 | 3,902 | 1,153 | 2,332 |
| +Systematic review, type | 28,922 | 21,398 | 5,755 | 6,763 |
| +Review, type | 314,176 | 201,162 | 39,859 | 111,234 |
NOTE: Search strategies: treatment: "treatment", prevention: "prevention OR screening [ti] OR screening [tw]", Not trial or treatment: "NOT (trial* OR treatment)."
Abbreviation: RCT, randomized controlled trial.
It is difficult to accurately separate the literature on associations from the literature on treatments and interventions—sometimes they intermingled in the same article. Subsets searches in Table 1 should be reviewed with caution given the non-perfect sensitivity and specificity of PubMed searches. However, clearly articles with relevance to treatment far outnumber articles with relevance to prevention across all subsets. Table 1 also provides search results with the string "NOT (trial* OR treatment)" to further exclude articles on clinical trials and treatmentrelated research (which can be either randomized or observational, e.g., predictive tools for treatment response). As shown, studies with "cohort" or "case control" are split between the literature with and without trials/treatment implications.
Knowledge management: studies, data, analyses
KM efforts can take many different forms, depending on whether one is tapping into published information; retrieving unpublished information; developing databases with raw data; or allowing a live stream of all collected data and analyses (Table 2). Published information is only a fraction of the total raw data that have been collected for or repurposed for research purposes and of the analyses that have been formally conducted, probed, or contemplated (4).
Table 2.
Different approaches to KM and KS
| Knowledge management |
| Published data: optimization of search engines, curation and cleaning, harmonization |
| Unpublished data: detection, registration, cleaning |
| Deposition of raw datasets in public: documentation, access control, ease of use, credit, independence |
| Live stream information |
| Knowledge synthesis |
| Same-level of information |
| Systematic reviews of published information |
| Meta-analyses of published information |
| Meta-analyses including also retrieved unpublished data |
| Field synopses of many meta-analyses |
| Collaborative meta-analyses of previously collected individual-level data |
| Collaborative meta-analyses of prospectively collected data from existing studies |
| Prospective consortia and meta-analyses thereof |
| Multiple levels of information |
| Cross-design synthesis and multilevel evidence appraisals |
| Modeling with real or simulated data |
| Meta-research (research on research) |
Efforts of KM for published data concentrate usually at search optimization, curation, cleaning, and harmonization. Published data may often be a selected, even distorted, subset of the whole information chosen based on statistical significance and/or other selection filters (5). Although some journals have begun efforts to publish "null" results (6, 7), these remain underrepresented in the published literature. If so, KM targeting past published data may yield largely misleading results. Empirical meta-research evaluations on the credibility of research findings in different research fields and with different methods and technologies may help anchor some credibility estimates. They may help decide whether a field is severely biased that it is a wasteful effort to collect, clean, and use published information. Conversely, the KM process may indicate that a systematic synthesis can indeed yield reliable results.
Unpublished data and analyses results are notoriously difficult to unearth. Some investigators may claim that unpublished data can be ignored, as they have not passed peer review. However, the seal of peer review is not a perfect discriminant. Registration of protocols and analyses would have helped to understand the depth of the problem. Nevertheless, publication of these documents is limited. For example, it is well documented that many clinical trials protocols and/or analyses are never published. Of those that are published, half or more of the originally considered outcomes are not reported, whereas many others are reported with analyses and results that deviate from the originally intended analyses (8–10). For observational epidemiology, these problems are probably more common (11). However, a priori registration of protocols is difficult for such studies, given their exploratory and iterative nature. Instead, it has been argued that registration should focus on study datasets (4, 12), that is, information on what variables have been collected and measured. This allows an understanding and assessment how many registered datasets could have undergone specific analyses.
Public deposition of raw datasets has attracted increasing attention over time, with several successful efforts for laboratory research, for example, genomic sequencing databases and functional databases (13). For microarray, macromolecular, and protein data, most high-impact journals have policies that require delineating some plan of making raw data, protocols, and analysis codes publicly available, routinely or after request to the authors; public deposition of such information is often a prerequisite for publication, but these policies are not necessarily enforced (14, 15).
There are several challenges related to deposition of raw datasets (Table 2). Some datasets may be deposited with poor documentation that hinders their usage by an outsider or may lead to erroneous data readings and misleading inferences. In addition, some public databases have minimum requirements when depositing data. Even if investigators are required to adhere to data-sharing policies (either from funding agencies or journal requirements), they often enter the minimal amount of required information. There are different modes of data access: open-to-all, cursory approval–based, and access to select investigators passing stringent standards of recognized expertise. Striking a balance between credit and independence is also challenging. Original investigators could (or should) be credited for analyses conducted on their data. However, it may also be advisable to keep further analyses separate from them: subsequent investigators who then use these published data should feel free to repeat and challenge the original analyses.
Finally, the live stream information model suggests that data, protocols, and analyses are readily available and visible to a wider circle, even the full public, as they accumulate, change, and evolve. This practice has been piloted in experiments trying to replicate the finding of bacteria with arsenic-containing DNA (16). Other fields, including cancer epidemiology, may also learn from it.
Knowledge synthesis of same-level information
There is a wide variety of KS methods (Table 2), but systematic reviews and meta-analyses are the most common. The majority of such reviews still depend on published information. Meta-analyses of published data are popular in many disciplines, especially those where unadjusted estimates and plain 2 × 2 tables are convenient. Some efforts may also be made to retrieve and include unpublished information, but success in this endeavor varies. For fields where there are many meta-analyses, field synopses have emerged (17) with simultaneous compilation of tens to millions of meta-analyses on the same field, as in several examples of applications in human genome epidemiology, for example, AlzGene, SzGene, and PDGene (18–20).
Other KS efforts involve investigators who control existing primary data. Such collaborative meta-analyses use a central secretariat to collect, query, clean, and synthesize individual-level data or statistics derived from individual-level data analyses procured by the original investigators of each included study. The advantages (potential standardization or harmonization of data and analyses, consistency of adjustments, multivariable models, interactions, and other complex models) and disadvantages (cost, effort, inability to fully standardize post hoc, selective availability of information, political difficulties) of this approach versus meta-analyses of the published literature have been extensively discussed previously (21, 22).
There is increasing interest in collaborative meta-analyses that use prospectively collected measurements from existing studies. This is the dominant paradigm in meta-analyses of GWAS (23, 24) that have led to a massive increase in the number of discovered genetic variants with strong statistical support (25). These meta-analyses may avoid the potential for selective reporting bias that threatens collaborative meta-analyses of previously collected data. Consortia working in this framework are common in human genome epidemiology but less common in other fields. Finally, one can envision prospective meta-analyses, where not only specific measurements but also the primary studies are designed prospectively, with the plan to eventually combine them. Such examples currently exist mostly from randomized trials (26). Nevertheless, the concept can conceivably be applied to future designed case–control studies, cohort studies, and biobanks (27), with prospective standardization of their designs and data collection and analyses procedures (28).
Landscape of KS methods used in cancer epidemiology
Table 3 shows the landscape of practiced KS methods for the PubMed subset of "Cancer NOT (trial* OR treatment)". The large majority of systematic reviews (85.5%) and all but 13 meta-analyses addressed human data. More effort is needed to systematically appraise evidence from animal studies (29, 30), which can be informative and influential for judging biologic plausibility and for other preclinical inferences.
Table 3.
Systematic reviews and meta-analyses in different fields of cancer (excluding trials and treatment)
| Search terms | All articles, N (all) | n (SR) | n (MA) | n (SR)/n (MA) | n (all)/n (MA) |
|---|---|---|---|---|---|
| Gene/genome/genetic | 268,597 | 1,999 | 920 | 2.2 | 291 |
| Epigenetic/methylation/mutation | 115,763 | 497 | 159 | 3.1 | 728 |
| Immune/allergy/asthma | 29,046 | 107 | 25 | 4.3 | 1162 |
| Hormone | 53,679 | 148 | 51 | 2.9 | 1,032 |
| Social/socioeconomic | 11,531 | 224 | 50 | 4.5 | 231 |
| Diet/dietary/nutrition/nutritional | 19,549 | 289 | 147 | 2.0 | 133 |
| Physical activity/exercise/obesity | 8,919 | 192 | 74 | 2.6 | 121 |
| Virus/bacteria/infection/infectious | 88,881 | 331 | 109 | 3.0 | 815 |
| Carcinogen | 30,286 | 201 | 88 | 2.3 | 344 |
| Radiation | 30,124 | 229 | 86 | 2.6 | 350 |
| Occupation/occupational | 16,839 | 344 | 177 | 1.9 | 95 |
| Smoking/smoke/tobacco | 20,660 | 413 | 232 | 1.8 | 89 |
| Alcohol | 17,921 | 170 | 83 | 2.0 | 215 |
| Biomarker | 72,709 | 373 | 97 | 3.8 | 750 |
NOTE: On the basis of PubMed searches conducted on August 29, 2012. The search resulted in 1,295,958 items overall, of which 6,763 were tagged by PubMed as systematic reviews and 2,332 as meta-analyses; when limited to studies tagged as human, there were 941,360 items overall, 5,780 systematic reviews, and 2,319 meta-analyses. All searches use as a backbone the search strategy "Cancer NOT (trial* OR treatment)" so as to avoid capturing articles on clinical trials and treatments. Some articles from case-control and cohort studies where treatment or treatment-related questions are discussed would be excluded by this strategy, but this latter group accounts generally for few meta-analysis, for example, a search with "Cancer AND (trial* OR treatment) AND (case-control or cohort) AND meta-analysis [type]" yields an additional 451 items (besides the 2,332 identified with "Cancer NOT (trial* OR treatment) AND meta-analysis [type]"); in a sample of 20 of the 451, only 10 are related to cancer epidemiology. Conversely, in detailed scrutiny of a sample of 127 items, among those identified with the search "Cancer NOT (trial* OR treatment) AND meta-analysis [type]", 114 are indeed meta-analyses of cancer epidemiology topics (positive predictive value 90%). Thus, the provided estimate of 2,332 metaanalysis articles in cancer epidemiology seems to be quite accurate, with <250 falsely identified articles and roughly equal number of falsely nonidentified articles. All searches in the table, with the exception of Gene/Genome/Genetic and Epigenetic/Methylation/Mutation, use the string "NOT (Gene OR Genome OR Genetic)", so as to exclude items that deal primarily with genetics, for example, immune response genes or hormone-related genes.
Abbreviations: N (all), total number of articles retrieved; n (SR), number of articles retrieved with type: systematic review; n (MA), number of articles retrieved with type: meta-analysis.
The fields of genetics or epigenetics dominate almost a third of the literature. Correspondingly, the same distribution applies to systematic reviews, whereas these 2 disciplines account for about half of the existing meta-analyses. The literatures on biomarkers, hormones, and infectious agents are extensive but have relatively fewer meta-analyses (n = 51–109 in each). Conversely, other concentrations with smaller shares of the total literature instead have as many or more published meta-analyses, in particular smoking, occupational, and nutritional fields.
The number of systematic reviews is 2- to 3-fold larger than the number of meta-analyses in most areas, except for biomarkers, immune/allergy/asthma, and social/socioeconomic factors, where the ratio is even larger. This may reflect the difficulty of conducting quantitative syntheses (e.g., for social and socioeconomic factors with extreme heterogeneity of definitions and measurements) or less established traditions for conducting meta-analyses. The ratio of all published articles per published meta-analysis is 556 overall; despite their emerging popularity, metaanalyses are still a small portion of the literature. Moreover, there is large variability in this ratio across different fields. It is smaller [n (all)/n (MA) = 89–133] for smoking, occupational, nutritional, and lifestyle areas; modestly high [n (all)/n (MA) = 215–350] for alcohol, social, genetics, carcinogens, and radiation; and very high [n (all)/n (MA) = 728–1,162] in epigenetics, biomarkers, immune factors, hormones, and infectious agents.
A more detailed examination of a sample of meta-analyses published in 1992, 2002, and 2012 shows the evolution of the application of these methods over time. "Cancer NOT (trial* OR treatment) AND meta-analysis [type]" yields 25 items in 1992, 49 in 2002, and 232 in the first 8 months of 2012 alone. On closer examination, 20 of the 25 meta-analysis–tagged articles published in 1992 are indeed meta-analyses related to cancer epidemiology, and the same applies to 44 of 49 tagged articles in 2002 and 50 of the 53 latest indexed articles in 2012.
Besides the geometric increase in the number of published meta-analysis articles each year, the areas represented have changed over time. The advent of metaanalyses on genetics and epigenetics is impressive. In 1992, there was only one quantitative review on leukemia cytogenetics. In 2002, of the 44 meta-analyses, 8 (18%) assessed genetic variants, 1 (2%) genomic hybridization, and 1 (2%) microarrays. In 2012, of the 50 most recently indexed published meta-analyses, 25 (50%) were on genetic variants, 2 (4%) on epigenetics, and another one on gene–menopause interaction. No other field in 2012 had such staggering increase in the number of meta-analyses (smoking, n = 3; alcohol, n = 3; biomarkers, n = 2; infectious agents, n = 2; dietary, n = 2; social, n = 1; occupational, n = 1; diagnostic tests, n = 3, other, n = 3 among 50 meta-analyses examined).
Moreover, there have been an increasing number of genes and genetic variants examined in meta-analyses over time. All genetic meta-analyses in 2002 focused on specific genes. Conversely, meta-analyses in 2012 included also genome-wide association meta-analyses, consortium analyses examining a number of variants, and field synopses. There is also a discernible change in the types of meta-analyses conducted over time, in particular about the use of consortium approaches and use of individual-level data (31). In the examined samples, there were 2 meta-analyses using individual-level data in 1992 (among 20), 5 in 2002 (among 44), and 9 among the most recent 50 meta-analyses in 2012. All the meta-analyses with individual-level data in 1992 and 2002 combined information that had been already collected in existing studies. One meta-analysis combined data from publicly available data on microarray experiments, whereas all the other meta-analyses created collaborative structures where investigators contributed their data and participated in the final manuscripts. These analyses pertained to nutritional factors (n = 4), hormones (n = 1), or smoking and alcohol (n = 1). Conversely, in 2012, the 9 meta-analyses using individual-level data targeted a very different set of risk factors: genetic factors (n = 6), gene expression data (n = 1), biomarkers (n = 1) and endometriosis (n = 1). The 6 meta-analyses of genetic factors were done by consortia conducting with genotype data generated prospectively for the project. The gene expression meta-analysis used data from publicly available databases, and the other 2 meta-analyses were done by investigators contributing previously collected data.
Caveats in current meta-analyses in cancer epidemiology
Despite the increase in the number and proportion of meta-analyses with individual-level data over time, these still represent the minority. Most currently published metaanalyses in cancer epidemiology continue to depend on published summary data. Many of these meta-analyses focus on genetic variants, often targeting a single or a few candidate genes and variants thereof. Interestingly, among the 50 published meta-analyses from 2012, 12 were done in China focusing on specific candidate genes from the era preceding GWAS. With one exception (32), all of these Chinese meta-analyses concluded that the examined candidate genes are significantly associated with the phenotypes of interest, although the P values were always very modest. On the basis of previous experience on candidate gene associations (33, 34), the credibility of these associations is very low. Another 3 meta-analyses from China addressed genetic variants previously highlighted from GWAS and also included primary data that the authors generated in their own sample and claimed replication of the genetic effects. Including also other fields beyond genetics, overall, 19 of the 50 (38%) meta-analyses in 2012 were from China and 17 of these 19 concluded with significant, favorable results. Previous empirical evaluations suggest that studies from China in different fields have frequent or even ubiquitous "positive" findings (35, 36).
The very high prevalence of "positive" meta-analyses, at the face of what should be mostly null associations, is worrisome. Apparently automation has allowed the massive production of potentially unreliable meta-analyses. The problem seems to be most acute for genetic epidemiology, which carries a lion’s share in currently published meta-analyses, but may extend also in other disciplines.
Knowledge synthesis: multiple-level information
Besides KS involving the same type of information combined across different studies, KS may also try to synthesize multiple levels of information and/or simulated rather than real data. Cross-design synthesis approaches can combine data from different types of designs and umbrella reviews try to compile information on different aspects of questions of interest, for example, incidence, prevalence, associations, predictive performance, and clinical treatment effects, if pertinent (37). The IARC monographs combine basic and epidemiologic data to arrive at a systematic approach to classification of carcinogens (38). The HuGENet Venice criteria attempt to do this for genetic associations (39) and Boffetta and colleagues recently proposed a merging of IARC monograph and Venice methods appraising evidence on gene–environment interactions (40).
As knowledge progresses from discovery to health-related applications, mixed methods become increasingly used to examine the evidence of validity and use of the information. Examples of KS using a mixture of methods for more advanced translation steps include the US Preventive Services Task Force documents for clinical preventive services (41) such as prostate and breast cancer screening (T2 translation stage, "does it work"), the CDC Community Guide for Preventive Services (ref. 42; T3 translation stage, "how does it work in community settings"), and CISNet, an NCI-funded consortia that evaluate using modeling and empirical data the impact of different interventions on real-world population outcomes (T4 translation stage), for example, as in a recent modeling article on contribution of screening and survival differences to racial disparities in colorectal cancer rates (43).
Knowledge synthesis: meta-research
Meta-research (research on research) may allow obtaining wide views on evidence concerning multiple research questions across one or more fields. It may help understand general patterns of study design, reporting, and biases. For example, meta-research evaluations have documented the problems of selective reporting and excess significance biases in cancer epidemiology studies and their meta-analyses (44–50). One may list here also efforts to reproduce published results. Such efforts, ranging from "forensic bioinformatics" to "reproducibility checks," have shown major reproducibility problems in several research fields such as -omics signatures (51) or preclinical data on drug targets (52).
Knowledge translation: using science to influence research, policy, and practice
KM and KS are not sufficient to move promising applications and interventions into practice. KT is a proactive process that involves communicating and disseminating synthesized information to influence policy, guideline development, practice, and research across the translation continuum. This is the most "messy" component of knowledge integration; it requires the "buy-in" from stakeholders with different perspectives, for example, see the recently discussed dissemination and implementation agenda for NIH (53).
Many forces affect the diffusion, adoption, and implementation of evidence-based recommendations into policy and practice and often operate independently from KS. They include public and private investments in research and development, policy and legal frameworks, oversight and regulation, product marketing, coverage and reimbursements, consumer advocacy, provider awareness, access, and health care services development and implementation. Deverka and colleagues (54) showed that for cancer genomic applications, different stakeholders hold disparate views of the synthesized knowledge presented to them. For example, payers generally require a higher level of evidence of clinical use than genomic researchers or test developers. Issues around differential access and implementation may contribute to the "lost in translation" phenomenon (55).
One aspect of KT may involve convening stakeholders around KS to address differences in evidentiary thresholds that drive decision making. This convening function of "knowledge brokering" links researchers and policy makers to facilitate interactions and forge partnerships to use evidence from existing knowledge and define areas for future research. In fields with rapidly changing landscape such as cancer genomics, KT knowledge brokering may need more robust proactive stakeholder engagement earlier in the decision-making process rather than later (1).
Conclusions and future prospects
The landscape of knowledge integration in cancer epidemiology has changed substantially over time and it continues to change. New methods are used more widely for managing, synthesizing, and translating information. Table 4 summarizes some possibilities that may enhance knowledge integration efforts in the future.
Table 4.
Suggestions for the future of knowledge integration
| Knowledge management |
| Methods for mining published and unpublished data |
| Registration of observational datasets and, when appropriate, protocols |
| Availability of raw data and analysis codes |
| Facilitation of repeatability and reproducibility checks, replication culture |
| Consideration of live stream information |
| Knowledge synthesis |
| Facilitation of consortia with prospective measurements |
| Optimization of multiconsortial space, competition, and communication |
| Prospective study networks |
| Knowledge translation |
| Anticipatory rather than post hoc brokering |
KM may benefit from more proactive steps rather than waiting to handle selectively reported, fragmented published data. Registration of observational datasets (4, 56), more systematic availability of raw data and analysis codes (57, 58), facilitation of repeatability and reproducibility checks (59), building a replication culture (60), and even consideration of live streaming of information may accelerate science, allow prompt recognition of false positives and dead ends, and facilitate translation of interesting observations that can be repeated and validated. The optimal way and implementation mode to achieve these changes needs carefully study.
In KS, the paradigm of large-scale international collaboration with prospective data collection has become dominant in human genome epidemiology (29), but it can also permeate more broadly many other fields. New epidemiologic studies and biobanks (25) may also be designed with the outlook that they will form part of a larger prospective network, rather than isolated proprietary experiments. For many diseases and subfields, it is possible that their several consortia with overlapping purposes may continue to co-exist. This may not necessarily be a drawback, as this may promote competition and independent replication across consortia. Regardless, it is important to have wide views of the information that is or could become available. This would help avoiding having to fund yet another study when there are hundreds available that can easily address the same question (4) or to prioritize studies and data collection in fields where the wider map of global evidence seems to have a dearth of data.
Finally, KT could benefit by wider spread and brokering of sound evidence from more reliable KM and KS efforts. It may be easier to set upfront goals, expectations, and rules of engagement and make all stakeholders aware of them, rather than wait for debates to be settled post hoc. A fascinating aspect of science is that not everything can be anticipated, but this does not mean that we cannot try to have some upfront planning and more transparency in protocols, analysis plans, and results.
Footnotes
Disclosure of Potential Confiicts of Interest
No potential conflicts of interest were disclosed.
References
- 1.Khoury MJ, Gwinn M, Dotson WD, Schully SD. Knowledge integration at the center of genomic medicine. Genet Med. 2012;14:643–7. doi: 10.1038/gim.2012.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Philippi S. Data and knowledge integration in the life sciences. Brief Bioinform. 2008;9:451. doi: 10.1093/bib/bbn046. [DOI] [PubMed] [Google Scholar]
- 3.Best A, Terpstra JL, Moor G, Riley B, Norman CD, Glasgow RE. Building knowledge integration systems for evidence-informed decisions. J Health Organ Manag. 2009;23:627–41. doi: 10.1108/14777260911001644. [DOI] [PubMed] [Google Scholar]
- 4.Ioannidis JP. The importance of potential studies that have not existed and registration of observational data sets. JAMA. 2012;308:575–6. doi: 10.1001/jama.2012.8144. [DOI] [PubMed] [Google Scholar]
- 5.Ioannidis JP. Why most published research findings are false. PLoS Med. 2005;2:e124. doi: 10.1371/journal.pmed.0020124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kheifets L, Olsen J. Should epidemiologists always publish their results? Yes, almost always. Epidemiology. 2008;19:532–3. doi: 10.1097/EDE.0b013e318177813d. [DOI] [PubMed] [Google Scholar]
- 7.Ioannidis JP. Journals should publish all "null" results and should sparingly publish "positive" results. Cancer Epidemiol Biomarkers Prev. 2006;15:186. doi: 10.1158/1055-9965.EPI-05-0921. [DOI] [PubMed] [Google Scholar]
- 8.Dwan K, Altman DG, Cresswell L, Blundell M, Gamble CL, Williamson PR. Comparison of protocols and registry entries to published reports for randomised controlled trials. Cochrane Database Syst Rev. 2011;19:MR000031. doi: 10.1002/14651858.MR000031.pub2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kirkham JJ, Dwan KM, Altman DG, Gamble C, Dodd S, Smyth R, et al. The impact of outcome reporting bias in randomised controlled trials on a cohort of systematic reviews. BMJ. 2010;340:c365. doi: 10.1136/bmj.c365. [DOI] [PubMed] [Google Scholar]
- 10.Dwan K, Altman DG, Arnaiz JA, Bloom J, Chan AW, Cronin E, et al. Systematic review of the empirical evidence of study publication bias and outcome reporting bias. PLoS One. 2008;3:e3081. doi: 10.1371/journal.pone.0003081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kavvoura FK, Liberopoulos G, Ioannidis JP. Selection in reported epidemiological risks: an empirical assessment. PLoS Med. 2007;4:e79. doi: 10.1371/journal.pmed.0040079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lash TL, Vandenbroucke JP. Should preregistration of epidemiologic study protocols become compulsory? Refiections and a counterproposal. Epidemiology. 2012;23:184–8. doi: 10.1097/EDE.0b013e318245c05b. [DOI] [PubMed] [Google Scholar]
- 13.National Center for Biotechnology Information, National Library of Medicine [cited 2012 Sep 6]; Available from: http://www.ncbi.nlm. nih.gov./
- 14.Alsheikh-Ali AA, Qureshi W, Al-Mallah MH, Ioannidis JP. Public availability of published research data in high-impact journals. PLoS One. 2011;6:e24357. doi: 10.1371/journal.pone.0024357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Savage CJ, Vickers AJ. Empirical study of data sharing by authors publishing in PLoS journals. PLoS One. 2009;4:e7078. doi: 10.1371/journal.pone.0007078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.RRResearch Available from: http://rrresearch.fieldofscience.com/2010/12/arsenic-associated-bacteria-nasas.html. Last accessed October 1, 2012.
- 17.Khoury MJ, Bertram L, Boffetta P, Butterworth AS, Chanock SJ, Dolan SM, et al. Genome-wide association studies, field synopses, and the development of the knowledge base on genetic variation and human diseases. Am J Epidemiol. 2009;170:269–79. doi: 10.1093/aje/kwp119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bertram L, McQueen MB, Mullin K, Blacker D, Tanzi RE. Systematic meta-analyses of Alzheimer disease genetic association studies: the AlzGene database. Nat Genet. 2007;39:17–23. doi: 10.1038/ng1934. [DOI] [PubMed] [Google Scholar]
- 19.Allen NC, Bagade S, McQueen MB, Ioannidis JP, Kavvoura FK, Khoury MJ, et al. Systematic meta-analyses and field synopsis of genetic association studies in schizophrenia: the SzGene database. Nat Genet. 2008;40:827–34. doi: 10.1038/ng.171. [DOI] [PubMed] [Google Scholar]
- 20.Lill CM, Roehr JT, McQueen MB, Kavvoura FK, Bagade S, Schjeide BM, et al. Comprehensive research synopsis and systematic meta-analyses in Parkinson's disease genetics: the PDGene database. PLoS Genet. 2012;8:e1002548. doi: 10.1371/journal.pgen.1002548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ioannidis JP, Rosenberg PS, Goedert JJ, O'Brien TR. International Meta-analysis of HIV Host Genetics. Commentary: meta-analysis of individual participants' data in genetic epidemiology. Am J Epidemiol. 2002;156:204–10. doi: 10.1093/aje/kwf031. [DOI] [PubMed] [Google Scholar]
- 22.Blettner M, Sauerbrei W, Schlehofer B, Scheuchenpflug T, Friedenreich C. Traditional reviews, meta-analyses and pooled analyses in epidemiology. Int J Epidemiol. 1999;28:1–9. doi: 10.1093/ije/28.1.1. [DOI] [PubMed] [Google Scholar]
- 23.Zeggini E, Ioannidis JP. Meta-analysis in genome-wide association studies. Pharmacogenomics. 2009;10:191–201. doi: 10.2217/14622416.10.2.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gögele M, Minelli C, Thakkinstian A, Yurkiewich A, Pattaro C, Pramstaller PP, et al. Methods for meta-analyses of genome-wide association studies: critical assessment of empirical evidence. Am J Epidemiol. 2012;175:739–49. doi: 10.1093/aje/kwr385. [DOI] [PubMed] [Google Scholar]
- 25.Hindorff LA, Sethupathy P, Junkins HA, Ramos EM, Mehta JP, Collins FS, et al. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc Natl Acad Sci U S A. 2009;106:9362–7. doi: 10.1073/pnas.0903103106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Province MA, Hadley EC, Hornbrook MC, Lipsitz LA, Miller JP, Mulrow CD, et al. The effects of exercise on falls in elderly patients. A preplanned meta-analysis of the FICSIT Trials. Frailty and injuries: cooperative studies of intervention techniques. JAMA. 1995;273:1341–7. [PubMed] [Google Scholar]
- 27.Manolio TA, Weis BK, Cowie CC, Hoover RN, Hudson K, Kramer BS, et al. New models for large prospective studies: is there a better way? Am J Epidemiol. 2012;175:859–66. doi: 10.1093/aje/kwr453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fortier I, Burton PR, Robson PJ, Ferretti V, Little J, L'Heureux F, et al. Quality, quantity and harmony: the DataSHaPER approach to integrating data across bioclinical studies. Int J Epidemiol. 2010;39:1383–93. doi: 10.1093/ije/dyq139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Macleod M. Why animal research needs to improve. Nature. 2011;477:511. doi: 10.1038/477511a. [DOI] [PubMed] [Google Scholar]
- 30.Sena ES, >van der Worp HB, Bath PM, Howells DW, Macleod MR. Publication bias in reports of animal stroke studies leads to major overstatement of efficacy. PLoS Biol. 2010;8:e1000344. doi: 10.1371/journal.pbio.1000344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Seminara D, Khoury MJ, O'Brien TR, Manolio T, Gwinn ML, Little J, et al. The emergence of networks in human genome epidemiology: challenges and opportunities. Epidemiology. 2007;18:1–8. doi: 10.1097/01.ede.0000249540.17855.b7. [DOI] [PubMed] [Google Scholar]
- 32.Zhang LQ, Zhou JN, Wang J, Liang GD, Li JY, Zhu YD, et al. Absence of association between N-acetyltransferase 2 acetylator status and colorectal cancer susceptibility: based on evidence from 40 studies. PLoS One. 2012;7:e32425. doi: 10.1371/journal.pone.0032425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Siontis KC, Patsopoulos NA, Ioannidis JP. Replication of past candidate loci for common diseases and phenotypes in 100 genome-wide association studies. Eur J Hum Genet. 2010;18:832–7. doi: 10.1038/ejhg.2010.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ioannidis JP, Tarone R, McLaughlin JK. The false-positive to falsenegative ratio in epidemiologic studies. Epidemiology. 2011;22:450–6. doi: 10.1097/EDE.0b013e31821b506e. [DOI] [PubMed] [Google Scholar]
- 35.Pan Z, Trikalinos TA, Kavvoura FK, Lau J, Ioannidis JP. Local literature bias in genetic epidemiology: an empirical evaluation of the Chinese literature. PLoS Med. 2005;2:e334. doi: 10.1371/journal.pmed.0020334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vickers A, Goyal N, Harland R, Rees R. Do certain countries produce only positive results? A systematic review of controlled trials. Control Clin Trials. 1998;19:159–66. doi: 10.1016/s0197-2456(97)00150-5. [DOI] [PubMed] [Google Scholar]
- 37.Ioannidis JP. Integration of evidence from multiple meta-analyses: a primer on umbrella reviews, treatment networks and multiple treatments meta-analyses. CMAJ. 2009;181:488–93. doi: 10.1503/cmaj.081086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.International Agency for Research on Cancer . Preamble to the IARC Monographs (amended January 2006) International Agency for Research on Cancer; Lyon, France: 2009. [Google Scholar]
- 39.Ioannidis JP, Boffetta P, Little J, O'Brien TR, Uitterlinden AG, Vineis P, et al. Assessment of cumulative evidence on genetic associations: interim guidelines. Int J Epidemiol. 2008;37:120–32. doi: 10.1093/ije/dym159. [DOI] [PubMed] [Google Scholar]
- 40.Boffetta P, Winn DM, Ioannidis JP, Thomas DC, Little J, Smith GD, et al. Recommendations and proposed guidelines for assessing the cumulative evidence on joint effects of genes and environments on cancer occurrence in humans. Int J Epidemiol. 2012;41:686–704. doi: 10.1093/ije/dys010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Agency for Healthcare Research and Quality US Preventive Services Task Force. [cited 2012 Sep 1]; Available from: http://www.ahrq.gov/clinic/uspstfix.htm.
- 42.The guide to community preventive services [cited 2012 Sep 1]; Available from: http://www.thecommunityguide.org/index.html.
- 43.Lansdorp-Vogelaar I, Kuntz KM, Knudsen AB, van Ballegooijen M, Zauber AG, Jemal A. Contribution of screening and survival differences to racial disparities in colorectal cancer rates. Cancer Epidemiol Biomarkers Prev. 2012;21:728–36. doi: 10.1158/1055-9965.EPI-12-0023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kyzas PA, Loizou KT, Ioannidis JP. Selective reporting biases in cancer prognostic factor studies. J Natl Cancer Inst. 2005;97:1043–55. doi: 10.1093/jnci/dji184. [DOI] [PubMed] [Google Scholar]
- 45.Kyzas PA, Denaxa-Kyza D, Ioannidis JP. Quality of reporting of cancer prognostic marker studies: association with reported prognostic effect. J Natl Cancer Inst. 2007;99:236–43. doi: 10.1093/jnci/djk032. [DOI] [PubMed] [Google Scholar]
- 46.Kyzas PA, Denaxa-Kyza D, Ioannidis JP. Almost all articles on cancer prognostic markers report statistically significant results. Eur J Cancer. 2007;43:2559–79. doi: 10.1016/j.ejca.2007.08.030. [DOI] [PubMed] [Google Scholar]
- 47.Schoenfeld J, Ioannidis JPA. Is everything we eat associated with cancer? A systematic cookbook review. Am J Clin Nutr. 2012 Nov 28; doi: 10.3945/ajcn.112.047142. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 48.Altman DG, McShane LM, Sauerbrei W, Taube SE. Reporting recommendations for tumor marker prognostic studies (REMARK): explanation and elaboration. PLoS Med. 2012;9:e1001216. doi: 10.1371/journal.pmed.1001216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mallett S, Royston P, Waters R, Dutton S, Altman DG. Reporting performance of prognostic models in cancer: a review. BMC Med. 2010;8:21. doi: 10.1186/1741-7015-8-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mallett S, Timmer A, Sauerbrei W, Altman DG. Reporting of prognostic studies of tumour markers: a review of published articles in relation to REMARK guidelines. Br J Cancer. 2010;102:173–80. doi: 10.1038/sj.bjc.6605462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Baggerly KA, Coombes KR. Deriving chemosensitivity from cell lines: forensic bioinformatics and reproducible research in high-throughput biology. Ann Appl Stat. 2009;3:1309–34. [Google Scholar]
- 52.Begley CG, Ellis LM. Drug development: raise standards for preclinical cancer research. Nature. 2012;483:531–3. doi: 10.1038/483531a. [DOI] [PubMed] [Google Scholar]
- 53.Glasgow RE, Vinson C, Chambers D, Khoury MJ, Kaplan RM, Hunter C. National Institutes of Health approaches to dissemination and implementation science: current and future directions. Am J Public Health. 2012;102:1274–81. doi: 10.2105/AJPH.2012.300755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Deverka PA, Schully SD, Ishibe N, Carlson JJ, Freedman A, Goddard KA, et al. Stakeholder assessment of the evidence for cancer genomic tests: insights from three case studies. Genet Med. 2012;14:656–62. doi: 10.1038/gim.2012.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lenfant C. Shattuck lecture–clinical research to clinical practice–lost in translation? N Engl J Med. 2003;349:868–74. doi: 10.1056/NEJMsa035507. [DOI] [PubMed] [Google Scholar]
- 56.Andre F, McShane LM, Michiels S, Ransohoff DF, Altman DG, Reis-Filho JS, et al. Biomarker studies: a call for a comprehensive biomarker study registry. Nat Rev Clin Oncol. 2011;8:171–6. doi: 10.1038/nrclinonc.2011.4. [DOI] [PubMed] [Google Scholar]
- 57.Donoho DL, Maleki A, Rahman IU, Shahram M, Stodden V. Reproducible research in computational harmonic analysis. Comput Sci Eng. 2009;11:8–18. [Google Scholar]
- 58.Baggerly K. Disclose all data in publications. Nature. 2010;467:401. doi: 10.1038/467401b. [DOI] [PubMed] [Google Scholar]
- 59.Ioannidis JP, Khoury MJ. Improving validation practices in "omics" research. Science. 2011;334:1230–2. doi: 10.1126/science.1211811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.The Reproducibility Initiative [cited 2012 Sep 1]; Available from: https://www.scienceexchange.com/reproducibility.


