Abstract
Remarkably different conformations can result when DNA binds with stereoisomeric compounds containing differing absolute configurations of substituents about chiral carbon atoms. Furthermore, the biochemical functions of covalent adducts with DNA are strongly affected by the stereochemistry of the ligands. Such stereochemical effects are manifested by DNA covalent adducts derived from metabolites of the non-planar fjord region environmental chemical carcinogen benzo[c]phenanthrene. To analyze these phenomena, an extensive conformational investigation for R and S stereoisomeric adducts to deoxyadenosine, derived from trans addition of enantiomeric anti diol epoxide metabolites of benzo[c]phenanthrene, has been carried out. We have surveyed the potential energy surface of the two adducts by varying systematically at 5° intervals in combination, the three important torsion angles that govern conformational flexibility of the carcinogen bulk with respect to the linked nucleoside. We carried out a grid search by creating 373, 248 structures for each isomer, and evaluated their molecular mechanical energies. This has permitted us to map the potential energy surface of each adduct in these three variables, and to delineate their low energy regions. The maps have a symmetric relationship which stems from the near mirror-image stereochemistry in the R and S isomers. This produces near mirror-image low energy structures in the nucleoside adducts. The limited sets of stereoisomer-dependent conformational domains delineated are determined by steric effects. Moreover, these features have been experimentally demonstrated to play governing structural roles in such carcinogen-damaged DNA duplexes: opposite orientations in the stereoisomer pairs computed for the nucleosides are observed by high-resolution NMR in the similarly modified DNA double helices, and are likely to play important roles in their interactions with enzymes involved in DNA transactions, and hence their biological activities.
Keywords: Conformational search, Potential energy surface, Benzo[c]phenanthrene, Environmental carcinogen, Fjord region polycyclic aromatic hydrocarbon, Stereochemistry and steric effects
2. INTRODUCTION
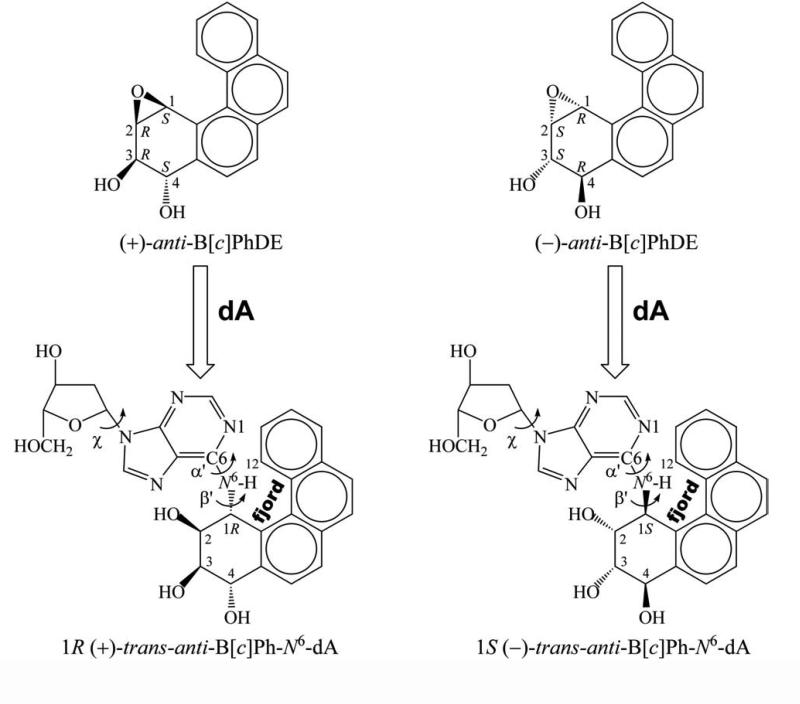
The biochemical functions of covalent adducts to DNA derived from metabolically activated polycyclic aromatic hydrocarbons (PAHs) are strongly affected by the stereochemistry of the ligands (1-5). Very different conformations can result when DNA binds with stereoisomeric compounds containing differing absolute configurations of substituents about chiral carbon atoms (6). Such stereochemical effects are manifested by DNA adducts derived from metabolites of the environmental chemical carcinogen benzo[c]phenanthrene. Benzo[c]phenanthrene (B[c]Ph) is representative of a class of polycyclic aromatic hydrocarbons (PAHs) containing a crowded fjord region which manifests deviation from planarity of the aromatic ring system (7). Metabolic activation of B[c]Ph produces diol epoxides, including a pair of enantiomers (3R,4S-dihydroxy-1S,2R-epoxy-1, 2, 3, 4-tetrahydrobenzo[c]phenanthrene and the corresponding 3S,4R,1R,2S isomer), known as (+)- and (−)-anti-B[c]PhDE, respectively (Figure 1) (8, 9). DNA can react with these metabolites to form covalently modified duplexes containing these bulky substituents (10). The stereoisomeric 1R (+) and 1S (−)-trans-anti-B[c]Ph-N6-dA adducts (Figure 1) are among these. Thermal melting studies revealed that these R and S adducted DNA duplexes have the same stabilities as the unmodified one (11). In addition, it has been found that they are not repaired by the nucleotide excision repair machinery (2) which removes bulky DNA adducts (12); thus these unrepaired adducts have the chance to encounter the DNA replication machinery. Mutations caused by the unfaithful replication of DNA adducts can contribute to cancer initiation via alteration in function of proto-oncogenes and tumor suppressor genes (13-15). Site specific mutagenesis studies show that the 1R (+) and 1S (−)-trans-anti-B[c]Ph-N6-dA adducts have different mutagenic specificities due to the different configurations of the chiral carbon at the linkage point (3).
Figure 1.
Structures of (+) and (−)-anti-B[c]PhDE and of 1R (+) and 1S (−)-trans-anti-B[c]Ph-N6-dA nucleoside adducts. Torsion angles χ, α’, β’ are defined as follows: χ, O4’-C1’-N9-C4; α’, N1-C6-N6-C1 (B[c]Ph); β’, C6- N6-C1(B[c]Ph)-C2(B[c]Ph). The fjord region encompasses the C1-C12 area.
High resolution NMR solution structures of adenine and guanine-B[c]PhDE adducts are available (16-18). These NMR structures reveal intercalation of the B[c]Ph moieties into the B-DNA duplex with opposite orientations of insertion with respect to the 5’ direction of the modified strand, in R and S isomers. The R isomer is 5’-directed while the S isomer is 3’-directed in adenine adducts (16, 17). In guanine adducts, the R isomer is 3’-directed while the S isomer is 5’-directed (18). Distorted Watson-Crick base pairing is retained in both cases.
The opposite orientation phenomenon of R versus S adducts has been extensively documented for the planar, non-hindered bay region PAH benzo[a]pyrene (6, 19-22) in both trans and cis opened guanine and adenine DNA adducts. However, the analogous adducts derived from the fjord region PAH B[c]Ph metabolites, which have remarkably different structural features (6) and biochemical functions (2-5, 23) due to the non-planar topology of the aromatic ring system (24), have not been fully investigated (25). As part of an effort to understand the origins of the structural motifs adopted by both bay and fjord region PAH–DNA adducts, we present results of an extensive computational investigation of the 1R (+) and 1S (−)-trans-anti-B[c]Ph-N6-dA adducts (Figure 1) on the nucleoside level. We have surveyed the potential energy surface of these two adducts by varying systematically at 5° intervals in combination, three key sources of conformational flexibility, the torsion angles χ, α’ and β’ (see Figure 1) of these two adducts. Thus, we created 373, 248 structures for each adduct, and evaluated each of their energies with AMBER 5.0 and the Cornell et al. force field (26). This has permitted us to map the potential energy surface of each adduct in these three variables, and to delineate the low energy regions and their relationship in the R/S adduct pair.
Our analyses reveal that the conformational preferences manifested on the nucleoside level, governed by stereochemistry, determine the orientations at the duplex DNA level, in these non-planar fjord region adducts. This work thus extends and generalizes a series of computational studies for R and S adducts of diol epoxide metabolites derived from both the non-planar fjord region benzo[c]phenanthrene and the planar bay region benzo[a]pyrene. The results show that favored nucleoside conformations govern observed conformational domains at the duplex DNA levels, with steric hindrance effects dominating the potential energy surface in all cases.
3. MATERIALS AND METHODS
3.1. Creating starting conformations
Coordinates of the high resolution NMR solution structure of the 1S (−)-trans-anti-B[c]Ph-N6-dA adduct in a double stranded DNA 11-mer sequence (17), were employed as the basis for the modified nucleoside structures. The modified nucleoside was excised from the duplex 11-mer of the 1S (−)-trans-anti-B[c]Ph-N6-dA adduct (17) and the 3’ and 5’ phosphorus atoms were replaced with hydrogen atoms. For the 1S (−)-trans-anti-B[c]Ph-N6-dA nucleoside adduct, this served as the starting model. To create a starting model for the 1R (+)-trans-anti-B[c]Ph-N6-dA, we constructed a mirror-image of the B[c]Ph in the 1S (−)-trans-anti-B[c]Ph-N6-dA adduct (by inverting the sign of the x coordinates) and then added this moiety to the nucleoside. The conformational details of these structures are given in Table A1, Appendix.
The present study employed fixed conformations for the OH-containing benzylic ring and the fjord region twist. The NMR solution structures in DNA duplexes suggest that these parameters can have some variability (16-18); however, these were not varied in the present comprehensive survey of the potential energy surface for the key torsion angles χ, α’, and β’ which govern the carcinogen–base orientation, and are the subject of another study (25).
Starting structures for the energy calculations were created with a torsion driver, a program which rotates the torsion angles χ, α’ and β’ (Figure 1) to chosen values and then computes the coordinates of the resulting structures. The starting structures uniformly surveyed the potential energy surface of the molecule with conformers in which χ, α’ and β’ sampled their 360° conformation space at 5° intervals, in combination, giving a total of (360°/5° = 72)3 = 373, 248 conformers for each adduct.
3.2. Energy computation
Energies of each of the 373, 248 structures for each adduct were computed with the molecular mechanics program AMBER 5.0 (27), using the Cornell et al. force field (26) with the parm96 parameter set. Since there are no negatively charged phosphates in the nucleoside, Na+ counterions were not needed. A sigmoidal distance-dependent dielectric function (28) was employed in the Coulombic term of the force field to model the dielectric effects of solvent water.
Parameters added to the AMBER 5.0 force field for the B[c]Ph-N6-dA adducts are the same as those given previously (21), except for the partial charges for the modified nucleosides; these were computed with Gaussian 94 (29) at the 6-31G* basis set level which is compatible with the rest of the Cornell et al. force field and parm96 parameter set; the least squares charge fitting algorithm RESP (30) provided with AMBER 5.0 was then used to fit the charge to each atomic center. Partial charges, atom types and topology information are given in Table A2, Appendix.
Computations were carried out at the Department of Energy's National Energy Research Supercomputer Center (Berkeley, CA), the National Science Foundation National Partnership for Advanced Computational Infrastructure (San Diego, CA) and on our own SGI workstations.
3.3. Statistical weights and thermodynamic quantities
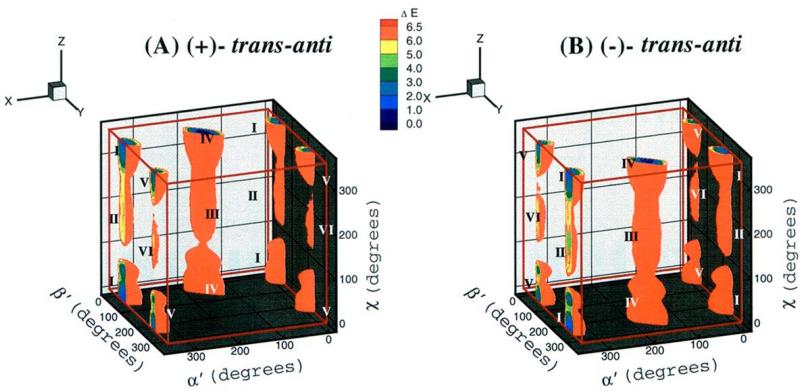
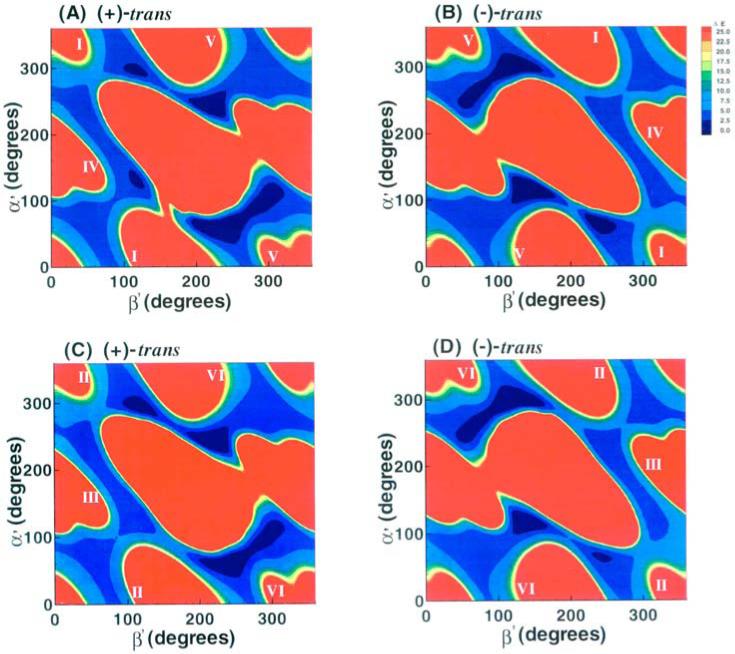
The computed energies were used to construct both three-dimensional energy surfaces as a function of the χ, α’, β’ torsion angles and two-dimensional energy contour slices through them; slices were made at 5° intervals of χ as a function of α’ and β’. The program TECPLOT was used for this purpose. These plots revealed six low-energy wells or domains (Figure 2). This permitted grouping the full data set into six regions, each of which encompassed one of the six low-energy domains:
Region 1: χ = 0°–105° and 240°–355°; α’ = 270°–355° and 0°–85°; β’ for 1R (+) adduct = 0°–155°; β’ for
 S (−) adduct = 200°–355°
S (−) adduct = 200°–355°Region 2: χ = 110°–235°; α’ = 270°–355° and 0°–85°; β’ for 1R (+) adduct = 0°–155°; β’ for 1S (−) adduct = 200°–355°
Region 3: χ =110°–235°; α’ = 90°–265°; β’ for 1R (+) adduct = 0°–155°; β’ for
 S (−) adduct = 200°–355°
S (−) adduct = 200°–355°Region 4: χ = 0°–105° and 240°–355°; α’ = 90°–265°; β’ for 1R (+) adduct = 0°–155°; β’ for 1S (−) adduct = 200°–355°
Region 5: χ = 0°–105° and 240°–355°; α’ = 0°–355°; β’ for 1R (+) adduct = 160°–355°; β’ for 1S (−) adduct = 0°–195°
Region 6: χ = 110°–235°; α’ = 0°–355°; β’ for 1R (+) adduct = 160°–355°; β’ for 1S (−) adduct = 0°–195°
Figure 2.
Three-dimensional χ, α’ and β’ energy topographies to 6.5 kcal/mol. I-VI denote the 6 low energy domains. (A), 1R(+)-trans-anti-B[c]Ph-N6-dA adduct; (B), 1S (−)-trans-anti-B[c]Ph-N6-dA adduct.
With this grouping, we computed fractional statistical weights as well as conformational free energies enthalpies, and entropies as in previous work (19-22).
The fractional statistical weight Pi of each conformer was computed from the relationship:
where Ei is the relative energy of a given conformer in kcal/mol with respect to the lowest energy structure, R is the universal gas constant, 1.987 × 10−3 kcal/mol·K, T =300 K, and N is 373, 248, the total number of conformers for each adduct.
Using the above grouping, we computed the combined fractional statistical weight, Wj, for each region (containing one low energy well) by summing the individual statistical weights, Pi, of each point in the region:
where nj is the number of conformers in each one of the six regions (j = 1, 2, ..., 6).
The conformational free energy of each well, Gj, was then computed from the relationship:
To obtain the conformational entropy of each well, Sj, we first compute pi, the fractional statistical weight of each conformer within its own well or region:
Then,
Conformational enthalpies for each well, Hj, are obtained from:
Computation of these thermodynamic parameters for each domain treats these as individual chemical species, which appears justifiable on the basis of the apparently high energy barriers between the domains (Figures 2 and 3). The program Insight II from Accelrys, Inc. was employed for computer graphic manipulation and visualization.
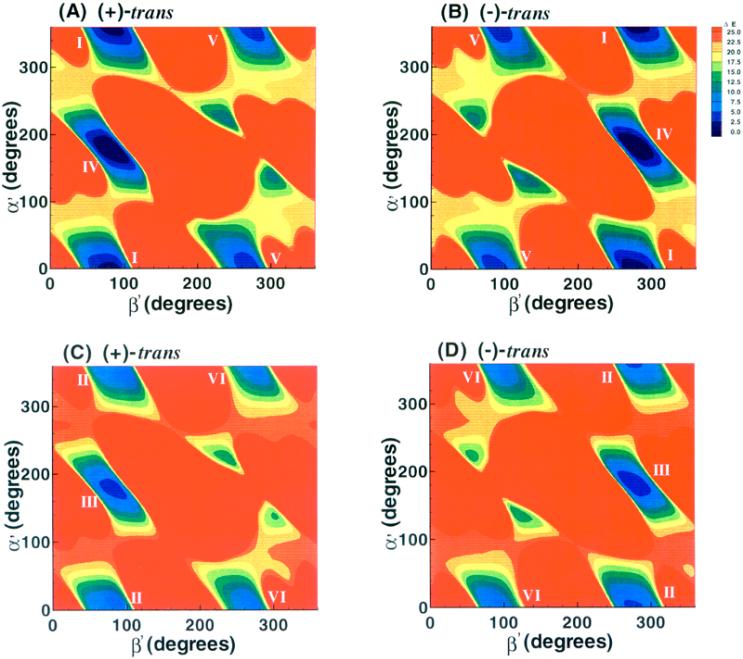
Figure 3.
α’, β’ energy contour maps to 25 kcal/mol. (A), 1R (+)-trans-anti-B[c]Ph-N6-dA adduct, χ = 10° (syn); (B), 1S (−)-trans-anti-B[c]Ph-N6-dA adduct, χ = 10° (syn); (C), 1R (+)-trans-anti-B[c]Ph-N6-dA adduct, χ = 200° (anti); (D), 1S (−)-trans-anti-B[c]Ph-N6-dA adduct, χ = 200° (anti).
4. RESULTS AND DISCUSSION
4.1. Symmetries in potential energy surfaces
The 3-dimensional energy topographies in χ, α’ and β’ to 6.5 kcal/mol are shown in Figure 2, for the 1R (+) and 1S (−)-trans-anti-B[c]Ph-N6-dA adducts. Each adduct has six low energy domains. Syn glycosidic torsion angle χ is found in domains I, IV and V, and domains II, III and VI are in the anti region. The ranges in the values of χ, α’ and β’ that characterize the low energy domains are given in Table 1.
Table 1.
Domains of Low-Energy Wells in 1R (+) and 1S (–)-trans-anti-B[c]Ph-N6-dA Adducts 1
| χ domain | α’ domain | β’ domain(deg) | ||
|---|---|---|---|---|
| (+) | (–) | |||
| domain | (deg) | (deg) | Adduct | Adduct |
| I | 10 + 100/– 130 | 0 + 40/– 45 | 90 + 35/–55 | 270 + 55/– 35 |
| II | 200 + 40/– 90 | 0 ± 30 | 90 + 25/– 45 | 270 + 45/– 25 |
| III | 200 + 40/– 90 | 180 + 40/– 35 | 90 + 25/– 50 | 270 + 50/– 25 |
| IV | 10 + 100/– 130 | 180 + 45/– 50 | 90 + 40/– 60 | 270 + 60/– 40 |
| V | 10 + 100/– 130 | 0 + 45/– 40 | 270 + 35/– 45 | 90 + 45/– 35 |
| VI | 200 + 40/– 90 | 0 + 35/– 30 | 270 ± 25 | 90 ± 25 |
Energy contour maps in the α’ and β’ plane (Figure 3) were created from slices through the approximate centers of the 3-dimensional plots in the syn (10°) and anti (200°) domains of χ. The 2-dimensional 1R (+) and 1S (−)-trans-anti adduct maps are symmetrical: the map for the 1R (+) adduct can be essentially transformed into that of the 1S (−) adduct by inverting the sign of the torsion angles α’ and β’, corresponding to an ~180° rotation about a central symmetry axis positioned perpendicular to the map plane at α’, β’ = 180°, 180°. Thus, a given energy feature in a specific α’, β’ region of the 1R (+) adduct map, has a corresponding feature in the 1S (−) adduct map in the −α’, −β’ region. This torsion angle symmetry is a hallmark of mirror-image pairs of molecules, although the symmetry is not exact in our nucleoside adduct pair.
The α’, β’ energy contour maps at 5° intervals of χ for each adduct reveal the same symmetries as in Figure 3. Table A3, Appendix, gives the number of conformers in 1 kcal/mol shells and Table 2 gives computed thermodynamic parameters and statistical weights. Differences in population between the structures of the 1R (+) and 1S (−) adduct stem from the symmetry breaking effect of the sugar residues and C4’-C5’ atoms. We note that domain IV (discussed below) is the most preferred domain, and that favorable conformational entropy plays the key part in stabilizing it.
Table 2.
Computed Statistical Weights and Thermodynamics Parameters for Low-Energy Domains in 1R (+) and 1S (–)-trans-anti-B[c]Ph-N6-dA Adducts a
| W% | G | H | TS | |||||
|---|---|---|---|---|---|---|---|---|
| (kcal/mol) | (kcal/mol) | (kcal/mol) | ||||||
| Domain | (+) | (–) | (+) | (–) | (+) | (–) | (+) | (–) |
| I | 9.440 | 12.593 | –1.537 | –1.687 | 2.464 | 2.287 | 4.001 | 3.974 |
| II | 0.044 | 0.082 | 1.666 | 1.313 | 5.756 | 5.391 | 4.090 | 4.078 |
| III | 0.338 | 0.812 | 0.448 | –0.052 | 4.345 | 3.856 | 3.896 | 3.909 |
| IV | 88.430 | 84.785 | –2.871 | –2.824 | 0.955 | 0.997 | 3.826 | 3.821 |
| V | 1.741 | 1.714 | –0.529 | –0.498 | 3.240 | 3.249 | 3.769 | 3.747 |
| VI | 0.007 | 0.014 | 2.760 | 2.376 | 6.613 | 6.226 | 3.853 | 3.849 |
Statistical weights, W, are given in percents of the population. G, H, and S are conformational free energy, enthalpy, and entropy, respectively. T = 300 K
4.2. Near mirror-image symmetries in low energy conformations
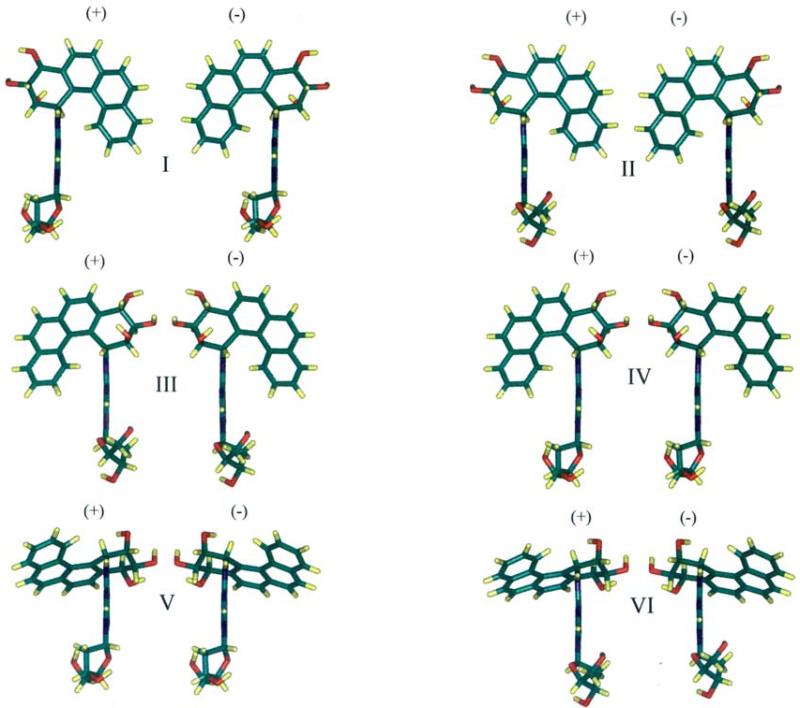
Color views of representative structures of each of the six low energy domains of the 1R (+) and 1S (−)-trans-anti adducts are shown in Figure 4. The 1R (+) adduct structure is nearly a mirror-image of the 1S (−) one in each domain, with the mirror-image symmetry broken by the sugars, whose O4’ atoms point in the same rather than the opposite directions in each pair.
Figure 4.
Color views of representative structures for 1R (+) and 1S (−)-trans-anti-B[c]Ph-N6-dA adducts. In each pair the left structure is the 1R (+)-trans-anti-B[c]Ph-N6-dA adduct and the right is the 1S (−) adduct. χ, α’, and β’ values and energies, ΔE, relative to the global minimum are the following. Domain I: 1R (+) adduct 10°, 0°, 90°; 1S (−) adduct 10°, 0°, 270°; ΔE +1.85, −1.36 kcal/mol. Domain II: 1R (+) adduct 200°, 0°, 90°; 1S (−) adduct 200°, 0°, 270°; ΔE +5.14, −4.73 kcal/mol. Domain III: 1R (+) adduct 200°, 180°, 90°; 1S (−) adduct: 200°, 180°, 270°; ΔE +3.87, −3.20 kcal/mol. Domain IV: 1R (+) adduct 10°, 180°, 90°; 1S (−) adduct 10°, 180°, 270°; ΔE +0.55, −0.22 kcal/mol. Domain V: 1R (+) adduct 10°, 0°, 270°; 1S (−) adduct 10°, 0°, 90°; ΔE +2.55, −2.28 kcal/mol. Domain VI: 1R (+) adduct 200°, 0°, 270°; 1S (−) adduct 200°, 0°, 90°; ΔE +6.01, −5.47 kcal/mol. The view is edge-on along the adenine with C8H directed toward the viewer.
4.3. Structural domain preferences
Syn and Anti Conformations
Domains I (syn) and II (anti) are essentially the same except for the difference in glycosidic torsion angles. The stabilization of domain I relative to domain II probably stems from the fact that the sugar residue has favorable interactions with both aromatic rings of the adenine in the syn conformation; when the adenine residue is in the anti orientation, the sugar contacts only its 5-membered ring. This characteristic is intrinsic to the deoxyadenosine moiety and is not significantly influenced by the B[c]Ph residue. Syn domains IV and V are favored over anti domains III and VI, respectively, for the same reason. The syn domain preference in nucleosides is overcome in DNA duplexes through maintenance of Watson-Crick base pairing, which requires the anti domain in B-DNA, as discussed below.
α’ and β’ Regions
Table 1 reveals that α’ is centered at 0° or 180° in each of the low energy domains, and Table 2 shows that the α’=180° region is favored more. The preference for these two α’ regions stems from the maximal overlap of the adenine aromatic ring π orbitals with the N6 lone pair p orbital when α’ = ~0° or ~180°.
With these α’ values, the preferred β’ are the 90° region for the 1R (+) isomer and the 270° (− 90°) region for the 1S (−) isomer; other combinations of α’ and β’ produced crowded structures. In normal B-DNA duplexes, the α’ = ~180° region is required to form the Watson-Crick hydrogen bond that involves the free hydrogen atom on N6-dA (6).
4.4. Steric hindrance produces opposite orientations in R and S isomers
Energy contour maps in the α’, β’ plane which considered only the van der Waals component of the total energy helped to elucidate the origin of the opposite orientations (Figure 5). For each adduct, these maps contain the same overall regions below 25 kcal/mol, with the same symmetry effects as those employing the full energy potential (Figure 3). The energy boundaries above 25 kcal/mol are determined by repulsive steric components of the Lennard-Jones potential employed to compute van der Waals energies (31), and hence steric repulsions are predominantly responsible for the different energy landscapes in the 1R (+) and 1S (−)-trans adducts and for the symmetries between them. The energy regions below 25 kcal/mol contain differing details in the total energy maps (Figure 3) than in the van der Waals component maps (Figure 5), due to modulating effects of electrostatic, torsional and other terms in the total energy.
Figure 5.
α’, β’ van der Waals energy component maps to 25 kcal/mol. (A), 1R (+)-trans-anti-B[c]Ph-N6-dA adduct, χ = 10° (syn); (B), 1S(−)-trans-anti-B[c]Ph-N6-dA adduct, χ = 10° (syn); (C), 1R (+)-trans-anti-B[c]Ph-N6-dA adduct, χ = 200° (anti); (D), 1S(−)-trans-anti-B[c]Ph-N6-dA adduct, χ = 200° (anti).
To further examine the nature of the steric repulsions, we rotated each structure shown in Figure 4 into that preferred by its stereoisomer, a 180° rotation around β’ in each case. For domains I and II of both 1R (+) and 1S (−) adducts, after rotation, the N1 of the purine is in a crowded region with the C4A, C4B and H3 of the B[c]Ph moiety; for domains III and IV, the rotated structures show crowding between N7 of the adenine and C4A, C4B and H3 of the B[c]Ph; the rotated structures of domains V and VI, however, show no crowding. After rotation, domains V and VI are close to allowed domains I and II, respectively. The crowding between atoms on the adenine and the benzylic ring of the B[c]Ph residues caused by rotation of β’ from the region preferred by one stereoisomer into the region preferred by the other has been termed “primary” steric hindrance (6).
4.5. Nucleoside conformations govern solution structures of DNA duplexes
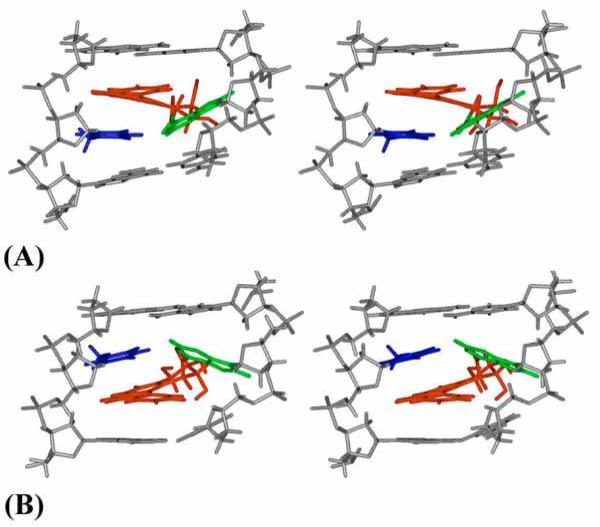
The footnote to Table 1 summarizes the χ, α’, β’ values adopted by the 1R (+) and 1S (−)-trans-anti-B[c]Ph-N6-dA adducts (16, 17) in double stranded DNA, determined from the analysis of high resolution NMR solution data. Both of the B[c]Ph NMR solution structures adopt α’, β’ combinations in the vicinity of domain IV, which is most favored for each adduct on the nucleoside level (Table 2). In the observed domain IV, the B[c]Ph moiety is placed on the 3’-side of the modified adenine in the 1S (−)-trans-anti adduct, and on the 5’-side in the 1R (+)-trans-anti-isomeric adduct in nucleosides and duplexes. However, in the duplexes distorted Watson-Crick base pairing is maintained. This is permitted by the 180° region adopted by torsion angle α’ (Figures 4 and 6), but also requires the anti domain for the glycosidic torsion angle χ, found on the nucleoside level in Domain III. The torsion angle β’ adopts values of ~ −90° for the 1S (−) and ~ +90° for the 1R (+)-trans-anti isomer (Table 1). Thus, the α’, β’ combination, computed in nucleosides and observed in duplexes, gives rise to the opposite orientations of the B[c]Ph, relative to the modified deoxyadenosyl residues.
Figure 6.
Stereo views of central modified duplex 5-mer NMR solution structures. The (A) 1R (+)-trans-anti-B[c]Ph-N6-dA (16) and (B) 1S (−)-trans-anti-B[c]Ph-N6-dA (17) adducts. B[c]Ph is red, modified adenine is green, and the partner thymine is blue. Backbone, sugar, and other nucleotide atoms are in gray.
These results are consistent with our previous studies on R and S benzo[c]phenanthrene diol epoxide guanine adducts (25), and on a number of R and S nucleoside adducts of diol epoxide metabolites derived from the planar bay region PAH, benzo[a]pyrene (19-22). In all cases symmetric potential energy surfaces are manifested in nucleosides and opposite orientations are observed in NMR solution structures of DNA duplexes (18, 32-35). Thus, our findings regarding opposite orientations of S and R isomers appear to be general, encompassing both planar bay and non-planar fjord region PAHs. The particular type of conformational theme adopted by the aromatic moiety in DNA duplexes, namely (1) at the helix exterior in a groove; (2) inserted into the helix with displacement of the covalently modified base to the helix exterior; or (3) intercalation with maintenance of Watson–Crick base pairing, is governed by a subtle interplay of hydrogen bonding, hydrophobic and steric forces involving both the aromatic ring of the carcinogen and the DNA (6). These are determined by the specific stereochemistry and adduction site of the differing isomers. Modulating conformational effects, stemming from flexibility in benzylic ring conformation and non-planarity in the aromatic ring system, observed in the NMR solution structures of these sterically hindered fjord region adducts (16-18) are considered in another investigation (25). However, opposite orientations are adopted by R and S stereoisomers irrespective of the specific conformational theme. It is quite remarkable that the opposite orientations in all polycyclic aromatic R and S adducted duplexes observed so far can be fully explained by the nucleoside conformations. These structural distinctions may play a role in producing differential mutagenic specificities observed in R and S isomeric adducts (3, 4, 36)
5. BIOLOGICAL IMPLICATIONS
Our comprehensive survey of the potential energy surface for a stereoisomeric pair of small molecules, namely carcinogen-linked nucleosides, defined the energetic and structural features which govern the orientation of the carcinogen with respect to its covalently bound base. The computations delineated limited sets of allowed conformational domains that are stereoisomer-dependent and determined by steric effects. These features have been experimentally demonstrated to play governing structural roles in such carcinogen-damaged DNA duplexes: opposite orientations in the stereoisomer pairs computed for the nucleosides are observed by high-resolution NMR in the similarly modified DNA double helices. It is plausible that the favored conformational domains would occur within enzymes that process these lesions. Observed differences in biological activities such as mutagenicity in the stereoisomer pair likely result from differential treatment by polymerases, which stem from the opposite orientations. Finally, this study underscores the power of the conformational searches to define conformations of small, biologically important molecules of limited flexibility, reveals their applicability to relevant macromolecules, and points to functional implications of the structural findings. Analogous approaches in defining conformational features, the role of stereochemistry, and their relation to biological activity in small ligands that may interact with proteins can be useful techniques in computer aided drug discovery.
ACKNOWLEDGEMENTS
This research is supported by NIH grant CA-28038 to S.B., NIH grant CA-46533 to D.J.P., and NIH grant CA-20851 to N.E.G.
8. APPENDIX
Table A1.
Conformational Features of Modified Nucleosides
| Dihedral Angle | 1R (+)-trans-anti-B[c]Ph-N6-A (deg) | 1S (–)-trans-anti-B[c]Ph-N6-A (deg) |
|---|---|---|
| C1-C2-C3-C4 | 54.30 | −54.30 |
| C2-C3-C4-C4A | −48.71 | 48.71 |
| C3-C4-C4A-C1A | 27.41 | −27.41 |
| C4-C4A-C1A-C1 | −9.14 | 9.14 |
| C4A-C1A-C1-C2 | 14.28 | −14.28 |
| C1A-C1-C2-C3 | −38.00 | 38.00 |
| C1-C2-O2-HO2 | 51.30 | 51.30 |
| C2-C3-O3-HO3 | −71.79 | 71.79 |
| C3-C4-O4-HO4 | −62.15 | 62.15 |
| C4B-C6B-C8B-C12 | 15.78 | −15.78 |
| C1-C4B-C6B-C8B | 9.15 | −9.15 |
| O4N-C1N-C2N-C3N | 37.16 | 37.16 |
| C1N-C2N-C3N-C4N | −36.55 | −36.55 |
| C2N-C3N-C4N-O4N | 23.63 | 23.63 |
| C3N-C4N-O4N-C1N | −0.44 | −0.44 |
| C4N-O4N-C1N-C2N | −23.22 | −23.22 |
| O4N-C4N-C5N-O5N | −56.54 | −56.54 |
| C4N-C5N-O5N-H5N | 2.40 | 2.40 |
| C2N-C3N-O3N-H3N | −39.79 | −39.79 |
| C3N-C4NC5N-O5N | 62.26 | 62.26 |
Table A2.
Partial Charges, Topology, and Atom Type Assignments
| Charges | |||||
|---|---|---|---|---|---|
| Atom | Type | Topology | 1R(+)-B[c]Ph | 1S(–)-B[c]Ph | |
| dA: | HO5 | HO | M | 0.369402 | 0.384554 |
| O5′ | OH | M | −0.597676 | −0.640506 | |
| C5′ | CT | M | 0.063023 | 0.215272 | |
| H5′1 | H1 | E | 0.077803 | 0.032584 | |
| H5′2 | H1 | E | 0.121727 | 0.075972 | |
| C4′ | CT | M | 0.032341 | 0.003844 | |
| H4′ | H1 | E | 0.115023 | 0.093768 | |
| O1′ | OS | S | −0.356514 | −0.350765 | |
| C1′ | CT | B | 0.054737 | −0.002370 | |
| H1′ | H2 | E | 0.127963 | 0.199964 | |
| N9 | N* | S | 0.039077 | −0.029234 | |
| C8P | CK | B | −0.016961 | 0.051096 | |
| H8P | H5 | E | 0.173580 | 0.192984 | |
| N7 | NB | S | −0.427606 | −0.485960 | |
| C5P | CB | S | 0.024748 | 0.071670 | |
| C4P | CB | S | 0.375799 | 0.411124 | |
| N3 | NC | S | −0.688109 | −0.737523 | |
| C2P | CQ | B | 0.591722 | 0.627065 | |
| H2P | H5 | E | 0.046343 | 0.040777 | |
| N1 | NC | S | −0.644863 | −0.654923 | |
| C6P | CA | S | 0.483293 | 0.463303 | |
| N6 | N2 | B | −0.562482 | −0.546219 | |
| HN6 | H | E | 0.333586 | 0.329206 | |
| C3′ | CT | M | 0.205377 | 0.337776 | |
| H3′ | H1 | E | 0.126271 | 0.093759 | |
| C2′ | CT | B | −0.130653 | −0.267874 | |
| H2′1 | HC | E | 0.083002 | 0.118957 | |
| H2′2 | HC | E | 0.067844 | 0.075082 | |
| O3′ | OH | M | −0.651333 | −0.673750 | |
| HO3 | HO | M | 0.408315 | 0.413234 | |
| B[c]Ph: | C1Z | CT | B | −0.012501 | 0.009626 |
| H1Z | H1 | E | 0.148079 | 0.128628 | |
| C2Z | CT | 3 | 0.009910 | 0.067960 | |
| H2Z | H1 | E | 0.156274 | 0.147212 | |
| O2Z | OH | S | −0.600130 | −0.610844 | |
| HO2Z | HO | E | 0.373496 | 0.372272 | |
| C3Z | CT | 3 | 0.201143 | 0.200388 | |
| H3Z | H1 | E | 0.111411 | 0.094092 | |
| O3Z | OH | S | −0.657781 | −0.659476 | |
| HO3Z | HO | E | 0.426531 | 0.419514 | |
| C4Z | CT | 3 | 0.156935 | 0.226752 | |
| H4Z | H1 | E | 0.037687 | 0.015203 | |
| O4Z | OH | S | −0.631642 | −0.644431 | |
| HO4Z | HO | E | 0.411246 | 0.411158 | |
| C4A | CA | S | 0.020305 | 0.002818 | |
| C5Z | CA | B | −0.126848 | −0.150585 | |
| H5Z | HA | E | 0.145944 | 0.149816 | |
| C6Z | CA | B | −0.234776 | −0.201667 | |
| H6Z | HA | E | 0.161741 | 0.155844 | |
| C6A | CA | S | 0.064061 | 0.065413 | |
| C7Z | CA | B | −0.125625 | −0.164495 | |
| H7Z | HA | E | 0.130481 | 0.137207 | |
| C8Z | CA | B | −0.267977 | −0.215210 | |
| H8Z | HA | E | 0.155668 | 0.144279 | |
| C8A | CA | S | 0.179590 | 0.148056 | |
| C9Z | CA | B | −0.209005 | −0.214245 | |
| H9Z | HA | E | 0.147059 | 0.152141 | |
| C10 | CA | B | −0.136028 | −0.150137 | |
| H10 | HA | E | 0.136226 | 0.138240 | |
| C11 | CA | B | −0.110064 | −0.060934 | |
| H11 | HA | E | 0.138868 | 0.108273 | |
| C12 | CA | B | −0.236757 | −0.301061 | |
| H12 | HA | E | 0.167586 | 0.194036 | |
| C8B | CA | S | 0.065726 | 0.089404 | |
| C6B | CA | S | −0.011029 | 0.010791 | |
| C4B | CA | E | −0.030583 | −0.058906 | |
Table A3.
Number of Conformations in 1 kcal/mol Shells in 1R (+) and 1S (–)-trans-anti-B[c]Ph-N6-dA Adducts, to 5 kcal/mol
| 0-1 kcal/mol | 1-2 kcal/mol | 2-3 kcal/mol | 3-4 kcal/mol | 4-5 kcal/mol | Total | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Domain | (+) | (–) | (+) | (–) | (+) | (–) | (+) | (–) | (+) | (–) | (+) | (–) |
| I | 0 | 0 | 81 | 127 | 430 | 462 | 629 | 621 | 780 | 766 | 1920 | 1976 |
| II | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3 | 110 | 3 | 110 |
| III | 0 | 0 | 0 | 0 | 0 | 1 | 93 | 297 | 429 | 485 | 522 | 783 |
| IV | 211 | 188 | 395 | 395 | 530 | 520 | 555 | 560 | 785 | 784 | 2476 | 2447 |
| V | 0 | 0 | 0 | 0 | 113 | 93 | 306 | 312 | 489 | 455 | 908 | 860 |
| VI | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Total | 211 | 188 | 476 | 522 | 1073 | 1076 | 1583 | 1790 | 2486 | 2600 | 5829 | 6176 |
REFERENCES
- 1.Chary P, Lloyd RS. In vitro replication by prokaryotic and eukaryotic polymerases on DNA templates containing site-specific and stereospecific benzo[a]pyrene-7, 8-dihydrodiol-9, 10-epoxide adducts. Nucleic Acids Res. 1995;23:1398–1405. doi: 10.1093/nar/23.8.1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Buterin T, Hess MT, Luneva N, Geacintov NE, Amin S, Kroth H, Seidel A, Naegeli H. Unrepaired fjord region polycyclic aromatic hydrocarbon-DNA adducts in ras codon 61 mutational hot spots. Cancer Res. 2000;60:1849–1856. [PubMed] [Google Scholar]
- 3.Ponten I, Sayer JM, Pilcher AS, Yagi H, Kumar S, Jerina DM, Dipple A. Factors determining mutagenic potential for individual cis and trans opened benzo[c]phenanthrene diol epoxide-deoxyadenosine adducts. Biochemistry. 2000;39:4136–4144. doi: 10.1021/bi991719q. [DOI] [PubMed] [Google Scholar]
- 4.Ramos LA, Ponten I, Dipple A, Kumar S, Yagi H, Sayer JM, Kroth H, Kalena G, Jerina DM. Site-specific mutagenesis in Escherichia coli by N2-deoxyguanosine adducts derived from the highly carcinogenic fjord-region benzo[c]phenanthrene 3, 4-diol 1, 2-epoxides. Chem Res Toxicol. 2002;15:1619–1626. doi: 10.1021/tx020073r. [DOI] [PubMed] [Google Scholar]
- 5.Rechkoblit O, Zhang Y, Guo D, Wang Z, Amin S, Krzeminsky J, Louneva N, Geacintov NE. Translesion synthesis past bulky benzo[a]pyrene diol epoxide N2-dG and N6-dA lesions catalyzed by DNA bypass polymerases. J Biol Chem. 2002;277:30488–30494. doi: 10.1074/jbc.M201167200. [DOI] [PubMed] [Google Scholar]
- 6.Geacintov NE, Cosman M, Hingerty BE, Amin S, Broyde S, Patel DJ. NMR solution strucutures of stereoisomeric covalent polycyclic aromatic carcinogen-DNA adducts: principles, patterns, and diversity. Chem Res Toxicol. 1997;10:111–146. doi: 10.1021/tx9601418. [DOI] [PubMed] [Google Scholar]
- 7.Hirshfeld FL. The structure of overcrowded aromatic compounds. Part VII. Out-of-plane deformation in benzo[c] phenanthrene and 1, 12-dimethyl-benzo[c]phenanthrene. J Chem Soc. 1963:2126–2135. [Google Scholar]
- 8.Levin W, Wood AW, Chang RL, Ittah Y, Croisy-Delcey M, Yagi H, Jerina DM, Conney AH. Exceptionally high tumor-initiating activity of benzo[c]phenanthrene bay-region diol-epoxides on mouse skin. Cancer Res. 1980;40:3910–3914. [PubMed] [Google Scholar]
- 9.Thakker DR, Levin W, Yagi H, Yeh HJ, Ryan DE, Thomas PE, Conney AH, Jerina DM. Stereoselective metabolism of the (+)-(S,S)- and (−)-(R,R)-enantiomers of trans-3, 4-dihydroxy-3, 4-dihydrobenzo[c]-phenanthrene by rat and mouse liver microsomes and by a purified and reconstituted cytochrome P-450 system. J Biol Chem. 1986;261:5404–5413. [PubMed] [Google Scholar]
- 10.Agarwal SK, Sayer JM, Yeh HJC, Pannell LK, Hilton BD, Pigott MA. Chemical characterization of DNA adducts derived from the configurationally isomeric benzo[c]phenanthrene-3, 4-diol-1, 2-epoxides. J Am Chem Soc. 1987:2497–2504. [Google Scholar]
- 11.Laryea A, Cosman M, Lin JM, Liu T, Agarwal R, Smirnov S, Amin S, Harvey RG, Dipple A, Geacintov NE. Direct synthesis and characterization of site-specific adenosyl adducts derived from the binding of a 3, 4-dihydroxy-1, 2-epoxybenzo[c]phenanthrene stereoisomer to an 11-mer oligodeoxyribonucleotide. Chem Res Toxicol. 1995;8:444–454. doi: 10.1021/tx00045a017. [DOI] [PubMed] [Google Scholar]
- 12.Wood RD. DNA damage recognition during nucleotide excision repair in mammalian cells. Biochimie. 1999;81:39–44. doi: 10.1016/s0300-9084(99)80036-4. [DOI] [PubMed] [Google Scholar]
- 13.Weinberg RA. How cancer arises. Sci Am. 1996;275:62–70. doi: 10.1038/scientificamerican0996-62. [DOI] [PubMed] [Google Scholar]
- 14.Garner RC. The role of DNA adducts in chemical carcinogenesis. Mutat Res. 1998;402:67–75. doi: 10.1016/s0027-5107(97)00283-2. [DOI] [PubMed] [Google Scholar]
- 15.Gibbs WW. Untangling the roots of cancer. Sci Am. 2003;289:56–65. doi: 10.1038/scientificamerican0703-56. [DOI] [PubMed] [Google Scholar]
- 16.Cosman M, Fiala R, Hingerty BE, Laryea A, Lee H, Harvey RG, Amin S, Geacintov NE, Broyde S, Patel D. Solution conformation of the (+)-trans-anti-[BPh]dA adduct opposite dT in a DNA duplex: intercalation of the covalently attached benzo[c]phenanthrene to the 5′-side of the adduct site without disruption of the modified base pair. Biochemistry. 1993;32:12488–12497. doi: 10.1021/bi00097a029. [DOI] [PubMed] [Google Scholar]
- 17.Cosman M, Laryea A, Fiala R, Hingerty BE, Amin S, Geacintov NE, Broyde S, Patel DJ. Solution conformation of the (−)-trans-anti-benzo[c]phenanthrene-dA ([BPh]dA) adduct opposite dT in a DNA duplex: intercalation of the covalently attached benzo[c]phenanthrenyl ring to the 3′-side of the adduct site and comparison with the (+)-trans-anti-[BPh]dA opposite dT stereoisomer. Biochemistry. 1995;34:1295–1307. doi: 10.1021/bi00004a024. [DOI] [PubMed] [Google Scholar]
- 18.Lin C, Huang X, Kolbanovskii A, Hingerty BE, Amin S, Broyde S, Geacintov NE, Patel DJ. Molecular topology of polycyclic aromatic carcinogens determines DNA adduct conformation: a link to tumorigenic activity. J Mol Biol. 2001;306:1059–1080. doi: 10.1006/jmbi.2001.4425. [DOI] [PubMed] [Google Scholar]
- 19.Xie XM, Geacintov NE, Broyde S. Stereochemical origin of opposite orientations in DNA adducts derived from enantiomeric anti-benzo[a]pyrene diol epoxides with different tumorigenic potentials. Biochemistry. 1999;38:2956–2968. doi: 10.1021/bi9825605. [DOI] [PubMed] [Google Scholar]
- 20.Xie XM, Geacintov NE, Broyde S. Origins of conformation differences between cis and trans DNA adducts derived from enantiomeric anti-benzo[a]pyrene diol epoxides. Chem Res Toxicol. 1999;12:597–609. doi: 10.1021/tx990021a. [DOI] [PubMed] [Google Scholar]
- 21.Tan J, Geacintov NE, Broyde S. Principles governing conformations in stereoisomeric adducts of bay region benzo[a]pyrene diol epoxides to adenine in DNA: steric and hydrophobic effects are dominant. J Am Chem Soc. 2000;122:3021–3032. [Google Scholar]
- 22.Tan J, Geacintov NE, Broyde S. Conformational determinants structures in stereoisomeric cis-opened anti-benzo[a]pyrene diol epoxide adducts to adenine in DNA. Chem Res Toxicol. 2000;13:811–822. doi: 10.1021/tx000094q. [DOI] [PubMed] [Google Scholar]
- 23.Frank EG, Sayer JM, Kroth H, Ohashi E, Ohmori H, Jerina DM, Woodgate R. Translesion replication of benzo[a]pyrene and benzo[c]phenanthrene diol epoxide adducts of deoxyadenosine and deoxyguanosine by human DNA polymerase ι. Nucleic Acids Res. 2002;30:5284–5292. doi: 10.1093/nar/gkf643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wu M, Yan S, Patel DJ, Geacintov NE, Broyde S. Relating repair susceptibility of carcinogen-damaged DNA with structural distortion and thermodynamic stability. Nucleic Acids Res. 2002;30:3422–3432. doi: 10.1093/nar/gkf427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu M, Yan S, Patel DJ, Geacintov NE, Broyde S. Cyclohexene ring and fjord region twist inversion in stereoisomeric DNA adducts of enantiomeric benzo[c]phenanthrene diol epoxides. Chem Res Toxicol. 2001;14:1629–1642. doi: 10.1021/tx010152n. [DOI] [PubMed] [Google Scholar]
- 26.Cornell WD, Cieplak P, Bayly CI, Gould IR, Merz KM, Ferguson DM, Spellmeyer DC, Fox T, Caldwell JW, Kollman PA. A second generation force field for the simulation of proteins and nucleic acids. J Am Chem Soc. 1995;117:5179–5197. [Google Scholar]
- 27.Case D, Pearlman D, Caldwell J, Cheatham T, Ross W, Simmerling C, Darden T, Merz K, Stanton R, Cheng A, Vincent J, Crowley M, Ferguson D, Radner R, Seibel G, Singh UC, Weiner P, Kollman P. AMBER 5.0. University of California; San Francisco, CA: 1997. [Google Scholar]
- 28.Hingerty BE, Ritchie RH, Ferrell TL, Turner JE. Dielectric effects in biopolymers: the theory of inonic saturation revisited. Biopolymers. 1985;24:427–439. [Google Scholar]
- 29.Frisch MJ, Trucks GW, Schlegel HB, Gill P, Johnsone BG, Robb MA, Cheeseman JR, Keith TA, Petersson GA, Montgomery JA, Raghavachari K, Al-Laham MA, Zakrzewski VG, Ortiz JV, Foresman JB, Cioslowski J, Stefanov BB, Nanayakkara A, Challacombe M, Peng CY, Ayala PY, Chen W, Wong MW, Andres JL, Reploge ES, Gomperts R, Martin RL, Fox DJ, Binkley JS, Defrees DJ, Baker J, Stewart JP, Head-Gordon M, Gonzalez C, Pople JA. Gaussian 94 (Revision A.1) Pittsburgh PA: 1995. [Google Scholar]
- 30.Bayly CI, Cieplak P, Cornell WD, Kollman PA. A well-behaved electrostatic potential based method using charge restraints for deriving atomic charges: the RESP model. J Phys Chem. 1993;97:10269–10280. [Google Scholar]
- 31.Atkins PW. Physical Chemistry. 4th ed. W. H. Freeman Co.; New York: 1990. [Google Scholar]
- 32.Cosman M, de los Santos C, Fiala R, Hingerty BE, Singh SB, Ibanez V, Margulis LA, Live D, Geacintov NE, Broyde S. Solution conformation of the major adduct between the carcinogen (+)- anti-benzo[a]pyrene diol epoxide and DNA. Proc Natl Acad Sci USA. 1992;89:1914–1918. doi: 10.1073/pnas.89.5.1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cosman M, de los Santos C, Fiala R, Hingerty BE, Ibanez V, Luna E, Harvey R, Geacintov NE, Broyde S, Patel DJ. Solution conformation of the (+)-cis-anti-[BP]dG adduct in a DNA duplex: intercalation of the covalently attached benzo[a]pyrenyl ring into the helix and displacement of the modified deoxyguanosine. Biochemistry. 1993;32:4145–4155. doi: 10.1021/bi00067a001. [DOI] [PubMed] [Google Scholar]
- 34.Cosman M, Hingerty BE, Luneva N, Amin S, Geacintov NE, Broyde S, Patel DJ. Solution conformation of the (−)-cis-anti-benzo[a]pyrenyl-dG adduct opposite dC in a DNA duplex: intercalation of the covalently attached BP ring into the helix with base displacement of the modified deoxyguanosine into the major groove. Biochemistry. 1996;35:9850–9863. doi: 10.1021/bi9605346. [DOI] [PubMed] [Google Scholar]
- 35.de los Santos C, Cosman M, Hingerty BE, Ibanez V, Margulis LA, Geacintov NE, Broyde S, Patel DJ. Influence of benzo[a]pyrene diol epoxide chirality on solution conformations of DNA covalent adducts: the (−)-trans-anti-[BP]G.C adduct structure and comparison with the (+)-trans-anti-[BP]G.C enantiomer. Biochemistry. 1992;31:5245–5252. doi: 10.1021/bi00138a002. [DOI] [PubMed] [Google Scholar]
- 36.Ponten I, Kroth H, Sayer JM, Dipple A, Jerina DM. Differences between the mutational consequences of replication of cis- and trans-opened benzo[a]pyrene 7, 8-diol 9, 10-epoxidedeoxyguanosine adducts in M13mp7L2 constructs. Chem Res Toxicol. 2001;14:720–726. doi: 10.1021/tx0002684. [DOI] [PubMed] [Google Scholar]